Abstract
Every human suffers through life a number of papillomaviruses (PVs) infections, most of them asymptomatic. A notable exception are persistent infections by Human papillomavirus 16 (HPV16), the most oncogenic infectious agent for humans and responsible for most infection-driven anogenital cancers. Oncogenic potential is not homogeneous among HPV16 lineages, and genetic variation within HPV16 exhibits some geographic structure. However, an in-depth analysis of the HPV16 evolutionary history was still wanting. We have analyzed extant HPV16 diversity and compared the evolutionary and phylogeographical patterns of humans and of HPV16. We show that codivergence with modern humans explains at most 30% of the present viral geographical distribution. The most explanatory scenario suggests that ancestral HPV16 already infected ancestral human populations and that viral lineages co-diverged with the hosts in parallel with the split between archaic Neanderthal-Denisovans and ancestral modern human populations, generating the ancestral HPV16A and HPV16BCD viral lineages, respectively. We propose that after out-of-Africa migration of modern human ancestors, sexual transmission between human populations introduced HPV16A into modern human ancestor populations. We hypothesize that differential coevolution of HPV16 lineages with different but closely related ancestral human populations and subsequent host-switch events in parallel with introgression of archaic alleles into the genomes of modern human ancestors may be largely responsible for the present-day differential prevalence and association with cancers for HPV16 variants.
Keywords: divergence, evolutionary medicine, Hominin evolution, host-switch, human papillomavirus, infection and cancer, sexually transmitted infection, variant, virus-host coevolution
Introduction
Virus lifestyle shapes viral population structure through essential virus life traits, such as infectivity, immunogenicity, latency, transmission rate, mutation rate, virion productivity, and duration of the infection (Sharp 2002). The interaction between the virus and the host’s immune system strongly influences viral evolution: immune memory reduces the number of susceptible hosts, and differential immune recognition and response against different pathogen phenotypes may favour selection of certain viral lineages (Holmes 2008). Finally, the evolutionary history of the host constrains further the population structure of the virus. Thus, for viruses and hosts with a long shared evolutionary history, the virus population is shaped by population-level processes of genetic drift and natural selection, in both the virus and the host populations (Holmes 2008).
Papillomaviruses (PVs) have a long common history with amniotes (Bravo and Félez-Sánchez 2015). PVs are double-stranded DNA viruses, with a circular genome of about 8 kb. More than 300 PVs have been described and above 200 of them have been retrieved from humans (Bzhalava et al. 2015). Human PVs (HPVs) infect dividing epithelia, and virtually all humans are hosts to a variable number of HPVs infections at cutaneous (Antonsson et al. 2003) but often also at mucosal sites (Ma et al. 2014). The viral infection does not kill the target cell and in most cases PVs can complete their life cycle and be maintained as a chronic, asymptomatic infection with viral genomes remaining episomally as plasmids in the infected cell (Doorbar et al. 2012). In some cases, they can cause productive, wart-like lesions or even be involved in certain cancers (Doorbar et al. 2012). The fine balance between viral replication and host immune tolerance suggests a long coexistence between PVs and their hosts, and indeed ancestral PVs probably infected the ancestral amniotes (García-Vallvé et al. 2005) and possibly all bony vertebrates (López-Bueno et al. 2016). The evolution of PVs in mammals likely started with an initial adaptive radiation event that generated a handful of viral crown groups, linked to the evolution of mammalian fur and skin glands (Gottschling et al. 2011; Bravo and Félez-Sánchez 2015). This diversification was followed by limited host-linked evolution and encompassed also incomplete lineage sorting and host-switch events (Gottschling et al. 2007; Gottschling et al. 2011). At shallower evolutionary timescale, it has been commonly assumed that HPV16 has co-diverged with modern human populations, based on certain geographical structure of HPV16 variant distribution (Bernard 1994; Cornet et al. 2012), but this hypothesis has never been rigorously tested.
Infections by anogenital HPVs are very common: >80% of all sexually active adults are infected by one of these HPVs at least once in their lifetime (Einstein et al. 2009; Chesson et al. 2014), and the time point prevalence of cervical HPVs infections in sexually active healthy women is estimated to be around 12% (Bruni et al. 2010) or even higher in younger women (Molden et al. 2016). At least 12 evolutionarily related HPVs are carcinogenic to humans (Bouvard et al. 2009), causing different fractions of anogenital and oropharyngeal cancers (Moscicki et al. 2012). HPV16 is not only the most prevalent HPV in infection-associated cancers of the cervix, vulva, vagina, anus, penis, and oropharynx (Moscicki et al. 2012) but also the most prevalent HPV infection of the genital tract occurring asymptomatically in healthy individuals (Bruni et al. 2010). Within the HPV16 lineage, distribution of genetic diversity presents certain geographic structure, with different HPV16 variants showing differential prevalence in different geographical regions (Cornet et al. 2012). Furthermore, the oncogenic potential is not homogeneous for all HPV16 variants (Schiffman et al. 2010) and varies in association with different human populations (Villa et al. 2000; Berumen et al. 2001; Burk et al. 2003; Xi et al. 2006; Zuna et al. 2011; Cornet et al. 2013). Such differential connection between viral phylogeography and oncogenic potential might indicate that the evolution of HPV16 lineage has been at least partly shaped by the differential host immune response.
In this study, we have analyzed human and virus genomic data to infer the evolutionary history of HPV16. The common understanding about the evolution of HPVs is that the modern repertoire of diverse HPVs has coevolved with modern humans as a host population, so that the current geographic distribution of HPVs reflects modern human migration dispersal patterns (Ho et al. 1993; Bernard 1994). Here, we show for the first time that differential coevolution of HPV16 lineages with different but closely related ancestral human populations together with recent host-switch events may be largely responsible for the present-day differential prevalence and association with cancers for HPV16 variants. The most parsimonious interpretation of our results requires that the ancestor of HPV16 already infected the ancestral human populations >500 thousands years ago (kya). The split between Neanderthals/Denisovans and modern human ancestor populations was mirrored by a split in the viral populations, namely HPV16A, carried by ancestral human populations, and HPV16BCD, carried by the populations of modern human ancestors in Africa. After the last out-of-Africa migration event 60–120 kya, the modern human ancestor populations that left Africa carried a reduced sample of the viral diversity in the continent: the HPV16B lineage remained in African populations but was lost by lineage sorting in those leaving the continent. The viral lineage HPV16CD gave rise in allopatry to the HPV16C variant in Africa and to the HPV16D variant outside Africa. Later, the interbreeding events between Neanderthal and Denisovan populations with modern human ancestor populations lead to a host-switch through sexual transmission of the HPV16A virus lineage from archaic populations into the modern human ancestors. The HPV16A lineage thus transmitted expanded rapidly in the new host populations and became dominant in Eurasia and in the Americas.
Results
HPV16 Phylogeography
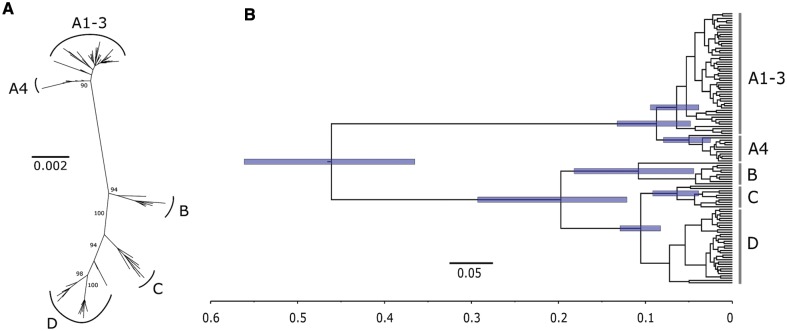
We inferred the phylogenetic relationship among all available, non-recombinant HPV16 genomes (n = 118) under maximum likelihood (ML) framework for both the full-genome alignment (supplementary material S1, Supplementary Material online) and for the coding region alignment (supplementary material S2, see also supplementary table S1, Supplementary Material online). Phylogenetic inference recovered all previously reported HPV16 variant lineages, and we retained for subsequent analysis HPV16A1-3, A4, B, C, and D lineages, all supported by >98% bootstrap values for the full-genome alignment (fig. 1A and supplementary fig. S1, Supplementary Material online). Pairwise distances under ML were significantly larger (2.28 times, 95% confidence interval [CI], 1.88–2.69; Wilcoxon–Mann–Whitney test P = 2.2e–16) for the full-genome than for the selection-filtered alignment (supplementary fig. S2, Supplementary Material online).
Fig. 1.
Phylogenetic tree of the 118 HPV16 complete genomes. (A) Unrooted ML phylogenetic tree inferred using the complete coding region alignment after excluding a total of 26 positively selected and 115 negatively selected codons. The final alignment encompassed 118 sequences, 6,093 bp and 316 distinct alignment patterns. Bootstrap support after 500 replicates is given for branches leading to the A1–3, A4, B, C, and D variant lineages. Scale bar in substitutions per nucleotide site. (B) Maximum clade credibility tree inferred for selection-filtered HPV16 genome coding region alignment under the HHS scenario, 9.6 × 10−9 [5.5 × 10−9–13.6 × 10−9] subs/site/yr as substitution rate prior and enforcing two time point calibrations: archaic Hominin divergence at 500 kya (95% CI, 400–600 kya) and recent modern human migration out-of-Africa at 90 kya (95% CI, 60–120 kya). Scale bar in millions of years ago. Node bars indicate the 95% probability intervals for the corresponding node age.
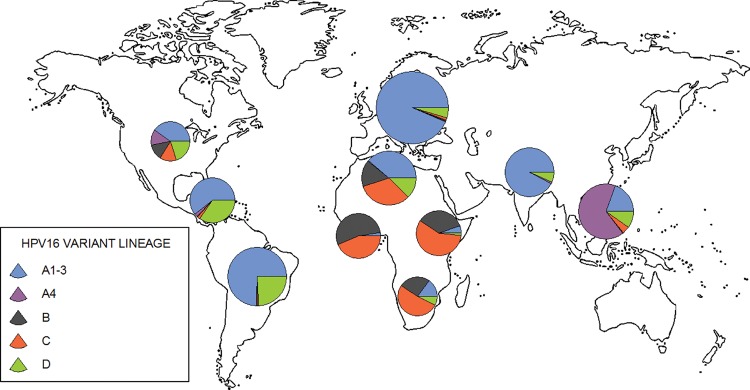
To assess HPV16 phylogeography, we analyzed a total of 1,719 HPV16 isolate cases (alignments available as supplementary materials S3–S4, Supplementary Material online) by complementing the 118 HPV16 full-genome sequences with 1,601 partial HPV16 isolate sequences spanning the LCR, the E6 oncogene, and the L2 capsid gene. A total of 1,680 global HPV16 isolate sequences with known geographical origin could be unambiguously assigned to a precise HPV16 lineage using an evolutionary placement algorithm (supplementary table S2, Supplementary Material online). Phylogeography of the 1,680 isolate cases showed that the HPV16A1-3 lineage predominated in Europe, South Asia, and Central/South America, and was also present in all other continental subgroups, albeit with very low prevalence in sub-Saharan Africa (fig. 2). HPV16A4 was the most prevalent lineage in East Asia and was also present in North America, but was virtually absent elsewhere. Variants B and C were largely restricted to Africa and were especially prevalent in Sub-Saharan Africa, although they were also observed in North America. Variant D was present in all continents, displaying low prevalence in Sub-Saharan Africa, and the highest frequency in Central/South America.
Fig. 2.
Phylogeographic distribution of the 1,680 HPV16 sequences encompassing the LCR, E6 and L2 genome loci. Each sequence was assigned to a specific HPV16 variant lineage (see the color coding for A1–3, A4, B, C, and D variants). The size of the pie charts is proportional to the number of sequences from the corresponding geographic region (supplementary table S3, supplementary material online).
To explain the evolution and diversity of HPV16 and to infer its origins, we explored the differential fit to the data of two alternative scenarios: 1) the recent-out-of-Africa (ROOA) model of exclusive codivergence for HPV16 with modern human populations and 2) the Hominin-host-switch (HHS) model, including a viral transmission between archaic and ancestral modern human populations. On the one hand, the ROOA scenario required exclusive codivergence of HPV16 lineages with modern human dispersals. On the other hand, the HHS scenario implied an ancestral virus-host codivergence episode during the split between Homo sapiens and H. neanderthalensis lineages, followed by more recent transmission events between Neanderthals/Denisovans and modern human ancestors (fig. 3). Both scenarios are plausible given the topology of the phylogenetic relationships between HPV16 lineages (fig. 1) and the impossibility to root the HPV16 tree using extant HPVs sequences.
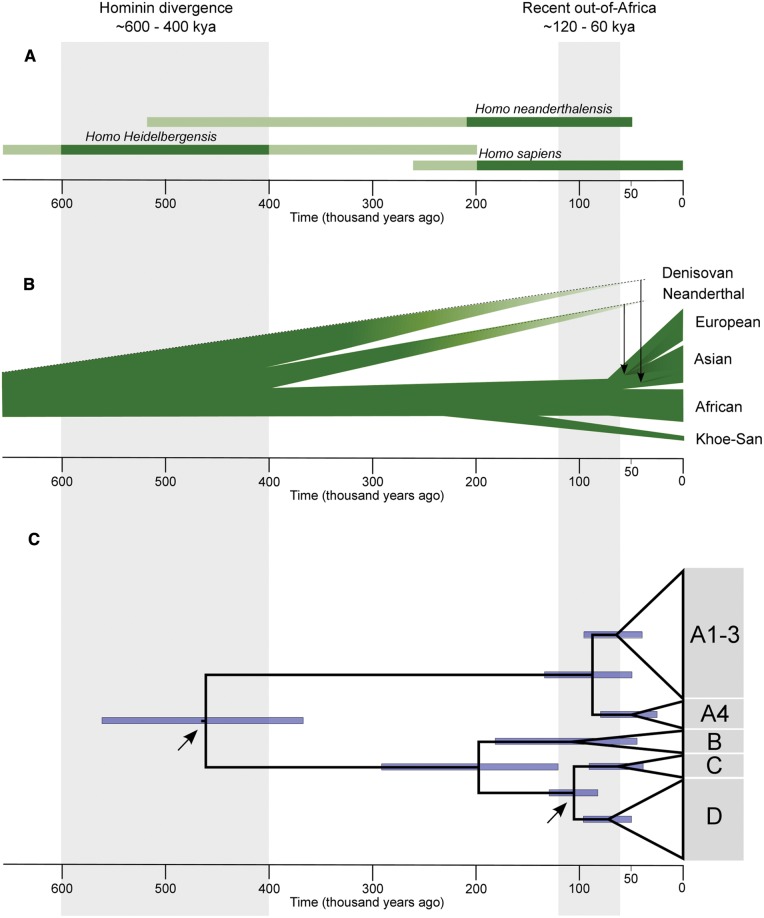
Fig. 3.
Timeline of divergence for archaic and modern human ancestors, and for HPV16. (A) Dated and classification-confirmed Hominin fossil taxa (light green represents uncertain classification, as reviewed by Scally and Durbin 2012). (B) Phylogenetic relationship of modern human, Neanderthal and Denisovan populations. Vertical arrows indicate the proposed interbreeding and subsequent gene flow from the Neanderthal and Denisovan populations to the ancestors of present-day modern humans. The evolutionary relationships between Neanderthals and Denisovans are still unresolved (Sawyer et al. 2015; Stringer and Barnes 2015). (C) Phylogenetic relationships for 118 selection-filtered HPV16 coding genome sequences using maximum clade credibility tree under the Hominin host-switch scenario. Blue bars indicate the 95% probability intervals for the corresponding node age. Arrows indicate the nodes used for calibration.
To test the explanatory power of the ROOA model, we analyzed the correlation between geography-based population structure of modern humans and of HPV16 by comparing genetic distances between geographically defined human metapopulations with the genetic distances between HPV16 isolates. We used nine previously described metapopulation groups (supplementary table S3, Supplementary Material online), excluding North America due to lack of human genome data, and tested the correlation between the FST distance matrices for humans and for HPV16 (supplementary fig. S3, Supplementary Material online). We observed indeed correlation between both matrices, but geographical structure across different human genome loci explained <30% of the worldwide HPV16 geographical structure (table 1, see also supplementary table S4, Supplementary Material online). Notably, most of the European and sub-Saharan African ascertained autosomal human genome loci variability showed significant but limited (<30%) correlation with the HPV16 LCR-E6 variability. Very interestingly, after excluding the sites under selection in the HPV16 E6 gene only the correlation with the mitochondrial genome variability and most of the sub-Saharan African but not of the European ascertained autosomal genome data remained significant, and accounted for barely 15% of the global distribution of HPV16 diversity. Nevertheless, it is important to note that after controlling for false discovery rate none of the human genome and of HPV16 correlations showed to be significant.
Table 1.
Mantel’s Permutation Test for Similarity of FST Distance Matrices between HPV16 and Human Genome Sequence Data Sets.
| Selection-filtered |
Including positions under selection |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Taxa | Loci | Region/SNPs | n | Correlation (%)a | P-valueb | FDRc | Correlation (%)a | P-valueb | FDRc |
| mtDNA | 1 | 16 kb genome | 875 | 12.4 | 0.037 | 0.017 | 15.8 | 0.026 | 0.018 |
| NRY | 1 | 5,000 kb | 551 | 8.5 | 0.062 | 0.028 | 11.8 | 0.044 | 0.036 |
| Europe | chr1 | 6,353 | 773 | 8.8 | 0.062 | 0.029 | 12.7 | 0.033 | 0.026 |
| Europe | chr2 | 5,918 | 773 | 6.3 | 0.091 | 0.040 | 9.6 | 0.054 | 0.040 |
| Europe | chr3 | 5,325 | 773 | 8.8 | 0.063 | 0.032 | 12.7 | 0.037 | 0.030 |
| Europe | chr4 | 4,211 | 773 | 8.9 | 0.063 | 0.030 | 12.7 | 0.036 | 0.029 |
| Europe | chr5 | 4,743 | 773 | 5.6 | 0.104 | 0.047 | 8.7 | 0.061 | 0.047 |
| Europe | chr6 | 4,736 | 773 | 6.3 | 0.095 | 0.041 | 9.5 | 0.055 | 0.042 |
| Europe | chr7 | 4,162 | 773 | 5.2 | 0.112 | 0.048 | 8.1 | 0.062 | 0.048 |
| Europe | chr8 | 3,750 | 773 | 8.0 | 0.070 | 0.036 | 11.6 | 0.045 | 0.037 |
| Europe | chr9 | 3,222 | 773 | 8.8 | 0.064 | 0.033 | 12.7 | 0.038 | 0.033 |
| Europe | chr10 | 4,223 | 773 | 9.2 | 0.059 | 0.026 | 13.1 | 0.035 | 0.028 |
| Europe | chr11 | 3,850 | 773 | 5.9 | 0.099 | 0.046 | 9.2 | 0.058 | 0.046 |
| Europe | chr12 | 3,339 | 773 | 5.1 | 0.116 | 0.049 | 8.0 | 0.066 | 0.049 |
| Europe | chr13 | 2,234 | 773 | 5.8 | 0.098 | 0.045 | 9.2 | 0.057 | 0.045 |
| Europe | chr14 | 2,307 | 773 | 5.9 | 0.097 | 0.043 | 9.3 | 0.056 | 0.043 |
| Europe | chr15 | 2,346 | 773 | 6.2 | 0.096 | 0.042 | 9.3 | 0.054 | 0.041 |
| Europe | chr16 | 2,636 | 773 | 7.5 | 0.078 | 0.038 | 11.1 | 0.045 | 0.038 |
| Europe | chr17 | 2,109 | 773 | 6.8 | 0.084 | 0.039 | 10.1 | 0.045 | 0.039 |
| Europe | chr18 | 2,243 | 773 | 9.8 | 0.056 | 0.025 | 13.9 | 0.033 | 0.025 |
| Europe | chr19 | 1,175 | 773 | 3.9 | 0.139 | 0.050 | 6.5 | 0.085 | 0.050 |
| Europe | chr20 | 2,067 | 773 | 9.9 | 0.055 | 0.023 | 13.8 | 0.031 | 0.021 |
| Europe | chr21 | 973 | 773 | 10.0 | 0.055 | 0.024 | 14.1 | 0.031 | 0.022 |
| Europe | chr22 | 1,141 | 773 | 7.7 | 0.073 | 0.037 | 11.6 | 0.042 | 0.034 |
Note.—HPV16 sequence data (1,192 bp) encompassing LCR (735 bp) and E6 (457 bp) regions for 1,101 worldwide isolates were used in the correlations. mitochondrial DNA (mtDNA); nonrecombining part of the Y chosomosome (NRY); autosomal SNP data stratified by chromosome and ascertained using a European (Europe) ancestry data (autosomal SNP data ascertained using a sub-Saharan ancestry data is provided in supplementary table S8, supplementary material online). For both datasets nine geographic metapopulations were defined: North/East/West/Southern Africa, South Asia, East Asia, Europe, Central and South America (supplementary table S2 and S5. supplementary material online).
R2 of the correlation coefficient.
Mantel-test after 10,000 permutations.
False discovery rate controlling procedure (q = 0.05).
Time Inference for HPV16 Diversification
To estimate divergence times for HPV16 diversification under the two alternative evolutionary scenarios, we applied a Bayesian Markov chain Monte Carlo (MCMC) inference. First, we tested whether we could accurately identify the root of the HPV16 genome diversity by performing phylogenetic inference incorporating the most closely related HPVs, members of Alphapapillomaviruses species 9. The long evolutionary distances between these viruses and HPV16 prevented unambiguous identification of the root of HPV16 lineages, with either ML (RAxML) or Bayesian (Phylobayes) approaches (supplementary fig. S4, Supplementary Material online). In all reconstructions, the preferred topology was (A,(B,(C,D))), which corresponded to the HHS scenario. Nevertheless, the likelihood values for the HPV16 trees obtained after forcing our two alternative scenarios, namely, the HHS and ROOA models, differed by <0.01%, and neither the Shimodaira–Hasegawa test nor the Robinson–Foulds split method showed significant differences between both topologies. When introducing time into the Bayesian (BEAST) phylogenetic analyses without imposing any prior on tree topology, the same (A,(B,(C,D))) topology (i.e., the HHS model) was systematically the preferred one for both HPV16 complete and HPV16 selection-filtered coding region alignments (supplementary fig. S4, Supplementary Material online). The combined difference in topology and branch length between trees obtained with the full-length and with the selection-filtered alignments was very small (as assessed by a K-score < 0.004) (supplementary table S5, Supplementary Material online). We resorted then to dated Bayesian MCMC inference for identifying the more plausible between the two alternative evolutionary scenarios. For the HHS scenario, we used the archaic Hominin divergence at 500 kya (95% CI, 400–600 kya, see references in Materials and Methods) to calibrate the time for the most recent common ancestor (tmrca) of extant HPV16 lineages, as well as the recent modern human migration out-of-Africa at 90 kya (95% CI, 60–120 kya, see references in Materials and Methods) to calibrate the tmrca of lineages HPV16C and D. For the ROOA scenario, we used the recent migration out-of-Africa with the same 90-kya settings as above but in this case to calibrate the tmrca of all HPV16 lineages. Both strict and relaxed clock were tested for all MCMC calibrations and the best log-likelihood value using Bayes Factor (2lnBF) was obtained for relaxed clock (2lnBF > 38). The demographic models of exponential growth or of Bayesian skyline for the population function of the coalescent tree prior yielded in both cases growth rates consistently above zero, excluding thus constant population size as a model. In addition, Bayesian skyline plot estimating changes in effective population size through time showed a significant increase of the HPV16 population in the last 12 kya (supplementary fig. S5, Supplementary Material online). For final analyses, we used the evolutionary models and priors showing the best performance, namely, relaxed log-normal molecular clock and a Bayesian skyline coalescent model (2lnBF > 83). Comparison of divergence time estimates between the HHS and ROOA scenarios for HPV16 evolution is presented in table 2. For the analyses based on the positions evolving neutrally, the log-marginal likelihoods showed the highest value and strongest support (2lnBF = 8.9) for the HHS scenario with a substitution rate 18.4 × 10 − 9 (95% CI, 14.3 ×10−9–22.1 × 10−9) subs/site/yr. For the analyses that included the positions under selection, the HHS model was again the preferred model albeit with a substitution rate two times higher. The log-marginal likelihood value for the HHS scenario including the positions under selection did not significantly differ from that for the ROOA model (2lnBF = 1.2) although the inferred evolutionary rate in the case of ROOA model needed to be around five times higher. Maximum clade credibility tree inferred for selection-filtered HPV16 genome coding region alignment under the HHS scenario and enforcing the two HHS model time point calibrations is presented in fig 1B. Furthermore, to estimate whether our time point calibrations had an impact on the divergence time estimates, we also performed MCMC analysis either by imposing the HHS model topology or without imposing any prior on tree topology and without using any time point calibrations (table 3). Depending on substitution rate priors, divergence times of extant HPV16 lineages showed to be between 260 kya and 4.8 Ma. In all cases, the preferred model was the HHS scenario. Although HPV16A was thus always the basal clade, divergence within HPV16A showed to be lower than within HPV16BCD (supplementary fig. S6, Supplementary Material online).
Table 2.
Substitution Rate and Divergence Time Estimates with Highest Probability Density (HPD) in Thousands of Years Ago (kya) Inferred for HPV16 Complete (Full) or Selection-Filtered Genome Alignment (Neutral), under the HHS or the ROOA Scenarios, and Considering Different Priors.
| Model | Calibration | Rate uniform prior | Data | ×10−9 (subs/site/yr) |
Log marginal |
2lnBF | tmrca ABCD (kya) |
tmrca A1–4 (kya) |
tmrca BCD (kya) |
tmrca CD (kya) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ×10−9 (subs/site/yr) | µ | (95% HPD) | likelihood | mean | (95% HPD) | mean | (95% HPD) | mean | (95% HPD) | mean | (95% HPD) | ||||
| HHS | HHS 2 calib | PVs estímate [13.2–24.7] a | Neutral | 18.4 | (14.5–22.1) | −12476.94 | Ref | 461 | (365–561) | 87 | (48–133) | 197 | (121–293) | 105 | (82–129) |
| HHS | HHS 2 calib | Human mtDNA [17.6–32.3]b | Neutral | 20.3 | (16–25.1) | −12480.34 | 6.8 | 456 | (358–553) | 78 | (42–123) | 184 | (109–273) | 101 | (76–124) |
| HHS | HHS 2 calib | PVs estimate [5.5–13.6]c | Neutral | 12.8 | (11.1–14.3) | −12484.91 | 15.9 | 484 | (396–580) | 130 | (80–190) | 250 | (158–360) | 122 | (100–144) |
| HHS | HHS 2 calib | Mammal genome [2.0–2.4]d | Neutral | 3.6 | (3.1–4.1) | −12567.19 | 180.5 | 571 | (488–655) | 423 | (284–578) | 408 | (267–575) | 180 | (158–201) |
| HHS | HHS 2 calib | 4.5-4.5E–11 | Neutral | 20.2 | (14.0–26.8) | −12482.18 | 10.5 | 457 | (357–5559) | 79 | (40–128) | 185 | (105–280) | 101 | (75–128) |
| HHS | HHS 1 calib 500 kya | 4.5-4.5E–11 | Neutral | 13.6 | (8.1–20.3) | −12482.41 | 10.9 | 490 | (389–587) | 125 | (55–206) | 311 | (157–471) | 197 | (95–312) |
| HHS | HHS 1 calib 90 kya | 4.5-4.5E–11 | Neutral | 32.1 | (17.0–49.6) | −12483.21 | 12.5 | 210 | (92–346) | 54 | (23–91) | 134 | (72–203) | 85 | (53–115) |
| ROOA | 90 kya | 4.5-4.5E–11 | Neutral | 80.5 | (40.3–128) | −12481.41 | 8.9 | 85 | (54–115) | 22 | (9–38) | 54 | (24–87) | 34 | (14–56) |
| HHS | HHS 2 calib | PVs estímate [13.2–24.7]a | Full | 22.9 | (20.3–25.3) | −19313.69 | 4.7 | 476 | (387–568) | 147 | (101–200) | 246 | (164–343) | 127 | (106–148) |
| HHS | HHS 2 calib | Human mtDNA [17.6–32.3]b | Full | 27.5 | (23.5–31.0) | −19311.33 | Ref | 459 | (364–551) | 120 | (79–166) | 216 | (140–300) | 116 | (95–138) |
| HHS | HHS 2 calib | PVs estimate [5.5–13.6]c | Full | 14.4 | (13.1–15.8) | −19333.81 | 44.9 | 519 | (431–607) | 240 | (167–324) | 330 | (212–455) | 152 | (8131–173) |
| HHS | HHS 2 calib | Mammal genome [2.0–2.4]d | Full | 4.6 | (4.0–5.2) | −19502.91 | 383.1 | 650 | (571–727) | 616 | (518–705) | 411 | (320–498) | 214 | (8189–232) |
| HHS | HHS 2 calib | 4.5-4.5E–11 | Full | 33.9 | (24.4–43.4) | −19314.23 | 5.8 | 446 | (346–544) | 95 | (53–142) | 186 | (110–271) | 105 | (80–132) |
| HHS | HHS 1 calib 500 kya | 4.5-4.5E–11 | Full | 22.0 | (13.2–31.2) | −19311.87 | 1.1 | 490 | (390–586) | 156 | (78–240) | 320 | (176–473) | 209 | (111–319) |
| HHS | HHS 1 calib 90 kya | 4.5-4.5E–11 | Full | 55.0 | (31.8–82.9) | −19311.82 | 1 | 199 | (99–322) | 63 | (31–100) | 130 | (73–193) | 84 | (54–115) |
| ROOA | 90 kya | 4.5-4.5E–11 | Full | 130.6 | (71.8–206.0) | −19311.94 | 1.2 | 84 | (54–115) | 27 | (12–44) | 55 | (27–88) | 36 | (17–58) |
Note.—Likelihood values in bold are the best-supported models, taken as reference (Ref) for the path sampling Bayes factor (2lnBF) comparison.
Table 3.
Substitution Rate and Divergence Time Estimates with Highest Probability Density (HPD) in Thousands of Years Ago (kya) Inferred for HPV16 Complete (Full) or Selection-Filtered Genome Alignment (Neutral) Either under the HHS Scenario or without Imposing Any Prior on Tree Topology, and without Using Any Time Point Calibrations, and Using Instead Only Substitution Rate Priors.
| Data | Model | Calibration | Uniform prior | ×10−9 (subs/site/yr) |
tmrca ABCD (kya) |
tmrca A1-4 (kya) |
tmrca BCD (kya) |
tmrca CD (kya) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ×10−9 (subs/site/yr) | µ | (95% HPD) | Mean | (95% HPD) | Mean | (95% HPD) | Mean | (95% HPD) | Mean | (95% HPD) | |||
| Full | No topology | No calib | PVs estimate [13.2–24.7]a | 18.0 | (12.5–23.8) | 581 | (298–911) | 188 | (109–278) | 391 | (221–589) | 261 | (146–392) |
| Neutral | No topology | No calib | 18.2 | (12.4–24.4) | 340 | (168–573) | 90 | (46–141) | 231 | (121–355) | 156 | (83–247) | |
| Neutral | HHS | No calib | 18.2 | (12.4–24.2) | 361 | (178–592) | 92 | (50–147) | 228 | (123–353) | 145 | (81–222) | |
| Full | No topology | No calib | Human mtDNA [17.6–32.3]b | 23.6 | (16.4–31.1) | 440 | (223–693) | 142 | (85–214) | 296 | (167–442) | 198 | (112–296) |
| Neutral | No topology | No calib | 24.0 | (16.4–31.7) | 258 | (124–420) | 68 | (37–107) | 175 | (94–269) | 118 | (61–186) | |
| Neutral | HHS | No calib | 23.9 | (16.4–31.6) | 273 | (136–444) | 70 | (37–108) | 173 | (95–271) | 110 | (60–167) | |
| Full | No topology | No calib | PVs estimate [5.5–13.6]c | 9.0 | (5.2–12.8) | 1181 | (532–2037) | 384 | (201–627) | 803 | (400–1330) | 536 | (267–870) |
| Neutral | No topology | No calib | 9.1 | (5.2–13.2) | 698 | (296–1228) | 185 | (85–313) | 185 | (86–312) | 317 | (145–539) | |
| Neutral | HHS | No calib | 9.1 | (5.2–13.1) | 742 | (324–1297) | 189 | (90–320) | 469 | (220–791) | 298 | (141–493) | |
| Full | No topology | No calib | Mammal genome [2.0–2.4]d | 2.1 | (1.8–2.4) | 4819 | (2914–7086) | 1564 | (1077–2133) | 3249 | (2108–4458) | 2177 | (1466–3015) |
| Neutral | No topology | No calib | 2.1 | (1.8–2.5) | 2863 | (1576–4415) | 755 | (457–1086) | 1919 | (1206–2741) | 1301 | (784–1916) | |
| Neutral | HHS | No calib | 2.1 | (1.8–2.4) | 3010 | (1686–4582) | 767 | (475–1123) | 1900 | (1187–2738) | 1208 | (761–1691) | |
Variation within the HPV16 E6 Oncogene
To investigate geographical differences in HPV16 genetic diversity, we estimated summary statistics within each geographic HPV16 population using all 1,123 available HPV16 nearly complete E6 sequences (table 4). Overall, HPV16 E6 genetic diversity was highest outside sub-Saharan Africa. Both Tajima’s D and Fu’s-Li’s D statistics as well as the more robust R2 index showed statistically large negative values for European isolates, suggesting past selective sweeps and/or population expansion, for HPV16 population in Europe only. HPV16 E6 haplotype diversity was similar in sub-Saharan Africa, Europe, and South America and significantly higher in North Africa and Central America than in sub-Saharan Africa (table 4). East Asian and Central American HPV16 isolates showed higher average number of pairwise differences compared with sub-Saharan African isolates, even after accounting for intralineage diversity. Furthermore, to estimate the geographical origin of the HPV16 E6 gene haplotype diversity, we constructed a median-joining haplotype network for the E6 gene sequences (table 5, see also supplementary fig. S7, Supplementary Material online). Two of the most common HPV16A1-3 lineage E6 haplotypes were observed in Eurasian isolate background, and only 1–3% of these common haplotypes originated from sub-Saharan African isolates. In contrast, the most frequent HPV16B and C variant E6 haplotypes were observed in >92% of African isolate background. The most common HPV16D E6 haplotype displayed the highest frequency (58%) in the American isolate background and the lowest (4–5%) in North and sub-Saharan African isolates. The second most common HPV16D E6 haplotype showed the highest frequency (58%) in North African isolates, intermediate frequencies in Eurasian and in American isolates (18% and 21%, respectively) and very small (3%) of the sub-Saharan African isolate background.
Table 4.
Summary Statistics of the HPV16 E6 Gene Sequences (n = 1,123) with Geographic Origin of the Sample.
| Metapopulation | n | bp | S | h | Hd | (95% CI)a | (95% CI)b | MPD | (95% CI)a | (95% CI)b | WIMP | Tajima’s D | P-valuec | (95% CI)b | P-valueb | Fu’s-Li’s D* | (95% CI)b | P-valueb | R2 | (95% CI)b | P-valueb |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sub-Saharan Africa | 213 | 456 | 34 | 32 | 0.754 | (0.712 –0.796) | (0.439 –0.892) | 3.049 | (2.739 –3.359) | (0.815 –7.389) | 1.459 | –1.427 | >0.1 | (–1.642 to 2.053) | 0.047 | –3.459 | (–2.178 to 1.523) | 0.003 | 0.044 | (0.033 –0.148) | 0.100 |
| North Africa | 221 | 456 | 30 | 26 | 0.861 | (0.839 –0.883) | (0.653 –0.918) | 4.985 | (4.52 –5.45) | (1.543 –12.592) | 0.969 | –0.196 | >0.1 | (–1.598 to 2.097) | 0.519 | –3.288 | (–1.959 to 1.311) | 0.466 | 0.081 | (0.035 –0.14) | 0.533 |
| Europe | 161 | 456 | 31 | 25 | 0.734 | (0.688 –0.78) | (0.16 –0.838) | 1.62 | (1.401 –1.839) | (0.194 –4.219) | 1.187 | –2.043 | <0.05 | (–1.62 to 2.05) | 0.001 | –3.825 | (–1.958 to 1.447) | 0.002 | 0.026 | (0.028 –0.164) | 0.017 |
| South Asia | 139 | 456 | 8 | 6 | 0.523 | (0.455 –0.591) | (0.056 –0.797) | 0.998 | (0.831 –1.165) | (0.043 –2.744) | 0.464 | –0.724 | >0.1 | (–1.509 to 2.026) | 0.253 | –0.454 | (–2.302 to 1.297) | 0.281 | 0.062 | (0.026 –0.193) | 0.302 |
| East Asia | 207 | 456 | 31 | 37 | 0.643 | (0.567 –0.719) | (0.495 –0.894) | 3.425 | (3.079 –3.771) | (0.853 –8.014) | 1.712 | 1.211 | >0.1 | (–1.593 to 2.11) | 0.091 | –2.848 | (–2.415 to 1.606) | 0.014 | 0.054 | (0.032 –0.146) | 0.180 |
| Central America | 50 | 456 | 21 | 14 | 0.889 | (0.851 –0.927) | (0.589 –0.925) | 4.551 | (3.641 –5.461) | (1.197 –10.979) | 1.803 | –0.237 | >0.1 | (–1.683 to 2.033) | 0.475 | –1.027 | (–2.568 to 1.511) | 0.167 | 0.103 | (0.052 –0.177) | 0.503 |
| South America | 132 | 402 | 18 | 17 | 0.78 | (0.744 –0.816) | (0.486 –0.893) | 3.184 | (2.774 –3.594) | (0.772 –7.859) | 0.947 | –0.096 | >0.1 | (–1.613 to 1.896) | 0.526 | –0.850 | (–2.371 to 1.543) | 0.185 | 0.087 | (0.035 –0.156) | 0.542 |
Note.—Number of samples (n), base pair (bp), Segregating sites (S), haplotypes (h), haplotype diversity (Hd), mean pairwise difference (MPD), weighted intralineage mean pairwise difference (WIMP) (Hurles et al. 2002), Tajima’s D, Fu’s and Li’s D* and R2 statistics were calculated using DnaSP (Librado and Rozas 2009).
Calculcated from empirical distribution.
Calculated from a coalescent simulation with 1,000 replications and no recombination.
Calculated assuming beta distribution.
Table 5.
HPV16 E6 Gene Major Haplotype Frequencies within Each Lineage and Geographical Background.
| Origin | A1–3a |
A1–3b |
A4 |
B |
C |
D |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | |
| America | 50 | 19.5 | 40 | 19.0 | 2 | 1.5 | 1 | 1.2 | 4 | 2.6 | 45 | 57.7 | 8 | 21.1 |
| Eurasia | 177 | 68.9 | 118 | 56.2 | 127 | 98.5 | 1 | 1.2 | 7 | 5.0 | 26 | 33.3 | 7 | 18.4 |
| North Africa | 27 | 10.5 | 46 | 21.9 | — | — | 4 | 4.7 | 53 | 37.6 | 4 | 5.0 | 22 | 57.9 |
| Sub-Saharan Africa | 3 | 1.2 | 6 | 2.9 | — | — | 79 | 92.9 | 77 | 54.6 | 3 | 3.8 | 1 | 2.6 |
Major haplotype including HPV16 E6 350G allele.
Major haplotype including HPV16 E6 350T allele.
Discussion
HPV16 is the most oncogenic virus for humans, and epidemiological evidence suggests that its oncogenic potential is associated with high viral loads and with infection persistence (Doorbar et al. 2012; Moscicki et al. 2012). Viral genotypic variation may underlie strong phenotypic variation so that different HPV16 variants may display differential persistence and viral load and therefore differential carcinogenicity. Several reports have communicated differential oncogenic potential for different HPV16 variant lineages (Villa et al. 2000; Schiffman et al. 2010; Zuna et al. 2011; Freitas et al. 2014). However, the differential risks for different variants seem to depend on the host population (Cornet et al. 2013; Qmichou et al. 2013), and to vary with the specific individual genetic background (Xi et al. 2006; Lopera et al. 2014). In the present study, we have addressed the origin, diversification, and dispersal of HPV16. We have used the largest available collection of HPV16 full-length genomes and partial sequences to generate estimates of the worldwide phylogeography and divergence time of extant HPV16 lineages. Our results support a scenario of codivergence of HPV16 with separate but closely related ancestral Hominin populations with subsequent host-switch events, likely through sexual transmission between archaic and modern human ancestral populations in the recent human evolutionary history. We have further compared the genetic diversity patterns of HPV16 with those estimated for modern human populations using genome data. Altogether, our results suggest that virus-host coevolution and host switch events have been fundamental factors that have shaped HPV16 evolution.
Purifying selection can mask the ancient origin of recently sampled pathogens (Wertheim and Kosakovsky Pond 2011), and our results point in the same direction. Results based on only HPV16 genome positions evolving neutrally showed significantly better fit to the data for our HHS model while analyses including positions under selection still preferred the HHS model but could not differentiate the two alternative scenarios, namely, the ROOA and the HHS model, tested in this study (table 2). More importantly, our MCMC inference showed that irrespective of the evolutionary rate prior used, divergence times for extant HPV16 lineages largely predate the estimated recent out-of-Africa migration of modern human ancestors 60–120 kya. Furthermore, although HPV16A is the oldest lineage, the tmrca for the HPV16BCD variants (197 kya, 95% HPD 121–291 kya) was older than the tmrca for the HPV16A lineage (88 kya, 95% HPD 50–134 kya), and the HPV16A lineage encompasses less genetic diversity than the sister HPV16BCD lineages. Taken together, the lower divergence within HPV16A, the good match for the tmrca of this lineage with the out-of-Africa migration of modern humans and the delayed expansion of HPV16A reinforce the HHS scenario: the exceptional interbreeding events between archaic and modern human populations resulted in a strong bottleneck for the transmission of HPV16A, so that only a small fraction of the HPV16A diversity available in the Neandertal/Denisovan populations underwent effectively a horizontal transfer toward populations of ancestral modern humans.
Mounting evidence suggests that virus-host codivergence is not the only essential driving force for PV evolution. At higher taxonomic levels virus-host co-phylogeny events explain only around 30% of the PV evolutionary history, while additional events of lineage sorting, lineage duplication and host switch have played major roles during the evolution of PVs (Gottschling et al. 2011). In the present study, we have quantified for the first time that codivergence between modern humans and HPV16 explains <30% of the current geographical distribution of the viral extant diversity, largely based on the virtual absence of HPV16B and HPV16C lineages outside Africa and on the enrichment of HPV16A and HPV16D outside Africa (fig. 2). The contribution of virus-host codivergence to HPV16 evolution, regardless of excluding the viral coding region sites under selection, showed to be consistent only for mitochondrial genome variability and less consistent using European ascertained autosomal single nucleotide polymorphisms (SNPs) compared with sub-Saharan African ascertained autosomal SNPs. It is tempting to speculate that this difference in consistency might be partly due to the SNPs introgressed from archaic populations into the genomes of non-African modern humans. However, the underlying biological complexities and limitations of the available data to compare the human genome and of HPV16 genetic diversity restrained us from further estimations of local adaptation between virus and host genomes. Nevertheless, under the ROOA scenario, one would have expected a consistently high correlation between human and viral phylogeography. Instead, several lines of evidence argue further against this ROOA scenario for HPV16. First, the HHS scenario is preferred over ROOA in terms of HPV16 lineage phylogeny (supplementary fig. S6, Supplementary Material online). Second, the hypothesis of exclusive codivergence of all HPV16 lineages with the ancestral dispersals of modern humans does not explain the largely predominant role of HVP16A (fig. 2), the most basal HPV16 lineage, in all continents and in all indigenous populations, except in sub-Saharan Africa (Picconi et al., 2003; Cornet et al. 2012; Mendoza et al. 2013; Qmichou et al. 2013; Tan et al. 2013; Lopera et al. 2014). Moreover, for indigenous populations in South America, for instance, the increased presence of HPV16A lineage variants has been proposed to reflect the influence of recent European occupation (Picconi et al. 2003). However, such a rapid selective sweep of the putative pre-Columbian HPV16A genetic diversity would require strong selection forces for the viral dynamics in very short time scale, which are not compatible with our current understanding of PV evolution (Bravo and Félez-Sánchez 2015). Third, the geographical dichotomy for the HPV16 E6 gene haplotype backgrounds consistently supported our HHS model over ROOA: most common HPV16A lineage haplotypes were observed mostly in Eurasian populations while most common HPV16B, C, and D lineage haplotypes were observed mainly in African populations (table 5). And fourth, while human genetic diversity is highest in sub-Saharan Africa (1000 Genomes Project Consortium et al. 2015), our results of HPV16 E6 variability showed consistently higher genetic diversity estimates outside sub-Saharan Africa (table 4).
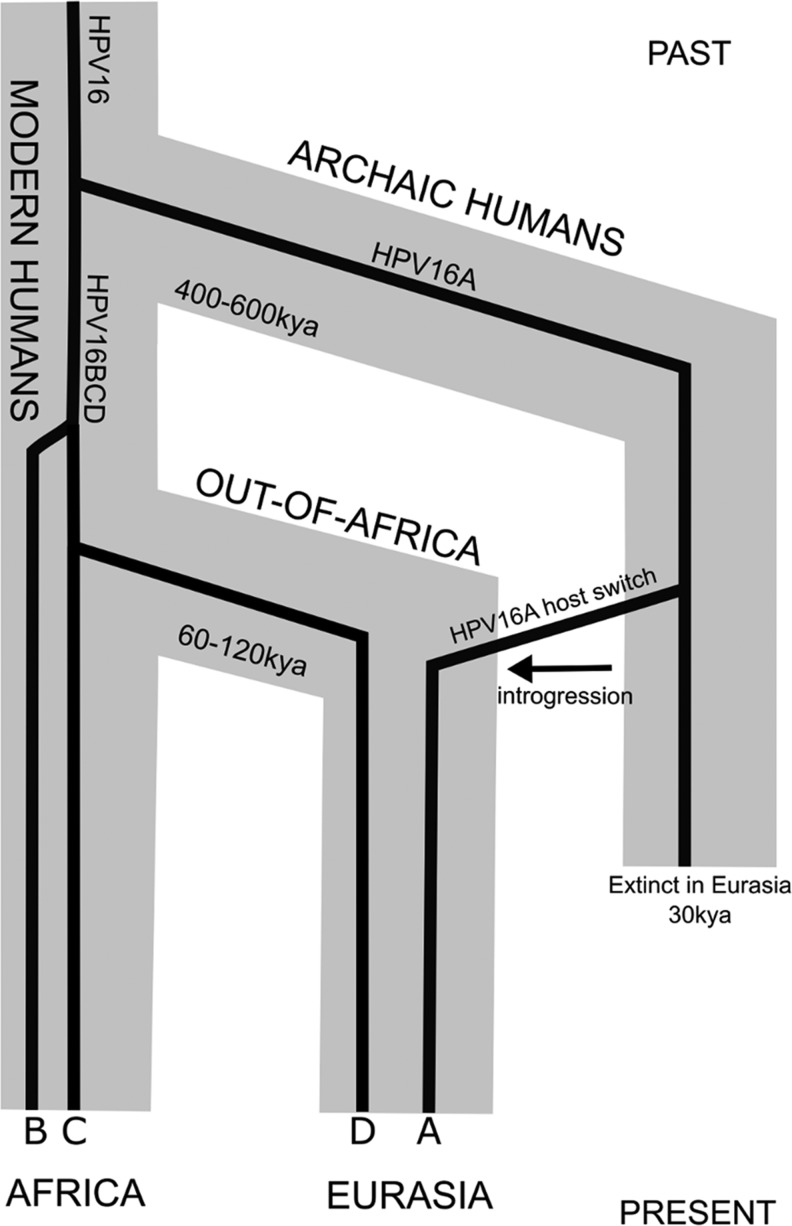
After rejecting the ROOA model as explanatory framework for HPV16 evolution, we have explored the HHS model as alternative scenario, implying a host switch of an ancestral HPV16 lineage between archaic and modern human ancestral populations (fig. 4). Indeed, recent genomic studies have confirmed the admixture of modern human ancestors with Neanderthals and Denisovans through repeated interbreeding after migration to Europe and Asia, respectively (Reich et al. 2010; Lazaridis et al. 2014; Qin and Stoneking 2015; Vernot and Akey 2015). Such interbreeding events lead to adaptive introgression of genetic material from our closest evolutionary relatives into the genome of a subset of modern human ancestors. We propose here that in parallel to the introgression of genetic material, sexual intercourse between Neanderthals/Denisovans and modern human ancestors also prompted sexual transmission of PVs, chiefly of the HPV16A lineage (fig. 4). Our results on HPV16 phylogeography, genetic diversity, and tmrca of the different HPV16 lineages sustain this HHS scenario, in which the divergence of extant HPV16 lineages (ca. 460 kya, table 2) predates the recent out-of-Africa migration of modern human ancestors (fig. 3), and thus, ancestral HPV16 lineages probably already infected the ancestral, closely related Hominin groups. The host split between the ancestral archaic (Neanderthal/Denisovan) and the modern human ancestor populations was probably mirrored by a viral split resulting in the ancestral HPV16A lineage and of the ancestral HPV16BCD lineage, respectively (fig. 3B and C). Among the recurrent out-of-Africa expansions of ancestral Hominin groups, which may have occurred some 460 kya, Neanderthals/Denisovans may have carried essentially the ancestral HPV16A. Evolution of HPV16 genomes in ancestral Hominin populations remaining in Africa, instead, would have lead to HPV16B and CD lineages (fig 4). The virtual absence of HPV16B outside Sub-Saharan Africa is parsimoniously explained if in the last out-of-Africa expansion, the modern human ancestors that left Africa probably lost the ancestral HPV16B lineage by a lineage sorting event (Johnson et al. 2003). Similar pattern of virus extinction after host population bottleneck has been observed in non-human primates (Kapusinszky et al. 2015). After the modern human dispersal, the HPV16CD ancestor generated in allopatry the HPV16C lineage in the populations remaining in Africa, and the HPV16D lineage in the populations outside Africa (fig. 4). During their expansion in Europe and in Asia, modern human ancestors experienced limited admixture with Neanderthal and Denisovan populations and were exposed to the HPV16A lineage, most likely through sexual contact. After the adaptive introgression of Neanderthal-Denisovan alleles, anatomically modern humans of non-African ancestry carry nowadays in average between 2% and 4% of their genomes of archaic human origin (Meyer et al. 2012; Prüfer et al. 2014). Remarkably, the horizontal transfer of genetic loci upon interbreeding of Neanderthal/Denisovan ancestry into the genome of modern human ancestors did not occur at random (Vernot and Akey 2014; Kuhlwilm et al. 2016; Vernot et al. 2016). Instead, genes involved in keratinocyte differentiation and innate immunity, which are loci directly involved in host–PV interaction, are particularly enriched in introgressed genomic loci, displaying >60% of Neanderthal/Denisovan ancestry in present day Eurasians (table 6) (Abi-Rached et al. 2011; Sankararaman et al. 2014; Vernot and Akey 2014; Vernot et al. 2016). Such genetic changes possibly altered the HPV16-human interplay, as this virus exclusively infects keratinocytes in particular epithelia and requires keratinocyte differentiation to accomplish the virus life cycle and virion production (Doorbar et al. 2012). Hence, we speculate that the adaptive introgression in modern humans of archaic alleles involved in keratinocyte differentiation and innate immunity may have elicited changes in the ecological niche of HPV16 that significantly enhanced the adaptive value of HPV16A and lead to increased prevalence of this viral lineage in modern human populations with Eurasian origin (figs. 2, 3B and C, and 4).
Fig. 4.
Cartoon timeline depicting interbreeding and subsequent gene flow of archaic alleles from Neanderthals and Denisovans into modern humans and the proposed sexual transmission of HPV16A lineage to the ancestors of modern human populations in Eurasia. Viral lineages HPV16A, B, C, and D are labeled at the bottom.
Table 6.
Global Prevalence of HPV16A Lineages (Upper Panel) and Proportion of Nonrandomly Introgressed Archaic Alleles into Modern Human Ancestors (Lower Panel).
| Human papillomavirus 16A lineages | Geography |
Ref. | ||
|---|---|---|---|---|
| Sub-Saharan Africa | West Eurasia | East Eurasia | ||
| HPV16A1-3 | <5% | 93% | 25-91% | This study |
| HPV16A4 | Absent | ∼1% | 60% | This study |
| Host’s genome loci directly involved in PVs life-cycle | ||||
| Neanderthal ancestry loci | Absent | 64% | 62% | Sankararaman et al. (2014) |
| Loci involved in keratinocyte differentiation | Absent | 40–70% | 40–66% | Vernot and Akey (2014) |
| HLA I loci (innate immunity) | <7% | 52–59% | 72–82% | Abi-Rached et al. (2011) |
| Toll-like receptor loci (innate immunity) | Absent | 15–39% | 17–51% | Dannemann et al. (2015) |
| APOBEC3A deletion (innate immunity) | <1% | 7% | 14-93% | Kidd et al. (2007) |
Note.—A selection of host’s genome loci is listed, directly involved in the Papillomavirus life-cycle through keratinocyte differentiation and innate immunity.
Taken together, our HHS evolutionary model provides explanatory potential for the three central flaws of the alternative ROOA scenario for the evolution of HPV16, namely, 1) the distant phylogenetic relationship between HPV16A and HPV16BCD clades (fig. 1A and B), which predates the most recent out-of-Africa migration of modern human ancestors (table 3); 2) the global increased prevalence of HPV16A outside Africa (fig. 2), and 3) the increased viral genetic diversity outside sub-Saharan Africa (table 4). In agreement with our HHS model, recent studies of parasites evolution have concluded that the divergence of at least five present day human parasites (lice, tapeworm, follicle mites, a protozoan, and bedbug) predates modern human origin. The most likely explanation for the presence of these closely related ancient pairs of parasite taxa involves host switch from an archaic to modern human ancestors (Ashford 2000; Reed et al. 2004). Furthermore, evolutionary studies of sexually transmitted herpes simplex-virus suggest ancient codivergence and later cross-species transmission of this pathogen between ancestors of chimpanzees and extinct Hominin groups (Wertheim et al. 2014).
Despite the explanatory power of our results, our study suffers from a number of limitations. Ideally, the global host and pathogen sequence data should be analyzed at the level of full genomes from the same individuals, but such data set is not available. Furthermore, there is hitherto no evidence of the presence of any PV sequences from ancient human samples. Indeed, we analyzed the currently available Neanderthal and Denisovan preassembly sequence data, and we could not find any significant traces of any known HPVs in these data sets. Unfortunately, the epithelial tropism of these viruses likely prevents retrieving viral sequences from fossil bones, the common reservoir for ancient DNA studies. We have also tried to identify sequence traces of PV DNA from the metagenomic sequence data extracted from tissue remains of the European Copper Age glacier mummy (Maixner et al. 2014) but could not find any traces of PV DNA in any of these sample data sets. However, future studies encompassing larger global sample of individuals with full human and HPV genome sequence data and/or well-preserved ancestral human samples containing PVs DNA will allow to test and refute or validate our hypothesis.
We have presented here the most comprehensive study of the evolution and diversity of HPV16, the most oncogenic infectious agent for humans. Our phylogeographic analyses confirm the limited contribution of virus-host codivergence to the evolution of the HPV16 lineage: most of the geography-based diversity within HPV16 cannot be explained alone by co-speciation with modern humans. Instead, our results suggest that ancestral HPV16 already infected the ancestor of H. sapiens and H. neanderthalensis half a million years ago, and that two main HPV16 lineages codiverged with either human lineage. When a population of modern humans migrated out of Africa some 60–120 kya, they carried with them a subset of the HPV16 diversity evolved in that continent. When modern humans encountered and interbred with Neanderthal/Denisovan populations in Europe and in Asia, a transfer of sexually transmitted pathogens occurred, in parallel with the genomic introgression. Gene alleles involved in keratinocyte differentiation and in innate immunity evolved among Neanderthals/Denisovans and transferred to modern humans, may have facilitated niche colonization and expansion of HPV16A in modern human populations with archaic admixture. This evolutionary advantage led to the enhanced prevalence of HPV16A lineage among modern humans with introgressed archaic genetic background. Indeed, our results show that sexually transmitted pathogens may have been effectively transferred between different Hominin populations in recent events. We propose that the repertoire of the HVP16 variants coevolved with archaic humans, the diversity of keratinocyte differentiation and innate immune genes that have undergone adaptive introgression, and eventually the interaction between viral and host genotypes may be largely responsible for the differential association of certain HPV16 variants with cancer risk in certain modern human populations.
Materials and Methods
HPV16 Complete Genomes and Partial Sequences
The full-genome sequences for 118 HPV16 isolates used for the analysis were retrieved from GenBank. Sequences were aligned with MUSCLE (Edgar 2004), at amino acid level for the coding region and at nucleotide level for the UTR, and tested for putative recombination break points using GARD algorithm (Kosakovsky Pond et al. 2006). The final full-genome alignment encompassed 118 full-length HPV16 genomes, 7,926 bp and 638 distinct alignment patterns (alignment available as supplementary material S1, Supplementary Material online). Best substitution model for the HPV16 full-genome data, manually curated and filtered using Gblocks (Castresana 2000), was inferred using jModelTest 2.0 (Darriba et al. 2012). Phylogenetic inference was performed under ML framework with RAxML v8.0.16 using the general time reversible (GTR) model of nucleotide substitution with Γ-rate heterogeneity parameter (GTR + Γ4) and 500 bootstrap replicates (Stamatakis 2014). Besides the unpartitioned analysis, four partition schemes were also explored: 1) partitions of noncoding and coding regions; 2) the same as before, but further partitioning the coding region into three codon positions; 3) eight partitions corresponding to the regulatory region and to the E6, E7, E1, E2, E5, L2, L1 genes; and 4) the same as before, but further partitioning each coding regions into three codon positions. Among all partition schemes explored, the unpartitioned scheme rendered the best AIC scores.
In order to focus on the HPV16 genome positions evolving under neutrality and to filter out the possible bias arising from the presence of positions undergoing natural selection, we identified and removed a total of 26 positively selected and 115 negatively selected codons from the full-genome alignment (supplementary table S1, Supplementary Material online). Each coding region of the HPV16 genome was separately assessed for codon-based selection using random effects likelihood (REL) model with ML fit (Pond and Muse 2005) and Bayes Factor of 20 (i.e., P-value = 0.05) as a level of significance, except the L2 gene region, which was assessed for selection using ML reconstruction of ancestral codons with a P-value = 0.05 as a level of significance, due to computational limitations of the REL model with the L2 alignment (Kosakovsky Pond and Frost 2005). The final coding region alignment manually curated and filtered for sites under selection, encompassed 118 HPV16 sequences, 6,093 bp and 316 distinct alignment patterns (alignment available as supplementary material S2, Supplementary Material online).
To define the root of the HPV16 complete genome ML tree, the five main lineages (i.e., A1–3, A4, B, C, and D) were identified and the two most divergent members of each lineage were retained. These 10 HPV16 variant genomes were aligned, separately for each E1, E2, L2, and L1 coding region, with the genomes of all other members of Alphapapillomaviruses species 9: HPV31, HPV33, HPV35, HPV52, HPV58, and HPV67. The concatenated coding regions genome alignment was submitted to ML phylogenetic inference following the same protocol described above. The procedure was replicated using Bayesian inference with Phylobayes (Lartillot and Philippe 2004), both at the amino acid and at nucleotide level. The differential likelihood of our two alternative hypothesis for the location of the root in the HPV16 lineage namely, ROOA and HHS models, was further assessed using Shimodaira and Hasegawa (1989) and Robinson and Fouls (1981) comparison tests.
Phylogenetic relationships of partial HPV16 sequences spanning the LCR, E6 and L2 loci from 1,601 HPV16 isolates retrieved worldwide (supplementary table S6, Supplementary Material online) were placed in the genetic landscape of HPV16 complete genome variability using an Evolutionary Placement Algorithm on RAxML v8.0.16 with the –f v command and the GTR + Γ4 model (Berger and Stamatakis 2011). The algorithm provides likelihood weights for placing partial sequences into the different nodes in the reference tree (supplementary table S2, Supplementary Material online), which in our case was the best ML tree obtained for the 118 complete HPV16 genomes (supplementary fig. 1, Supplementary Material online). Partial HPV16 sequences were introduced into the initial genome alignment using MAFFT (Katoh and Standley 2013). The final alignment consisted of 1,719 isolates (118 complete genomes, 1,082 isolates of 1,338 bp partial sequences spanning the LCR and E6 loci and 519 isolates of 665 bp partial sequences spanning the LCR, E6 and L2 loci, see available alignments in supplementary materials S3–S4, Supplementary Material online). For each partial sequence, we integrated the likelihood weights for all nodes using 0.7 as a likelihood cutoff value to confidently assign each sample into one of the five main HPV16 variant lineages (supplementary table S2, Supplementary Material online). Finally, we stratified the HPV16 variant isolate data into 10 geographical population groups: North/East/West/Southern Africa, South Asia, East Asia, Europe, Central and South America using the United Nations standard geographical criteria (supplementary table S3, Supplementary Material online). To compare the genetic variability patterns observed for HPV16 isolates within each geographic population group we calculated the summary statistics of the available 1,123 HPV16 E6 sequences (table 4). Further to visualize the genetic variability of the 1,123 HPV16 E6 456bp nearly complete gene sequences within each geographic background a haplotype-specific median joining network was constructed using NETWORK 4.6.1.3 (www.fluxus-technology.com) with equal weight for variable positions.
Bayesian MCMC Inference
Evolutionary analysis for HPV16 lineages was estimated using both the complete 7,926 bp genome alignment and the selection-filtered 6,093 bp coding region alignment of 118 HPV16 variants (supplementary materials S1–S2,Supplementary Material online), and using Bayesian MCMC analysis as implemented in BEAST v.1.8.1 (Drummond et al. 2012). Bayesian MCMC inferences were performed with the GTR + Γ4 model. Genealogies were estimated using both strict and relaxed molecular clocks with an uncorrelated log-normal distribution of rates. Two credible PVs substitution rate estimates from the literature were used as uniform priors: 9.6 × 10−9 [5.5 × 10−9–13.6 × 10−9] subs/site/yr (Shah et al. 2010) and 19.5 × 10−9 [13.2 × 10−9–24.7 × 10−9] subs/site/yr (Rector et al. 2007). We also assessed the suitability of two additional plausible substitution rates, namely, the values inferred for mammalian genomes 2.2 × 10−9 [2.0 × 10−9 − 2.4 × 10−9] subs/site/yr (Kumar and Subramanian 2002) and for human mitochondrial genomes 25.3 × 10−9 [17.6 × 10−9–32.3 × 10−9] subs/site/yr (Fu et al. 2014). In addition, a noninformative uniform prior between 4.5 × 10−11 and 4.5 subs/site/yr covering all plausible PVs rate for both molecular clock models was used (table 2). Two recently revised evolutionary time point estimates for human populations were used as prior normal distribution for calibration of the HPV16 variant phylogenetic tree (table 2 and fig. 3C): first, the archaic divergence of modern humans and Neanderthals/Denisovans was set at 500 kya (95% CI, 400–600 kya) (Reich et al. 2010; Scally and Durbin 2012; Buck and Stringer 2014; Mendez et al. 2016); second, modern humans migration out-of-Africa was set at 90 kya (95% CI, 60–120 kya) (Scally and Durbin 2012; Liu et al. 2015). All evolutionary related parameters were estimated with the following demographic models: 1) constant population size, 2) exponential growth, and 3) Bayesian skyline coalescent models (Pybus et al. 2000). Two independent runs were performed for each parameter combination with 100 million generations and subsampling every 10,000 generations. The resulting tree and log-files were combined for further analysis after removing a 10% burn-in from each run. Statistical confidence in parameter estimates was assessed by reporting marginal posterior means and their associated 95% HPD intervals (95% HPD). Efficient mixing of the chains in the Bayesian MCMC analysis was assessed with effective sample size values >200 for all parameters. Convergence of each of the simulations was visually confirmed before data were merged for further analysis. The combined Bayesian phylogenetic inferences were analyzed using Tracer v.1.6 and TreeAnnotator v.1.8.1 and visualized using Figtree v.1.4.2. Best model estimates to explain the HPV16 variant diversity were selected using two independent path sampling runs with at least 1 million iterations per path step and until no significant chances were observed in log marginal likelihoods for Bayes Factor (2lnBF) estimates (Baele et al. 2012, 2013) as implemented in BEAST v.1.8.1 (Drummond et al. 2012). The support of a particular model was assessed with 2lnBF < 2 indicating no support, 2lnBF = 2–6 indicating positive model support, 2lnBF = 6–10 indicating strong positive support and 2lnBF > 10 indicating definite distinction between competing models.
To determine the best topology for the HPV16 viral lineages, the maximum clade credibility trees were also inferred for both complete and selection-filtered coding region HPV16 genome alignments, without imposing any prior on tree topology, using BEAST v.1.8.1 (Drummond et al. 2012), and further assessing the distance between the obtained trees using K tree score (Soria-Carrasco et al. 2007). Wilcoxon–Mann–Whitney U test was used to determine the significance of intertaxa pairwise differences between the complete and selection filtered coding region HPV16 ML trees. Summary statistics, including Tajima’s D, Fu’s and Li’s D* and R2 were calculated using DnaSP v.5.10.1. (Librado and Rozas 2009).
Human Genome Data
To compare the phylogeography of HPV16 with the global variability of the human genome, we compiled human mitochondrial, Y chromosome, and autosomal genotype data of the same 938 nonrelated individuals of the CEPH Human Genome Diversity Panel representing 51 human populations worldwide (Cann et al. 2002). In order to systematically compare the host and pathogen genome diversity distribution, we stratified the human population genetic data in nine subcontinental metapopulation groups (supplementary table S3, Supplementary Material online) using the same geographical classification presented for HPV16 isolate data in supplementary table S6 (Supplementary Material online). This geographical grouping of human population genetic data reflected, as far as possible, the metapopulation groups from which the viral isolates were sampled. All mitochondrial complete genome sequences were manually aligned using MUSCLE (Edgar 2004) and 875 complete mitochondrial DNA genomes available from (Lippold et al. 2014) were used in this study. Y chromosome nonrecombining region sequences from 551 males were retrieved (Lippold et al. 2014) and all variable loci data were included in this study. Autosomal single nucleotide polymorphism (SNP) loci data ascertained using either a European or sub-Saharan African individual as previously reported (Reich et al. 2010) were used in this study. All autosomal SNP markers were previously genotyped in 934 CEPH diversity panel individuals (Green et al. 2010; Reich et al. 2010). The African and European ascertained data sets consisted of 12,162 and 111,970 SNPs, respectively, covering all autosomal chromosomes of the human genome. All autosomal SNP genotype quality control and data mining were performed using PLINK software (Purcell et al. 2007). Only SNPs with MAF > 0.01 were used in further analysis. Phasing for each genotype data sets per chromosome were performed using BEAGLE software (Browning and Browning 2007). Genetic FST distances (WRIGHT 1951) between the nine geographical population groups defined above, and excluding North America, were estimated for 1,101 available HPV16 isolate sequences encompassing LCR (735bp) and E6 (457 bp) genome regions (supplementary fig. S3, Supplementary Material online) as well as for each of the human genome marker data sets (mitochondrial, Y chromosome, and autosomal SNPs). Mantel’s permutation test for matrix similarity was implemented for pairwise combinations of FST distance matrices with 10,000 permutations. To search for possible traces of HPVs, including all known HPV16 variants, in archaic humans we retrieved all available Neanderthal and Denisovan pre-assembly sequence data from http://cdna.eva.mpg.de/neandertal/altai/AltaiNeandertal and http://www.ebi.ac.uk/ena/data/view/PRJEB3092, respectively. Reads were mapped against all known HPVs genomes using bowtie2 (version: 2.2.5) with –very-sensitive command for the alignment. Further details of data processing are available from the authors upon request.
Supplementary Material
Supplementary figures S1–S7, tables S1–S6 and materials S1–S4 are available at Molecular Biology and Evolution online.
Author Contributions
Designed and conceived the project: VNP, IGB. Retrieved data and performed the phylogenetic inference: VNP, CMO. Performed the modern and archaic human genome data analyses and Bayesian MCMC inference: VNP. Wrote the manuscript: VNP, IGB. All authors read and approved the final article.
Supplementary Material
Acknowledgments
This research was financially supported by The European Society of Clinical Microbiology and Infectious Diseases (ESCMID Research Grant 2015 VNP), Aaltonen Foundation (VNP), Otto A. Malmi Foundation (VNP), The São Paulo Research Foundation (FAPESP grant #2011/24035-2 and #2012/23290-1, CMO) and the Europen Research Council (Contract #647916, IGB). No agency had any role in the interpretation of the results or in the preparation of the article. Alexander Herbig is warmly thanked for the help in the analysis of the metagenomic data of the European Copper Age glacier mummy. Xavier Bosch, Samuel Alizon and Yannis Michalakis are thanked for helpful comments and review of an initial version of the article.
References
- 1000 Genomes Project Consortium, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA.2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, Babrzadeh F, Gharizadeh B, Luo M, Plummer FA, et al. 2011. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonsson A, Karanfilovska S, Lindqvist PG,, Hansson BG. 2003. General acquisition of human papillomavirus infections of skin occurs in early infancy. J Clin Microbiol. 41:2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashford RW. 2000. Parasites as indicators of human biology and evolution. J Med Microbiol. 49:771–772. [DOI] [PubMed] [Google Scholar]
- Baele G, Lemey P, Bedford T, Rambaut A, Suchard MA, Alekseyenko AV. 2012. Improving the accuracy of demographic and molecular clock model comparison while accommodating phylogenetic uncertainty. Mol Biol Evol. 29:2157–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baele G, Li WLS, Drummond AJ, Suchard MA, Lemey P. 2013. Accurate model selection of relaxed molecular clocks in bayesian phylogenetics. Mol Biol Evol. 30:239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SA, Stamatakis A. 2011. Aligning short reads to reference alignments and trees. Bioinformatics 27:2068–2075. [DOI] [PubMed] [Google Scholar]
- Bernard HU. 1994. Coevolution of papillomaviruses with human populations. Trends Microbiol. 2:140–143. [DOI] [PubMed] [Google Scholar]
- Berumen J, Ordoñez RM, Lazcano E, Salmeron J, Galvan SC, Estrada RA, Yunes E, Garcia-Carranca A, Gonzalez-Lira G, Madrigal-de la Campa A. 2001. Asian-American variants of human papillomavirus 16 and risk for cervical cancer: a case-control study. J Natl Cancer Inst. 93:1325–1330. [DOI] [PubMed] [Google Scholar]
- Bouvard V, Baan R, Straif K, Grosse Y, Secretan B, Ghissassi FE, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, et al. 2009. A review of human carcinogens—Part B: biological agents. Lancet Oncol. 10:321–322. [DOI] [PubMed] [Google Scholar]
- Bravo IG, Félez-Sánchez M. 2015. Papillomaviruses: viral evolution, cancer and evolutionary medicine. Evol Med Public Heal. 2015:32–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruni L, Diaz M, Castellsagué X, Ferrer E, Bosch FX, de Sanjosé S. 2010. Cervical human papillomavirus prevalence in 5 continents: meta-analysis of 1 million women with normal cytological findings. J Infect Dis. 202:1789–1799. [DOI] [PubMed] [Google Scholar]
- Buck LT, Stringer CB. 2014. Homo heidelbergensis. Curr Biol. 24:R214–R215. [DOI] [PubMed] [Google Scholar]
- Burk RD, Terai M, Gravitt PE, Brinton LA, Kurman RJ, Barnes WA, Greenberg MD, Hadjimichael OC, Fu L, McGowan L, et al. 2003. Distribution of human papillomavirus types 16 and 18 variants in squamous cell carcinomas and adenocarcinomas of the cervix. Cancer Res. 63:7215–7220. [PubMed] [Google Scholar]
- Bzhalava D, Eklund C, Dillner J. 2015. International standardization and classification of human papillomavirus types. Virology 476:341–344. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand M-F, Morel V, Piouffre L, Bodmer J, Bodmer WF, Bonne-Tamir B, Cambon-Thomsen A, et al. 2002. A human genome diversity cell line panel. Science 296:261–262. [DOI] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Chesson HW, Dunne EF, Hariri S, Markowitz LE. 2014. The estimated lifetime probability of acquiring human papillomavirus in the United States. Sex Transm Dis. 41:660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet I, Gheit T, Franceschi S, Vignat J, Burk RD, Sylla BS, Tommasino M, Clifford GM. 2012. Human papillomavirus type 16 genetic variants: phylogeny and classification based on E6 and LCR. J Virol 86:6855–6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet I, Gheit T, Iannacone MR, Vignat J, Sylla BS, Del Mistro A, Franceschi S, Tommasino M, Clifford GM. 2013. HPV16 genetic variation and the development of cervical cancer worldwide. Br J Cancer. 108:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M, Andrús AM, Kelso J, Rambaut A. 2016. Neandertal- and Denisovan-like Haplotypes Contributes to Adaptive Variation in Human Toll-like Receptors. Am J Hum Genet. 98:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9:772.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. 2012. The biology and life-cycle of human papillomaviruses. Vaccine 1–16. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einstein MH, Schiller JT, Viscidi RP, Strickler HD, Coursaget P, Tan T, Halsey N, Jenkins D. 2009. Clinician’s guide to human papillomavirus immunology: knowns and unknowns. Lancet Infect Dis. 9:347–356. [DOI] [PubMed] [Google Scholar]
- Freitas LB, Chen Z, Muqui EF, Boldrini NAT, Miranda AE, Spano LC, Burk RD. 2014. Human papillomavirus 16 non-European variants are preferentially associated with high-grade cervical lesions. PLoS One 9:e100746.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PLF, Aximu-Petri A, Prüfer K, de Filippo C, et al. 2014. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514:445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Vallvé S, Alonso A, Bravo IG. 2005. Papillomaviruses: different genes have different histories. Trends Microbiol. 13:514–521. [DOI] [PubMed] [Google Scholar]
- Gottschling M, Göker M, Stamatakis A, Bininda-Emonds ORP, Nindl I, Bravo IG. 2011. Quantifying the phylodynamic forces driving papillomavirus evolution. Mol Biol Evol. 28:2101–2113. [DOI] [PubMed] [Google Scholar]
- Gottschling M, Stamatakis A, Nindl I, Stockfleth E, Alonso A, Bravo IG. 2007. Multiple evolutionary mechanisms drive papillomavirus diversification. Mol Biol Evol. 24:1242–1258. [DOI] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH-Y, et al. 2010. A draft sequence of the Neandertal genome. Science 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho L, Chan SY, Burk RD, Das BC, Fujinaga K, Icenogle JP, Kahn T, Kiviat N, Lancaster W, Mavromara-Nazos P. 1993. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J Virol. 67:6413–6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes EC. 2008. Evolutionary history and phylogeography of human viruses. Annu Rev Microbiol. 62:307–328. [DOI] [PubMed] [Google Scholar]
- Hurles ME, Nicholson J, Bosch E, Renfrew C, Sykes BC, Jobling MA. 2002. Y chromosomal evidence for the origins of oceanic-speaking peoples. Genetics 160:289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KP, Adams RJ, Page RDM, Clayton DH. 2003. When do parasites fail to speciate in response to host speciation? Syst Biol. 52:37–47. [DOI] [PubMed] [Google Scholar]
- Kapusinszky B, Mulvaney U, Jasinska AJ, Deng X, Freimer N, Delwart E. 2015. Local virus extinctions following a host population bottleneck. J Virol 89:8152–8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30:772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Newman TL, Tuzun E, Kaul R., Eichler EE. 2007. Population stratification of a common APOBEC gene deletion polymorphism PLoS Genet. 3:e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 22:1208–1222. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. 2006. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 23:1891–1901. [DOI] [PubMed] [Google Scholar]
- Kuhlwilm M, Gronau I, Hubisz MJ, de Filippo C, Prado-Martinez J, Kircher M, Fu Q, Burbano HA, Lalueza-Fox C, de la Rasilla M, et al. 2016. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature 530:429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Subramanian S. 2002. Mutation rates in mammalian genomes. Proc Natl Acad Sci U S A. 99:803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. 2004. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol. 21:1095–1109. [DOI] [PubMed] [Google Scholar]
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics [Internet] 25:1451–1452. Available from: http://www.ncbi.nlm.nih.gov/pubmed/19346325 [DOI] [PubMed] [Google Scholar]
- Lippold S, Xu H, Ko A, Li M, Renaud G, Butthof A, Schröder R, Stoneking M. 2014. Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences. Investig Genet. 5:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Martinón-Torres M, Cai Y, Xing S, Tong H, Pei S, Sier MJ, Wu X, Edwards RL, Cheng H, et al. 2015. The earliest unequivocally modern humans in southern China. Nature 526:696–699. [DOI] [PubMed] [Google Scholar]
- Lopera EA, Baena A, Florez V, Montiel J, Duque C, Ramirez T, Borrero M, Cordoba CM, Rojas F, Pareja R, et al. 2014. Unexpected inverse correlation between Native American ancestry and Asian American variants of HPV16 in admixed Colombian cervical cancer cases. Infect Genet Evol. 28:339–348. [DOI] [PubMed] [Google Scholar]
- López-Bueno A, Mavian C, Labella AM, Castro D, Borrego JJ, Alcami A, Alejo A. 2016. Concurrence of iridovirus, polyomavirus, and a unique member of a new group of fish papillomaviruses in lymphocystis disease-affected gilthead Sea Bream. J Virol. 90:8768–8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Madupu R, Karaoz U, Nossa CW, Yang L, Yooseph S, Yachimski PS, Brodie EL, Nelson KE, Pei Z. 2014. Human papillomavirus community in healthy persons, defined by metagenomics analysis of human microbiome project shotgun sequencing data sets. J Virol. 88:4786–4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maixner F, Thomma A, Cipollini G, Widder S, Rattei T, Zink A. 2014. Metagenomic analysis reveals presence of Treponema denticola in a tissue biopsy of the Iceman. PLoS One 9:e99994.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez FL, Poznik GD, Castellano S, Bustamante CD. 2016. The divergence of Neandertal and modern human Y chromosomes. Am J Hum Genet. 98:728–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza L, Picconi MA, Mirazo S, Mongelós P, Giménez G, Basiletti J, Arbiza J. 2013. Distribution of HPV-16 variants among isolates from Paraguayan women with different grades of cervical lesion. Int J Gynecol Obstet. 122:44–47. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molden T, Feiring B, Ambur OH, Christiansen IK, Hansen M, Laake I, Meisal R, Myrvang E, Jonassen CM, Trogstad L. 2016. Human papillomavirus prevalence and type distribution in urine samples from Norwegian women aged 17 and 21 years: a nationwide cross-sectional study of three non-vaccinated birth cohorts. Papillomavirus Res. 2:153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscicki A-B, Schiffman M, Burchell A, Albero G, Giuliano AR, Goodman MT, Kjaer SK, Palefsky J. 2012. Updating the natural history of human papillomavirus and anogenital cancers. Vaccine 30 Suppl 5:F24–F33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picconi MA, Alonio LV, Sichero L, Mbayed V, Villa LL, Gronda J, Campos R, Teyssié A. 2003. Human papillomavirus type-16 variants in Quechua aboriginals from Argentina. J Med Virol. 69:546–552. [DOI] [PubMed] [Google Scholar]
- Pond SK, Muse SV. 2005. Site-to-site variation of synonymous substitution rates. Mol Biol Evol. 22:2375–2385. [DOI] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus OG, Rambaut A, Harvey PH. 2000. An integrated framework for the inference of viral population history from reconstructed genealogies. Genetics 155:1429–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin P, Stoneking M. 2015. Denisovan ancestry in East Eurasian and native American populations. Mol Biol Evol. 32:2665–2674. [DOI] [PubMed] [Google Scholar]
- Qmichou Z, Khyatti M, Berraho M, Ennaji MM, Benbacer L, Nejjari C, Benjaafar N, Benider A, Attaleb M, El Mzibri M. 2013. Analysis of mutations in the E6 oncogene of human papillomavirus 16 in cervical cancer isolates from Moroccan women. BMC Infect Dis. 13:378.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rector A, Lemey P, Tachezy R, Mostmans S, Ghim S-J, Van Doorslaer K, Roelke M, Bush M, Montali RJ, Joslin J, et al. 2007. Ancient papillomavirus-host co-speciation in Felidae. Genome Biol. 8:R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DL, Smith VS, Hammond SL, Rogers AR, Clayton DH. 2004. Genetic analysis of lice supports direct contact between modern and archaic humans. PLoS Biol. 2:e340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468:1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DF, Foulds LR. 1981. Comparison of phylogenetic trees. Math Biosci. 53:131–147. [Google Scholar]
- Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S, Renaud G, Viola B, Hublin J-JJ, Gansauge MM-T, Shunkov MV, Derevianko AP, Pru fer K, Kelso J, Pa a bo S. 2015. Nuclear and mitochondrial DNA sequences from two Denisovan individuals. Proc Natl Acad Sci U S A.112:2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A, Durbin R. 2012. Revising the human mutation rate: implications for understanding human evolution. 13:745–753. [DOI] [PubMed] [Google Scholar]
- Schiffman M, Rodriguez AC, Chen Z, Wacholder S, Herrero R, Hildesheim A, Desalle R, Befano B, Yu K, Safaeian M, et al. 2010. A population-based prospective study of carcinogenic human papillomavirus variant lineages, viral persistence, and cervical neoplasia. Cancer Res. 70:3159–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SD, Doorbar J, Goldstein RA. 2010. Analysis of host-parasite incongruence in papillomavirus evolution using importance sampling. Mol Biol Evol. 27:1301–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM. 2002. Origins of human virus diversity. Cell 108:305–312. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. 1999. Letter to the editor multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 16:1114–1116. [Google Scholar]
- Soria-Carrasco V, Talavera G, Igea J, Castresana J. 2007. The K tree score: quantification of differences in the relative branch length and topology of phylogenetic trees. Bioinformatics 23:2954–2956. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer CB, Barnes I. 2015. Deciphering the Denisovans. Proc Natl Acad Sci U S A. 112:15542–15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SE, Garland SM, Rumbold AR, Zardawi I, Taylor-Thomson D, Condon JR, Tabrizi SN. 2013. Investigating a cluster of vulvar cancers in young women: distribution of human papillomavirus and HPV-16 variants in vulvar dysplastic or neoplastic biopsies. Sex Health. 10:18–25. [DOI] [PubMed] [Google Scholar]
- Vernot B, Akey JM. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science 343:1017–1021. [DOI] [PubMed] [Google Scholar]
- Vernot B, Akey JM. 2015. Complex history of admixture between modern humans and Neandertals. Am J Hum Genet. 96:448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernot B, Tucci S, Kelso J, Schraiber JG, Wolf AB, Gittelman RM, Dannemann M, Grote S, McCoy RC, Norton H, et al. 2016. Excavating Neandertal and Denisovan DNA from the genomes of Melanesian individuals. Science 352:235–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa LL, Sichero L, Rahal P, Caballero O, Ferenczy A, Rohan T, Franco EL. 2000. Molecular variants of human papillomavirus types 16 and 18 preferentially associated with cervical neoplasia. J Gen Virol. 81:2959–2968. [DOI] [PubMed] [Google Scholar]
- Wertheim JO, Kosakovsky Pond SL. 2011. Purifying selection can obscure the ancient age of viral lineages. Mol Biol Evol. 28:3355–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim JO, Smith MD, Smith DM, Scheffler K, Kosakovsky Pond SL. 2014. Evolutionary origins of human herpes simplex viruses 1 and 2. Mol Biol Evol. 31:2356–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. 1951. The genetical structure of populations. Ann Eugen. 15:323–354. [DOI] [PubMed] [Google Scholar]
- Xi LF, Kiviat NB, Hildesheim A, Galloway DA, Wheeler CM, Ho J, Koutsky LA. 2006. Human papillomavirus type 16 and 18 variants: race-related distribution and persistence. J Natl Cancer Inst. 98:1045–1052. [DOI] [PubMed] [Google Scholar]
- Zuna RE, Tuller E, Wentzensen N, Mathews C, Allen RA, Shanesmith R, Dunn ST, Gold MA, Wang SS, Walker J, et al. 2011. HPV16 variant lineage, clinical stage, and survival in women with invasive cervical cancer. Infect Agent Cancer. 6:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.