Enterococcus faecalis wound infections remain poorly understood. We demonstrate in vivo that E faecalis infection results in 2 different states: colonization or infection. We show that E faecalis infection modulates host immunity, resulting in persistent infection and delayed healing.
Keywords: Enterococcus faecalis, wound infection, immune evasion, multiple peptide resistant factor, persistence
Abstract
Enterococcus faecalis is one of the most frequently isolated bacterial species in wounds yet little is known about its pathogenic mechanisms in this setting. Here, we used a mouse wound excisional model to characterize the infection dynamics of E faecalis and show that infected wounds result in 2 different states depending on the initial inoculum. Low-dose inocula were associated with short-term, low-titer colonization whereas high-dose inocula were associated with acute bacterial replication and long-term persistence. High-dose infection and persistence were also associated with immune cell infiltration, despite suppression of some inflammatory cytokines and delayed wound healing. During high-dose infection, the multiple peptide resistance factor, which is involved in resisting immune clearance, contributes to E faecalis fitness. These results comprehensively describe a mouse model for investigating E faecalis wound infection determinants, and suggest that both immune modulation and resistance contribute to persistent, nonhealing wounds.
Wound infections affect between 7% and 15% of hospitalized people globally [1]. Enterococcus faecalis is one of the most frequently isolated bacterial species across all types of wounds, including diabetic foot ulcers, burns, and surgical sites [2–4]. In surgical site infections, E faecalis is the third most commonly isolated organism [5, 6]. E faecalis infections are increasingly difficult to treat due to their intrinsic and acquired resistance to a range of antibiotics [7]. Despite the high frequency of E faecalis in wound infections, little is known about its pathogenic strategies in this niche.
Bacterial biofilms, which are often polymicrobial in nature, are a major factor in wound healing and are associated with a poorer prognosis [8–10]. Moreover, biofilm formation promotes survival and persistence of infecting microbes because it facilitates defense against the host immune response [11]. E faecalis encodes several factors that contribute to biofilm formation, including 2 sortase enzymes, SrtC and SrtA, which polymerize and attach endocarditis- and biofilm-associated pili to the cell wall, respectively [12–14]. These pili aid in the attachment of E faecalis to surfaces, which is required in the initial stages of biofilm formation in vitro and in vivo during catheter-associated urinary tract infection [15, 16]. Other biofilm-associated factors that are attached to the cell wall by SrtA include Ace, aggregation substance, and Esp [17–20].
In addition to initial adhesion and colonization, E faecalis must also overcome host defenses to establish infection. E faecalis can modulate and evade the host immune response in a number of settings [21–24]. Biofilm formation, along with expression of the SrtA substrate aggregation substance, can promote E faecalis survival within macrophages and neutrophils [25, 26]. The multiple peptide resistance factor (MprF) protein of E faecalis confers resistance to antimicrobial peptides via electrostatic repulsion [27, 28], and is important for surviving both neutrophil-mediated clearance and within epithelial cells and macrophages in a variety of gram-positive bacteria [29–31].
Previously, a mouse wound excisional model was developed to study wound healing processes [32–34]. This model has been used to examine bacterial factors required for wound infection by Pseudomonas aeruginosa, Acinetobacter baumannii, and Staphylococcus aureus [35–38]. In the current study, we characterized the dynamics of E faecalis infection in the model. We establish the minimal doses of E faecalis required for colonization and infection of wounds. We also demonstrate a role for the innate immune defense factor MprF in wound infection, and show that modulation of early inflammatory responses and delayed wound healing are coupled with persistence of E faecalis within wounds.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Strains used are shown in Supplementary Table 1. For mouse infections, E faecalis was grown statically at 37°C for 15–18 hours in Brain Heart Infusion medium (BHI; Neogen, Lansing, Michigan) without antibiotics. Clinical strains isolated from patient wounds were provided by Tan Tock Seng Hospital, Singapore.
Genetic Manipulation
Construction of E faecalis OG1RF ΔmprF1 and ΔmprF2 were previously described [27]. OG1RF ΔmprF1/2 was made by subcloning the ΔmprF1 deletion construct from pJRS213-ΔmprF1 into pGCP213 to create pGCP213-ΔmprF1 and transforming the plasmid into OG1RF ΔmprF2. Chromosomal deletions were constructed as described previously.
Mouse Wound Excisional Model
Mouse wound infections were modified from a previous study [39]. Male C57BL/6 mice (7–8 weeks old, 22 to 25 g; InVivos, Singapore) were anesthetized with 3% isoflurane and the dorsal hair trimmed. Following trimming, Nair cream (Church and Dwight Co) was applied and the fine hair removed via shaving with a scalpel. This 2-step shaving method ensured the wound dressing remains for >5 days without detachment. The skin was then disinfected with 70% ethanol. A 6-mm biopsy punch (Integra Miltex, New York) was used to create a full-thickness wound and 10 μL of the respective bacteria inoculum applied. The wound site was then sealed with a transparent dressing (Tegaderm, 3M, St Paul Minnesota). At indicated time points, mice were euthanized and a 1 × 1 cm piece of skin surrounding the wound site was excised into sterile phosphate buffered saline (PBS). Excised wounds were homogenized and viable bacteria enumerated by plating onto BHI agar with (Supplementary Table 1) and without antibiotics to check for contamination. Mice with contaminants were excluded from our datasets and subsequent analysis. For coinfection experiments, competitive index (CI) was determined with the following formula:
Histology
Wound tissues were excised as described above and fixed in 4% paraformaldehyde in 1 × PBS (pH 7.4) for 24 hours at 4°C. Samples were then submerged in 15% and 30% sucrose gradient for 24 hours each, embedded in Optimal Cutting Temperature (OCT) embedding media (Sakura, California), and frozen in liquid nitrogen. Thin sections (10 μm) were then obtained with a Leica CM1860 UV cryostat (Leica Biosystems, Ernst-Leitz Strasse, Germany) and stained with hemotoxylin and eosin (H&E). Images of H&E stained sections were acquired using an Axio Scan.Z1 slide scanner (Carl Zeiss, Göttingen, Germany) fitted with a 20×/0.8 Apochrome objective.
Gene Probe and Fluorescence In Situ Hybridization
E faecalis was detected with the oligonucleotide probe 5′-GGTGTTGTTAGCATTTCG/Cy3/-3′ (IDT Technologies, Iowa). Probe 5′-GCTGCCTCCCGTAGGAGT/Alexa Fluor 488/-3′ (IDT Technologies, Iowa) targets the 16S rRNA of organisms in the domain of Bacteria [40]. Cryosectioned tissue sections were dehydrated in a graded ethanol series (70% and 80%) for 3 minutes each. Tissue sections were then immersed in a 0.2% Sudan Black (Sigma-Aldrich, Iowa) solution (prepared in 96% ethanol) for 20 minutes and washed 3 times with a 0.02% Tween solution (prepared in 1 × PBS). A total of 25 μL of 25% formamide hybridization buffer (20 mM Tris-HCl [pH 8.0], 5M NaCl, 0.1% sodium dodecyl sulfate, and 25% formamide) containing 100 pmol of the labeled probe (50 μg/mL stock) was added to the sections and incubated overnight at 48°C. Slides were then immersed in 50 mL of wash buffer (0.5M EDTA and 5M NaCl, 20mM Tris-HCl [pH 8.0]) for 30 minutes in a 46°C water bath. After washing, slides were plunged into ice cold water for 5 seconds and left to dry.
Confocal Laser Scanning Microscopy
Hybridized samples were mounted using Citifluor (Citifluor, London) and imaged using an Elyra PS.1 LSM780 inverted laser scanning confocal microscope (Carl Zeiss, Göttingen, Germany) fitted with a 63×/1.4 Plan-Apochromat oil immersion objective using the Zeiss Zen Black 2012 SP2 software suite. Laser power and gain were kept constant between experiments.
Scanning Electron Microscopy
Excised skin samples were fixed using 2.5% glutaraldehyde (prepared in 0.1M phosphate buffer (PB) pH 7.4) for 48 hours at 4°C and then washed 3 times with 0.1M PB. Fixed samples were then dehydrated with a graded ethanol series (once with 30%, 50%, 70%, 80%, 90%, and twice with 100% for 15 minutes at each step) together with gentle agitation. Samples were then subjected to amyl acetate immersion for 30 minutes. Samples were next critical point dried with the Bal-Tec CPD-030 Critical Point Dryer (Bal-Tec AG, Balzers, Liechtenstein) overnight and deposited onto scanning electron microscopy (SEM) specimen stubs using NEM Tape (Nisshin Em. Co. Ltd, Tokyo, Japan). Samples were then sputter coated with gold using a Bal-Tec SCD 005 sputter coater (Bal-Tec AG, Balzers, Liechtenstein). Samples were viewed using a JSM-6360LV scanning electron microscope (JEOL, Tokyo, Japan).
Cytokine Luminex MAP Analysis
Luminex multi-analyte profiling (MAP) analysis was performed using the Bio-Plex Pro™ Mouse Cytokine 23-plex Assay (Bio-Rad, California) as previously described [41].
Flow Cytometry
Skin was cut into pieces and incubated in RPMI containing 10% serum, 0.2 mg/mL collagenase IV (Roche, Basel, Switzerland) and 20000 U/mL of DNAse I (Roche, Basel, Switzerland) for 1 hour at 37°C. Cells were then passed through a 19 G syringe and filtered through a 100 μm cell strainer (BD Biosciences, New Jersey) to obtain a homogenous cell suspension, which was stained with the fluorochrome or biotin-conjugated monoclonal antibodies listed in Supplementary Table 2. Multiparameter analyses of cell suspensions were performed on an LSR II (BD Biosciences, San Jose). Data were analyzed with FlowJo software (TreeStar, Oregon).
Statistical Analyses
Statistical analyses were performed with GraphPad Prism software (Version 6.05 for Windows, California) and are described in the respective figure legends. Principal component analysis was performed in R (Version 3.3.2) with the packages factoextra (Version 1.0.4) and FactoMineR (Version 1.34).
Ethics Statement
All procedures were approved and performed in accordance with the Institutional Animal Care and Use Committee in Nanyang Technological University (ARF SBS/NIEA0198Z).
RESULTS
Minimum Colonization Dose for E faecalis in Wounds is 10 CFU
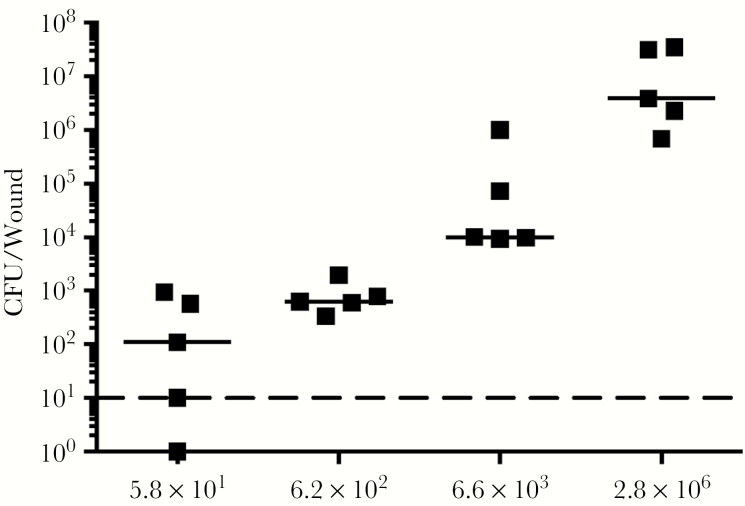
To investigate the colonization and infection dynamics of E faecalis OG1RF in wounds, we first determined the colonization dose required to colonize >50% of excisional wounds in C57BL/6 mice (CD50). We examined a range of infection inocula, from 101 to 106 colony forming units (CFU), at 24 hours postinoculation (hpi) and determined the CD50 to be 5.8 × 101 CFU, which resulted in 50% of the mice displaying recoverable CFU (Figure 1). The CD90 was 6.2 × 102 CFU. In general, we observed that the median recoverable CFU for all inocula at 24 hpi was similar to the initial inoculum. At inocula of 102 we observed no visible, macroscopic signs of inflammation (Supplementary Figure 1). By contrast, at 24 hpi, inocula of 106 resulted in visible, macroscopic inflammation with redness and accompanied by presence of serous exudates at the wound site of all infected mice (Supplementary Figure 1). Henceforth, we defined 106 as the infectious dose (ID90). These findings suggest that the initial bacterial inoculum can result in wounds of 2 different states: colonization or infection.
Figure 1.
The CD50 of Enterococcus faecalis wound infection is 101 colony forming units (CFU). Male C57BL/6 mice were wounded and infected with E faecalis OG1RF with inocula of 5.8 × 101 (circle), 6.2 × 102 (square), 6.6 × 103 (triangle), or 2.8 × 106 (inverted triangle) CFU. Wounds were harvested at 24 hours postinoculation and the recovered bacteria enumerated. Each point represents one mouse, and the solid horizontal lines indicate the median. The horizontal dashed line indicates the limit of detection; n = 5.
E faecalis Infection is Associated With High Titer Persistence in Wounds
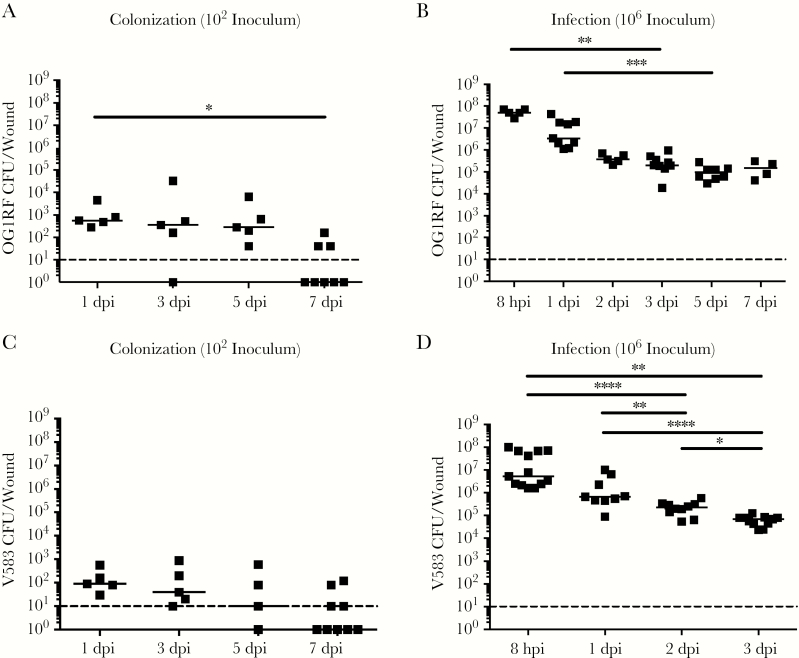
To further investigate the differences between low-inoculum colonization and high-inoculum infection dynamics, we inoculated mice with the CD90 colonization dose of 102 CFU, or the infection dose of 106 CFU, and monitored the mice for 7 days postinoculation (dpi). We observed that, regardless of the initial infection inoculum, viable bacteria were recovered at all time points (24 hpi to 7 dpi). However, after a 102 CFU inoculation, E faecalis persisted at 102 CFU and only decreased at 7 dpi (Figure 2A). By contrast, when mice were inoculated with 106 CFU, we observed a rapid increase to 108 CFU by 8 hpi, followed by a decrease at 3 dpi to 105 CFU, which was maintained throughout the course of the experiment (Figure 2B). Consistent with this, at inocula of 106 we observed visible inflammation and wound exudates only at 8 and 24 hpi, which resolved after 2 dpi (data not shown).
Figure 2.
Colonization and infection dynamics of Enterococcus faecalis in wounds. Wounds were harvested at the indicated time points postinoculation and the colony forming units (CFU) enumerated. Mice were inoculated with (A) 102 CFU of OG1RF, (B) 106 CFU of OG1RF, (C) 102 CFU of V583, or (D) 106 CFU of V583. Each dot represents 1 mouse, and the solid horizontal lines indicate the median, N (biological replicates) = 2, n (technical replicates) = ≥5. Statistical analysis was performed using Kruskall–Wallis test with Dunn’s post-test to correct for multiple comparisons. * P < .05, ** P < .01. Abbreviations: hpi, hours postinfection; dpi, days postinfection.
E faecalis wound infection dynamics were not strain specific because the clinical blood isolate E faecalis V583 [42] displayed similar infection kinetics to that of strain OG1RF (Figure 2C and D). In addition, clinical E faecalis wound isolates inoculated at the infection dose resulted in similar high-titer wound infections at 8 hpi (Supplementary Figure 2). Together, these results demonstrate and confirm that the initial inoculum can result in the following states: colonization in the absence of increased E faecalis titers and overt inflammation, or infection associated with acute bacterial replication and overt inflammation.
MprF Contributes to E faecalis Fitness During Wound Infection
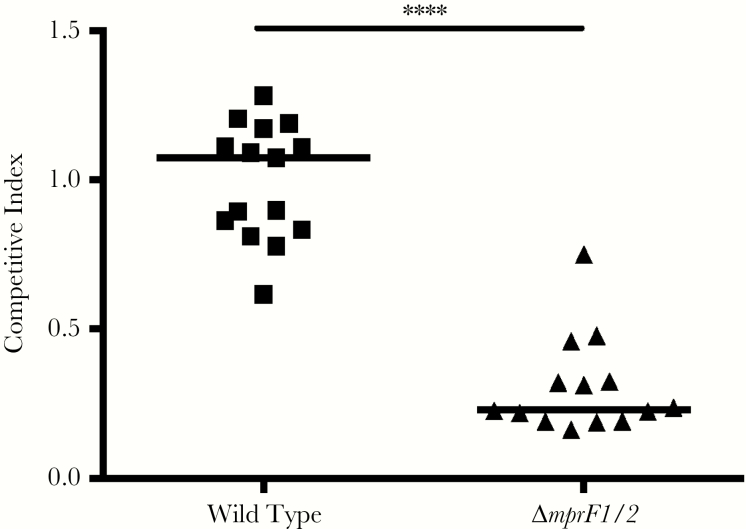
To determine E faecalis factors involved in wound colonization and infection, we examined the fitness of previously described biofilm factors as well as factors involved in immune defense (Figure 3, Supplementary Figure 3). In single species and competitive infections, we found that a ΔsrtAC double mutant strain defective in biofilm formation was not attenuated in fitness during coinfection at 8 hpi or 3 dpi (Supplementary Figure 3B–D). Because we observed overt inflammation after high-dose E faecalis infection in wounds, we predicted that resistance to host immune killing may be important for its survival. E faecalis encodes 2 paralogues of MprF [27, 28]. To address the contribution of these gene products to fitness in wounds, we coinfected mice with wild-type E faecalis OG1X and OG1RF Δmprf1/2, and found that OG1RF Δmprf1/2 was significantly less fit during coinfection at 3 dpi, but not at 8 hpi (Figure 3, Supplementary Figure 3A). OG1RF Δmprf1/2 was not attenuated in single species infection (Supplementary Figure 3D). Together, these results suggest that traditional biofilm-associated factors may be less important for E faecalis wound infection than its ability to resist immune defense mechanisms.
Figure 3.
Multiple peptide resistance factor (MprF) contributes to fitness during Enterococcus faecalis wound infection. Wounds were infected with a 1:1 ratio of E faecalis strains OG1X:wild type OG1RF or OG1X:OG1RF ΔmprF1/2, at 106 colony forming units per inoculum, harvested at 3 days postinfection, and the recovered bacteria enumerated on selective media for each strain. Each dot represents 1 mouse, and the solid horizontal lines indicate the median, N = 3, n = 5. Statistical analysis was performed using Mann–Whitney U test. **** P < .0001. Abbreviations: N, biological replicates: n, technical replicates
E faecalis Forms Microcolonies on the Wound Surface
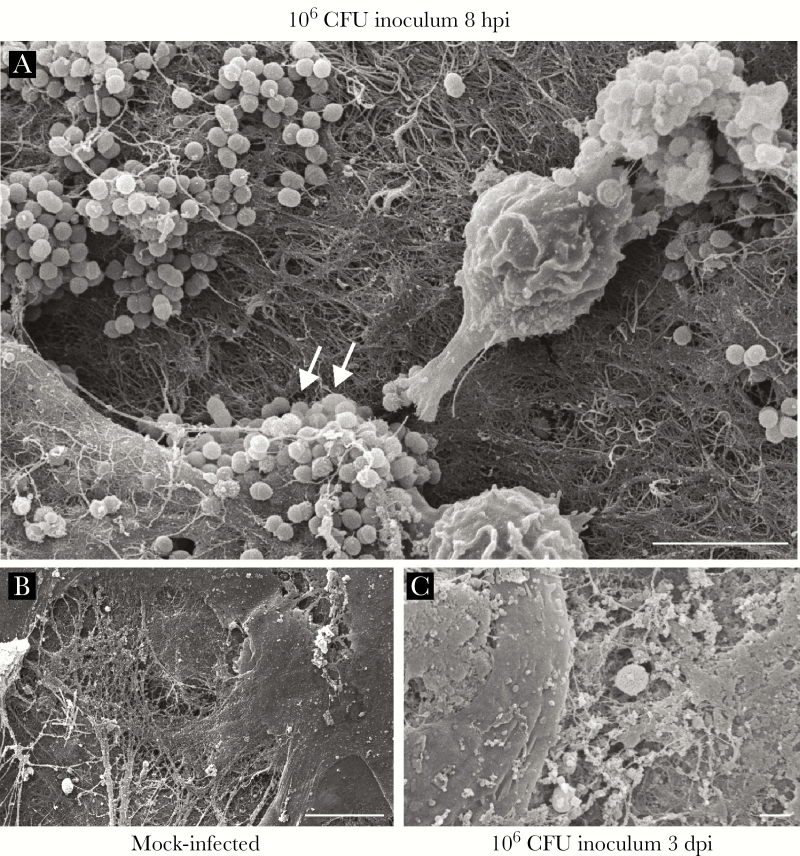
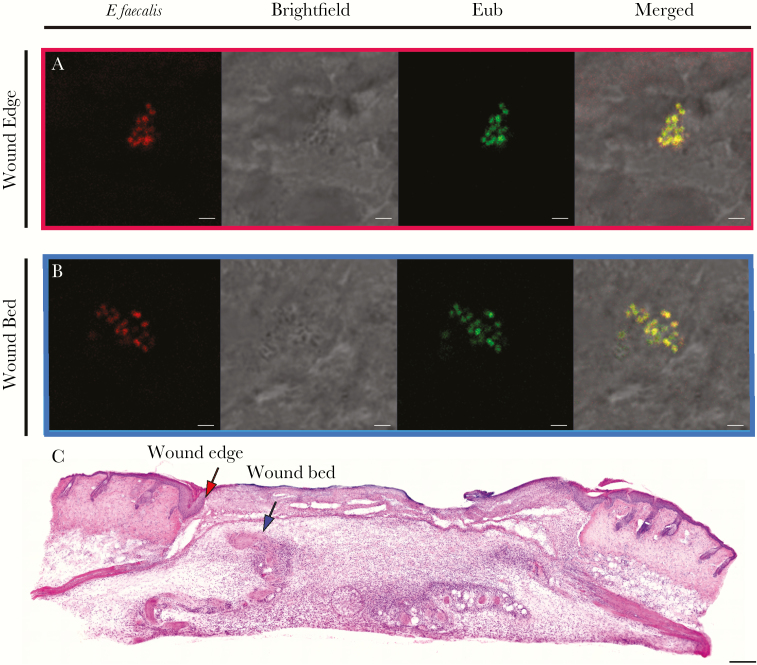
Because we observed marked changes in the CFU recovered from infected wounds over time, we hypothesized that the spatial distribution of E faecalis may also vary across time during infection. To address this question, we performed SEM at 8 hpi and 3 dpi, which represent the peak of infection and the onset of stable colonization, respectively (Figure 2B). At 8 hpi, we observed E faecalis microcolonies on infected wounds that appeared to be encased within a matrix, indicative of early biofilm development (Figure 4A). By contrast, at 3 dpi we were unable to detect E faecalis on the surface of the wounds (Figure 4B). Because the CFU burden was still high at 3 dpi, we reasoned that E faecalis may instead be embedded within the tissue. To determine the spatial localization of subsuperficial E faecalis in 3 dpi wounds, we performed fluorescence in situ hybridization (FISH) on 3 dpi wound samples. Using FISH probes specific for E faecalis, we observed E faecalis microcolonies at the wound edge (Figure 5A and C) and in the wound bed (Figure 5B and C). These results suggest that E faecalis form biofilm-like microcolonies within wounds and can be encapsulated or internalized within the host tissues. We postulate that both properties may contribute to protection from the host immune response and persistence within wounds.
Figure 4.
Enterococcus faecalis forms microcolonies in acutely infected wounds. Mice were wounded and infected with 106 CFU E faecalis OG1RF or mock infected with phosphate buffered saline. Wounds were harvested at the indicated postinfection time points for scanning electron microscopy. E faecalis microcolonies encapsulated by fibrous material were visible at 8 hours postinfection (hpi) (white arrows, A), but not in mock-infected wounds (B) or in infected wounds at 3 days postinfection (dpi) (C). Bar represents 5 μm. Images shown are representative of 3 independent experiments.
Figure 5.
Enterococcus faecalis is present at the wound edge and in the wound bed. Male C57BL/6 mice were wounded and infected with 106 CFU of E faecalis OG1RF. Wounds were harvested at 3 days postinfection, cryosectioned, and subjected to (A,B) fluorescence in situ hybridization or (C) H&E staining. A, E faecalis-specific probe or probe specific for the domain bacteria (Eub) were used for fluorescence in situ hybridization. The brightfield channel is represented in grey scale. Red and blue arrows (C) correspond to the red and blue boxes (A,B) and represent the wound edge and wound bed, respectively. A,B, Bar represents 2 μm. Images shown are representative of 3 independent experiments.
High-Titer E faecalis Infection Alters Wound Healing and Delays Wound Closure
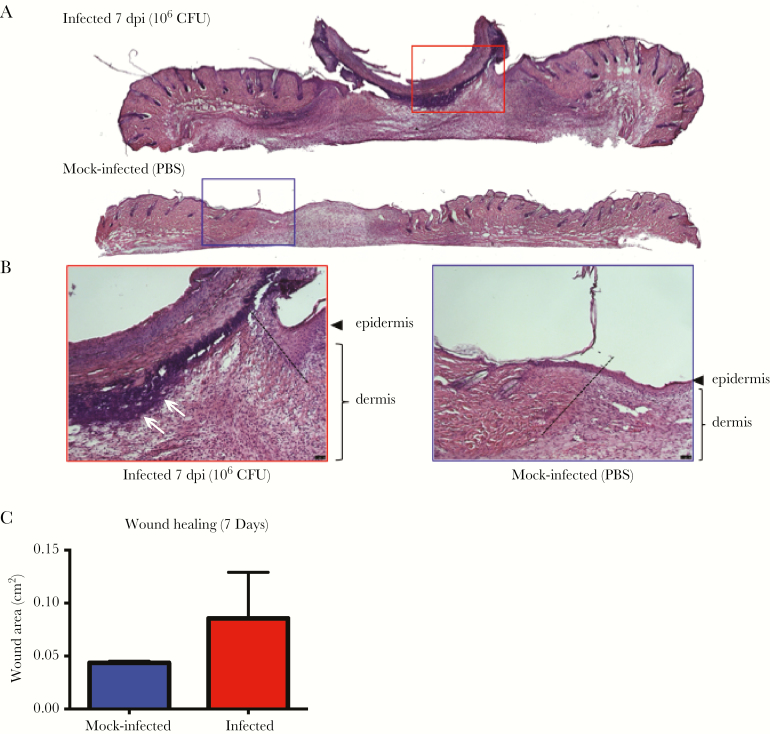
Infection of wounds by P aeruginosa and S aureus correlate with delayed re-epithelization and wound healing [43, 44]. To determine whether E faecalis similarly affects wound healing, we performed histology on skin tissue obtained from wounds of infected mice at 7 dpi. Hemotoxylin and eosin (H&E) staining revealed a hyperthickened epidermis, indicative of nonprogressive wound healing, with delayed closure in the infected tissues, which was not seen in the wounded, mock-infected controls (Figure 6A and B). Moreover, we also observed large numbers of polymorphonuclear leukocytes in H&E stained infected samples as late as 7 dpi as compared to mock-infected controls (Figure 6A). In addition, granulation tissue, which is indicative of dermal healing, was not properly formed in infected wounds, whereas healing was visible in mock-infected controls (Figure 6A and B). Long-term persistence of E faecalis also resulted in delayed wound closure (Figure 6C). These observations show that high-titer E faecalis infection negatively affects the wound healing process and delays the onset of wound closure.
Figure 6.
Enterococcus faecalis infection alters wound healing dynamics. Wounds were harvested at 7 days postinfection (dpi) and subjected to H&E staining. A,B, Red and blue boxes represent the wound edge for the infected wounds and mock-infected controls, respectively. B, Higher magnification images of the boxes depicted in (A) and dashed line indicate the wound edge. Clusters of polymorphonuclear leukocytes are present at the 7 dpi wound (white arrows) but absent from the mock-infected wound. Bar represents 20 μm. Images are representative observations from 3 independent samples examined. C, Wound area at 7 dpi. Blue and red bar graphs represent the mean area of the mock infected and infected wounds respectively. Error bars represent the standard deviation. Measurements were made from 4 independent mice from each group.
E faecalis Can Persist Within Wounds While Escaping Immune Clearance
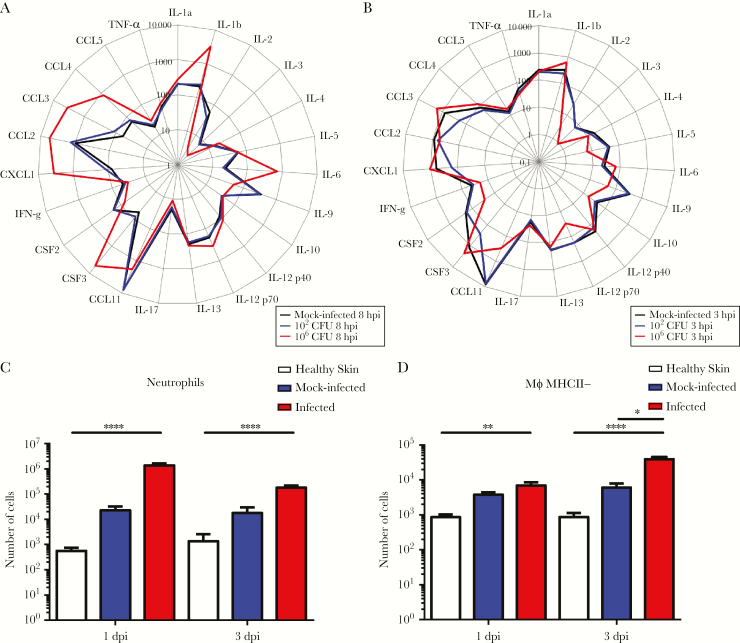
We next hypothesized that E faecalis might escape host detection during wound infection, contributing to its ability to persist and delay wound healing. Therefore, to examine the host immune response to E faecalis infection, we first performed cytokine, growth factor, and chemokine analysis on supernatants from wound homogenates inoculated with either 102 or 106 CFU, or PBS. At both 8 hpi and 3 dpi, wounds inoculated with 102E faecalis CFU had cytokine and growth factor levels similar to the mock-infected controls (Figure 7A and B). By contrast, wounds infected with 106 CFU displayed significantly higher levels of the inflammatory cytokine IL-1b, as well as growth factors/chemokines CSF3, CXCL1, CCL2, CCL3, and CCL4, compared to the mock-infected controls at 8 hpi (Figure 7A, Supplementary Figure 4A), when macroscopic inflammation was observed. At 3 dpi, when E faecalis wound titers resolved to 105 CFU (Figure 2B), we observed significantly lower levels of IL-2, IL-5, IL-10, IL12-p70, CCL11, IFN-γ, and CSF2 compared to both 102 CFU-inoculated and mock-infected controls (Figure 7B, Supplementary Figure 4B). Reduced cytokine and chemokine levels during steady-state infection suggest that E faecalis can modulate the host immune response in wounds to promote persistence.
Figure 7.
Enterococcus faecalis modulates the soluble and cellular host response at the wound site. Mice were wounded and infected with 102 colony forming units (CFU) or 106 CFU of E faecalis, or mock infected with phosphate buffered saline. At the indicated times, wounds were processed into single cell suspensions and subjected to (A, B) cytokine analysis, shown in pg/mL. Total number of (C) neutrophils (CD45+ MHCII− CD11b+ Ly6G+) and (D) macrophages (CD45+ MHCII− CD11b+ Ly6C− CD64+) infiltrating into and accumulating in the skin analyzed by flow cytometry. N = 2, n = 5. Error bars represent the standard error of mean. Statistical analysis was performed using Kruskal–Wallis test with Dunn’s post-hoc test. * P < .05, ** P < .01, *** P < .001, **** P < .0001. Abbreviations: CCL, chemokine ligand; CSF, colony stimulating factor; CXC, C-X-C motif chemokine; IL, interleukin; N, biological replicates; n, technical replicates; TNF, tumor necrosis factor; IFN, interferon.
To gain further insight into the spectrum of soluble factors that were most associated with E faecalis immune modulation during infection, we performed principal component analysis (PCA) (Supplementary Figure 4C). The PCA profiles of wounds infected with 106 CFU at 8 hpi and 3 dpi were distinct and clustered separately, confirming that high inoculum infection results in a temporally distinct inflammatory profile (Supplementary Figure 4C). Differences in IL-1β, IL-2, IL-12p70, and CCL11 specifically explained the variation between the PCA profiles and best represented differences between all sample groups. Among these, IL-2, IL-12p70, and CCL11 were significantly decreased in the 106 CFU infected group when compared to the mock-infected controls, suggesting that down-regulation of these cytokines in particular may be associated with an attenuated immune response (Supplementary Figure 4B).
To complement the analysis of soluble immune effectors, we performed flow cytometry to quantify the immune cell types present during infection (Supplementary Figure 5). Nearly all immune cell types examined were significantly increased in the infected wounds compared to the healthy skin at 1 and 3 dpi (Supplementary Figure 6); and neutrophil infiltration correlated with neutrophil-related chemokine expression in the infected wounds compared to the mock-infected controls (Figure 7A and C). Notably, at 3 dpi, there were significantly more MHCII− macrophages in the infected wounds compared to mock-infected controls (Figure 7D). Despite the presence of significant immune infiltrates at 3 dpi, the E faecalis bacterial burden in the wounds persisted at >105 CFU.
Taken together, these data demonstrate that high-titer inocula, resulting in high-titer wound infection, is associated with an acute inflammatory response concomitant with the peak of infection. The resolution of acute high-titer infection to a lower steady-state infection at 3 dpi corresponds to a suppression of cytokine and chemokine levels but also the presence of immune cellular infiltrate, suggesting a complex immunomodulatory program that is insufficient to resolve acute E faecalis wound infection.
DISCUSSION
Surgical site infections are prevalent and can extend the average hospital stay by 5 to 17 days [1]. Despite the prevalence and clinical importance of E faecalis wound infection, we know nothing of its pathogenic mechanisms in this infection setting. Here, we established a modified mouse wound excisional model to study the infection dynamics of E faecalis in surgical site infections.
We show that acute high-titer E faecalis wound infection associated with ≥106 CFU is associated with a robust cellular host immune response and visible signs of inflammation, along with delayed wound healing, whereas inflammation is suppressed or absent in lower-titer infections. Our observations are consistent with reports showing that bacterial counts of ≥106 perturb healing in humans [45, 46]. However, despite an early robust inflammatory response, E faecalis can persist in the local wound site regardless of the inoculum load. Consistent with this, we observed E faecalis in both the wound bed and at the epidermal wound edge at 3 dpi, suggesting that E faecalis reservoirs within host cells may promote persistence in this niche.
Consistent with reports that most wound infections involve biofilms, we observed the presence of microcolonies at the surface of E faecalis infected wounds at 8 hpi. However, a sortase null mutant, deficient in the surface display of a variety of biofilm-associated factors, was not attenuated in wounds. Together, these findings suggest that E faecalis wound-associated microcolonies or biofilms require other bacterial or host factors for their development, and that currently understood biofilm factors are less crucial in this niche. Instead, we found that a factor involved in resistance to host immune defenses is important for E faecalis survival in wounds. We observed that an MprF null strain was not attenuated at 8 hpi when acute bacterial replication is occurring, but displayed a fitness defect at 3 dpi when the bacterial burden resolves to a steady state of 105 CFU. Interestingly, the only immune cell type we observed to be significantly greater in infected wounds compared to mock-infected wounds was MHCII− macrophages and we only observed this at 3 dpi. Based on these findings, it is tempting to speculate that MprF may contribute to E faecalis survival in the face of macrophage infiltration. Consistent with this, we and others have shown that E faecalis can survive within and suppress immune activation of macrophages [23, 47, 48].
Importantly, we showed that E faecalis wound infection resulted in immunomodulation. At 3 dpi, IL-2, IL-5, IL-10, IL-12p70, CCL11, IFN-γ, and CSF2 levels were significantly lower in infected wounds compared to mock-infected wounds, suggesting active immune suppression at the cytokine and chemokine level [49, 50]. However, we still observed significant immune cell infiltrate at 3 dpi, indicating that immune modulation may be insufficient to limit a full inflammatory response. Nevertheless, the proinflammatory cellular infiltrate was not able to clear E faecalis from the wounds. Thus, it is tempting to speculate that E faecalis wound infection includes an active immune evasion or immune suppression component, which contributes to high-titer infection and long-term persistence, leading to the development of a chronic, nonhealing wound. Further, even modest E faecalis-mediated immune suppression may provide an advantage for coinfecting organisms commonly found with E faecalis in polymicrobial wound infections [10, 39]. Given the widespread prevalence of enterococcal wound infections, further studies into factors that promote E faecalis pathogenesis in wounds and its consequences on wound healing are critical.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgments. We would like to thank Pei Yi Choo for her assistance with the FISH imaging. We would also like to thank Milton Kwek and Declan Lunny for their help and advice regarding the histology, sectioning, and staining of skin samples.
Financial support. This work was supported by the National Research Foundation and Ministry of Education Singapore under its Research Centre of Excellence Program, by the National Research Foundation under its Singapore NRF Fellowship program (NRF-NRFF2011-11), and by the Ministry of Education Singapore under its Tier 2 program (MOE2014-T2-1–129).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. World Health Organization. Global guidelines for the prevention of surgical site infections. World Health Organization, 2016. [PubMed] [Google Scholar]
- 2. Dowd SE, Sun Y, Secor PR et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 2008; 8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev 2001; 14:244–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giacometti A, Cirioni O, Schimizzi AM et al. Epidemiology and microbiology of surgical wound infections. J Clin Microbiol 2000; 38:918–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. National Nosocomial Infections SurveillanceSystem. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1992 through June 2004, issued October 2004. Am J Infect Control 2004; 32:470–85. [DOI] [PubMed] [Google Scholar]
- 6. Gjødsbøl K, Christensen JJ, Karlsmark T, Jørgensen B, Klein BM, Krogfelt KA. Multiple bacterial species reside in chronic wounds: a longitudinal study. Int Wound J 2006; 3:225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hollenbeck BL, Rice LB. Intrinsic and acquired resistance mechanisms in enterococcus. Virulence 2012; 3:421–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hurlow J, Couch K, Laforet K, Bolton L, Metcalf D, Bowler P. Clinical Biofilms: a challenging frontier in wound care. Adv Wound Care 2015; 4:295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Metcalf DG, Bowler PG. Biofilm delays wound healing: A review of the evidence. Burns Trauma 2015; 1:5–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tay WH, Chong KK, Kline KA. Polymicrobial-host interactions during infection. J Mol Biol 2016; 428:3355–71. [DOI] [PubMed] [Google Scholar]
- 11. Hall‐Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol 2009; 11:1034–43. [DOI] [PubMed] [Google Scholar]
- 12. Kline KA, Kau AL, Chen SL et al. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol 2009; 191:3237–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nielsen HV, Flores-Mireles AL, Kau AL et al. Pilin and sortase residues critical for endocarditis- and biofilm-associated pilus biogenesis in Enterococcus faecalis. J Bacteriol 2013; 195:4484–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nallapareddy SR, Singh KV, Sillanpää J et al. Endocarditis and biofilm-associated pili of Enterococcus faecalis. J Clin Invest 2006; 116:2799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nielsen HV, Guiton PS, Kline KA et al. The metal ion-dependent adhesion site motif of the Enterococcus faecalis EbpA pilin mediates pilus function in catheter-associated urinary tract infection. MBio 2012; 3:e00177–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Flores-Mireles AL, Pinkner JS, Caparon MG, Hultgren SJ. EbpA vaccine antibodies block binding of Enterococcus faecalis to fibrinogen to prevent catheter-associated bladder infection in mice. Sci Transl Med 2014; 6:254ra127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nallapareddy SR, Qin X, Weinstock GM, Höök M, Murray BE. Enterococcus faecalis adhesin, ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. Infect Immun 2000; 68:5218–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Süssmuth SD, Muscholl-Silberhorn A, Wirth R, Susa M, Marre R, Rozdzinski E. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect Immun 2000; 68:4900–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dunny GM, Leonard BA, Hedberg PJ. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J Bacteriol 1995; 177:871–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shankar V, Baghdayan AS, Huycke MM, Lindahl G, Gilmore MS. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect Immun 1999; 67:193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Varahan S, Iyer VS, Moore WT, Hancock LE. Eep confers lysozyme resistance to Enterococcus faecalis via the activation of the extracytoplasmic function sigma factor SigV. J Bacteriol 2013; 195:3125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thurlow LR, Thomas VC, Fleming SD, Hancock LE. Enterococcus faecalis capsular polysaccharide serotypes C and D and their contributions to host innate immune evasion. Infect Immun 2009; 77:5551–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zou J, Shankar N. Enterococcus faecalis infection activates phosphatidylinositol 3-kinase signaling to block apoptotic cell death in macrophages. Infect Immun 2014; 82:5132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Park SY, Shin YP, Kim CH et al. Immune evasion of Enterococcus faecalis by an extracellular gelatinase that cleaves C3 and iC3b. J Immunol 2008; 181:6328–36. [DOI] [PubMed] [Google Scholar]
- 25. Baldassarri L, Bertuccini L, Creti R et al. Glycosaminoglycans mediate invasion and survival of Enterococcus faecalis into macrophages. J Infect Dis 2005; 191:1253–62. [DOI] [PubMed] [Google Scholar]
- 26. Rakita RM, Vanek NN, Jacques-Palaz K et al. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect Immun 1999; 67:6067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kandaswamy K, Liew TH, Wang CY et al. Focal targeting by human β-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci U S A 2013; 110:20230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bao Y, Sakinc T, Laverde D et al. Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS One 2012; 7:e38458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Camejo A, Buchrieser C, Couvé E et al. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog 2009; 5:e1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Peschel A, Jack RW, Otto M et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J Exp Med 2001; 193:1067–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thedieck K, Hain T, Mohamed W et al. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol 2006; 62:1325–39. [DOI] [PubMed] [Google Scholar]
- 32. Kirchner LM, Meerbaum SO, Gruber BS et al. Effects of vascular endothelial growth factor on wound closure rates in the genetically diabetic mouse model. Wound Repair Regen 2003; 11:127–31. [DOI] [PubMed] [Google Scholar]
- 33. Cross SE, Naylor IL, Coleman RA, Teo TC. An experimental model to investigate the dynamics of wound contraction. Br J Plast Surg 1995; 48:189–97. [DOI] [PubMed] [Google Scholar]
- 34. Stiernberg J, Norfleet AM, Redin WR, Warner WS, Fritz RR, Carney DH. Acceleration of full-thickness wound healing in normal rats by the synthetic thrombin peptide, TP508. Wound Repair Regen 2000; 8:204–15. [DOI] [PubMed] [Google Scholar]
- 35. Turner KH, Everett J, Trivedi U, Rumbaugh KP, Whiteley M. Requirements for Pseudomonas aeruginosa acute burn and chronic surgical wound infection. PLoS Genet 2014; 10:e1004518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thompson MG, Black CC, Pavlicek RL et al. Validation of a novel murine wound model of Acinetobacter baumannii infection. Antimicrob Agents Chemother 2014; 58:1332–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watters C, DeLeon K, Trivedi U et al. Pseudomonas aeruginosa biofilms perturb wound resolution and antibiotic tolerance in diabetic mice. Med Microbiol Immunol 2013; 202:131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. DeLeon S, Clinton A, Fowler H, Everett J, Horswill AR, Rumbaugh KP. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect Immun 2014; 82:4718–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Keogh D, Tay WH, Ho YY et al. Enterococcal metabolite cues facilitate interspecies niche modulation and polymicrobial infection. Cell Host Microbe 2016; 20:493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kempf VA, Trebesius K, Autenrieth IB. Fluorescent in situ hybridization allows rapid identification of microorganisms in blood cultures. J Clin Microbiol 2000; 38: 830–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rousseau M, Goh HMS, Holec S et al. Bladder catheterization increases susceptibility to infection that can be prevented by prophylactic antibiotic treatment. JCI Insight 2016; 1: e88178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bourgogne A, Garsin DA, Qin X et al. Large scale variation in Enterococcus faecalis illustrated by the genome analysis of strain OG1RF. Genome Biol 2008; 9:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goldufsky J, Wood SJ, Jayaraman V et al. Pseudomonas aeruginosa uses T3SS to inhibit diabetic wound healing. Wound Repair Regen 2015; 23:557–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schierle CF, De la Garza M, Mustoe TA, Galiano RD. Staphylococcal biofilms impair wound healing by delaying reepithelialization in a murine cutaneous wound model. Wound Repair Regen 2009; 17:354–9. [DOI] [PubMed] [Google Scholar]
- 45. Bendy R Jr, Nuccio P, Wolfe E et al. Relationship of quantitative wound bacterial counts to healing of decubiti: Effect of topical gentamicin. Antimicrob Agents Chemother 1964; 10:147–55. [PubMed] [Google Scholar]
- 46. Robson MC, Heggers JP. Delayed wound closure based on bacterial counts. J Surg Oncol 1970; 2:379–83. [DOI] [PubMed] [Google Scholar]
- 47. Tien BYQ, Goh HMS, Chong KKL et al. Enterococcus faecalis promotes innate immune suppression and polymicrobial catheter-associated urinary tract infection [published online ahead of print 11 Sep, 2017]. Infect Immun doi: 10.1128/IAI.00378-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zou J, Shankar N. The opportunistic pathogen Enterococcus faecalis resists phagosome acidification and autophagy to promote intracellular survival in macrophages. Cell Microbiol 2016; 18:831–43. [DOI] [PubMed] [Google Scholar]
- 49. Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen 2008; 16:585–601. [DOI] [PubMed] [Google Scholar]
- 50. Leoni G, Neumann PA, Sumagin R, Denning TL, Nusrat A. Wound repair: role of immune-epithelial interactions. Mucosal Immunol 2015; 8:959–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.