Summary
This manuscript explores the potential of developing a novel immunotherapeutic approach for the prevention/treatment of parainfluenza virus 3 infections by characterizing the cellular immune response to all 7 encoded viral antigens and establishing a hierarchy of immunogenicity
Keywords: Parainfluenza virus 3, virus-specific T cells, immunotherapy
Abstract
Parainfluenza virus type 3 (PIV3) infections are a major cause of morbidity and mortality in immunocompromised individuals, with no approved therapies. Our group has demonstrated the safety and efficacy of adoptively transferred virus-specific T cells for the prevention and treatment of a broad range of viral infections including BK virus, cytomegalovirus, adenovirus, human herpesvirus 6, and Epstein-Barr virus. However, this approach is restricted to well-characterized viruses with known immunogenic/protective T-cell target antigens, precluding extension to PIV3. We now characterize the cellular immune response to all 7 PIV3-encoded antigens in 17 healthy donors and define a hierarchy of immunogenicity based on the frequency of responding donors and the magnitude of specific cells. We show that reactive populations of both CD4+ and CD8+ T cells are capable of producing Th1-polarized effector cytokines and killing PIV3-expressing targets. Furthermore, we confirm the clinical relevance of these cells by demonstrating a direct correlation between the presence of PIV3-specific T cells and viral control in allogeneic hematopoietic stem cell transplant recipients. Taken together, our findings support the clinical use of PIV3-specific T cells produced with our Good Manufacturing Practice–compliant manufacturing process, in immunocompromised patients with uncontrolled infections.
(See the editorial commentary by Waghmare et al on pages 147–9.)
The adoptive transfer of in vitro expanded virus-specific T cells (VSTs) has been shown to effectively treat drug-refractory viral infections in allogeneic hematopoietic stem cell transplant (HSCT) recipients. However, this strategy has been restricted to immunologically well-characterized viruses such as cytomegalovirus (CMV), Epstein-Barr virus (EBV), BK virus, human herpesvirus 6 (HHV-6), and adenovirus (AdV) [1–8]. The purpose of the current study was to explore the potential of targeting the respiratory virus parainfluenza virus type 3 (PIV3) using this approach.
Parainfluenza virus types 1, 2, 3, and 4 are paramyxoviruses that primarily affect young children, where the pathogenic spectrum comprises upper respiratory tract infections (URTIs) and lower respiratory tract infections (LRTIs) and includes syndromes such as the common cold, croup, bronchitis, bronchiolitis, and pneumonia [9, 10]. Whereas infections in immunocompetent individuals are generally well-controlled via a variety of innate and adaptive immune mechanisms [11–15], PIV3 infections in the immunocompromised host (eg, HSCT recipients) are frequent (up to 18%), associated with substantial morbidity (eg, decreased lung function, multiorgan failure, and graft loss) and mortality rates as high as 60% [16–20]. Currently there are no approved treatments, and the use of the nucleoside analogue ribavirin (with or without intravenous immunoglobulin) has had limited effect [21–23].
To establish whether such a virus can be targeted immunotherapeutically, it is important to demonstrate that protective T cells are present in the peripheral blood of healthy seropositive individuals. Furthermore, if such cells exist, it is important to know which virally encoded antigens they recognize, so that the most effective T cells can be prepared for adoptive transfer. To address these questions, we now comprehensively profile the cellular immune response against PIV3 in both healthy adults and patients with documented infections.
METHODS
Donors and Cell Lines
Blood samples were obtained from healthy volunteers and HSCT recipients with informed consent using Baylor College of Medicine institutional review board–approved protocols (H-7634, H-36346, H-35436, and H-25064). Peripheral blood mononuclear cells (PBMCs), which were isolated by Ficoll gradient, were used to generate phytohemagglutinin (PHA) blasts and VSTs. PHA blasts were generated from PBMCs (2 × 106/well of a 24-well tissue culture-treated plate) using PHA (5 μg/mL; Sigma-Aldrich, St Louis, Missouri), and cells were maintained in VST medium (RPMI 1640; [HyClone Laboratories, Logan, Utah], 45% Click’s medium [Irvine Scientific, Santa Ana, California], 2 mM GlutaMAX TM-I [Life Technologies, Grand Island, New York], and 5% human AB serum [Valley Biomedical, Winchester, Virginia]) supplemented with interleukin (IL) 2 (100 U/mL; NIH, Bethesda, Maryland), which was replenished every 3 days.
VST Generation
Pepmixes
For stimulation, we used custom-ordered peptide libraries (15 mers overlapping by 11 amino acids) spanning the PIV3 proteins M, HN, N, F, PC, PP, and L (Genemed Synthesis, San Antonio, Texas). Control pepmixes spanning AdV (Hexon and Penton) were purchased from JPT Technologies (Berlin, Germany). Lyophilized pepmixes for PIV3 antigens were reconstituted at 10 mg/mL in dimethyl sulfoxide (DMSO; Sigma-Aldrich), and lyophilized pepmixes for the control virus AdV antigens were reconstituted at 200 μg/mL in DMSO and stored at –80°C.
VST Activation
Fifteen million fresh or frozen PBMCs were pelleted and pulsed for 30 minutes at 37°C with pepmixes, individually or pooled (100–500 ng/peptide/15 × 106 PBMCs). After incubation, cells were resuspended in VST media supplemented with 400 U/mL IL-4 and 10 ng/mL IL-7 (R&D Systems, Minneapolis, Minnesota) and plated in a 24-well plate (2 × 106/well) or transferred to a G-Rex10 device (Wilson Wolf, Minneapolis, Minnesota) (15 × 106/G-Rex10). Media and cytokines were replenished on day 7, and cultures were split when they reached a density of >3 × 106 cells/well or >50 × 106 cells/G-Rex10. On days 9–11, VSTs were harvested, counted, and used for phenotypic and functional studies.
VST Expansion
Ten million PIV3 specific T cells (PIV3-STs) were restimulated on days 9–11 of culture at a 1:1 ratio with irradiated (30 Gy) autologous pepmix-pulsed PHA blasts or PBMCs. Cultures were resuspended in a total of 20 mL VST media supplemented with IL-4 and IL-7 and transferred to a 24-well tissue culture plate (2 × 106 cells/well). On days 13 and 17, cultures were replenished with fresh media supplemented with IL-2 (100 U/mL). On days 19–21, VSTs were harvested and used for further studies.
Flow Cytometry
Immunophenotyping
PIV3-STs were surface stained with monoclonal antibodies to CD3, CD56, and CD45RO (Becton Dickinson [BD], Franklin Lakes, New Jersey) and CD4, CD8, CD16, and CD62L (Beckman Coulter, Pasadena, California). For staining, cells were washed once with phosphate-buffered saline (PBS) (Sigma-Aldrich) and pelleted, and antibodies were added in saturating amounts (2–5 μL). After 15 minutes’ incubation at 4°C in the dark, cells were washed twice and analyzed. Approximately 30000 live cells were acquired using a Gallios™ flow cytometer equipped with Gallios software.
Intracellular Cytokine Staining
VSTs were harvested, resuspended at a concentration of 5 × 106/mL in VST media, and plated at 100 μL/well in a 96-well plate. The cells were then stimulated with test or control pepmix in the presence of brefeldin A (1 μg/mL), monensin (1 μg/mL), and CD28 and CD49d (1 μg/mL) (BD) overnight. Subsequently, VSTs were washed with PBS, pelleted, and surface stained with CD8 and CD3 (5 μL/antibody/tube). After 15 minutes’ incubation at 4°C in the dark, they were washed, pelleted, fixed, and permeabilized with Cytofix/Cytoperm solution (BD) for 20 minutes at 4°C in the dark. After washing with PBS containing fetal bovine serum (FBS) and saponin (BD), cells were incubated with 20 μL interferon gamma (IFN-γ) and tumor necrosis factor alpha (TNF-α) antibodies (BD) for 30 minutes at 4°C in the dark. Cells were then washed twice with cold PBS containing FBS and saponin, and at least 20000 live cells from each population were analyzed with a Gallios™ flow cytometer equipped with Gallios software.
Functional Studies
Enzyme-Linked Immunospot Assay
Enzyme-linked immunospot (ELIspot) analysis was used to quantitate the frequency of T cells that secreted IFN-γ and granzyme B in response to PIV3 antigen exposure. Fresh PBMCs were resuspended at 5 × 106/mL in VST media, and 100 μL (5 × 105 cells) was added to each ELIspot well. Expanded PIV3-STs were resuspended at 2 × 106/mL in VST media and 100 µL (2 × 105 cells) was added to each well. Antigen-specific activity was measured after direct stimulation of the cells with each of the individual PIV3 pepmixes (M, HN, N, F, PC, PP, and L) and AdV pepmixes (Hexon and Penton). PHA (1 μg/mL) was used as a positive control while unstimulated cells served as a negative control and each condition was run in duplicate. After 20–24 hours of incubation, plates were developed per manufacturer instructions, dried at room temperature, and then sent to Zellnet Consulting (New York, New York) for quantification. Spot-forming cells (SFC) and input cell numbers were plotted as a box-and-whisker plot, where the whisker ends were defined as 10th and 90th percentile, and data points outside this range were represented as dots.
Multiplex
The cytokine profile was assessed using the Milliplex High Sensitivity Human Cytokine Panel (Millipore, Billerica, Massachusetts). A total of 1 × 105 VSTs were stimulated using pepmix (M, HN, N, F, PC, PP, and L) overnight. Subsequently, supernatant was collected, plated in duplicate samples, and incubated overnight at 4°C with antibody-immobilized beads, then washed and incubated for 1 hour at room temperature with the biotinylated detection antibodies. Finally, streptavidin-phycoerythrin was added for 30 minutes at room temperature. Samples were washed and analyzed using xPONENT software.
Chromium Release Assay
We measured the cytotoxic specificity of the PIV3-STs in a standard 4-hour chromium 51 (Cr51) release assay, using effector:target (E:T) ratios of 40:1, 20:1, 10:1, and 5:1. VSTs were used as effectors and the targets were PHA blasts pulsed with pepmixes. Autologous PHA blasts alone were used as specificity controls. The percentage of specific lysis was calculated as [(experimental release – spontaneous release) / (maximum release – spontaneous release)] × 100.
RESULTS
PIV3
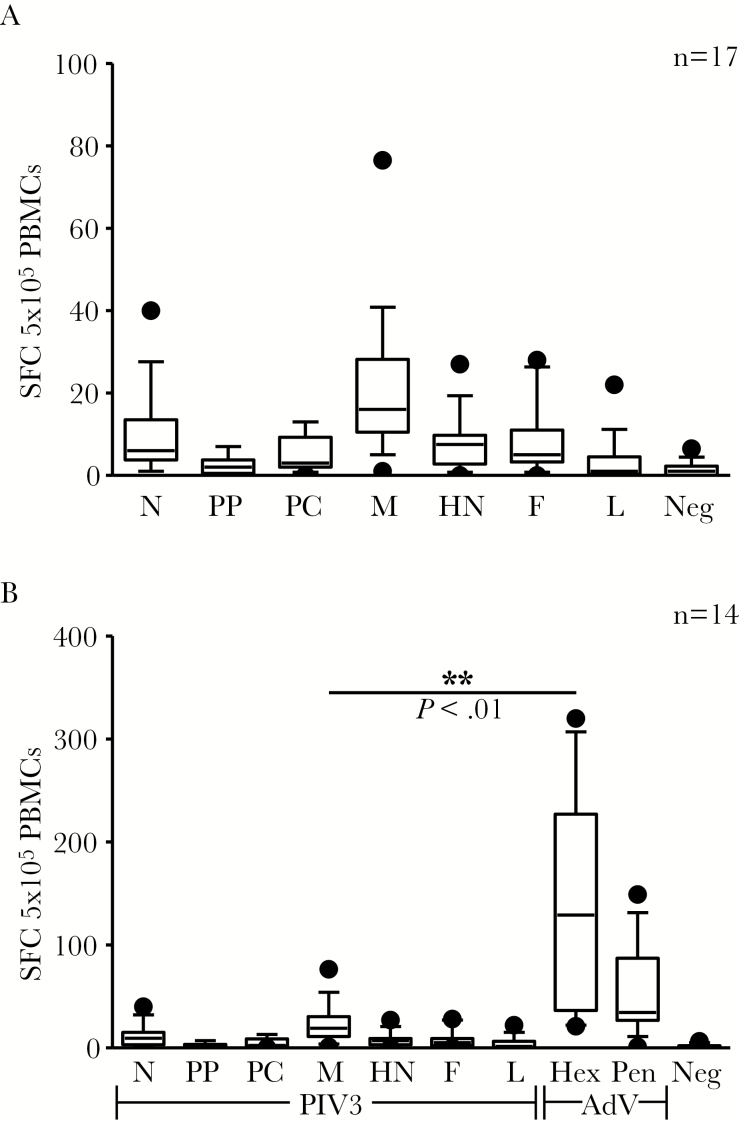
PIV3, a negative-sense, single-stranded RNA virus of the family Paramyxoviridae, encodes for 7 viral antigens: nucleocapsid (N), phosphoprotein (PP), protein C (PC), matrix (M), hemagglutinin-neuraminidase (HN), fusion (F), and large protein (L) [9, 24, 25] (Supplementary Figure 1A and Supplementary Table 1). To characterize the cellular immune response to this virus, we assessed the T-cell activity directed against all 7 viral antigens by exposing PBMCs from 17 healthy donors to peptide libraries (15 mers overlapping by 11aa) and evaluating the frequency of IFN-γ–producing antigen-specific T cells by ELIspot assay. In general, the frequency of circulating virus-specific T cells was low (mean ± SEM) (N: 9.1 ± 2.5 SFC/5 × 105 PBMCs; PP: 2.3 ± 0.6; PC: 4.6 ± 1.1; M: 20.3 ± 4.2; HN: 7.8 ± 1.6; F: 7.9 ± 2.0; L: 3.2 ± 1.3 [n = 17]; Figure 1A)—substantially lower than against AdV (139.8 ± 26.6 and 50.7 ± 9.8 SFC/5 × 105; Hexon and Penton, respectively [n = 14]; Matrix vs Hexon, P = .0013; Figure 1B).
Figure 1.
Frequency of parainfluenza virus type 3 (PIV3)–specific T cells in healthy donors. A, Frequency of PIV3-specific T cells in peripheral blood as determined by interferon-γ enzyme-linked immunospot assay (n = 17). B, Frequency of PIV3 and adenovirus (Hexon and Penton)–reactive T cells in peripheral blood in 14 individuals. Data points outside the 10th to 90th percentile are represented as dots. Results are reported as spot-forming cells per 5 × 105 peripheral blood mononuclear cells plated. Abbreviations: AdV, adenovirus; F, fusion; Hex Pen, Hexon and Penton; HN, hemagglutinin-neuraminidase; L, large protein; M, matrix; N, nucleocapsid; PBMCs, peripheral blood mononuclear cells; PC, protein C; PIV3, parainfluenza virus 3; PP, phosphoprotein; SFC, spot-forming cells.
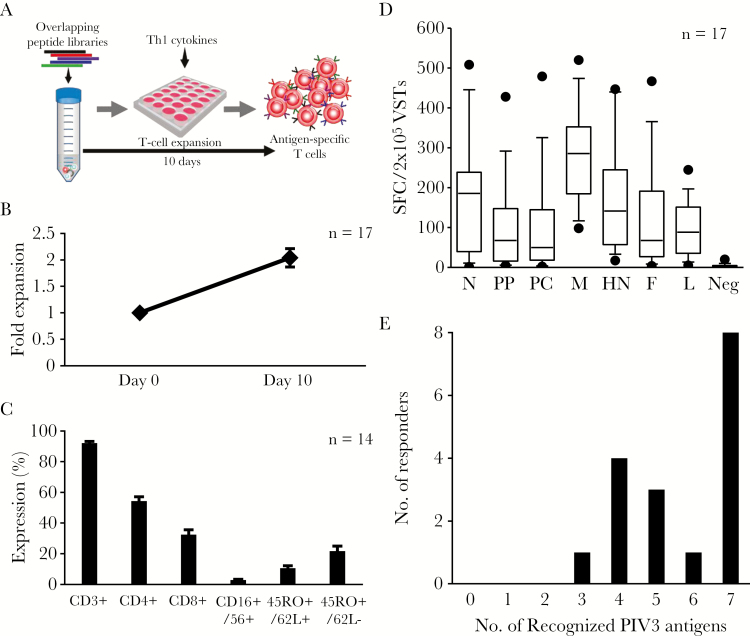
To investigate whether the paucity of reactive T cells in peripheral blood was due to the limited immunogenicity of the virus or simply reflected a number of circulating T cells below the ELIspot detection threshold, we performed a single in vitro stimulation designed to selectively amplify PIV3-specific T cells [4]. Thus, we exposed PBMCs to a mastermix of the PIV3 peptide libraries followed by a 9- to 11-day expansion period (Figure 2A), resulting in a mean ± SEM 2.0 ± 0.2-fold increase in total cell numbers (n = 17) (Figure 2B). The expanded cells were predominantly CD3+ T cells (mean ± SEM: 92.1% ± 1.14% [n = 14]), with a mixture of helper (CD4+) (mean ± SEM: 54.6% ± 2.6%) and cytotoxic (CD8+) cells (mean ± SEM: 31.6% ± 3%), a subset of which expressed central and effector memory markers (CD45RO+/CD62L+ [mean ± SEM: 10.5% ± 1.6%] and CD45RO+/CD62L– [mean ± SEM: 21.7% ± 3.2%], respectively; Figure 2C). To next assess whether these cells represented a PIV3-enriched population, we repeated our IFN-γ ELIspot and, as shown in Figure 2D, we were able to detect increased activity against all viral antigens (fold enrichment of reactive T cells is summarized in Supplementary Figure 2), allowing us to establish a hierarchy of immunodominance based on the frequency of responding donors and magnitude of reactive cells (Table 1).
Figure 2.
Activation and expansion of PIV3-specific T cells. A, Schematic of the T-cell expansion protocol. B, Fold expansion achieved over a 9- to 11-day period based on cell counting using Trypan blue exclusion (n = 17). C, Phenotype of the expanded lines (mean ± standard error of the mean) (n = 14). D, Specificity of the expanded cells as determined by interferon-γ enzyme-linked immunospot assay. Data points outside the 10th to 90th percentile are represented as dots. E, Number of antigens recognized by the donors screened. Only lines with a total of ≥30 spot-forming cells/2 × 105 input cells for a given antigen were considered to be positive. Abbreviations: F, fusion; HN, hemagglutinin-neuraminidase; L, large protein; M, matrix; N, nucleocapsid; PC, protein C; PIV3, parainfluenza virus 3; PP, phosphoprotein; SFC, spot-forming cell; VST, virus-specific T cell.
Table 1.
Parainfluenza Virus 3 Antigen Hierarchy of Immunodominance
| Antigen | Responding Donors |
SFC/2 × 105 VSTs (Mean ± SEM) |
Median SFC/2 × 105 VSTs (Range) |
|---|---|---|---|
| M | 17 | 283 ± 29 | 286 (98–520) |
| HN | 16 | 183 ± 32 | 172 (37–447) |
| N | 14 | 196 ± 38 | 200 (39–509) |
| F | 13 | 157 ± 37 | 80 (31–467) |
| PC | 13 | 134 ± 37 | 86 (34–480) |
| L | 13 | 117 ± 17 | 93 (44–245) |
| PP | 10 | 140 ± 35 | 125 (34–428) |
Abbreviations: F, fusion; HN, hemagglutinin-neuraminidase; L, large protein; M, matrix; N, nucleocapsid; PC, protein C; PP, phosphoprotein; SEM, standard error of the mean; SFC, spot-forming cell; VST, virus-specific T cell.
M was recognized in all 17 donors and induced the highest frequency of specific cells (mean ± SEM: 283 ± 29 SFC/2 × 105), followed, in descending order, by HN (n = 16; mean ± SEM: 183 ± 32), N (n = 14; mean ± SEM: 196 ± 38), F (n = 13; mean ± SEM: 157 ± 37), PC (n = 13; mean ± SEM: 134 ± 37), L (n = 13; mean ± SEM: 117 ± 17), and PP (n = 10; mean ± SEM: 140 ± 35). Notably, all donors recognized ≥3 antigens and more than half of the donors recognized ≥6 antigens (Figure 2E).
Functional Characterization of PIV3 VSTs
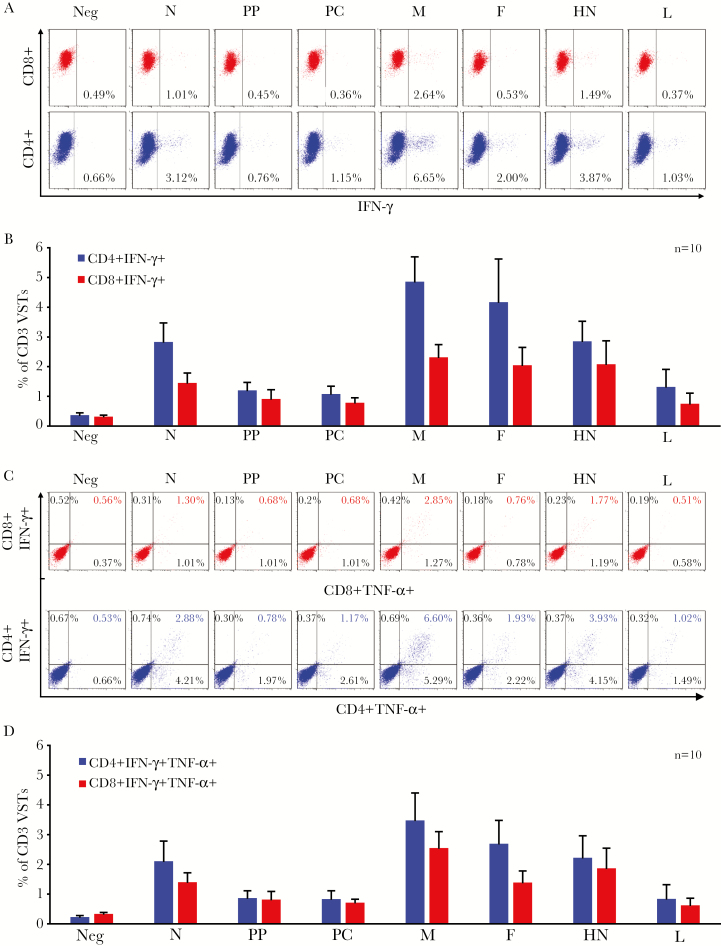
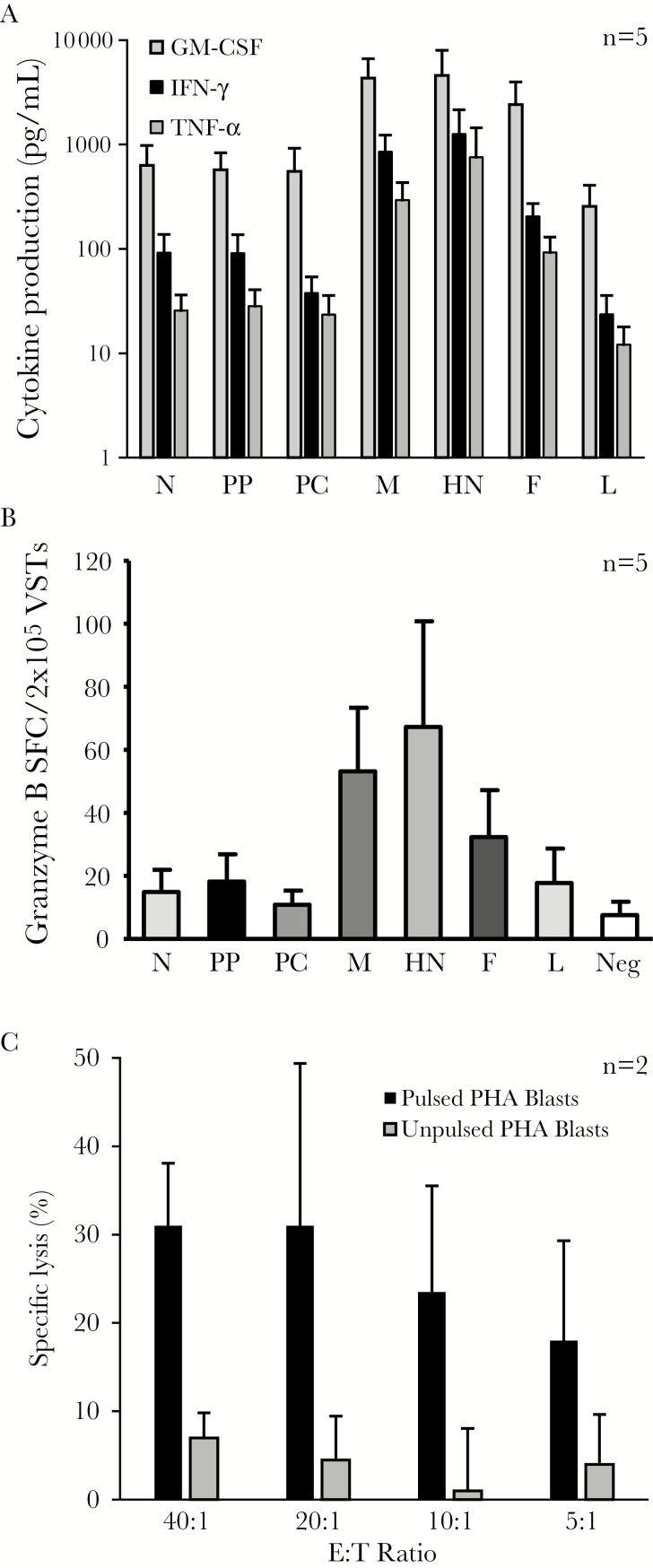
To evaluate whether PIV3 specificity was mediated predominantly by CD4+ or CD8+ T cells, we performed intracellular cytokine staining (ICS), gating on CD4+ and CD8+ IFN-γ–producing cells. For all 7 antigens, T-cell activity was detected predominantly in the CD4+ compartment, with a minor CD8 response. Figure 3A shows fluorescence-activated cell sorting data from a representative donor, and Figure 3B summarizes the results for all 10 VST lines studied. As the production of multiple proinflammatory cytokines and effector molecules has been shown to correlate with enhanced cytolytic function and improved in vivo activity, we additionally evaluated the production of the Th1 cytokines TNF-α and granulocyte-macrophage colony-stimulating factor (GM-CSF) in response to antigenic stimulation [4, 26, 27]. ICS analysis demonstrated that the majority of IFN-γ–producing cells also produced TNF-α (detailed results from 1 donor, Figure 3C; summary results for 10 donors, Figure 3D. Furthermore, reactive T cells additionally produced GM-CSF, as measured by Luminex array (Figure 4A), as well as the effector molecule granzyme B (mean ± SEM) (N: 14 ± 7 SFC/2 × 105; PP: 18 ± 8; PC: 11 ± 4, M: 53 ± 17, HN: 67 ± 32, F: 32 ± 14; and L: 17.8 ± 11.4 [n = 5]) (Figure 4B). Thus, our expanded PIV3-specific T-cell lines were polyclonal, Th1-polarized, and polyfunctional. Finally, to investigate the cytolytic potential of these VSTs in vitro, we co-cultured PIV3-STs with Cr51-labeled, peptide-loaded autologous PHA blasts. As shown in Figure 4C, PIV3-loaded targets were specifically recognized and lysed by our expanded PIV3-STs at all E:T ratios tested, whereas control target cells were not killed (n = 2).
Figure 3.
Parainfluenza virus 3–specific T cells are polyclonal and polyfunctional. A, Interferon gamma (IFN-γ) production, as assessed by intracellular cytokine staining (ICS) from CD4 helper (bottom row) and CD8 cytotoxic T cells (top row) on day 10 after antigenic stimulation in 1 representative donor (dot plots were gated on CD3+ cells). B, Summary results of IFN-γ production for 10 donors screened (mean ± standard error of the mean [SEM]). C, Dual IFN-γ and tumor necrosis factor alpha production as evaluated by ICS from CD4 helper (bottom row) and CD8 cytotoxic (top row) T cells on day 10 after stimulation in 1 representative donor (dot plots gated on CD3+ cells). D, Summary results for 10 donors screened (mean ± SEM). Abbreviations: F, fusion; HN, hemagglutinin-neuraminidase; IFN-γ, interferon gamma; L, large protein; M, matrix; N, nucleocapsid; PC, protein C; PP, phosphoprotein; TNF-α, tumor necrosis factor alpha; VST, virus-specific T cell.
Figure 4.
Parainfluenza virus (PIV3)–specific T cells are Th1-polarized and cytolytic. A, Cytokine profile of PIV3-specific T cells as measured by multiplex bead array. B, Production of granzyme-B by enzyme-linked immunospot assay. Results are reported as mean ± standard error of the mean spot-forming cells/2 × 105 input cells. C, Cytolytic potential of PIV3-specific T-cell lines as evaluated by standard 4-hour chromium 51 release assay using PIV3 pepmix-pulsed phytohemagglutinin blasts as targets (n = 2). Results are presented as percentage of specific lysis (mean ± standard deviation). Abbreviations: E:T, effector:target ratio; F, fusion; GM-CSF, granulocyte-macrophage colony-stimulating factor; HN, hemagglutinin-neuraminidase; IFN-γ, interferon gamma; L, large protein; M, matrix; N, nucleocapsid; PC, protein C; PHA, phytohemagglutinin; PP, phosphoprotein; SFC, spot-forming cell; TNF-α, tumor necrosis factor alpha; VST, virus-specific T cell.
Detection of PIV3-Specific T Cells in HSCT recipients
To assess the in vivo relevance of PIV3-specific T cells in the control of viral infection, we investigated whether allogeneic HSCT recipients had circulating T cells directed against our identified immunodominant target antigens in their peripheral blood during a PIV3 infection.
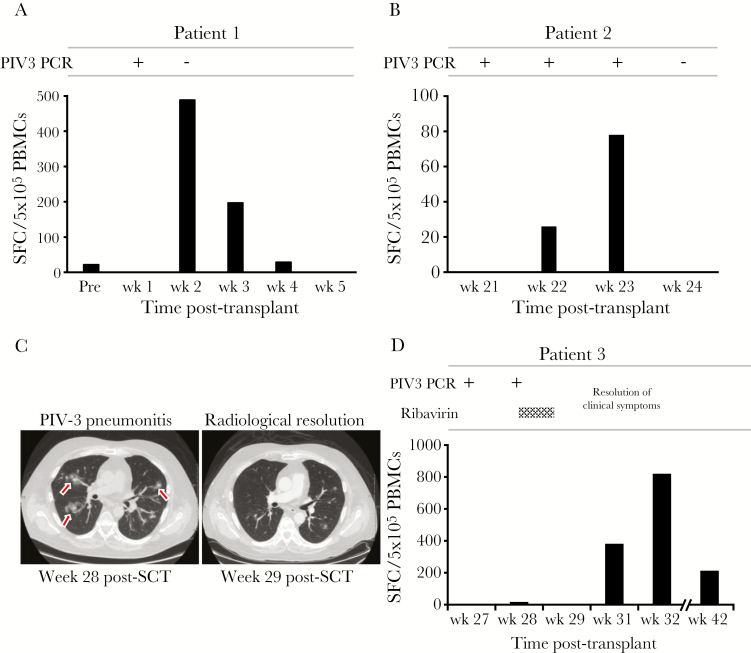
Patient 1 is a 20-month-old girl with acute lymphoblastic leukemia (ALL) who received a matched related donor transplant from her sister. Week 1 posttransplant, she developed a cough, congestion, and a runny nose. Viral screening (reverse- transcription polymerase chain reaction [RT-PCR]) of her nasal secretions was positive for PIV3, but no infiltrates were present on chest radiography. By week 2 posttransplant, her cough and congestion had improved with no antiviral therapy, and repeat screening was negative for PIV3. We analyzed the circulating frequency of PIV3-specific T cells over the course of her infection. Prior to transplant, she had a weak but detectable response against PIV3 antigens F and N (22 SFC/5 × 105 PBMCs), which became undetectable following her myeloablative conditioning regimen with cytarabine, cyclophosphamide, and total body irradiation. However, within a week of viral exposure, PIV3-specific T cells had expanded in vivo (489 SFC/5 × 105 PBMCs), representing a 22-fold increase in reactive cells, as seen in Figure 5A. Thereafter, the reactive population declined, coincident with viral clearance. Importantly, over this same time frame the response against influenza virus, which was not active, remained static, confirming that this robust expansion in PIV3-reactive T cells represented a virus-driven response.
Figure 5.
Detection of parainfluenza virus 3 (PIV3)–specific T cells in the peripheral blood of hematopoietic stem cell (HSCT) recipients who controlled PIV3 infection. Peripheral blood mononuclear cells (PBMCs) isolated from 3 HSCT recipients during an acute PIV3 infection were tested for specificity against our panel of PIV3 antigens, using interferon-γ enzyme-linked immunospot assay as a readout. A, B, and D, Results from 3 patients in whom PIV3 infection was rapidly controlled, coincident with a detectable rise in PIV3-specific T cells. C, Axial computed tomography scan of the lungs (patient 3) displaying multifocal patchy ground-glass nodular infiltrates that resolved within 1 week. Abbreviations: PBMC, peripheral blood mononuclear cell; PCR, polymerase chain reaction; PIV3, parainfluenza virus 3; SCT, stem cell transplant; SFC, spot-forming cell.
Patient 2 is a 4-year-old boy with history of high-risk ALL who underwent an umbilical cord blood transplant. On week 21 post-HSCT, he presented with a runny nose, congestion, and nonproductive cough, confirmed to be associated with PIV3 by RT-PCR on nasal wash. He was followed as an outpatient with weekly PCR analysis of nasal secretions, and no antiviral therapy was administered. By week 24, the virus was not detected and his symptoms resolved. To determine whether T-cell immunity played a role in viral clearance, we again assessed the PIV3-directed T-cell immune response during the infection (weeks 22 and 23 post-HSCT), and as shown in Figure 5B, there was robust cellular activity detected during the period of active infection.
Patient 3 is a 60-year-old man with high-risk acute myeloid leukemia who received an allogeneic HSCT, complicated by grade 3/4 lower gastrointestinal and skin graft-vs-host disease, requiring tacrolimus and prednisone. At week 27 post-HSCT, the patient developed rhinorrhea and mild cough, and a nasal wash was positive for PIV3. He was initially followed as an outpatient but developed worsening dyspnea and was hospitalized at week 28 with multifocal pneumonitis on a computed tomography (CT) scan of his chest. A subsequent bronchoscopy and bronchoalveolar lavage revealed the presence of only PIV3. He received empiric antibiotic therapy, 4 days of inhaled ribavirin, and tapering of prednisone, and within 1 week his symptoms had improved and his radiological abnormalities had resolved as confirmed by repeat CT (Figure 5C). The T-cell immune response, measured during infection, showed a several-fold rise in the frequency of PIV3-specific T cells (Figure 5D), coincident with clinical improvement.
In contrast, patient 4, a 4-year-old girl with very high-risk ALL, and patient 5, a 3-year-old boy with an immune dysregulation syndrome, developed possible PIV3 LRTIs (RT-PCR positive for PIV3 from nasal wash, LRTI symptoms, and new pulmonary infiltrates on radiography [28]), at weeks 25 and 3 post-HSCT, respectively, requiring supplemental oxygen and prolonged hospitalization. Following 3 weeks of inhaled ribavirin, PIV3 became undetectable in patient 4. However, patient 5 required 13 weeks of inhaled ribavirin therapy, interrupted once for a 1-week trial of DAS-181 that was discontinued due to transaminitis. During this time, he remained persistently hypoxic and his nasal wash was consistently positive for PIV3 until week 17 posttransplant. Furthermore, in contrast to the previous examples, neither of these patients developed immune responses to PIV3 (patient 4: 12 SFC/5 × 105 PBMCs [week 26 post-HSCT] vs 12 SFC 5 × 105 PBMCs [week 34 post-HSCT]; patient 5: 6 SFC/5 × 105 PBMCs [week 5 post-HSCT] vs 6 SFC 5 × 105 PBMCs (week 13 post-HSCT]) (Supplementary Figure 3A and 3B).
DISCUSSION
In this study, we characterized the T-cell immune response to the 7 antigens encoded by PIV3 and established a hierarchy of immunogenicity based on the profile of T-cell activity detected in 17 healthy virus-exposed individuals. Overall, M, HN, N, and F were the most frequently recognized antigens with activity detected in 17, 16, 14, and 13 individuals, respectively. Moreover, we detected endogenous T-cell activity to M, HN, N, and F in the peripheral blood of 3 HSCT recipients with active PIV3-associated URTIs (patients 1 and 2) and LRTI (patient 3) who cleared their infections, while the absence of reactive T cells contributed to prolonged possible LRTI in 2 other patients. These results demonstrate the in vivo relevance of PIV3-reactive T cells in mediating antiviral activity. Finally, we show that PIV3-specific T cells, which produce multiple effector molecules and kill antigen-loaded targets, can be readily expanded ex vivo using current Good Manufacturing Practice (GMP)–compliant manufacturing methodologies, providing rationale for a future clinical trial of adoptively transferred T cells to treat PIV3 infections in immunocompromised patients. Whether the induced cells also cross-reactively recognize other PIV serotypes is yet to be determined.
PIV is a small, negative-stranded enveloped RNA virus from the Paramyxoviridae family, which can be divided into 4 serotypes (PIV1–4) based on antigenic and genetic determinants. PIV3 is most frequently detected (52%), followed by PIV1 (26%), PIV2 (12%), and PIV4 (4%) [29]. In rare cases, PIV has been linked to febrile seizures, acute encephalitis, Guillain-Barré syndrome, and meningitis, but it is most commonly associated with childhood respiratory tract infections/disease such as croup (laryngotracheobronchitis) (PIV1 and PIV2) and bronchiolitis and pneumonia (PIV3) [30–32]. Indeed, PIV3 is a common cause of acute respiratory infection–associated hospitalization in children [10, 29]. In immunocompetent adults, PIV3-associated symptoms are generally mild and self-limiting, but in immunocompromised individuals (eg, recipients of stem cell and solid organ transplantation, HIV/AIDS [33], and primary immune deficiencies), PIV3 infections can cause significant morbidity and mortality. For example, nosocomial outbreaks of PIV3 have been linked to respiratory failure and mortality rates of up to 60% in HSCT recipients with severe disease [34–36], while in lung transplant recipients, PIV3-associated bronchiolitis and pneumonia have been linked with acute and chronic lung allograft rejection [37, 38].
The therapeutic options for PIV3 infections are limited. Ribavirin is a synthetic guanosine analogue with broad- spectrum activity against a range of DNA and RNA viruses. However, in immunocompromised hosts, it neither reduces viral shedding nor decreases the mortality associated with PIV3-related pneumonia and hence is not recommended for the treatment of PIV3 infections [23, 39]. DAS 181 is an inhaled sialidase fusion protein preventing viral attachment to the host cell, with demonstrated success in case reports [40, 41]. In a study of 16 HSCT recipients with PIV-associated URTIs and probable/proven pneumonia, administration of DAS 181 led to a complete response (defined as symptomatic improvement or resolution of cough/dyspnea/fever and/or hypotension) in 9 patients [42]. However, a prior study noted that prolonged administration (>7 days) was associated with a decline in lung function (as measured by forced expiratory volume in 1 second [FEV1]), hypersensitivity pneumonitis, and development of neutralizing antibodies to DAS 181 [43]. A randomized controlled trial of DAS 181 is currently underway in HSCT recipients. Given the safe and effective use of adoptively transferred T cells to treat other clinically problematic viruses [1, 4–6], we set out to identify appropriate PIV3 antigens against which an effector T-cell response might be effectively generated.
The identification of immunogenic viral antigens and the demonstration that reactive cells provide protective immunity in vivo are essential precursors for the development of an immune-based therapy. Though the immune response to PIV3 had not been previously studied, the clinical importance of reactive T cells was inferred by the numerous clinical studies reporting persistent and often fatal PIV infections occurring in patients with compromised T-cell immunity, which could not be compensated by antibody (intravenous immunoglobulin) therapy [23, 39]. Thus, we sought to characterize the T-cell response against all 7 encoded PIV3 antigens to identify those that will induce protective immunity.
Unlike other viruses (eg, AdV) that our group has immunologically profiled, the circulating frequency of PIV3-specific T cells in peripheral blood was low (PIV3: range, 3–127 SFC/5 × 105 [n = 17] vs AdV: range, 49–408 SFC/5 × 105 [n = 14]) [44]. However, with a single in vitro stimulation, we were able to selectively expand reactive T cells from all 17 healthy donors tested, with detectable activity against ≥3 antigens in all donors. Recognition was mediated predominantly by antigen-specific CD4+ T cells, similar to the profile of activity detected against AdV. Both CD4+ and CD8+ PIV3-reactive populations produced Th1-polarized effector molecules, including IFN-γ, TNF-α and granzyme B, and were able to kill antigen-loaded targets, alluding to the potential clinical benefit associated with the adoptive transfer of T cells reactive against the dominant target antigens M, HN, N, and F. Finally, to assess the clinical significance of these findings, we monitored 5 HSCT recipients with active PIV3 infections. Three patients successfully controlled the virus within 2–4 weeks, and clearance correlated with an elevation in T-cell activity against M, HN, N, and F (Figure 5). However, the 2 patients who lacked detectable PIV3-specific T-cell activity developed possible LRTIs with radiographic pneumonia and required prolonged inhaled ribavirin treatment (3 and 13 weeks, respectively) (Supplementary Figure 3).
Taken together, this evidence suggests that PIV3-specific T cells with specificity for M, HN, N, and F support viral control in vivo. Furthermore, given that our in vitro manufacturing processes are GMP-compliant, we conclude that adoptive T-cell transfer for PIV3 is feasible, either as a single virus-specific product or incorporated into our multivirus product [1], which currently targets AdV, EBV, CMV, BK virus, and HHV-6, to support broad-spectrum antiviral protection in immunocompromised patients.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Financial support. This work was supported by the Flow Cytometry and Cell and Vector Production shared resources in the Dan L. Duncan Comprehensive Cancer Center (support grant P30 CA125123). R. J. A. and P. I. A.-H. are supported by the National Institutes of Health (grant numbers T32 DK060445-11 and T32 HL92332-12, respectively). J. F. V. is supported by a Mentored Research Scholars Grant in Applied and Clinical Research (grant number MRSG-14-197-01-LIB) from the American Cancer Society.
Potential conflicts of interest. A. M. L., J. F. V., I. T., and P. I. A.-H. have filed for intellectual property and submitted a patent application. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Papadopoulou A, Gerdemann U, Katari UL et al. Activity of broad-spectrum T cells as treatment for AdV, EBV, CMV, BKV, and HHV6 infections after HSCT. Sci Transl Med 2014; 6:242ra83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Leen AM, Christin A, Myers GD et al. Cytotoxic T lymphocyte therapy with donor T cells prevents and treats adenovirus and Epstein-Barr virus infections after haploidentical and matched unrelated stem cell transplantation. Blood 2009; 114:4283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heslop HE, Brenner MK, Rooney CM. Donor T cells to treat EBV-associated lymphoma. N Engl J Med 1994; 331:679–80. [DOI] [PubMed] [Google Scholar]
- 4. Gerdemann U, Katari UL, Papadopoulou A et al. Safety and clinical efficacy of rapidly-generated trivirus-directed T cells as treatment for adenovirus, EBV, and CMV infections after allogeneic hematopoietic stem cell transplant. Mol Ther 2013; 21:2113–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Leen AM, Bollard CM, Mendizabal AM et al. Multicenter study of banked third-party virus-specific T cells to treat severe viral infections after hematopoietic stem cell transplantation. Blood 2013; 121:5113–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Doubrovina E, Oflaz-Sozmen B, Prockop SE et al. Adoptive immunotherapy with unselected or EBV-specific T cells for biopsy-proven EBV+ lymphomas after allogeneic hematopoietic cell transplantation. Blood 2012; 119:2644–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fuji S, Kapp M, Grigoleit GU, Einsele H. Adoptive immunotherapy with virus- specific T cells. Best Pract Res Clin Haematol 2011; 24:413–9. [DOI] [PubMed] [Google Scholar]
- 8. Gerdemann U, Keukens L, Keirnan JM et al. Immunotherapeutic strategies to prevent and treat human herpesvirus 6 reactivation after allogeneic stem cell transplantation. Blood 2013; 121:207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Henrickson KJ. Parainfluenza viruses. Clin Microbiol Rev 2003; 16:242–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abedi GR, Prill MM, Langley GE et al. Estimates of parainfluenza virus- associated hospitalizations and cost among children aged less than 5 years in the United States, 1998–2010. J Pediatric Infect Dis Soc 2016; 5:7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bose S, Kar N, Maitra R, DiDonato JA, Banerjee AK. Temporal activation of NF-kappaB regulates an interferon-independent innate antiviral response against cytoplasmic RNA viruses. Proc Natl Acad Sci U S A 2003; 100:10890–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lewandowska-Polak A, Brauncajs M, Paradowska E et al. Human parainfluenza virus type 3 (HPIV3) induces production of IFNγ and RANTES in human nasal epithelial cells (HNECs). J Inflamm (Lond) 2015; 12:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spriggs MK, Murphy BR, Prince GA, Olmsted RA, Collins PL. Expression of the F and HN glycoproteins of human parainfluenza virus type 3 by recombinant vaccinia viruses: contributions of the individual proteins to host immunity. J Virol 1987; 61:3416–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mostafa HH, Vogel P, Srinivasan A, Russell CJ. Non-invasive imaging of sendai virus infection in pharmacologically immunocompromised mice: NK and T cells, but not neutrophils, promote viral clearance after therapy with cyclophosphamide and dexamethasone. PLoS Pathog 2016; 12:e1005875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tremonti LP, Lin JS, Jackson GG. Neutralizing activity in nasal secretions and serum in resistance of volunteers to parainfluenza virus type 2. J Immunol 1968; 101:572–7. [PubMed] [Google Scholar]
- 16. Cortez KJ, Erdman DD, Peret TC et al. Outbreak of human parainfluenza virus 3 infections in a hematopoietic stem cell transplant population. J Infect Dis 2001; 184:1093–7. [DOI] [PubMed] [Google Scholar]
- 17. Shah DP, Shah PK, Azzi JM, Chemaly RF. Parainfluenza virus infections in hematopoietic cell transplant recipients and hematologic malignancy patients: a systematic review. Cancer Lett 2016; 370:358–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sydnor ER, Greer A, Budd AP et al. An outbreak of human parainfluenza virus 3 infection in an outpatient hematopoietic stem cell transplantation clinic. Am J Infect Control 2012; 40:601–5. [DOI] [PubMed] [Google Scholar]
- 19. Toupin M, Hamadah A, Madore S, Padmore R, Allan DS. Impact of parainfluenza virus type 3 infection on engraftment after hematopoietic SCT. Bone Marrow Transplant 2012; 47:451–2. [DOI] [PubMed] [Google Scholar]
- 20. Erard V, Chien JW, Kim HW et al. Airflow decline after myeloablative allogeneic hematopoietic cell transplantation: the role of community respiratory viruses. J Infect Dis 2006; 193:1619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chemaly RF, Hanmod SS, Rathod DB et al. The characteristics and outcomes of parainfluenza virus infections in 200 patients with leukemia or recipients of hematopoietic stem cell transplantation. Blood 2012; 119:2738–45; quiz 969. [DOI] [PubMed] [Google Scholar]
- 22. Elizaga J, Olavarria E, Apperley J, Goldman J, Ward K. Parainfluenza virus 3 infection after stem cell transplant: relevance to outcome of rapid diagnosis and ribavirin treatment. Clin Infect Dis 2001; 32:413–8. [DOI] [PubMed] [Google Scholar]
- 23. Nichols WG, Corey L, Gooley T, Davis C, Boeckh M. Parainfluenza virus infections after hematopoietic stem cell transplantation: risk factors, response to antiviral therapy, and effect on transplant outcome. Blood 2001; 98:573–8. [DOI] [PubMed] [Google Scholar]
- 24. Durbin AP, Karron RA. Progress in the development of respiratory syncytial virus and parainfluenza virus vaccines. Clin Infect Dis 2003; 37:1668–77. [DOI] [PubMed] [Google Scholar]
- 25. Moscona A. Entry of parainfluenza virus into cells as a target for interrupting childhood respiratory disease. J Clin Invest 2005; 115:1688–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kannanganat S, Ibegbu C, Chennareddi L, Robinson HL, Amara RR. Multiple-cytokine-producing antiviral CD4 T cells are functionally superior to single- cytokine-producing cells. J Virol 2007; 81:8468–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lachmann R, Bajwa M, Vita S et al. Polyfunctional T cells accumulate in large human cytomegalovirus-specific T cell responses. J Virol 2012; 86:1001–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seo S, Xie H, Campbell AP et al. Parainfluenza virus lower respiratory tract disease after hematopoietic cell transplant: viral detection in the lung predicts outcome. Clin Infect Dis 2014; 58:1357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fry AM, Curns AT, Harbour K, Hutwagner L, Holman RC, Anderson LJ. Seasonal trends of human parainfluenza viral infections: United States, 1990–2004. Clin Infect Dis 2006; 43:1016–22. [DOI] [PubMed] [Google Scholar]
- 30. Olivares F, Salinas M, Soto A, Dabanch J, Fica A. Severe acute disseminated encephalomyelitis associated with parainfluenza 3 infection: case report [in Spanish]. Rev Chilena Infectol 2015; 32:476–81. [DOI] [PubMed] [Google Scholar]
- 31. Arisoy ES, Demmler GJ, Thakar S, Doerr C. Meningitis due to parainfluenza virus type 3: report of two cases and review. Clin Infect Dis 1993; 17:995–7. [DOI] [PubMed] [Google Scholar]
- 32. Branche AR, Falsey AR. Parainfluenza virus infection. Semin Respir Crit Care Med 2016; 37:538–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cohen AL, Sahr PK, Treurnicht F et al. parainfluenza virus infection among human immunodeficiency virus (HIV)-infected and HIV-uninfected children and adults hospitalized for severe acute respiratory illness in South Africa, 2009–2014. Open Forum Infect Dis 2015; 2:ofv139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hodson A, Kasliwal M, Streetly M, MacMahon E, Raj K. A parainfluenza-3 outbreak in a SCT unit: sepsis with multi-organ failure and multiple co-pathogens are associated with increased mortality. Bone Marrow Transplant 2011; 46:1545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewis VA, Champlin R, Englund J et al. Respiratory disease due to parainfluenza virus in adult bone marrow transplant recipients. Clin Infect Dis 1996; 23:1033–7. [DOI] [PubMed] [Google Scholar]
- 36. Maziarz RT, Sridharan P, Slater S et al. Control of an outbreak of human parainfluenza virus 3 in hematopoietic stem cell transplant recipients. Biol Blood Marrow Transplant 2010; 16:192–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Khalifah AP, Hachem RR, Chakinala MM et al. Respiratory viral infections are a distinct risk for bronchiolitis obliterans syndrome and death. Am J Respir Crit Care Med 2004; 170:181–7. [DOI] [PubMed] [Google Scholar]
- 38. Billings JL, Hertz MI, Savik K, Wendt CH. Respiratory viruses and chronic rejection in lung transplant recipients. J Heart Lung Transplant 2002; 21:559–66. [DOI] [PubMed] [Google Scholar]
- 39. Chakrabarti S, Collingham KE, Holder K, Oyaide S, Pillay D, Milligan DW. Parainfluenza virus type 3 infections in hematopoetic stem cell transplant recipients: response to ribavirin therapy. Clin Infect Dis 2000; 31:1516–8. [DOI] [PubMed] [Google Scholar]
- 40. Waghmare A, Wagner T, Andrews R et al. Successful treatment of parainfluenza virus respiratory tract infection with DAS181 in 4 immunocompromised children. J Pediatric Infect Dis Soc 2015; 4:114–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen YB, Driscoll JP, McAfee SL et al. Treatment of parainfluenza 3 infection with DAS181 in a patient after allogeneic stem cell transplantation. Clin Infect Dis 2011; 53:e77–80. [DOI] [PubMed] [Google Scholar]
- 42. Salvatore M, Satlin MJ, Jacobs SE et al. DAS181 for treatment of parainfluenza virus infections in hematopoietic stem cell transplant recipients at a single center. Biol Blood Marrow Transplant 2016; 22:965–70. [DOI] [PubMed] [Google Scholar]
- 43. Zenilman JM, Fuchs EJ, Hendrix CW et al. Phase 1 clinical trials of DAS181, an inhaled sialidase, in healthy adults. Antiviral Res 2015; 123:114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Leen AM, Sili U, Vanin EF et al. Conserved CTL epitopes on the adenovirus hexon protein expand subgroup cross-reactive and subgroup-specific CD8+ T cells. Blood 2004; 104:2432–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.