Infection with influenza B viruses of both the B/Yamagata/16/88-like and the B/Victoria/2/87-like lineage induced hemagglutinin (HA)–specific antibodies with antibody-dependent cellular cytotoxicity (ADCC) activity in humans. Only antibodies directed to the HA stalk were capable of mediating ADCC and displayed lineage cross-reactivity.
Keywords: Influenza B virus, hemagglutinin, antibodies, natural killer cells, antibody-dependent cellular cytotoxicity
Abstract
Influenza A virus (IAV) and influenza B virus (IBV) cause substantial morbidity and mortality during annual epidemics. Two distinct lineages of IBV are distinguished, based on variation in hemagglutinin (HA): B/Victoria/2/87-like (B/Vic) and B/Yamagata/16/88-like (B/Yam). Here, we show that, in humans, primary IBV infection with either lineage induces HA-specific antibody-dependent cellular cytotoxicity (ADCC)–mediating antibodies. IBV infection induced antibodies specific to the HA head and stalk, but only HA stalk–specific antibodies mediated ADCC efficiently and displayed cross-reactivity with IBV of both lineages. This corresponds to recent findings that 2 points of contact between the effector and target cell (ie, HA and sialic acid, respectively, and the fragment crystallizable [Fc] domain and Fcγ receptor IIIα, respectively) are required for efficient ADCC activity and that antibodies specific for the receptor-binding site located in the head domain of HA therefore fail to mediate ADCC. Potentially, ADCC-mediating antibodies directed to the HA stalk of IBV contribute to cross-protective immunity to IBV of both lineages.
(See the editorial commentary by Ramilo, on pages 1–2.)
Influenza viruses cause 3–5 million cases of severe illness and half a million deaths worldwide per year during annual epidemics [1]. Cocirculating influenza A viruses (IAV; subtypes H1N1 and H3N2) and influenza B virus (IBV) are responsible for this morbidity and mortality. Like IAV, IBV displays antigenic drift by accumulating mutations in the gene encoding hemagglutinin (HA), resulting in viral escape from recognition by neutralizing antibodies induced by previous infections or vaccinations [2–4]. However, antigenic drift of IBV is less pronounced than that of IAV [5].
IBV possesses 2 major surface glycoproteins, HA and neuraminidase (NA). Based on antigenic and genetic variation of HA, 2 distinct lineages of IBV that cocirculate globally are distinguished, the B/Victoria/2/87-like (B/Vic) lineage and the B/Yamagata/16/88-like (B/Yam) lineage [6–8]. The earliest IBV isolate originates from the 1940s [9], and it is currently thought that the 2 lineages diverged in the 1970s [2, 6, 10, 11]. Although the IBV lineages are often compared to the different IAV subtypes, the homology of the HA amino acid sequence between the 2 lineages of IBV is higher than between the 2 subtypes of seasonal IAV [7].
The primary protective measure against IBV infection is vaccination. Currently used seasonal influenza vaccines aim to induce virus-neutralizing antibodies directed to the variable head domain of HA and are mainly trivalent inactivated vaccines, which contain IBV of a single lineage, in addition to influenza A(H1N1) and A(H3N2) components. However, IBV lineage mismatches between circulating IBV strains and the strains used for trivalent inactivated vaccine production frequently occur [12–14]. Notably, in the 10 seasons between 2001–2002 and 2010–2011, the IBV lineage selected for inclusion in the seasonal TIV only matched the dominant circulating IBV lineage 5 times, with the lineage of 46% and 58% of strains not represented in the vaccine in the United States and Europe, respectively [12, 14]. Vaccine effectiveness is significantly higher against a vaccine-matched IBV strain than against a strain from the opposing lineage, proving that vaccine-induced antibodies cross-neutralize poorly and that the 2 lineages are antigenically distinct [14–16]. The difficulty of predicting which IBV lineage will circulate in the coming season, combined with limited cross-reactivity in the case of a vaccine mismatch, forms the basis for the rationale to include IBVs of both lineages in quadrivalent seasonal vaccines.
Virus-specific antibodies are induced upon natural infection with or vaccination against IBV. Antibodies directed to antigenic sites in or around the receptor-binding site located in the head domain of HA have the capacity to directly neutralize IBV. In addition to direct neutralization, virus-specific antibodies can have other modes of action, in particular those of the immunoglobulin G1 (IgG1) and IgG3 subclasses [17]. After antibody binding to antigens displayed on virus-infected cells, the fragment crystallizable (Fc) domain of these antibodies engages Fc receptors on host effector cells that subsequently kill virus-infected cells through antibody-dependent cellular cytotoxicity (ADCC) [18, 19], antibody-dependent phagocytosis [20, 21], or complement-dependent cytotoxicity [22]. Specifically, ADCC is predominantly induced by interaction between the Fc region of virus-specific antibodies and Fcγ receptor IIIα (FcγRIIIα; also known as CD16), present on natural killer (NK) cells. IAV-specific monoclonal antibodies directed to the conserved HA stalk domain were shown to have protective efficacy through Fcγ receptor–dependent mechanisms in mice [19, 23, 24].
Here, we analyzed the presence of functional HA-specific ADCC-mediating antibodies in serum samples obtained from children after primary infection with IBV from either lineage. Using recombinant full-length HA from both lineages in solid-phase ADCC assays, we demonstrated that primary IBV infection induces HA-specific ADCC-mediating antibodies and that these antibodies cross-reacted with HA from IBV of the opposing lineage. By using both the HA1 subunit of HA and a chimeric HA protein that contains only the stalk region of IBV, we showed that cross-lineage reactive antibodies were directed to the HA stalk. Furthermore, IBV HA head–specific antibodies present in human sera were not capable of mediating ADCC.
Methods
Ethics Statement
Serum samples were obtained from healthy volunteers and subjects enrolled in another study (ie, the PIENTER 2 study) [25]. The Medical Ethical Committee of University Hospital, Erasmus Medical Center (MEC-2015–095), approved the collection of blood specimens from healthy volunteers; all participants gave informed consent and permission for use of materials. The work described has been carried out in accordance with the code of ethics of the World Medical Association (ie, the Declaration of Helsinki).
Study Subjects
Healthy Volunteers
Pilot experiments, assay setup, and end point titrations were performed with serum samples from healthy coworkers from the Erasmus Medical Center Department of Viroscience. Coworkers had received trivalent influenza vaccine at least once in previous years.
PIENTER 2 Participants
Serum samples were collected during a nationwide, cross-sectional, population-based study performed in the Netherlands from February 2006 to June 2007 (ie, the PIENTER 2 study) [25, 26]. For the study described here, 41 serum samples were selected from children 1–7 years of age who did not receive influenza vaccination (1:1 ratio of samples to children). The numbers of samples from children who were 1, 2, 3, 4, 5, 6, and 7 years of age were 3, 5, 5, 5, 6, 10, and 7, respectively. HI titers against multiple representative IAV and IBV strains (circulating or in vaccines between 1999–2000 and 2006–2007) were determined in the scope of another study [25]. Based on HI data, 18 subjects were categorized as B/Vic responders, and 23 were categorized as B/Yam responders (Tables 1 and 2). B/Vic and B/Yam responders were defined as individuals with HI antibodies against viruses of the corresponding lineage and the absence of antibodies against the opposing lineage.
Table 1.
Number of Sera From Responders to Influenza B Virus From the B/Victoria-Like Lineage (B/Vic) and Influenza B Virus From the B/Yamagata-Like Lineage (B/Yam), by Age
| Responders by Age, No. | |||||||
|---|---|---|---|---|---|---|---|
| Variable | 1 y | 2 y | 3 y | 4 y | 5 y | 6 y | 7 y |
| B/Vic responders (n = 18) | 1 | 2 | 2 | 2 | 2 | 5 | 4 |
| B/Yam responders (n = 23) | 2 | 3 | 3 | 3 | 4 | 5 | 3 |
Table 2.
Responses to Influenza A(H1N1) Virus, Influenza A(H3N2) Virus, Influenza B Virus From the B/Victoria-Like Lineage (B/Vic), and Influenza B Virus From the B/Yamagata-Like Lineage (B/Yam) Among Individuals With Responses to B/Vic or B/Yam
| Response,a No. (%) | ||||
|---|---|---|---|---|
| Variable | A(H1N1) | A(H3N2) | B/Vic | B/Yam |
| B/Vic responders (n = 18) | 7 (39) | 14 (78) | 15 (83) | 0 (0) |
| B/Yam responders (n = 23) | 11 (48) | 21 (91) | 0 (0) | 23 (100) |
B/Vic or B/Yam responders were defined on the basis of the presence of HI serum antibodies against viruses of the corresponding lineage and the absence of antibodies against viruses of the opposing lineage. The highest HI titers obtained with representative A(H1N1), A(H3N2), B/Vic, and B/Yam strains from the relevant seasons are shown.
Abbreviation: Hi, hemagglutination inhibition.
Defined as individuals with an HI titer of ≥40 to the specified influenza A or B virus.
Detection of HA-Specific Antibodies by Enzyme-Linked Immunosorbent Assay (ELISA)
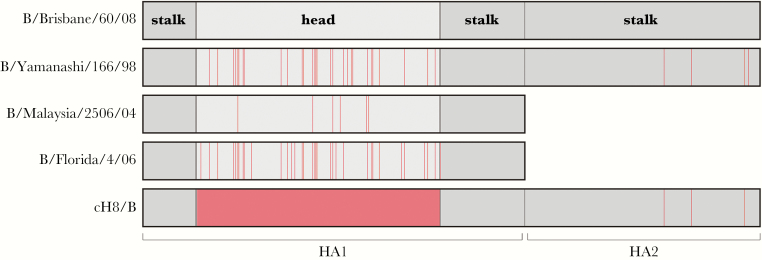
Ninety-six–well enzyme immunoassay/radioimmunoassay plates (Greiner) were coated overnight at 4°C with 50 ng/well of full-length HA derived from B/Yamanashi/166/98 (B/Yam-lineage) or B/Brisbane/60/08 (B/Vic-lineage), the HA1 subunit derived from B/Florida/4/06 (B/Yam-lineage) or B/Malaysia/2506/04 (B/Vic-lineage) or chimeric HA, containing the stalk of IBV B/Yamagata/16/88 and the head domain of the IAV A/mallard/Sweden/24/02 (cH8/B) [27] in 0.05 M carbonate/bicarbonate buffer (pH 9.6; Sigma Aldrich; Figure 1). Plates were blocked for 1 hour at room temperature, using phosphate-buffered saline (PBS) with 0.05% Tween (PBST; pH 7.2; Thermo Fisher Scientific) supplemented with 2% (w/v) milk powder (Semper). A 5-fold dilution series of serum samples was prepared in blocking buffer starting at 1:40 and incubated on the coated plates for 1 hour at room temperature. A positive control (serum from an individual with a strong response to IBV) and negative controls (pooled sera from influenza virus–naive children; positive and negative controls were on antigen-coated plates) were included. In addition, mock-coated plates were used as negative control. Plates were washed with PBST and incubated with anti-human IgG horseradish peroxidase for 1 hour at room temperature. Subsequently, plates were washed with PBST, and 3,3ʹ,5,5ʹ-tetramethylbenzidine peroxidase substrate (Svanova Biotech) was added. Reactions were stopped with 1.8 M H2SO4, and absorbance was measured at 450 nm by using a SpectraMax Plus 384 Microplate Reader (Molecular Devices). End point titers were determined as the reciprocal of the highest dilution at which the OD450 was below background (defined as 3 times the OD450 of PBS controls) and used for subsequent analysis.
Figure 1.
Schematic overview of hemagglutinin (HA) antigens used in the enzyme-linked immunosorbent assay and antibody-dependent cellular cytotoxicity assay. B/Brisbane/60/08 was used as a reference sequence, and mutations relative to B/Brisbane/60/08 in the various antigens are shown in red. The cH8/B antigen contains the globular head of an influenza A virus of the H8 subtype, and the stalk in from B/Yamagata/16/88.
ADCC Assay
We measured the presence of ADCC-mediating antibodies in serum following interaction with purified full-length recombinant HA from representative IBVs, B/Yamanashi/166/98 (B/Yam) and B/Brisbane/60/08 (B/Vic, made available by Protein Sciences). To determine the head or stalk specificity of the ADCC-mediating antibodies, assays were performed with the HA1 subunit from B/Florida/4/06 (B/Yam) and B/Malaysia/2506/04 (B/Vic) or with chimeric HA consisting of the head domain of an H8 IAV (A/mallard/Sweden/24/02) and the stalk of IBV B/Yamagata/16/88 (cH8/B) (Figure 1). An ADCC assay was performed as described previously [28]. High-binding, 96-well, flat-bottomed plates (Immunolon) were coated overnight at 4°C (200 ng/well), blocked, washed with a set dilution (1:160) of serum, and incubated at 37°C. Appropriate positive and negative controls were included. After 2 hours, the continuous NK cell-line NK92.05-CD16 [29] was added (1 × 105 cells/well), and plates were incubated for 5 hours at 37°C in the presence of Golgistop, Golgiplug, and a human monoclonal antibody (labeled with V450) directed to the degranulation marker CD107a (all from BD Biosciences). Subsequently, cells were stained with fixable Live/Dead (Molecular Probes) and CD56 (labeled with PE; BD Biosciences) and acquired on a FACS Canto II (BD Biosciences); percentages of degranulating CD107a+ cells were determined within the respective Live-stained CD56+ NK cell populations.
Determination of ADCC End Point Titers
ADCC end point titers were determined as described previously [28]. Sera were tested as outlined above in a 4-fold serial dilution series (from 1:10 to 1:163840), and the percentages of CD107a+ NK92.05-CD16 cells were calculated by subtracting the background values obtained with mock-coated plates from the values obtained with HA-coated plates. The highest serum dilution still resulting in a population in which >5% of cells were CD107a+ NK92.05-CD16s, which was chosen as a cutoff, was recorded as the end point ADCC serum antibody titer. This cutoff represents the background value obtained with the negative serum pool, multiplied by ±3.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism. CD107a+ percentages obtained with different antigens were compared by performing a nonparametric Wilcoxon rank test or a Mann-Whitney test on paired or unpaired serum samples, respectively. A nonparametric Friedman test involving paired samples with multiple comparisons was performed to compare the ELISA and ADCC data obtained with the different antigens in a side-by-side experiment.
RESULTS
ADCC End Point Titration and Selection of Optimal Serum Dilution
Sera obtained from 2 coworkers positive for IBV-specific HI antibodies were analyzed for the presence of ADCC-mediating antibodies against full-length HA from both IBV lineages (B/Brisbane/60/08 and B/Yamanashi/166/98). A serum pool obtained from seronegative children and intravenous immunoglobulin (IVIG) were used as negative and positive controls, respectively. End point B/Vic- and B/Yam-specific ADCC titers were determined by creating 4-fold serial dilutions of serum samples; an arbitrary cutoff (end point) for presence of ADCC-mediating antibodies was set at 5% CD107a+ NK92.05-CD16 cells (Supplementary Figure 1A and 1B). At a 1:10 dilution and occasionally at dilutions of 1:40 and 1:160, lower percentages of CD107a+ cells were observed than at higher dilutions, indicative of a prozonal effect. B/Vic- and B/Yam-specific ADCC-mediating antibodies were in general equally present and detected with similar CD107a+ percentages at a set dilution and end point titers (Supplementary Figure 1A and 1B). Since, at a 1:160 serum dilution, CD107a+ NK cells were readily detected with minimal prozonal effects, this dilution was chosen for subsequent experiments.
First-Time IBV Infection of Children Induces ADCC-Mediating Antibodies
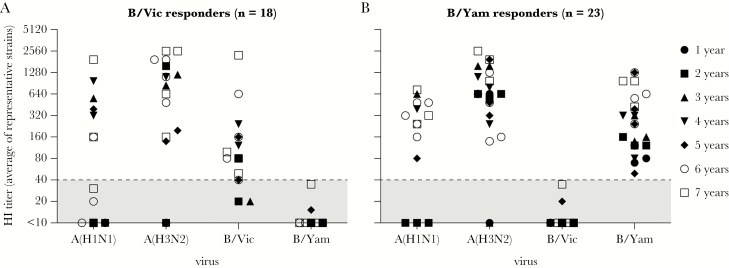
Sera from 41 pediatric subjects aged 1–7 years with serological evidence of first-time exposure to IBV of either lineage were selected (Table 1). Based on HI data that were generated with various epidemic IBV strains that these subjects had potentially been exposed to [25], sera from subjects with evidence of infection with the B/Vic (n = 18) or B/Yam (n = 23) lineage were identified (B/Vic or B/Yam responders, respectively; Table 2). All subjects had detectable HI titers against ≥1 epidemic IBV strains from a single lineage, but not all subjects reached HI titers of ≥40. In addition, most subjects had A(H1N1)- and A(H3N2)-specific HI antibodies, indicating that they had experienced 1 or multiple IAV infections (Figure 2A and 2B show the highest HI titers obtained with representative IAV and IBV strains).
Figure 2.
Influenza A virus (IAV)– and influenza B virus (IBV)–specific hemagglutination inhibition (HI) responses in selected subjects. HI titers against representative influenza A(H1N1), influenza A(H3N2), influenza B/Vic, and influenza B/Yam viruses circulating in the relevant seasons were determined for 41 selected subjects in the scope of another study. Subjects were categorized as B/Vic (n = 18; A) or B/Yam (n = 23; B) responders. A, HI antibodies against B/Vic were detected in all responders; 15 of 18 subjects reached HI titers of ≥40. HI titers of ≥40 specific for B/Yam were not detected. B, HI antibodies against B/Yam were detected in 23 of 23 subjects; all reached HI titers of ≥40. HI titers of ≥40 specific for B/Vic were not detected.
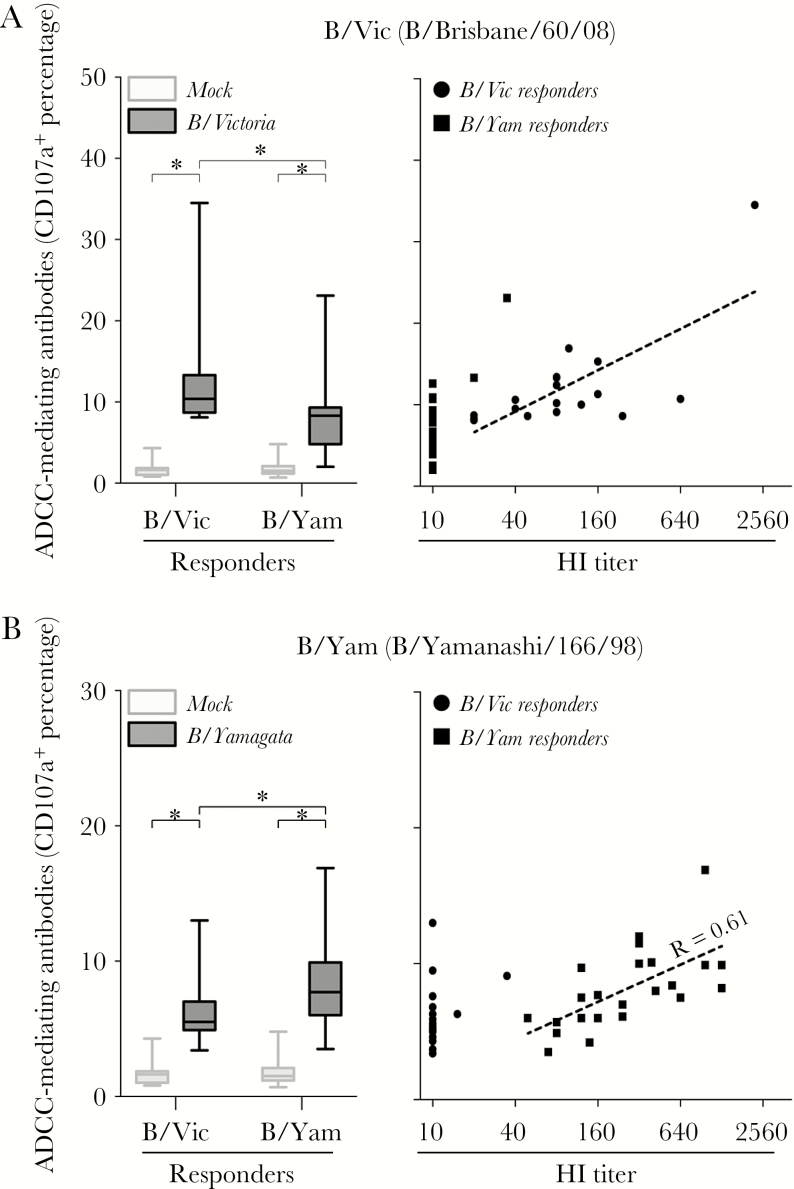
The presence of ADCC-mediating antibodies directed to the HA of either IBV lineage in B/Vic and B/Yam responders was determined by a solid-phase ADCC assay, using representative full-length HAs from both IBV lineages (ie, B/Brisbane/60/08 [B/Vic] and B/Yamanashi/166/98 [B/Yam]) as antigens. Significant levels of HA-specific ADCC-mediating antibodies reactive with B/Vic and B/Yam HA were detected in B/Vic responders and B/Yam responders, respectively (P < .0001), indicating that infection induced ADCC-mediating antibodies specific for HA of the corresponding lineage (Figure 3A and 3B). In both B/Vic and B/Yam responders, limited correlation was observed between ADCC and HI antibody titers directed to IBV of the homologous lineage (the slopes were significantly different from 0; Figure 3A and 3B). Interestingly, ADCC activity to HA of the heterologous IBV lineage was also detected; the ADCC activity to HA of B/Yam was detected in B/Vic responders (P < .0001; Figure 3A), and the ADCC activity to HA of B/Vic was detected in B/Yam responders (P < .0001; Figure 3B). ADCC activity to HA of the heterologous lineage was significantly lower than to that of the corresponding lineage for both B/Vic (P = .0022; Figure 3A) and B/Yam (P = .0136; Figure 3B) responders. Notably, these HA-specific cross-reactive ADCC-mediating antibodies were detected in the absence of HI antibodies against the heterologous lineage.
Figure 3.
Hemagglutinin (HA)–specific antibody-dependent cellular cytotoxicity (ADCC)–mediating antibodies after primary influenza B virus (IBV) infection. ADCC responses against a representative B/Vic antigen (B/Brisbane/60/08; A) and a representative B/Yam antigen (B/Yamanashi/166/98; B) were determined in both B/Vic and B/Yam responder sera. A, Statistically significantly higher CD107a+ percentages were detected after stimulation with B/Vic in both the B/Vic and B/Yam responders as compared to the phosphate-buffered saline (PBS) control (left panel). B/Vic-specific responses were significantly higher in the B/Vic responders as compared to the B/Yam responders. Correlation between B/Vic hemagglutination inhibition (HI) titers and ADCC percentages (right panel). B, Statistically significantly higher CD107a+ percentages were detected after stimulation with B/Yam in both the B/Vic and B/Yam responders as compared to the PBS control (left panel). B/Yam-specific responses were significantly higher in the B/Yam responders as compared to the B/Vic responders. Correlation between B/Yam HI titers and ADCC percentages (right panel). CD107a+ percentages obtained with different antigens were compared by performing a nonparametric Wilcoxon rank test or Mann-Whitney test on paired or unpaired serum samples, respectively.
IBV Infection Induces HA Head–Specific and HA Stalk–Specific Antibodies
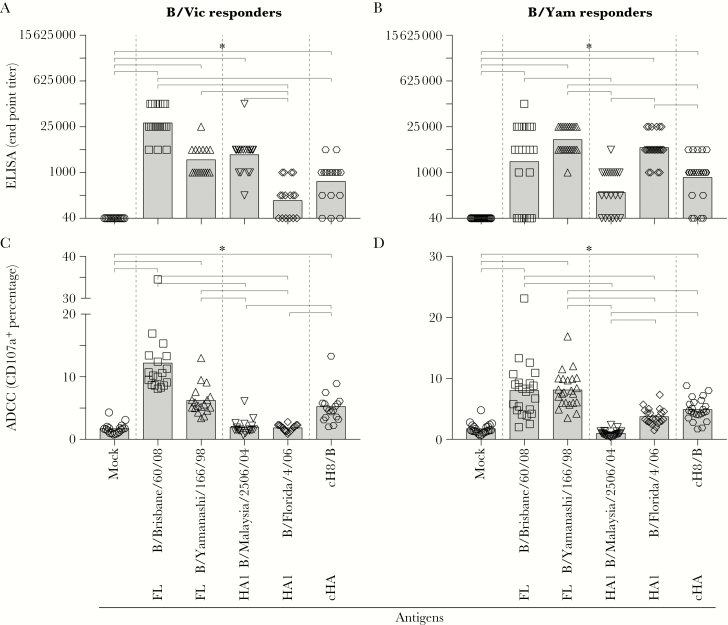
To determine whether IBV infection induced antibodies specific to the HA head and/or stalk, we performed an ELISA to assess the presence of serum antibodies to full-length HA and the HA1 subunit of both lineages or a chimeric HA (representing the IBV stalk only). Representative full-length HAs (B/Brisbane/60/08 [B/Vic] and B/Yamanashi/166/98 [B/Yam]) and representative HA1 subunits (B/Malaysia/2506/04 [B/Vic] and B/Florida/04/06 [B/Yam]) from both IBV lineages were used as antigens. The chimeric HA consisted of the head domain of an irrelevant IAV (H8 subtype) and the stalk domain of IBV B/Yamagata/16/88 (cH8/B) (Figure 1). Since humans presumably do not possess antibodies to IAV of the H8 subtype, cH8/B-reactive antibodies are likely specific for the IBV HA stalk.
In both B/Vic and B/Yam responders, antibodies recognizing the corresponding full-length HA were detected by ELISA (P < .0001 for both comparisons; Figure 4A and 4B). Antibodies reactive with heterologous full-length HAs were also detected in both responder groups (P < .0001 for both comparisons), indicating the presence of antibodies that react across both IBV lineages. However, when the HA1 subunit was used as antigen, antibodies reactive with HA1 of the corresponding lineage (P < .0001 for both comparisons) were predominantly detected, and reactivity with HA1 of the opposing lineage was not detected (P > .9999 and P = .4509 for B/Vic and B/Yam responders, respectively). HA stalk–specific antibodies were detected in both B/Vic and B/Yam responders (P = .0288 and P = .0137, respectively), indicating that HA stalk–specific antibodies were likely responsible for the cross-reactivity observed against the full-length HA of the opposing IBV lineage. ELISA titers obtained with the HA1 subunit of the corresponding IBV lineage correlated with HI titers but not with those obtained with HA1 of the opposing lineage or cH8/B in both the B/Vic and B/Yam responders (data not shown).
Figure 4.
Detection of antibody-dependent cellular cytotoxicity [ADCC]–mediating hemagglutinin (HA) stalk–specific and HA head–specific antibodies after primary influenza B virus (IBV) infection. A and B, Antibodies against either the full-length (FL) HA of both lineages, the HA1 of both lineages, or the HA stalk (cHA, chimeric HA) after a primary IBV infection were detected by enzyme-linked immunosorbent assay (ELISA). End point titers were determined by creating 5-fold serial dilutions of serum samples, and a cutoff (end point) for the presence of specific antibodies was set at 3 times the background level. Statistically significantly higher end point titers were detected against the full-length HA of both lineages, the homologous HA1 subunit, and the HA stalk as compared to the phosphate-buffered saline (PBS) control. C and D, Presence of ADCC-mediating antibodies against either the full-length HA of both lineages, the HA1 of both lineages, or the HA stalk after a primary IBV infection. Statistically significantly higher CD107a+ were detected upon stimulation with the full-length HA of both lineages and the HA stalk as compared to the PBS control. Stimulation with the HA1 subunit only, homologous or heterologous, did not lead to significant differences as compared to the PBS control. A nonparametric Friedman test on paired samples with multiple comparisons was performed to compare the ELISA and ADCC data obtained with the different antigens.
HA Stalk–Specific Antibodies Are Responsible for ADCC
To resolve the specificity of cross-reactive ADCC, ADCC reactivity to the HA stalk, HA1 subunits, or full-length HAs was determined. Concordant with results of ELISA and the ADCC assays (Figure 3), ADCC activity cross-reactive with full-length HA of the opposing lineage was detected in both B/Vic and B/Yam responders (P < .0001 for all comparisons between full-length HA and mock; Figure 4C and 4D). Although antibodies reactive with the homologous HA1 were detected by ELISA, these antibodies failed to mediate ADCC to the HA1 of the corresponding and opposing IBV lineage in both B/Vic responders (P > .9999 and P > .9999, respectively) and B/Yam responders (P = .2023 and P = .3494, respectively), although limited reactivity with the B/Yam HA1 subunit was detected in B/Yam responders. In contrast, ADCC-mediating antibodies were readily detected to cH8/B in both B/Vic (P < .0001) and B/Yam (P = .0035) responders, indicating that ADCC activity was predominantly specific for the HA stalk. HA stalk–specific ADCC activity correlated significantly with ELISA end point titers and OD450 values measured with cH8/B (data not shown). In conclusion, infection of pediatric subjects with IBV of the B/Vic or B/Yam lineage induced both HA head–specific and HA stalk–specific antibodies, but only HA stalk–specific antibodies were capable of mediating ADCC across the 2 lineages of IBV.
DISCUSSION
Here, we assessed whether human infections with IBV induced ADCC-mediating antibodies and determined the cross-reactivity of these antibodies with IBV of the 2 antigenically distinct IBV lineages. To this end, serum samples obtained from children 1–7 years of age with serological evidence of having experienced an infection with IBV of either lineage were tested for the presence of IBV-specific ADCC activity. Infection of pediatric subjects with IBV of either the B/Vic or B/Yam lineage induced ADCC-mediating antibodies that were specific for HA of the corresponding lineage and that displayed cross-reactivity with HA of IBV from the opposing lineage. IBV infection induced both HA head–specific and HA stalk–specific binding antibodies as demonstrated by ELISA, but interestingly, only HA stalk–specific antibodies were capable of mediating ADCC.
First time infection with 2009 pandemic influenza A(H1N1) virus was previously shown to induce ADCC-mediating antibodies in children [28, 30], using paired preinfection and postinfection serum samples. Here, we used selected serum samples from young children who tested positive by HI for the presence of antibodies to IBV belonging to one of the phylogenetic lineages, B/Yam or B/Vic. These serum samples, obtained after primary IBV infection, were tested for the presence of ADCC-mediating antibodies. Similar to infection with IAV [28, 30], infection with IBV induced ADCC activity, which correlated with HI antibody titers to a limited extent.
The selected serum samples were obtained from 41 pediatric subjects, selected for presence of serum antibodies to IBV of a single lineage. Eighteen of these subjects were identified to be infected with an IBV of the B/Vic lineage, and 23 were infected with IBV of the B/Yam lineage. Of note, it has been shown that HI antibodies to IBV of one lineage fail to react with an IBV of the opposing lineage [7, 25]. Since the HI assay only detects antibodies directed to epitopes located in or near the receptor binding site of HA, we performed ELISA with full-length HA, the HA1 subunit, or chimeric HA cH8/B to demonstrate that primary IBV infection induced antibodies reactive with both the HA head and stalk. Cross-lineage reactive antibodies were detected by ELISA and proved to be mainly directed to the HA stalk. To determine whether HA-specific ADCC-mediating antibodies cross-reacted with IBV from the opposing lineage, sera from B/Vic responders were tested for B/Yam-specific ADCC activity, and sera from B/Yam responders were tested for B/Vic-specific ADCC activity. Of interest, cross-lineage ADCC activity was observed both in B/Vic responders and B/Yam responders and was found to be exclusively mediated by stalk-specific antibodies. Notably, the full-length HA and HA1 subunit used in these assays did not fully antigenically match the IBV strain to which subjects were exposed, owing to the availability of antigens and variation in the IBV strains. Representative antigens, antigenically close to the viruses the subjects were infected with, were therefore chosen.
Interestingly, HA head–specific antibodies present in human sera failed to mediate ADCC after binding to their epitopes located in the HA head domain of IBV of the corresponding (or opposing) lineage. This corresponds to recent findings, showing that monoclonal antibodies that interfere with binding of HA to sialic acid molecules on NK cells are not capable of mediating ADCC. It has been hypothesized that the interaction between Fc and FcγRIIIα alone is not enough to mediate FcγRIIIα activation and that the additional interaction, between HA and sialic acid, is required to possibly stabilize the immunological synapse between target and effector cell for efficient ADCC [31–33]. Here we show in human polyclonal sera that antibodies to the stalk region and not those specific for the HA head are responsible for ADCC. Notably, the HA1 subunit used in the assays described here also contains part of the stalk, potentially explaining the ADCC reactivity detected against the HA1 from the B/Yam lineage in the B/Yam responders. Furthermore, we show that, in the solid-phase ADCC assay and with the use of NK92.05-CD16 cells, 2 points of contact between effector cells and HA are still required for ADCC activity.
The induction of HA stalk–specific antibodies has gained considerable interest as universal vaccination approach against IAV [34–37]. Monoclonal antibodies directed to the HA stalk have broad reactivity and are capable of inducing protection from challenge with IAV of various subtypes in animal models. This protection is predominantly conferred by Fcγ receptor–dependent mechanisms [19, 38, 39]. Monoclonal antibodies recognizing conserved IBV epitopes have also been identified [40, 41]. One of these is broadly reactive, of the IgG1 subclass (46B8), and proved capable of neutralizing IBV from both lineages [42]. 46B8 mediated ADCC in vitro and was capable of protecting mice from lethal IBV challenge. An Fc-defective variant of 46B8 did not confer protection from challenge, demonstrating that Fcγ receptor–dependent mechanisms were crucial in this model. Novel universal IBV vaccination approaches focus on the induction of HA stalk–specific antibodies; sequential vaccination of mice with chimeric HAs consisting of the globular head domains from irrelevant IAVs and stalks from IBVs induced HA stalk–specific antibodies was performed, conferring protection from challenge with multiple IBVs from both lineages [27]. Notably, ADCC was identified as the most likely correlate of protection in these experiments.
Collectively, the data presented here show that initial IBV infection of children induces both HA head–specific and HA stalk–specific antibodies. The HA head–specific antibodies (as detected by HI and ELISA) bind to IBV of the corresponding lineage only, whereas HA stalk–specific antibodies recognized IBV of the corresponding and opposing lineages. Interestingly, HA head–specific antibodies did not mediate ADCC upon binding to HA, but HA stalk–specific antibodies did. This could be explained by the hypothesis that 2 points of contact between target and effector cell are required for efficient ADCC and that HA head–specific antibodies potentially disrupt the interaction between HA and sialic acid. Altogether, this study demonstrates that human infection with IBV induces ADCC-mediating antibodies mainly directed to the HA stalk of IBV, which display cross-reactivity with IBV of both lineages. These ADCC-mediating antibodies might afford a certain degree of cross-protection from infection with antigenically distinct IBV, but the minimal requirements for protection remain to be determined.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Presented in part: Annual FLUCOP Meeting, Sienna, Italy, April 2017.
Notes
Acknowledgments. We thank Kerry S. Campbell, for supplying the NK92.06-CD16 cell line, and Manon Cox of Protein Sciences, for supplying recombinant proteins.
Disclaimer. The funding source had no role in study design or collection, analysis, and interpretation of data.
Financial support. This work was supported by Innovative Medicines Initiative (FLUCOP grant 115672–3), the National Institute of Allergy and Infectious Diseases (grant U19 AI109946), and the European Union (FLUNIVAC grant 602604 to G. F. R. and R. D. d. V.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Lambert LC, Fauci AS. Influenza vaccines for the future. N Engl J Med 2010; 363:2036–44. [DOI] [PubMed] [Google Scholar]
- 2. Chen R, Holmes EC. The evolutionary dynamics of human influenza B virus. J Mol Evol 2008; 66:655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koel BF, Burke DF, Bestebroer TM, et al. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 2013; 342:976–9. [DOI] [PubMed] [Google Scholar]
- 4. Smith DJ, Lapedes AS, de Jong JC, et al. Mapping the antigenic and genetic evolution of influenza virus. Science 2004; 305:371–6. [DOI] [PubMed] [Google Scholar]
- 5. Bedford T, Suchard MA, Lemey P, et al. Integrating influenza antigenic dynamics with molecular evolution. Elife 2014; 3:e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rota PA, Wallis TR, Harmon MW, Rota JS, Kendal AP, Nerome K. Cocirculation of two distinct evolutionary lineages of influenza type B virus since 1983. Virology 1990; 175:59–68. [DOI] [PubMed] [Google Scholar]
- 7. van de Sandt CE, Bodewes R, Rimmelzwaan GF, de Vries RD. Influenza B viruses: not to be discounted. Future Microbiol 2015; 10:1447–65. [DOI] [PubMed] [Google Scholar]
- 8. Yamashita M, Krystal M, Fitch WM, Palese P. Influenza B virus evolution: co-circulating lineages and comparison of evolutionary pattern with those of influenza A and C viruses. Virology 1988; 163:112–22. [DOI] [PubMed] [Google Scholar]
- 9. Francis T., Jr A new type of virus from epidemic influenza. Science 1940; 92:405–8. [DOI] [PubMed] [Google Scholar]
- 10. Lindstrom SE, Hiromoto Y, Nishimura H, Saito T, Nerome R, Nerome K. Comparative analysis of evolutionary mechanisms of the hemagglutinin and three internal protein genes of influenza B virus: multiple cocirculating lineages and frequent reassortment of the NP, M, and NS genes. J Virol 1999; 73:4413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen JM, Guo YJ, Wu KY, et al. Exploration of the emergence of the Victoria lineage of influenza B virus. Arch Virol 2007; 152:415–22. [DOI] [PubMed] [Google Scholar]
- 12. Ambrose CS, Levin MJ. The rationale for quadrivalent influenza vaccines. Hum Vaccin Immunother 2012; 8:81–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Heikkinen T, Ikonen N, Ziegler T. Impact of influenza B lineage-level mismatch between trivalent seasonal influenza vaccines and circulating viruses, 1999–2012. Clin Infect Dis 2014; 59:1519–24. [DOI] [PubMed] [Google Scholar]
- 14. Belshe RB. The need for quadrivalent vaccine against seasonal influenza. Vaccine 2010; 28(Suppl 4):D45–53. [DOI] [PubMed] [Google Scholar]
- 15. Beran J, Wertzova V, Honegr K, et al. Challenge of conducting a placebo-controlled randomized efficacy study for influenza vaccine in a season with low attack rate and a mismatched vaccine B strain: a concrete example. BMC Infect Dis 2009; 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beyer WEP, Palache AM, Boulfich M, Osterhaus ADME. Rationale for two influenza B lineages in seasonal vaccines: A meta-regression study on immunogenicity and controlled field trials. Vaccine 2017; 35:4167–76. [DOI] [PubMed] [Google Scholar]
- 17. Bruhns P, Iannascoli B, England P, et al. Specificity and affinity of human Fcgamma receptors and their polymorphic variants for human IgG subclasses. Blood 2009; 113:3716–25. [DOI] [PubMed] [Google Scholar]
- 18. Jegaskanda S, Laurie KL, Amarasena TH, et al. Age-associated cross-reactive antibody-dependent cellular cytotoxicity toward 2009 pandemic influenza A virus subtype H1N1. J Infect Dis 2013; 208:1051–61. [DOI] [PubMed] [Google Scholar]
- 19. DiLillo DJ, Tan GS, Palese P, Ravetch JV. Broadly neutralizing hemagglutinin stalk-specific antibodies require FcγR interactions for protection against influenza virus in vivo. Nat Med 2014; 20:143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ana-Sosa-Batiz F, Vanderven H, Jegaskanda S, et al. Influenza-Specific Antibody-Dependent Phagocytosis. PLoS One 2016; 11:e0154461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Henry Dunand CJ, Leon PE, Huang M, et al. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 2016; 19:800–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Co MD, Terajima M, Thomas SJ, et al. Relationship of preexisting influenza hemagglutination inhibition, complement-dependent lytic, and antibody-dependent cellular cytotoxicity antibodies to the development of clinical illness in a prospective study of A(H1N1)pdm09 Influenza in children. Viral Immunol 2014; 27:375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. El Bakkouri K, Descamps F, De Filette M, et al. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J Immunol 2011; 186:1022–31. [DOI] [PubMed] [Google Scholar]
- 24. Impagliazzo A, Milder F, Kuipers H, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 2015; 349:1301–6. [DOI] [PubMed] [Google Scholar]
- 25. Bodewes R, de Mutsert G, van der Klis FR, et al. Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin Vaccine Immunol 2011; 18:469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van der Klis FR, Mollema L, Berbers GA, de Melker HE, Coutinho RA. Second national serum bank for population-based seroprevalence studies in the Netherlands. Neth J Med 2009; 67:301–8. [PubMed] [Google Scholar]
- 27. Ermler M, Kirkpatrick E, Sun W, et al. Chimeric hemagglutinin constructs induce broad protection against influenza B virus challenge in the mouse model. J Virol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. de Vries RD, Nieuwkoop NJ, Pronk M, et al. Influenza virus-specific antibody dependent cellular cytoxicity induced by vaccination or natural infection. Vaccine 2017; 35:238–47. [DOI] [PubMed] [Google Scholar]
- 29. Binyamin L, Alpaugh RK, Hughes TL, Lutz CT, Campbell KS, Weiner LM. Blocking NK cell inhibitory self-recognition promotes antibody-dependent cellular cytotoxicity in a model of anti-lymphoma therapy. J Immunol 2008; 180:6392–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mesman AW, Westerhuis BM, Ten Hulscher HI, et al. Influenza virus A(H1N1)2009 antibody-dependent cellular cytotoxicity in young children prior to the H1N1 pandemic. J Gen Virol 2016; 97:2157–65. [DOI] [PubMed] [Google Scholar]
- 31. He W, Tan GS, Mullarkey CE, et al. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc Natl Acad Sci U S A 2016; 113:11931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cox F, Kwaks T, Brandenburg B, et al. HA antibody-mediated FcγRIIIa activity is both dependent on FcR engagement and interactions between HA and sialic acids. Front Immunol 2016; 7:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leon PE, He W, Mullarkey CE, et al. Optimal activation of Fc-mediated effector functions by influenza virus hemagglutinin antibodies requires two points of contact. Proc Natl Acad Sci U S A 2016; 113:E5944–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Vries RD, Altenburg AF, Rimmelzwaan GF. Universal influenza vaccines: a realistic option?Clin Microbiol Infect 2016; 22(Suppl 5):120–4. [DOI] [PubMed] [Google Scholar]
- 35. Wiersma LC, Rimmelzwaan GF, de Vries RD. Developing universal influenza vaccines: hitting the nail, not just on the head. Vaccines (Basel) 2015; 3:239–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. de Vries RD, Altenburg AF, Rimmelzwaan GF. Universal influenza vaccines, science fiction or soon reality?Expert Rev Vaccines 2015; 14:1299–301. [DOI] [PubMed] [Google Scholar]
- 37. Krammer F, Palese P. Influenza virus hemagglutinin stalk-based antibodies and vaccines. Curr Opin Virol 2013; 3:521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 2011; 333:850–6. [DOI] [PubMed] [Google Scholar]
- 39. DiLillo DJ, Palese P, Wilson PC, Ravetch JV. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J Clin Invest 2016; 126:605–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dreyfus C, Laursen NS, Kwaks T, et al. Highly conserved protective epitopes on influenza B viruses. Science 2012; 337:1343–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yasugi M, Kubota-Koketsu R, Yamashita A, et al. Human monoclonal antibodies broadly neutralizing against influenza B virus. PLoS Pathog 2013; 9:e1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chai N, Swem LR, Reichelt M, et al. Two escape mechanisms of influenza A virus to a broadly neutralizing stalk-binding antibody. PLoS Pathog 2016; 12:e1005702. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.