Abstract
Human immunodeficiency virus type 1 (HIV-1) drug resistance genotyping is recommended to help in the selection of antiretroviral therapy and to prevent virologic failure. There are several ultrasensitive assays able to detect HIV-1 drug-resistance minority variants (DRMVs) not detectable by standard population sequencing–based HIV genotyping assays. Presence of these DRMVs has been shown to be clinically relevant, but its impact does not appear to be uniform across drug classes. In this review, we summarize key evidence for the clinical impact of DRMVs across drug classes for both antiretroviral treatment–naive and antiretroviral treatment–experienced patients, and highlight areas where more supporting evidence is needed.
Keywords: HIV-1 drug resistance, drug resistant minority variants, treatment failure
Human immunodeficiency virus type 1 (HIV-1) drug resistance genotyping is recommended to help in the selection of antiretroviral therapy (ART) and to prevent virologic failure (VF). Genotypic tests for HIV drug resistance that are based on standard population sequencing fail to detect drug-resistant minority variants (DRMVs) present in less than 15%–25% of the total viral population [1, 2]. More sensitive techniques have been developed, such as allele-specific polymerase chain reaction (ASPCR) [3], oligonucleotide ligation assay [4], SNaPshot assay [5], and ultra-deep sequencing (UDS) [6, 7], to detect and quantify DRMVs. The lower limit of detection of minority variants differs widely between assays, with an upper range of 2%–5% for the HIV-SNaPshot assay [5] and certain UDS assays [8], and a lower range of <0.01% has been reported for ASPCR [9].
DRMVs have been shown to be clinically relevant, but their impact does not appear to be uniform across drug classes. The clinical relevance of DRMVs is related to the genetic barrier to resistance to specific drugs and can be classified based on 3 levels of evidence. The best evidence that DRMVs may adversely affect response to ART lies in the assessment of resistance mutations active against the nonnucleoside reverse transcriptase inhibitors (NNRTIs) and CCR5 antagonists. These drugs have a low genetic barrier to resistance and a single mutation can dramatically impact drug susceptibility. A moderate amount of evidence has accumulated that DRMVs against nucleoside reverse transcriptase inhibitors (NRTIs) and the integrase strand transfer inhibitor (INSTI) raltegravir may affect their clinical efficacy. Finally, in the case of the protease inhibitors (PIs), elvitegravir and dolutegravir, there is little evidence so far that DRMVs increase the risk of VF, although few studies have been performed and more research is needed. In this review, we review key evidence for both ART-naive and ART-experienced patients and highlight areas where more supporting evidence is needed.
NNRTIs AND NRTIs
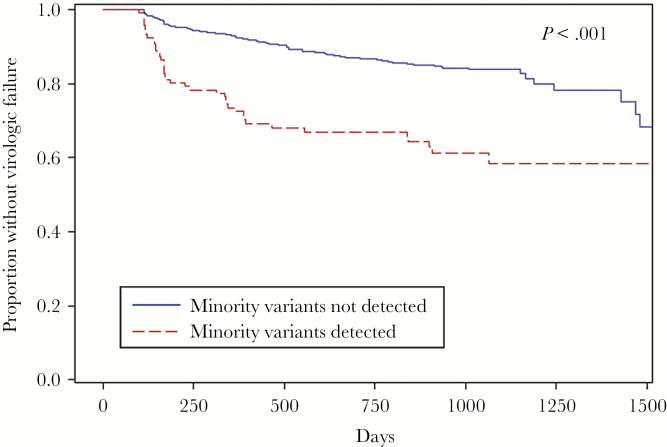
There have been several studies evaluating the effects of baseline low-frequency NNRTI and NRTI resistance mutations on the rates of treatment failure for ART-naive individuals. A pooled analysis was performed of 10 studies involving 985 ART-naive participants and included only individuals with no detectable pre-ART NNRTI and NRTI resistance by standard genotyping [10]. The pooled analysis showed that 14% of participants harbored either NNRTI or NRTI DRMVs by ultrasensitive assays and the presence of DRMVs at baseline was associated with more than twice the risk of VF. The increased risk of VF was most evident early after therapy initiation (Figure 1) and was mediated primarily by NNRTI-resistant minority variants (hazard ratio [HR], 2.6). The effect of the minority variants was detectable even after controlling for medication adherence [11]. In addition, the risk of VF was detected in a dose-dependent manner and even at some of the lowest DRMV frequencies (<0.5% and 10–99 mutant copies/mL) [10]. These results are supported by several subsequent studies. In a Europe-wide case-control study of 260 participants initiating an NNRTI-based regimen, DRMVs were detected by UDS in 21% of subjects and the presence of DRMVs was associated with an increased risk of VF (odds ratio [OR], 2.75) [12]. The strength of the minority variant (MV) effect was similar between DRMVs to NNRTIs (OR, 2.41) or NRTIs (OR, 2.27), but the effect of the NRTI-resistant MVs did not reach statistical significance, possibly due to the smaller numbers of NRTI MVs detected. In addition, a direct dose-effect relationship between the mutational load of NNRTI DRMVs and risk of VF was found.
Figure 1.
Kaplan-Meier curves for the proportion of patients without virologic failure on first-line nonnucleoside reverse transcriptase inhibitor (NNRTI)–based combination antiretroviral therapy regimen by the presence of HIV-1 nucleoside reverse transcriptase inhibitor and NNRTI-resistant minority variants. Adapted from [10].
There is also strong evidence that NNRTI-resistant MVs are commonly found in those failing an NNRTI-based ART regimen and that these mutations increase in the risk of VF. In the AIDS Clinical Trials Group (ACTG) study 398, NNRTI-resistant MVs were more commonly detected in treatment-experienced individuals and were associated with an increased risk of virologic failure [13]. In addition, a number of other studies have demonstrated the detection of NNRTI and NRTI DRMVs in treatment-experienced individuals that may affect downstream treatment efficacy [14–19].
NNRTIs IN AFRICAN STUDIES
Several studies have now demonstrated the selection of DRMVs in African patients treated with single-dose nevirapine (sdNVP) [20–22]. One of the largest studies was the Optimal Combination Therapy After Nevirapine Exposure (OCTANE) Trial 1 of 232 women treated with sdNVP with DRMVs evaluated by ASPCR. Exposure to sdNVP was associated with an increased detection of NNRTI DRMVs and these MVs were associated with a significantly increased risk of VF (HR, 2.71) after restarting an NNRTI-based regimen [23]. The risk of VF appeared to be mitigated by a longer interval between sdNVP exposure and start of combination ART [24]. This finding is likely due to the continued decay of the proportion of drug-resistant variants after sdNVP in light of the fact that DRMV frequency was associated with the extent of risk for the primary endpoint (VF or death) [23]. Interestingly, in the parallel OCTANE Trial 2 of African women without prior sdNVP exposure, no significant association was detected between NNRTI DRMV detection and risk of treatment failure [25]. A subsequent study by the ANRS team also showed that while an ultrasensitive assay could detect more DRMVs in treatment-naive African patients, these mutations were not associated with an increased risk of VF [26]. The differential effects of DRMVs seen in these African patients could be related to differences in their NRTI backbone, HIV subtype differences, or extent of multiple linked mutations. However, these hypotheses deserve further exploration.
CCR5 ANTAGONIST
HIV-1 requires the use of either CCR5 or CXCR4 as a co-receptor to enter target cells. Maraviroc, a CCR5 antagonist that blocks HIV-1 entry, was approved for clinical use against R5-tropic virus. Several phenotypic and genotypic assays have been developed to assess HIV-1 co-receptor tropism [27]. Historically, a phenotypic HIV tropism assay has been used in the United States, but minority populations of CXCR4-using (R4-tropic) virus can be a cause of VF [28–31]. In a retrospective reanalysis of treatment-naive patients, UDS showed the same ability as an improved version of the phenotypic assay in predicting the response to Maraviroc [32]. A genotypic tropism assay with UDS is now available in the United States (HIV-1 Coreceptor Tropism with Reflex to Ultradeep Sequencing, Quest Diagnostics). This test includes initial population sequencing followed by UDS analysis of those samples that showed only R5-tropic virus by population sequencing. In a retrospective analysis of 327 treatment-experienced patients who received maraviroc in the Maraviroc versus Optimized Therapy in Viremic Antiretroviral Treatment-Experienced Patients and A4001029 trials, this test had greater sensitivity than population sequencing alone to detect minority non-R5 and was equivalent to the phenotypic test for predicting maraviroc responders from nonresponders [33].
INTEGRASE INHIBITORS
Integrase strand transfer inhibitors (INSTIs) have become a key component of ART, but the clinical impact of INSTI DRMVs remains understudied. The concern surrounding raltegravir (RAL) DRMVs is two-fold. First, there have been several case reports of RAL DRMVs that led to subsequent treatment failure [34, 35]. In addition, RAL DRMVs can be found in a large subset of treatment-naive patients [36] and may be found at a higher rate in treatment-naive individuals [37]. However, those findings have not been replicated in other studies and RAL DRMVs have not yet been found to definitively impact VF rates in larger studies of treatment-naive [6, 38, 39] or treatment-experienced individuals [40]. The clinical impact of DRMVs on elvitegravir (EVG) and dolutegravir (DTG) has not been carefully studied. However, there is evidence that ultrasensitive assays can detect EVG and DTG DRMVs that would affect the predicted susceptibility profiles for these drugs, especially in INSTI-experienced patients [41, 42].
PROTEASE INHIBITORS
The use of more sensitive genotyping methods has significantly increased the number of PI DRMVs detected in treatment-naive patients, but these DRMVs have not been shown to affect the efficacy of first-line PI therapy [43, 44]. For example, a retrospective analysis of 123 baseline clinical samples from the ADVANZ and ADVANZ-3 studies found that NNRTI DRMVs increased the risk of VF for an EFV-based regimen, but PI DRMVs had no significant effect on VF for a PI-based regimen [14]. For treatment-experienced patients, PI DRMVs have been detected in a high proportion of individuals, but these mutations have also not been clearly linked to increased risk of VF [43, 45]. Several factors can contribute to this lack of association. PIs have a high genetic barrier to resistance and thus, multiple mutations are required to confer significant resistance [46, 47]. In addition, the prevalence of transmitted PI-resistance mutations is low and viruses with multiple PI-resistance mutations are rarely transmitted [48, 49]. Of note, in an analysis of the CASTLE study, the presence of NRTI DRMVs was associated with virologic failure of the lopinavir-based ART regimen [44], although this effect was not found in other studies [50].
CONCLUSIONS
The detection of DRMVs of HIV-1 has been shown to be clinically significant mainly in 3 settings: detection of NNRTI-resistant minority variant prior to the initiation of a first-line NNRTI-based regimen outside of Africa, detection of NNRTI-resistant minority variants after exposure to sdNVP, and detection of CXCR4-using variants prior to initiation of maraviroc-containing regimens. However, there are a number of scenarios where the clinical impact of DRMVs remains controversial and additional studies are needed. These include first-line NNRTI-based regimens in an African setting, as well as DRMVs against NRTIs, PIs, and INSTIs.
During the past decade, UDS technologies have continued to evolve and their sequencing costs have been greatly reduced, making it an increasingly cost-efficient technique, especially in regions where demand for HIV genotyping is high [8, 51]. Adoption of these platforms may improve access to HIV genotyping worldwide, and additional studies on the clinical impact of DRMVs are needed to guide the interpretation of these minority variants in the clinical setting.
Financial support. N. S.-A. is supported by a predoctoral fellowship from Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III, Spain (award number FI14/00391). J. Z. L. is supported by a grant from the National Institute of Allergy and Infectious Diseases (grant number AI100699).
Supplement sponsorship. This work is part of a supplement sponsored by the National Institute of Allergy and Infectious Disease, NIH, and the Centers for Disease Control and Prevention.
Potential conflicts of interest. J. Z. L. has consulted for United BioPharma, Gilead Sciences, and Merck, and received research support from Gilead Sciences and Merck. J. R. A. has consulted for Gilead Sciences, Merck, Viiv, Janssen, and Teva. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Grant RM, Kuritzkes DR, Johnson VA et al. Accuracy of the TRUGENE HIV-1 genotyping kit. J Clin Microbiol 2003; 41:1586–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schuurman R, Demeter L, Reichelderfer P, Tijnagel J, de Groot T, Boucher C. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J Clin Microbiol 1999; 37:2291–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Paredes R, Marconi VC, Campbell TB, Kuritzkes DR. Systematic evaluation of allele-specific real-time PCR for the detection of minor HIV-1 variants with pol and env resistance mutations. J Virol Methods 2007; 146:136–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ellis GM, Vlaskin TA, Koth A et al. Simultaneous and sensitive detection of human immunodeficiency virus type 1 (HIV) drug resistant genotypes by multiplex oligonucleotide ligation assay. J Virol Methods 2013; 192:39–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jakobsen MR, Tolstrup M, Søgaard OS et al. Transmission of HIV-1 drug-resistant variants: prevalence and effect on treatment outcome. Clin Infect Dis 2010; 50:566–73. [DOI] [PubMed] [Google Scholar]
- 6. Li JZ, Chapman B, Charlebois P et al. ; ACTG A5262 Study Team Comparison of Illumina and 454 deep sequencing in participants failing raltegravir-based antiretroviral therapy. PLoS One 2014; 9:e90485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gibson RM, Meyer AM, Winner D et al. Sensitive deep-sequencing-based HIV-1 genotyping assay to simultaneously determine susceptibility to protease, reverse transcriptase, integrase, and maturation inhibitors, as well as HIV-1 coreceptor tropism. Antimicrob Agents Chemother 2014; 58:2167–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dudley DM, Chin EN, Bimber BN et al. Low-cost ultra-wide genotyping using Roche/454 pyrosequencing for surveillance of HIV drug resistance. PLoS One 2012; 7:e36494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Paredes R, Lalama CM, Ribaudo HJ et al. ; AIDS Clinical Trials Group (ACTG) A5095 Study Team Pre-existing minority drug-resistant HIV-1 variants, adherence, and risk of antiretroviral treatment failure. J Infect Dis 2010; 201:662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li JZ, Paredes R, Ribaudo HJ et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA 2011; 305:1327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li JZ, Paredes R, Ribaudo HJ et al. Relationship between minority NNRTI resistance mutations, adherence, and the risk of virologic failure. AIDS 2011; 26:185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cozzi-Lepri A, Noguera-Julian M, Di Giallonardo F et al. ; CHAIN Minority HIV-1 Variants Working Group Low-frequency drug-resistant HIV-1 and risk of virological failure to first-line NNRTI-based ART: a multicohort European case-control study using centralized ultrasensitive 454 pyrosequencing. J Antimicrob Chemother 2015; 70:930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Halvas EK, Wiegand A, Boltz VF et al. Low frequency nonnucleoside reverse-transcriptase inhibitor-resistant variants contribute to failure of efavirenz-containing regimens in treatment-experienced patients. J Infect Dis 2010; 201:672–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Casadellà M, Manzardo C, Noguera-Julian M et al. ; ADVANZ and ADVANZ-3 Investigators Clinical value of ultradeep HIV-1 genotyping and tropism testing in late presenters with advanced disease. AIDS 2015; 29:1493–504. [DOI] [PubMed] [Google Scholar]
- 15. Casadellà M, Noguera-Julian M, Sunpath H et al. Treatment options after virological failure of first-line tenofovir-based regimens in South Africa: an analysis by deep sequencing. AIDS 2016; 30:1137–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Charpentier C, Lee GQ, Rodriguez C et al. Highly frequent HIV-1 minority resistant variants at baseline of the ANRS 139 TRIO trial had a limited impact on virological response. J Antimicrob Chemother 2015; 70:2090–6. [DOI] [PubMed] [Google Scholar]
- 17. Kyeyune F, Gibson RM, Nankya I et al. Low-frequency drug resistance in HIV-infected Ugandans on antiretroviral treatment is associated with regimen failure. Antimicrob Agents Chemother 2016; 60:3380–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohamed S, Penaranda G, Gonzalez D et al. Comparison of ultra-deep versus Sanger sequencing detection of minority mutations on the HIV-1 drug resistance interpretations after virological failure. AIDS 2014; 28:1315–24. [DOI] [PubMed] [Google Scholar]
- 19. Nishizawa M, Matsuda M, Hattori J et al. Longitudinal detection and persistence of minority drug-resistant populations and their effect on salvage therapy. PLoS One 2015; 10:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Johnson JA, Li JF, Morris L et al. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis 2005; 192:16–23. [DOI] [PubMed] [Google Scholar]
- 21. Flys T, Nissley DV, Claasen CW et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J Infect Dis 2005; 192:24–9. [DOI] [PubMed] [Google Scholar]
- 22. Coovadia A, Hunt G, Abrams EJ et al. Persistent minority K103N mutations among women exposed to single-dose nevirapine and virologic response to nonnucleoside reverse-transcriptase inhibitor-based therapy. Clin Infect Dis 2009; 48:462–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boltz VF, Zheng Y, Lockman S et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proc Natl Acad Sci U S A 2011; 108:9202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lockman S, Shapiro RL, Smeaton LM et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med 2007; 356:135–47. [DOI] [PubMed] [Google Scholar]
- 25. Boltz VF, Bao Y, Lockman S et al. ; OCTANE/A5208 Team Low-frequency nevirapine (NVP)-resistant HIV-1 variants are not associated with failure of antiretroviral therapy in women without prior exposure to single-dose NVP. J Infect Dis 2014; 209:703–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Derache A, Iwuji CC, Danaviah S et al. Prevalence and impact of pretreatment drug resistance in the ANRS 12249 TASP TRIAL [abstract 43]. In: 24th Conference on Retroviruses and Opportunistic Infections, Seattle, WA, 2017. [Google Scholar]
- 27. Rose JD, Rhea AM, Weber J, Quiñones-Mateu ME. Current tests to evaluate HIV-1 coreceptor tropism. Curr Opin HIV AIDS 2009; 4:136–42. [DOI] [PubMed] [Google Scholar]
- 28. Archer J, Braverman MS, Taillon BE et al. Detection of low-frequency pretherapy chemokine (CXC motif) receptor 4 (CXCR4)-using HIV-1 with ultra-deep pyrosequencing. AIDS 2009; 23:1209–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Archer J, Rambaut A, Taillon BE, Harrigan PR, Lewis M, Robertson DL. The evolutionary analysis of emerging low frequency HIV-1 CXCR4 using variants through time—an ultra-deep approach. PLoS Comput Biol 2010; 6:e1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsibris AMN, Korber B, Arnaout R et al. Quantitative deep sequencing reveals dynamic HIV-1 escape and large population shifts during CCR5 antagonist therapy in vivo. PLoS One 2009; 4:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Westby M, Lewis M, Whitcomb J et al. Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir. J Virol 2006; 80:4909–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Swenson LC, Mo T, Dong WW et al. Deep V3 sequencing for HIV type 1 tropism in treatment-naive patients: a reanalysis of the MERIT trial of maraviroc. Clin Infect Dis 2011; 53:732–42. [DOI] [PubMed] [Google Scholar]
- 33. Kagan RM, Johnson EP, Siaw M et al. A genotypic test for HIV-1 tropism combining Sanger sequencing with ultradeep sequencing predicts virologic response in treatment-experienced patients. PLoS One 2012; 7:e46334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stefic K, Salmona M, Capitao M et al. Unravelling the dynamics of selection of multiresistant variants to integrase inhibitors in an HIV-1-infected child using ultra-deep sequencing. J Antimicrob Chemother 2017; 72:850–4. [DOI] [PubMed] [Google Scholar]
- 35. Codoñer FM, Pou C, Thielen A et al. Dynamic escape of pre-existing raltegravir-resistant HIV-1 from raltegravir selection pressure. Antiviral Res 2010; 88:281–6. [DOI] [PubMed] [Google Scholar]
- 36. Charpentier C, Laureillard D, Piketty C et al. High frequency of integrase Q148R minority variants in HIV-infected patients naive of integrase inhibitors. AIDS 2010; 24:867–73. [DOI] [PubMed] [Google Scholar]
- 37. Liu J, Miller MD, Danovich RM et al. Analysis of low-frequency mutations associated with drug resistance to raltegravir before antiretroviral treatment. Antimicrob Agents Chemother 2011; 55:1114–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mukherjee R, Jensen ST, Male F et al. Switching between raltegravir resistance pathways analyzed by deep sequencing. AIDS 2011; 25:1951–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Armenia D, Vandenbroucke I, Fabeni L et al. Study of genotypic and phenotypic HIV-1 dynamics of integrase mutations during raltegravir treatment: a refined analysis by ultra-deep 454 pyrosequencing. J Infect Dis 2012; 205:557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nguyen HL, Charpentier C, Nguyen N et al. Longitudinal analysis of integrase N155H variants in heavily treated patients failing raltegravir-based regimens. HIV Med 2013; 14:85–91. [DOI] [PubMed] [Google Scholar]
- 41. Gibson RM, Weber J, Winner D, Miller MD, Quiñones-Mateu ME. Contribution of human immunodeficiency virus type 1 minority variants to reduced drug susceptibility in patients on an integrase strand transfer inhibitor-based therapy. PLoS One 2014; 9:e104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fonager J, Larsson JT, Hussing C, Neess Engsig F, Nielsen C, Fischer TK. Identification of minority resistance mutations in the HIV-1 integrase coding region using next generation sequencing. J Clin Virol 2015; 73:95–100. [DOI] [PubMed] [Google Scholar]
- 43. Lataillade M, Chiarella J, Yang R et al. Virologic failures on initial boosted-PI regimen infrequently possess low-level variants with major PI resistance mutations by ultra-deep sequencing. PLoS One 2012; 7:e30118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lataillade M, Chiarella J, Yang R et al. Prevalence and clinical significance of HIV drug resistance mutations by ultra-deep sequencing in antiretroviral-naive subjects in the CASTLE study. PLoS One 2010; 5:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fisher R, van Zyl GU, Travers SA et al. Deep sequencing reveals minor protease resistance mutations in patients failing a protease inhibitor regimen. J Virol 2012; 86:6231–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tang MW, Shafer RW. HIV-1 Antiretroviral resistance scientific principles and clinical applications. Drugs 2012; 72:1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wensing AM, Calvez V, Günthard HF et al. 2017 update of the drug resistance mutations in HIV-1. Top Antivir Med 2017; 24:132–3. [PMC free article] [PubMed] [Google Scholar]
- 48. Hofstra LM, Sauvageot N, Albert J et al. SPREAD Program Transmission of HIV drug resistance and the predicted effect on current first-line regimens in Europe. Clin Infect Dis 2016; 62:655–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rhee SY, Blanco JL, Jordan MR et al. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med 2015; 12:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Metzner KJ, Rauch P, von Wyl V et al. Efficient suppression of minority drug-resistant HIV type 1 (HIV-1) variants present at primary HIV-1 infection by ritonavir-boosted protease inhibitor-containing antiretroviral therapy. J Infect Dis 2010; 201:1063–71. [DOI] [PubMed] [Google Scholar]
- 51. Ekici H, Rao SD, Sönnerborg A, Ramprasad VL, Gupta R, Neogi U. Cost-efficient HIV-1 drug resistance surveillance using multiplexed high-throughput amplicon sequencing: implications for use in low- and middle-income countries. J Antimicrob Chemother 2014; 69:3349–55. [DOI] [PubMed] [Google Scholar]