Abstract
Gene expression levels change as an individual ages and responds to environmental conditions. With the exception of humans, such patterns have principally been studied under controlled conditions, overlooking the array of developmental and environmental influences that organisms encounter under conditions in which natural selection operates. We used high-throughput RNA sequencing (RNA-Seq) of whole blood to assess the relative impacts of social status, age, disease, and sex on gene expression levels in a natural population of gray wolves (Canis lupus). Our findings suggest that age is broadly associated with gene expression levels, whereas other examined factors have minimal effects on gene expression patterns. Further, our results reveal evolutionarily conserved signatures of senescence, such as immunosenescence and metabolic aging, between wolves and humans despite major differences in life history and environment. The effects of aging on gene expression levels in wolves exhibit conservation with humans, but the more rapid expression differences observed in aging wolves is evolutionarily appropriate given the species’ high level of extrinsic mortality due to intraspecific aggression. Some expression changes that occur with age can facilitate physical age-related changes that may enhance fitness in older wolves. However, the expression of these ancestral patterns of aging in descendant modern dogs living in highly modified domestic environments may be maladaptive and cause disease. This work provides evolutionary insight into aging patterns observed in domestic dogs and demonstrates the applicability of studying natural populations to investigate the mechanisms of aging.
Keywords: RNA-Seq, transcriptome, senescence, immunosenescence, Canis lupus, canid
Introduction
In contrast to genetic changes occurring across generations, alterations of gene expression allow immediate and acutely sensitive responses to changing conditions. Gene expression can be affected by multiple factors such as age (Göring et al. 2007; Hong et al. 2008; López-Otín et al. 2013; Rahman et al. 2013; Peters et al. 2015), social stressors (Weaver et al. 2006; Cole et al. 2007; McGowan et al. 2009; Cole et al. 2011, 2012; Tung et al. 2012; Murphy et al. 2013; Powell et al. 2013), and infectious disease status (Whitney et al. 2003; Cobb et al. 2005; Ramilo et al. 2007; Blankley et al. 2014; Mejias and Ramilo 2014). Aging is associated with a multitude of gene expression changes in blood including cellular senescence and dysfunctional immune responses (e.g., immunosenescence and inflammaging) (Boren and Gershwin 2004; Kirkwood 2005; Baylis et al. 2013). In contrast, chronic stress due to adverse social environments activates immune pathways and can affect antiviral responses, susceptibility to infectious agents, wound healing abilities, and individual fitness (Sapolsky 2005; Weaver et al. 2006; Miller et al. 2009; Chen et al. 2011; Cole et al. 2011, 2012; Tung et al. 2012; Murphy et al. 2013; Powell et al. 2013).
Investigations of gene expression patterns in natural populations of nonmodel species can reveal which environmental factors are most influential on the regulation of specific molecular pathways. However, with the exception of baboons (Runcie et al. 2013; Tung et al. 2015), the transcriptome-wide effects of intrinsic and environmental variables on gene expression in vertebrates have only been studied in humans and species in captivity. Knowledge regarding the generality of aging effects on gene expression patterns across species is further limited by short lifespan biases of model species and unintended artificial selection on aging rates (Ricklefs 2010). Thus, despite a long history of research on the diverse ecology, social structures, and life histories of wild animal populations (e.g., Wilson 2000), the impacts of aging and environmental factors on interindividual variation at a functional genomic level in natural animal populations remain poorly understood (Ricklefs 2010).
To address this knowledge gap, we used RNA sequencing (RNA-Seq) to quantify genome-wide expression levels in wild gray wolves (Canis lupus) from Yellowstone National Park (YNP), USA. Gray wolves serve as an excellent model for studying the influences of age and social factors on gene expression levels, as wolves live in highly cooperative societies characterized by social hierarchies (Mech 1999; Packard 2003). Further, the divergence time of approximately 80 My between wolves and more extensively studied primate and murine models (dos Reis et al. 2012) affords the opportunity to assess the long-term evolutionary conservation of expression patterns. We take advantage of the exceptional observational database available for the YNP wolf population, in which factors potentially affecting gene expression such as social status, age, and disease presence have been extensively documented since the wolf reintroduction in 1995–1996 (vonHoldt et al. 2008, 2010; Almberg et al. 2009; MacNulty, Smith, Vucetich et al. 2009; Smith et al. 2010; Almberg et al. 2012; Smith et al. 2012; Stahler et al. 2013; Cubaynes et al. 2014).
Wolf packs consist of an adult pair of breeding wolves (denoted alpha), their offspring (denoted subordinates), and additional adult wolves, which may breed but are considered subordinate to alphas (denoted beta) (Mech 1999, 2003; Packard 2003). The breeding pair is considered dominant to other individuals, and social hierarchy is most commonly demonstrated by nonaggressive behavior and submissive postures resulting in unchanged or slightly lower glucocorticoid levels in subordinates (Mech 1999; Packard 2003; Smith et al. 2012; Molnar et al. 2015). This social system is in contrast to hierarchies in which dominance is maintained by displacements and threats toward lower ranking individuals, such as in wild baboons and macaques, which result in higher levels of stress in low-ranking individuals (Sapolsky 2004, 2005). Given the widespread impacts that social stress can have on individual health and the immune system as detected in blood (Sapolsky 2005; Weaver et al. 2006; Miller et al. 2009; Chen et al. 2011; Cole et al. 2011, 2012; Tung et al. 2012; Murphy et al. 2013; Powell et al. 2013), we aimed to compare influences of social rank on gene expression levels in blood of wolves, which could be collected with minimum handling, to those observed in primate social systems.
Active disease infection also impacts gene expression variation (Whitney et al. 2003; Cobb et al. 2005; Ramilo et al. 2007; Blankley et al. 2014; Mejias and Ramilo 2014). Sarcoptic mange is an infectious disease frequently observed in YNP gray wolves, induced by the mite Sarcoptes scabiei and analogous to human scabies (Almberg et al. 2012, 2015). Sarcoptes mites are known to elicit gene expression changes in humans and trigger allergic reactions causing skin inflammation, itching, and hair loss in wolves (Arlian et al. 2004, 2006; Almberg et al. 2012). Therefore, we hypothesized that presence of mange in YNP wolves results in expression signatures of immunosuppression and inflammatory response (Arlian et al. 2004, 2006; Velavan and Ojurongbe 2011; Gregori et al. 2012).
Finally, aging is also expected to be a factor affecting gene expression in gray wolves. We predict that rapid aging observed at the physical level in wolves (MacNulty, Smith, Vucetich et al. 2009; Stahler et al. 2013; Cubaynes et al. 2014) would be mirrored by gene expression changes with age. Although the molecular impact of age has been well described in humans (Göring et al. 2007; Hong et al. 2008; López-Otín et al. 2013; Rahman et al. 2013; Peters et al. 2015), the relative influence and specific pathways altered by age in a wild population confronted with multiple environmental challenges is unknown. In baboons, the only wild species assessed to date, the impact of age on gene expression was found to be modest, and the molecular pathways altered by age were not evaluated (Runcie et al. 2013; Tung et al. 2015). Therefore, to assess conservation of patterns across the mammalian phylogeny, and because impacts of aging on gene expression of specific pathways are virtually unknown for wild populations, we compared our age results to humans (Göring et al. 2007; Hong et al. 2008).
To investigate natural variation in gene expression levels, we use a linear mixed model to simultaneously assess the effects of social rank (alpha vs. all subordinates), age, disease status (infected with mange vs. not infected), and sex on genome-wide expression in whole blood while controlling for genetic relatedness. As these factors can also influence cell-type abundance in blood (Linton and Dorshkind 2004; Cole et al. 2011, 2012; Baylis et al. 2013; Powell et al. 2013), we perform transcript origin analysis (TOA) (Cole et al. 2011) and transcriptome representation analysis (TRA) (Powell et al. 2013) to infer the cellular origins of differentially expressed transcripts. Among the variables studied, we find that gene expression variation among wild wolves is primarily explained by age, with multiple age-related genes and processes conserved between wolves and humans. In contrast, we observe a surprising absence of expression differences with other variables such as rank, indicating that impacts of rank in primate models cannot be extended to the effects of social hierarchy in other vertebrate species.
Results
Sequence Data
To assess transcriptome variation across gray wolves in their natural environment, we quantified gene expression levels from whole blood samples of 27 wolves using RNA-Seq. A total of 1.26 billion 100 base-pair reads of sufficient quality (Phred score > 20) and length (> 25 bp after trimming) were generated (mean of 46.69 million reads per individual; supplementary table S1, Supplementary Material online). On average, 87.5% of these reads aligned to the dog genome (CanFam3.1.74) and up to 35% of the uniquely mapped reads aligned to annotated protein-coding regions (supplementary table S1, Supplementary Material online). Of the 19,856 protein-coding genes annotated in the dog genome, we obtained 13,558 genes (68.3%) with sufficient read depth across individuals (≥10 reads in at least 75% of cDNA libraries) to be included in expression analyses. The number of detected genes in whole blood of wolves (N = 13,558) was similar to that found in lymphocytes of humans (N = 12,758) (Göring et al. 2007) and whole blood of baboons (N = 10,409) (Tung et al. 2015).
Association of Gene Expression Levels with Intrinsic and Environmental Factors
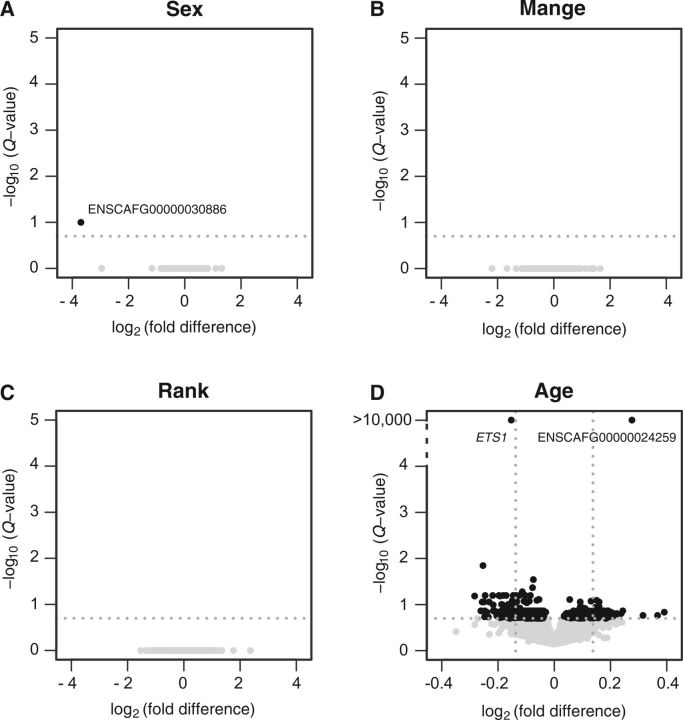
We assessed the effects of intrinsic and environmental factors on transcriptome-wide gene expression using a significance cut-off of false discovery rate (FDR) = 0.2 given the limited sample size. Similar to other studies that evaluated effects of sex on blood gene expression in mammals (e.g., Whitney et al. 2003; Tung et al. 2015), we observed minimal expression differences between male and female wolves, with only one gene significantly associated with sex, ENSCAFG00000030886 (fold difference [FD] = 0.08; Q value = 0.1; fig. 1A andsupplementary table S2, Supplementary Material online). This protein-coding gene belongs to the thymosin β4 family but has not been formally named. Further, despite the presence of physical symptoms characteristic of mange in seven of the sampled wolves, we did not identify any genes differentially expressed in wolves affected by mange (fig. 1B). Finally, we predicted that social rank would be broadly associated with gene expression levels, comparable to effects of social conditions in humans and nonhuman primates (Cole et al. 2007, 2012; Tung et al. 2012). However, we did not identify any genes in which expression levels were significantly associated with social rank (fig. 1C).
Fig. 1.
Difference in gene expression according to sex, mange, social rank, and age in wolf blood. (A–D) Black dots represent genes for which expression is significantly associated with a variable, whereas gray dots illustrate nonsignificant genes. The significance threshold (Q value = 0.2) is delimited by the horizontal line y = −log10(0.2). FD is expression of females relative to males (A), mange-affected wolves relative to nonaffected individuals (B), alpha wolves relative to subordinates (C), and difference per year of age (D) for 13,558 genes expressed in wolf whole blood. (D) Vertical lines depict the thresholds used in the TOA (table 3).
In contrast, controlling for rank, sex, and mange infection status, we detected 625 genes associated with age, comprising 214 genes up-regulated and 411 genes down-regulated with age with diverse functions (fig. 1D andtable 1 and supplementary table S2, Supplementary Material online). These age-related gene expression changes may have been induced by intracellular changes in gene transcription or by age-related changes in proportions of blood cell-types (Linton and Dorshkind 2004; Baylis et al. 2013). To evaluate potential changes in blood cell composition with age, we performed TOA (table 2), which infers the cellular origin of transcripts (Cole et al. 2011), in parallel with TRA (table 3), which assesses changes in cell abundance based on genes previously identified to have highly cell-type-specific expression (Powell et al. 2013). In accordance with observations of immunosenescence in humans, genes down-regulated with age were largely associated with B cells (P < 0.001; table 2), which appear to partly be due to a lower prevalence of B cells in older wolves (TRA; FD per year = 0.98; P = 0.001; table 3). Further, consistent with development of more pro-inflammatory phenotypes in older individuals (reviewed in Boren and Gershwin 2004; Baylis et al. 2013), genes up-regulated with age were significantly associated with innate immunity cells including dendritic cells (P < 0.001; table 2) and natural killer cells (P = 0.004; table 2). TRA indicated a slightly increased proportion of monocytes in older individuals (FD per year = 1.01; P < 0.001; table 3).
Table 1.
Biological Processes and Genes Associated with Age in Gray Wolves.
| Enriched GO Terma | GO P Value | Gene Symbol | Ensembl Gene ID | Annual FD | Q Valueb |
|---|---|---|---|---|---|
| Regulation of metabolic process | 2.25E-5 | ETS1 | ENSCAFG00000010304 | 0.900 | <0.001 |
| CDK4 | ENSCAFG00000000280 | 0.910 | 0.137 | ||
| RNA metabolic process | 1.91E-5 | DKC1 | ENSCAFG00000019620 | 0.948 | 0.175 |
| NOP56 | ENSCAFG00000006651 | 0.958 | 0.198 | ||
| Chromatin modification | 2.97E-3 | SIRT6 | ENSCAFG00000019123 | 0.896 | 0.200 |
| HIRA | ENSCAFG00000014619 | 0.974 | 0.137 | ||
| Immune response | 6.28E-6 | DLA-DQA1 | ENSCAFG00000000812 | 0.945 | 0.137 |
| DLA-DRA | ENSCAFG00000000803 | 0.950 | 0.146 | ||
| DLA-88 | ENSCAFG00000000487 | 0.945 | 0.180 | ||
| DLA-DOB | ENSCAFG00000000819 | 0.883 | 0.175 | ||
| Lymphocyte activation | 1.55E-9 | IL7 | ENSCAFG00000008373 | 0.907 | 0.180 |
| IL27RA | ENSCAFG00000016504 | 0.919 | 0.146 | ||
| CD5 | ENSCAFG00000016332 | 0.938 | 0.157 | ||
| CXCR5 | ENSCAFG00000012439 | 0.898 | 0.175 | ||
| Lipid metabolic process | 1.17E-2 | LEP | ENSCAFG00000001672 | 1.078 | 0.151 |
| LIAS | ENSCAFG00000015991 | 1.035 | 0.183 | ||
| LIPH | ENSCAFG00000013246 | 1.113 | 0.184 |
aA comprehensive gene list and full GO results are presented in supplementary tables S2–S4, Supplementary Material online.
b Q values of genes significantly associated with age after controlling for relatedness, social rank, mange presence, and sex using linear mixed effects models.
Table 2.
Cell Types Inferred to Mediate Age-Related Gene Expression Differences.
| Cell Type |
P Value |
|
|---|---|---|
| FD > 1.10a | FD < 0.90b | |
| CD14+ monocytes | 0.035 | 0.998 |
| BDCA4+ dendritic cells | <0.001* | 0.973 |
| CD56+ NK cells | 0.004* | 0.338 |
| CD4+ T cells | 0.762 | 0.424 |
| CD8+ T cells | 0.460 | 0.768 |
| CD19+ B cells | 0.126 | <0.001* |
Note.—NK, natural killer. FD, fold difference per year of age.
aTOA results of up-regulated genes (N = 137 genes).
bTOA results of down-regulated genes (N = 111 genes).
*Significant (P < 0.01) overrepresentation of genes predominantly expressed by a specific cell type.
Table 3.
Leukocyte Prevalence Differences with Age in Wolf Blood.
| Cell Types | Genes Z > 6a | FD | P Valueb |
|---|---|---|---|
| CD14+ monocytes | 337 | 1.01 | <0.001* |
| BDCA4+ dendritic cells | 250 | 1.00 | 0.912 |
| CD56+ NK cells | 349 | 1.00 | 0.043 |
| CD4+ T cells | 62 | 1.00 | 0.606 |
| CD8+ T cells | 45 | 1.01 | 0.220 |
| CD19+ B cells | 87 | 0.98 | 0.001* |
Note.—FD, fold difference in cell prevalence per year of age; NK, natural killer.
aGenes diagnostic of a cell type are defined by average expression in a cell type > 6 standard deviations above the mean (Genes Z > 6) (Powell et al. 2013).
bTRA results of cell-type differences with age.
*Significant (P < 0.01) difference in cell-type prevalence.
To assess the conservation of aging processes across species, we compared the 625 age-related genes in wolf leukocytes to a catalog of age-related genes in human lymphocytes (Göring et al. 2007; Hong et al. 2008). Wolf age-related genes were significantly enriched with human age-related genes, as we found that of the 420 age-related wolf genes represented in the human study, 255 (60.7%) genes were associated with age in humans (hypergeometric test; P = 0.046). The majority of these overlapping genes were associated with age in the same direction (60.8% of overlapping genes, N = 155 concordant genes, χ2 = 11.863, df = 1, P = 0.001).
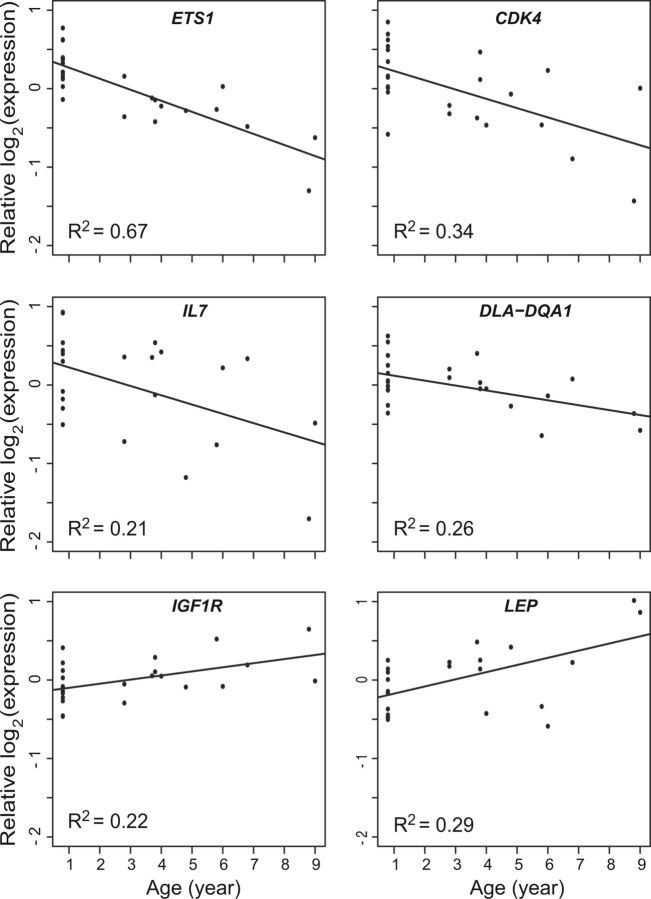
A diversity of genes and biological processes we found to be associated with age in wolves (table 1) has previously been associated with aging in humans (Boren and Gershwin 2004; Kirkwood 2005; Baylis et al. 2013). For example, we observed significant age-related down-regulation of ETS1 (FD per year = 0.900; Q value < 0.001; fig. 1D and 2 and table 1 and supplementary table S2, Supplementary Material online), a transcription factor involved in a variety of functions including regulation of the immune response (Garrett-Sinha 2013) and cellular senescence (Ohtani et al. 2001). Similarly, CDK4, which is known to be inhibited by p16, a key protein activated during cellular senescence (Krishnamurthy et al. 2004; Liu et al. 2009; López-Otín et al. 2013), was down-regulated with age (FD = 0.909; Q value = 0.137; fig. 2 and table 1 and supplementary table S2, Supplementary Material online). Additionally, we observed significant age-related expression differences in genes associated with telomere maintenance (e.g., DKC1, a component of the telomerase holoenzyme; FD = 0.948; Q value = 0.2; and NOP56; FD = 0.958; Q value = 0.198; table 1 and supplementary table S2, Supplementary Material online) (Poncet et al. 2008), DNA repair and genomic stability (e.g., SIRT6; FD = 0.896; Q value = 0.2) (Mostoslavsky et al. 2006; Di Mauro and David 2009), and epigenetic regulation (genes down-regulated with age were enriched for the gene ontology (GO) terms “chromatin organization,” P = 0.005 and “histone modification,” P = 0.011; table 1 and supplementary table S3, Supplementary Material online).
Fig. 2.
Expression of a subset of genes significantly associated with age in gray wolves. Relative expression represents the normalized, log 2-transformed expression levels of wolves after controlling for relatedness using linear mixed effects models. A comprehensive gene list is available in supplementary table S2, Supplementary Material online.
Paralleling intracellular senescence, aging has been found to dysregulate the propensity of the adaptive immune system to respond to novel infectious disease (a phenomenon known as “immunosenescence”) (Baylis et al. 2013; López-Otín et al. 2013). In agreement with previous work, our results suggest reduced activity of B and T cells with age, as interleukin and interleukin-related genes, required for differentiation and proliferation of lymphocytes, were down-regulated in older wolves (IL7, IL21R, IL27RA; Q values < 0.2; fig. 2 and table 1 and supplementary table S2, Supplementary Material online). In addition, we observed age-related down-regulation of multiple MHC class II genes (Dog Leukocyte Antigen genes DLA-DQA1, DLA-DRA, DLA-88, and DLA-DOB; Q values < 0.2; fig. 2 and table 1 and supplementary table S2, Supplementary Material online), whose expression is likely limited to antigen-presenting cells (B cells, monocytes) and activated T cells. GO analyses further support immunosenescence, as 30% of terms enriched in the genes down-regulated with age are explicitly related to immune system processes (e.g., lymphocyte activation, antigen receptor-mediated signaling pathway, T- and B-cell activation and proliferation, and interleukin-2 production; supplementary table S3, Supplementary Material online).
In contrast to the abundance of immunity-related GO terms enriched in the genes down-regulated with age in wolves, GO terms enriched in genes up-regulated with age were dominated by biological processes related to lipid metabolism (supplementary table S4, Supplementary Material online) and included leptin (LEP), lipoic acid synthetase, and lipase member H (LIPH; Q values < 0.2; fig. 2 and table 1 and supplementary table S2, Supplementary Material online). We also observed an age-related increase in expression of insulin-like growth factor 1 receptor (IGF1R; FD = 1.038; Q value = 0.188; fig. 2 and supplementary table S2, Supplementary Material online), which has been found to regulate lifespan (Holzenberger et al. 2003).
Discussion
We examined factors that we predicted to influence gene expression in gray wolves based on gene expression patterns observed in humans and other primates, which included sex, mange presence, social rank, and age. The negligible effect of sex on gene expression in blood is consistent with similar findings in humans and macaques (Whitney et al. 2003; Tung et al. 2015). However, the lack of detectable differentially expressed genes in mange-infected wolves was unexpected, given the presence of visible symptoms of mange such as hairless patches with skin lesions (Almberg et al. 2015). This result may partly be due to limited statistical power but may also be caused by the immunosuppressive properties that S. scabiei evoke in their hosts by impacting IL10 signaling (Arlian et al. 2006). The possibility of altered IL10 signaling in wolves with mange is supported by the presence of IL10RA in the top five genes associated with mange in our data set, although this result is not significant at our Q-value threshold of 0.2.
The absence of a significant effect of rank on gene expression was also unexpected but likely reflects a less hierarchically based social system in wolves compared to many primates species. In wolves, the term alpha denotes the breeding pair of a pack (which are typically the parents of other pack members) and signifies little to no differences in stress levels, resource access, or received aggression from other wolves (Mech 1999; Cubaynes et al. 2014; Molnar et al. 2015). Additionally, wolf rank and age are positively correlated (Pearson correlation r = 0.357; P = 0.068) and may have effects on the same biological pathways (Snyder-Mackler et al. 2014), potentially limiting our statistical power to detect rank effects independent of aging.
Overall, we found that age has a much broader impact on gene regulation than sex, mange infection status, or rank in a social top-predator exposed to naturally occurring abiotic and biotic challenges. Wolves show signatures of senescence consistent with aging in model organisms and humans (Baylis et al. 2013; Beirne et al. 2014), which supports the hypothesis that aging effects on cellular processes are evolutionarily conserved through mammal phylogeny (Palacios et al. 2011; Nussey et al. 2013). Theory predicts that higher levels of extrinsic mortality should lead to an increased rate of senescence because a relatively larger amount of energy is expected to be dedicated to reproduction than to somatic maintenance and repair (Kirkwood 1977, 2005). The short time period over which age-related expression differences manifest in wolves (spanning a few years compared to decades in humans) (Göring et al. 2007) is evolutionarily appropriate given the high extrinsic mortality in this species, due mainly to interpack strife (Cubaynes et al. 2014), and rapid physical senescence (MacNulty, Smith, Mech et al. 2009; MacNulty, Smith, Vucetich et al. 2009; Stahler et al. 2013).
Stress related to changes in rank, physical injuries, and disease (Almberg et al. 2015) may augment the expression signature of aging in wolves or result in expression differences unique from that observed in humans. Genes associated with age in wolves were significantly enriched for human age-related genes, yet we detected 165 age-related genes in wolves that were not found to change with age in humans (Göring et al. 2007). Further, 100 genes associated with age in both wolves and humans showed opposite trends of expression levels with age between the two species (Göring et al. 2007). A longitudinal study, and comparison to gene expression in captive wolves, would be valuable in decomposing the specific factors that have long-lasting impacts on gene expression in wolves.
We identified age-related changes in the expression of genes that may be critical in the progression of the aging process, such as up-regulation of IGF1R, the primary receptor of the IGF1 protein product. IGF1 signaling is thought to be an evolutionarily conserved regulator of the trade-off between growth and survival, as the IGF1 pathway promotes growth during early development but can increase the rate of aging later in life (Rollo 2002; Salminen and Kaarniranta 2010). The existence of a trade-off in IGF1 signaling is supported by studies across domestic dog breeds, in which plasma IGF1 protein levels across breeds positively correlate with body size but negatively correlate with lifespan (Sutter et al. 2007; Hoopes et al. 2012). This mechanism has been suggested to be the reason that small dog breeds can have twice the lifespan of larger breeds (Kraus et al. 2013). The rapid development of yearling wolves (MacNulty, Smith, Mech et al. 2009; MacNulty, Smith, Vucetich et al. 2009) likely requires early IGF1R expression. However, the function of up-regulated IGF1R we observe in old wolves is unknown and is potentially nonadaptive. IGF1R is down-regulated with age in humans (Göring et al. 2007), which may be an important contributor to human longevity. Consequently, up-regulation of IGF1R may be a leading driver of the rapid senescence of wolves and other short-lived species.
We also observed increased expression of LEP in older wolves, concordant with increased leptin protein levels observed in aging humans, dogs, and model species (Mobbs 1998; Ishioka et al. 2002, 2007; Ma et al. 2002). Leptin is produced by adipocytes and reduces reflex appetite (Sánchez-Rodrı´guez et al. 2000). Its increase with age, which is often disproportionately more than would follow from an increase in adipose tissue, is thought to indicate a leptin-resistant state in elderly individuals and can contribute to obesity (Ahren et al. 1997; Li et al. 1997; Sánchez-Rodrı´guez et al. 2000; Ma et al. 2002). Because the weight of wolves is strongly correlated with age (Pearson correlation r = 0.768; P = 2.967 × 10−5; supplementary table S1, Supplementary Material online), we do not have the statistical power to test whether LEP expression increases with age more than expected by weight gain alone. However, the overall weight gain and increased expression of LEP with age in wolves supports the hypothesis that wild wolves undergo metabolic aging (MacNulty, Smith, Mech et al. 2009).
Although we find age-related gene expression patterns shared by humans and wolves, their impacts on health and fitness may be vastly different for each species. For example, metabolic aging and obesity are recognized as having negative health impacts in older humans (e.g., Alexander et al. 2003). In contrast, increased weight in wolves enhances reproductive success in females of all ages (Stahler et al. 2013) and dispatching ungulate prey in males and females (MacNulty, Smith, Mech et al. 2009). Further, male wolves exhibit increasing aggressiveness with age, which may facilitate dominance and breeding opportunities within a pack and enhance success during interpack territorial encounters (Cassidy KA, Mech LD, MacNulty DR, Stahler DR, Smith DW, personal communication). Thus, although senescence does occur in wolves, some age-related changes may also allow older wolves to maintain fitness and, consequently, can be maintained by selection (e.g., Schwarz et al. 2016). In dogs, which live in human-dominated environments, such age-related changes may be maladaptive and result in morbidity. Likewise, gene expression patterns that evolved under a hunter-gatherer lifestyle in humans may lead to health problems for individuals with a western lifestyle and diet (Carrera-Bastos et al. 2011; Farooqui 2015). Consequently, for both modern humans and their domestic companions, such legacy effects may result in disease in aging individuals.
Our results establish the ancestral baseline for gene expression in dogs and show that transcriptome-wide gene expression analysis applied to natural populations has the potential to reveal novel functional and evolutionary insights into the mechanisms and drivers of aging. In general, our study suggests that aging is the critical factor affecting gene expression levels in a wild wolf population and provides a new paradigm for investigating the aging process in natural systems.
Materials and Methods
Sampling and Phenotypic Traits
Whole blood was collected from 27 wild gray wolves in YNP, USA. Sampling was conducted during winters 2011, 2012, and 2013 during annual wolf captures according to US national guidelines for handling animals and with all required permits held by the National Park Service (Smith et al. 2012). Individual animal information is provided in supplementary table S1, Supplementary Material online.
Age and Sex
For each of the 27 wolves, sex was determined during handling. Males were coded as 0 and females as 1. Ages were estimated from an assumed birth date of April 15 (mean whelp date in study area) (Stahler et al. 2013) and were continuously coded for the linear models (supplementary table S1, Supplementary Material online and supplementary fig. S2, Supplementary Material online). Age was determined by either capturing pups (N = 13), which are easy to distinguish due to body and tooth size, or recapturing individuals that had known birth years. The known ages (years) of the 21 wolves included in the analyses were 0.8 (N = 13); 2.8 (N = 1); 3.7 (N = 1); 3.8 (N = 3); 4.8 (N = 1); 5.8 (N = 1); 6.8 (N = 1); and 8.8 (N = 1). For the five individuals included in the expression analyses that were not first identified as pups, tooth wear was used to estimate the age of adults (Gipson et al. 2000). The estimated ages (years) of the five wolves used in the analyses were 2.8 (N = 1); 4 (N = 1); 4.8 (N = 1); 6 (N = 1); and 9 (N = 1). Estimated ages of the two older wolves (6 and 9 years) were corroborated by the fact that they had been monitored by NPS for multiple consecutive years (5 and 6 years of monitoring, respectively).
Social Status
Social rankings of the 27 individuals were determined from behavioral observations and reproductive status and were categorized, specific to the time of sampling, as alpha or nonalpha (supplementary table S1, Supplementary Material online). Individuals that were the only or primary breeders in a pack or behaviorally dominant to all pack members (excluding their mates) were identified as alpha wolves. In well-known and larger packs, individuals that were subordinate to the leading pair but dominant to other pack members were ranked as beta. Individuals showing neither reproductive nor antagonistic behavior were classified in the subordinate category (Mech 1999; Packard 2003; Smith et al. 2012). Because age and social rank are highly correlated in YNP wolves (Pearson correlation r = 0.614; P = 6.55 × 10−5), we focused on the distinction between alpha wolves (scored as 1) and nonalpha animals (scored as 0; betas and subordinates), which significantly reduced the magnitude of this correlation (N = 27; P < 0.01; psych R package; paired.r function to test for a significant decrease in correlation) (Revelle 2015).
Mange Infection Status
The severity of mange infection in each wolf was assessed based on the presence of physical symptoms (hairlessness and scratching lesions) at the time of capture and scored according to the percentage of body surface affected (0: no physical symptoms; 1: < 5% of the body affected by lesions; 2: 6–50%; and 3: > 50%) (Pence et al. 1983; Almberg et al. 2012). As only one individual presented severe symptoms of mange infection, we classified mange as a binary variable (0: no symptom; 1: visible symptoms; supplementary table S1, Supplementary Material online). Continuous coding of mange severity led to qualitatively identical null findings (results not shown).
RNA Extraction, Library Preparation, and Sequencing
Whole blood was preserved in PAXgene Blood RNA tubes (PreAnalytiX, Qiagen, Hombrechtikon, Switzerland) and stored at −80 °C. Total RNA extraction was performed following the manufacturer’s instructions of PAXgene Blood RNA kit (PreAnalytiX, Qiagen, Hombrechtikon, Switzerland). RNA quality was assessed with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). All samples had a RNA integrity number (RIN) > 7. Total RNA was treated with the Globin-Zero kit (Epicentre, Illumina, Madison, WI) and purified with a modified Qiagen RNeasy MinElute (Qiagen Inc., Valencia, CA) cleanup procedure or ethanol precipitation (supplementary table S1, Supplementary Material online). cDNA libraries were synthesized using a strand-specific kit from Epicentre (ScriptSeq v2 Library Prep kit, Illumina, Madison, WI) with ScriptSeq Index PCR primers (Epicentre, Illumina, Madison, WI). cDNA libraries were quantified with the KAPA SYBR Fast qPCR library quantification kit (Kapa Biosystems Inc., Wilmington, MA) and pooled at five to six samples per lane. Single-end 100 bp sequencing was performed on Illumina's HiSeq2000 platform (supplementary table S1, Supplementary Material online) at the Broad Stem Cell Research Center’s biosequencing core, UC Los Angeles, CA (BSCRC) and the Vincent J. Coates Genomics Sequencing Laboratory (GSL), UC Berkeley, CA. The data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al. 2002) and are accessible through GEO series accession number GSE80440 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80440).
Mapping and Expression Quantification
Quality filtering and adaptor trimming was performed using Trim Galore! v.0.3.1 (www.bioinformatics.babraham.ac.uk/projects/trim_galore/). A moderate coverage (25x) genome assembly of the gray wolf is available (Freedman et al. 2014). However, this assembly is based on mapping reads to the domestic dog. Therefore, given the availability of the well-annotated dog genome (Canisl. familiaris; Ensembl CanFam3.1.74) (Lindblad-Toh et al. 2005; Hoeppner et al. 2014), as well as the short divergence time between domestic dogs and wolves (< 29 Ka) (Thalmann et al. 2013; Freedman et al. 2014; Skoglund et al. 2015; Fan et al. 2016), we used the domestic dog genome as a reference. Splice-aware mapping to the domestic dog genome (Ensembl CanFam3.1.74, unmasked) was implemented with TopHat2 v.2.0.10 (Engström et al. 2013; Kim et al. 2013). We allowed up to three mismatches, a total of 3-bp length of gaps, and an edit distance no more than 3 bp between the reads and the reference sequence to account for the divergence between the domestic dog and the gray wolf. Only reads that uniquely mapped to the reference were kept in subsequent analyses. Only uniquely mapped reads were kept in subsequent analyses. The expression level of each gene was quantified using the union mode of the python script htseq-count (HTSeq 0.6.1) (Anders et al. 2015). Individual expression profiles were filtered to only include protein-coding genes with at least ten reads in at least 75% of cDNA libraries (N = 13,558 protein-coding genes). Genome assembly and GTF files of the domestic dog genome (CanFam3.1.74) used as reference in this study can be retrieved at http://dec2013.archive.ensembl.org/Canis_familiaris/Info/Index.
Data Preprocessing
To identify outlier samples, we used the adjacency function in the WGCNA R package (Langfelder and Horvath 2008) to build a Euclidean distance-based sample network from the log 2-transformed read counts. Samples were designated as outliers if their standardized connectivity deviated by more than three standard deviations from the mean (Horvath 2011). After removing two individual outliers from the data (supplementary table S1, Supplementary Material online), we used conditional quantile normalization (cqn package in R) to normalize for sequencing depth (using trimmed mean of M values) (Robinson et al. 2010), GC-content, and gene length (Hansen et al. 2012), averaged from the transcripts in the canFam3.1.74 gtf file. We used principal component analysis of the normalized, log 2-transformed expression data to assess general impacts of technical, phenotypic, or environmental variables on gene expression variance. We identified significant effects of technical factors on overall gene expression (supplementary fig. S1A, Supplementary Material online) including effects of sequencing core (correlated with PC1 [Pearson correlation r = 0.59, P = 1.892 × 10−3] and PC2 [Pearson correlation r = −0.519, P = 7.809 × 10−3] of the normalized gene expression data), library preparation procedure (column purifications vs. ethanol precipitation: correlated with PC1 [Pearson correlation r = 0.791, P = 2.557 × 10−6]), barcode bleed (Kircher et al. 2012) of RNA-Seq from other samples sequenced in the same lane (Johnston R, unpublished data, March 2015; correlated with PC2 [Pearson correlation r = −0.566, P = 3.178 × 10−3] and PC6 [Pearson correlation r = −0.486, P = 1.387 × 10−2]), and year of sampling (correlated with PC1 [Pearson correlation r = 0.623, P = 8.86 × 10−4]). To control for these effects, we regressed out sequencing core, library preparation procedure, and sequencing lane composition for each gene prior to the main analyses (supplementary fig. S1B, Supplementary Material online and supplementary table S1, Supplementary Material online). Year of sampling, which significantly correlated with library preparation procedure (Pearson correlation r = 0.721, P = 4.853 × 10−5), was not significantly associated with PC1-12 after regressing out the effect of library preparation (supplementary fig. S1B, Supplementary Material online). For the subset of samples in which time of day of collection was available (supplementary table S1, Supplementary Material online), we did not detect a significant effect of time of day on overall gene expression (supplementary fig. S1, Supplementary Material online).
Linear Mixed Effects Models
For each gene, we modeled the fixed effects of age, sex, social rank, and mange infection status on the residuals of the normalized, log 2-transformed gene expression levels of the 25 samples using linear mixed models in gemma v.0.94 (Zhou and Stephens 2012) as done previously (Tung et al. 2012). To fit a random effect controlling for kinship, we calculated pairwise relatedness between individuals using the triadic maximum likelihood approach in Coancestry (Wang 2011), based on genotypes obtained from 24 microsatellite loci (supplementary table S5, Supplementary Material online). Allele frequencies for the 24 markers were estimated using a comprehensive data set of 371 YNP gray wolves (vonHoldt et al. 2008, 2010; vonHoldt BM, unpublished data).
All significance levels for the linear models were adjusted for multiple hypothesis testing using the Q value method in R (Storey and Tibshirani 2003), with the null distribution constructed based on 100 random permutations. Because of the difficulty of obtaining samples from animals in the wild for RNA analyses, our sample size limited the statistical power of the linear model analysis. Therefore, we set a relatively liberal FDR threshold of 0.2 to identify candidate genes associated with rank, age, sex, and mange. The FDR threshold of 0.2 corresponded to a minimum detectably significant FD of approximately 2.2% for age and approximately 7.8% for sex (supplementary table S2, Supplementary Material online). Q values of all genes meeting the 0.2 FDR threshold are presented in supplementary table S2, Supplementary Material online.
GO Analysis
To assess which biological functions were significantly enriched in the genes affected by our variables of interest (defined as genes associated at Q value < 0.2), we used g:ProfileR (Reimand et al. 2007) based on the Canisfamiliaris gene annotation (Ensembl 79). Queries were composed of genes up- or down-regulated with each variable of interest and the background set included all genes tested in our analysis (N = 13,558). We set the minimal overlap between the GO term and the query, as well as the minimal number of genes within a functional category, to 3. All GO analyses were corrected for multiple testing using the Benjamini–Hochberg FDR method (Benjamini and Hochberg 1995).
Differential White Blood Cell Counts
To assess the impacts of age, rank, mange, and sex on the relative abundance of lymphocytes, neutrophils, monocytes, basophils, and eosinophils, blood smears were prepared for eleven of the 27 individuals during the collection of PAXgene-preserved blood (supplementary table S6, Supplementary Material online). Subsequently, the histological slides were fixed and stained according to standard procedures (i.e., fixation using the methanol and staining according to the Wright–Giemsa method). White blood cell (WBC) differentials were determined by quantifying the abundance of lymphocytes, neutrophils, monocytes, basophils, and eosinophils relative to a total number of white cells fixed at NtotalWBC = 100. WBC counts were calculated as the average of two replicated slides. For each WBC type, counts were correlated in a linear model with each explanatory variable (age, sex, rank, or mange). No significant result was obtained. Therefore, we assessed effects of age on cell population with TRA (Powell et al. 2013), which is more sensitive to small differences in cell prevalence than blood smear cell counts because TRA assesses gene expression of multiple cell markers.
TOA and TRA
TOA was used to assess the extent to which differentially expressed genes were predominately characteristic of one or more subtypes of circulating leukocytes (Cole et al. 2011). Differential expression for TOA was defined by the FD, with differentially expressed genes defined by <0.90 and >1.10 FD per year of age. Thresholds of FD were selected to achieve the largest biological effect size possible while still yielding an input list with at least 50 differentially expressed genes. TRA was used to assess the prevalence of cell-type-specific RNAs (Powell et al. 2013) identified a priori based on highly cell-type-diagnostic transcripts in humans (Cole et al. 2011; Powell et al. 2013).
Supplementary Material
Acknowledgments
We would like to acknowledge Emily Almberg and Erin Stahler for their contribution to data collection. We are thankful to Devaughn Fraser for her significant help in the blood smear cell counts and to Ryan Harrigan for his comments on the manuscript. This work was supported by the National Science Foundation (grant numbers DEB 1257716 to R.K.W. and B.M.V. and DEB 1245373 to D.R.S. and D.W.S.); the National Institutes of Health (grant numbers P30 AG017265 to S.W.C., S10 RR029668 and S10 RR027303 to work with the Vincent J. Coates GSL at UC Berkeley); the Austrian Academy of Sciences (to P.C.); the Max Kade Foundation (to P.C.); the Howard Hughes Medical Institute (to R.A.J.); the National Park Service (to D.R.S. and D.W.S.); the Yellowstone Park Foundation (to D.R.S. and D.W.S.); the Tapeats Fund (to D.R.S. and D.W.S.); the Perkin-Prothro Foundation (to D.R.S. and D.W.S.); and an anonymous donor (to D.R.S. and D.W.S.).
References
- Ahren B, Mansson S, Gingerich RL, Havel PJ. 1997. Regulation of plasma leptin in mice: influence of age, high-fat diet, and fasting. Regul Integr Comp Physiol. 273:R113–R120. [DOI] [PubMed] [Google Scholar]
- Alexander CM, Landsman PB, Teutsch SM, Haffner SM. 2003. NCEP-defined metabolic syndrome, diabetes, and prevalence of coronary heart disease among NHANES III participants age 50 years and older. Diabetes 52:1210–1214. [DOI] [PubMed] [Google Scholar]
- Almberg ES, Cross PC, Dobson AP, Smith DW, Hudson PJ. 2012. Parasite invasion following host reintroduction: a case study of Yellowstone's wolves. Philos Trans R Soc Lond Ser B Biol Sci. 367:2840–2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almberg ES, Cross PC, Dobson AP, Smith DW, Metz MC, Stahler DR, Hudson PJ. 2015. Social living mitigates the costs of a chronic illness in a cooperative carnivore. Ecol Lett. 18:660–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almberg ES, Mech LD, Smith DW, Sheldon JW, Crabtree RL. 2009. A serological survey of infectious disease in Yellowstone National Park’s canid community. PLoS One 4:e7042.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W. 2015. HTSeq: a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlian LG, Morgan MS, Neal JS. 2004. Extracts of scabies mites (Sarcoptidae: Sarcoptes scabiei) modulate cytokine expression by human peripheral blood mononuclear cells and dendritic cells. J Med Entomol. 41:69–73. [DOI] [PubMed] [Google Scholar]
- Arlian LG, Morgan MS, Paul CC. 2006. Evidence that scabies mites (Acari: Sarcoptidae) influence production of interleukin-10 and the function of T-regulatory cells (Tr1) in humans. J Med Entomol. 43:283–287. [DOI] [PubMed] [Google Scholar]
- Baylis D, Bartlett D, Patel H, Roberts H. 2013. Understanding how we age: insights into inflammaging. Longev Healthspan. 2:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beirne C, Delahay R, Hares M, Young A. 2014. Age-related declines and disease-associated variation in immune cell telomere length in a wild mammal. PLoS One 9:e108964.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 57:289–300. [Google Scholar]
- Blankley S, Berry MPR, Graham CM, Bloom CI, Lipman M, O'Garra A. 2014. The application of transcriptional blood signatures to enhance our understanding of the host response to infection: the example of tuberculosis. Philos Trans R Soc Lond Ser B Biol Sci. 369:20130427.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boren E, Gershwin ME. 2004. Inflamm-aging: autoimmunity, and the immune-risk phenotype. Autoimmun Rev. 3:401–406. [DOI] [PubMed] [Google Scholar]
- Carrera-Bastos P, Fontes-Villalba M, O’Keefe JH, Lindeberg S, Cordain L. 2011. The western diet and lifestyle and diseases of civilization. Res Rep Clin Cardiol. 2:15–35. [Google Scholar]
- Chen E, Miller GE, Kobor M, Cole SW. 2011. Maternal warmth buffers the effects of low early-life socioeconomic status on pro-inflammatory signaling in adulthood. Mol Psychiatry. 16:729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb JP, Mindrinos MN, Miller-Graziano C, Calvano SE, Baker HV, Xiao W, Laudanski K, Brownstein BH, Elson CM, Hayden DL, et al. 2005. Application of genome-wide expression analysis to human health and disease. Proc Natl Acad Sci U S A. 102:4801–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Conti G, Arevalo JMG, Ruggiero AM, Heckman JJ, Suomi SJ. 2012. Transcriptional modulation of the developing immune system by early life social adversity. Proc Natl Acad Sci U S A. 109:20578–20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JMG, Cacioppo JT. 2011. Transcript origin analysis identifies antigen-presenting cells as primary targets of socially regulated gene expression in leukocytes. Proc Natl Acad Sci U S A. 108:3080–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole SW, Hawkley LC, Arevalo JM, Sung CY, Rose RM, Cacioppo JT. 2007. Social regulation of gene expression in human leukocytes. Genome Biol. 8:R189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubaynes S, MacNulty DR, Stahler DR, Quimby KA, Smith DW, Coulson T. 2014. Density-dependent intraspecific aggression regulates survival in northern Yellowstone wolves (Canis lupus). J Anim Ecol. 83:1344–1356. [DOI] [PubMed] [Google Scholar]
- Di Mauro T, David G. 2009. Chromatin modifications: the driving force of senescence and aging? Aging 1:182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PCJ, Yang Z. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc R Soc Lond Ser B Biol Sci. 279:3491–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P, Steijger T, Sipos B, Grant G, Kahles A, Rätsch G, Goldman N, Hubbard T, Harrow J, Guigó R, et al. 2013. Systematic evaluation of spliced alignment programs for RNA-seq data. Nat Methods. 10:1185–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Silva P, Gronau I, Wang S, Armero AS, Schweizer RM, Ramirez O, Pollinger J, Galaverni M, Ortega Del-Vecchyo D, et al. 2016. Worldwide patterns of genomic variation and admixture in gray wolves. Genome Res. 26:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farooqui AA. 2015. Effect of long term consumption of high calorie diet and calorie restriction on human health. High calorie diet and the human brain. Cham (Switzerland: ): Springer International Publishing; p. 1–28. [Google Scholar]
- Freedman AH, Schweizer RM, Gronau I, Han E, Ortega-Del Vecchyo D, Silva PM, Galaverni M, Fan Z, Marx P, Lorente-Galdos B, et al. 2014. Genome sequencing highlights genes under selection and the dynamic early history of dogs. PLoS Genet. 10:e1004016.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett-Sinha L. 2013. Review of Ets1 structure, function, and roles in immunity. Cell Mol Life Sci. 70:3375–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson PS, Ballard WB, Nowak RM, Mech LD. 2000. Accuracy and precision of estimating age of gray wolves by tooth wear. J Wildl Manage. 64:752–758. [Google Scholar]
- Göring HHH, Curran JE, Johnson MP, Dyer TD, Charlesworth J, Cole SA, Jowett JBM, Abraham LJ, Rainwater DL, Comuzzie AG, et al. 2007. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 39:1208–1216. [DOI] [PubMed] [Google Scholar]
- Gregori S, Goudy KS, Roncarolo MG. 2012. The cellular and molecular mechanisms of immuno-suppression by human type 1 regulatory T cells. Front Immunol. 3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KD, Irizarry RA, Wu Z. 2012. Removing technical variability in RNA-seq data using conditional quantile normalization. Biostatistics 13:204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeppner MP, Lundquist A, Pirun M, Meadows JRS, Zamani N, Johnson J, Sundström G, Cook A, FitzGerald MG, Swofford R, et al. 2014. An improved canine genome and a comprehensive catalogue of coding genes and non-coding transcripts. PLoS One 9:e91172.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421:182–187. [DOI] [PubMed] [Google Scholar]
- Hong M-G, Myers AJ, Magnusson PKE, Prince JA. 2008. Transcriptome-wide assessment of human brain and lymphocyte senescence. PLoS One 3:e3024.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopes B, Rimbault M, Liebers D, Ostrander E, Sutter N. 2012. The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm Genome. 23:780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S. 2011. Integrated weighted correlation network analysis of mouse liver gene expression data. Weighted network analysis: applications in genomics and systems biology. New York: Springer Book; p. 421. [Google Scholar]
- Ishioka K, Hosoya K, Kitagawa H, Shibata H, Honjoh T, Kimura K, Saito M. 2007. Plasma leptin concentration in dogs: effects of body condition score, age, gender and breeds. Res Vet Sci. 82:11–15. [DOI] [PubMed] [Google Scholar]
- Ishioka K, Soliman MM, Sagawa M, Nakadomo F, Shibata H, Honjoh T, Hashimoto A, Kitamura H, Kimura K, Saito M. 2002. Experimental and clinical studies on plasma leptin in obese dogs. J Vet Med Sci. 64:349–353. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Sawyer S, Meyer M. 2012. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 40:e3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood TBL. 1977. Evolution of ageing. Nature 270:301–304. [DOI] [PubMed] [Google Scholar]
- Kirkwood TBL. 2005. Understanding the odd science of aging. Cell 120:437–447. [DOI] [PubMed] [Google Scholar]
- Kraus C, Pavard S, Promislow DEL. 2013. The size–life span trade-off decomposed: why large dogs die young. Am Nat. 181:492–505. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J, Torrice C, Ramsey MR, Kovalev GI, Al-Regaiey K, Su L, Sharpless NE. 2004. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 114:1299–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9(1): 13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Matheny M, Nicolson M, Türner N, Scarpace PJ. 1997. Leptin gene expression increases with age independent of increasing adiposity in rats. Diabetes 46:2035–2039. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DBKM, Clamp MCJ, Kulbokas EJ, 3rd, Zody MCME, Xie XBM, Wayne RK, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–819. [DOI] [PubMed] [Google Scholar]
- Linton PJ, Dorshkind K. 2004. Age-related changes in lymphocyte development and function. Nat Immunol. 5:133–139. [DOI] [PubMed] [Google Scholar]
- Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Ibrahim JG, Thomas NE, Sharpless NE. 2009. Expression of p16INK4a in peripheral blood T-cells is a biomarker of human aging. Aging Cell 8:439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, Kroemer G. 2013. The hallmarks of aging. Cell 153:1194–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XH, Muzumdar R, Yang XM, Gabriely I, Berger R, Barzilai N. 2002. Aging is associated with resistance to effects of leptin on fat distribution and insulin action. J Gerontol A Biol Sci Med Sci. 57:B225–B231. [DOI] [PubMed] [Google Scholar]
- MacNulty DR, Smith DW, Mech LD, Eberly LE. 2009. Body size and predatory performance in wolves: is bigger better? J Anim Ecol. 78:532–539. [DOI] [PubMed] [Google Scholar]
- MacNulty DR, Smith DW, Vucetich JA, Mech LD, Stahler DR, Packer C. 2009. Predatory senescence in ageing wolves. Ecol Lett. 12:1347–1356. [DOI] [PubMed] [Google Scholar]
- McGowan P, Sasaki A, D'Alessio AC, Dymov S, Labonté B, Szyf M, Turecki G, Meaney MJ. 2009. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 12:342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mech LD. 1999. Alpha status, dominance, and division of labor in wolf packs. Can J Zool. 77:1196–1203. [Google Scholar]
- Mech LD. 2003. Wolf social ecology In: Mech LD, Boitani L, editors. Wolves—behavior, ecology, and conservation. Chicago (IL: ): The University of Chicago Press; p. 1–34. [Google Scholar]
- Mejias A, Ramilo O. 2014. Transcriptional profiling in infectious diseases: ready for prime time? J Infect. 68:S94–S99. [DOI] [PubMed] [Google Scholar]
- Miller GE, Chen E, Fok AK, Walker HA, Lim A, Nicholls EF, Cole SW, Kobor M. 2009. Low early-life social class leaves a biological residue manifested by decreased glucocorticoid and increase proinflammatory signaling. Proc Natl Acad Sci U S A. 106:14716–14721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobbs CV. 1998. Leptin and aging In: Mobbs CV, Hof PR, editors. Functional endocrinology of aging. Basel (Switzerland: ): Karger; p. 228–240. [Google Scholar]
- Molnar B, Fattebert J, Palme R, Ciucci P, Betschart B, Smith DW, Diehl P-A. 2015. Environmental and intrinsic correlates of stress in free-ranging wolves. PLoS One 10:e0137378.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. 2006. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124:315–329. [DOI] [PubMed] [Google Scholar]
- Murphy MLM, Slavich GM, Rohleder N, Miller GE. 2013. Targeted rejection triggers differential pro- and anti-inflammatory gene expression in adolescents as a function of social status. Clin Psychol Sci. 1:30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussey DH, Froy H, Lemaitre J-F, Gaillard J-M, Austad SN. 2013. Senescence in natural populations of animals: widespread evidence and its implications for bio-gerontology. Ageing Res Rev. 12:214–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani N, Zebedee Z, Huot TJG, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. 2001. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature 409:1067–1070. [DOI] [PubMed] [Google Scholar]
- Packard JM. 2003. Wolf behavior: reproductive, social and intelligent In: Mech LD, Boitani L, editors. Wolves—behavior, ecology, and conservation. Chicago (IL) and London: The University of Chicago Press; p. 35–65. [Google Scholar]
- Palacios MG, Winkler DW, Klasing KC, Hasselquist D, Vleck CM. 2011. Consequences of immune system aging in nature: a study of immunosenescence costs in free-living Tree Swallows. Ecology 92:952–966. [DOI] [PubMed] [Google Scholar]
- Pence DB, Windberg LA, Pence BC, Sprowls R. 1983. The epizootiology and pathology of sarcoptic mange in coyotes, Canis latrans, from South Texas. J Parasitol. 69:1100–1115. [PubMed] [Google Scholar]
- Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, Reinmaa E, Sutphin GL, Zhernakova A, Schramm K, et al. 2015. The transcriptional landscape of age in human peripheral blood. Nat Commun. 6:8570.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet D, Belleville A, t'Kint de Roodenbeke C, Roborel de Climens A, Ben Simon E, Merle-Beral H, Callet-Bauchu E, Salles G, Sabatier L, Delic J, et al. 2008. Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B-chronic lymphocytic leukemia. Blood 111:2388–2391. [DOI] [PubMed] [Google Scholar]
- Powell ND, Sloan EK, Bailey MT, Arevalo JMG, Miller GE, Chen E, Kobor MS, Reader BF, Sheridan JF, Cole SW. 2013. Social stress up-regulates inflammatory gene expression in the leukocyte transcriptome via beta-adrenergic induction of myelopoiesis. Proc Natl Acad Sci U S A. 110:16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman AA, Karim AN, Hamid ANA, Harun R, Wan Ngah WZ. 2013. Senescence-related changes in gene expression of peripheral blood mononuclear cells from octo/nonagenarians compared to their offspring. Oxid Med Cell Longev. 2013:14.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, et al. 2007. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood 109:2066–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J, Kull M, Peterson H, Hansen J, Vilo J. 2007. g: profiler—a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 35:W193–W200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelle W. 2015. Psych: procedures for personality and psychological research. Evanston (IL: ): Northwestern University. [Google Scholar]
- Ricklefs RE. 2010. Life-history connections to rates of aging in terrestrial vertebrates. Proc Natl Acad Sci U S A. 107:10314–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollo CD. 2002. Growth negatively impacts the life span of mammals. Evol Dev. 4:55–61. [DOI] [PubMed] [Google Scholar]
- Runcie DE, Wiedmann RT, Archie EA, Altmann J, Wray GA, Alberts SC, Tung J. 2013. Social environment influences the relationship between genotype and gene expression in wild baboons. Philos Trans R Soc Lond Ser B Biol Sci. 368:20120345.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Kaarniranta. K. 2010. Insulin/IGF-1 paradox of aging: regulation via AKT/IKK/NF-κB signaling. Cell Signal. 22:573–577. [DOI] [PubMed] [Google Scholar]
- Sánchez-Rodrı´guez M, Garcı´a-Sánchez A, Retana-Ugalde R., Mendoza-Núñez VcM 2000. Serum leptin levels and blood pressure in the overweight elderly. Arch Med Res. 31:425–428. [DOI] [PubMed] [Google Scholar]
- Sapolsky RM. 2004. Social status and health in humans and other animals. Annu Rev Anthropol. 33:393–418. [Google Scholar]
- Sapolsky RM. 2005. The influence of social hierarchy on primate health. Science 308:648–652. [DOI] [PubMed] [Google Scholar]
- Schwarz F, Springer SA, Altheide TK, Varki NM, Gagneux P, Varki A. 2016. Human-specific derived alleles of CD33 and other genes protect against postreproductive cognitive decline. Proc Natl Acad Sci U S A 113:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Ersmark E, Palkopoulou E, Dalén L. 2015. Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr Biol. 25:1515–1519. [DOI] [PubMed] [Google Scholar]
- Smith DW, Stahler DR, Albers E, McIntyre R, Metz M, Irving J, Raymond R, Anton C, Cassidy-Quimby K, Bowersock N. 2010. Yellowstone wolf project, Annual Report. Yellowstone National Park (WY: ): National Park Service. [Google Scholar]
- Smith DW, Stahler DR, Stahler E, Metz M, Quimby K, McIntyre R, Ruhl C, Martin H, Kindermann R, Bowersock N, et al. 2012. Yellowstone wolf project, Annual report. Yellowstone National Park (WY: ): National Park Service. [Google Scholar]
- Snyder-Mackler N, Somel M, Tung J. 2014. Shared signatures of social stress and aging in peripheral blood mononuclear cell gene expression profiles. Aging Cell 13:954–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahler DR, MacNulty DR, Wayne RK, vonHoldt B, Smith DW. 2013. The adaptive value of morphological, behavioural and life-history traits in reproductive female wolves. J Anim Ecol. 82:222–234. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter NB, Bustamante CD, Chase K, Gray MM, Zhao K, Zhu L, Padhukasahasram B, Karlins E, Davis S, Jones PG, et al. 2007. A single IGF1 allele is a major derterminant of small size in dogs. Science 316:112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann O, Shapiro B, Cui P, Schuenemann VJ, Sawyer SK, Greenfield DL, Germonpré MB, Sablin MV, López-Giráldez F, Domingo-Roura X, et al. 2013. Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science 342:871–874. [DOI] [PubMed] [Google Scholar]
- Tung J, Barreiro LB, Johnson ZP, Hansen KD, Michopoulos V, Toufexis D, Michelini K, Wilson ME, Gilad Y. 2012. Social environment is associated with gene regulatory variation in the rhesus macaque immune system. Proc Natl Acad Sci U S A. 109:6490–6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J, Zhou X, Alberts SC, Stephens M, Gilad Y. 2015. The genetic architecture of gene expression levels in wild baboons. eLife 4: e04729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velavan T, Ojurongbe O. 2011. Regulatory T cells and parasites. J Biomed Biotechnol. 2011:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, Stahler DR, Smith DW, Earl DA, Pollinger JP, Wayne RK. 2008. The genealogy and genetic viablity of reintroduced Yellostone grey wolves. Mol Ecol. 17:252–274. [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Stahler DR, Bangs EE, Smith DW, Jimenez MD, Mack CM, Niemeyer CC, Pollinger JP, Wayne RK. 2010. A novel assessment of population structure and gene flow in grey wolf populations of the Northern Rocky Mountains of the United States. Mol Ecol. 19:4412–4427. [DOI] [PubMed] [Google Scholar]
- Wang J. 2011. COANCESTRY: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resour. 11:141–145. [DOI] [PubMed] [Google Scholar]
- Weaver ICG, Meaney MJ, Szyf M. 2006. Maternal care effects on hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 103:3480–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney AR, Diehn M, Popper SJ, Alizadeh AA, Boldrick JC, Relman DA, Brown PO. 2003. Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A. 100:1896–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson EO. 2000. Sociobiology: the new synthesis. Cambridge and London: The Belknap Press of Harvard University Press. [Google Scholar]
- Zhou X, Stephens M. 2012. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 44:821–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.