Abstract
Transgenic mice have provided invaluable information about gene function and regulation. However, because of marked differences between rodents and primates, some areas of human biology such as early embryonic development, aging, and maternal–fetal interactions would be best studied in a nonhuman primate model. Here, we report that gene transfer into rhesus monkey (Macaca mulatta) preimplantation embryos gives rise to transgenic placentas that express a reporter transgene (eGFP). Blastocysts resulting from culture of in vitro fertilized ova were transduced with a self-inactivating lentiviral vector and transferred into recipient females. One twin and one singleton pregnancy were produced from a single stimulation cycle, and one live rhesus monkey was born from each pregnancy. Placentas from all conceptuses showed expression of the transgene as detected by reverse transcription–PCR, ribonuclease protection assay, direct epifluorescence, immunohistochemistry, and Western blot analysis. Integration in somatic tissues of the offspring was not detected. A maternal immune response to the xenogeneic placental antigen was shown by the presence of anti-GFP antibodies in peripheral blood of the recipient females by day 99 of gestation (term = 165 days). These results demonstrate that transgene expression during gestation is compatible with successful pregnancy in nonhuman primates and provides an approach that could be broadly applicable to the development of novel models for primate biomedical research.
Pronuclear injection of mouse zygotes is an efficient and practical means of producing transgenic mice. However, other mammalian species have not achieved similar success, and transgenic offspring in these species have proven laborious to produce. Production of transgenic monkeys faces significant difficulties because of constraints on resources and practical limitations on nonhuman primate assisted reproductive technologies and embryology. Because of these difficulties, we have explored alternative methodologies to optimize the production of genetically altered rhesus monkey preimplantation embryos. Oncoretroviruses using internal promoters to drive transgene expression have achieved transgene insertion in mice (1), cattle (2, 3), and monkeys (4). However, offspring resulting from these studies show significant transgene silencing, and transgenes delivered by these vectors generally remain nonfunctional in these animals. This problem has been addressed, in part, by the production of self-inactivating (SIN) retroviral vectors (5). SIN vectors have a deletion within the U3 region of the 3′ long terminal repeat (LTR) so that during the viral life cycle the deletion is transferred to the 5′ LTR. Cis-acting elements within this region have been shown to contribute to vector silencing, and mutation of these sequences ameliorates the silencing event (6).
Alternative integrating vectors may provide useful tools for transgene delivery while avoiding vector silencing. Lentiviral vectors based on HIV-1 (7, 8) can retain expression in multipotent cells (hematopoietic and embryonic stem cells) and their differentiated derivatives (9, 10). Other elements that have been incorporated into lentiviral vectors may enhance transgene expression by posttranscriptional mechanisms. The inclusion of an intron in transgene vectors (11–13) and other posttranscriptional regulatory elements such as the woodchuck hepatitis virus posttranscriptional regulatory element have been demonstrated to increase transgene expression (14). Thus, these vectors may be well suited for transgene delivery to preimplantation primate embryos.
We chose a SIN lentiviral vector (15, 16) for delivery of transgenes into preimplantation rhesus monkey embryos because of the stability and longevity of expression in other multipotent cell types. For these studies, we used a vesicular stomatitis virus G protein pseudotyped vector. The vector used the intron containing human elongation factor-1α (EF1α) promoter, directing expression of enhanced green fluorescent protein (eGFP) (SIN-EF-GFP-W) (17). We established several pregnancies from the nonsurgical transfer of rhesus blastocysts injected with lentiviral vectors and obtained two live rhesus infants from these trials. Placentas from all pregnancies as well as other extraembryonic tissues (i.e., amnion and umbilical cord) showed expression of eGFP. The efficient expression of placental transgenes in vivo should provide opportunities to gain insights into primate placental development and function during pregnancy.
Materials and Methods
Lentiviral Vector Preparation.
Human embryonic kidney 293T cells and human fibrosarcoma HT1080 cells (ATCC CCL-121) were grown in DMEM (Life Technologies, Grand Island, NY) supplemented with 10% heat-inactivated FBS/2 mM l-glutamine/50 units/ml penicillin/50 μg/ml streptomycin (Life Technologies). Vesicular stomatitis virus envelope glycoprotein G (VSV-G)-pseudotyped SIN-EF-GFP-W vector particles were prepared by transiently transfecting the SIN-EF-GFP-W transfer vector plasmid (15 μg) (17), the packaging plasmid pCMVΔR8.91 (10 μg) (12), and the VSV-G envelope plasmid pMD.G (5 μg) (11) into subconfluent 293T cells by calcium phosphate coprecipitation as described (17). Vector-conditioned medium was collected 24 h posttransfection, centrifuged at 2,000 × g to remove cellular debris, and filtered through a 0.45-μm-pore-sized filter (Nalgene) before being aliquoted and frozen at −80°C. To determine vector titer, an aliquot of the vector preparation was thawed and serial dilutions were added in the presence of 6 μg/ml polybrene (Sigma) to 2 × 105 HT1080 cells that had been seeded in 12-well plates 8 h earlier. Fresh medium was added after 4 h of transduction, and 72 h later the relative end-point vector titer (transducing units/ml; TU/ml) was determined by flow cytometric analysis (17).
In Vitro Fertilization, Embryo Culture, and Transfer.
Mature female rhesus monkeys were hyperstimulated with 60 units/day human recombinant follicle-stimulating hormone (Ares-Serono, Randolph, MA) for 7–9 days, followed by 1,000 units of human recombinant chorionic gonadotropin (Ares-Serono) 27 h before laparoscopic oocyte retrieval. Semen preparation from electroejaculated males and in vitro fertilization were done as described (18). Embryos were cultured in sequential human embryo culture medium G1.1 5% defined FBS (HyClone) and G2.2 2% serum (IVF Science Scandinavia, Gothenburg, Sweden) (19). Day 7–8 blastocysts were injected with 1 × 106 green fluorescent protein (GFP) TU/ml of vector-conditioned medium supplemented with 8 μg/ml polybrene in the blastocoel by using an Eppendorf Transjector micromanipulator. After 18–24 h of culture, blastocysts were transferred to naturally cycling female rhesus monkeys via nonsurgical, transcervical uterine transfer (20). Pregnancy was confirmed by assay for serum CG and monitored by ultrasound examination. All procedures were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and under the approval of the University of Wisconsin Graduate School Animal Care and Use Committee.
Cell Culture.
Cytotrophoblasts were isolated by enzymatic digestion of the placenta. The placentas of both live births were dispersed by trypsin/DNase I digestion as described (21). Isolated placental cells were incubated in DMEM supplemented with 25 mM Hepes and 10% FBS. Cells were imaged under phase-contrast microscopy, and epifluorescence images were captured by using 1-sec exposures.
Transgene Detection.
Tissues were collected and stored at 4°C in RNA Later (Ambion, Austin, TX) until homogenization. Total RNA and genomic DNA were extracted by using RNA-STAT 60 (Tel-Test, Friendswood, TX). One microgram of total RNA was reverse-transcribed by using the Geneamp kit (Perkin–Elmer). PCR using primers that spanned the EF1α promoter (22) intron (EF1α.exon1.up: CGC AAC GGG TTT GCC GCC AGA ACA C; GFPintron.down: G TCG TCC TTG AAG AAG ATG GTG CGC) was performed on reverse transcription (RT) reactions to amplify transgene cDNA. Genomic DNA was amplified by using the same primers supplemented with 2X Master Amp Enhancer (1.5 mM MgCl2) (Epicentre Technologies, Madison, WI). Ribonuclease protection assays were performed essentially as we have described (23). A 300-bp fragment of eGFP was PCR-amplified (5′-GTG ACC ACC CTG ACC TAC GGC GTG C-3′ and 5′-CTT GTC GGC CAT GAT ATA GAC GTT G-3′) and subcloned into a vector containing flanking T7 and T3 promoters (pBlue eGFP300). pBlue eGFP300 and pTRI′-actin-125-Human (internal control template) (Ambion) were linearized, and 32P-CTP-labeled antisense RNA probes were transcribed in vitro by using T7 RNA polymerase (Ambion). RNase protection assays were performed by using an RPA III kit (Ambion).
Immunochemistry.
Tissue was collected and fixed in 2% paraformaldehyde for 4 h and then infused with 9% sucrose for 4 h and 20% sucrose overnight. Tissue was then embedded in OCT, frozen in liquid nitrogen, and stored at −80°C until sectioning. Ten-micrometer frozen sections were cut and fixed for 5 min in acetone. Endogenous peroxidase activity was quenched by a 10-min incubation with 0.5% hydrogen peroxide, and slides were blocked with 2% BSA/Tris-buffered saline (20 mM Tris, 500 mM NaCl). Immunostaining was achieved as we have described (24) by using a mouse mAb against GFP (3 μg/ml; CLONTECH). Bound primary antibody was incubated with a biotinylated horse anti-mouse antibody and subsequently bound with Vectastain ABC peroxidase complex (Vector Laboratories). Localization of positive cells was done by using the NovaRed substrate (Vector Laboratories).
Western Blots.
Total isolated protein (250 μg) was fractionated on a 10% polyacrylamide gel, blotted onto a poly(vinylidene difluoride) membrane, blocked with 0.2% nonfat dry milk TTBS (Tris-buffered saline, 0.1% Tween 20) overnight, and probed with a rabbit anti-GFP peptide antibody (CLONTECH) directly conjugated by horseradish peroxidase (1:500). The ECL Plus kit (Amersham Pharmacia) was used to detect antibodies against eGFP. To identify maternal antibodies directed against eGFP, 500 ng of recombinant eGFP protein (CLONTECH) was fractionated on a 10% polyacrylamide gel, blotted, and cut into individual strips. Each strip was blocked and incubated with individual rhesus serum samples (1:100) for 2 h. Secondary rabbit anti-monkey IgG (Sigma), directly conjugated with alkaline phosphatase, was incubated with the individual strips. Positive binding was visualized by enhanced chemiluminescence by using the Immuno-Star chemiluminescent system (Bio-Rad).
Results
Oocytes were collected from gonadotropin-stimulated female rhesus monkeys, fertilized in vitro, and cultured for 7–9 days to the blastocyst stage. Recombinant, self-inactivating lentiviral vector was microinjected into the blastocoel of 14 embryos produced from a single stimulation cycle. The embryos were cultured for an additional 18–24 h to allow reexpansion of the blastocyst.
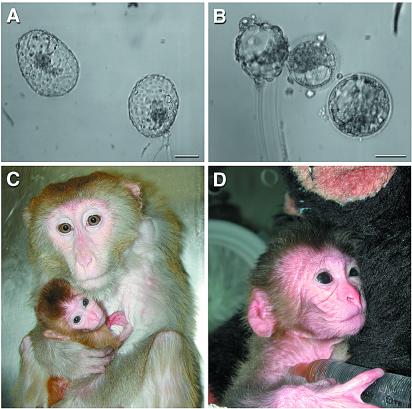
The embryos then were transferred to six naturally cycling females via nonsurgical, transcervical blastocyst transfer (20). Two females that each received two blastocysts (Fig. 1 A and B) on days 4 and 6 after the luteinizing hormone peak became pregnant with twins and a single fetus, respectively. The female carrying the singleton pregnancy was supplemented with 4.8 mg of progesterone injected daily, and the other female received none. The twin pregnancy was delivered by cesarean section on day 153 postinsemination, with one male infant, animal A (Fig. 1C), born alive. A female fetus, animal B, apparently had died in utero and was severely autolysed. The surviving male weighed 326 g and had a monodiscoid placenta (macaques usually have two discs) weighing 130 g. The singleton pregnancy, another male, animal C (Fig. 1D), was delivered by cesarean section on day 159 postinsemination. He weighed 532 g and had a bidiscoid placenta weighing 168 g. Two other pregnancies ensued after transfer of injected embryos from an independent stimulation. Both females carrying these pregnancies received supplementary progesterone. When the pregnancies were determined not to be viable by ultrasound examination of the uterus, the progesterone was removed and, 1 to 2 days later, the uterine contents were expelled naturally and collected. Histopathological examination showed that the placenta was forming but fetal tissues were not identified.
Figure 1.
SIN lentiviral vector-transduced rhesus monkey blastocysts and infants. In vitro produced blastocysts that contributed to the twin and singleton infants, respectively (A and B). Shown are the surviving twin (infant A) with dam (C) and male singleton (infant C) in a perinatal incubator holding a 10% dextrose syringe with nipple (D). He subsequently was returned to his dam as well. Embryos were exposed to greater than 1 sec of continuous epifluorescent excitation at 488 nm but were not yet expressing robust amounts of eGFP. (Bars = 100 μm.)
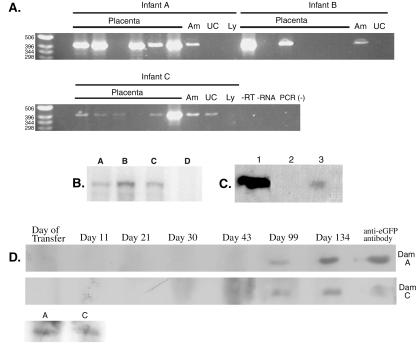
Transgene expression first was assayed by RT-PCR in all available tissues (Fig. 2A). Primers were designed to span intron 1 of the EF1α promoter so that transcribed mRNA and integrated vector DNA could be distinguished easily. Expression of eGFP mRNA could be detected in the chorionic disk of all placentas by RT-PCR and ribonuclease protection assay (Fig. 2B), with some placental biopsies having low or nondetectable levels of eGFP mRNA and some biopsies showing very robust expression. The amnion of all three animals showed transgene expression. The umbilical cord showed transgene expression only in animal C.
Figure 2.
Transgene expression and the subsequent maternal humoral response. (A) RT-PCR in the three animals; results are shown for six placental biopsies along with amnion (Am), umbilical cord (UC), and cord lymphocytes (Ly). (B) Ribonuclease protection assay for eGFP mRNA. Lanes: A–C, placental RNA from animals A, B, and C; D, nontransgenic placenta. (C) Western blot for GFP. Lanes: 1, 100 ng recombinant eGFP; 2, control placenta; 3, 250 μg of total protein from placenta A. (D) Maternal serum samples from dam A and C as well as fetal cord blood (A and C, bottom blot) were assayed for antibodies directed against placental eGFP as detected by Western blot analysis. A positive control is shown that uses a mouse monoclonal anti-GFP antibody.
Integrated vector DNA was assayed by using the same primers as for the RT-PCR experiments. As expected, the amplified fragment was approximately 1 kb longer because of the inclusion of the intron (data not shown). Placental biopsies that had detectable levels of eGFP also showed genomic integration of the transgene. Transgene insertion in cells collected from the infants (peripheral blood lymphocytes and hair follicles) was not detectable by PCR analysis in the limited samples available.
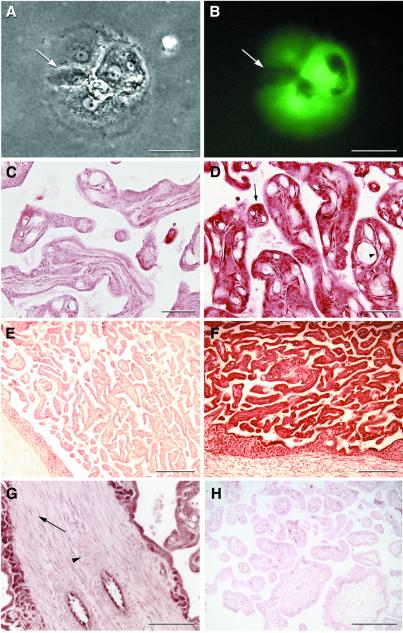
eGFP protein expression was detected in the placenta of all three fetuses. Directly after the delivery of the infants, the placentas from both live births were dispersed by trypsin/DNase I treatment, plated, and cultured to determine placental transgene expression. Cytotrophoblasts obtained from the dispersed placentas expressed robust amounts of eGFP as seen by direct epifluorescence immediately after the dispersion. Fluorescence was sustained during culture as cytotrophoblasts migrated, fused, and differentiated into syncytia (Fig. 3 A and B) (21). Nonexpressing trophoblasts could be seen fusing with transgenic colonies and ultimately sharing cytoplasmic eGFP.
Figure 3.
Transgene protein expression in placenta. Phase-contrast (A) and epifluorescent images (B) of cultured trophoblasts from placenta A are shown. Arrow indicates a nonexpressing trophoblast that has fused recently with a transgenic syncytium. (C and E) Bright-field negative controls for D and F. Anti-GFP immunohistochemistry of placenta A (D) frozen sections reveals widespread eGFP expression. Arrow indicates an expressing trophoblast, and arrowhead shows an expressing fetal villous endothelial cell. (F) Low-power view of placenta C showing eGFP expression throughout the trophoblastic shell. (G) Immunohistochemical localization of eGFP in placenta A showing the absence of expression in placental stroma and vascular smooth muscle. Arrow shows nonexpressing stromal cells, and arrowhead shows nonexpressing vascular smooth muscle cells. (H) Photomicrograph of a nontransgenic control placenta probed with the same anti-GFP antibody. [Bars = 50 μm (A and B), 40 μm (C and D), 200 μm (E, F, and H), and 100 μm (G).]
To investigate the expression pattern of eGFP in situ, we performed immunohistochemical studies on frozen tissue sections by using an anti-GFP mAb. Indirect immunostaining of placental sections revealed trophoblast staining in villi of animal A (Fig. 3 C and D) and in animal C (Fig. 3 E and F), which is consistent with the expression in cultured dispersed trophoblasts. However, not all trophoblasts showed expression. Expression was seen in fetal villous endothelial cells (Fig. 3D), but villous stroma and vessel smooth muscle within the placenta did not express eGFP (Fig. 3G). Placental eGFP protein expression also was shown by Western blot of total placental protein (Fig. 2C).
Pregnancy induces tolerance to paternal antigens by unknown mechanisms. A pregnant female produces a nonproductive cellular (25) and humoral (26) immune response to these antigens. We were interested to see whether the surrogate female carrying a transgenic fetus would produce an immune response to the transgene. We were able to detect rhesus antibodies directed against eGFP in peripheral blood in surrogate females carrying transgenic fetuses. Antibodies specific for recombinant eGFP were first detected by approximately day 99 in both pregnant animals and continued throughout pregnancy (Fig. 2D). Anti-GFP IgG, presumably from passive transport of maternal IgG across the placenta, also was detected in umbilical cord serum from both live infants (Fig. 2D).
Discussion
In widely used rodent models, organization and development of embryonic and extraembryonic tissues vary significantly from that of the human, whereas early development, implantation, and placentation are very similar in human and nonhuman primates (27, 28). To better understand human placental development and function, we wished to use the rhesus monkey as an in vivo model for molecular studies of these events. To this end, we have produced two live rhesus monkeys that expressed a transgene in the placenta and extraembryonic membranes. Expression of the transgene was shown by mRNA and protein production. This demonstrates that expression of transgenic protein is compatible with successful primate pregnancy after transduction of preimplantation embryos. Furthermore, the efficiency of transduction in blastocysts with a SIN Lentiviral vector as well as the efficiency of obtaining live births from transduced embryos indicates that these methods will be powerful tools for studying primate placental physiology in vivo.
Blastocyst-stage genetic manipulation has not been reported previously in primates. A significant limitation on the feasibility of this approach has been the difficulties with the transfer of blastocyst-stage embryos in rhesus monkeys, the most widely used nonhuman primate model. Only recently have parameters of successful implantation (e.g., timing of embryo transfer) been examined (20, 29) and the birth of rhesus monkeys after transcervical transfer of blastocyst-stage embryos been achieved. We have shown in the current studies that transcervical blastocyst transfer has proven to be a reliable means of obtaining live rhesus infants even after extensive manipulation in vitro such as micromanipulation, viral genetic modification, and fluorescent imaging. We propose that nonsurgical blastocyst transfer may be valuable for further genetic studies such as the production of rhesus embryonic stem cell (30) chimeras to study primate cell fates during development.
A recent study using an oncoretroviral vector in rhesus oocytes demonstrated integration of vector DNA, but the live infant did not express the transgene protein in extraembryonic or fetal/neonatal cells (4). The most current lentiviral vectors used in our studies may represent a significant improvement over the original retroviruses used in previous reports. The lentiviral vectors used in this study incorporate SIN features that allow transcription to be driven primarily by internal promoters and have incorporated posttranscriptional regulatory elements, including an intron in the transgene cassette and the woodchuck hepatitis virus posttranscriptional regulatory element (17). Viral promoters (e.g., cytomegalovirus IE) are used to overexpress transgenes because of their small size and high expression in vitro. However, viral promoters are problematic in vivo and often are not appropriate for developmental studies. The use of mammalian promoters appropriate for the development context of interest would be desirable. The human EF1α promoter is a strong mammalian promoter that is expressed well in tissues studied here. Further studies with trophoblast-specific and developmentally regulated promoters are possible for use in vivo in the primate placenta (e.g., chorionic gonadotropin-α, placental GH, or HLA-G).
The success of rhesus pregnancies in which a nonmammalian transgene is expressed in the placenta is of general interest in the context of maternal–fetal immune interactions. Although it expresses paternal antigens, the fetus is not eliminated by the maternal immune system during mammalian gestation. The placenta undoubtedly plays a central role here, although the precise mechanisms by which this maternal tolerance of the fetal semiallograft is generated remain poorly understood. The hemochorial placenta of the rhesus macaque has close homology with the human, including organization into chorionic villi, a syncytial trophoblast layer in direct contact with maternal blood, and extravillous trophoblasts that invade the maternal endometrium and play a role in vascular reorganization in response to pregnancy (28). Nonprimate species have significantly different organization of the maternal–fetal interface.
Although it has long been recognized that antibodies directed against paternal MHC alleles are present in maternal serum (26), these antibodies most likely are produced in response to fetal lymphocytes found in maternal circulation. The placenta does not express these classical MHC class I alleles in the trophoblasts exposed to the maternal immune system (see below); thus, an antipaternal response is not equivalent to an antiplacental response. We therefore were surprised to identify antibodies against eGFP in the serum of rhesus monkey surrogate dams carrying transduced embryos. Because at this time we have not yet detected expression of eGFP in lymphocytes of the offspring, the antibodies generated most likely indicate a response against eGFP expressed in the placental trophoblasts. The presence of this humoral antibody response during the middle third of pregnancy in the rhesus indicates that transgene expression was initiated well before term and was sustained throughout the remainder of gestation, persisting in the placenta at delivery. Based on these results, we propose that the detection of fetal transgene-specific antibodies in maternal peripheral circulation may be a reliable preterm indicator of transgenesis in nonhuman primates. The importance of the maternal immune response in pregnancy is still controversial. In mice, maternal B cells directed toward fetal-specific MHC class I molecules are deleted during gestation (31). Although we did not directly measure GFP-specific B cell numbers, total anti-GFP antibody titer was sustained over the ≈2 months of pregnancy where antibodies were detectable. Thus, there was no evidence for B cell deletion in this primate model.
This model now provides a way to investigate directly the importance of the expression of selected MHC loci during pregnancy. Although the specific role(s) of MHC class I and II molecules in regulating the maternal immune response are not well defined, their likely importance at the maternal–fetal barrier is evidenced by the dearth of classical MHC class I and II molecule expression in the primate placenta and the preferential expression of nonpolymorphic MHC class I molecules: these include HLA-G (32, 33) and HLA-E (34) in the human placenta and Mamu-AG (24, 35, 36) and Mamu-E (37) in the rhesus placenta. Overexpression of classical polymorphic MHC alleles (or of antisense nonpolymorphic mRNAs) in the placenta, or other immune modulators or cytokines, could aid in the understanding of the role(s) of the maternal immune system at the maternal–fetal interface. Given the robust transgene expression in the placenta, these methods could be very useful for studying placental physiology outside the arena of immune regulation. Placental dysfunction contributes significantly to maternal and fetal morbidity, and the molecular bases of the placental components of pathologies of pregnancy (e.g., preeclampsia, intrauterine growth restriction) are not well understood. Accordingly, therapies directed at these problems are lacking, at least in part, because of inadequate models for physiological studies. The rhesus monkey would be an excellent model for studying the molecular basis of human placental dysfunction and exploring novel therapeutic approaches in this crucial organ.
Acknowledgments
We thank Ares Advanced Technology for generously providing r-hFSH and r-hCG for these studies. We are grateful to Didier Trono for providing initial lentiviral vectors and Drs. K. M. Downs and R. R. Magness for critical review of the manuscript. We also thank Fritz Wegner and Steve Jacoris for performing hormone assays and the veterinary, pathology, and animal care staff for technical support. This work was supported by National Institutes of Health Grants HD26458 and RR14040 (to T.G.G.), RR00167 (Wisconsin Regional Primate Research Center), and HL65519 (to R.G.H.). This is publication 40-029 of the Wisconsin Regional Primate Research Center.
Abbreviations
- SIN
self-inactivating
- EF1α
elongation factor-1α
- eGFP
enhanced green fluorescent protein
- RT
reverse transcription
References
- 1.Soriano P, Cone R D, Mulligan R C, Jaenisch R. Science. 1986;234:1409–1413. doi: 10.1126/science.3024318. [DOI] [PubMed] [Google Scholar]
- 2.Haskell R E, Bowen R A. Mol Reprod Dev. 1995;40:386–390. doi: 10.1002/mrd.1080400316. [DOI] [PubMed] [Google Scholar]
- 3.Chan A W, Homan E J, Ballou L U, Burns J C, Bremel R D. Proc Natl Acad Sci USA. 1998;95:14028–14033. doi: 10.1073/pnas.95.24.14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan A W S, Chong K Y, Martinovich C, Simerly C, Schatten G. Science. 2001;291:309–312. doi: 10.1126/science.291.5502.309. [DOI] [PubMed] [Google Scholar]
- 5.Hawley R G, Covarrubias L, Hawley T, Mintz B. Proc Natl Acad Sci USA. 1987;84:2406–2410. doi: 10.1073/pnas.84.8.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osborne C S, Pasceri P, Singal R, Sukonnik T, Ginder G D, Ellis J. J Virol. 1999;73:5490–5496. doi: 10.1128/jvi.73.7.5490-5496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 8.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 9.Miyoshi H, Smith K A, Mosier D E, Verma I M, Torbett B E. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- 10.Hamaguchi I, Woods N B, Panagopoulos I, Andersson E, Mikkola H, Fahlman C, Zufferey R, Carlsson L, Trono D, Karlsson S. J Virol. 2000;74:10778–10784. doi: 10.1128/jvi.74.22.10778-10784.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brinster R L, Allen J M, Behringer R R, Gelinas R E, Palmiter R D. Proc Natl Acad Sci USA. 1988;85:836–840. doi: 10.1073/pnas.85.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi T, Huang M, Gorman C, Jaenisch R. Mol Cell Biol. 1991;11:3070–3074. doi: 10.1128/mcb.11.6.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buchman A R, Berg P. Mol Cell Biol. 1988;8:4395–4405. doi: 10.1128/mcb.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zufferey R, Donello J E, Trono D, Hope T J. J Virol. 1999;73:2886–2892. doi: 10.1128/jvi.73.4.2886-2892.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zufferey R, Dull T, Mandel R J, Bukovsky A, Quiroz D, Naldini L, Trono D. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyoshi H, Blömer U, Takahashi M, Gage F H, Verma I M. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramezani A, Hawley T S, Hawley R G. Mol Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- 18.Bavister B D, Boatman D E, Leibfried L, Loose M, Vernon M W. Biol Reprod. 1983;28:983–999. doi: 10.1095/biolreprod28.4.983. [DOI] [PubMed] [Google Scholar]
- 19.Gardner D K, Vella P, Lane M, Wagley L, Schlenker T, Schoolcraft W B. Fertil Steril. 1998;69:84–88. doi: 10.1016/s0015-0282(97)00438-x. [DOI] [PubMed] [Google Scholar]
- 20.Wolfgang, M. J., Eisele, S. G., Knowles, L., Browne, M. A., Schotzko, M. L. & Golos T. G. (2001) J. Med. Prim., in press. [DOI] [PubMed]
- 21.Golos T G, Handrow R R, Durning M, Fisher J M, Rilling J K. Endocrinology. 1992;131:89–99. doi: 10.1210/endo.131.1.1612035. [DOI] [PubMed] [Google Scholar]
- 22.Mizushima S, Nagata S. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khorram O, Garthwaite M, Grosen E, Golos T G. Fertil Steril. 2001;75:174–179. doi: 10.1016/s0015-0282(00)01658-7. [DOI] [PubMed] [Google Scholar]
- 24.Slukvin I I, Lunn D P, Watkins D I, Golos T G. Proc Natl Acad Sci USA. 2000;97:9104–9109. doi: 10.1073/pnas.97.16.9104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tafuri A, Alferink J, Möller P, Hämmerling G J, Arnold B. Science. 1995;270:630–633. doi: 10.1126/science.270.5236.630. [DOI] [PubMed] [Google Scholar]
- 26.Innes A, Cunningham C, Power D A, Catto G R D. Am J Reprod Immunol. 1989;19:146–150. doi: 10.1111/j.1600-0897.1989.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 27.O'Rahilly R, Muller F. Developmental Stages in Human Embryos. Carnegie Institution of Washington; 1987. [Google Scholar]
- 28.Benirschke K, Kaufmann P. Pathology of the Human Placenta. New York: Springer; 1990. [Google Scholar]
- 29.Nusser K D, Mitalipov S, Widmann A, Gerami-Naini B, Yeoman R R, Wolf D P. Hum Reprod. 2001;16:130–137. doi: 10.1093/humrep/16.1.130. [DOI] [PubMed] [Google Scholar]
- 30.Thomson J A, Kalishman J, Golos T G, Durning M, Harris C P, Becker R A, Hearn J P. Proc Natl Acad Sci USA. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aït-Azzouzene D, Gendron M C, Houdayer M, Langkopf A, Burki K, Nemazee D, Kanellopoulos-Langevin C. J Immunol. 1998;161:2677–2683. [PubMed] [Google Scholar]
- 32.McMaster M T, Librach C L, Zhou Y, Lim K H, Janatpour M J, DeMars R, Kovats S, Damsky C, Fisher S J. J Immunol. 1995;154:3771–3778. [PubMed] [Google Scholar]
- 33.Yelavarthi K K, Fishback J L, Hunt J S. J Immunol. 1991;146:2847–2854. [PubMed] [Google Scholar]
- 34.Wei X H, Orr H T. Hum Immunol. 1990;29:131–142. doi: 10.1016/0198-8859(90)90076-2. [DOI] [PubMed] [Google Scholar]
- 35.Boyson J E, Iwanaga K K, Golos T G, Watkins D I. J Immunol. 1997;159:3311–3321. [PubMed] [Google Scholar]
- 36.Slukvin I I, Boyson J E, Watkins D I, Golos T G. Biol Reprod. 1998;58:728–738. doi: 10.1095/biolreprod58.3.728. [DOI] [PubMed] [Google Scholar]
- 37.Boyson J E, McAdam S N, Gallimore A, Golos T G, Liu X, Gotch F M, Hughes A L, Watkins D I. Immunogenetics. 1995;41:59–68. doi: 10.1007/BF00182314. [DOI] [PubMed] [Google Scholar]