This review provides a perspective on early nitrate-elicited signaling events with an emphasis on calcium, phospholipase C, and the potential role of phosphate modifications in nitrate signaling.
Keywords: Gene expression, nitrate, nitrogen, phospholipase C, phosphorylation, primary response, signaling
Abstract
Nitrogen (N) is an essential macronutrient that impacts many aspects of plant physiology, growth, and development. Besides its nutritional role, N nutrient and metabolites act as signaling molecules that regulate the expression of a wide range of genes and biological processes. In this review, we describe recent advances in the understanding of components of the nitrate signaling pathway. Recent evidence posits that in one nitrate signaling pathway, nitrate sensed by NRT1.1 activates a phospholipase C activity that is necessary for increased cytosolic calcium levels. The nitrate-elicited calcium increase presumably activates calcium sensors, kinases, or phosphatases, resulting in changes in expression of primary nitrate response genes. Consistent with this model, nitrate treatments elicit proteome-wide changes in phosphorylation patterns in a wide range of proteins, including transporters, metabolic enzymes, kinases, phosphatases, and other regulatory proteins. Identifying and characterizing the function of the different players involved in this and other nitrate signaling pathways and their functional relationships is the next step to understand N responses in plants.
Introduction
Nitrogen (N) is an essential mineral nutrient that plants require the most in quantitative terms (Frink et al., 1999). It is a critical constituent of proteins and nucleic acids and is therefore indispensable for life. In order to sustain the high crop productivity demanded by modern agriculture, fields are subjected to massive applications of N fertilizers, which contribute 30–50% of their total yield (Crawford and Glass, 1998; Stewart et al., 2005). Global demand for N fertilizers in 2014 was 113147000 tonnes. This amount has been projected to grow at approximately 1.4% per year, reaching 119418000 tonnes by 2018 (FAO, 2015). From these large amounts of N fertilizer added to crops every year, only 25–50% is taken up by plants. Excess N leaches into water streams, becoming an important promoter of hypoxic zones and eutrophication (Hirel et al., 2011; Robertson and Vitousek, 2009). A significant fraction of N is also converted to N oxide gases that contribute to global warming (Crutzen et al., 2008; Davidson, 2009). Improving N use efficiency in plants is critical for efficient sustainable agriculture.
The main source of N in well-aerated soils is nitrate (NO3–) (Crawford and Forde, 2002). Nitrate is also the most abundant N resource available in agricultural lands (Owen and Jones, 2001). Nitrate concentrations in agricultural soils typically range between 1 and 5 mM (Owen and Jones, 2001). This supply is not steady because the predominantly negative charge of ground particles makes the nitrate ion highly mobile in the soil solution, varying its concentration and potentially causing its depletion by run-off (Miller and Cramer, 2004). Biotic factors such as plant absorption and microbial denitrification also contribute to soil nitrate depletion (Crawford and Glass, 1998).
Plants evolved sophisticated mechanisms to cope with variable N concentrations in the soil. The root architecture adjusts to this fluctuating environment: lateral root elongation is stimulated by exogenous nitrate application, favoring root colonization of nitrate-rich soil patches (Gojon et al., 2009; Zhang and Forde, 1998). However, plants subjected to long-term N treatments have a different behavior: plants grown under N-sufficient conditions develop fewer lateral roots than plants grown under low N conditions, a strategy that allows N foraging only when this nutrient is scarce (Gifford et al., 2008). This local root nitrate acquisition needs to be coordinated with systemic signals in order to coordinate N supply and demand within the plant (Ruffel et al., 2011). Besides its role as a nutrient, nitrate is a local and systemic signal that coordinates its uptake with plant growth and development (Alvarez et al., 2012; Ruffel et al., 2014; Ruffel et al., 2011). Nitrate induces changes in the transcription of genes involved in N acquisition, nitrate assimilation, production of reducing equivalents needed for N metabolism, C metabolism, and an array of other functions (Redinbaugh and Campbell, 1991; Scheible et al., 2004; Vidal and Gutiérrez, 2008; Wang et al., 2000; Wang et al., 2003). Consequently, nitrate has a myriad of effects on plant development: it induces seed germination, regulates root growth and architecture, controls shoot growth, and delays flowering (Alboresi et al., 2005; Castro Marín et al., 2011; Drew and Saker, 1975; Liu et al., 2010; Rahayu et al., 2005; Remans et al., 2006; Vidal et al., 2014; Walch-Liu et al., 2000; Walch-Liu et al., 2006; Yuan et al., 2016).
Inside the cell, nitrate is reduced by Nitrate Reductase (NR) and the nitrite product is further reduced to ammonia by Nitrite Reductase (NiR). The ammonia generated is incorporated into the amino acid glutamate to generate glutamine by the GS/GOGAT cycle (Krapp, 2015; Marschner, 2012; Xu et al., 2012). Subsequent reactions transfer the assimilated N into other amino acids or biomolecules (Krapp, 2015). Since incorporation of inorganic N into amino acids requires a carbon (C) skeleton, a fine balance between N and C must be attained by the plant (Alvarez et al., 2012; Ruffel et al., 2014). Early reports established that nitrate acquisition varies according to the availability of photosynthates, imposing diurnal oscillations over nitrate uptake (Clement et al., 1978; Delhon et al., 1996). Molecular and computational studies proved that the expression of many nitrate-controlled genes is also subjected to regulation by C metabolites (Gutiérrez et al., 2007b; Palenchar et al., 2004). Girin et al. (2007) identified the N- and-C-dependent transcriptional control of a nitrate transporter, which was localized to a 150 bp sequence that is sensitive to nitrate, N metabolites, and sucrose (Girin et al., 2007). Furthermore, N metabolites control transcription of the circadian clock master regulator CCA1, which, in turn, regulates the transcription of genes involved in N assimilation, establishing a connection between the circadian clock and N nutrition (Gutiérrez et al., 2008).
Nitrate signaling has been actively investigated in recent years. In this review, we will discuss recent advances, with an emphasis on early nitrate-elicited changes in calcium levels and protein modification by phosphorylation. For discussion of other aspects of N responses in plants, we recommend a number of recent reviews (Alvarez et al., 2012; Gutiérrez, 2012; Krapp, 2015; Krouk, 2016; Krouk et al., 2011; Medici and Krouk, 2014; O’Brien et al., 2016; Ruffel et al., 2014; Vidal et al., 2014; Vidal et al., 2015).
Nitrate sensing and primary response
The dual-affinity transporter and sensor (transceptor) Nitrate Transporter 1.1 (NRT1.1/NPF6.3) triggers nitrate-dependent changes in gene expression. Besides its nitrate uptake function, NRT1.1 regulates the expression of key nitrate assimilatory genes. Its affinity changes according to the phosphorylation status of residue T101 (Ho et al., 2009; Hu et al., 2009). Under low nitrate conditions, CBL-Interacting Protein Kinase 23 (CIPK23) phosphorylates this residue, shifting NRT1.1 into a high-affinity nitrate carrier (Liu and Tsay, 2003). This change also triggers a weak up-regulation of the Nitrate Transporter 2.1 (NRT2.1) high-affinity nitrate transporter (Ho et al., 2009) (Fig. 1).
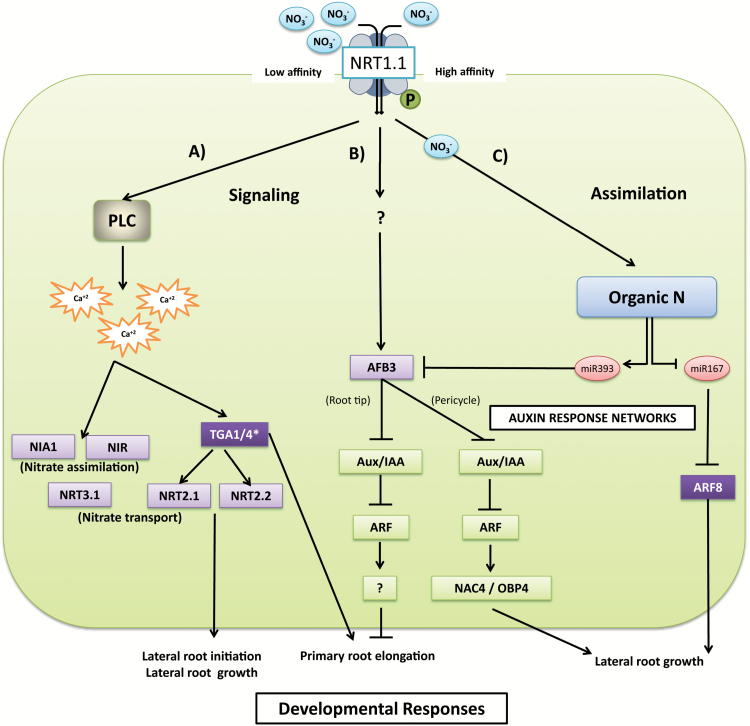
Fig. 1.
Summary of nitrate signaling and assimilation. Nitrate is sensed and transported by the NRT1.1 transceptor, changing its affinity by modifications of phosphorylation status and triggering a signaling pathway. Under low nitrate conditions, CIPK23 phosphorylates NRT1.1, changing it into a high-affinity transporter. Nitrate sensing elicits changes in the phosphorylation status of NRT1.1, and activates, through PLC, calcium influx, which acts as a second messenger (A). This cascade mediates changes in the expression of transcription factors (TGA1/4*) and genes involved in nitrate transport (NRT2.1, NRT2.2 and NRT3.1) and nitrate assimilation (NIA1 and NiR). On the other hand, AFB3 is regulated by nitrate in a PLC- and calcium-independent pathway (B). ABF3 modulates the expression of NAC4 and OBP4, with subsequent effects on root remodeling. Finally, nitrate assimilation (C) produces organic N, which induces miR393 and represses miR167, regulating the abundance of AFB3 and ARF8, respectively. Nitrate-responsive genes are depicted in lilac, transcription factors in purple, and microRNAs in pink. For clarity purposes, the cell nucleus is not shown. *TGA1 and TGA4 are redundant regulatory factors that mediate nitrate responses in Arabidopsis roots. However, the connection between TGA4 and the PLC–calcium pathway has not been experimentally validated. Other relevant transcription factors, such as HRS1 and NLP7, were not included in this figure because their connection with calcium signaling is currently unknown. (This figure is available in colour at JXB online.)
The primary transcriptional response to a signal corresponds to changes in gene expression that occur independent of de novo protein translation (Herschman, 1991). The nitrate response is characterized by rapid and often transient changes in gene expression that can be detected as early as minutes after nitrate treatments (Gowri et al., 1992; Krouk et al., 2010; Medici and Krouk, 2014). Nitrate response genes are characterized by biological functions such as metabolism, energy, N and sulfur (S) metabolism, amino acid metabolism, ammonium assimilation, the pentose phosphate pathway, glycolysis, and gluconeogenesis (Gutiérrez et al., 2007a). The first studies on the primary nitrate response reported a cycloheximide-independent transcript accumulation of the nitrate assimilation enzymes NR, glutamine synthetase (GS2) and ferredoxin-dependent glutamate synthase (Fd-GOGAT) (Gowri et al., 1992; Redinbaugh and Campbell, 1993). With the advent of technologies to evaluate the transcriptome on a routine basis, a more detailed characterization of both primary and secondary nitrate responses has been obtained. An early transcriptomic study using the Affymetrix ATH1 chip showed large effects on gene expression 20 minutes after nitrate treatments. This change was larger in roots, with 1176 affected transcripts, compared with shoots, with only 183 affected transcripts (Wang et al., 2003). Another study reported induction of the nitrate transporters NRT1.1, NRT2.1, Nitrate Transporter 2.2 (NRT2.2) and Nitrate Transporter 2.4 (NRT2.4) in N-starved seedlings after nitrate addition. All nitrate assimilation genes were also up-regulated (Scheible et al., 2004). To assess the effects of nitrate without the confounding presence of downstream N metabolites, a subsequent transcriptomic study was performed in NR-null double mutant plants. Using this genetic background, 595 nitrate-responsive genes were identified, with over-representation of the following functional categories: energy, metabolism, glycolysis, and gluconeogenesis (Wang et al., 2004). A subsequent meta-analysis of transcriptomic data obtained under various nitrate treatments found genes and pathways that were affected in all studies (C and carbohydrate metabolism, N and S metabolism, pentose phosphate pathway, and others), as well as transcripts and functions that were affected only under certain conditions (protein synthesis and others). Comparison of the transcriptomic data of the NR-null double mutants with these datasets suggested that N metabolites regulated changes in expression that were context-dependent (i.e. that responded to nitrate only under certain conditions) (Gutiérrez et al., 2007a).
N availability alters hormonal signaling pathways in order to control plant growth (Krouk, 2016). Ristova et al. (2016) used combinatorial treatments of hormones (auxin, cytokinin, and abscisic acid) and N (nitrate and ammonium) to dissect N–hormone interactions. Individual treatments had distinct effects upon root growth, whereas combinations of treatments showed an array of interactions that ranged from pronounced (auxin and nitrate) to non-significant (nitrate and ammonium). A strong interaction between auxin and both nitrate and ammonium was found. Another strong and complex interaction between cytokinin, abscisic acid, nitrate, and ammonium suggested the existence of integration mechanisms for these signals. Measurements of genome-wide expression changes and subsequent GeneCloud analysis (Krouk et al., 2015) showed that specific signals or their combinations target specific regulatory modules. Certain genes showed ‘logic gate’ expression changes, with up-regulation occurring only under combined treatments. From all the ATH1 probes tested in this work, approximately 42% responded to composite signals, indicating that most gene expression changes are regulated by multiple inputs (Ristova et al., 2016).
The importance of NRT1.1 in nitrate responses was first established by a Serial Analysis of Gene Expression (SAGE) approach in Arabidopsis thaliana (Muños et al., 2004). In this study, transcript abundance in roots of the loss-of-function chl1-5 mutant differed by roughly 400 transcripts compared with wild-type plants when grown in the presence of ammonium nitrate. Transcripts of the Nitrate Transporter 1.5 (NRT1.5/NPF7.3) gene as well as the amino acid transporter At4g38250 were overexpressed. Interestingly, this mutant also showed a strong up-regulation of the high-affinity nitrate transporter NRT2.1. These results suggested that NRT1.1 is required for normal regulation of expression of nitrate transporters and other genes (Muños et al., 2004). Additional evidence supporting a role for NRT1.1 in nitrate sensing came from experiments with the loss-of-function mutants chl1-5 and nrg1: both alleles altered the nitrate-dependent induction of Nitrate reductase 1 (NIA1), Nitrite Reductase 1 (NiR), and NRT2.1 gene expression (Wang et al., 2009). The function of NRT1.1 as a sensor was further supported by analysis of the chl1-9 mutant, which retains NRT2.1 biphasic gene expression response but is deficient in nitrate transport (Ho et al., 2009). Interestingly, recent evidence indicates that nitrate-dependent gene induction and developmental responses are controlled by independent signaling pathways that are triggered downstream of NRT1.1 (Bouguyon et al., 2015).
Nitrate signaling and calcium
One model for nitrate signaling proposes that nitrate is sensed by NRT1.1, eliciting the immediate production of second messengers, which would consequently trigger changes in gene expression (Medici and Krouk, 2014; Wang et al., 2009). An attractive second messenger in this pathway is calcium (Ca2+). Support for the role of Ca2+ in nitrate signaling comes from early experiments in barley and corn, in which the expression of nitrate-responsive genes was shown to be altered by either EGTA or LaCl3 pre-treatment, pointing to a potential role of Ca2+ as a second messenger (Sakakibara et al., 1997; Sueyoshi et al., 1999).
Ca2+ is a key second messenger required for signal transduction in plants and other organisms (Dodd et al., 2010; Steinhorst and Kudla, 2014). Intracellular Ca2+ levels are in the micromolar range and are subject to tight regulation. In contrast, the cell exterior and intracellular compartments, such as the vacuole and endoplasmic reticulum (ER), have higher Ca2+ concentrations, typically in the millimolar range. Consequently, these different Ca2+ concentrations generate an electrochemical gradient across these compartments (Hashimoto and Kudla, 2011). This gradient allows a fast influx of Ca2+, facilitated by Ca2+ channels, into the cytoplasm. Ca2+ channels are located in the plasma membrane and in internal reserve compartments such as the vacuole and ER. Three types of Ca2+ channels have been identified in animals: voltage-dependent calcium channels (VDCCs), receptor-operated calcium channels (ROCCs), and mechanical-stimulation-gated channels (Hamilton et al., 2015; Sukharev and Sachs, 2012; Tsien and Tsien, 1990). VDCCs and ROCCs have been found in plants (Nagata et al., 2004; Sanders et al., 2002). Electrophysiological and genetic approaches have been used to demonstrate that mechanosensitive calcium channels are also present in plants (Hamilton et al., 2015).
Different biotic and environmental perturbations can cause specific spatiotemporal changes in cytosolic Ca2+ concentration (Dodd et al., 2010). Different types of stimuli trigger unique changes in free cytosolic Ca2+ that differ in frequency, amplitude, and localization. The ‘Ca2+ signature’ differs according to the identity of the Ca2+ elicitor and its intensity. Ca2+ sensor proteins, such as calmodulin kinases (CaM), CaM-related proteins, Ca2+-dependent protein kinases (CDPK), and calcineurin-like proteins (CBL), perceive cytosolic Ca2+ changes and transduce the signal to downstream signaling cascades that trigger changes in enzyme activity, cytoskeleton orientation, phosphorylation, and gene expression (Dodd et al., 2010; Hashimoto and Kudla, 2011).
It was recently shown that Ca2+ has a role in plant nitrate signal transduction and is important for nitrate-dependent regulation of gene expression in A. thaliana plants. Using aequorin reporter lines, it was shown that nitrate treatment causes a rapid increase in cytoplasmic Ca2+ levels in roots as well as in whole seedlings (Riveras et al., 2015). This response was inhibited when plants were pre-treated with either the Ca2+ channel blocker LaCl3 or the chelating agent EGTA. In addition, this Ca2+ increase was abolished in the NRT1.1 mutants chl1-5 and chl1-9, demonstrating that this response requires a functional transceptor. A concomitant increase in inositol 1, 4, 5-triphosphate (IP3) in response to nitrate treatments suggested that activity of a PLC is also involved in this pathway (Riveras et al., 2015).
PLCs are membrane-associated enzymes that break phospholipids, causing lipid membrane remodeling and generating multiple second messengers (Tuteja and Sopory, 2008). In plants, there are two classes of PLCs, which differ according to their substrate specificity: phosphatidylinositol-specific (PI-PLC) and non-specific (NPC). Plant NPCs share homology with bacterial PLCs. NPCs can have a preference for either acetylcholine (PC-PLC), phosphatidylethanolamine (PE-PLC), or phosphatidylserine (PS-PLC) (Rupwate and Rajasekharan, 2012).
PI-PLCs are the most extensively studied PLC class. They hydrolyze phosphatidylinositol 4,5-bisphosphate (PIP2) from the plasma membrane, producing IP3 and diacylglycerol (DAG) (Rupwate and Rajasekharan, 2012; Singh et al., 2015). The mechanism of action of PI-PLCs has been widely studied in animals and is based on activation of PI-PLCs by the α subunit of G protein (Gαq), which is dissociated in response to a stimulus (Munnik and Testerink, 2009). Once PI-PLC is activated, it splits a membrane phospholipid into IP3 and DAG. Subsequently, IP3 is released into the cytoplasm, where it can bind to IP3 receptors, promoting Ca2+ influx from intracellular reserves. The released Ca2+ can act as a second messenger (Hashimoto and Kudla, 2011; van Leeuwen et al., 2007). DAG remains membrane bound and can activate protein kinase C (PKC), which transduces the signal to effector molecules (Munnik and Testerink, 2009). DAG can also be converted to diacylglycerolpyrophosphate (DGPP), through the action of diacylglycerol kinase (DGK). DGPP can be used as a substrate to generate phosphatidic acid (PA) through phospholipase D (PLD) activity.
This mode of action of PI-PLCs appears to be present in plants but with some inconsistencies in comparison to animal systems: biochemical studies have shown that the plasma membrane PIP2 levels of plants are much lower than in animals (Delage et al., 2013). This was recently confirmed by experiments using radioactivity and biosensors: only small quantities of PIP2 were found in plasma membrane of plants by FRAP (fluorescence recovery after photobleaching) analysis and co-labeling experiments (Delage et al., 2013; Munnik and Testerink, 2009; van Leeuwen et al., 2007). Furthermore, plant cells lack homologs of animal IP3 receptors and protein kinase C signaling pathway components (Chen et al., 2011; Munnik and Testerink, 2009). Despite these differences from the animal model, products generated from IP3 and DAG, in particular inositol hexaphosphate (IP6) and phosphatidic acid (PA), respectively, can act as second messengers in plants (Arisz et al., 2009; Chen et al., 2011).
PI-PLC proteins have three minimal domains: the EF-hand in the N-terminal; the XY catalytic domains required for phosphotesterase activity; and a C2 domain on the C-terminal, which is responsible for Ca2+-dependent phospholipid binding (Singh et al., 2015; Tasma et al., 2008). In addition, a PH (pleckstrin homology) domain is found only in animals, and is responsible for the recognition of PIP2, which facilitates PLC membrane attachment (Singh et al., 2015). This domain has not been found in plants, and it is not known whether and how PIP2 binding occurs in plants (Pokotylo et al., 2014). PI-PLCs are also subclassified according to their subcellular localization as membrane-bound or cytosolic (Singh et al., 2015). Enzymatic activity for both groups is dependent on Ca2+ availability, which can also regulate substrate availability (Chen et al., 2011; Singh et al., 2015). Despite the absence of the PIP2 binding domain, it has been reported that the activity of PI-PLCs requires cell membrane association, either transiently or permanently, in order to associate with PIP2 (Helling et al., 2006; Lomasney et al., 1996).
Nine PI-PLC genes (AtPLC1–AtPLC9) have been identified in the A. thaliana genome (Mueller-Roeber and Pical, 2002; Tasma et al., 2008). Many of these have been characterized as multifunctional enzymes because of their role in modulating plant physiology under different biotic and abiotic stresses, nutritional deficiencies, or environmental conditions (Rupwate and Rajasekharan, 2012; Singh et al., 2015). In addition, it has been shown that three PLCs and most NPCs are implicated in growth and development processes in A. thaliana (Singh et al., 2015). A significant role for NtPLC3 in pollen tube growth has been reported in Nicotiana tabacum: NtPLC3 accumulates in the pollen tube plasma membrane during elongation. Growth of tobacco pollen tubes was blocked by treatment with U73122, a PI-PLC inhibitor; it was also reduced by NtPLC3 overexpression (Helling et al., 2006).
Expression analysis for various PI-PLC genes in A. thaliana showed that PLC isoforms are differentially expressed in diverse plant organs, as well as in response to biotic and abiotic stresses (Arisz et al., 2009; Helling et al., 2006; Hunt et al., 2004; Liu et al., 2006; Munnik and Testerink, 2009; Pokotylo et al., 2014; Tasma et al., 2008). AtPLC1, AtPLC2, AtPLC3, AtPLC4, AtPLC5, and AtPLC9 are specifically expressed in roots (Pokotylo et al., 2014; Tasma et al., 2008). Sequence homology, predicted phylogenetic relationships, and expression patterns suggest that some AtPLC pairs (AtPLC1–ATPLC3, AtPLC4–AtPLC5, and AtPLC8–AtPLC9) might be related to each other. AtPLC8 and AtPLC9 are proposed to lack enzymatic activity because they have a large deletion in the Y catalytic domain (Tasma et al., 2008). However, it has been shown that AtPLC9 has a role in stress responses. plc9 loss-of-function mutant plants have a heat-resistant phenotype (Zheng et al., 2012). Interestingly, it has also been shown that AtPLC4 and AtPLC5 expression is induced by nitrate: several previous studies showed that their mRNA levels were up-regulated after KNO3 treatments (Alvarez et al., 2014; Canales et al., 2014; Riveras et al., 2015; Vidal et al., 2013b; Wang et al., 2003; Wang et al., 2004).
A direct link between nitrate signaling and PI-PLCs has been established in Arabidopsis. Nitrate treatment triggered an increase in IP3 and cytoplasmatic Ca2+, which was not observed in plants pre-treated with the PLC inhibitor U73122. This suppression of nitrate-dependent increases in cytoplasmatic Ca2+ and IP3 was also observed in two different NRT1.1 mutants, chl1-5 and chl1-9, demonstrating that this response is NRT1.1-dependent. Furthermore, U73122 pre-treatment also affected the induction of canonical nitrate-responsive genes [NRT2.1, TGACG Sequence-specific Binding Protein 1 (TGA1), NiR, Nitrate Transporter 3.1 (NRT3.1), and NIA1], which resembled the expression levels observed for nrt1.1 mutants under the same conditions. These responses were not observed when plants were pre-treated with the non-functional analog U73343 (Riveras et al., 2015; Thompson et al., 1991).
These results suggest that NRT1.1 as well as phospholipase activity are required for nitrate-dependent increases in cytoplasmic Ca2+ levels and IP3 (Fig. 1A). However, it was also observed that expression of the nitrate-sensitive gene Auxin Signaling F-Box 3 (AFB3) was not affected (Fig 1B). This indicates that as well as the Ca2+ and PI-PLC-dependent pathway downstream of NRT1.1, there is another PI-PLC-independent pathway that controls the expression of nitrate-responsive genes (Riveras et al., 2015).
The role of phosphorylation in nitrate signaling
Phosphorylation is arguably one of the most important and well-documented post-translational protein modifications. Activation or inactivation of protein function, including components in signaling pathways, are mediated by phosphorylation/autophosphorylation cycles. Protein phosphorylation can impact the ability of proteins to interact to form hetero- or homodimers, protein stabilization or degradation, and change in localization, among other functions (Huber, 2007; Hunter, 1995; Olsen et al., 2006).
The role of protein phosphorylation in nitrate signaling was initially suggested through experiments with protein kinase and phosphatase inhibitors (Sakakibara et al., 1997; Sueyoshi et al., 1999). Kinase and phosphatase inhibition had distinct effects on the expression of genes coding for nitrogen-assimilatory enzymes, such as NR, NiR, and GS2, in maize leaves (Sakakibara et al., 1997). Inhibition of Ca2+/calmodulin-dependent protein kinases by treatment with the antagonists W-7 and trifluoperazine had no effect on NR and NiR mRNA levels in barley leaves (Sueyoshi et al., 1999). However, pre-treatments with W-7 had an inhibitory effect on the nitrate-elicited response of the NR (34%), NiR (34%), GS2 (63%), and Fd-GOGOAT (59%) genes. This implies that calmodulin-dependent and -independent protein kinases are at least partially involved in the nitrate signaling pathway (Sakakibara et al., 1997). Similarly, the importance of other groups of protein kinases has been proposed. It has been reported that the tyrosine kinase inhibitors genistein, quercetin, and curcumin compromise nitrate response in barley leaves (Sueyoshi et al., 1999). On the other hand, involvement of type 1 and type 2A serine/threonine protein phosphatases in nitrate signaling has been found using the inhibitors okadaic acid and calyculin A (Sueyoshi et al., 1999). Strong down-regulation of nitrate-induced accumulation of NR and NiR mRNAs in both maize and barley leaves suggests that alteration of protein dephosphorylation also has an impact on nitrate signaling (Sakakibara et al., 1997; Sueyoshi et al., 1999).
The function of central components involved in nitrate signaling and transport are modulated by phosphorylation/dephosphorylation. Nitrate transport activity across the plasma membrane and the tonoplast is regulated by phosphorylation (Liu and Tsay, 2003; Migocka et al., 2013). NRT1.1 can switch its affinity for nitrate depending on the phosphorylation status of a key threonine residue, T101. This residue is located in the intracellular side, between the second and third transmembrane helices of NRT1.1 (Ho et al., 2009; Hu et al., 2009; Liu and Tsay, 2003). Under low-nitrate conditions, CBL-Interacting Protein Kinase 23 (CIPK23) phosphorylates this residue, shifting NRT1.1 into a high-affinity nitrate carrier (Liu and Tsay, 2003) (Fig. 1). In addition, the nitrate primary response is highly reduced and a weak up-regulation of the NRT2.1 high-affinity transporter also occurs under low-nitrate conditions (Filleur et al., 2001; Ho et al., 2009). Recently, it was found that the calcium sensor Calcineurin B-Like Protein 1 (CBL1) and the ABA-sensitive Protein Phosphatase 2C (ABI2) are also important players in nitrate signaling in A. thaliana (Léran et al., 2015). The activity of ABI2 prevents full phosphorylation of CIPK23, inhibiting its kinase activity toward its substrate, NRT1.1. Under abiotic stress conditions, ABA inactivates ABI2, releasing its influence over CIPK23 and enhancing phosphorylation of NRT1.1, resulting in a net decrease in nitrate uptake. This finding suggest a mechanism by which plant stress signaling and nutrient uptake are coordinated by ABI2 (Léran et al., 2015).
Mutations mimicking or eliminating T101 phosphorylation abolish the dual-affinity transport activity of NRT1.1 and lock the transporter in either the high-affinity or low-affinity mode, respectively (Liu and Tsay, 2003). Recently, two independent groups unraveled the three-dimensional structure of NRT1.1 by X-ray crystallography (Parker and Newstead, 2014; Sun et al., 2014). The structural analysis showed a phosphorylation-dependent dimerization switching mechanism for the dual-affinity transporter. When T101 of NRT1.1 is dephosphorylated, the transporter forms homodimers and works as a low-affinity transporter. On the other hand, phosphorylated T101 turns the protein into a high-affinity transporter by structurally decoupling the dimer (Sun et al., 2014; Sun and Zheng, 2015). This finding is consistent with the idea that the non-phosphorylated form of NRT1.1 is the predominantly active form in nitrate signaling (Ho et al., 2009). A more recent report showed that phosphorylation of NRT1.1 has different signaling functions: the dephosphorylated form is critical for the primary root response, represented by the short-term induction of NRT2.1. The phosphorylated form triggers the induction of auxin transport, repressing lateral root emergence under low-nitrate conditions (Bouguyon et al., 2015). Therefore, differential phosphorylation of NRT1.1 not only affects the affinity of the transporter, but can also affect the output targets of the primary nitrate response.
Besides its role in nitrate acquisition and the primary response, phosphorylation also affects N metabolism. The first step of nitrate reduction is catalyzed by NR, an enzyme that is subjected to post-translational control by phosphorylation and a subsequent inhibitory interaction with 14-3-3 proteins (Bachmann et al., 1996b; Kaiser et al., 2002). This regulation was first observed as a change in NR activity during light/dark cycles in spinach leaves. Experiments using 32P labeling and kinase assays established that these changes were dependent on the phosphorylation status of NR (Huber et al., 1992; Mackintosh, 1992). Site-specific phosphorylation was assessed with peptide-antibodies raised against serine 543 and phospho-serine 543 of NR, showing that phosphorylation at this site was necessary but not sufficient for 14-3-3 binding (Weiner and Kaiser, 2001). Later studies showed that, in the presence of Mg2+ and/or other divalent cations, 14-3-3 proteins bind phospho-NR, inactivating it (Athwal and Huber, 2002; Bachmann et al., 1996a). This inhibition/activation switch is triggered by several environmental and intrinsic factors, such as photosynthesis, sugars, anoxia, and pH. Nitrate does not have any effect on this post-translational modification. However, it does exert transcriptional control over the NR gene (Kaiser et al., 2002).
A full-scale proteomic study performed in Arabidopsis seedlings under nitrate deprivation conditions showed that nitrate starvation and resupply affects both protein abundance and phosphorylation modifications. Changes in abundance were observed for 170 proteins, and 36 were reported to modify their phosphorylation status. Nitrate deprivation increased the abundance of stress-responsive proteins and proteins that play roles in catabolism and proteolysis; it also down-regulated biosynthetic proteins. After a 48-hour starvation, a down-regulation of enzymes such as NiR, Carbamoyl Phosphate Synthase (CARB), Arginosuccinate Synthase (ACC), and Carbonic Anhydrase (CA2) was observed (Wang et al., 2012). Engelsberger and Schulze (2012) assessed phosphorylation changes triggered by nitrate and ammonia treatments after N starvation. In general, transient phosphorylation changes were observed predominantly in the AHA1 and 2 subunits from the P-type plasma membrane ATPase and in proteins involved in N assimilation, amino acid biosynthesis, nucleotide metabolism, and tetrapyrrole synthesis. In general, protein phosphorylation status changed upon N resupply, with short-term responses centered in plasma membrane-associated proteins, medium-term responses with cytosolic proteins, and long-term responses with nuclear/cell interior proteins. This phosphorylation response behaved in the manner of ‘waves’ from the cell membrane to the interior. The most rapid phosphorylation change observed by the authors was in the high-affinity transporter NRT2.1, which was dephosphorylated within 3 minutes of N supply. Changes in the phosphorylation status of the ammonium transporters AMT1.1 and AMT1.3 were also observed (Engelsberger and Schulze, 2012).
A relationship between Ca2+ signaling and phosphorylation was previously established in animal models (Gresset et al., 2010). This study showed that the activity of some PI-PLCs was modulated by phosphorylation of the X/Y linker, which connects the two catalytic domains. This post-translational modification regulated PI-PLC activity, establishing a link between phosphorylation and phospholipase activation (Gresset et al., 2010). Phosphorylation sites have already been identified in many plant PI-PLCs by either mass spectrometry or bioinformatics predictions (Durek et al., 2010). A phosphoproteomic study in nitrogen-starved Arabidopsis seedlings showed that proteins from the phosphatidylinositol pathway are phosphorylated upon nitrate resupply. This effect was not observed when the seedlings were provided with ammonium as a N source, establishing that N-dependent phosphorylation of this pathway is nitrate-specific (Engelsberger and Schulze, 2012). Further characterization of plant PI-PLCs involved in nitrate signaling, as well as their modulation by phosphorylation, would provide a better understanding of the underlying mechanism that connects Ca2+, phosphorylation, and nitrate signaling.
Nitrate-elicited changes in gene expression
From the evidence discussed above, it can be concluded that nitrate-triggered changes in phosphorylation status and intracellular Ca2+ levels affect the transcription of at least some nitrate-responsive genes. Thousands of N-regulated genes have been identified from transcriptomic studies, generating mounting evidence pointing to either endogenous or exogenous N signals controlling a broad range of responses in different processes, including metabolism, growth, and development (Alvarez et al., 2012; Ruffel et al., 2014; Wang et al., 2004; Wang et al., 2003). Underlying N regulation of metabolism and development are transcription factors, which play important roles in regulating the expression of nitrate-responsive genes, including sentinel genes such as NRT1.1/NPF6.3, NRT2.1, NRT2.2, NIA1, NIA2, and NIR. So far, only a handful of transcription factors mediating nitrate responses have been identified: these are Arabidopsis Nitrate Regulated 1 (ANR1) (Zhang and Forde, 1998), NIN-like Protein 6 (NLP6) (Konishi and Yanagisawa, 2013), NIN-like Protein 7 (NLP7) (Marchive et al., 2013), LOB Domain-Containing proteins (LBD37/38/39) (Rubin et al., 2009), Squamosa Promoter Binding Protein-Like 9 (SPL9) (Krouk et al., 2010), Basic Leucine-Zipper 1 (bZIP1) (Para et al., 2014), NAC Domain Containing Protein 80 (NAC4) (Vidal et al., 2013b), TGA1/TGA4 (Alvarez et al., 2014), Teosinte Branched1/Cycloidea/Proliferating Cell Factor 20 (TCP20) (Guan et al., 2014), and Nitrate Regulatory Gene (NRG2) (Xu et al., 2016). Interaction with target gene promoters has been experimentally verified for only TGA1, NLP6/7, bZIP1, and TCP20.
NLP7 has been described as an important regulator of early nitrate-dependent gene expression changes. Its subcellular localization is nitrate responsive: in the presence of nitrate, NLP7 moves from the cytoplasm into the nucleus. Different nlp7 mutant alleles treated with nitrate show a misregulation of up to 58% of nitrate-induced genes. In one study using chromatin immunoprecipitation coupled to a whole-genome tiling array (ChIP-chip), 851 genes were identified as being bound by NLP7 in the presence of nitrate, further supporting a role for NLP7 as a main regulator (Marchive et al., 2013). Recently, a genetic screen led to the identification of NRG2 as a new regulatory factor of the nitrate response: a mutation in NRG2 disrupted the induction of nitrate-responsive genes after nitrate treatment. Interestingly, NRG2 interacts physically with NLP7 in the nucleus (Xu et al., 2016). TCP20 was identified by yeast one-hybrid screens that used the nitrate enhancer DNA fragments of NIA1 and NRT2.1 as baits. Expression of over 100 nitrate-responsive genes is controlled by this transcription factor. Electrophoretic mobility shift experiments demonstrated that TCP20 binds to the promoters of NIA1, NRT2.1, and NRT1.1. Root growth assays of loss-of-function mutants revealed an impairment in nitrate-induced lateral root growth (root foraging) and an impairment in systemic root growth. Therefore, this transcription factor has a critical role in nitrate-induced transcriptional changes, systemic signaling, and root foraging (Guan et al., 2014). The transcription factors TGA1 and TGA4 were identified through an integrative bioinformatics approach. Transcript levels of TGA1 and TGA4 vary in response to nitrate treatments of Arabidopsis roots. Experiments with the ATH1 Affymetrix microarray showed that the tga1/tga4 double mutant and wild-type plants had different nitrate responses, demonstrating that these transcription factors regulate genes involved in nitrate transport and metabolic functions. ChIP analysis indicated that these transcription factors control the expression of the high-affinity nitrate transporters NRT2.1 and NRT2.2 by binding directly to their promoters. Moreover, mutations of either tga1/tga4 or nrt2.1/nrt2.2 show similar alterations in nitrate-dependent lateral root growth. This suggests that TGA1/TGA4 and NRT2.1/NRT2.2 function in the same nitrate signaling pathway, regulating lateral root density in Arabidopsis (Alvarez et al., 2014).
In recent years, a novel technique named TARGET (Transient Transformation System for Genome-Wide Transcription Factor Target Discovery) was developed for analysis of direct transcriptional control. Briefly, this method consists of the transfection of protoplasts with a construct containing red fluorescent protein (RFP) and the transcription factor of interest fused to the glucocorticoid receptor (GR). RFP-containing protoplasts are then selected by fluorescence-activated cell sorting (FACS). The effect of the transcription factor on gene induction is subsequently assessed by dexamethasone treatment, which allows entry of the transcription factor into the nucleus. Experiments are performed either with or without the translation repressor cycloheximide, allowing discrimination between direct and indirect transcription factor targets (Bargmann et al., 2013).
This approach enabled the discovery of downstream targets for the transcription factor Hypersensitivity to Low Pi-Elicited Primary Root Shortening 1 (HRS1), which is strongly induced by nitrate. Interestingly, GO and semantic gene enrichment analysis (Krouk et al., 2015) of targets of HRS1 showed that the terms ‘phosphate’ and ‘cell division’ are up-regulated by this transcription factor. Further analysis showed that HRS1 is involved in both nitrate and phosphate signaling, integrating both pathways (Medici et al., 2015).
The downstream targets of the master transcription factor bZIP1, which integrates light and N sensing, were also identified using TARGET and ChIP-Seq (Obertello et al., 2010; Para et al., 2014). A total of 1308 bZIP1 primary targets were found by Para et al. (2014). They were categorized according to their interaction with bZIP1 and by the downstream regulation exerted by this transcription factor, as type I, poised (bound by bZIP1, but not regulated by it); type II, stable (bound and regulated by bZIP); and type III, transient (not bound, but regulated by bZIP). Interestingly, N-related biological processes were enriched in the type III class. Further analysis revealed that the canonical early N response genes NRT2.1, NIN-like protein 3 (NLP3), and LBD39 belong to this category. These results support the conclusion that the early N response follows a ‘hit-and-run’ transcription model, in which a transcription factor rapidly and transiently activates a large number of targets, facilitating the spread of the signal (Para et al., 2014).
Post-transcriptional mechanisms have also been found to be involved in nitrate-elicited gene expression changes. A study focusing on cell-specific root nitrogen responses (Birnbaum et al., 2003) showed that exogenous application of nitrate causes an increase in organic N, triggering the down-regulation of miR167, which targets the auxin response factor ARF8. This mechanism has the net effect of an up-regulation of ARF8 in pericycle cells in response to nitrate, controlling N-dependent lateral root initiation (Gifford et al., 2008) (Fig. 1C). The authors also reported that, from the initial ~6000 nitrate-responsive transcripts found, only 771 responded across all five cell types studied, and 87% were differentially regulated in one or a few cell types, indicating that N responses vary within root cell types (Gifford et al., 2008). It has been shown that the miR393/AFB3 regulatory module regulates nitrate-mediated root branching by an incoherent feed-forward loop in which exogenous nitrate induces early induction of AFB3. However, at later time points, miR393 is also induced, targeting the AFB3 mRNA and decreasing its abundance (Vidal et al., 2010). Subsequently, AFB3/miR393 control the expression of the transcription factor NAC4 (Vidal et al., 2013b) (Fig 1b).
Many early transcriptomic studies that address plant responses to nitrate relied on the Affymetrics Arabidopsis ATH1 Genome Array. This methodology has inherent limitations, in that it allows researchers to analyze only genes that are represented within the array. Vidal et al. (2013a) used next-generation sequencing technology to explore mRNA and small RNA components of the transcriptome. Besides the 13411 nitrate-responsive genes that are present in the array, this study identified 3022 additional nitrate-induced genes. Furthermore, not only coding RNAs were induced by nitrate, but also small non-coding RNAs such as miR5640, which targets Phosphoenol Pyruvate Carboxylase 3 (AtPPC3), an enzyme that has a role in carbon balance in plants. Interestingly, it was found that antisense transcripts for TCP transcription factor 23 (TCP23) are induced by nitrate, suggesting that additional post-transcriptional regulatory mechanisms may be important for nitrate responses (Vidal et al., 2013a).
Future perspectives
The dual-affinity transceptor NRT1.1 and its modification by phosphorylation at the T101 residue have been well characterized. However, the effect of differential phosphorylation on other factors, and what role it plays in nitrate signaling, needs to be further examined. In addition, many questions about N-dependent changes in phosphorylation remain. Are all nitrate-elicited phosphorylation changes dependent on the NRT1.1 transceptor? How is the sensed nitrate signal relayed to the immediate targets in the signaling pathway? Which kinases/phosphatases are responsible for the phosphorylation status switches of the different components? Given the limitations of current proteomics experiments, additional studies evaluating rapid and transient phosphorylation changes in response to nitrate and other N-nutrient/metabolites are necessary to assess the breath and impact of this particular post-translational modification.
Characterization of the early Ca2+-elicited response described in this review has just begun: the Arabidopsis genome has nine PLC genes and two of them are induced by nitrate. Uncovering which PLCs are relevant for N signaling, and understanding the biochemical mechanisms and post-translational modifications that link them to nitrate sensing, is essential to better understand nitrate signaling in plants. Additional elements of this pathway, such as IP3-derived signals, remain to be characterized in plants. Furthermore, nitrate signaling also has a PLC and Ca2+-independent component, as evidenced by AFB3 expression (Fig. 1B), suggesting the possibility of other second messenger(s) being involved in nitrate responses. The identity of this second messenger (or messengers) is still an open question.
Finally, elucidating the different nitrate-sensing mechanisms and understanding their spatiotemporal cross-talk, at the cell-specific, organ-specific, and organism level, will be essential to provide a holistic understanding of N-nutrient/metabolite sensing and responses in plants.
Acknowledgements
This work was supported by grants from the Howard Hughes Medical Institute, Fondo de Desarrollo de Areas Prioritarias (FONDAP) Center for Genome Regulation (15090007), Millennium Nucleus Center for Plant Systems and Synthetic Biology (NC130030), and Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) 1141097 to RAG.
References
- Alboresi A, Gestin C, Leydecker MT, Bedu M, Meyer C, Truong HN. 2005. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant, Cell & Environment 28, 500–512. [DOI] [PubMed] [Google Scholar]
- Alvarez JM, Riveras E, Vidal EA, et al. 2014. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. The Plant Journal 80, 1–13. [DOI] [PubMed] [Google Scholar]
- Alvarez JM, Vidal EA, Gutiérrez RA. 2012. Integration of local and systemic signaling pathways for plant N responses. Current Opinion in Plant Biology 15, 185–191. [DOI] [PubMed] [Google Scholar]
- Arisz SA, Testerink C, Munnik T. 2009. Plant PA signaling via diacylglycerol kinase. Biochimica et Biophysica Acta 1791, 869–875. [DOI] [PubMed] [Google Scholar]
- Athwal GS, Huber SC. 2002. Divalent cations and polyamines bind to loop 8 of 14-3-3 proteins, modulating their interaction with phosphorylated nitrate reductase. The Plant Journal 29, 119–129. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Athwal GS, Wu K, Ferl RJ, Huber SC. 1996a. 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Letters 398, 26–30. [DOI] [PubMed] [Google Scholar]
- Bachmann M, Huber JL, Liao PC, Gage DA, Huber SC. 1996b. The inhibitor protein of phosphorylated nitrate reductase from spinach (Spinacia oleracea) leaves is a 14-3-3 protein. FEBS Letters 387, 127–131. [DOI] [PubMed] [Google Scholar]
- Bargmann BO, Marshall-Colon A, Efroni I, Ruffel S, Birnbaum KD, Coruzzi GM, Krouk G. 2013. TARGET: a transient transformation system for genome-wide transcription factor target discovery. Molecular Plant 6, 978–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN. 2003. A gene expression map of the Arabidopsis root. Science 302, 1956–1960. [DOI] [PubMed] [Google Scholar]
- Bouguyon E, Brun F, Meynard D, et al. 2015. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants 1, 15015. [DOI] [PubMed] [Google Scholar]
- Canales J, Moyano TC, Villarroel E, Gutiérrez RA. 2014. Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Frontiers in Plant Science 5, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro Marín I, Loef I, Bartetzko L, Searle I, Coupland G, Stitt M, Osuna D. 2011. Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta 233, 539–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen GQ, Snyder CL, Greer MS, Weselake RJ. 2011. Biology and biochemistry of plant phospholipases. Critical Reviews in Plant Sciences 30, 239–258. [Google Scholar]
- Clement CR, Hopper MJ, Jones LHP, Leafe EL. 1978. The uptake of nitrate by Lolium perenne from flowing nutrient solution: II. Effect of light, defoliation, and relationship to CO2 flux. Journal of Experimental Botany 29, 1173–1183. [Google Scholar]
- Crawford NM, Forde BG. 2002. Molecular and developmental biology of inorganic nitrogen nutrition. The Arabidopsis Book 1, e0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM, Glass ADM. 1998. Molecular and physiological aspects of nitrate uptake in plants. Trends in Plant Science 3, 389–395. [Google Scholar]
- Crutzen PJ, Mosier AR, Smith KA, Winiwarter W. 2008. N2O release from agro-biofuel production negates global warming reduction by replacing fossil fuels. Atmospheric Chemistry and Physics 8, 389–395. [Google Scholar]
- Davidson EA. 2009. The contribution of manure and fertilizer nitrogen to atmospheric nitrous oxide since 1860. Nature Geoscience 2, 659–662. [Google Scholar]
- Delage E, Puyaubert J, Zachowski A, Ruelland E. 2013. Signal transduction pathways involving phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: convergences and divergences among eukaryotic kingdoms. Progress in Lipid Research 52, 1–14. [DOI] [PubMed] [Google Scholar]
- Delhon P, Gojon A, Tillard P, Passama L. 1996. Diurnal regulation of NO3– uptake in soybean plants IV. Dependence on current photosynthesis and sugar availability to the roots. Journal of Experimental Botany 47, 893–900. [Google Scholar]
- Dodd AN, Kudla J, Sanders D. 2010. The language of calcium signaling. Annual Review of Plant Biology 61, 593–620. [DOI] [PubMed] [Google Scholar]
- Drew MC, Saker LR. 1975. Nutrient supply and growth of seminal root system in barley: II. Localized, compensatory increases in lateral root growth and rates of nitrate uptake when nitrate supply is restricted to only part of the root system. Journal of Experimental Botany 26, 79–90. [Google Scholar]
- Durek P, Schmidt R, Heazlewood JL, Jones A, MacLean D, Nagel A, Kersten B, Schulze WX. 2010. PhosPhAt: the Arabidopsis thaliana phosphorylation site database. An update. Nucleic Acids Research 38, D828–D834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelsberger WR, Schulze WX. 2012. Nitrate and ammonium lead to distinct global dynamic phosphorylation patterns when resupplied to nitrogen-starved Arabidopsis seedlings. The Plant Journal 69, 978–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO 2015. World fertilizer trends and outlook to 2018. Rome: Food and Agriculture Organization of the United Nations. [Google Scholar]
- Filleur S, Dorbe MF, Cerezo M, Orsel M, Granier F, Gojon A, Daniel-Vedele F. 2001. An Arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Letters 489, 220–224. [DOI] [PubMed] [Google Scholar]
- Frink CR, Waggoner PE, Ausubel JH. 1999. Nitrogen fertilizer: retrospect and prospect. Proceedings of the National Academy of Sciences of the United States of America 96, 1175–1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford ML, Dean A, Gutiérrez RA, Coruzzi GM, Birnbaum KD. 2008. Cell-specific nitrogen responses mediate developmental plasticity. Proceedings of the National Academy of Sciences of the United States of America 105, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girin T, Lejay L, Wirth J, Widiez T, Palenchar PM, Nazoa P, Touraine B, Gojon A, Lepetit M. 2007. Identification of a 150 bp cis-acting element of the AtNRT2.1 promoter involved in the regulation of gene expression by the N and C status of the plant. Plant, Cell & Environment 30, 1366–1380. [DOI] [PubMed] [Google Scholar]
- Gojon A, Nacry P, Davidian JC. 2009. Root uptake regulation: a central process for NPS homeostasis in plants. Current Opinion in Plant Biology 12, 328–338.m [DOI] [PubMed] [Google Scholar]
- Gowri G, Kenis JD, Ingemarsson B, Redinbaugh MG, Campbell WH. 1992. Nitrate reductase transcript is expressed in the primary response of maize to environmental nitrate. Plant Molecular Biology 18, 55–64. [DOI] [PubMed] [Google Scholar]
- Gresset A, Hicks SN, Harden TK, Sondek J. 2010. Mechanism of phosphorylation-induced activation of phospholipase C-gamma isozymes. Journal of Biological Chemistry 285, 35836–35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan P, Wang R, Nacry P, Breton G, Kay SA, Pruneda-Paz JL, Davani A, Crawford NM. 2014. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proceedings of the National Academy of Sciences of the United States of America 111, 15267–15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA. 2012. Systems biology for enhanced plant nitrogen nutrition. Science 336, 1673–1675. [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Gifford ML, Poultney C, Wang R, Shasha DE, Coruzzi GM, Crawford NM. 2007a Insights into the genomic nitrate response using genetics and the Sungear Software System. Journal of Experimental Botany 58, 2359–2367. [DOI] [PubMed] [Google Scholar]
- Gutiérrez RA, Lejay LV, Dean A, Chiaromonte F, Shasha DE, Coruzzi GM. 2007b Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biology 8, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez RA, Stokes TL, Thum K, et al. 2008. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proceedings of the National Academy of Sciences of the United States of America 105, 4939–4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton ES, Schlegel AM, Haswell ES. 2015. United in diversity: mechanosensitive ion channels in plants. Annual Review of Plant Biology 66, 113–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Kudla J. 2011. Calcium decoding mechanisms in plants. Biochimie 93, 2054–2059. [DOI] [PubMed] [Google Scholar]
- Helling D, Possart A, Cottier S, Klahre U, Kost B. 2006. Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. The Plant Cell 18, 3519–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschman HR. 1991. Primary response genes induced by growth factors and tumor promoters. Annual Review of Biochemistry 60, 281–319. [DOI] [PubMed] [Google Scholar]
- Hirel B, Tetu T, Lea PJ, Dubois F. 2011. Improving nitrogen use efficiency in crops for sustainable agriculture. Sustainability 3, 1452–1485. [Google Scholar]
- Ho CH, Lin SH, Hu HC, Tsay YF. 2009. CHL1 functions as a nitrate sensor in plants. Cell 138, 1184–1194. [DOI] [PubMed] [Google Scholar]
- Hu HC, Wang YY, Tsay YF. 2009. AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. The Plant Journal 57, 264–278. [DOI] [PubMed] [Google Scholar]
- Huber JL, Huber SC, Campbell WH, Redinbaugh MG. 1992. Reversible light/dark modulation of spinach leaf nitrate reductase activity involves protein phosphorylation. Archives of Biochemistry and Biophysics 296, 58–65. [DOI] [PubMed] [Google Scholar]
- Huber SC. 2007. Exploring the role of protein phosphorylation in plants: from signalling to metabolism. Biochemical Society Transactions 35, 28–32. [DOI] [PubMed] [Google Scholar]
- Hunt L, Otterhag L, Lee JC, et al. 2004. Gene-specific expression and calcium activation of Arabidopsis thaliana phospholipase C isoforms. New Phytologist 162, 643–654. [DOI] [PubMed] [Google Scholar]
- Hunter T. 1995. Protein kinases and phosphatases: the yin and yang of protein phosphorylation and signaling. Cell 80, 225–236. [DOI] [PubMed] [Google Scholar]
- Kaiser WM, Weiner H, Kandlbinder A, Tsai CB, Rockel P, Sonoda M, Planchet E. 2002. Modulation of nitrate reductase: some new insights, an unusual case and a potentially important side reaction. Journal of Experimental Botany 53, 875–882. [DOI] [PubMed] [Google Scholar]
- Konishi M, Yanagisawa S. 2013. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nature Communications 4, 1617. [DOI] [PubMed] [Google Scholar]
- Krapp A. 2015. Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Current Opinion in Plant Biology 25, 115–122. [DOI] [PubMed] [Google Scholar]
- Krouk G. 2016. Hormones and nitrate: a two-way connection. Plant Molecular Biology 91, 599–606. [DOI] [PubMed] [Google Scholar]
- Krouk G, Carré C, Fizames C, Gojon A, Ruffel S, Lacombe B. 2015. GeneCloud reveals semantic enrichment in lists of gene descriptions. Molecular Plant 8, 971–973. [DOI] [PubMed] [Google Scholar]
- Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. 2010. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biology 11, R123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G, Ruffel S, Gutiérrez RA, Gojon A, Crawford NM, Coruzzi GM, Lacombe B. 2011. A framework integrating plant growth with hormones and nutrients. Trends in Plant Science 16, 178–182. [DOI] [PubMed] [Google Scholar]
- Léran S, Edel KH, Pervent M, et al. 2015. Nitrate sensing and uptake in Arabidopsis are enhanced by ABI2, a phosphatase inactivated by the stress hormone abscisic acid. Science Signaling 8, ra43. [DOI] [PubMed] [Google Scholar]
- Liu HT, Huang WD, Pan QH, Weng FH, Zhan JC, Liu Y, Wan SB, Liu YY. 2006. Contributions of PIP(2)-specific-phospholipase C and free salicylic acid to heat acclimation-induced thermotolerance in pea leaves. Journal of Plant Physiology 163, 405–416. [DOI] [PubMed] [Google Scholar]
- Liu J, An X, Cheng L, Chen F, Bao J, Yuan L, Zhang F, Mi G. 2010. Auxin transport in maize roots in response to localized nitrate supply. Annals of Botany 106, 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KH, Tsay YF. 2003. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. The EMBO Journal 22, 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomasney JW, Cheng HF, Wang LP, Kuan Y, Liu S, Fesik SW, King K. 1996. Phosphatidylinositol 4,5-bisphosphate binding to the pleckstrin homology domain of phospholipase C-delta1 enhances enzyme activity. Journal of Biological Chemistry 271, 25316–25326. [DOI] [PubMed] [Google Scholar]
- MacKintosh C. 1992. Regulation of spinach-leaf nitrate reductase by reversible phosphorylation. Biochimica et Biophysica Acta 1137, 121–126. [DOI] [PubMed] [Google Scholar]
- Marchive C, Roudier F, Castaings L, Bréhaut V, Blondet E, Colot V, Meyer C, Krapp A. 2013. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nature Communications 4, 1713. [DOI] [PubMed] [Google Scholar]
- Marschner H. 2012. Marschner’s mineral nutrition of higher plants. London: Academic Press. [Google Scholar]
- Medici A, Krouk G. 2014. The primary nitrate response: a multifaceted signalling pathway. Journal of Experimental Botany 65, 5567–5576. [DOI] [PubMed] [Google Scholar]
- Medici A, Marshall-Colon A, Ronzier E, et al. 2015. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nature Communications 6, 6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migocka M, Warzybok A, Papierniak A, Kłobus G. 2013. NO3⁻/H⁺ antiport in the tonoplast of cucumber root cells is stimulated by nitrate supply: evidence for a reversible nitrate-induced phosphorylation of vacuolar NO3⁻/H⁺ antiport. PLoS One 8, e73972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Cramer MD. 2004. Root nitrogen acquisition and assimilation. Plant and soil 274, 1–36. [Google Scholar]
- Mueller-Roeber B, Pical C. 2002. Inositol phospholipid metabolism in Arabidopsis. Characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiology 130, 22–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Testerink C. 2009. Plant phospholipid signaling: “in a nutshell”. Journal of Lipid Research 50Suppl, S260–S265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muños S, Cazettes C, Fizames C, Gaymard F, Tillard P, Lepetit M, Lejay L, Gojon A. 2004. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. The Plant Cell 16, 2433–2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata T, Iizumi S, Satoh K, et al. 2004. Comparative analysis of plant and animal calcium signal transduction element using plant full-length cDNA data. Molecular Biology and Evolution 21, 1855–1870. [DOI] [PubMed] [Google Scholar]
- O’Brien JA, Vega A, Bouguyon E, Krouk G, Gojon A, Coruzzi G, Gutiérrez RA. 2016. Nitrate transport, sensing, and responses in plants. Molecular Plant 9, 837–856. [DOI] [PubMed] [Google Scholar]
- Obertello M, Krouk G, Katari MS, Runko SJ, Coruzzi GM. 2010. Modeling the global effect of the basic-leucine zipper transcription factor 1 (bZIP1) on nitrogen and light regulation in Arabidopsis. BMC Systems Biology 4, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. 2006. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648. [DOI] [PubMed] [Google Scholar]
- Owen AG, Jones DL. 2001. Competition for amino acids between wheat roots and rhizosphere microorganisms and the role of amino acids in plant N acquisition. Soil Biology & Biochemistry 33, 651–657. [Google Scholar]
- Palenchar PM, Kouranov A, Lejay LV, Coruzzi GM. 2004. Genome-wide patterns of carbon and nitrogen regulation of gene expression validate the combined carbon and nitrogen (CN)-signaling hypothesis in plants. Genome Biology 5, R91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Para A, Li Y, Marshall-Colon A, et al. 2014. Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 111, 10371–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JL, Newstead S. 2014. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature 507, 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokotylo I, Kolesnikov Y, Kravets V, Zachowski A, Ruelland E. 2014. Plant phosphoinositide-dependent phospholipases C: variations around a canonical theme. Biochimie 96, 144–157. [DOI] [PubMed] [Google Scholar]
- Rahayu YS, Walch-Liu P, Neumann G, Römheld V, von Wirén N, Bangerth F. 2005. Root-derived cytokinins as long-distance signals for NO3–-induced stimulation of leaf growth. Journal of Experimental Botany 56, 1143–1152. [DOI] [PubMed] [Google Scholar]
- Redinbaugh MG, Campbell WH. 1991. Higher-plant responses to environmental nitrate. Physiologia Plantarum 82, 640–650. [Google Scholar]
- Redinbaugh MG, Campbell WH. 1993. Glutamine synthetase and ferredoxin-dependent glutamate synthase expression in the maize (Zea mays) root primary response to nitrate (evidence for an organ-specific response). Plant Physiology 101, 1249–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remans T, Nacry P, Pervent M, Girin T, Tillard P, Lepetit M, Gojon A. 2006. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiology 140, 909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristova D, Carré C, Pervent M, et al. 2016. Combinatorial interaction network of transcriptomic and phenotypic responses to nitrogen and hormones in the Arabidopsis thaliana root. Science Signaling 9, rs13. [DOI] [PubMed] [Google Scholar]
- Riveras E, Alvarez JM, Vidal EA, Oses C, Vega A, Gutiérrez RA. 2015. The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiology 169, 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GP, Vitousek PM. 2009. Nitrogen in agriculture: balancing the cost of an essential resource. Annual Review of Environment and Resources 34, 97–125. [Google Scholar]
- Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. 2009. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. The Plant Cell 21, 3567–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffel S, Gojon A, Lejay L. 2014. Signal interactions in the regulation of root nitrate uptake. Journal of Experimental Botany 65, 5509–5517. [DOI] [PubMed] [Google Scholar]
- Ruffel S, Krouk G, Ristova D, Shasha D, Birnbaum KD, Coruzzi GM. 2011. Nitrogen economics of root foraging: transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proceedings of the National Academy of Sciences of the United States of America 108, 18524–18529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupwate SD, Rajasekharan R. 2012. Plant phosphoinositide-specific phospholipase C: an insight. Plant Signaling & Behavior 7, 1281–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara H, Kobayashi K, Deji A, Sugiyama T. 1997. Partial characterization of the signaling pathway for the nitrate-dependent expression of genes for nitrogen-assimilatory enzymes using detached maize leaves. Plant and Cell Physiology 38, 837–843. [Google Scholar]
- Sanders D, Pelloux J, Brownlee C, Harper JF. 2002. Calcium at the crossroads of signaling. The Plant Cell 14Suppl, S401–S417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible WR, Morcuende R, Czechowski T, et al. 2004. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiology 136, 2483–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Bhatnagar N, Pandey A, Pandey GK. 2015. Plant phospholipase C family: Regulation and functional role in lipid signaling. Cell Calcium 58, 139–146. [DOI] [PubMed] [Google Scholar]
- Steinhorst L, Kudla J. 2014. Signaling in cells and organisms—calcium holds the line. Current Opinion in Plant Biology 22, 14–21. [DOI] [PubMed] [Google Scholar]
- Stewart WM, Dibb DW, Johnston AE, Smyth TJ. 2005. The contribution of commercial fertilizer nutrients to food production. Agronomy Journal 97, 1–6. [Google Scholar]
- Sueyoshi K, Mitsuyama T, Sugimoto T, Kleinhofs A, Warner RL, Oji Y. 1999. Effects of inhibitors for signaling components on the expression of the genes for nitrate reductase and nitrite reductase in excised barley leaves. Soil Science and Plant Nutrition 45, 1015–1019. [Google Scholar]
- Sukharev S, Sachs F. 2012. Molecular force transduction by ion channels: diversity and unifying principles. Journal of Cell Science 125, 3075–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Bankston JR, Payandeh J, Hinds TR, Zagotta WN, Zheng N. 2014. Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature 507, 73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zheng N. 2015. Molecular mechanism underlying the plant NRT1.1 dual-affinity nitrate transporter. Frontiers in Physiology 6, 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasma IM, Brendel V, Whitham SA, Bhattacharyya MK. 2008. Expression and evolution of the phosphoinositide-specific phospholipase C gene family in Arabidopsis thaliana. Plant Physiology and Biochemistry 46, 627–637. [DOI] [PubMed] [Google Scholar]
- Thompson AK, Mostafapour SP, Denlinger LC, Bleasdale JE, Fisher SK. 1991. The aminosteroid U-73122 inhibits muscarinic receptor sequestration and phosphoinositide hydrolysis in SK-N-SH neuroblastoma cells. A role for Gp in receptor compartmentation. Journal of Biological Chemistry 266, 23856–23862. [PubMed] [Google Scholar]
- Tsien RW, Tsien RY. 1990. Calcium channels, stores, and oscillations. Annual Review of Cell Biology 6, 715–760. [DOI] [PubMed] [Google Scholar]
- Tuteja N, Sopory SK. 2008. Plant signaling in stress: G-protein coupled receptors, heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signaling & Behavior 3, 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen W, Vermeer JE, Gadella TW, Jr, Munnik T. 2007. Visualization of phosphatidylinositol 4,5-bisphosphate in the plasma membrane of suspension-cultured tobacco BY-2 cells and whole Arabidopsis seedlings. The Plant Journal 52, 1014–1026. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Álvarez JM, Moyano TC, Gutiérrez RA. 2015. Transcriptional networks in the nitrate response of Arabidopsis thaliana. Current Opinion in Plant Biology 27, 125–132. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Araus V, Lu C, Parry G, Green PJ, Coruzzi GM, Gutiérrez RA. 2010. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America 107, 4477–4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Gutiérrez RA. 2008. A systems view of nitrogen nutrient and metabolite responses in Arabidopsis. Current Opinion in Plant Biology 11, 521–529. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Canales J, Gutiérrez RA. 2014. Nitrogen control of developmental phase transitions in Arabidopsis thaliana. Journal of Experimental Botany 65, 5611–5618. [DOI] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Krouk G, Katari MS, Tanurdzic M, McCombie WR, Coruzzi GM, Gutiérrez RA. 2013a Integrated RNA-seq and sRNA-seq analysis identifies novel nitrate-responsive genes in Arabidopsis thaliana roots. BMC Genomics 14, 701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal EA, Moyano TC, Riveras E, Contreras-Lopez O, Gutiérrez RA. 2013b Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proceedings of the National Academy of Sciences of the United States of America 110, 12840–12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Ivanov II, Filleur S, Gan Y, Remans T, Forde BG. 2006. Nitrogen regulation of root branching. Annals of Botany 97, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Liu P, Neumann G, Bangerth F, Engels C. 2000. Rapid effects of nitrogen form on leaf morphogenesis in tobacco. Journal of Experimental Botany 51, 227–237. [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM. 2000. Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. The Plant Cell 12, 1491–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. 2003. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiology 132, 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Tischner R, Gutiérrez RA, Hoffman M, Xing X, Chen M, Coruzzi G, Crawford NM. 2004. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiology 136, 2512–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Xing X, Wang Y, Tran A, Crawford NM. 2009. A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiology 151, 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Bian Y, Cheng K, Zou H, Sun SS, He JX. 2012. A comprehensive differential proteomic study of nitrate deprivation in Arabidopsis reveals complex regulatory networks of plant nitrogen responses. Journal of Proteome Research 11, 2301–2315. [DOI] [PubMed] [Google Scholar]
- Weiner H, Kaiser WM. 2001. Antibodies to assess phosphorylation of spinach leaf nitrate reductase on serine 543 and its binding to 14-3-3 proteins. Journal of Experimental Botany 52, 1165–1172. [PubMed] [Google Scholar]
- Xu G, Fan X, Miller AJ. 2012. Plant nitrogen assimilation and use efficiency. Annual Review of Plant Biology 63, 153–182. [DOI] [PubMed] [Google Scholar]
- Xu N, Wang R, Zhao L, et al. 2016. The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. The Plant Cell 28, 485–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S, Zhang ZW, Zheng C, et al. 2016. Arabidopsis cryptochrome 1 functions in nitrogen regulation of flowering. Proceedings of the National Academy of Sciences of the United States of America 113, 7661–7666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang HM, Forde BG. 1998. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 279, 407–409. [DOI] [PubMed] [Google Scholar]
- Zheng SZ, Liu YL, Li B, Shang ZL, Zhou RG, Sun DY. 2012. Phosphoinositide-specific phospholipase C9 is involved in the thermotolerance of Arabidopsis. Plant Journal 69, 689–700. [DOI] [PubMed] [Google Scholar]