Abstract
Pre-mRNA splicing is an essential step in gene expression in most eukaryote genes. Here we present the feasibility of a genetically encoded luciferase reporter to monitor the pre-mRNA splicing process in living cells and animals. We showed that the splicing activity change induced by isoginkgetin could be readily visualized in vitro both in a dose and time dependent manner. Moreover, the pre-mRNA splicing process could be also obviously detected in mice by bioluminescence imaging and confirmed by RT-PCR. Our work provided a reporter system that allows high-throughput screening of chemical libraries to identify potential compounds leading to aberrant patterns of splicing.
OCIS codes: (170.0170) Medical optics and biotechnology; (170.3880) Medical and biological imaging; (170.6280) Spectroscopy, fluorescence and luminescence
1. Introduction
In most eukaryote genes, the protein-coding regions (exons) are interrupted by noncoding sequences (introns), which must be removed in the nucleus by splicing from pre-mRNA before nuclear export and translation into protein. Pre-mRNA splicing is an essential step in gene expression that is mediated by a highly dynamic multi-small nuclear ribonucleoprotein (multi-snRNP) called spliceosome [1]. The spliceosome complex is composed of four snRNP particles (U1, U2, U4/U6 and U5) and more than 100 snRNP or non-snRNP associated proteins that are necessary to complex assembly and splicing catalysis [2]. Splicing depends on the recognition of consensus splice sites at the boundaries between exons and introns, and occurs in two sequential chemical reactions. First, U1 snRNP interacts with 5′ splice site and U2 snRNP recognizes the branch point adenosine, forming a lariat complex. A subsequent ATP-dependent step stabilizes the branch point/U2 snRNP interaction, leading to the integration of the U4/U5/U6 tri-snRNP complex [3, 4]. The multiple conformational rearrangements of this complex are followed by the production of catalytically active spliceosome, wherein U1 and U4 snRNP are released. Finally, after the cleavage of the 5′ and 3′ splice site and the release of lariat products, exon ligation takes place [5, 6].
The gene expression at the mRNA levels is typically monitored in vitro by biochemical approaches including Northern blotting, reverse-transcriptase polymerase chain reaction (RT-PCR) analysis, RNA protection assays [7], exon junction microarrays [8] and autoradiography [9]. However, these methods generally require the lysis of cells and thus cannot fully reflect the characteristics of mRNA in living cells, which is critical to assess the mRNA processing pattern in vivo. Several radionuclides or reporter protein based probes, such as radiolabeled antisense oligonucleotides (RASONs) [10, 11] or ribozymes [12, 13], have been extensively used to evaluate the endogenous transcribed mRNA. RASONs are designed to be complementary to a small segment of interest mRNA. Nevertheless, RASONs are fundamentally limited by their nonspecific binding to serum proteins, poor cell membrane permeability and high background activity [14]. The ribozyme-based RNA reporter refers to the employment of the reporter gene systems and antisense RNA binding to achieve signal amplification and targeting specificity, respectively. In this context, the designed ribozyme would splice the reporter onto the target RNA in-frame with the guidance of an attached antisense sequence, leading to a fusion RNA containing the reporter, whose activity can be detected by sensitive cooled charge-coupled device (CCD) cameras [15, 16]. However, the splicing efficiency primarily dependents on the high specificity in the binding domains between ribozymes and target mRNA. Moreover, the exon context surrounding the ribozyme sequence would greatly influence its folding, and thus hinder its catalytic activity [17]. Therefore, development of promising imaging probes to detect and quantify mRNA splicing events in living subjects remains a challenge.
In the present study, we described a genetically encoded luciferase reporter for noninvasive monitoring the pre-mRNA splicing processes in living cells and animals. We show that this splicing reporter is able to provide quantitative and real-time measurements of the extent of splicing activity in response to extracellular stimuli. Our data may open a novel avenue for imaging of pre-mRNA splicing in vivo and the high-throughout screening of modulators of the splicing machinery with this reporter system, which would shed new insights into the treatment of human diseases caused by splicing defects.
2. Materials and methods
2.1. Luciferase reporter construction
To get an intron-containing luciferase reporter (Luc-intron), a 68-nucleotide chimeric intron was firstly inserted at nucleotide position 834 of a firefly luciferase gene pGL3-control (Promega) by mutagenesis. Then the intron-containing luciferase was amplified and inserted into the multiple cloning sites (MCS) of a lentivirus vector pHBLV-CMVIE-ZsGreen-Puro (Hanbio Inc. China). An intronless luciferase gene was also inserted into the MCS of the same lentivirus vector as a control, named Luc-control. Both Luc-intron and Luc-control reporter were transcribed from a cytomegalovirus (CMV) promoter. The sequence of the chimeric intron is as follow: 5′-GTA AGT ATC AAG AAT TCA GAA GGA ATT ACT AGT TCT TAC TGA TGC TCA CTT TGC CTT TCT CTC CAC AG-3′.
2.2. Cell culture and generation of stable cell line
Human non-small-cell lung cancer A549 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, HyClone), supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% Penicillin-Streptomycin Solution (HyClone) at 37 °C in an incubator with 5% CO2. To obtain the stable cell lines, HEK293 cells were transfected with the two luciferase constructs using Lipofectamine 2000 (Invitrogen) and virus supernatants were harvested 48 h later. Then the virus was infected with A549 cells and the integration was selected using puromycin for 2-3 weeks. The final stable cell lines were named A549-luc-intron cells and A549-luc-control cells, respectively. The luciferase assays were used to validate the two stable cell lines.
2.3. Luciferase assay
To determine the luciferase activity of cells, cells were plated in 24-well plates, and cultured for 24 h in an incubator with 5% CO2. Then the cells were lysed with cell lysis buffer (CLB, TransGen Biotech, China) and performed luciferase assays using Glomax-20/20 Luminometer (Promega). For the drug treatment experiment, isoginkgetin (Chembest, China) was dissolved in DMSO. Then the A549-luc-intron cells or A549-luc-control cells were treated with different concentrations of isoginkgetin as indicated. 24 h later, the cells were harvested for luciferase assay using a Promega luciferase assay system as described above.
2.4. Cell viability assay
To determine the cell viability treated by isoginkgetin, Cell Counting Kit-8 (CCK-8, Dojindo Laboratories, Japan) was performed. Cells were treated with different concentrations of isoginkgetin for different times. In one experiment, cells were treated with isoginkgetin (10 or 40 μM) for different times (6, 18, 24, 48 h). In the other experiment, cells were treated with isoginkgetin (40 μM) for 24 h, and then cultured for different times (2, 6, 8, 24 h) with the new medium. At the end of each experiment, 50 μl CCK-8 was added in each well for 3 h, and then the absorbance at 450 nm was measured by GloMax Discover and Explorer Detection Systems (Promega).
2.5. Reverse transcription PCR (RT-PCR)
Total RNA was isolated with Trizol (Invitrogen) according to the manufacturer’s protocols. The RT-PCR was preformed using a RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher) according to the manufacturer’s instructions. Specifically, 1 μg of total RNA was incubated with oligo (dT) primer at 65 °C for 5 min, and then added with components containing M-MLV, dNTP mix, RNase inhibitor and reaction buffer, followed by 60 min at 42 °C. Finally the reaction was terminated by heating at 70 °C for 5 min. The product of the first cDNA was then used for PCR in 50 μl total volume. All primer sequences are available upon request.
2.6. In vitro bioluminescence imaging assay
To determine the linear relationship between luciferase activity and cell numbers, a dilution series of A549-luc-control cells or A549-luc-intron cells were seeded in a 24-well plate. Then D-luciferin (150 μg/ml) was added to the cells and in vitro bioluminescence imaging assay was performed using a Xenogen Lumina II system. The luminescence signal for each well was determined and showed as average values.
2.7. In vivo bioluminescence imaging
All animal experiments were performed according to the Guild for the Care and Use of Laboratory Animals approved by Xidian University. A549-luc-intron cells (1 × 108) were collected in PBS and injected into the right flank of nude mice (n = 5) to allow the tumor growth. When the tumor volume is approximately 100 mm3, the xenograft mouse model was injected intraperitoneally (i.p.) with isoginkgetin of 15 mg/kg. For the bioluminescence imaging of the nude mice, in vivo Imaging System Lumina II (Xenogen) was used. At 0 h and 24 h after treatment, the mice were anaesthetized with 2% isoflurane and injected i.p. 200 μl of D-luciferin (150 mg/kg) (TransGen Biotech, China) for bioluminescence imaging. The regions of interest (ROI) of constant area were expressed as p/sec/cm2/sr using the Living Imaging Software 4.1 (Xenogen).
2.8. Statistical analysis
The results are showed as the means ± SD and were analyzed by unpaired Student’s t-test or two-way ANOVA. P values < 0.05 were considered statistically significant.
3. Results and discussion
3.1. Validation and characterization of splicing reporter
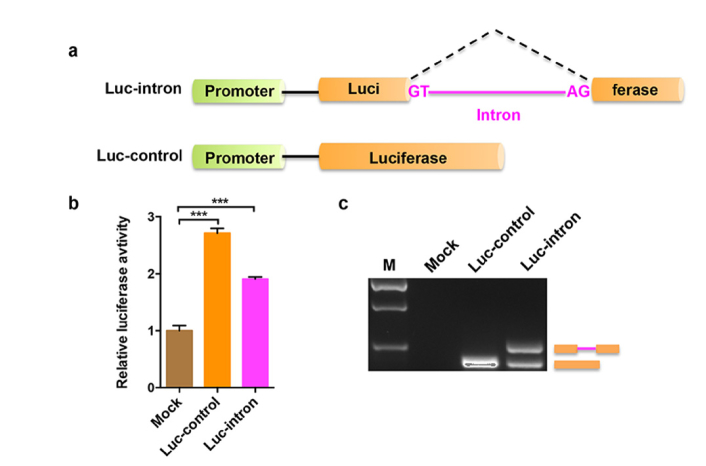
In order to monitor the pre-mRNA splicing, we designed a reporter Luc-intron in which the open reading frame (ORF) of firefly luciferase gene was interrupted by a chimeric intron (Fig. 1(a)). According to the conserved sequence principles of pre-mRNA splicing, the 5 ' splice site of GT and 3′ splice site of AG are at the both ends of the chemic intron for the conservative camp. The intron was inserted at nucleotide position 834 of a firefly luciferase gene, which is controlled by a CMV promoter. We also created a control reporter (Luc-control) identical to the splicing reporter except that there is no interruption by chimeric intron. To characterize the splicing reporters, Luc-intron or Luc-control reporters were co-transfected with pRL-TK renilla luciferase control plasmid into A549 cells, respectively. Then a dual luciferase reporter assay was performed to determine the luciferase expression (Fig. 1(b)). The results showed that the luciferase activity was restored after the intron was spliced out, and the luciferase signal from Luc-intron reporter was slightly lower than that from Luc-control.
Fig. 1.
Characterization of the splicing reporter. (a) Schematic diagram of the intron-containing (Luc-intron) and intronless (Luc-control) luciferase reporters. Both luciferase genes are under the control of a CMV promoter. GT and AG represent the 5′ and 3′ splice site of intron, respectively. (b) The A549 cells were transfected with the Luc-intron or Luc-control plasmid, together with an internal control plasmid pRL-TK that expresses renilla luciferase. Mock indicates that the cells were added only transfection reagent. Dual luciferase reporter assay was performed 24 h after transfection. Error bars represent the standard deviations for three independent experiments. ***p < 0.001 (c) RT-PCR analysis of total RNA isolated from the cells treated by the same method as described in (b). M indicates DNA marker.
To further validate the splicing reporters, equal amounts of Luc-intron or Luc-control construct was transfected into cells and RT-PCR was performed to analyze the total RNA from each group. Consistent with the luciferase assay, the results confirmed that transcripts from Luc-intron were spliced, and gene expression of the spliced Luc-intron is a little lower than that of Luc-control (Fig. 1(c)). Therefore, the splicing reporter is successfully constructed and should be able to detect changes in mRNA splicing.
3.2. Correlation between bioluminescence and cell number
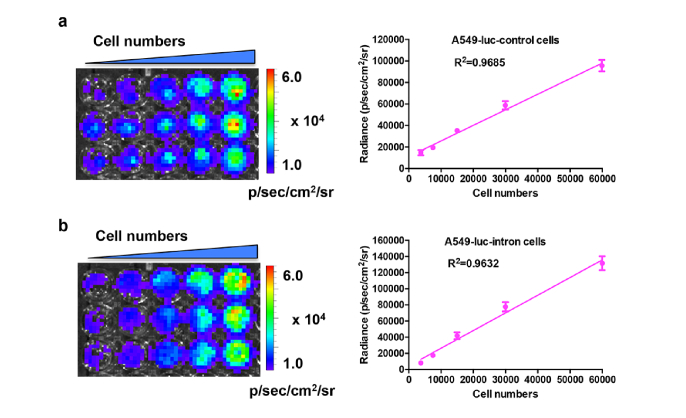
Stable cell lines (A549-luc-intron and A549-luc-control) expressing each reporter were generated using a lentivirus system. To assess the luciferase sensitivity of these two cell lines, different numbers of cells were diluted and in vitro bioluminescence imaging assay was performed. As shown in Fig. 2(a), the bioluminescence signal increased according to the increase of cells numbers. A high correlation (R2 = 0.9685) was observed between bioluminescence intensity and cell numbers. For the other cell line, as the numbers of A549-luc-intron cells was increased, the bioluminescence signal was also accordingly increased (Fig. 2(b)). The analysis of bioluminescence intensities in the region of interest (ROI) indicated a high correlation (R2 = 0.9632) between intensity and cell number. The results suggested that these two cell lines exhibited great luciferase sensitivity and can be used in further experiment.
Fig. 2.
Correlation between bioluminescence signal and cell numbers in cells. The A549-luc-control cells (a) or A549-luc-intron cells (b) were seeded with different cell numbers in a 24-well plate. Then the in vitro bioluminescence imaging was performed following the addition of D-luciferin (150 μg/ml) substrate to the cells. The correlation curves were plotted for each cell line and the square of the correlation coefficients were 0.9685 and 0.9632, respectively.
3.3. Measuring splicing activity in a dose dependent manner
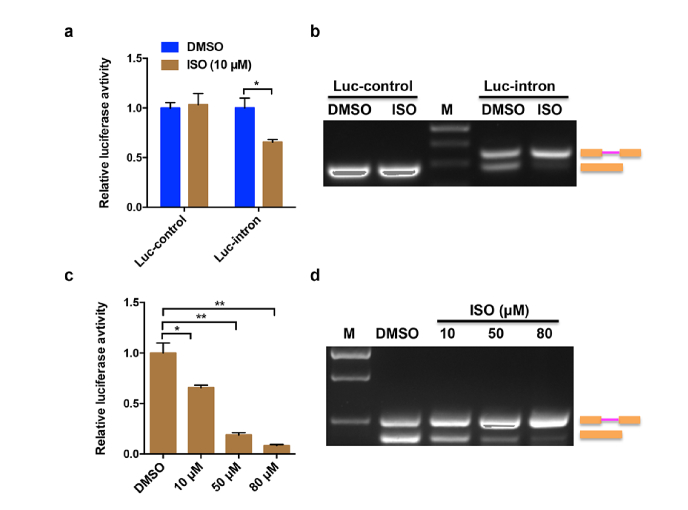
To verify the feasibility of Luc-intron reporter for detecting splicing change upon extrinsic intervention, a naturally occurring agent, isoginkgetin (ISO), is utilized to treat the A549-luc-intron or A549-luc-control cells. ISO has been found to be a general cell-permeable inhibitor of pre-mRNA splicing by blocking spliceosome-mediated splicing process at the pre-spliceosome/A complex stage [18]. As expected, ISO decreased significantly the luciferase activity of Luc-intron reporter but has no effect on Luc-control reporter (Fig. 3(a)). To verify that the observed luciferase activity changes were due to the inhibition of pre-mRNA splicing, RT-PCR assays were performed to analyze the total RNA extracted from DMSO and ISO treated A549-luc-intron or A549-luc-control cells, respectively. As shown in Fig. 3(b), comparing with the DMSO group, the exposure to ISO resulted in the increased accumulation of unspliced pre-mRNA in A549-luc-intron cells but not in A549-luc-control cells.
Fig. 3.
Measuring of splicing activity at different doses of ISO treatment. (a) The A549-luc-control cells or A549-luc-intron cells were treated with ISO (10 μM) or DMSO for 24 h. Then the luciferase activities were measured. (b) RT-PCR analysis of total RNA isolated from the cells treated with the same method described in (a). Sizes of unspliced and spliced products were indicated. (c) The A549-luc-intron cells were treated with DMSO or different doses of ISO (10, 50, 80 μM) for 24 h. Then the luciferase activities were measured. (d) RT-PCR analysis of total RNA isolated from the cells treated with the different doses of ISO used in (c). Error bars represent the standard deviations for three independent experiments. *p < 0.05, **p < 0.01.
To further test whether the splicing reporter is dose dependent, the A549-luc-intron cells were treated with different concentrations of ISO (10 μM, 50 μM, 80 μM). The results showed that the luciferase activities were decreased with the increased concentrations of ISO (Fig. 3(c)). When the concentration of ISO reached 80 μM, the decrease was the most significant. Consistent with this, RT-PCR results confirmed that the effect of ISO treatment was also dose-dependent, leading to an increased shift from spliced mRNA to unspliced pre-mRNA (Fig. 3(d)).
3.4. In vitro real-time monitor the splicing activity
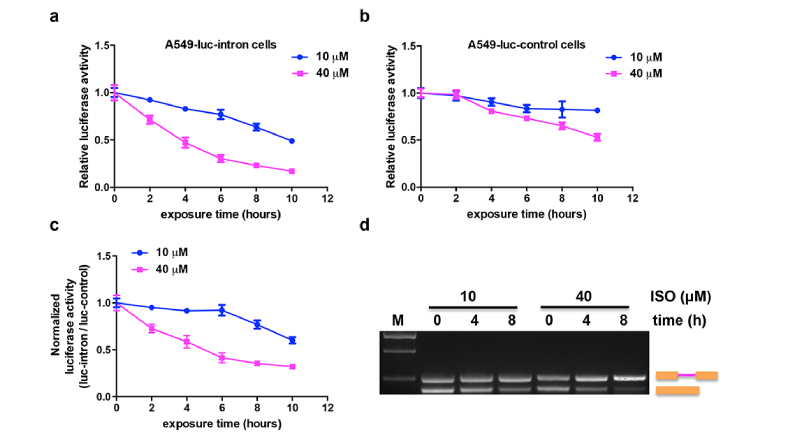
To further explore the efficacy of Luc-intron reporter in monitoring splicing change in real-time, A549-luc-intron cells were treated with different concentrations of ISO (10 μM and 40 μM) for different time points. As shown in Fig. 4(a), the luciferase activity was consistently decreased with the increase of exposure time to ISO. And the 40 μM ISO resulted in much more decrease of luciferase signal than 10 μM did. We also did the same experiment with A549-luc-control cells. Unexpectedly, we found that both concentrations of ISO led to a slight or moderate decrease of the luciferase activity over the time course (Fig. 4(b)). Specifically, the luciferase activity from 10 μM ISO treated group was decreased about 18% at 10 h time point, and the decrease was not statistically significant compared to that at 0 h time point. In comparison, the 40 μM group induced an approximately 50% decrease of luciferase activity at the end time point of treatment. The reduction of luciferase activity in A549-luc-control cells may be attributed to the fact that ISO is also an anti-tumor agent and could inhibit cell growth [19].
Fig. 4.
In vitro real time monitor splicing activity induced by ISO. Luciferase activity versus time for A549-luc-intron cells (a) or A549-luc-control cells (b) treated with 10 μM or 40 μM of ISO. (c) The normalization of luciferase activity from A549-luc-intron cells to that from A549-luc-control cells treated with 10 μM or 40 μM ISO at the same period of time. (d) RT-PCR analysis of total RNA isolated from A549-luc-intron cells treated with 10 or 40 μM ISO for 0, 4 or 8 h.
To minimize the effect of cell growth arrest induced by ISO, we normalized the luciferase activity change in A549-luc-intron cells with that in A549-luc-control cells. A ratiometric analysis (Luc-intron/Luc-control) revealed a less reduction in luciferase activity than that in A549-luc-intron cells (Fig. 4(c)). Without normalization with Luc-control, the lowest reduction in luciferase activity at 10 h time point induced by 10 μM ISO was 48.9% of that at 0 h time point, but after normalization, the reduction level was relieved (60% of 0 h). Similar effect has also been observed in the 40 μM ISO treated cells (16.9% before normalization v.s. 32% after normalization). The normalization curve reflected that the observed decrease of luciferase activity occurred was due to the inhibition of splicing but not cell growth arrest induced by ISO. To further confirm this result, RT-PCR was performed and demonstrated that treatment with 40 μM ISO resulted in a greater increase of pre-mRNA accumulation with 8 h than did treatment with 10 μM ISO (Fig. 4(d)).
3.5. Monitor the inhibition of cell proliferation and splicing by ISO in a reversible fashion
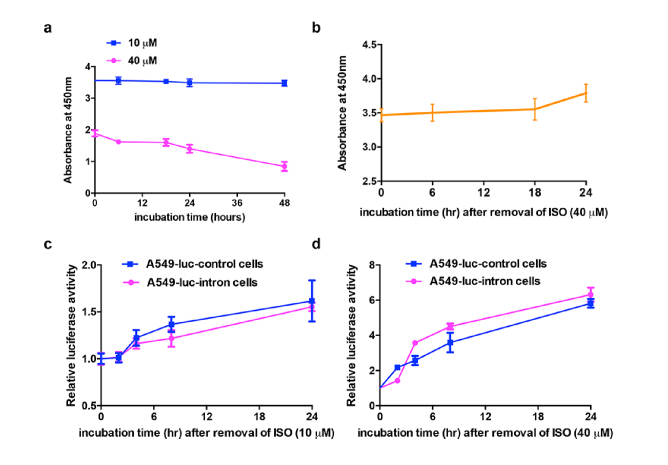
As a general inhibitor of pre-mRNA splicing is frequently accompanied with anti-proliferative activity [20], we verified that whether ISO could induce cell growth arrest. Therefore, we determined the cell viability in A549-luc-control cells treated by different concentrations of ISO. As shown in Fig. 5(a), the absorbance values had no obvious change with the increasing incubation time of 10 μM ISO, whereas the values were decreased in cells treated with 40 μM ISO for the same period of time, suggesting that ISO exerted an anti-proliferative effect on cell growth when it reached a certain concentration. These results were consistent with our previous findings that luciferase activity in A549-luc-control cells treated by either 10 μM or 40 μM ISO was decreased to a certain extent.
Fig. 5.
Monitor the inhibition of cell proliferation and splicing by ISO in a reversible fashion. (a) CKK-8 assay for the A549 cells treated with 10 μM or 40 μM of ISO over time. (b) The A549 cells were treated with 40 μM of ISO for 24 h. Then the ISO was removed and the fresh medium was added for CKK-8 assay at the indicated time. (c, d) The luciferase activity from A549-luc-intron cells or A549-luc-control cells exposed to fresh medium after 24 h treatment with (c) 10 μM or (d) 40 μM of ISO.
Then we attempted to figure out whether the inhibitory of cell growth by ISO (40 μM) is reversible or not. To this end, the ISO was removed after exposure to A549-luc-control cells for 24 h, and then the cells were cultured with fresh medium for a period of time. The results showed that the cell growth was restored with the increasing time in new medium (Fig. 5(b)). Consistent with this, the luciferase activity from A549-luc-control and A549-luc-intron cells were also recovered after removal of 10 μM (Fig. 5(c)) or 40 μM (Fig. 5(d)) ISO that exposure for 24 h. Taken together, removal of ISO restored both the cellular proliferation and luciferase activity, suggesting that ISO is a reversible inhibitor of pre-mRNA splicing.
3.6. In vivo imaging of splicing by ISO
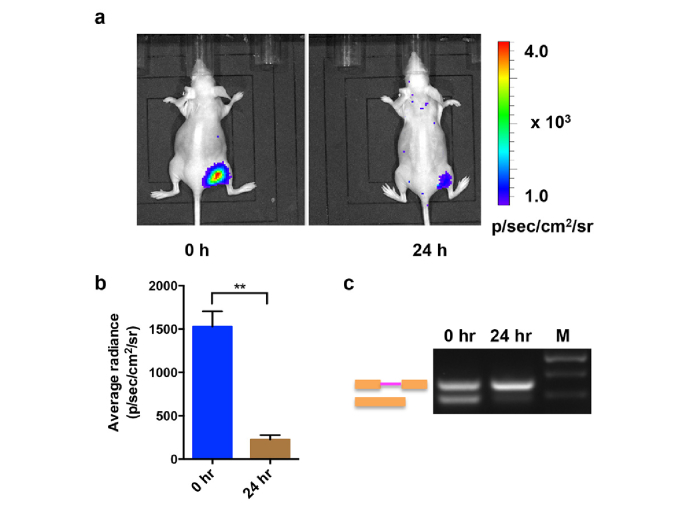
To demonstrate the feasibility of the splicing reporter in living subjects, we established a xenograft mice model by implanting A549-luc-intron cells in the right flank of nude mice (n = 5). The mice were imaged before and after ISO treatment by intraperitoneal injection of 15 mg/kg ISO. As shown in Fig. 6(a), a significant decrease in the bioluminescence signal was observed in the tumors after ISO treatment. A quantitative ROI analysis revealed a 6.8-fold reduction in the luminescence intensity after the injection of ISO for 24 h (Fig. 6(b)). To confirm that reduced the bioluminescent signal was due to the inhibition of pre-mRNA splicing, RT-PCR was performed to analyze the total RNA isolated from tumors of the xenograft mice receiving ISO treatment at 0 h and 24 h time point. The results demonstrated ISO treatment led to a greater increase in pre-mRNA accumulation and a decrease in spliced mRNA accordingly at 24 h than that at 0 h (Fig. 6(c)). Thus, these results indicate that the present splicing reporter is applicable to in vivo imaging of pre-mRNA splicing activity upon stimulation with exogenous chemical compounds.
Fig. 6.
In vivo imaging of splicing by ISO. (a) The xenograft mice were intraperitoneally injected with 15 mg/kg ISO. The bioluminescence imaging was performed at the 0 h time point and 24 h time point after ISO treatment. A representative image was shown. (b) The quantification of the bioluminescence signal from the region of interest in mice treated by ISO at 0 or 24 h as described in (a). (c) RT-PCR analysis of total RNA isolated from tumors of the xenograft mice receiving ISO treatment at 0 h and 24 h time point.
Gene expression in eukaryotes is precisely controlled both at the transcriptional and post-transcriptional levels by such mechanisms as DNA methylation, mRNA processing and mRNA translation. Pre-mRNA splicing is a fundamental process of gene regulation at the post-transcriptional level. An image approach that could visualize these processes would offer more reliable and accurate information on the expression levels of the gene of interest. In this study, we have demonstrated that the pre-mRNA splicing processes can be monitored with the Luc-intron genetically encoded luciferase reporter in living cells and living animals. Although we did not image an endogenous mRNA, our work represents an important proof-of-concept toward imaging gene expression especially gene splicing in living subjects.
Numerous reporter gene systems were developed to visualize ribozyme- or spliceosome-mediated RNA splicing in mammalian cells and live animals. Sumitaka et al. [21] characterized a β-lactamase-based reporter in which the self-splicing Tetrahymena ribozyme was inserted into the coding region of the mRNA of a bacterial enzyme, TEM-1 β-lactamase (Bla). In this system, ribozyme self-splicing leads to uninterrupted Bla mRNA, which is translated into the reporter enzyme Bla. The splicing activity of ribozyme is signaled by the ratio of blue to green fluorescence in cells treated by a CCF2/AM substrate. In the following studies, Sumitaka et al. expanded this concept and developed a split β-lactamase reporter system to quantify the trans-splicing activity of Tetrahymena ribozyme in single mammalian cells by flow cytometry and fluorescence microscopy [22]. This split β-lactamase reporter was also extended to image a mutant p53 mRNA both in single cells and noninvasively in living mice [15, 16]. Compared to the ribozyme-mediated imaging of RNA splicing in vivo, Nasim et al. [23] devised a double reporter in which splicing produces luciferase signal whereas unspliced and spliced RNA produced β-galactosidase activity, providing a convenient assay for detecting splicing efficiency. Bhaumik et al. [24] designed a split luciferase reporter system based on the mammalian spliceosome to monitor pre-mRNA splicing in vivo. This spliceosome-mediated RNA trans-splicing (SMaRT) technique is relied on the employment of engineered pre-trans-splicing molecules (PTM), which is complementary to intronic sequences in the target mRNA. However, the visualization of pre-mRNA splicing in living subjects by RNA trans-splicing is limited to the luciferase sequence of the reporter system. Thus, the same group provided a generalizable strategy for imaging levels of any pre-mRNA by excluding the ATG start codon in the PTM that encoding a full-length luciferase gene [25]. Moreover, the coding domain of this PTM could be easily exchanged to encode a reporter protein for other imaging modality such as PET or MRI, which is more robust and clinically relevant.
In comparison with the aforementioned reporter systems, our splicing reporter is entirely based on luciferase expression as readout. By using lenti-virus system to establish stable cell lines expressing reporter at identical genetic loci, we excluded the possible effects at the transcriptional level owing to different genomic contexts. Given the fundamental role of splicing in gene expression, exposure to splicing inhibitors, ISO for example, would affect the stability of the reporter or steps in its expression other than splicing, and cause toxicity to cells. To overcome this problem, we also constructed a control luciferase reporter Luc-control that was identical to the splicing reporter except for its lack of an intron. By comparing the luciferase activities of these two reporters from cells treated by ISO at different doses or different time points, we normalized the possible effects on luciferase signal due to the toxicity of ISO in cells. We found that the decrease of luciferase signal from ISO-treated A549-luc-intron cells was alleviated after normalization with the luciferase signal from A549-luc-control cells treated by the same means. Therefore, the combination of these two reporter systems could monitor merely the splicing pattern change excluding cell toxicity induced by extracellular stimuli.
4. Conclusion
Our study offers a valuable reporter system to noninvasively monitor the pre-mRNA splicing process in living subjects. In principle, this strategy can be potentially applied in the high-throughput screening of modulators of splicing and splicing-dependent processes in cells. This would be an important advancement in the field of RNA imaging and splicing related drug development. Further studies are ongoing to optimize the design of our reporter system towards this direction.
Funding
National Natural Science Foundation of China (No. 81571721, 81772010), and Natural Science Basis Research Plan in Shaanxi Province of China (Program No. 2016JM8016).
Disclosures
No potential conflicts of interest were disclosed.
References and links
- 1.Staley J. P., Guthrie C., “Mechanical devices of the spliceosome: motors, clocks, springs, and things,” Cell 92(3), 315–326 (1998). 10.1016/S0092-8674(00)80925-3 [DOI] [PubMed] [Google Scholar]
- 2.Wahl M. C., Will C. L., Lührmann R., “The spliceosome: design principles of a dynamic RNP machine,” Cell 136(4), 701–718 (2009). 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 3.Black D. L., Chabot B., Steitz J. A., “U2 as well as U1 small nuclear ribonucleoproteins are involved in premessenger RNA splicing,” Cell 42(3), 737–750 (1985). 10.1016/0092-8674(85)90270-3 [DOI] [PubMed] [Google Scholar]
- 4.Konarska M. M., Sharp P. A., “Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes,” Cell 49(6), 763–774 (1987). 10.1016/0092-8674(87)90614-3 [DOI] [PubMed] [Google Scholar]
- 5.Padgett R. A., Konarska M. M., Grabowski P. J., Hardy S. F., Sharp P. A., “Lariat RNA’s as intermediates and products in the splicing of messenger RNA precursors,” Science 225(4665), 898–903 (1984). 10.1126/science.6206566 [DOI] [PubMed] [Google Scholar]
- 6.Ruskin B., Krainer A. R., Maniatis T., Green M. R., “Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro,” Cell 38(1), 317–331 (1984). 10.1016/0092-8674(84)90553-1 [DOI] [PubMed] [Google Scholar]
- 7.Blackwell T. K., Walker A. K., “Transcription mechanisms,” WormBook 2006, 1–16 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson J. M., Castle J., Garrett-Engele P., Kan Z., Loerch P. M., Armour C. D., Santos R., Schadt E. E., Stoughton R., Shoemaker D. D., “Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays,” Science 302(5653), 2141–2144 (2003). 10.1126/science.1090100 [DOI] [PubMed] [Google Scholar]
- 9.Krainer A. R., Maniatis T., Ruskin B., Green M. R., “Normal and mutant human beta-globin pre-mRNAs are faithfully and efficiently spliced in vitro,” Cell 36(4), 993–1005 (1984). 10.1016/0092-8674(84)90049-7 [DOI] [PubMed] [Google Scholar]
- 10.Liu M., Wang R. F., Zhang C. L., Yan P., Yu M. M., Di L. J., Liu H. J., Guo F. Q., “Noninvasive imaging of human telomerase reverse transcriptase (hTERT) messenger RNA with 99mTc-radiolabeled antisense probes in malignant tumors,” J. Nucl. Med. 48(12), 2028–2036 (2007). 10.2967/jnumed.107.042622 [DOI] [PubMed] [Google Scholar]
- 11.Qin G., Zhang Y., Cao W., An R., Gao Z., Li G., Xu W., Zhang K., Li S., “Molecular imaging of atherosclerotic plaques with technetium-99m-labelled antisense oligonucleotides,” Eur. J. Nucl. Med. Mol. Imaging 32(1), 6–14 (2005). 10.1007/s00259-004-1700-0 [DOI] [PubMed] [Google Scholar]
- 12.Cech T. R., “Self-splicing of group I introns,” Annu. Rev. Biochem. 59(1), 543–568 (1990). 10.1146/annurev.bi.59.070190.002551 [DOI] [PubMed] [Google Scholar]
- 13.Sullenger B. A., Cech T. R., “Ribozyme-mediated repair of defective mRNA by targeted, trans-splicing,” Nature 371(6498), 619–622 (1994). 10.1038/371619a0 [DOI] [PubMed] [Google Scholar]
- 14.Gauchez A. S., Du Moulinet D’Hardemare A., Lunardi J., Vuillez J. P., Fagret D., “Potential use of radiolabeled antisense oligonucleotides in oncology,” Anticancer Res. 19(6B), 4989–4997 (1999). [PubMed] [Google Scholar]
- 15.So M. K., Gowrishankar G., Hasegawa S., Chung J. K., Rao J., “Imaging target mRNA and siRNA-mediated gene silencing in vivo with ribozyme-based reporters,” ChemBioChem 9(16), 2682–2691 (2008). 10.1002/cbic.200800370 [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa S., Gowrishankar G., Rao J., “Detection of mRNA in mammalian cells with a split ribozyme reporter,” ChemBioChem 7(6), 925–928 (2006). 10.1002/cbic.200600061 [DOI] [PubMed] [Google Scholar]
- 17.Woodson S. A., Cech T. R., “Alternative secondary structures in the 5′ exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA,” Biochemistry 30(8), 2042–2050 (1991). 10.1021/bi00222a006 [DOI] [PubMed] [Google Scholar]
- 18.O’Brien K., Matlin A. J., Lowell A. M., Moore M. J., “The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing,” J. Biol. Chem. 283(48), 33147–33154 (2008). 10.1074/jbc.M805556200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon S. O., Shin S., Lee H. J., Chun H. K., Chung A. S., “Isoginkgetin inhibits tumor cell invasion by regulating phosphatidylinositol 3-kinase/Akt-dependent matrix metalloproteinase-9 expression,” Mol. Cancer Ther. 5(11), 2666–2675 (2006). 10.1158/1535-7163.MCT-06-0321 [DOI] [PubMed] [Google Scholar]
- 20.Salton M., Misteli T., “Small Molecule Modulators of Pre-mRNA Splicing in Cancer Therapy,” Trends Mol. Med. 22(1), 28–37 (2016). 10.1016/j.molmed.2015.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasegawa S., Jackson W. C., Tsien R. Y., Rao J., “Imaging Tetrahymena ribozyme splicing activity in single live mammalian cells,” Proc. Natl. Acad. Sci. U.S.A. 100(25), 14892–14896 (2003). 10.1073/pnas.2036553100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasegawa S., Choi J. W., Rao J., “Single-cell detection of trans-splicing ribozyme in vivo activity,” J. Am. Chem. Soc. 126(23), 7158–7159 (2004). 10.1021/ja049144u [DOI] [PubMed] [Google Scholar]
- 23.Nasim M. T., Eperon I. C., “A double-reporter splicing assay for determining splicing efficiency in mammalian cells,” Nat. Protoc. 1(2), 1022–1028 (2006). 10.1038/nprot.2006.148 [DOI] [PubMed] [Google Scholar]
- 24.Bhaumik S., Walls Z., Puttaraju M., Mitchell L. G., Gambhir S. S., “Molecular imaging of gene expression in living subjects by spliceosome-mediated RNA trans-splicing,” Proc. Natl. Acad. Sci. U.S.A. 101(23), 8693–8698 (2004). 10.1073/pnas.0402772101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walls Z. F., Puttaraju M., Temple G. F., Gambhir S. S., “A generalizable strategy for imaging pre-mRNA levels in living subjects using spliceosome-mediated RNA trans-splicing,” J. Nucl. Med. 49(7), 1146–1154 (2008). 10.2967/jnumed.107.047662 [DOI] [PubMed] [Google Scholar]