Abstract
Objective. Gout is associated with dyslipidaemia. Association of the apolipoprotein A1-C3-A4 gene cluster with gout has previously been reported in a small study. To investigate a possible causal role for this locus in gout, we tested the association of genetic variants from APOA1 (rs670) and APOC3 (rs5128) with gout.
Methods. We studied data for 2452 controls and 2690 clinically ascertained gout cases of European and New Zealand Polynesian (Māori and Pacific) ancestry. Data were also used from the publicly available Atherosclerosis Risk in Communities study (n = 5367) and the Framingham Heart Study (n = 2984). Multivariate adjusted logistic and linear regression was used to test the association of single-nucleotide polymorphisms with gout risk, serum urate, triglyceride and high-density lipoprotein cholesterol (HDL-C).
Results. In Polynesians, the T-allele of rs670 (APOA1) increased (odds ratio, OR = 1.53, P = 4.9 × 10−6) and the G-allele of rs5128 (APOC3) decreased the risk of gout (OR = 0.86, P = 0.026). In Europeans, there was a strong trend to a risk effect of the T-allele for rs670 (OR = 1.11, P = 0.055), with a significant protective effect of the G-allele for rs5128 being observed after adjustment for triglycerides and HDL-C (OR = 0.81, P = 0.039). The effect at rs5128 was specific to males in both Europeans and Polynesians. Association in Polynesians was independent of any effect of rs670 and rs5128 on triglyceride and HDL-C levels. There was no evidence for association of either single-nucleotide polymorphism with serum urate levels (P ⩾ 0.10).
Conclusion. Our data, replicating a previous study, supports the hypothesis that the apolipoprotein A1-C3-A4 gene cluster plays a causal role in gout.
Keywords: gout, hyperuricaemia, gene, association, apolipoprotein
Rheumatology key message
Replicated association of the apolipoprotein A1-C3-A4 gene cluster with gout implicates apolipoprotein metabolism as causal in gout.
Introduction
Gout typically presents as an acute autoinflammatory response to MSU crystals that form in individuals with hyperuricaemia (HU). While the aetiology of HU is becoming better understood, the genetic and biochemical factors predisposing individuals with HU to symptomatic gout are poorly understood [1]. Gout and HU are associated with an increased level of very low-density lipoprotein triglyceride (VLDL-Tg) [2], with association of VLDL-Tg with gout evident using an asymptomatic hyperuricaemic comparison group [3]. These findings implicate triglyceride and apolipoprotein metabolism in the aetiology of gout.
Hyperuricaemic-hypertriglyceridaemic patients have an increased ratio of apolipoprotein C3–C2 [4] along with increased VLDL levels [5]. The apolipoprotein molecules are encoded within a gene cluster of ∼60 kb on human chromosome 11q23 that encodes for APOA1, APOC3, APOA4 and APOA5 [6]. Among these apolipoproteins, APOA1 is a major contributor to reverse cholesterol transport in the liver by acting as the ligand for the ABCA1 cholesterol transporter and as an obligatory cofactor for lecithin-cholesterol acyltransferase [7]. The major C-allele at the -75 position (rs670) in the promoter region has been associated with increased levels of circulating high-density lipoprotein cholesterol (HDL-C) in Han Chinese [8]. APOC3, predominantly produced in the liver (reviewed in [9]), facilitates the exchangeable component of Tg-rich lipoproteins (chylomicron and VLDL) and is an inhibitor of lipoprotein and hepatic lipase [10]. In APOC3, the minor allele of rs5218 in the 3′ non-coding region (3238G/C) has repeatedly been associated with hypertriglyceridaemia [11–14]. However this candidate variant was not associated with gout in a small Spanish case–control sample set [4]. In contrast, supporting a causal role for APOA1 in gout, the C-allele of rs670 was associated with gout in the same Spanish sample set (odds ratio, OR = 1.99, P = 0.01) [4]. The aim of this study was to test for association of APOA1 variant rs670 and APOC3 variant rs5128 with gout.
Methods
Subjects
Demographic and clinical details of all study sample sets are summarized in supplementary Table S1, available at Rheumatology Online. The New Zealand (NZ) gout participants were mostly recruited from Auckland, Wellington and Christchurch, NZ. Additional cases of European ancestry were recruited by the Eurogout consortium within the European Crystal Network (n = 762) [15] and by the Arthritis Genomics Recruitment Initiative in Australasia (n = 83). All gout cases were clinically ascertained by the ARA preliminary classification criteria [16]. All variables except for biochemical measurements [serum urate (SU), serum triglyceride and HDL-C] and BMI were self-reported.
The NZ control group self-reported their lack of gouty arthritis. It comprised 457 European and 994 Polynesian individuals recruited primarily from the Auckland area, using the same protocol as the cases, and 1001 European controls sampled from the Otago region [17]. Apart from the Otago controls, the NZ participants were not requested to fast. On the basis of the self-reported ancestry of grandparents, the Polynesian participants were divided into three subgroups: Eastern Polynesian (EP)—NZ Māori and Cook Island Māori (489 cases, 626 controls), Western Polynesian (WP)—Tongan, Samoan and Niuean (412 cases, 298 controls) and a small mixed Eastern and Western ancestry group (EP/WP) (34 cases, 70 controls). The stratification of Polynesian subjects into EP and WP was because of genetic diversity between the two groups [18]. The NZ Multi-region Ethics Committee (105/10/130) and the following institutional committees in Europe and Australia granted ethical approval: Research and Ethics Committee, Repatriation General Hospital, South Australia (32/08); Research Ethics Committee, University of New South Wales; Ethikkommission, Technische Universität Dresden (EK 8012012); South East Scotland Research Ethics Committee (04/S1102/41); Commission Cantonale (VD) D’éthique de la Recherche sur l’être Humain, Université de Lausanne; Commissie Mensgebonden Onderzoek regio Arnhem—Nijmegen; Partners Health Care System Institutional Review Board. Written informed consent according to the Declaration of Helsinki was obtained from all participants.
Participants from the Atherosclerosis Risk in Communities (ARIC) study and the Framingham Heart Study (FHS) (Generation 3 only) cohorts were also used as additional controls for the gout risk analysis and for evaluating association with SU (approval number #834; The Genetic Basis of Gout). Subjects from ARIC and FHS who self-reported as taking diuretic medication, who were first-degree related, who were not of European ancestry, who were diagnosed with kidney disease or who reported physician-diagnosed gout, were excluded. The final ARIC and FHS data sets consisted of 5367 and 2984 people of European ancestry, respectively. For all ancestral groups (except the 1001 controls recruited from the Otago region of NZ, for whom SU data were not available), control participants were further stratified into a normouricaemia group (with a SU level <0.41 mmol/l) and a HU group with a SU level of ⩾0.41 mmol/l.
Genotyping
In the NZ samples, rs670 and rs5128 Taqman genotyping was done using a LightCycler 480 Real-Time PCR System (Roche Applied Science, IN, USA) in 384-well plates. The ARIC sample set was genotyped on the Affymetrix SNP 6 platform, and the FHS sample set had been genotyped by the Affymetrix SNP 5 platform and a custom-designed gene-centric 50K +500K array SNP platform. Both single-nucleotide polymorphisms (SNPs) were imputed in the ARIC data set using IMPUTE2 (ver. 2.3.0) [19] to a common reference panel from the 1000 Genomes project [Phase I integrated variant set release (ver. 3), March 2012, NCBI build 37 (hg19)]. Both SNPs were imputed in FHS generation 3 subjects using MACH1 v1.0.15 [20] with the HapMap2 CEU (release 22, build 36) sample set used as reference haplotypes. For the 1001 controls recruited from the Otago region of NZ, 640 participants were genotyped using the Affymetrix 6 platform and the remainder using the Illumina Omni2.5 platform. Rs5128 was genotyped by both platforms, whereas rs670 was imputed (IMPUTE ver. 2.2) to a common reference panel from the 1000 Genomes project [Phase I integrated variant set release (ver. 3), March 2012, NCBI build 37 (hg19)], followed by quality score (Q cor.9) filtering. The genotype distributions of all sample sets presented in Table 1 were in Hardy–Weinberg equilibrium (P > 0.01).
Table 1.
Association analysis of rs670 and rs5128 with risk of gout using all controls
| Case genotypes, n (freq) | Control genotypes, n (freq) | Unadjusted | Adjusteda | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | T Freq | CC | CT | TT | T Freq | Allelic OR (T-Allele), (95% CI) | Allelic P | Allelic OR (T-Allele), (95% CI) | Allelic P | |
|
| ||||||||||||
| rs670 (APOA1) | ||||||||||||
| European | 1204 (0.693) | 470 (0.270) | 64 (0.037) | 598 (0.172) | 6911 (0.708) | 2629 (0.269) | 227 (0.023) | 3083 (0.158) | 1.11 (1.01, 1.22) | 0.035 | 1.11 (1.00, 1.24) | 0.055 |
| 1.07 (0.93, 1.25) | 0.34 | |||||||||||
| 1.14 (1.00, 1.30) | 0.051 | |||||||||||
| Eastern Polynesian | 356 (0.739) | 87 (0.180) | 39 (0.081) | 165 (0.171) | 487 (0.780) | 132 (0.212) | 05 (0.008) | 142 (0.114) | 1.50 (1.20, 1.88) | 0.0004 | 1.61 (1.12, 2.15) | 0.001 |
| 1.74 (1.26, 2.42) | 0.001 | |||||||||||
| 1.56 (1.16, 2.10) | 0.003 | |||||||||||
| EP/WP | 20 (0.588) | 10 (0.294) | 04 (0.118) | 18 (0.265) | 51 (0.739) | 17 (0.246) | 01 (0.015) | 19 (0.138) | 2.11 (1.04, 4.28) | 0.039 | 1.97 (0.88, 4.39) | 0.097 |
| 1.98 (0.84, 4.67) | 0.12 | |||||||||||
| 2.23 (0.91, 5.43) | 0.078 | |||||||||||
| Western Polynesian | 266 (0.670) | 121 (0.305) | 10 (0.025) | 141 (0.178) | 221 (0.742) | 71 (0.238) | 06 (0.020) | 83 (0.139) | 1.34 (1.00, 1.81) | 0.053 | 1.38 (0.97, 1.95) | 0.074 |
| 1.49 (1.01, 2.21) | 0.045 | |||||||||||
| 1.47 (1.02, 2.12) | 0.039 | |||||||||||
|
| ||||||||||||
| CC | CG | GG | G Freq | CC | CG | GG | G Freq | Allelic OR (G-Allele), (95% CI) | Allelic P | Allelic OR (G-Allele), (95% CI) | Allelic P | |
|
| ||||||||||||
| rs5128 (APOC3) | ||||||||||||
| European | 1464 (0.849) | 245 (0.142) | 16 (0.009) | 277 (0.080) | 8165 (0.833) | 1542 (0.157) | 94 (0.010) | 1730 (0.088) | 0.90 (0.79, 1.03) | 0.13 | 0.91 (0.79, 1.06) | 0.22 |
| 0.81 (0.66, 0.99) | 0.039 | |||||||||||
| 0.89 (0.75, 1.06) | 0.20 | |||||||||||
| Eastern Polynesian | 192 (0.400) | 241 (0.502) | 47 (0.098) | 335 (0.349) | 268 (0.432) | 271 (0.436) | 82 (0.132) | 435 (0.350) | 0.99 (0.83, 1.19) | 0.95 | 0.98 (0.78, 1.24) | 0.91 |
| 0.93 (0.71, 1.20) | 0.57 | |||||||||||
| 0.96 (0.76, 1.22) | 0.74 | |||||||||||
| EP/WP | 15 (0.441) | 14 (0.412) | 05 (0.147) | 24 (0.353) | 26 (0.371) | 30 (0.429) | 14 (0.200) | 58 (0.414) | 0.79 (0.45, 1.40) | 0.42 | 0.83 (0.42, 1.63) | 0.60 |
| 1.04 (0.50, 2.17) | 0.92 | |||||||||||
| 0.74 (0.37, 1.50) | 0.40 | |||||||||||
| Western Polynesian | 163 (0.405) | 177 (0.439) | 63 (0.156) | 303 (0.376) | 82 (0.275) | 157 (0.527) | 59 (0.198) | 275 (0.461) | 0.71 (0.57, 0.87) | 0.002 | 0.65 (0.50, 0.84) | 0.001 |
| 0.62 (0.46, 0.82) | 0.001 | |||||||||||
| 0.66 (0.50, 0.86) | 0.003 | |||||||||||
aTop value adjusted for age and sex; middle value adjusted for age, sex, triglyceride and HDL-C; bottom value adjusted for age, sex and BMI. Polynesian sample sets were additionally adjusted for STRUCTURE ancestry estimate. WP: Western Polynesian; EP: Eastern Polynesian. HDL-C: high-density lipoprotein cholesterol; freq: frequency.
Statistical analyses
Associations with gout risk and blood metabolites (SU, serum triglyceride and HDL-C) were determined using logistic and linear regression, respectively, using STATA ver. 8.0 (StataCorp, College Station, TX, USA). All associations were adjusted for age and sex with additional adjustors being serum triglyceride, HDL-C and BMI. In Polynesians, a STRUCTURE [21] ancestral estimate was also included as an adjustor, as previously described [22], in order to account for admixture, primarily with Europeans. Meta-analysis of allele counts was performed in R within STATA, using rmeta to calculate combined ORs and to evaluate heterogeneity between studies, with a random effect model being used when PHet < 0.05. Using rmeta, the OR is a ratio of frequency ref/alt alleles in cases vs the frequency of ref/alt alleles in controls, whereas the OR derived from logistic regression measures the relationship between the binary dependent variable (gout) and independent variable (genotype) by estimating probabilities using a logistic function. Coefficients with P ⩽ 0.05 were considered to indicate a nominally significant association. Because there was little linkage disequilibrium between rs670 and rs5128 (r2 in NZ Europeans = 0.01, r2 in NZ Polynesians = 0.08), association analysis of haplotypes was not done; instead, epistatic interaction was tested by incorporating an interaction term in the logistic regression analysis.
Power calculations for the individual sample sets are presented in supplementary Table S2, available at Rheumatology Online. For rs670, using all controls, there was adequate power in the European sample set to detect an allelic effect size of OR > 1.2, and adequate power in the combined Polynesian sample set to detect an allelic effect size of OR > 1.3. For rs5128, power was adequate in both sample sets to detect an effect size of OR > 1.2.
Models of inheritance were estimated by formulating the genotype predictor in the logistic regression model in different ways: full genotype model (CC, CT, TT)—two ORs; additive model (0, 0.5, 1) or (0, 1, 2)—one OR (this should be approximately the same as the allelic OR but may differ if there is lack of fit to Hardy–Weinberg equilibrium in either cases or controls); dominant model (0, 1, 1)—one OR; or recessive model (0, 0, 1)—one OR. A model selection tool (Akaike Information Criterion [23]) was used to select the most likely model.
Results
Association of rs670 and rs5128 was first evaluated with serum triglyceride and HDL-C levels in the combined gout and non-gout European and Polynesian sample sets (supplementary Table S3, available at Rheumatology Online). Evidence for association of the G-allele of rs5128 (APOC3) with serum triglyceride and both variants with HDL-C (C-allele of rs5128 and T-allele of rs670) was observed in both ancestral groups, with a consistent direction of association. For example, for rs5128 in serum triglyceride the effect size was, per copy of the minor G allele, 0.150 mmol/l in Europeans and 0.149 mmol/l in Polynesians, a direction of effect consistent with previous reports (ref. [14] and citations therein). In Polynesians and Europeans, the minor T allele of rs670 was associated with increased HDL-C, an opposing effect to that previously observed in Han Chinese [8].
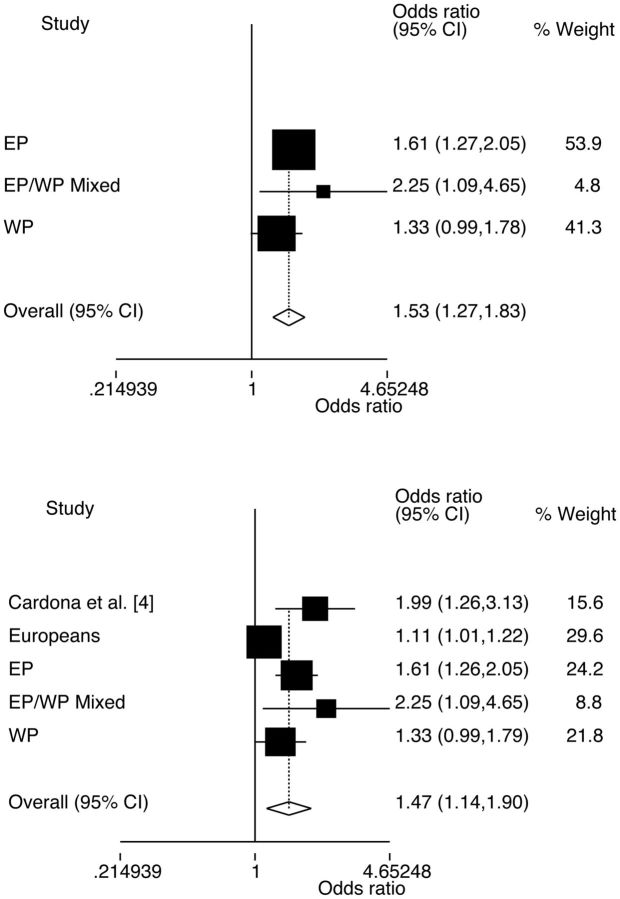
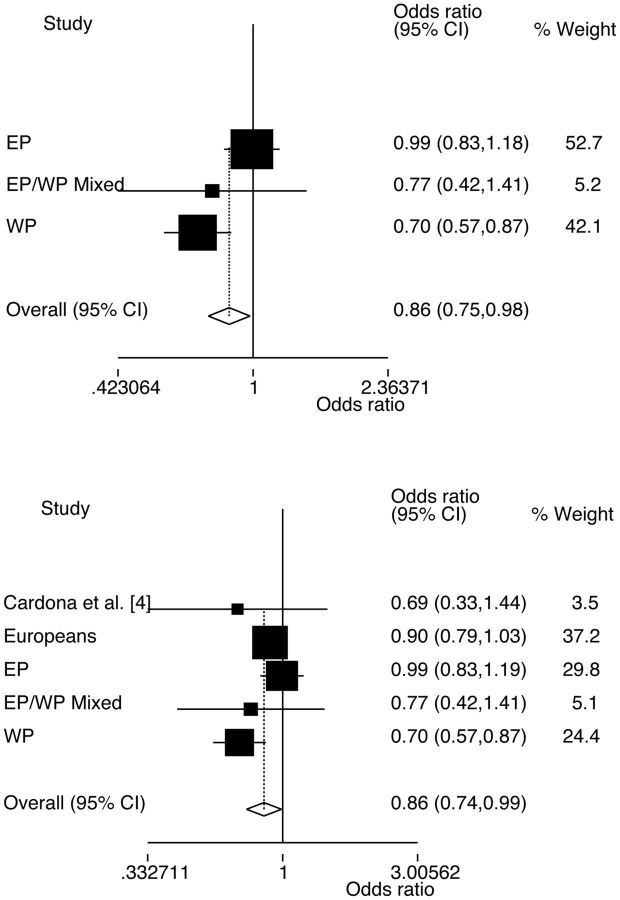
Increased risk of gout was associated with the minor T-allele of rs670 in the EP sample set (Table 1; OR = 1.61, P = 0.001). When the three Polynesian sample sets were combined by meta-analysis, increased risk of gout was associated with the T-allele (Fig. 1A; ORmeta = 1.53, Pmeta = 4.88 × 10−6). A strong trend towards association of the T-allele with gout risk was observed in Europeans (OR = 1.11, P = 0.055) (Table 1). Combined with the Polynesian sample sets and the previously published Cardona et al. [4] data in meta-analysis, there was also evidence for association of rs670 with the risk of gout (Fig. 1B; ORmeta = 1.47, Pmeta = 0.003). The minor G-allele of rs5128 was associated with reduced risk of gout in the WP sample set (Table 1; OR = 0.65, P = 0.001) as well as in meta-analysis of the combined Polynesian sample set (Fig. 2A; ORmeta = 0.86, Pmeta = 0.026). No evidence of association of the G-allele with gout risk was observed in Europeans (OR = 0.91, P = 0.22), although adjustment by serum triglyceride and HDL-C level provided nominal evidence for association (OR = 0.81, P = 0.039). When the European, Cardona et al. [4] and Polynesian samples were combined by meta-analysis for rs5128 there was evidence for association with gout (Fig. 2B; ORmeta = 0.86, Pmeta = 0.036). There was no evidence for epistatic interaction between rs670 and rs5128 in determining the risk of gout in Europeans or Polynesians (ORInteraction = 0.81, P = 0.40 and ORInteraction = 0.64, P = 0.11, respectively).
Fig. 1.
Meta-analysis of Polynesian, and Polynesian and European subgroups using all controls for rs670
Top, Polynesian subgroups: PHet = 0.34 and P for OR is 4.9 × 10−6. Bottom, Polynesian and European subgroups: PHet = 0.003 and P for OR is 0.003. WP: Western Polynesian; EP: Eastern Polynesian.
Fig. 2.
Meta-analysis of Polynesian, and Polynesian and European subgroups using all controls for rs5128
Top, Polynesian subgroups: PHet = 0.048 and P for OR is 0.026. Bottom, Polynesian and European subgroups: PHet = 0.15 and P for OR is 0.036. WP: Western Polynesian; EP: Eastern Polynesian.
To investigate the inheritance pattern for each of rs670 and rs5128, four models were estimated by conditional logistic regression (with strata defined by population): full genotype model, additive model (that should approximate the allelic OR), dominant model and recessive model. The Akaike Information Criteria [23] were used to select the most favourable model: a recessive model for rs670 and an additive model for rs5128. On this basis, ORs were calculated for rs670 under a recessive model (comparing the TT-genotype group to the combined CC/CT-genotype group) using all controls and adjusting for age and sex and, for Polynesians, ancestry. This revealed significant evidence for association in Europeans [OR = 1.62 (1.17, 2.23), P = 0.001] compared with OR = 1.11 (P = 0.055) under an additive model and strengthened evidence for association in EP [OR = 21.40 (6.51, 70.35), P = 4.4 × 10−7) compared with OR = 1.61, P = 0.001 under an additive model. For WP, evidence for association was not significant [OR = 1.72 (0.51, 5.80), P = 0.38] compared with OR = 1.38, P = 0.074 under an additive model with statistical evidence for association similar in the sample set of mixed Eastern and WP ancestry [OR = 6.81 (0.67, 68.82), P = 0.10] to the additive model (OR = 1.97, P = 0.097).
Association analyses were adjusted by serum triglyceride and HDL-C (in addition to age and sex), with no substantial changes in effect size or P-values in the Polynesian analyses (Table 1), indicating that the observed associations with gout were independent of serum triglyceride and HDL-C levels. However, in Europeans, the serum triglyceride and HDL-C adjustment reduced the evidence for association of rs670 with gout [OR = 1.11, P = 0.055 to OR = 1.07, P = 0.34, although a recessive model provided significant evidence for association; OR = 1.63 (1.06, 2.50), P = 0.026] and provided evidence for association of rs5128 with gout (OR = 0.81, P = 0.039). However, in Europeans, as was also observed in Polynesians, adjustment for BMI did not substantially influence the effect size or P-values at either SNP.
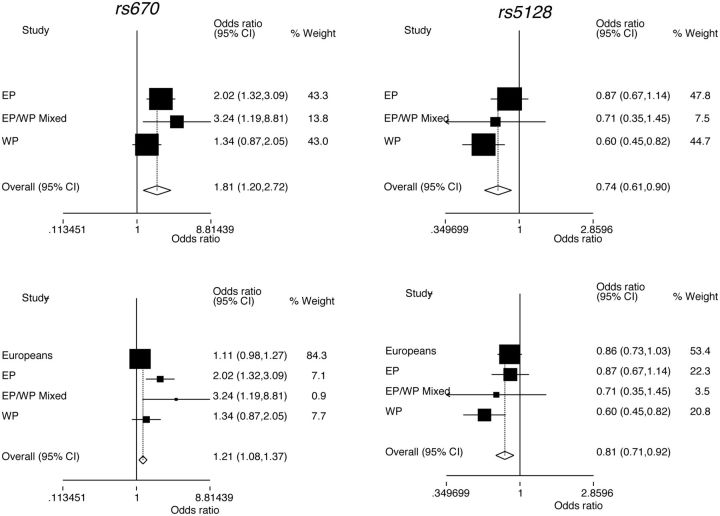
Analysis was conducted using HU controls (SU ⩾ 0.41 mmol/l) (supplementary Table S4, available at Rheumatology Online). Consistent with the unstratified controls analysis, for the T-allele of rs670 variant, increased gout risk was observed in the EP sample set (OR =1.84, P = 0.004) and in combined Polynesians (Fig. 3; OR = 1.81, P = 1.2 × 10−4). A recessive model was not used owing to the scarcity of the TT-genotype in the Polynesian HU controls. Evidence for a significant protective role of the G-allele of rs5128 was observed in the WP sample set (OR = 0.63, P = 0.007) and in combined Polynesians (Fig. 3; OR = 0.74, P = 0.002). In Europeans, there was no significant evidence for association of rs670 with gout using HU controls, although there was significant association at rs5128 after adjustment for serum triglyceride and HDL-C (supplementary Table S4, available at Rheumatology Online; OR = 0.77, P = 0.045). However, for rs670 under a recessive model, there was significant evidence for association in Europeans [OR = 1.66 (1.02, 2.70), P = 0.040]. Meta-analysis with Polynesians strengthened the evidence for association of rs5128 with gout (Fig. 3; OR = 0.81, P = 0.001), but weakened the evidence for association of rs670 with gout (OR = 1.21, P = 0.001).
Fig. 3.
Meta-analysis of Polynesian, and European and Polynesian sample sets using HU controls
Top, Polynesian sample set: for rs670 (APOA1) PHet = 0.18 and P for OR is 1.2 × 10−4 and for rs5128 (APOC3) PHet = 0.20 and P for OR is 0.002. Bottom, Polynesian and European sample set: for rs670 (APOA1) PHet = 0.011 and P for OR is 0.001 and for rs5128 (APOC3) PHet = 0.20 and P for OR is 0.001. WP: Western Polynesian; EP: Eastern Polynesian.
Using all controls, stratification by sex did not reveal significant association of rs670 with either sex in Europeans, using either an allelic or recessive model, with significant association observed in males and females in combined Polynesians using both models (Table 2). However, at rs5128, the association was restricted to males in both the European and combined Polynesian sample sets (Table 2; OR = 0.78, P = 0.027 and OR = 0.69, P = 0.001, respectively). Similar data were observed using HU controls for rs670 (OR = 1.56, P = 0.012 and OR = 2.86, P = 0.006 for combined Polynesian males and females, respectively, using the allelic model) and for rs5128 (OR = 0.75, P = 0.032 and OR = 0.66, P = 0.003 for European and combined Polynesian males, respectively).
Table 2.
Association analysis of rs670 and rs5128 with gout using all controls in males and females
| Case genotypes, n (freq) | Control genotypes, n (freq) | Unadjusted | Adjusted | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | T Freq | CC | CT | TT | T Freq | Allelic OR (T-Allele), (95% CI) | Allelic P | OR, (95% CI) | P-values | ||
|
| |||||||||||||
| rs670 (APOA1) | |||||||||||||
| Males | Combined Europeans | 1018 (0.694) | 393 (0.268) | 55 (0.037) | 503 (0.171) | 3471 (0.711) | 1299 (0.266) | 110 (0.022) | 1519 (0.156) | 1.12 (1.01, 1.25) | 0.039 | 1.09 (0.92, 1.28) | 0.056 |
| 1.58 (0.99, 2.54) | 0.31 | ||||||||||||
| Combined Polynesians | 536 (0.713) | 172 (0.229) | 44 (0.058) | 260 (0.173) | 344 (0.763) | 102 (0.226) | 05 (0.011) | 112 (0.124) | 1.41 (1.13, 1.77) | 3 × 10−3 | 1.63 (1.23, 2.16) | 1 × 10−3 | |
| 6.54 (2.34, 18.28) | 3 × 10−4 | ||||||||||||
| Females | Combined Europeans | 184 (0.684) | 76 (0.282) | 09 (0.033) | 94 (0.174) | 3440 (0.704) | 1330 (0.272) | 117 (0.024) | 1564 (0.160) | 1.11 (0.88, 1.40) | 0.36 | 1.00 (0.67, 1.50) | 0.99 |
| 1.82 (0.62, 5.31) | 0.27 | ||||||||||||
| Combined Polynesians | 106 (0.658) | 46 (0.286) | 09 (0.056) | 64 (0.199) | 413 (0.770) | 117 (0.218) | 06 (0.011) | 129 (0.120) | 1.80 (1.29, 2.51) | 5 × 10−4 | 1.67 (1.05, 2.67) | 0.031 | |
| 14.69 (2.83, 76.24) | 1 × 10−3 | ||||||||||||
|
| |||||||||||||
| CC | CG | GG | G Freq | CC | CG | GG | G Freq | Allelic OR (G-Allele), (95% CI) | Allelic P | Allelic OR (G-Allele), (95% CI) | Allelic P | ||
|
| |||||||||||||
| rs5128 (APOC3) | |||||||||||||
| Males | Combined Europeans | 1232 (0.846) | 210 (0.144) | 14 (0.010) | 238 (0.082) | 4093 (0.835) | 759 (0.155) | 52 (0.011) | 863 (0.088) | 0.92 (0.80, 1.07) | 0.30 | 0.78 (0.62, 0.97) | 0.027 |
| Combined Polynesians | 304 (0.402) | 363 (0.480) | 89 (0.112) | 541 (0.358) | 165 (0.366) | 213 (0.472) | 73 (0.162) | 359 (0.398) | 0.84 (0.71, 1.00) | 0.046 | 0.69 (0.55, 0.85) | 1 × 10−3 | |
| Females | Combined Europeans | 230 (0.865) | 34 (0.128) | 02 (0.007) | 38 (0.071) | 4072 (0.831) | 783 (0.160) | 42 (0.009) | 867 (0.088) | 0.79 (0.57, 1.11) | 0.18 | 0.94 (0.56, 1.60) | 0.83 |
| Combined Polynesians | 66 (0.410) | 69 (0.429) | 26 (0.161) | 121 (0.376) | 209 (0.391) | 243 (0.455) | 82 (0.154) | 407 (0.381) | 0.98 (0.76, 1.26) | 0.87 | 1.14 (0.80, 1.60) | 0.47 | |
Adjusted for age, triglyceride and HDL-C. Polynesians are additionally adjusted for STRUCTURE ancestry estimate. For rs670 in the adjusted column, the allelic OR is top and the recessive model OR is bottom. HDL-C: high-density lipoprotein cholesterol; freq: frequency.
Neither of the SNPs showed any evidence for association with SU in European and Polynesian non-gout controls (Table 3; all age- and sex-adjusted P > 0.10), even when rs670 was analysed under a recessive model (β = –0.0025, P = 0.61 and β = –0.0050, P = 0.84, respectively). Meta-analysis of European and Polynesian data also provided no support for association of the tested SNPs with SU (βmeta = –0.001 mmol/l, Pmeta = 0.62, PHet = 0.12 for rs670 and βmeta = 0.003 mmol/l, Pmeta = 0.097, PHet = 0.29 for rs5128).
Table 3.
Association analysis of rs670 and rs5128 with SU (mmol/l) in controls
| Number | Unadjusted β (95% CI) | P-values | Adjusted β (95% CI) | P-values | |
|---|---|---|---|---|---|
| rs670 (APO A1) | |||||
| European non-gout | 8755 | −0.0016 (−0.0050, 0.0019) | 0.37 | −0.0002 (−0.0030, 0.0025) | 0.87 |
| Polynesian non-gout | 821 | −0.0131 (−0.0265, 0.00004) | 0.057 | −0.0098 (−0.0219, 0.0023) | 0.11 |
| rs5128 (APO C3) | |||||
| European non-gout | 8764 | 0.0024 (−0.0019, 0.0068) | 0.28 | 0.0020 (−0.0015, 0.0055) | 0.26 |
| Polynesian non-gout | 821 | 0.0131 (0.0043, 0.0220) | 4 × 10−3 | 0.0068 (−0.0014, 0.0149) | 0.10 |
Adjusted for age and sex. The Polynesian sample set is additionally adjusted for STRUCTURE ancestry estimate and ancestral group. SU: serum urate.
Discussion
We demonstrated association of the minor T-allele of rs670 with increased risk of gout in Polynesians (Fig. 1; OR = 1.53, P = 4.9 × 10−6). This replicates the finding of Cardona et al. [4] in a small Spanish sample set of 68 cases and 165 controls (OR = 1.99, P = 0.0031). Additionally, there was a very strong trend to association in the European sample set (OR = 1.11, P = 0.055). Thus, these rs670 data are notable in that they represent the strongest replicated evidence to date for association of an apparently non-urate-controlling genetic variant with gout (the TLR4 gene is to our knowledge the only other non-urate-controlling gene having replicated association with gout [24, 25]). There was no evidence for association of rs670 with SU; therefore, these data suggest that this variant causes gout in the presence of HU. The relationship between hypertriglyceridaemia and gout is likely to be complex, and the interpretation of data such as these is complicated by the established observational association between HU/gout and hypertriglyceridaemia. There is some evidence that rs670 is associated with HDL-C, and rs5128 has been consistently associated with triglyceride levels, including in the data sets studied here with the triglyceride-raising G-allele of rs5128 protecting from gout in Polynesians (Table 1 and supplementary Table S3, available at Rheumatology Online). However, because the gout associations observed here were independent of HDL-C and triglyceride levels (Tables 2 and 3), the simplest interpretation is that the functional effects (marked by rs670 and rs5128) on apolipoprotein metabolism are directly causal of gout.
The association of rs5128 (APOC3) with gout was restricted to males in both Europeans and Polynesians (Table 2). As the existing literature is scant in the area of apolipoprotein metabolism and the risk of gout, it is not possible to speculate on a biological basis for this effect. Elucidating the basis for this male-specific effect will be important in understanding both the molecular basis of risk of gout at APOC3 and identifying differences in the pathogenesis of gout between men and women. A recessive model was the most favourable for rs670, and use of this model provided significant evidence for association with gout in Europeans using all controls and HU controls (OR = 1.62, P = 0.001 and OR = 1.66, P = 0.040, respectively).
The rs670 variant maps to the -75 nucleotide position in the promoter region of the APOA1 gene [26]. In the published literature, whether or not this variant influences APOA1 expression is unclear, with two studies yielding conflicting results [27, 28]. However the Genotype-Tissue Expression database (www.gtexportal.org; ver. 6) associates the gout risk T-allele of rs670 with increased expression of APOA1 in heart left-ventricle (P = 3.3 × 10−16) but in no other tissues. Thus, the association of rs670 with gout may be mediated by an influence on the expression of APOA1. There is evidence that APOA1 is involved in gout inflammatory pathways. APOA1 inhibits IL-1β production [29], which is a key factor for monocyte recruitment in gouty inflammation [30]. Moreover, the APOA1 mimetic peptide 4F can inhibit proinflammatory gene expression by altering the assembly of toll-like receptor-ligand complexes in cell membranes [31]. Finally, an increased level of MSU crystal-bound APOA1 has been reported in acute gout [32]. Chiang et al. [32] speculated that APOA1 could be involved in the resolution of acute gout attacks, whereas our genetic association data are consistent with an additional role for APOA1 in the initiation of gout. As far as we are aware, APOA1 has not been directly implicated in VLDL metabolism, although evidence exists that increased levels of triglyceride-enriched VLDL particles have a positive association with HDL-APOA1 catabolism (ref. [33] and citations therein).
We detected stronger association of rs670 (APOA1) in Polynesians than in Europeans. Our evidence suggests that this effect is independent of control of urate, although it will be necessary to retest association of these variants with urate levels in a larger Polynesian sample set. If the replicated association of rs670 with gout in Polynesians was due to contribution to the formation and accumulation of MSU crystals and/or inflammatory processes in gout, then this would indicate a pathogenic process, outside the inherent HU present in Polynesians [34], contributing to the increased prevalence of gout in NZ Polynesians (Māori and Pacific) [35]. It is possible that the pathogenic pathway is related to the presence of Tg-rich VLDL1 particles in Māori and Pacific people with gout [3].
The minor G-allele of rs5128 decreased the risk of gout by nearly 15% in Polynesian cases compared with normouricaemic controls (Fig. 1B) and by nearly 25% compared with HU controls (Fig. 2B). The G-allele is associated with increased expression of APOC3 in a range of non-immune tissues (www.gtexportal.org), meaning that the rs5128-associated effect on gout is likely to be mediated by an influence on the expression of APOC3. The association is genetically independent of rs670 (r2 between the two SNPs is 0.08 in Polynesians), and there was no epistasis apparent between the two variants. There was also evidence for association with gout in Europeans after adjustment by serum triglyceride and HDL-C levels, also with a protective effect of the minor allele. The lower frequency of the G-allele of rs5128 in gout cases compared with in non-gout controls (Table 1) is consistent in direction of association with the observation of Cardona et al. [4], in which they reported non-significant association (OR = 0.75, P = 0.42). The association with rs5128 in our study was independent of serum triglycerides and HDL-C. As is the case for rs670, a possible role for rs5128 in regulation of urate levels will need to be evaluated in a larger sample set, especially given the established association of rs5128 with triglyceride levels (supplementary Table S3, available at Rheumatology Online, and refs [11–14]) and the report that, in Europeans at least, there is evidence from a Mendelian randomization study that increased triglycerides are causal of increased serum urate [36]. However, we note that the triglyceride-raising G-allele of rs5128 is protective against gout.
APOC3 is a non-competitive inhibitor of lipoprotein lipase and thus reduces the lipolysis of Tg-rich lipoproteins, VLDL and chylomicron [37]. Animal studies show that upregulation of APOC3 can induce hypertriglyceridaemia due to delayed clearance of VLDL particles [38], and downregulation can ameliorate the hypertriglyceridaemia [39]. This delayed hydrolysis of VLDL can increase the life span of these particles in the circulation and can result in formation of Tg-rich VLDL particles (VLDL1), which are present in Polynesian gout cases but not in European gout cases in which over-production of VLDL has been observed [3]. The high frequency of the G-allele of rs5128 in Polynesians (>4-fold higher than in Europeans) might ultimately result in lower activity of lipoprotein lipase and thus promote the presence of Tg-rich VLDL1 particles in Polynesian gout cases. It is also possible that rs5128 could influence the inflammatory response; for example, APOC3 and APOC3-rich VLDL can each induce expression of adhesion molecules in endothelial cells and enhance binding and activation of monocytes [40].
Our findings confirm association of APOA1 with gout and are the first to associate APOC3 with gout, although the association in Polynesians does require replication. No evidence for association with urate points towards a role for APOA1 and APOC3 in either formation of MSU or, more likely, in the inflammatory response to these crystals. However, it is not possible, until larger association studies are undertaken with SU as outcome, to eliminate the possibility that the variants studied here are also associated with urate levels. Nevertheless, these data strongly support the value of further genetic, observational and clinical studies of gout in connection with the pathways involving the apolipoprotein A1-C3-A4 locus; such research may reveal dyslipidaemia-focused approaches that could be applied to the management of gout.
Supplementary Material
Acknowledgements
The authors would like to thank Jill Drake, Roddi Laurence, Christopher Franklin, Meaghan House and Gabrielle Sexton for recruitment. We thank Labtests (Auckland) for their assistance in recruitment. Matthew Brown, Linda Bradbury and The Arthritis Genomics Recruitment Initiative in Australia network are acknowledged. The European Crystal Network was formed after the first European Crystal Workshop in Paris, March 2010 (Prof. Frédéric Lioté, Paris and Prof. Alexander So, Lausanne, convenors). The Atherosclerosis Risk in Communities and Framingham Heart study analyses (project #834) were approved by the relevant Database of Genotype and Phenotype (www.ncbi.nim.nih/gov/dbgap) Data Access Committees. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung and Blood Institute contracts N01-HC-55015, N01-HC-55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. The authors thank the staff and participants of the ARIC study for their important contributions. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and National Institutes of Health Roadmap for Medical Research. The Framingham Heart Study and the Framingham SHARe project are conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with Boston University. The Framingham SHARe data used for the analyses described in this manuscript were obtained through Database of Genotype and Phenotype. This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or the National Heart, Lung, and Blood Institute.
Funding: This work was supported by the Health Research Council of New Zealand, Arthritis New Zealand, New Zealand Lottery Health, the University of Otago and Arthritis Australia.
Disclosure statement: The authors have declared no conflicts of interest.
Supplementary data
Supplementary data are available at Rheumatology Online.
References
- 1. Merriman TR, Choi HK, Dalbeth N. The genetic basis of gout. Rheum Dis Clin North Am 2014;40:279–90. [DOI] [PubMed] [Google Scholar]
- 2. Matsubara K, Matsuzawa Y, Jiao S. et al. Relationship between hypertriglyceridemia and uric acid production in primary gout. Metabolism 1989;38:698–701. [DOI] [PubMed] [Google Scholar]
- 3. Rasheed H, Hsu A, Dalbeth N. et al. The relationship of apolipoprotein B and very low density lipoprotein triglyceride with hyperuricemia and gout. Arthritis Res Ther 2014;16:495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cardona F, Tinahones FJ, Collantes E. et al. Contribution of polymorphisms in the apolipoprotein AI-CIII-AIV cluster to hyperlipidaemia in patients with gout. Ann Rheum Dis 2005;64:85–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tinahones F, Collantes E, C-Soriguer FJ. et al. Increased VLDL levels and diminished renal excretion of uric acid in hyperuricaemic–hypertriglyceridaemic patients. Br J Rheumatol 1995;34:920–4. [DOI] [PubMed] [Google Scholar]
- 6. Pennacchio LA, Olivier M, Hubacek JA. et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science 2001;294:169–73. [DOI] [PubMed] [Google Scholar]
- 7. Fielding C, Shore V, Fielding P. A protein cofactor of lecithin: cholesterol acyltransferase. Biochem Biophys Res Comm 1972;46:1493–8. [DOI] [PubMed] [Google Scholar]
- 8. Li Y, Yin R, Zhou Y. et al. Associations of the apolipoprotein A-I gene polymorphism and serum lipid levels in the Guangxi Hei Yi Zhuang and Han populations. Int J Mol Med 2008;21:753–64. [PubMed] [Google Scholar]
- 9. Karathanasis SK. Apolipoprotein multigene family: tandem organization of human apolipoprotein AI, CIII, and AIV genes. Proc Natl Acad Sci U S A 1985;82:6374–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McConathy W, Gesquiere J, Bass H. et al. Inhibition of lipoprotein lipase activity by synthetic peptides of apolipoprotein C-III. J Lipid Res 1992;33:995–1003. [PubMed] [Google Scholar]
- 11. Ordovas JM, Civeira F, Genest J., Jr et al. Restriction fragment length polymorphisms of the apolipoprotein AI, C-III, A-IV gene locus. Relationships with lipids, apolipoproteins, and premature coronary artery disease. Atherosclerosis 1991;87:75–86. [DOI] [PubMed] [Google Scholar]
- 12. Dammerman M, Sandkuijl LA, Halaas JL, Chung W, Breslow JL. An apolipoprotein CIII haplotype protective against hypertriglyceridemia is specified by promoter and 3' untranslated region polymorphisms. Proc Natl Acad Sci U S A 1993;90:4562–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zeng Q, Dammerman M, Takada Y. et al. An apolipoprotein CIII marker associated with hypertriglyceridemia in Caucasians also confers increased risk in a west Japanese population. Hum Genet 1995;95:371–5. [DOI] [PubMed] [Google Scholar]
- 14. Surguchov AP, Page GP, Smith L. et al. Polymorphic markers in apolipoprotein C-III gene flanking regions and hypertriglyceridemia. Arterioscler Thromb Vasc Biol 1996;16:941–7. [DOI] [PubMed] [Google Scholar]
- 15. Lioté F, Merriman T, Nasi S, So A. Workshop report: 4th European crystal network meeting. Arthritis Res Ther 2013;15:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wallace DJ, Klinenberg JR, Morhaim D. et al. Coexistent gout and rheumatoid arthritis. Arthritis Rheum 1979;22:81–6. [DOI] [PubMed] [Google Scholar]
- 17. Jones GT, Bown MJ, Gretarsdottir S. et al. A sequence variant associated with sortilin-1 (SORT1) on 1p13.3 is independently associated with abdominal aortic aneurysm. Hum Mol Genet 2013;22:2941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Phipps-Green AJ, Hollis-Moffatt JE, Dalbeth N. et al. A strong role for the ABCG2 gene in susceptibility to gout in New Zealand Pacific Island and Caucasian, but not Mãori, case and control sample sets. Hum Mol Genet 2010;19:4813–9. [DOI] [PubMed] [Google Scholar]
- 19. Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Willer CJ, Ding J. et al. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic Epidemiol 2010;34:816–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet 2000;67:170–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hollis-Moffatt JE, Phipps-Green AJ, Chapman B. et al. The renal urate transporter SLC17A1 locus: confirmation of association with gout. Arthritis Res Ther 2012;14:R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Akaike H. A new look at statistical model identification. IEEE Trans Automat Contr 1974;19:716–22. [Google Scholar]
- 24. Qing YF, Zhou JG, Zhang QB. et al. Association of TLR4 gene rs2149356 polymorphism with primary gouty arthritis in a case–control study. PLoS One 2013;8:e64845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rasheed H, McKinney C, Stamp LK. et al. The Toll-Like Receptor 4 (TLR4) variant rs2149356 and risk of gout in European and Polynesian sample sets. PLoS One 2016;11:e0147939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pagani F, Sidoli A, Giudici GA. et al. Human apolipoprotein A-I gene promoter polymorphism: association with hyperalphalipoproteinemia. J Lipid Res 1990;31:1371–7. [PubMed] [Google Scholar]
- 27. Jeenah M, Kessling A, Miller N, Humphries S. G to A substitution in the promoter region of the apolipoprotein AI gene is associated with elevated serum apolipoprotein AI and high density lipoprotein cholesterol concentrations. Mol Biol Med 1990;7:233–41. [PubMed] [Google Scholar]
- 28. Kamboh MI, Aston CE, Nestlerode CM. et al. Haplotype analysis of two APOA1/MspI polymorphisms in relation to plasma levels of apo A-I and HDL-cholesterol. Atherosclerosis 1996;127:255–62. [DOI] [PubMed] [Google Scholar]
- 29. Hyka N, Dayer J-M, Modoux C. et al. Apolipoprotein AI inhibits the production of interleukin-1β and tumor necrosis factor-α by blocking contact-mediated activation of monocytes by T lymphocytes. Blood 2001;97:2381–9. [DOI] [PubMed] [Google Scholar]
- 30. Dalbeth N, Haskard D. Mechanisms of inflammation in gout. Rheumatology 2005;44:1090–6. [DOI] [PubMed] [Google Scholar]
- 31. White CR, Smythies LE, Crossman DK. et al. Regulation of pattern recognition receptors by the apolipoprotein A-I mimetic peptide 4F. Arterioscler Thromb Vasc Biol 2012;32:2631–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chiang SL, Ou TT, Wu YJ. et al. Increased level of MSU crystal-bound protein apolipoprotein A-I in acute gouty arthritis. Scand J Rheumatol 2014;43:498–502. [DOI] [PubMed] [Google Scholar]
- 33. Vergès B, Adiels M, Boren J. et al. Interrelationships between the kinetics of VLDL subspecies and HDL catabolism in abdominal obesity: a multicenter tracer kinetic study. J Clin Endocinol Metab 2014;99:4281–90. [DOI] [PubMed] [Google Scholar]
- 34. Merriman TR. Population heterogeneity in the genetic control of serum urate. Semin Nephrol 2011;31:420–5. [DOI] [PubMed] [Google Scholar]
- 35. Winnard D, Wright C, Taylor WJ. et al. National prevalence of gout derived from administrative health data in Aotearoa New Zealand. Rheumatology 2012;51:901–9. [DOI] [PubMed] [Google Scholar]
- 36. Rasheed H, Hughes K, Flynn TJ, Merriman TR. Mendelian randomization provides no evidence for a causal role of serum urate in increasing serum triglyceride levels. Circ Cardiovasc Genet 2014;7:830–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zheng C. Updates on apolipoprotein CIII: fulfilling promise as a therapeutic target for hypertriglyceridemia and cardiovascular disease. Curr Opin Lipidol 2014;25:35–9. [DOI] [PubMed] [Google Scholar]
- 38. Waterworth DM, Ribalta J, Nicaud V. et al. ApoCIII gene variants modulate postprandial response to both glucose and fat tolerance tests. Circulation 1999;99:1872–7. [DOI] [PubMed] [Google Scholar]
- 39. Maeda N, Li H, Lee D. et al. Targeted disruption of the apolipoprotein C-III gene in mice results in hypotriglyceridemia and protection from postprandial hypertriglyceridemia. J Biol Chem 1994;269:23610–6. [PubMed] [Google Scholar]
- 40. Kawakami A, Aikawa M, Alcaide P. et al. Apolipoprotein CIII induces expression of vascular cell adhesion molecule-1 in vascular endothelial cells and increases adhesion of monocytic cells. Circulation 2006;114:681–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.