In chronic hepatitis C virus (HCV) infection, IFNL4 genetic variation influences hepatic interferon-stimulated gene (ISG) expression. Here, RNA sequencing analysis demonstrated that clinically unfavorable IFNL4 polymorphisms are associated with elevated ISG expression by peripheral blood mononuclear cells in chronic HCV.
Keywords: Hepatitis C, interferon, interferon-stimulated genes, IFNL4, peripheral blood mononuclear cells
Abstract
Polymorphisms at IFNL4 strongly influence spontaneous resolution and interferon therapeutic response in hepatitis C virus (HCV) infection. In chronic HCV, unfavorable alleles are associated with elevated interferon (IFN)-stimulated gene (ISG) expression in the liver, but extrahepatic effects are less well characterized. We used RNA sequencing (RNA-Seq) to examine whether IFNL4 genetic variation (rs368234815) modulates ISG expression in peripheral blood mononuclear cells (PBMC) during chronic HCV infection. ISG expression was elevated in unstimulated PBMC homozygous for the unfavorable ΔG IFNL4 variant; expression following IFN-α stimulation was comparable across genotypes. These findings suggest that lambda interferons may have broader systemic effects during HCV infection.
Type III interferons (IFN-III), which include IFNL1, IFNL2, IFNL3, and IFNL4, are antiviral cytokines thought to act primarily at epithelial surfaces. Like type I interferons (IFN-I), IFN-III induce hundreds of interferon-stimulated genes (ISGs). IFN-III play significant roles in the host response to hepatitis C virus (HCV) infection. IFN-III are induced by HCV infection in liver and in primary human hepatocytes [1]. Noncoding genetic polymorphisms proximal to (rs8099917) and intronic within (rs12979860) the IFNL4 gene are strongly associated with exogenous IFN-I therapeutic response and spontaneous clearance of HCV (reviewed in [2]). An additional variant, rs368234815, in IFNL4 similarly influences the outcome of acute HCV infection and response to IFN-I therapy [3]. This site has 2 different alleles: ΔG (encoding an apparent IFNL4 open reading frame [ORF]) or TT (frameshift variant that disrupts the IFNL4 ORF). Somewhat surprisingly, the ΔG genotype is associated with unfavorable clinical outcomes. These polymorphic sites are in strong linkage disequilibrium.
Most functional studies on the effects of IFN-III single nucleotide polymorphisms (SNPs) in HCV have focused on hepatocytes and hepatic tissue. IFNL4 SNPs (rs8099917 and rs12979860) are associated with altered viral kinetics and fibrosis progression in the liver of HCV patients [4, 5]. In cultured primary human hepatocytes, the rs12979860 SNP affects antiviral responses [6]. Less is known about the role of IFN-III genetic variation in nonhepatic tissue. We therefore sought to determine whether IFN-III genetic variation impacts functional responses in extrahepatic cells from chronic HCV patients. We treated patient peripheral blood mononuclear cells (PBMCs) ex vivo with IFN-I and assessed the impact of IFNL4 genotype on global transcriptome responses by RNA sequencing (RNA-Seq). Our data indicate that, in chronic HCV, IFNL4 genotype influences baseline ISG expression levels in PBMC, and thereby affects the ISG response to additional IFN-I stimulation. These studies demonstrate a functional link between IFNL4 genetic variation and the immune response in cells of extrahepatic origin.
METHODS
Patients and Cells
Blood samples were collected from consenting patients receiving care for chronic HCV at New York Presbyterian Hospital/Weill Cornell Medicine. PBMCs were isolated by Ficoll gradient centrifugation and cryopreserved in liquid nitrogen. Protocols were carried out under approval from institutional review boards of Weill Cornell Medicine and Rockefeller University.
Cell Treatment and Nucleic Acid Extraction
Cryopreserved PBMCs were thawed in RPMI supplemented with 10% fetal bovine serum (FBS) and incubated at 37°C overnight. The next day, cells were either mock treated or treated with 10 U/mL Intron A (recombinant IFN-α2b, Merck) for 4 hours. PBMC DNA and RNA were extracted with the DNeasy Blood and Tissue Kit (Qiagen) and the RNeasy Mini Kit (Qiagen), respectively. PBMC samples for targeted RNA expression (TREx) and quantitative reverse transcription polymerase chain reaction (qRT-PCR) analyses were thawed and treated independently from samples used in RNA-Seq experiments (same patient collection and RNA extraction procedure, different cryovial aliquots; specific samples are detailed in Supplementary Table 1).
IFNL4 SNP Genotyping
DNA samples were genotyped for the rs12979860 SNP using primers F: GGGACCGCTACGTAAGTCAC and R: CGCT TATCGCATACGGCTA and for the rs368234815 and rs1176 48444 variants using primers F: ACTGTGTGTGCTGTGCCTTC and R: GGACGAGAGGGCGTTAGAG. PCR amplicons were directly sequenced by Sanger method to determine genotype.
RNA-Seq Library Preparation
RNA-Seq libraries were prepared from PBMC total RNA samples with the Illumina TruSeq RNA Sample Preparation v2 kit. Libraries were pooled and sequenced in multiplex on the Illumina HiSeq 2500 platform, generating 13–15 million 100 nt single-end reads per library.
RNA-Seq Data Processing and Analysis
RNA-Seq reads were mapped to the human genome (hg19, supplemented with Ensembl v66 transcript annotations) using the Tophat (v2.0.3) software package [7]. Per gene read counts were quantified on Ensembl v66 annotations appended with IFNL4 transcript information.
Differential gene expression analysis was performed using the voom/limma (v3.22.4) tools [8], with linear models including factors for IFNL4 rs368234815 genotype, sex, and IFN-α stimulation. For defining the set of genes induced by IFN-α (PBMC ISG set), samples were grouped into 2 conditions (unstimulated and IFN-α–stimulated) and contrasted directly. For all other differential expression analyses, samples were grouped by IFNL4 rs368234815 SNP genotype and contrasted as indicated.
ISG set enrichment testing was performed with CAMERA [9], using linear model factors detailed above. Gene ontology (GO) term enrichment analyses were conducted with the GOrilla webtool (http://cbl-gorilla.cs.technion.ac.il/);P value ranked gene lists (differential expression for each IFNL4 rs368234815 genotype pairwise comparison, all expressed protein-coding genes) were used as input.
Targeted RNA Expression Assay
Targeted RNA expression (TREx) probes for 150 PBMC ISGs and 10 “housekeeping” normalization genes (Supplementary File 6) were designed with the Illumina DesignStudio webtool. TREx libraries were prepared with the Illumina TruSeq Targeted RNA Expression Kit and sequenced on the MiSeq platform, generating an average of 3.75 × 105 reads per sample (range 2.9 × 104 to 8.0 × 105). One patient sample set (p9) was removed from analysis due to insufficient read depth. Illumina MiSeq Reporter software (v2.5.1) was used to conduct pairwise differential expression analyses.
RESULTS
We obtained PBMCs from 23 treatment-naive patients chronically infected with HCV prior to their treatment with IFN-α (Supplementary Table 1). We genotyped PBMCs at IFNL4 SNPs rs12979860, rs368234815, and rs117648444 (Supplementary Table 1). Because rs368234815 has been shown to be most predictive of spontaneous clearance and treatment outcomes (reviewed in [10]) we focused our analyses on samples grouped by this variant. Of note, one patient was discordant for the rs12979860/rs368234815 haplotype (p18, rs12979860 CT, rs368234815 ΔG/ΔG). Of the 23 patients, 7 were assigned to the TT/TT group (homozygous for the clinically favorable allele), 9 to the TT/ΔG group, and 7 to the ΔG/ΔG group (homozygous for the unfavorable allele). Although most patients were infected with HCV genotype 1, this cohort also included several patients infected with other HCV genotypes (detailed in Supplementary Table 1).
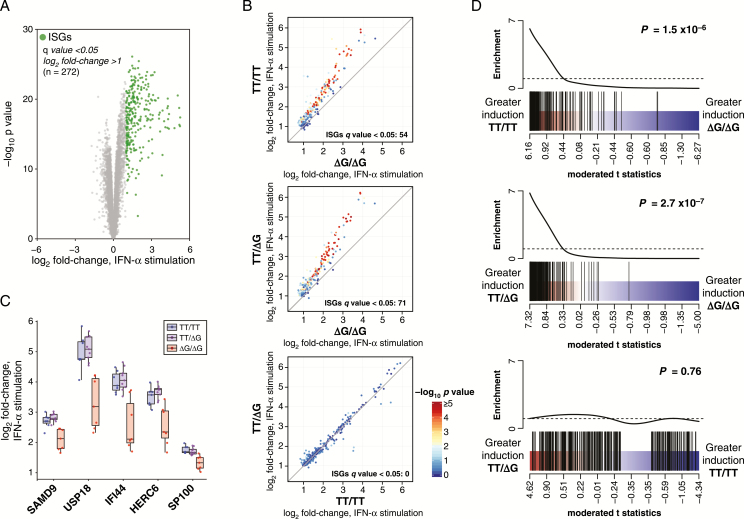
To characterize the integrative effects of IFN-III polymorphisms and IFN-I stimulation on PBMC gene expression patterns, we performed RNA-Seq on unstimulated and IFN-α–stimulated cells and conducted several complementary gene expression analyses. We first defined the specific ISGs induced by PBMC in response to IFN-α. We identified 272 genes as significantly upregulated (q value < 0.05, log2 fold-change >1) when comparing IFN-α stimulation to unstimulated cells, irrespective of IFNL4 genotype (Figure 1A; Supplementary File 1). This “PBMC ISG set” includes “classical” ISGs (eg, MX1, OAS1, STAT1, ISG15), as well as ISGs typically associated with leukocyte functions (eg, CD38, TNFSF13B, IL15, IL15RA).
Figure 1.
Interferon-stimulated gene (ISG) fold-change induction is attenuated in chronic hepatitis C virus (HCV) peripheral blood mononuclear cells (PBMCs) homozygous for ΔG/ΔG genotype at IFNL4 rs368234815 single nucleotide polymorphism (SNP). PBMC from individuals with chronic HCV were mock treated or stimulated with interferon-alpha (IFN-α) ex vivo and examined by RNA-Seq for gene expression fold-change analysis. A, Volcano plot depicting average fold-change in gene expression for PBMC samples (chronic HCV patients, n = 23) in response to IFN-α stimulation. PBMC IFN-stimulated gene (ISG) set (highlighted in green) was defined as q value < 0.05, log2 fold-change > 1, IFN-α stimulated versus unstimulated. B, PBMC ISG average fold-change expression (IFN-α simulated vs unstimulated) by IFNL4 rs368234815 SNP genotype (TT/TT, n = 7; TT/ΔG, n = 9; ΔG/ΔG, n = 7). Points are colored by –log10P value for indicated pairwise contrast. C, Individual sample fold-change expression (IFN-α simulated vs unstimulated) for the top 5 differentially induced ISGs in any pairwise contrasts (ranked by F test P value). D, Barcode plots representing PBMC ISG set enrichment in fold-change expression (IFN-α simulated vs unstimulated) by IFNL4 rs368234815 SNP genotype. PBMC ISGs (black bars) are ranked among all expressed genes by moderated t statistic for IFNL4 rs368234815 SNP genotype pairwise contrasts specified. P values for CAMERA gene set tests are indicated.
We next examined whether IFNL4 rs368234815 genotypes affected the magnitude of ISG induction in IFN-α–stimulated PBMC. We observed pronounced increases in ISG expression for all stimulated samples. However, in pairwise comparisons of IFNL4 rs368234815 genotypes (IFN-α–stimulated vs unstimulated), we found significant (q value < 0.05) differential induction of numerous ISGs across the genotype groups (Supplementary File 2). In general, we observed higher fold-change values for many ISGs in the TT/TT and TT/ΔG groups as compared to the ΔG/ΔG group (Figures 1B, 1C). No ISGs cleared significance thresholds in the TT/ΔG versus TT/TT comparison. To determine if differences in single gene fold-change values reflect a broader differential induction of the IFN-I response, we performed gene set enrichment testing for the PBMC ISG set across rs368234815 genotypes. As a set, ISGs were induced with lower fold-change values in ΔG/ΔG PBMC than other genotypes (Figure 1D); these differences were highly significant (P = 1.5 × 10−6, TT/TT vs ΔG/ΔG; P = 2.7 × 10−7, TT/ΔG vs ΔG/ΔG). These results indicate that in chronic HCV infection, the fold-change induction of an ex vivo IFN-I response is reduced in PBMC homozygous for IFNL4 rs368234815 ΔG/ΔG relative to other genotypes.
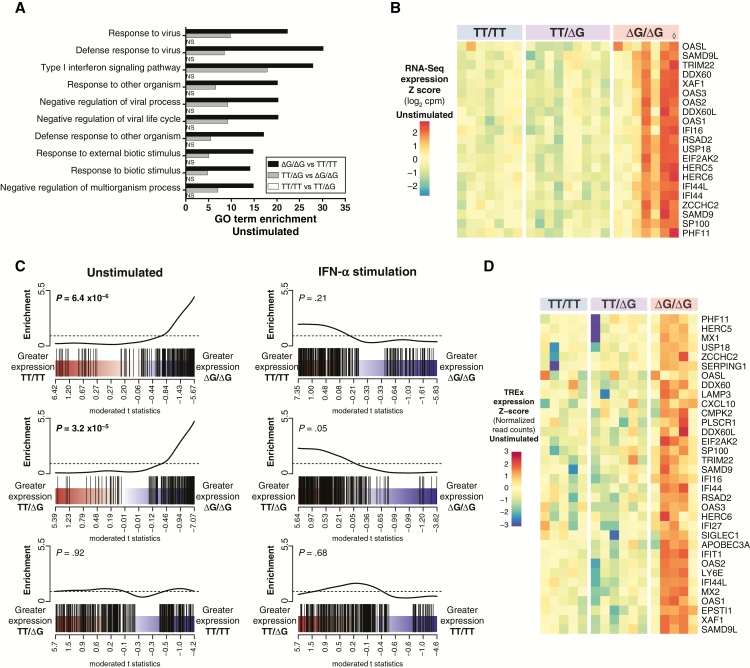
Because the above analysis was based on fold-change values (IFN-α–stimulated relative to unstimulated) across IFNL4 genotypes, we sought to determine whether our results could be explained by differences in underlying gene expression patterns for unstimulated cells, IFN-α–stimulated cells, or both. We conducted independent data analyses in which IFNL4 genotype groups were contrasted solely in unstimulated or in IFN-α–stimulated conditions. In the unstimulated condition, we first used GO term enrichment analysis to annotate differential gene expression across IFNL4 genotypes. We found numerous GO terms significantly enriched (q value < 0.01, Supplementary File 3) in TT/TT versus ΔG/ΔG (61 GO terms) and TT/ΔG versus ΔG/ΔG (73 GO terms) comparisons, but none in the TT/TT versus TT/ΔG contrast. The majority of significant GO terms were limited to innate antiviral immunity and IFN-I signaling (Supplementary File 3; Figure 2A), suggesting that genotype-associated differences in IFN signaling are evident in PBMC despite the absence of exogenous IFN-α stimulation.
Figure 2.
Interferon-stimulated gene (ISG) expression is elevated in unstimulated chronic hepatitis C virus (HCV) peripheral blood mononuclear cells (PBMCs) homozygous for ΔG/ΔG genotype at IFNL4 rs368234815 single nucleotide polymorphism (SNP), but reaches similar expression levels across all genotypes upon interferon-alpha (IFN-α) stimulation. PBMC from individuals with chronic HCV were mock treated or stimulated with IFN-α and subjected to RNA-Seq. Differential gene expression was analyzed for IFNL4 rs368234815 SNP genotype groups (TT/TT, n = 7; TT/ΔG, n = 9; ΔG/ΔG, n = 7) within the unstimulated condition, and within the IFN-α–stimulated condition. A, Top ranked (q value, TT/TT vs ΔG/ΔG) gene ontology (GO) terms enriched in unstimulated chronic HCV PBMC for indicated IFNL4 rs368234815 SNP pairwise contrasts. NS, not significant (q value > 0.01). B, Heatmap displaying unstimulated PBMC RNA-Seq expression values for ISGs differentially expressed in any IFNL4 genotype pairwise contrast (F test, q value < 0.05). Expression values scaled as per gene z score of log2 normalized RNA-Seq read counts per million reads (cpm). ◊ marks patient sample p18, discordant for rs12979860/rs368234815 haplotype (rs12979860 CT, rs368234815 ΔG/ΔG). C, Barcode plots representing PBMC ISG set expression enrichment in PBMC by IFNL4 rs368234815 SNP genotype. PBMC ISGs (black bars) are ranked among all expressed genes by moderated t statistic for IFNL4 rs368234815 SNP genotype pairwise contrasts specified. P values for CAMERA gene set tests are indicated. D, Heatmap displaying unstimulated PBMC ISG normalized expression values as measured by targeted RNA expression (TREx) assay in independent replicate samples (TT/TT, n = 5; TT/ΔG, n = 6; ΔG/ΔG, n = 5). ISGs were selected for heatmap display if differential expression was measured statistically significant in RNA-Seq samples (q value < 0.05) and/or TREx samples (q value < 0.1).
We next tested for differential gene expression in the unstimulated condition. We detected 37 individual genes, many of which were ISGs, differentially expressed across genotypes (F test q value < 0.05, Supplementary File 4; Figure 2B). Although many individual ISGs failed to clear significance thresholds, as a set, ISGs were expressed at significantly higher levels in unstimulated PBMC homozygous for IFNL4 rs368234815 ΔG/ΔG polymorphism than in other genotypes (Figure 2E) (P = 6.4 × 10−4, TT/TT vs ΔG/ΔG; P = 3.2 × 10−5, TT/ΔG vs ΔG/ΔG). These expression patterns indicate that in chronic HCV infection, unstimulated PBMC homozygous for IFNL4 rs368234815 ΔG/ΔG express many ISGs at elevated levels compared to other genotypes (further detailed in Supplementary Figure 1).
We conducted similar analyses in the IFN-α–stimulated condition. No significant GO terms were enriched in any pairwise contrast of rs368234815 genotypes. Furthermore, although we identified a small number of differentially expressed genes (q value < 0.05, Supplementary File 5), none of them are ISGs. Indeed, in PBMC ISG set enrichment testing, no significant enrichment by genotype group was detected in analysis of IFN-α–stimulated samples (Figure 2C).
To validate our findings with an orthogonal assay, we used the Illumina Targeted RNA Expression (TREx) platform to quantify mRNA levels for 150 ISGs and 10 “housekeeping” control genes in multiplex. Using independently prepared (same patients, different PBMC aliquots, independent IFN-α stimulations) PBMC RNA samples, we found that, consistent with our RNA-Seq results, unstimulated PBMC from the rs368234815 ΔG/ΔG group generally expressed ISGs at elevated levels compared to other genotypes; again, differences were not observed when comparing genotypes in the IFN-α–stimulated condition. Using the TREx statistical framework, we found that 21 ISGs were expressed at significantly higher levels in unstimulated PBMC from the ΔG/ΔG group as compared to the TT/TT or TT/ΔG groups (Figure 2D; Supplementary File 6). Additional validation experiments by quantitative reverse transcript polymerase chain reaction (qRT-PCR) for 3 selected ISGs (RSAD2, EIF2AK2, and OAS3) generated similar results (Supplementary Figure 1C).
DISCUSSION
Our results indicate that in chronic HCV infection, peripheral immune cells homozygous for IFNL4 rs368234815 ΔG/ΔG polymorphism express ISGs at elevated levels as compared to other genotypes. However, ΔG/ΔG cells maintain the potential to induce ISGs to levels comparable with other IFNL4 genotypes upon further IFN-α stimulation.
These data are largely consistent with studies on liver gene expression patterns, which associate elevated ISG expression with heterozygosity or homozygosity for unfavorable IFNL4 region alleles [11, 12]. Relatedly, a single unfavorable IFNL4 allele is sufficient to confer poor response to IFN therapy [3]. However, in PBMC, we only observed elevated ISG expression in the homozygous ΔG/ΔG group, but not the heterozygous TT/ΔG group. The basis for this discrepancy remains unclear, but could suggest a gene dosing effect in the context of PBMC stimulation. Such an effect would be consistent with a reported supra-additive model for CC homozygosity at rs12979860 in the spontaneous clearance of HCV [13].
Attenuated PBMC ISG induction in chronic HCV is associated with failure of IFN-based therapy [14]. Here, we link IFNL4 rs368234815 ΔG/ΔG genotype to ISG expression levels in PBMC, but the mechanisms contributing to this signature remain unclear. Because rs368234815 is in strong linkage disequilibrium with noncoding rs8099917 and rs12979860 SNPs, assigning a phenotype to a single site is challenging. We did not detect IFNL4 mRNA expression in any PBMC samples. Although the ΔG allele creates an ORF, a direct role for hepatic IFNL4 protein in HCV pathogenesis has not been established. If IFNL4 protein is produced by infected hepatocytes, then PBMCs may directly respond with increased ISG expression. While hematopoietic cells are not considered primary targets of IFNL action due to poor receptor expression [2], we did detect RNA expression of IFNLR1 and IL10R2 (Supplementary Figure 2). However, we cannot determine which PBMC subsets express the IFNL receptor, or if the observed mRNA correlates with receptor protein levels. Furthermore, we did not detect apparent differences in ISG expression levels from those ΔG/ΔG PBMC carrying a functionally impaired IFN-λ4-S70 variant (rs117648444) [15], although our sample size was insufficient to conduct a proper statistical comparison. Interestingly, intronic IFNL4 genetic variation (rs12979860) has been associated with postpartum PBMC ISG expression levels in healthy mothers after childbirth [16].
Alternatively, elevated ISG expression in unstimulated PBMC could be an indirect consequence of interferon activity in the infected liver. In this model, IFNL4 variation affects the production of paracrine factors (for example, IFN-I or perhaps signals from damaged cells) that act on PBMC systemically or locally in the liver. Direct and indirect mechanisms are not mutually exclusive, and evaluating their respective impact on PBMC gene expression will be experimentally challenging. Nonetheless, our results suggest that chronic HCV infection in patients with unfavorable IFNL4 SNPs may ultimately modulate IFN-I signaling not only in liver, but also PBMC and perhaps other tissue types.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank R. Peterson for PBMC database management. We acknowledge the support of C. Zhao, X. Wang, and W. Zhang at The Rockefeller University Genomics Resource Center. We thank E. Castillo, B. Flatley, and A. Webson for laboratory support. We further acknowledge the numerous primary research articles that could not be referenced due to space limitations.
Previous presentation. This study was presented in February 2016 at the Keystone Symposium: Cell Biology and Immunology of Persistent Infection in Banff, Alberta, Canada.
Financial support. This work was supported in part by the National Institutes of Health (grant numbers DK095031 to J. W. S., AI091707 to C. M. R., AI111825 to B. R. R. and C. M. R.); the John C. Whitehead Presidential Fellowship to B. R. R.; the American Cancer Society Institutional Research Grant (grant number 02-196 to UT Soutwestern Medical Center); The Rockefeller University Center for Clinical and Translational Science Award (grant number UL1RR024143); and by the Center for Basic and Translational Research on Disorders of the Digestive System through the generosity of the Leona M. and Harry B. Helmsley Charitable Trust. Additional funding was provided by the Greenberg Medical Research Institute, the Starr Foundation, the Ronald A. Shellow, M.D. Memorial Fund (C. M. R.), and the Troup Fund of the Kaleida Health Foundation (A. H. T). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the other funders.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Park H, Serti E, Eke O et al. . IL-29 is the dominant type III interferon produced by hepatocytes during acute hepatitis C virus infection. Hepatology 2012; 56:2060–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wack A, Terczyńska-Dyla E, Hartmann R. Guarding the frontiers: the biology of type III interferons. Nat Immunol 2015; 16:802–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Prokunina-Olsson L, Muchmore B, Tang W et al. . A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 2013; 45:164–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thompson AJ, Muir AJ, Sulkowski MS et al. . Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology 2010; 139:120–9.e18. [DOI] [PubMed] [Google Scholar]
- 5. Noureddin M, Wright EC, Alter HJ et al. . Association of IL28B genotype with fibrosis progression and clinical outcomes in patients with chronic hepatitis C: a longitudinal analysis. Hepatology 2013; 58:1548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sheahan T, Imanaka N, Marukian S et al. . Interferon lambda alleles predict innate antiviral immune responses and hepatitis C virus permissiveness. Cell Host Microbe 2014; 15:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014; 15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res 2012; 40:e133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O’Brien TR, Prokunina-Olsson L, Donnelly RP. IFN-λ4: the paradoxical new member of the interferon lambda family. J Interferon Cytokine Res 2014; 34:829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Honda M, Sakai A, Yamashita T et al. ; Hokuriku Liver Study Group Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology 2010; 139:499–509. [DOI] [PubMed] [Google Scholar]
- 12. Urban TJ, Thompson AJ, Bradrick SS et al. . IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology 2010; 52:1888–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shebl FM, Pfeiffer RM, Buckett D et al. . IL28B rs12979860 genotype and spontaneous clearance of hepatitis C virus in a multi-ethnic cohort of injection drug users: evidence for a supra-additive association. J Infect Dis 2011; 204:1843–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. He XS, Ji X, Hale MB et al. . Global transcriptional response to interferon is a determinant of HCV treatment outcome and is modified by race. Hepatology 2006; 44:352–9. [DOI] [PubMed] [Google Scholar]
- 15. Terczyńska-Dyla E, Bibert S, Duong FH et al. ; Swiss Hepatitis C Cohort Study Group Reduced IFNλ4 activity is associated with improved HCV clearance and reduced expression of interferon-stimulated genes. Nat Commun 2014; 5:5699. [DOI] [PubMed] [Google Scholar]
- 16. Price AA, Tedesco D, Prasad MR et al. . Prolonged activation of innate antiviral gene signature after childbirth is determined by IFNL3 genotype. Proc Natl Acad Sci U S A 2016; 113:10678–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.