Abstract
Background
Rapid diagnostic tests based on Plasmodium falciparum histidine-rich protein II (PfHRP-II) and P. falciparum lactate dehydrogenase (PfLDH) antigens are widely deployed for detection of P. falciparum infection; however, these tests often miss cases of low-level parasitemia, and PfHRP-II tests can give false-negative results when P. falciparum strains do not express this antigen.
Methods
We screened proteomic data for highly expressed P. falciparum proteins and compared their features to those of PfHRP-II and PfLDH biomarkers. Search criteria included high levels of expression, conservation in all parasite strains, and good correlation of antigen levels with parasitemia and its clearance after drug treatment. Different assay methods were compared for sensitive detection of parasitemia in P. falciparum cultures.
Results
Among potential new biomarkers, a P. falciparum homolog of insulin-degrading enzyme (PfIDEh) met our search criteria. Comparative enzyme-linked immunosorbent assays with monoclonal antibodies against PfLDH or PfIDEh showed detection limits of 100–200 parasites/µL and 200–400 parasites/µL, respectively. Detection was dramatically improved by use of real-time immuno–polymerase chain reaction (PCR), to parasitemia limits of 0.02 parasite/µL and 0.78 parasite/µL in PfLDH- and PfIDEh-based assays, respectively.
Conclusions
The ability of PfLDH- or PfIDEh-based immuno-PCR assays to detect <1 parasite/µL suggests that improvements of bound antibody sensor technology may greatly increase the sensitivity of malaria rapid diagnostic tests.
Keywords: insulin-degrading enzyme, lactate dehydrogenase enzyme, PfHRP-II, enzyme-linked immunosorbent assay, immuno-PCR
Summary
Rapid diagnostic tests with greater sensitivity and specificity are needed for malaria control and elimination. Results from immuno–polymerase chain reaction assays with PfLDH and PfIDEh antigens suggest that these improvements may be achievable by new developments in antibody sensor technology.
Malaria control programs including insecticide-treated nets, rapid diagnostic tests (RDTs), and antimalarial prophylaxis and treatment have made considerable progress in recent years [1, 2]. Malaria incidence and mortality, taking into account population growth, are estimated to have decreased by 41% and 62% between 2000 and 2015 [3]. To achieve further progress in anticipation of malaria eradication, the World Health Organization (WHO) has adopted the Global Technical Strategy for Malaria 2016–2030. Greater sensitivity and specificity of RDTs are needed to support this strategy, especially by improved detection of asymptomatic low-level parasitemias that remain reservoirs of transmission and malaria outbreaks [4–6].
WHO sponsors a program of RDT evaluations for reliability and performance (http://www.who.int/malaria/areas/diagnosis/rapid-diagnostic-tests/product-testing-round6/en/). These comparisons have shown that many RDTs perform comparably to expert detection of Plasmodium falciparum infections by microscopy, with the advantage that RDTs can be performed in minutes by village triage staff at points of care and do not require the labor, time, and training necessary for examination of blood films in a laboratory [7]. Nevertheless, improved RDTs are needed as they can miss very low, asymptomatic parasitemias; insufficient sensitivity is thought to be among the reasons that recent mass screening and treatment trials failed to achieve sustained reductions of parasite prevalence or disease incidence [8].
Various approaches are now being pursued for diagnostic tests of greater sensitivity and specificity [9]. Among these are high-sensitivity P. falciparum histidine-rich protein II (PfHRP-II) antigen–based RDTs with detection 8–16 times better than conventional RDTs [10], and polymerase chain reaction (PCR)–based assays that can detect very low parasitemias (≤1 parasite/µL), although the expertise and laboratory requirements for these assays remain high and samples can be prone to contamination or degradation [11]. To improve the practicability of nucleic acid detection, loop-mediated isothermal amplification (LAMP) tests have been developed for malaria diagnosis [12]. LAMP may prove practical for very low parasitemia detection depending on the affordability and reliability it can achieve in routine field settings [13].
Commonly used RDTs rely on detection of antigens such as PfHRP-II, P. falciparum lactate dehydrogenase (PfLDH), or aldolase [6]. PfHRP-II–based RDTs have greater sensitivity than PfLDH- or aldolase-based RDTs [14]. However, false-negative PfHRP-II tests can occur with P. falciparum strains that do not produce PfHRP-II antigen [15, 16] or from a prozone-like effect with high levels of antigen [17]. PfHRP-II antigen can remain in the blood for weeks after parasites are cleared by drug treatment, yielding persistently positive results of no use for assessments of clinical outcome [18].
Considering the above observations, we asked 2 questions: (1) Can different P. falciparum antigens be found that support diagnostic sensitivity levels similar to those of PfHRP-II, but with reliable detection of all P. falciparum strains and the rapid disappearance from blood after treatment that characterizes PfLDH—that is, can a new RDT candidate be found that has the advantages of both PfHRP-II and PfLDH? (2) Can the sensitivity of RDTs based on PfHRP-II, PfLDH, or a new candidate antigen be significantly improved below the current 100–200 parasites/µL limit [14], for better detection of asymptomatic infections (<1 parasite/µL) [1, 19]? Here, we describe a survey of proteomic and genomic databases for novel RDT candidates, identify a P. falciparum insulin-degrading enzyme homolog (PfIDEh) as a conserved, highly expressed protein with tandem repeats, and apply real-time immuno-PCR (i-PCR) with PfLDH or PfIDEh to obtain >100-fold better detection than by enzyme-linked immunosorbent assay (ELISA). Implications of these findings for new-generation RDTs are discussed.
METHODS
Computer Database Searches, Bioinformatics, and Analysis of PfIDEh Polymorphisms
Proteomic data available from 10 studies [20–29] were downloaded from PlasmoDB (http://plasmodb.org/plasmo/). Canonical repeats were detected by Finder programs (http://nihserver.mbi.ucla.edu/Repeats/ and http://www.ebi.ac.uk/Tools/pfa/radar/). Antigenicity was analyzed by MacVector 15.0 software (Apex). Polymorphic regions of the PfIDEh gene were sequenced by standard methods (Supplementary Materials).
Peptide Immunogens and Rabbit Antibodies
Peptide Synthesis, Keyhole Limpet Hemocyanin Coupling, and Rabbit Monoclonal Antibody Production
Peptides (Supplementary Table 1) were synthesized on a 433A Automated Peptide Synthesizer (Applied Biosystems, Foster City, California), confirmed by high-performance liquid chromatography and matrix-assisted laser desorption/ionization–time-of-flight mass spectrometry, and coupled via amide groups to keyhole limpet hemocyanin (KLH). A multiple antigen peptide was constructed from NTSDDDNTSDDDNTS of the PfIDEh repeat region 2 by Bio-Synthesis, Inc (Lewisville, Texas). Production of rabbit antisera and monoclonal antibodies (mAbs) against peptide sequences is described in the Supplementary Materials.
His-Tagged PfIDEh Recombinant Protein and Production of Mouse Monoclonal Antibodies
A codon-optimized version of PfIDEh (P. falciparum Dd2 line; GenBank: PFDG_00647) was designed with an added hexa-histidine tail sequence and expressed in Baculovirus-infected Sf9 cells (GenScript Corporation, www.genscript.com). BALB/c mice were immunized with 50 μg of purified recombinant PfIDEh (rc-PfIDEh) in complete Freund adjuvant followed by two 25-μg booster injections in incomplete Freund adjuvant at 2-week intervals; mAbs were produced from standard hybridoma fusions and purified by protein A/G affinity columns (GenScript Corporation).
DNA Vaccinations and Mouse Monoclonal Antibody Production
Five DNA fragments (L1–L5) from regions of the PfIDEh coding sequence (Figure 1A) were amplified by PCR (Supplementary Table 2) and cloned into VR2001-TOPO. Mice were vaccinated and used to produce mAbs by standard methods; binding specificities and association (ka) and dissociation (kd) rate constants of these antibodies were determined from immunoblot, immunoprecipitation, mass spectrometry, and surface plasmon resonance analyses (see Supplementary Materials for details).
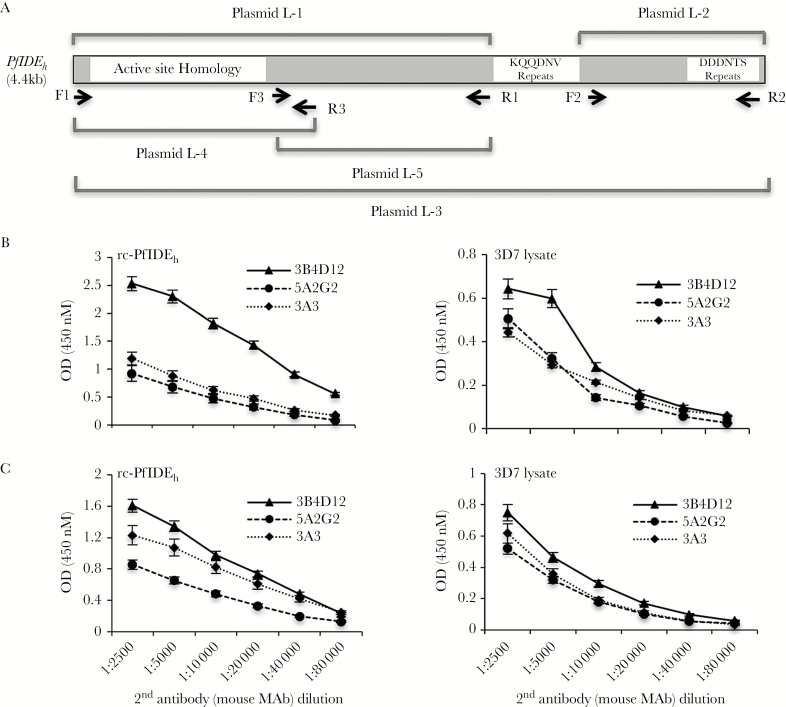
Figure 1.
Map of Plasmodium falciparum insulin-degrading enzyme homolog (PfIDEh) plasmid inserts and plots of enzyme-linked immunosorbent assay (ELISA) results with monoclonal antibody (mAb) pairs. A, Plasmid inserts were amplified from the PfIDEh sequence by indicated primer pairs. Schematic of the PfIDEh primary structure shows relative locations of the KQQDNV and DDDNTS repeats. B, Plots show signal levels obtained from horseradish peroxidase (HRP)–labeled 3B4D12, 5A2G2, and 3A3 mAbs in ELISAs of recombinant PfIDEh (rc-PfIDEh) or of PfIDEh captured by rabbit mAb AID-1-1-9 from 3D7 parasites. C, Plots show signal levels from HRP-labeled goat antimouse antibody applied to unlabeled 3B4D12, 5A2G2, and 3A3 mAbs in ELISAs of rc-PfIDEh or of PfIDEh captured by rabbit mAb AID-1-1-9 from 3D7 parasites.
Enzyme-Linked Immunosorbent Assay
For PfHRP-II ELISA, paired mouse mAbs (mouse immunoglobulin M MPFM-55A; horseradish peroxidase [HRP]–conjugated mouse immunoglobulin G MPFG-55A) were purchased from Immunology Consultants Laboratory (Portland, Oregon). For PfLDH ELISA, anti-PfLDH mouse mAbs MBS498007 and MBS498008 were purchased from Mybiosource (San Diego, California). For PfIDEh ELISA, mAbs generated in this study were used. HRP labeling was performed with a conjugation kit as recommended (Abcam, catalog number ab102890).
Immuno-PCR Detection of Captured PfLDH and PfIDEh
The i-PCR assays were carried out as described [30, 31] with modifications to optimize blocking conditions and decrease nonspecific signals. A 342-bp biotinylated DNA marker was prepared by PCR amplification using the pUC19 plasmid as the template and primer pair, pUC-bio (5ʹ-biotin-CCCGGATCCCAGCAATAAACCAGCCAGCC-3ʹ) and F1 (5ʹ-TATGCAGTGCTGCCATAACCATGA-3ʹ).
To capture PfLDH or PfIDEh for i-PCR, mouse mAb (MBS498007 or 3B4D12) was coated onto the wells of a microtiter plate overnight at 4°C (50 μL/well, 8 μg/mL). The wells were washed 5 times with 150 mL of Tris-buffered saline (20 mM Tris, 150 mM NaCl, pH = 7.4) and then treated for 1 hour at room temperature with 150 mL of Neptune block buffer (ImmunoChemistry Technologies, Bloomington, Minnesota) plus 200 ng/mL boiled (single-stranded) salmon sperm DNA (Thermo Fisher Scientific, catalog number AM9680). Two-fold serial dilutions of 3D7 parasite lysate in RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate; Thermo Fisher Scientific, catalog number 89900) or recombinant PfIDEh in 1× phosphate-buffered saline (PBS) buffer (10 mM PO43−, 137 mM NaCl, and 2.7 mM KCl, pH = 7.4) were added (30 μL/well) for 2 hours at 37°C. After 5 washes with 1× PBS containing 0.05% Tween 20, the wells containing captured antigen were incubated at 37°C for 30 minutes with streptavidin-labeled anti-PfLDH MBS498008 or anti-PfIDEh AID1-1-9 (Streptavidin Conjugation Kit, Abcam, catalog number ab102921) in Neptune Block buffer. Unbound streptavidin-labeled antibodies were cleared from the wells by additional 6 washes with 1× PBS containing 0.05% Tween 20 at room temperature.
For i-PCR detection, 30 μL of 0.5 ng/μL biotinylated DNA in Neptune Block buffer was added to each well. After 30 minutes of incubation at room temperature with the streptavidin-labeled antibodies, unbound biotinylated DNA was removed by 10 washes with 1× PBS containing 0.05% Tween 20. Bound DNA was released from the biotin-streptavidin complex by incubation with Bam HI (1 unit/well, 30 μL volume) for 2 hours at 37°C. Real-time PCR detection was performed on 6-μL samples in a Rotor-Gene Q instrument (Qiagen, Valencia, California) with cycling parameters: initial 5 minutes denaturation step at 95°C, followed by 40 cycles of 5 seconds at 95°C and 10 seconds at 60°C. All experiments were performed in duplicate. Statistical analyses were performed using Prism 7 software (GraphPad Software, La Jolla, California).
P. falciparum Cultures and PfHRP-II, PfLDH, and PfIDEh Assessments After Chloroquine Treatment
Plasmodium falciparum 3D7 parasites were cultivated in O+ human red blood cells at 2% hematocrit under standard in vitro conditions. Cultures were treated with 150 nM chloroquine (Sigma, St Louis, Missouri) at parasitemias of 8% (mixed stages). Infected erythrocytes and culture media were collected at the time of chloroquine treatment and at 24-hour intervals for 9 days.
RESULTS
P. falciparum Proteome Searches for Highly Expressed Conserved Proteins With Tandem Repeats
Sensitive PfHRP-II-based RDTs take advantage of the multiple binding sites for mAbs to tandem repeats (most commonly AHHAAD) that occur throughout much of the PfHRP-II protein [32]. These repeats increase mAb avidity and help to account for the greater sensitivity of PfHRP-II detection over PfLDH- and aldolase-based tests, even though PfLDH and aldolase are among the most abundant proteins in proteome databases [20]. Considering the variability and even absence of PfHRP-II expression by some P. falciparum strains, we surveyed proteomic data from 10 studies [20–29] for proteins that are conserved in all known parasite strains, contain tandem repeats, and are highly represented in asexual (ring, trophozoite, schizont) stage parasites. Relative representations were estimated from the number of spectra from each protein relative to the number from PfLDH in the parasitized erythrocytes. Only proteins with >2% representation relative to PfLDH and ≥3 canonical peptide repeats were included for further analysis. Proteins reported to be nonessential to the parasites in vitro [33] were also removed from consideration.
Eight P. falciparum proteins identified by the above selection criteria are listed in Supplementary Table 3 along with percentage representation of spectra relative to PfLDH, information on repeats, and predicted antigenicities. Pfg377 (PF3D7_1250100), leading the list with 33% representation relative to LDH, is an abundant osmiophilic body protein that contributes to maintenance of the parasitophorous vacuole and egress of female gametocytes [34]. Another 4 of the 8 proteins are well-described antigens: a mature parasite-infected erythrocyte surface antigen (MESA, PF3D7_0500800) [35], a glycophorin binding protein (GBP, PF3D7_1016300) [36], an asparagine-rich antigen (PF3D7_1110400) [37], and parasite liver stage antigen 3 (LSA 3; PF3D7_0220000) [38]. A P. falciparum homolog of insulin degrading enzyme (PfIDEh; PF3D7_1118300) shows conservation in other Plasmodium species but contains 2 regions of tandem repeats that are not found in the homologs of Plasmoidum vivax, Plasmodium knowlesi, or rodent malaria parasites (Supplementary Figure 1A). The 2 remaining candidates in Supplementary Table 3 are Plasmodium proteins of unknown function (PF3D7_0401800 and PF3D7_1120000).
Antibodies to Tandem Repeats and Selection of PfIDEh for Comparative Evaluation
Synthetic peptides of the repeat units listed in Supplementary Table 3 were KLH-coupled to immunize rabbits. Antisera against 4 KLH-peptides yielded bands on P. falciparum (3D7) immunoblots at relative molecular weights consistent with their corresponding proteins: PfIDEh, GBP, asparagine-rich antigen, and protein PF3D7_1120000 (Supplementary Figure 2A). Rabbit antisera against the KQQDNV repeats of PfIDEh yielded a distinct band at RMW ~175000. This band was confirmed in immunoblots of 4 additional P. falciparum strains from geographically distant regions (Fab9, South Africa; C2A, Thailand; D10, Papua New Guinea; 7G8, Brazil; Supplementary Figure 2B). Recognition of rc-PfIDEh on the immunoblots further confirmed the rabbit antisera against KQQDNV repeats (Supplementary Figure 2B).
To assess variation in the number of PfIDEh KQQDNV repeats, we examined sequences of 320 P. falciparum isolates from regions of Africa, Asia, the Americas, and Oceania. Results showed PfIDEh with 3–15 KQQDNV repeats among these parasites (Supplementary Table 4; Supplementary Figure 1B). A second region of DDDNTS repeats in PfIDEh was also surveyed; all 320 isolates contained 3 or 4 of these repeats (Supplementary Table 4; Supplementary Figure 1B). The overall sequence conservation among isolates, together with the abundant presence of PfIDEh in proteomic data and strong specific recognition of KQQDNV by rabbit antisera, led us to select PfIDEh for further evaluation as a potential diagnostic marker.
Monoclonal Antibodies (Rabbit and Mouse) for PfIDEh Detection
The KQQDNV repeats of PfIDEh are missing from the IDEh sequences of P. vivax, P. knowlesi, and rodent malaria parasites (Supplementary Figure 1A) and thus provide epitopes for P. falciparum–specific recognition. In addition, the multiple epitopes of these repeats may boost functional antibody binding (avidity) [39, 40]. Greater binding has also been reported with mAbs from rabbits relative to mAbs from mice [41]. We therefore obtained 6 KQQDNV-binding mAbs from rabbit lymphocytes, and, from these, selected mAb AID-1-1-9 based on its strong recognition of the peptide in ELISA experiments. Immunoprecipitation and mass spectroscopy confirmed recognition of PfIDEh by AID-1-1-9 (Supplementary Figure 2C; Supplementary Table 5).
For ELISA detection of PfIDEh, we required a second mAb against an epitope away from the KQQDNV region bound by rabbit mAb AID-1-1-9. For this purpose, we sought a mouse mAb to provide assay flexibility and low background signal. Two approaches were used: (1) immunize mice with rounds of rc-PfIDEh from baculovirus; and (2) immunize mice with plasmid DNA expressing different regions of PfIDEh, with or without boosting by baculovirus-produced rc-PfIDEh protein (Figure 1A). These approaches yielded 2 mouse mAbs (3B4D12, 5A2G2) from lymphocytes of mice immunized with rc-PfIDEh and 1 mAb (3A3) from lymphocytes of mice vaccinated by plasmid L-2 and boosted with PfIDEh. Each of these mAbs was conjugated with HRP and tested in ELISA plates coated with either:(1) rc-PfIDEh or (2) PfIDEh captured by mAb AID-1-1-9 from parasite lysate. The mAb 3B4D12 yielded strongest detection of both rc-PfIDEh and the captured PfIDEh (Figure 1B). To test if the different signal intensities from mAbs 3B4D12, 5A2G2, and 3A3 could be due to differential HRP labeling, we repeated the ELISA experiments with unlabeled mouse mAbs followed by an HRP-labeled goat antimouse antibody. Results again showed that the signals were strongest with mAb 3B4D12 (Figure 1C), suggesting that mAb binding, not HRP labeling efficiency, was the greater determinant of signal intensity.
Binding dissociation constants (KD) of mAbs AID-1-1-9 and 3B4D12 were determined against rc-PfIDEh by surface plasmon resonance (Supplementary Table 6). KDs of 2.7 × 10–9 M and 4.3 × 10–8 M were obtained for AID-1-1-9 and 3B4D12, respectively. These values compare to reported KDs of 2.5 × 10–8 M to 42 × 10–8 M for several mouse mAbs against PfLDH [42], and to the KD of 1.4 × 10–9 M for anti-PfLDH mouse mAb MBS498007 in our present study (Supplementary Table 6, determined against an rc-PfLDH fragment). All of these the KDs were weaker than the KD of MPFM-55A and other mAbs against the runs of repeats in the PfHRP-II antigen (KD 3.05 × 10–10 M [43]; 1.1 × 10–10 M, Supplementary Table 6).
ELISA Detection of PfHRP-II, PfLDH, and PfIDEh in P. falciparum Lysates
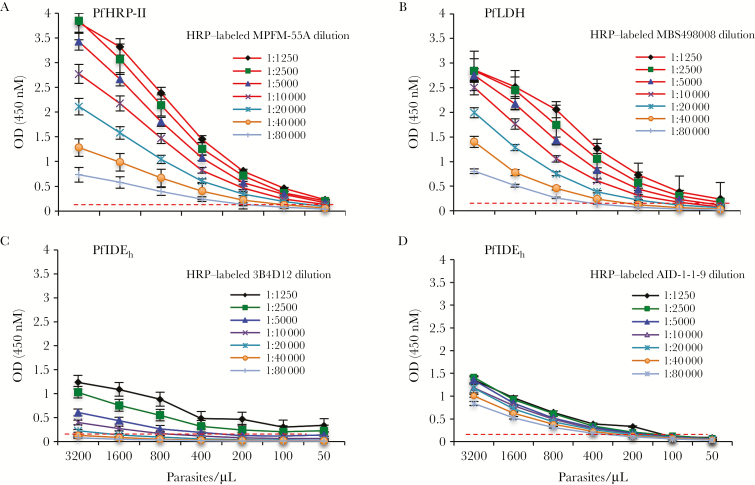
Figure 2 presents results from ELISA experiments to detect PfHRP-II, PfLDH, and PfIDEh in lysates from P. falciparum cultures. Capture of PfHRP-II by unlabeled mAb MPFG-55A, followed by detection with HRP-labeled MPFM-55A, yielded readily observable signals to parasite dilutions of 100–200 parasites/µL, depending on the concentration of HRP-labeled detection antibody (Figure 2A). Similar sensitivity of detection was obtained from PfLDH capture by mAb MBS498007 and detection by mAb MBS498008 (Figure 2B). Capture of PfIDEh with AID-1-1-9 mAb followed by detection with 3B4D12 was about 3-fold less sensitive (Figure 2C), whereas use of these mAbs in the opposite order—that is PfIDEh capture by mAb 3B4D12 followed by detection with AID-1-1-9—was about 2-fold less sensitive, with a detection lower limit of 200–400 parasites/µL (Figure 2D).
Figure 2.
Parasite detection in Plasmodium falciparum cultures by enzyme-linked immunosorbent assay based on P. falciparum histidine-rich protein II (PfHRP-II), P. falciparum lactate dehydrogenase (PfLDH), and P. falciparum insulin-degrading enzyme homolog (PfIDEh). A, PfHRP-II in 3D7 lysates was captured by unlabeled MPFG-55A and detected by horseradish peroxidase (HRP)–labeled MPFM-55A. Signal levels decreased with dilutions of HRP-labeled-MPFM-55A. B, PfLDH in 3D7 lysates was captured by MBS498007 and detected by HRP-labeled-MBS498008. Signal levels decreased with dilutions of HRP-labeled-MBS498008. C, PfIDEh in 3D7 lysates was captured by rabbit monoclonal antibody (mAb) AID-1-1-9 and detected by HRP-labeled mAb 3B4D12. Signal levels decreased with dilutions of HRP-labeled mAb 3B4D12. D, PfIDEh in 3D7 lysates was captured by mAb 3B4D12 and detected by HRP-labeled AID-1-1-9. Signal levels decreased with dilutions of HRP-labeled AID-1-1-9. Dashed line in the panels represents the absorbance signal cutoff. Results are the average ± standard deviation of 3 independent experiments.
High-Sensitivity Detection of Captured PfLDH or PfIDEh by Immuno-PCR
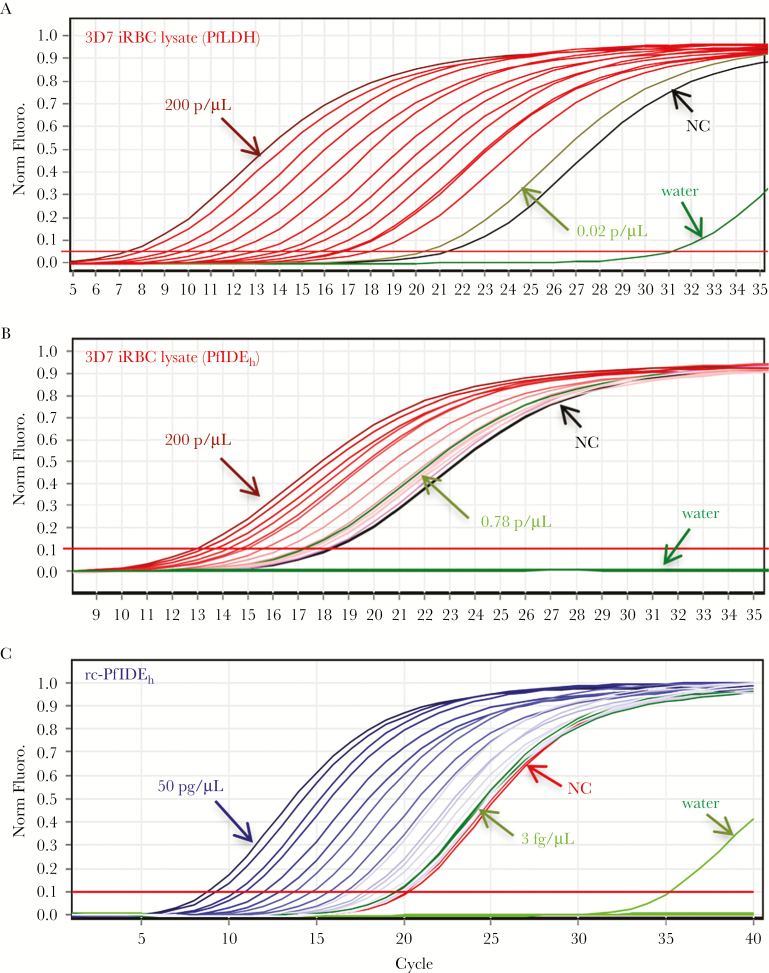
Considering the limited sensitivity of colorimetric signals from the HRP-conjugated mAbs in ELISA, we looked for methods to improve detection of captured PfLDH or PfIDEh antigen. Quantitative PCR (qPCR) of oligonucleotides conjugated to detection antibodies is reported to yield signals that are up to 100,000-fold more sensitive than those from colorimetric ELISA [44]. We therefore implemented i-PCR assays that combine mAb capture with the sensitivity of qPCR. In experiments on parasite lysates, PfLDH capture by MBS498007 followed by i-PCR of streptavidin-labeled MBS498008 yielded signals with a limit of detection (LOD) of 0.02 parasite/µL (Figure 3A). PfIDEh capture by mAb 3B4D12, followed by i-PCR of bound mAb AID-1-1-9, yielded an LOD of 0.78 parasite/µL (Figure 3B). Background signal with the detection antibodies likely explains much of the difference between LODs of these assays. We also obtained standard curves from serial dilutions of known rc-PfIDEh concentration (Figure 3C). Results showed a delta cycle threshold (∆Ct) of 20.79 from 3 fg/µL recombinant protein (0.017 pM, assuming predicted MW 174000), just above the calculated ∆Ct of 20.55 for the LOD [30].
Figure 3.
Immuno–polymerase chain reaction (i-PCR) detection of Plasmodium falciparum lactate dehydrogenase (PfLDH) and P. falciparum insulin-degrading enzyme homolog (PfIDEh) antigens. A, i-PCR curves of PfLDH in 2-fold serially diluted lysates from a culture of P. falciparum 3D7. MBS498007 was employed as the capture antibody and streptavidin-conjugated MBS498008 was used for i-PCR detection of PfLDH. Signal levels were proportional to the lysate dilution. Results from negative controls (NC; no added biotinylated DNA) and no lysate (water only) are shown in black and dark green, respectively. Final PCR products were run on 2% agarose gel to confirm the specific amplification of targeted region (161 bp). Late signals in water control samples (dark green) were from primer dimers, also confirmed by gel electrophoresis. Light-green curve shows the delta cycle threshold (∆Ct) of the tested sample concentration (0.02 parasite/μL) closest to the limit of detection (LOD), calculated as the value of the NC plus 3 standard deviations (SD). B, i-PCR curves of PfIDEh in 2-fold serially diluted lysates from a culture of P. falciparum 3D7. Mouse monoclonal antibody (mAb) 3B4D12 was employed as the capture antibody and streptavidin-conjugated rabbit mAb AID-1-1-9 was used for detection of PfIDEh. Signal levels were proportional to the lysate dilution. Results from negative controls of no added biotinylated DNA and no lysate (water only) are shown in black and dark green, respectively. Light-green curve shows the ∆Ct of the tested sample concentration (0.78 parasite/μL) closest to the LOD, which is calculated as the value of the NC plus 3 SD and is limited by the background signal from NC. C, i-PCR curves of 2-fold serially diluted recombinant PfIDEh (rc-PfIDEh) protein captured by 3B4D12 and detected by AID-1-1-9. Results from negative controls of no added biotinylated DNA and no lysate (water only) are shown in black and dark green, respectively. Light-green curve shows the ∆Ct of the tested rc-PfIDEh concentration (3 fg/μL) closest to the LOD, calculated as the value of the NC plus 3 SD. All experiments were performed in duplicate. Abbreviations: iRBC, infected red blood cell; Norm. Fluoro., normalized fluorescence.
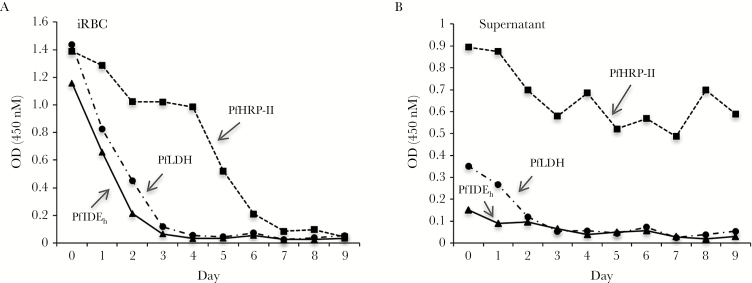
Clearance of PfHRP-II, PfLDH, and PfIDEh From Chloroquine-Treated P. falciparum Cultures
Parasitized erythrocytes and media were collected from P. falciparum cultures (3D7 line) before and after treatment with 150 ng/mL chloroquine. In comparative ELISAs over a 9-day period, PfHRP-II persisted in the samples until day 7 after treatment, whereas the levels of PfIDEh and PfLDH fell to near zero by day 3 (Figure 4A). Samples of culture supernatant included readily detectable amounts of PfHRP-II and PfLDH but only low levels of PfIDEh, suggesting little release of this protein into the medium from P. falciparum–infected cells (Figure 4B).
Figure 4.
Persistence of detectable Plasmodium falciparum insulin-degrading enzyme homolog (PfIDEh), P. falciparum lactate dehydrogenase (PfLDH), and P. falciparum histidine-rich protein II (PfHRP-II) after treatment of P. falciparum culture with chloroquine. A, Enzyme-linked immunosorbent assay (ELISA) optical density (OD) values from protein in pelleted infected erythrocytes. B, ELISA OD values from protein in supernatants. Monoclonal antibody pairs used to detect PfIDEh, PfLDH, and PfHRP-II were as follows: 3B4D12 and horseradish peroxidase (HRP)–labeled AID-1-1-9; MBS498007 and HRP-labeled MBS498008; and MPFG-55A and HRP-labeled MPFM-55A, respectively. Abbreviations: iRBC, infected red blood cell.
DISCUSSION
RDTs are increasingly important for detection of silent parasite carriage and local outbreaks in programs of malaria control and elimination. Detection of asymptomatic parasitemias as low as 1 parasite/µL in field settings, minimization of false-positive and false-negative results, differentiation of the Plasmodium species, fast results, and inexpensive and ready implementation are among desired properties of an ideal RDT. RDTs based on antigen detection in inexpensive lateral flow formats have some of these properties and are presently in wide use, but these can miss infections and have sensitivities generally limited to 100–200 parasites/µL [6, 14].
In this study, we identified a new P. falciparum–specific diagnostic marker, PfIDEh, and assessed it relative to the PfHRP-II and PfLDH antigens detected by many commercially available RDTs. In our comparisons of mAb-based capture assays, PfHRP-II- and PfLDH-based ELISAs showed detection limits of 100–200 parasites/µL, whereas the PfIDEh-based ELISA was about 2-fold less sensitive, with a limit of 200–400 parasites/µL. However, with use of i-PCR instead of colorimetric development to detect bound mAb, detection sensitivities improved greatly, to limits of 0.02 parasite/µL and 0.78 parasite/µL in PfLDH- and PfIDEh-based assays, respectively. These results show that the relatively poor sensitivities of ELISAs and related colorimetric development assays are attributable to insensitive sensor technology, not to inherent inability of the mAb to detect antigen at much lower levels. Indeed, the i-PCR limits of <1 parasite/µL show that mAb detection can serve for assays in the sensitivity range of LAMP and PCR-based detection.
PfHRP-II, presently the most commonly used antigen in RDTs for P. falciparum infection, contains multiple tandem repeats rich in alanine, histidine, and aspartic acid, comprising respectively, 37%, 34%, and 10% of the protein [32]. The mAbs against these repeats bind with very high avidity (KD = 3.05 × 10-10 M) ([43]). Strong antibody responses against tandem repeats have likewise been reported for other Plasmodium proteins and antigens of Leishmania and Trypanosoma cruzi [39, 45, 46] and are often targets of B-cell responses [46, 47]. In this study, we specifically targeted the tandem KQQDNV repeats to optimize detection of PfIDEh as a marker of P. falciparum infection. The functional binding affinity of a rabbit mAb against these repeats (KD = 2.7 × 10-9 M), however, was not as strong as that of mouse mAb against the PfHRP-II repeats, a difference that may arise from the much greater number of repeats in the PfHRP-II protein.
The different thresholds of parasitemia detection by i-PCR assays based on PfLDH or PfIDEh may be partly explained by the levels of these proteins in proteomic data. Amounts of antigen produced by different P. falciparum stages, particularly the ring and gametocytes stages that circulate in the bloodstream, may also affect the ability of ELISA or i-PCR assay to sense parasitemias.
Considering the exquisite sensitivities of i-PCR for PfIDEh as well as PfLDH, we did not pursue i-PCR assays with PfHRP-II as there are recent reports that some P. falciparum strains do not produce this antigen [16]. PfIDEh and PfLDH are proteins that are highly conserved in P. falciparum, and there is no evidence that either is deleted from any parasite strain. Additionally, unlike the PfHRP-II antigen that can persist in vitro or in vivo for weeks after parasites are killed by drug treatment, PfIDEh and PfLDH clear from P. falciparum cultures within a few days and are therefore less likely to yield false-positive results like those that confound outcome assessments with PfHRP-II–based RDTs [18].
The ability of PfLDH- or PfIDEh-based i-PCR to detect very low-level parasitemias suggests that technologically improved sensing of bound mAb may be able to greatly increase the sensitivity of antigen-based diagnostic tests. Additional proof of principle might be obtained by LAMP detection of the oligonucleotide-labeled secondary mAb in a suitable format, although the complexity and cost of such a hybrid i-PCR/LAMP system would likely be prohibitive in the field setting. Technologies involving various oligonucleotides, nanoparticles, or gold-based sensors may offer possibilities for greatly improved sensing [48–50]. If sufficiently improved detection can be applied with such technologies, a new generation of inexpensive, highly sensitive, and practical point-of-care RDTs may be possible for malaria elimination programs.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We dedicate this report to the memory of Dr Manoochehr Rasouli, who generously shared his expertise on antibody and ELISA methods. We thank Brian Brown, National Institutes of Health (NIH) Library Editing Service, for editing the manuscript.
Financial support. This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH.
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. malERA Consultative Group on Diagnoses and Diagnostics. A research agenda for malaria eradication: diagnoses and diagnostics. PLoS Med 2011; 8:e1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hemingway J, Shretta R, Wells TN et al. Tools and strategies for malaria control and elimination: what do we need to achieve a grand convergence in malaria? PLoS Biol 2016; 14:e1002380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. World Health Organization Global Malaria Programme. World malaria report. Available at: http://www.who.int/malaria/publications/world-malaria-report-2016/en/. Accessed 24 August 2017. [Google Scholar]
- 4. Slater HC, Ross A, Ouédraogo AL et al. Assessing the impact of next-generation rapid diagnostic tests on Plasmodium falciparum malaria elimination strategies. Nature 2015; 528:S94–101. [DOI] [PubMed] [Google Scholar]
- 5. Wu L, van den Hoogen LL, Slater H et al. Comparison of diagnostics for the detection of asymptomatic Plasmodium falciparum infections to inform control and elimination strategies. Nature 2015; 528:S86–93. [DOI] [PubMed] [Google Scholar]
- 6. Tietje K, Hawkins K, Clerk C et al. The essential role of infection-detection technologies for malaria elimination and eradication. Trends Parasitol 2014; 30:259–66. [DOI] [PubMed] [Google Scholar]
- 7. Najers JB, Hammer J.. Malaria: new patterns and perspectives. World Bank technical paper 1992; 183:19. [Google Scholar]
- 8. Cook J, Xu W, Msellem M et al. Mass screening and treatment on the basis of results of a Plasmodium falciparum-specific rapid diagnostic test did not reduce malaria incidence in Zanzibar. J Infect Dis 2015; 211:1476–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng Z, Cheng Z. Advances in molecular diagnosis of malaria. Adv Clin Chem 2017; 80:155–92. [DOI] [PubMed] [Google Scholar]
- 10. Plucinski MM, Rogier E, Dimbu PR, Fortes F, Halsey ES, Aidoo M. Estimating the added utility of highly sensitive histidine-rich protein 2 detection in outpatient clinics in sub-Saharan Africa. Am J Trop Med Hyg 2017; doi:10.4269/ajtmh.17-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hofmann N, Mwingira F, Shekalaghe S, Robinson LJ, Mueller I, Felger I. Ultra-sensitive detection of Plasmodium falciparum by amplification of multi-copy subtelomeric targets. PLoS Med 2015; 12:e1001788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lau YL, Lai MY, Fong MY, Jelip J, Mahmud R. Loop-mediated isothermal amplification assay for identification of five human Plasmodium species in Malaysia. Am J Trop Med Hyg 2016; 94:336–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oriero EC, Van Geertruyden JP, Nwakanma DC, D’Alessandro U, Jacobs J. Novel techniques and future directions in molecular diagnosis of malaria in resource-limited settings. Expert Rev Mol Diagn 2015; 15:1–8. [DOI] [PubMed] [Google Scholar]
- 14. Visser T, Daily J, Hotte N, Dolkart C, Cunningham J, Yadav P. Rapid diagnostic tests for malaria. Bull World Health Organ 2015; 93:862–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baker J, Ho MF, Pelecanos A et al. Global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: implications for the performance of malaria rapid diagnostic tests. Malar J 2010; 9:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng Q, Gatton ML, Barnwell J et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J 2014; 13:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luchavez J, Baker J, Alcantara S et al. Laboratory demonstration of a prozone-like effect in HRP2-detecting malaria rapid diagnostic tests: implications for clinical management. Malar J 2011; 10:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. World Health Organization. Malaria rapid diagnostic test performance: summary results of WHO product testing of malaria RDTs: rounds 1–6 (2008-2015). Available at: http://apps.who.int/iris/bitstream/10665/204119/1/9789241510042_eng.pdf?ua=1. Accessed 24 August 2017. [Google Scholar]
- 19. Ricks KM, Adams NM, Scherr TF, Haselton FR, Wright DW. Direct transfer of HRPII-magnetic bead complexes to malaria rapid diagnostic tests significantly improves test sensitivity. Malar J 2016; 15:399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Florens L, Washburn MP, Raine JD et al. A proteomic view of the Plasmodium falciparum life cycle. Nature 2002; 419:520–6. [DOI] [PubMed] [Google Scholar]
- 21. Lasonder E, Janse CJ, van Gemert GJ et al. Proteomic profiling of Plasmodium sporozoite maturation identifies new proteins essential for parasite development and infectivity. PLoS Pathog 2008; 4:e1000195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Florens L, Liu X, Wang Y et al. Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol Biochem Parasitol 2004; 135:1–11. [DOI] [PubMed] [Google Scholar]
- 23. Bowyer PW, Simon GM, Cravatt BF, Bogyo M. Global profiling of proteolysis during rupture of Plasmodium falciparum from the host erythrocyte. Mol Cell Proteomics 2011; 10:M110.001636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Acharya P, Pallavi R, Chandran S et al. A glimpse into the clinical proteome of human malaria parasites Plasmodium falciparum and Plasmodium vivax. Proteomics Clin Appl 2009; 3:1314–25. [DOI] [PubMed] [Google Scholar]
- 25. Oehring SC, Woodcroft BJ, Moes S et al. Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol 2012; 13:R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Khan SM, Franke-Fayard B, Mair GR et al. Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 2005; 121:675–87. [DOI] [PubMed] [Google Scholar]
- 27. Silvestrini F, Lasonder E, Olivieri A et al. Protein export marks the early phase of gametocytogenesis of the human malaria parasite Plasmodium falciparum. Mol Cell Proteomics 2010; 9:1437–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Solyakov L, Halbert J, Alam MM et al. Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Nat Commun 2011; 2:565. [DOI] [PubMed] [Google Scholar]
- 29. Treeck M, Sanders JL, Elias JE, Boothroyd JC. The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites’ boundaries. Cell Host Microbe 2011; 10:410–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Niemeyer CM, Adler M, Wacker R. Detecting antigens by quantitative immuno-PCR. Nat Protoc 2007; 2:1918–30. [DOI] [PubMed] [Google Scholar]
- 31. He X, Patfield SA. Immuno-PCR assay for sensitive detection of proteins in real time. Methods Mol Biol 2015; 1318:139–48. [DOI] [PubMed] [Google Scholar]
- 32. Wellems TE, Howard RJ Recombinant DNA clone containing a genomic fragment of PfHRP-II gene from Plasmodium falciparum US patent 5130416A. 1986. [Google Scholar]
- 33. Maier AG, Rug M, O’Neill MT et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell 2008; 134:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Suárez-Cortés P, Sharma V, Bertuccini L et al. Comparative proteomics and functional analysis reveal a role of Plasmodium falciparum osmiophilic bodies in malaria parasite transmission. Mol Cell Proteomics 2016; 15:3243–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Waller KL, Nunomura W, An X, Cooke BM, Mohandas N, Coppel RL. Mature parasite-infected erythrocyte surface antigen (MESA) of Plasmodium falciparum binds to the 30-kDa domain of protein 4.1 in malaria-infected red blood cells. Blood 2003; 102:1911–4. [DOI] [PubMed] [Google Scholar]
- 36. Ravetch JV, Kochan J, Perkins M. Isolation of the gene for a glycophorin-binding protein implicated in erythrocyte invasion by a malaria parasite. Science 1985; 227:1593–7. [DOI] [PubMed] [Google Scholar]
- 37. LaCount DJ, Vignali M, Chettier R et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 2005; 438:103–7. [DOI] [PubMed] [Google Scholar]
- 38. Prieur E, Druilhe P. The malaria candidate vaccine liver stage antigen-3 is highly conserved in Plasmodium falciparum isolates from diverse geographical areas. Malar J 2009; 8:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thuy NT, Goto Y, Lun ZR, Kawazu S, Inoue N. Tandem repeat protein as potential diagnostic antigen for Trypanosoma evansi infection. Parasitol Res 2012; 110:733–9. [DOI] [PubMed] [Google Scholar]
- 40. Hernan R, Heuermann K, Brizzard B. Multiple epitope tagging of expressed proteins for enhanced detection. Biotechniques 2000; 28:789–93. [DOI] [PubMed] [Google Scholar]
- 41. Rossi S, Laurino L, Furlanetto A et al. Rabbit monoclonal antibodies: a comparative study between a novel category of immunoreagents and the corresponding mouse monoclonal antibodies. Am J Clin Pathol 2005; 124:295–302. [DOI] [PubMed] [Google Scholar]
- 42. Lee GC, Jeon ES, Le DT et al. Development and evaluation of a rapid diagnostic test for Plasmodium falciparum, P. vivax, and mixed-species malaria antigens. Am J Trop Med Hyg 2011; 85:989–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leow CH, Jones M, Cheng Q, Mahler S, McCarthy J. Production and characterization of specific monoclonal antibodies binding the Plasmodium falciparum diagnostic biomarker, histidine-rich protein 2. Malar J 2014; 13:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sano T, Smith CL, Cantor CR. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science 1992; 258:120–122. [DOI] [PubMed] [Google Scholar]
- 45. Biswas S, Tomar D, Rao DN. Investigation of the kinetics of histidine-rich protein 2 and of the antibody responses to this antigen, in a group of malaria patients from India. Ann Trop Med Parasitol 2005; 99:553–62. [DOI] [PubMed] [Google Scholar]
- 46. Goto Y, Carter D, Guderian J, Inoue N, Kawazu S, Reed SG. Upregulated expression of B-cell antigen family tandem repeat proteins by Leishmania amastigotes. Infect Immun 2010; 78:2138–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Coppel RL, Cowman AF, Anders RF et al. Immune sera recognize on erythrocytes Plasmodium falciparum antigen composed of repeated amino acid sequences. Nature 1984; 310:789–92. [DOI] [PubMed] [Google Scholar]
- 48. Spengler M, Adler M, Niemeyer CM. Highly sensitive ligand-binding assays in pre-clinical and clinical applications: immuno-PCR and other emerging techniques. Analyst 2015; 140:6175–94. [DOI] [PubMed] [Google Scholar]
- 49. Rogier E, Plucinski M, Lucchi N et al. Bead-based immunoassay allows sub-picogram detection of histidine-rich protein 2 from Plasmodium falciparum and estimates reliability of malaria rapid diagnostic tests. PLoS One 2017; 12:e0172139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hemben A, Ashley J, Tothill IE. Development of an immunosensor for PfHRP 2 as a biomarker for malaria detection. Biosensors (Basel) 2017; 7 pii:E28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.