Abstract
Bivalves have evolved a range of complex shell forming mechanisms that are reflected by their incredible diversity in shell mineralogy and microstructures. A suite of proteins exported to the shell matrix space plays a significant role in controlling these features, in addition to underpinning some of the physical properties of the shell itself. Although, there is a general consensus that a minimum basic protein tool kit is required for shell construction, to date, this remains undefined. In this study, the shell matrix proteins (SMPs) of four highly divergent bivalves (The Pacific oyster, Crassostrea gigas; the blue mussel, Mytilus edulis; the clam, Mya truncata, and the king scallop, Pecten maximus) were analyzed in an identical fashion using proteomics pipeline. This enabled us to identify the critical elements of a “basic tool kit” for calcification processes, which were conserved across the taxa irrespective of the shell morphology and arrangement of the crystal surfaces. In addition, protein domains controlling the crystal layers specific to aragonite and calcite were also identified. Intriguingly, a significant number of the identified SMPs contained domains related to immune functions. These were often are unique to each species implying their involvement not only in immunity, but also environmental adaptation. This suggests that the SMPs are selectively exported in a complex mix to endow the shell with both mechanical protection and biochemical defense.
Keywords: biomineralization, shell matrix proteins, calcification, calcite, aragonite, evolution
Introduction
Bivalves are the second most bio-diverse extant molluscan class after the gastropods (Bieler et al. 2014) with more than 9000 living species and they are characterized by a tremendous array of shell architectures. These shells play a key role in protecting bivalves from predators, pathogens and to some extent from other environmental conditions, such as desiccation, wave action and iceberg damage (Checa 1993; Harper et al. 2012). The shells are formed by a biologically controlled process (biomineralization), which results in a composite material that is made of approximately 95% calcium carbonate (CaCO3) and between 1% and 5% organic components (Lowenstam and Weiner 1989). In nature, CaCO3 exists as different crystal polymorphs such as aragonite, calcite, and vaterite, which along with crystal size and organic matrix endow these biomineralized structures with their unique physio-chemical properties (Harper 2000). It is of great interest to material scientists that the aragonite and calcite shells formed by bivalves are very durable compared with the geological inorganic forms. For example, aragonite formed biologically by bivalves is 3000 times tougher than pure aragonite (Currey 1977). The CaCO3 crystals in shells are arranged in layers with a distinctive pattern to form complex microstructures. So far, more than 30 different microstructures of bivalve CaCO3 have been documented (Carter 1980).
In this regard, paleontological research has greatly aided our understanding of shell microstructure evolution. Bivalve fossil records date back to the Cambrian, although the first radiation event occurred in the Ordovician. There is substantive evidence that the primitive mineralogy was aragonite (Vendrasco et al. 2011) with clear patterns in the developing mineralogy associated with the seawater chemistry in which these organisms evolved (Wood and Zhuravlev 2012). The calcitic mineralogy is likely to have derived after the taxa evolved along with the changes in the global environment from the aragonitic-facilitating seas of the Ediacaran/early Cambrian to the subsequent calcitic seas (542–488 My) (Wood and Zhuravlev 2012). Modern bivalves secrete a range of both aragonite and calcite and the development of different mineralogies and structures is clearly a polyphyletic trait (Harper 2016). Why this complex pattern of evolution has occurred is still under debate, but may not be related to the different solubilities of the calcium polymorphs (Harper 2016).
It is commonly accepted that shell formation is biologically controlled through diverse molecules such as proteins, glycoproteins, lipids, and carbohydrates (Goulletquer and Wolowicz 1989). The majority of the research to date has focused on the shell matrix proteins (SMPs) and sugar moieties present in the shell matrix space. SMPs play a significant role in CaCO3 crystal nucleation, crystal growth and in generating the different crystal polymorphs even though they constitute only a very minor proportion of the shell matrix (Wheeler et al. 1981; Falini et al. 1996). SMPs are secreted in the mantle, an epithelial tissue lining the shell and are exported to the shell matrix space (Addadi et al. 2006).
Proteomic research investigating SMPs has been hampered by the lack of genomic resources in these non-model organisms. These data, namely, genome and transcriptome sequences, are needed to provide the cDNA transcripts or gene predictions, against which, the peptide fragments can be matched. Access to these sequences significantly enhances the chances of identifying meaningful functional information (Joubert et al. 2010). Initially, the mollusc SMPs were predicted from the genomic and transcriptomic data from mantle tissue (Miyamoto et al. 1996; Sudo et al. 1997) but this did not confirm their presence and incorporation into the shell matrix space. Next generation sequencing has enabled the rapid development of mantle transcriptome databases, with the resultant ESTs used as reference sequences to identify the peptides or proteins extracted from shell matrix space (Joubert et al. 2010; Berland et al. 2011). So far, SMPs from a variety of different mollusc species have been identified employing this approach, e.g., L. gigantea (Marie et al. 2013), H. asinina (Marie et al. 2010), H. cumingii (Berland et al. 2013), P. margaretifera, and P. maxima (Marie et al. 2012), C. gigas (Marie et al. 2011; Zhang et al. 2012), M. edulis (Marie et al. 2011; Liao et al. 2015) M. galloprovincialis (Gao et al. 2015) etc. Also, many groups have generated transcriptomic data from the mantle of different species, which could facilitate future shell proteomic studies (Jeong et al. 2007; Clark et al. 2010; Joubert et al. 2010; Shi et al. 2013; Artigaud et al. 2014; Freer et al. 2014). A preliminary outcome from analyses of these data is the recognition of the complexity of the molecular and structural diversity of the SMPs that are brought into play by the different species for growth and control of their intricately ordered shell structures, resulting in few shared proteins even with in a taxon (Jackson et al. 2010).
The analysis of specific bio-mineralizing proteins in divergent taxa such as sea urchins, gastropods, and bivalves has revealed the evolutionary divergence and independent parallel evolution of these genes (Jackson et al. 2010). In spite of having similar morphological traits, the SMPs even from closely related bivalve clades show very different protein sequences with extensive domain shuffling. For example, the PIF protein from P. margertifera, P. maxima P. penguin, and M. galloprovincialis show different primary amino acid sequences with diverse domain arrangements (Suzuki et al. 2009). However, some exceptions exist, such as the carbonic anhydrases, which are frequently found in the shell matrix and are highly conserved throughout the metazoans (Le Roy et al. 2014). Establishing a complete catalogue of proteins and their functions present in the shell matrix space is critical, not only understanding their role in an evolutionary context but also in terms of the constraints faced by the organism in their living environment and their provision of adaptive plasticity. With the latter becoming increasingly important with regard to understanding how shell microstructures impact on the plasticity of the shell and the ability of the organisms to cope with environmental change (Charrier et al. 2013).
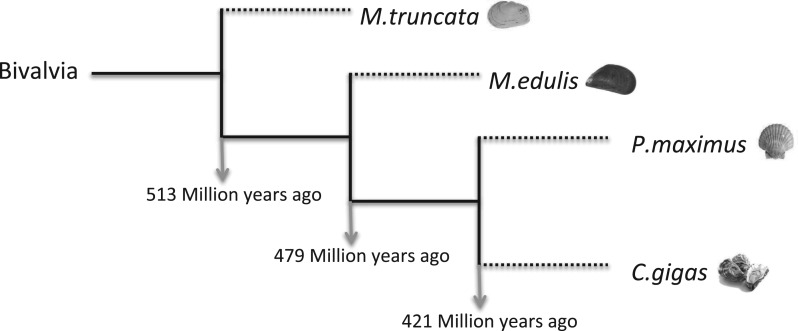
In this study, the SMPs identified from three model species namely, the Pacific oyster (Crassostrea gigas), the blue mussel (Mytilus edulis), and the king scallop (Pecten maximus) are presented. These data are supplemented with the recently published proteomic data of the soft shell clam (Mya truncata) (Arivalagan et al. 2016) to enable a comparison of the SMPs of highly divergent species with the aim of under covering an evolutionary conserved suite of shell proteins. These species are widely distributed in the North Sea and are of considerable economic importance in Europe. They differ in their evolutionary lineage, modus vivendi and shell structure. M. truncata (taxon: Heteroconchia) diverged from the other three species (taxon: Pteriomorphia) approximately 513 My ago (Plazzi and Passamonti 2010). Within the Pteriomorphia bivalves, C. gigas (clade: Ostreoida), and P. maximus (clade: Pectinoida) diverged from M. edulis (clade: Mytiloida) around 479 My ago. The divergence of the Ostreoida and Pectinoida is estimated to be 421 My ago (Ren et al. 2010) (fig. 1).
Fig. 1.
Phylogeny of four bivalve models and their corresponding divergence times based on a Bayesian analysis using mitochondrial genes (Plazzi and Passamonti 2010; Ren et al. 2010).
In terms of life style, C. gigas usually found in intertidal and sub-tidal zones, they settle on one another in large numbers and can even form reefs (Wrange et al. 2010). M. edulis lives in intertidal zones by attaching to any roughened, scarred or pitted surface by means of byssal threads (Seed 1969). P. maximus are free swimmers and live in relatively shallow waters. Unlike the other three models, M. truncata is burrower and lives 1–2 feet below the seabed and uses its siphon to filter microalgae. Also, the mineralogy and microstructure of these species differ significantly from one another. The C. gigas shell is purely calcite: the outer shell layers are calcite prisms, the middle shell layers are chalky calcite and inner shell layers are foliated calcite (Marie et al. 2011). In contrast, M. truncata shell is composed only of aragonite, with a granular microstructure in the outer layer, and cross-lamellar aragonite in the inner layer. Both P. maximus and M. edulis shells are a combination of both calcitic and aragonitic layers, but with differing organization. The M. edulis shell is composed of calcite prisms in the outer layer, with middle and inner layers of aragonite nacre. P. maximus has outer and inner layers of foliated calcite layers and a middle layer of cross-lamellar aragonite.
To date, the majority of the research on SMPs has been focused on their roles in the context of the biological control of shell formation, calcium biochemistry pathways, and crystal growth. However, the role of other proteins present in the shell matrix space has been ignored or at best considered as cellular contaminants. In this study, the shell matrix proteins of these four evolutionary divergent bivalves (C. gigas, M. edulis, M. truncata, and P. maximus) were extracted and analyzed in an identical fashion via proteomics pipeline. This enabled robust interrogation of the data to test the hypothesis that there is an ancient conserved “tool box” of calcification proteins and that proteins of varying functional domains are exported selectively to the shell matrix space to enhance successful species-specific adaptation.
Materials and Methods
Sample Preparation
Bivalve samples were collected at different locations in west coast of Scotland (supplementary fig. S1, Supplementary Material online). Proteomics experiment was carried out using the shells from the animals used for generating transcriptome data. Each shell was washed overnight with 5–10% sodium hypochlorite (NaOCl), to remove any organic impurities and the periostracum. Any remaining periostracum was removed by manual abrasion. The shells were then washed with water for an hour and air-dried. The shells were then cut and polished with a dremel tool. The cut shell pieces were washed briefly with NaOCl and then rinsed with water and air-dried. A mortar and pestle was used to grind the shell pieces into a powder, which was then graded using a 250 µm mesh.
Shell powders (3 g) were decalcified using 5% cold acetic acid for an hour and then left in 10% acetic acid overnight for the complete dissolution of CaCO3. The resulting liquid was centrifuged (14,000 rpm, 20 min, 4 °C) to separate the supernatant [acid soluble (ASM)] and the pellet [acid insoluble fractions (AIM)]. The AIM was washed five times with water, freeze-dried. The supernatant was concentrated and washed with water by centrifugation through a 10 kDa filter (Sartorius, VIVASPIN 20) to enrich the proteins from small molecules, such as peptides and other impurities. The filter containing the ASM proteins was then washed with water and freeze-dried. Prior to the digestion of the proteins, denaturation was performed, by treating both ASM and AIM samples with 30 µL of 8 M urea solution for an hour at 37 °C. Next 15 mM dithiothreitol (DTT) dissolved in 100 mM triethyl ammonium bicarbonate (TEAB) was added and an incubation at 37 °C performed for 1 h. Alkylation was performed by the further addition of 20 µL of 15 mM iodoacetamide, with the samples kept in the dark at room temperature for 1 h. After this alkylation step, trypsin was added to the AIM and ASM samples (8.5 and 5 µg, respectively) and incubated at 37 °C overnight. The samples were acidified with 10 µL of 10% formic acid and the digested peptides were desalted, by applying 40 µL of the digested matrix samples to 4 mm Empore™ SPE (Sigma-Aldrich, France) cartridges. The resulting peptides were quantified using the Bicinchoninic acid kit (Sigma-Aldrich, France).
Mass Spectrometry Analysis
One µg of the peptide sample was obtained by mixing both AIM and ASM samples and the mix was analyzed in a nano LC system (Dionex Ultimate 3000, France) coupled to a LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, France). The injected digest was enriched on a 5 mm C18 column (5 µm, 100 Å pore, 300 µm i.d., LC packing, France) and the peptides were separated on a 50 cm nano-column packed with C18 phase (3 µm, 75 µm i.d.) at a flow rate of 300 nL·min−1 using the following gradient: 1% solvent B (98% ACN, 0.1% formic acid) to 40% B in 180 min, 40% B to 60% B in 2 min, 60% B for 28 min. The mass spectrometer was operated in positive ion mode and the MS and MS/MS spectra acquired in the Orbitrap and linear ion trap, respectively. The parameters used for survey scans were 300–2000 m/z, resolution 30,000 AGC (automatic gain control) target 2 × 105 and maximum injection time 100 ms. CID fragmentation was performed for 20 of the most abundant precursors with the following parameters: minimum intensity 500, isolation window 2 Da, normalized collision energy 35%, AGC target 5000, and maximum injection time 100 ms, dynamic exclusion was enabled (repeat count 1, duration 80 s).
Data Analysis
MS/MS spectra were searched against the six-frame translated mantle transcriptomic databases of P. maximus, M. edulis, and C. gigas from British Antarctic Survey (Yarra et al. 2016) using in-house Mascot server (Matrix Science, London, UK; version 2.4.1) These data were supplemented by additional protein sequences of C. gigas from Uniprot (http://uniprot.org, last accessed April 2015). The following parameters were used for the database search: carbamido-methylation of cysteine as a fixed modification and oxidation of methionine and de-amidation of aspartic acid and asparagine as variable modifications. The mass tolerance for MS and MS/MS experiments was set as 10 ppm and 0.5 Da respectively. Protein identification results from Mascot were further validated using Scaffold software (version 3.6.5, Proteome Software Inc., Portland, USA). Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the Peptide Prophet algorithm. Protein identifications were validated if they could be established at greater than 95.0% probability (assigned by Protein Prophet algorithm) and matched at least two unique peptides, each containing minimum eight amino acids. False discovery rate was estimated to be <1% by the scaffold software.
BLAST2GO tool (version 3.3) was used to carry out sequence similarity protein searches against the NCBI database. Interspecies comparison of the SMPs from each species was carried out using locally installed NCBI BLAST tool version 2.4.0 (Blast-p) (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/, last accessed March 2016) with SEG filtering disabled. Circos (http://circos.ca/- version 0.69, last accessed March 2016) was used to represent the BLAST-p results. Conserved domains were detected using Simple Modular Architecture Research (SMART) domain prediction tool (http://smart.embl.de/, last accessed March 2016).
Results and Discussion
Although the literature is abundant with lists of genes and proteins identified from the mantle tissue and also the shell matrix space of different species, a robust comparison, in terms of the number and nature of the identified proteins, is difficult due to the different extraction and analytical techniques employed. Therefore, in this study, protein extraction, peptide generation, and data analysis were carried out under same conditions, thus enabling a much more reliable comparative study of SMPs from the four different species and investigation of a potentially ancient shell-building molecular tool kit. The shell proteome of M. truncata was recently published by our group (Arivalagan et al. 2016) wherein we employed the same analytical workflow as described in this work, which enables us to compare the shell proteome of the four species. It is still under debate as to whether the shell building mechanism is conserved in molluscs. Previous molecular phylogeny and paleontological studies have identified multiple origins of nacre evolution within the Mollusca (Jackson et al. 2010; Vendrasco et al. 2011) and this is merely one of the possible components of a mollusc shell. To date, the existence of an ancient conserved shell building molecular mechanism within the bivalves remains unexplored. In this study, we specifically chose to carry out an inter-species comparison of molluscs that exhibit a range of shell mineralogy and microstructures to gain insight into the conservation of SMPs.
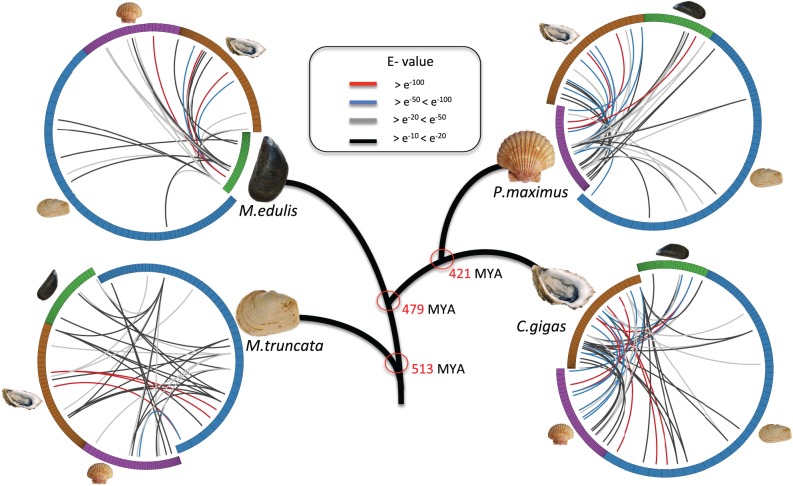
55, 46, and 46 different SMPs were identified in the shell matrices of C. gigas, M. edulis, and P. maximus, respectively (supplementary tables S1 and S2, Supplementary Material online) and in case of M. truncata 67 SMPs had been identified in a recent study (Arivalagan et al. 2016) (supplementary table S3, Supplementary Material online). It is worth to note that proteins reported are inferred from the peptides detected in MS/MS experiments and potentially represent a part of the shell forming proteome. The SMPs were searched against NCBI-nr database using BLAST-p program in order to find similar proteins with known functions. 24 similar proteins (≈48% of the identified SMPs) were retrieved in P. maximus, 16 in M. edulis (≈34%), 31 in M. truncata (≈46%) and 48 in C. gigas (≈87%). The higher number of similar proteins was found in C.gigas is due to the availability of an annotated genome. The number and extent of sequence similarities between the identified SMPs varies with specific-species comparisons (fig. 2). A high degree of matches was found between C. gigas and P. maximus and the least number of significant matches was obtained in comparisons of M. edulis to the other species. In general, the results are consistent with evolutionary divergence times except for M. edulis. This was surprising, as based solely on the principle of gene conservation over evolutionary time, it was expected that M. edulis would share greater number of similar proteins with C. gigas and P. maximus since they are closer in divergence times. However, M. truncata actually shared more proteins with C. gigas and P. maximus than M. edulis. This result cannot be explained purely by the differences in the crystal structure of the shells of the different species (Hedegaard and Wenk 1998; Chateigner et al. 2000), as the C. gigas shell is completely calcite, whereas that of M. truncata is completely aragonite. M. edulis, which is a mix of aragonite and calcite, would be expected to share significant protein homologies with both.
Fig. 2.
Circos representation of the sequence similarities of identified SMPs from the four bivalve models (cut-off E-value ≥ e−10). Colored lines represent similarity scores, with the corresponding E-values denoted in the figure.
Several other factors might be contributing to this surprising finding, which relate to environmental factors and the innate highly plastic adaptation strategies of Mytilus species. For example, M. edulis species exhibit a high degree of tolerance to freezing and to variations in salinity (Williams 1970). Several populations of M. edulis have shown rapid adaptation to changes in their environment (temperature, salinity, ocean acidification, availability of food, etc.) over only a few generations (Widdows 1978; Vuorinen et al. 2002) by altering not only their shell structure but also their life history traits, such as, shell size, life span and reproductive behavior. In addition, M. edulis has several hybridization zones with M. trossulus and M. galloprovincialis in the Atlantic Ocean, the Baltic, and North seas (Gardner 1996; Riginos and Cunningham 2005). These factors might have exerted selective pressure on the shell proteins in M. edulis to evolve several times in parallel.
In spite of the considerable differences in SMPs between the four species, a set of shared SMPs was identified between the four species, which indicate the existence of a shell-forming “molecular tool box” inherited from their common ancestors and potentially conserved among the class Bivalvia for the purpose of building a calcified shell (Marin et al. 2007). Although, BLAST sequence similarity analysis enabled the identification of longer cDNA transcripts within the transcriptome, matching the peptide fragments isolated in this study, the functional annotation of such transcripts was incomplete. In order to detect multiple domains within the same protein from divergent evolutionary sources, a domain prediction tool was used to provide additional annotation and further compare the SMPs in the context of their functions (Schultz et al. 1998, 2000). The final annotations were binned into different functional groups and are discussed in detail below.
Biocalcification—the Basic Tool Kit
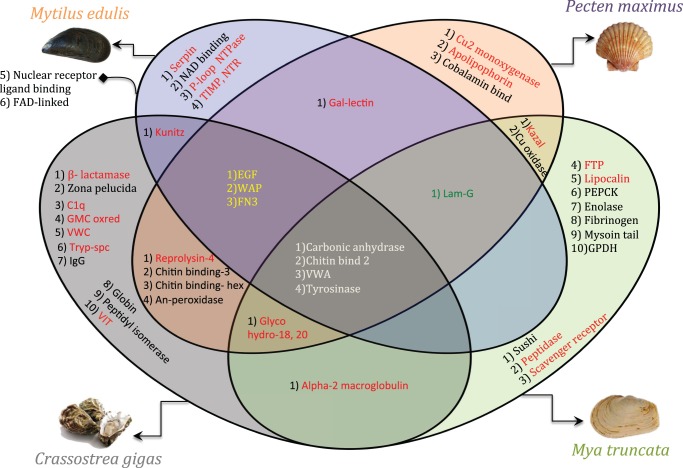
This study identified proteins with functional domains that are common to the shell matrix space of all four species (fig. 3). Thus, irrespective of shell morphology and microstructure, we suggest that these four domains are evolutionarily maintained and represent part of the “basic tool kit” for the construction of the CaCO3 molluscan exo-skeleton. These domains are tyrosinase, carbonic anhydrase, chitin binding-2, and Von Willebrand factor-A.
Fig. 3.
Venn diagram of the protein-domains identified from the four bivalve SMPs. Domains common to the four bivalve models are indicated in white. Domains in yellow and green are related to calcite and aragonite shell layers. Immunity-related domains are represented in red.
The main function of the tyrosinase domain in invertebrates is sclerotization, particularly, the arthropods. In this process, insoluble protein polymers (Nagai et al. 2007) are formed by transforming tyrosine to L-3,4-dihydroxyphenylalanine (L-DOPA), which is the same mechanism employed in the formation of periostracum in bivalves (Zhang et al. 2006). Tyrosinase has undergone multiple lineage-restricted expansions, even in the closely related bivalve super families Ostroidea (Crassostrea) and Pterioidea (Pinctada) clades (Aguilera et al. 2014). Similarly, in the shells of the four species under investigation, tyrosinases of different lengths and domain arrangements were identified (supplementary fig. S2, Supplementary Material online). C. gigas and P. maximus had paralogous tyrosinase domain-containing proteins and it has been hypothesized that these two species may have evolved these paralogs to perform two distinctive functions during shell formation (Nagai et al. 2007). In the case of P. maximus and M. edulis, one protein with two-tyrosinase domain repeats was identified, but the functional significance of such tyrosinase repeats is yet to be discovered.
Carbonic anhydrase (CA) belongs to the metallo-enzyme superfamily; it plays a key role in catalyzing the reverse hydration of carbon dioxide during calcium carbonate formation. Earlier experiments on these domain-containing proteins documented their role either in calcification (Marie et al. 2008) or as an inhibitor of the calcification process (Nacrein) (Miyamoto et al. 1996). In the present study, this domain differed in sequence length between species. However, in spite of this difference the conformation and activity of carbonic anhydrase in the biocalcification process is suggested to be highly conserved in metazoans (Le Roy et al. 2014).
Chitin binding 2 (ChBt-2) and Von Willebrand factor type-A (VWA) domains are extra cellular matrix domains usually found together in SMPs, such as PIF, the acidic matrix protein first identified in Pinctada fucata, Blue mussel shell protein (BMSP), etc. The main role of the ChBt-2 domain in the SMP is to interact with chitin as this is the key component of the mollusc organic matrix (Mann 1988). The VWA domain is usually found in glycoproteins and exhibits an adhesion function through protein–protein interactions (Whittaker and Hynes 2002). These domains are also components of hydrophilic glyco-proteins, which usually assemble to form silk-like protein complexes (Weiner 1979), which play a key role in chitin-scaffolding and in arranging CaCO3 crystals to such scaffolds.
Crystal Lattice Associated Proteins
As indicated earlier, all the four species have different layers of CaCO3 polymorphs in their shells. Apart from the basic tool kit, a subset of protein domains was identified that was common to the shells with similar crystalline layers and these domains potentially influence the specific nucleation or arrangement of particular polymorphs of CaCO3. Three domains were observed in shells containing a calcite layer: epidermal growth factor (EGF), fibronectin-3 domains (FN3), and whey acidic protein (WAP) domains. These were absent in M. truncata, which has a shell made entirely of aragonite entirely (fig. 3).
The EGF domain is a calcium-binding motif and comprises 45 amino acids arranged in two anti-parallel β-sheets with several cysteine residues (Stenflo et al. 2000). The EGF-like domain has been reported previously in shell matrix proteins, but the authors noted that this protein only occurred in the prismatic (calcitic) layers but not in the nacreous layer of Pinctada shells (Marie et al. 2011). EGF domains are always found in the SMPs as tandem repeats and this repeating design is favored in any system during evolution due to its stability against enzyme attack, with the repeating structures forming compact dimeric or tetrameric pseudo-symmetrical structures (Miller et al. 2001) which is thought to help in the compact arrangement of calcite crystals. Although the Lam-G domain is specific to shells with aragonite layers, intriguingly it exhibits a function similar to EGF (Panayotou et al. 1989). This may be due to the presence of 8 cysteine repeats that show homology to EGF, which contains six cysteine repeats (Beck et al. 1990), implying that these domains have an important role in specific crystal nucleation events and arrangements.
The extracellular matrix adhesive glycoprotein, fibronectin (Donaldson and Mahan 1983) is also only identified in shells made of calcite layers. Hanein et al (1993) demonstrated the affinity of fibronectin for calcite in the presence of water molecules (Hanein et al. 1993). Thus fibronectin might help in specific arrangement of calcite crystals in the hydrogel scaffold.
WAPs are highly conserved proteinase inhibitor domains found in a whole range of species, from arthropods to mammals. WAP domains in lustrin A and perlwapin in the nacre of gastropods H. laevigata and H. asinina play major roles in shell formation (Shen et al. 1997; Treccani et al. 2006; Marie et al. 2010). Treccani et al. (2006) proposed that WAP domains inhibit crystal growth in the fast growing c-axis and thus form a platy geometrical aragonite crystal. They also reported strong interactions between perlwapin and calcite crystals (Treccani et al. 2006). The WAP domains identified in previous proteomic studies were always discussed in relation to nacre or aragonite formation. However, WAP domains have also recently been observed in fully calcite eggshells (Mann K and Mann M 2015; Rose-Martel et al. 2015). Our observations support this latter finding and it is highly likely that WAP domains are also involved in calcite formation.
Immunomodulatory Proteins
In the context of shell formation, the identification and presence of a large number of proteins containing immunity-associated domains is puzzling. On the one hand their presence in the shell matrix space seems to reinforce the notion of the protective function of the shell through the use of additional biochemical pathways. How this actually contributes to the overall defense of the organism needs further investigation. Out of 47 separate functional domains identified in this study, 21 domains (≈45%) could be related to immunity function (fig. 3). This observation may raise questions with respect to this group of proteins potentially being contaminants from the mantle tissue. Proteins such as, actins, tubulins, and myosins, which are the most common and abundant proteins in cellular tissues, can be found in proteomics studies of the shell matrix space, but they are considered to be cellular contaminants (Marie et al. 2013; Ramos-Silva et al. 2013). However, none of these highly abundant structural proteins were identified in the shell extractions described here. This verifies the efficiency of the shell cleaning and protein extraction procedures. In addition, some of the domains with immunity functions were species-specific, which suggests environmental adaptation. These immunity-domain proteins are described in more detail below.
Proteases and Protease Inhibitors
Proteases are enzymes that perform proteolysis and are found to be highly expressed in haemocytes during microbial invasions (Gerdol et al. 2011). The peptidase domain was found to be unique to M. truncata SMPs, whilst the trypsin-like serine protease (Tryp-Spc) domain was unique to C. gigas. The metalloprotease domain reprolysin is a highly conserved disintegrin domain and is associated with the inflammatory response. This domain is present in both C. gigas and P. maximus. Although the disintegrin domain has been described earlier in the sea urchin genome (Angerer et al. 2006), this is the first report of its occurrence in bivalves. Out of the four gigasin family proteins identified in C. gigas, only gigasin-6 contains β-lactamase domain and could be classified as a protease in accordance with the earlier reports (Marie et al. 2011). This suggests that gigasin-6 might be an inactivated form of lactamase-related proteins and may exhibit protease functions.
Protease inhibitors are enzymes, which inhibit proteases released by pathogens. Several protease inhibitors such as metallo-proteinase inhibitors including the tissue inhibitor of metalloproteinase (TIMP), inter-alpha-trypsin inhibitor (VIT), and serine protease inhibitors containing serpin, kazal and kunitz domains were identified in this study. TIMP and serpin were unique to M. edulis and have been previously reported in the haemocytes and tissue of the gastropod Haliotis discus discus (Bathige et al. 2015), Crassostrea gigas (Montagnani et al. 2001), and the clam, Tegillarca granosa (Wang et al. 2012). Although, TIMP has been identified previously in C. gigas hemocytes and associated tissues, this protein was not identified in this study. This result may be due to the fact that even though many immunity proteins are secreted, only a few are selectively exported to the shell matrix space. The VIT domain was found uniquely in C. gigas, and has previously been identified in the haemocytes of B. glabrata gastropod mollusc (Mitta et al. 2005) with an immunity function. Serine protease inhibitors with domains such as kazal were present in both P. maximus, M. truncata, with kunitz domain proteins present in M. edulis and C. gigas. So far, SMPs containing both kazal and kunitz domains have not been reported in the same species. Therefore, the presence of either one of these serine protease inhibitors in the shell matrix space might indicate an adaptation trait.
Alpha-2-macroglobulin (A2M) was identified in both C. gigas and M. truncata, and belongs to a class of protease inhibitor that inhibits wide variety of endopeptidases, which was first discovered in humans (Family I39, MEROPS peptidase database) (Rawlings et al. 2016). However, the mode of action of A2M in invertebrates is similar to that of found in humans and has been shown to interact with a wide variety of proteases and thus may be used as a defense against several infections caused by bacteria, parasites and pathogens (Armstrong et al. 1996). A2M is the part of the ancient innate immune system and this idea is supported by homology to complement factors C1, C3 and C5 (Armstrong et al. 1998).
Pattern Recognizing Receptors and Associated Proteins
Pattern recognizing receptors (PRR) are proteins involved in the initial sensing of infections and are found in plants and animals. They are present in epithelial, endothelial, and fibroblasts, which recognize the pathogens through pathogen associated molecular patterns (PAMPS) (Takeuchi and Akira 2010). Lipid binding proteins, lectins, toll-like receptors (TLR), nod-like receptors (NLR) etc. all fall under this category and are associated with the innate immune response (Gordon 2002).
With the exception of M. edulis, we detected at least one lipid-binding domain in the other species. C. gigas had a complement component (C1q) domain-containing protein, which belonged to the C1q protein family. A bacterial challenge experiment using Listonella anguillarum in the scallop Chlamys farreri showed significant increases in the levels of C1q proteins in haemocytes, kidney and gills, confirming their participation in immunity related functions (Zhang et al. 2008; Gerdol et al. 2011). Lipocalin and the scavenger receptor (SR) are lipopolysaccharide-binding domains, which were uniquely found in M. truncata. The SR domain recognizes gram positive bacteria and is involved in the immune signaling cascade (Fabriek et al. 2009). Lipocalins are multifunctional domains and have been shown to be recruited for various biological functions such as coloration, complement fixation, etc. as well as regulation of the inflammatory response (Greene et al. 2001). P. maximus SMPs had matches to an apolipophorin-III, a lipid-binding domain, and an immune stimulating protein factor. Previous experiments have shown the former domain to participate in the detoxification of lipopolysaccharide endotoxins, involvement in interactions with gram positive bacteria and also lysozyme activity (Weers and Ryan 2006).
The Gal-lectin proteins identified in P. maximus and M. edulis, and the FTP domains found in M. truncata SMPs are lectins that recognize carbohydrate moieties. Lectins are glycoproteins used for pattern recognition and defense responses (Bachére et al. 1995). The increased expression of lectins in hemocytes or tissues during disease or in stressed conditions has previously been reported in bivalves (Kang et al. 2006).
In M. edulis, a P-loop containing NTPase domain was identified, which has not been reported to date in any bivalve shells. This domain along with leucine rich repeats are usually found in the metazoan nucleotide oligomerization domain (NOD)-like receptors (Koonin and Aravind 2002). The P-loop NTPase mediates self-oligomerization in the presence of ATP. This domain shows a high specificity for pathogens and parasites and thus significant involvement in the innate immunity in molluscs (Yeretssian et al. 2008).
Glycohydro-18 and 20 domains are functional domains of chitinase or chitobiase and were found in proteins of C. gigas, P. maximus, and M. truncata. The main role of these domains is to degrade chitin polymer to dimers and subsequently to monomers (Joshi et al. 1989). Chitinase and chitinase-like proteins are triggered by PRRs when they recognize the chitin cell wall of pathogens, particularly fungi (Vega and Kalkum 2012). Interestingly, chitinase is also involved in the remodeling of shells (Yonezawa et al. 2016).
Immunity Cascades and SMPs
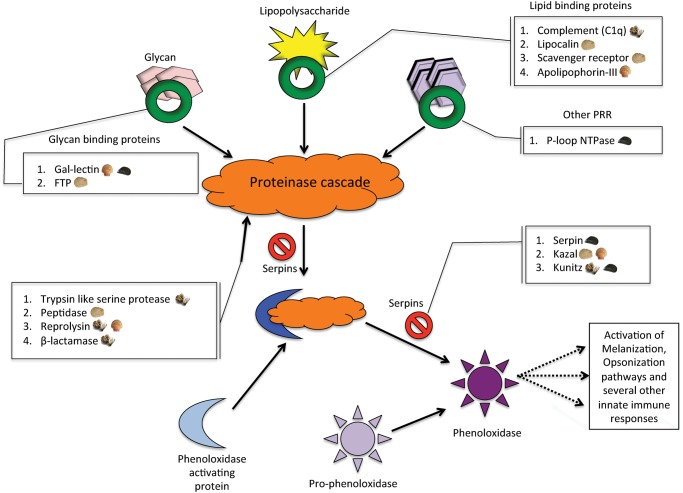
Phenoloxidase is an important precursor in the several biochemical cascades of innate immunity pathways in invertebrates such as melanization, phagocytosis, capsulation, opsonization etc. (Söderhäll and Cerenius 1998; Cannon et al. 2004). In the phenoloxidase pathway (fig. 4), organic molecules such as glycans, lipopolysaccharides are assembled by lectins, lipid binding proteins and PRRs along with proteinases to form a proteinase cascade. These bind to the phenoloxidase activating protein to form a complex that converts prophenoloxidase to phenoloxidase. Serpins act as an inhibitor and regulate this cascade. Intriguingly, all the identified SMPs containing immunity domains in the four species are part of the phenoloxidase pathway indicating selective export of these SMPs to the shell and the critical importance of this immune response pathway to bivalve molluscs.
Fig. 4.
Representation of the phenoloxidase oxidation pathway in arthropods adapted from Jiravanichpaisal et al. (2006). The boxed annotations show the identified functional domains and their corresponding protein families in this activation pathway.
Proteins with No Predicted Functions
In addition to biomineralization and immunity-related proteins, there were also proteins identified which had domains with no predicted functions. For example, C. gigas had two unique proteins, one with six repeats of a Von Willebrand factor-C (VWC) domain and another with a glucose-methanol-choline (GMC) oxidoreductase domain. Previously, single VWC domain containing proteins have been discussed in the context of immunity in crustaceans and arthropods (Sheldon et al. 2007; Chen et al. 2011), so the presence of a protein with six repeats of the VWC domain may indicate an immune function. Similarly the GMC domain is a highly conserved domain and has been shown to play a significant role in the development and immunity of insects (Iida et al. 2007). However, the role of these domains in the immune response of bivalves has not been demonstrated to date and they could also represent novel proteins, with, as yet, unknown functions.
Many of the proteins identified contain one or more low complexity domains (LCDs) in addition to the functional domains noted above. These LCDs confer the proteins with enhanced flexibility. This can be advantageous as during the transport of proteins across the cell membrane, proteins with flexible confirmations pass more readily than the folded proteins (Wright and Dyson 1999). As the SMPs are secreted and exported from the mantle, low complexity regions may aid in their transportation via epithelial cell membranes of the mantle. The intrinsic plasticity conferred by the LCDs could also allow a single protein to recognize several biological targets without sacrificing its specificity (Wright and Dyson 1999). Low complexity regions tend to evolve rapidly via mitotic slippage or meiotic recombination events (Marcotte et al. 1999) and could be an environmental adaptation response (Verstrepen et al. 2005).
Conclusions
Our data describes for the first time, the components of an evolutionary conserved bivalve “shell forming proteome tool kit” that are involved in shell calcification processes. This investigation takes advantage of the development of a standardized extraction and analytical proteomic pipeline, which was applied to four divergent bivalve species (C. gigas, M. edulis, P. maximus, and M. truncata), exhibiting different shell mineralogies and microstructures. This work enabled the identification of SMPs that are specific to aragonite and calcite mineralogies. Proteins specific to a single species were hypothesized to be adaptative to their modus vivendi, especially as these were most often identified in the blue mussel, M. edulis that inhabits highly variable environments and is subject to frequent inter-species hybridization events. Of significant note was the fact that numerous immunity-related domains were identified, which may aid in defense against pathogens. Thus shells are endowed with both embedded biochemical defense mechanisms and mechanical protection. This newly uncovered complexity of SMPs may underpin the evolutionary processes, which led to the successful diversification of the bivalves. Work is now in progress on the different elements of this basic tool kit to determine how these conserved proteins interact to produce the myriad of shell complexity observed in extant bivalves.
Supplementary Material
Supplementary figures S1 and S2 and tables S1–S3 are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
This work was supported by funding from the CACHE (Calcium in a Changing Environment) initial training network (ITN) under the European Union Seventh Framework Programme, reference grant agreement number 605051. We acknowledge E. Dufour (UMR 7209, MNHN) for shell sample preparation. We thank G. Bolbach and L. Matheron (IBPS-FR3631, Paris) for proteomic analysis and discussions.
References
- Addadi L, Joester D, Nudelman F, Weiner S. 2006. Mollusk shell formation: a source of new concepts for understanding biomineralization processes. Chem Eur J. 12:980–987. [DOI] [PubMed] [Google Scholar]
- Aguilera F, McDougall C, Degnan BM. 2014. Evolution of the tyrosinase gene family in bivalve molluscs: independent expansion of the mantle gene repertoire. Acta Biomater. 10:3855–3865. [DOI] [PubMed] [Google Scholar]
- Angerer L, Hussain S, Wei Z, Livingston BT. 2006. Sea urchin metalloproteases: a genomic survey of the BMP-1/tolloid-like, MMP and ADAM families. Dev Biol. 300:267–281. [DOI] [PubMed] [Google Scholar]
- Arivalagan J, Marie B, Sleight VA, Clark MS, Berland S, Marie A. 2016. Shell matrix proteins of the clam, Mya truncata: roles beyond shell formation through proteomic study. Mar Genomics 27:69–74. [DOI] [PubMed] [Google Scholar]
- Armstrong PB, Melchior R, Quigley JP. 1996. Humoral immunity in long-lived arthropods. J Insect Physiol. 42:53–64. [Google Scholar]
- Armstrong PB, Melchior R, Swarnakar S, Quigley JP. 1998. α2-Macroglobulin does not function as a C3 homologue in the plasma hemolytic system of the American horseshoe crab, Limulus. Mol Immunol. 35:47–53. [DOI] [PubMed] [Google Scholar]
- Artigaud S, Thorne MAS, Richard J, Lavaud R, Jean F, Flye-Sainte-Marie J, Peck LS, Pichereau V, Clark MS. 2014. Deep sequencing of the mantle transcriptome of the great scallop Pecten maximus. Mar Genomics 15:3–4. [DOI] [PubMed] [Google Scholar]
- Bachére E, Mialhe E, Noel D, Boulo V, Morvan A, Rodriguez J. 1995. Knowledge and research prospects in marine mollusc and crustacean immunology. Aquaculture 132:17–32. [Google Scholar]
- Bathige SDNK, Umasuthan N, Godahewa GI, Whang I, Kim C, Park H-C, Lee J. 2015. Three novel clade B serine protease inhibitors from disk abalone, Haliotis discus discus: molecular perspectives and responses to immune challenges and tissue injury. Fish Shellfish Immunol. 45:334–341. [DOI] [PubMed] [Google Scholar]
- Beck K, Hunter I, Engel J. 1990. Structure and function of laminin: anatomy of a multidomain glycoprotein. FASEB J. 4:148–160. [DOI] [PubMed] [Google Scholar]
- Berland S, Ma Y, Marie A, Andrieu J-P, Bedouet L, Feng Q. 2013. Proteomic and profile analysis of the proteins laced with aragonite and vaterite in the freshwater mussel Hyriopsis cumingii shell biominerals. Protein Pept Lett. 20:1170–1180. [DOI] [PubMed] [Google Scholar]
- Berland S, Marie A, Duplat D, Milet C, Sire JY, Bédouet L. 2011. Coupling proteomics and transcriptomics for the identification of novel and variant forms of mollusk shell proteins: a study with P. margaritifera. ChemBioChem 12:950–961. [DOI] [PubMed] [Google Scholar]
- Bieler R, Mikkelsen PM, Collins TM, Glover E. a, González VL, Graf DL, Harper EM, Healy J, Kawauchi GY, Sharma PP, et al. 2014. Investigating the bivalve tree of life—an exemplar-based approach combining molecular and novel morphological characters. Invertebr Syst. 28:32–115. [Google Scholar]
- Cannon JP, Haire RN, Rast JP, Litman GW. 2004. The phylogenetic origins of the antigen-binding receptors and somatic diversification mechanisms. Immunol Rev. 200:12–22. [DOI] [PubMed] [Google Scholar]
- Carter JG. 1980. Guide to bivalve shell microstructures In: Rhoads DC, Lutz RA. editors. Skeletal Growth of Aquatic Organisms: Biological Records of Environmental Change (Topics in Geobiology). New York: Plenum Publishing Corporation; p. 645–673. [Google Scholar]
- Charrier M, Marie A, Guillaume D, Bédouet L, Le Lannic J, Roiland C, Berland S, Pierre JS, Le Floch M, Frenot Y, et al. 2013. Soil calcium availability influences shell ecophenotype formation in the sub-antarctic land snail, Notodiscus hookeri. PLoS One 8:e84527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner D, Hedegaard C, Wenk HR. 2000. Mollusc shell microstructures and crystallographic textures. J Struct Geol. 22:1723–1735. [Google Scholar]
- Checa A. 1993. Non-predatory shell damage in recent deep-endobenthic bivalves from Spain. Palaeogeogr Palaeoclimatol Palaeoecol. 100:309–331. [Google Scholar]
- Chen YH, Jia XT, Zhao L, Li CZ, Zhang S, Chen YG, Weng SP, He JG. 2011. Identification and functional characterization of Dicer2 and five single VWC domain proteins of Litopenaeus vannamei. Dev Comp Immunol. 35:661–671. [DOI] [PubMed] [Google Scholar]
- Clark MS, Thorne MA, Vieira FA, Cardoso JC, Power DM, Peck LS. 2010. Insights into shell deposition in the Antarctic bivalve Laternula elliptica: gene discovery in the mantle transcriptome using 454 pyrosequencing. BMC Genomics 11:362.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currey JD. 1977. Mechanical Properties of Mother of Pearl in Tension. Proc R Soc Lond B 196:443–463. [Google Scholar]
- Donaldson DJ, Mahan JT. 1983. Fibrinogen and fibronectin as substrates for epidermal cell migration during wound closure. J Cell Sci. 62:117–127. [DOI] [PubMed] [Google Scholar]
- Fabriek BO, Bruggen R, Van Deng DM, Ligtenberg AJM, Nazmi K, Schornagel K, Vloet RPM, Dijkstra CD, Van Den Berg TK. 2009. The macrophage scavenger receptor CD163 functions as an innate immune sensor for bacteria. Blood 113:887–892. [DOI] [PubMed] [Google Scholar]
- Falini G, Albeck S, Weiner S, Addadi L. 1996. Control of aragonite or calcite polymorphism by mollusk shell macromolecules. Science 271:67–69. [Google Scholar]
- Freer A, Bridgett S, Jiang J, Cusack M. 2014. Biomineral proteins from Mytilus edulis mantle tissue transcriptome. Mar Biotechnol. 16:34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Liao Z, Wang X-X, Bao L-F, Fan M-H, Li X-M, Wu C-W, Xia S-W. 2015. Layer-by-layer proteomic analysis of Mytilus galloprovincialis shell. PLoS One 10:e0133913.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JP. 1996. The Mytilus edulis species complex in southwest England: effects of hybridization and introgression upon interlocus associations and morphometric variation. Mar Biol. 125:385–399. [Google Scholar]
- Gerdol M, Manfrin C, De Moro G, Figueras A, Novoa B, Venier P, Pallavicini A. 2011. The C1q domain containing proteins of the Mediterranean mussel Mytilus galloprovincialis: a widespread and diverse family of immune-related molecules. Dev Comp Immunol. 35:635–643. [DOI] [PubMed] [Google Scholar]
- Gordon S. 2002. Pattern recognition receptors: doubling up for the innate immune response. Cell 111:927–930. [DOI] [PubMed] [Google Scholar]
- Goulletquer P, Wolowicz M. 1989. The shell of Cardium edule, Cardium glaucum and Ruditapes philippinarum: organic content, composition and energy value, as determined by different methods. J Mar Biol Assoc UK. 69:563–572. [Google Scholar]
- Greene LH, Chrysina ED, Irons LI, Papageorgiou AC, Acharya KR, Brew K. 2001. Role of conserved residues in structure and stability: tryptophans of human serum retinol-binding protein, a model for the lipocalin superfamily. Protein Sci. 10:2301–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D, Geiger B, Addadi L. 1993. Fibronectin adsorption to surfaces of hydrated crystals. An analysis of the importance of bound water in protein-substrate interactions. Langmuir. 9:1058–1065. [Google Scholar]
- Harper EM. 2000. Are calcitic layers an effective adaptation against shell dissolution in the Bivalvia? J Zool. 251:179–186. [Google Scholar]
- Harper EM. 2016. Unanswered questions in the evolution of biomineralisation In: Grupe G, McGlynn GC, editors. Isotopic landscapes in bioarchaeology. Springer, Berlin/Heidelberg: p. 1–13. [Google Scholar]
- Harper EM, Clark MS, Hoffman JI, Philipp EER, Peck LS, Morley SA. 2012. Iceberg scour and shell damage in the Antarctic bivalve Laternula elliptica. PLoS One 7:e46341.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedegaard C, Wenk H-R. 1998. Microstructure and texture patterns of mollusc shells. J Mollus Stud. 64:133–136. [Google Scholar]
- Iida K, Cox-Foster DL, Yang X, Ko W-Y, Cavener DR. 2007. Expansion and evolution of insect GMC oxidoreductases. BMC Evol Biol. 11:7–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DJ, McDougall C, Woodcroft B, Moase P, Ross RA, Kube M, Reinhardrt R, Rokhsar DS, Montagnani C, Joubert C, et al. 2010. Parallel evolution of nacre building gene sets in molluscs. Mol Biol Evol. 27:591–608. [DOI] [PubMed] [Google Scholar]
- Jeong MS, Park JS, Song SH, Jang SB. 2007. Characterization of antibacterial nanoparticles from the scallop, Patinopecten yessoensis. Biosci Biotechnol Biochem. 71:2242–2247. [DOI] [PubMed] [Google Scholar]
- Jiravanichpaisal P, Lee BL, Söderhäll K. 2006. Cell-mediated immunity in arthropods: hematopoiesis, coagulation, melanization and opsonization. Immunobiology 211:213–236. [DOI] [PubMed] [Google Scholar]
- Joshi S, Kozlowski M, Richens S, Comberbach DM. 1989. Chitinase and chitobiase production during fermentation of genetically improved Serratia liquefaciens. Enzyme Microb Technol. 11:289–296. [Google Scholar]
- Joubert C, Piquemal D, Marie B, Manchon L, Pierrat F, Zanella-Cléon I, Cochennec-Laureau N, Gueguen Y, Montagnani C. 2010. Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: focus on biomineralization. BMC Genomics 11:613.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang YS, Kim YM, Park K, Il, Kim Cho S, Choi KS, Cho M. 2006. Analysis of EST and lectin expressions in hemocytes of Manila clams (Ruditapes philippinarum) (Bivalvia: Mollusca) infected with Perkinsus olseni. Dev Comp Immunol. 30:1119–1131. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Aravind L. 2002. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 9:394–404. [DOI] [PubMed] [Google Scholar]
- Liao Z, Bao L, Fan M, Gao P, Wang X, Qin C-L, Li X-M. 2015. In-depth proteomic analysis of nacre, prism, and myostracum of Mytilus shell. J Proteomics 122:26–40. [DOI] [PubMed] [Google Scholar]
- Lowenstam HA, Weiner S. 1989. On biomineralization. Oxford University Press, Oxford. [Google Scholar]
- Mann K, Mann M. 2015. Proteomic analysis of quail calcified eggshell matrix: a comparison to chicken and turkey eggshell proteomes. Proteome Sci. 13:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann S. 1988. Molecular recognition in biomineralization. Nature 332:119–124. [Google Scholar]
- Marcotte EM, Pellegrini M, Yeates TO, Eisenberg D. 1999. A census of protein repeats. J Mol Biol. 293:151–160. [DOI] [PubMed] [Google Scholar]
- Marie B, Jackson DJ, Ramos-Silva P, Zanella-Cléon I, Guichard N, Marin F. 2013. The shell-forming proteome of Lottia gigantea reveals both deep conservations and lineage-specific novelties. FEBS J. 280:214–232. [DOI] [PubMed] [Google Scholar]
- Marie B, Joubert C, Tayalé A, Zanella-Cléon I, Belliard C, Piquemal D, Cochennec-Laureau N, Marin F, Gueguen Y, Montagnani C. 2012. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc Natl Acad Sci U S A. 109:20986–20991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Luquet G, Bédouet L, Milet C, Guichard N, Medakovic D, Marin F. 2008. Nacre calcification in the freshwater mussel Unio pictorum: carbonic anhydrase activity and purification of a 95 kDa calcium-binding glycoprotein. ChemBioChem 9:2515–2523. [DOI] [PubMed] [Google Scholar]
- Marie B, Marie A, Jackson DJ, Dubost L, Degnan BM, Milet C, Marin F. 2010. Proteomic analysis of the organic matrix of the abalone Haliotis asinina calcified shell. Proteome Sci. 8:54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marie B, Ramos-Silva P, Marin F, Marie A. 2013. Proteomics of CaCO3 biomineral-associated proteins: how to properly address their analysis. Proteomics 13:3109–3116. [DOI] [PubMed] [Google Scholar]
- Marie B, Le Roy N, Zanella-Cléon I, Becchi M, Marin F. 2011. Molecular evolution of mollusc shell proteins: insights from proteomic analysis of the edible mussel mytilus. J Mol Evol. 72:531–546. [DOI] [PubMed] [Google Scholar]
- Marie B, Trinkler N, Zanella-Cleon I, Guichard N, Becchi M, Paillard C, Marin F. 2011. Proteomic identification of novel proteins from the calcifying shell matrix of the manila clam Venerupis Philippinarum. Mar Biotechnol. 13:955–962. [DOI] [PubMed] [Google Scholar]
- Marie B, Zanella-cléon I, Guichard N, Becchi M, Marin F. 2011. Novel proteins from the calcifying shell matrix of the Pacific oyster Crassostrea gigas. Mar Biotechnol. 13:1159–1168. [DOI] [PubMed] [Google Scholar]
- Marin F, Luquet G, Marie B, Medakovic D. 2007. Molluscan shell proteins: primary structure, origin, and evolution. Curr Top Dev Biol. 80:209–276. [DOI] [PubMed] [Google Scholar]
- Miller J, McLachlan AD, Klug A. 2001. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. J Trace Elem Exp Med. 14:157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitta G, Galinier R, Tisseyre P, Allienne JF, Girerd-Chambaz Y, Guillou F, Bouchut A, Coustau C. 2005. Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Dev Comp Immunol. 29:393–407. [DOI] [PubMed] [Google Scholar]
- Miyamoto H, Miyashita T, Okushima M, Nakano S, Morita T, Matsushiro A. 1996. A carbonic anhydrase from the nacreous layer in oyster pearls. Proc Natl Acad Sci U S A. 93:9657–9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montagnani C, Le Roux F, Berthe F, Escoubas J. 2001. Cg-TIMP, an inducible tissue inhibitor of metalloproteinase from the Pacific oyster Crassostrea gigas with a potential role in wound healing and defense mechanisms. FEBS Lett. 500:64–70. [DOI] [PubMed] [Google Scholar]
- Nagai K, Yano M, Morimoto K, Miyamoto H. 2007. Tyrosinase localization in mollusc shells. Comp Biochem Physiol B Biochem Mol Biol. 146:207–214. [DOI] [PubMed] [Google Scholar]
- Panayotou G, End P, Aumailley M, Timpl R, Engel J. 1989. Domains of laminin with growth-factor activity. Cell 56:93–101. [DOI] [PubMed] [Google Scholar]
- Plazzi F, Passamonti M. 2010. Towards a molecular phylogeny of Mollusks: bivalves’ early evolution as revealed by mitochondrial genes. Mol Phylogenet Evol. 57:641–657. [DOI] [PubMed] [Google Scholar]
- Ramos-Silva P, Marin F, Kaandorp J, Marie B. 2013. Biomineralization toolkit: the importance of sample cleaning prior to the characterization of biomineral proteomes. Proc Natl Acad Sci U S A. 110:E2144–E2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Finn R. 2016. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 44:D343–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren J, Liu X, Jiang F, Guo X, Liu B. 2010. Unusual conservation of mitochondrial gene order in Crassostrea oysters: evidence for recent speciation in Asia. BMC Evol Biol. 10:394.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riginos C, Cunningham CW. 2005. Local adaptation and species segregation in two mussel (Mytilus edulis× Mytilus trossulus) hybrid zones. Mol Ecol. 14:381–400. [DOI] [PubMed] [Google Scholar]
- Rose-Martel M, Smiley S, Hincke MT. 2015. Novel identification of matrix proteins involved in calcitic biomineralization. J. Proteomics 116:81–96. [DOI] [PubMed] [Google Scholar]
- Le Roy N, Jackson DJ, Marie B, Ramos-Silva P, Marin F. 2014. The evolution of metazoan α-carbonic anhydrases and their roles in calcium carbonate biomineralization. Front Zool. 11:1–16.24401080 [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A. 95:5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seed R. 1969. The ecology of Mytilus edulis L. (Lamellibranchiata) on exposed rocky shores - I. Breeding and settlement. Oecologia 3:277–316. [DOI] [PubMed] [Google Scholar]
- Sheldon TJ, Miguel-Aliaga I, Gould AP, Taylor WR, Conklin D. 2007. A novel family of single VWC-domain proteins in invertebrates. FEBS Lett. 581:5268–5274. [DOI] [PubMed] [Google Scholar]
- Shen X, Belcher AM, Hansma PK, Stucky GD, Morse DE. 1997. Molecular cloning and characterization of lustrin-A, a matrix protein from shell and pearl nacre of Haliotis rufescens. J Biol Chem. 272:32472–32481. [DOI] [PubMed] [Google Scholar]
- Shi Y, Yu C, Gu Z, Zhan X, Wang Y, Wang A. 2013. Characterization of the pearl oyster (Pinctada martensii) mantle transcriptome unravels biomineralization genes. Mar Biotechnol. 15:175–187. [DOI] [PubMed] [Google Scholar]
- Söderhäll K, Cerenius L. 1998. Role of the prophenoloxidase-activating system in invertebrate immunity. Curr Opin Immunol. 10:23–28. [DOI] [PubMed] [Google Scholar]
- Stenflo J, Stenberg Y, Muranyi A. 2000. Calcium-binding EGF-like modules in coagulation proteinases: function of the calcium ion in module interactions. Biochim Biophys Acta 1477:51–63. [DOI] [PubMed] [Google Scholar]
- Sudo S, Fujikawa T, Nagakura T, Ohkubo T, Sakagushi K, Tanaka M, Nakashima K, Takahashi T. 1997. Structures of mollusc shell framework proteins. Nature 387:565–567. [DOI] [PubMed] [Google Scholar]
- Suzuki M, Saruwatari K, Kogure T, Yamamoto Y, Nishimura T, Kato T, Nagasawa H. 2009. An acidic matrix protein, Pif, is a key macromolecule for nacre formation. Science 325:1388–1390. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. 2010. Pattern recognition receptors and inflammation. Cell 140:805–820. [DOI] [PubMed] [Google Scholar]
- Treccani L, Mann K, Heinemann F, Fritz M. 2006. Perlwapin, an abalone nacre protein with three four-disulfide core (whey acidic protein) domains, inhibits the growth of calcium carbonate crystals. Biophys J. 91:2601–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega K, Kalkum M. 2012. Chitin, chitinase responses, and invasive fungal infections. Int J Microbiol. 2012:920459.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendrasco MJ, Checa AG, Kouchinsky AV. 2011. Shell microstructure of the early bivalve Pojetaia and the independent origin of nacre within the mollusca. Palaeontology 54:825–850. [Google Scholar]
- Verstrepen KJ, Jansen A, Lewitter F, Fink GR. 2005. Intragenic tandem repeats generate functional variability. Nat Genet. 37:986–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuorinen I, Antsulevich AE, Maximovich NV. 2002. Spatial distribution and growth of the common mussel Mytilus edulis L. in the archipelago of SW-Finland, northern Baltic Sea. Boreal Environ Res. 7:41–52. [Google Scholar]
- Wang Q, Bao Y, Huo L, Gu H, Lin Z. 2012. A novel tissue inhibitor of metalloproteinase in blood clam Tegillarca granosa: Molecular cloning, tissue distribution and expression analysis. Fish Shellfish Immunol. 33:645–651. [DOI] [PubMed] [Google Scholar]
- Weers PMM, Ryan RO. 2006. Apolipophorin III: role model apolipoprotein. Insect Biochem Mol Biol. 36:231–240. [DOI] [PubMed] [Google Scholar]
- Weiner S. 1979. Aspartic acid-rich proteins: major components of the soluble organic matrix of mollusk shells. Calcif Tissue Int. 29:163–167. [DOI] [PubMed] [Google Scholar]
- Wheeler AP, George JW, Evans CA. 1981. Control of calcium carbonate nucleation and crystal growth by soluble matrx of oyster shell. Science 212:1397–1398. [DOI] [PubMed] [Google Scholar]
- Whittaker CA, Hynes RO. 2002. Distribution and evolution of von Willebrand/integrin A domains: widely dispersed domains with roles in cell adhesion and elsewhere. Mol Biol Cell 13:3369–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdows J. 1978. Physiological indices of stress in Mytilus edulis. J Mar Biol Assoc UK. 58:125–142. [Google Scholar]
- Williams RJ. 1970. Freezing tolerance in Mytilus edulis. Comp Biochem Physiol. 35:145–161. [Google Scholar]
- Wood R, Zhuravlev AY. 2012. Escalation and ecological selectively of mineralogy in the Cambrian Radiation of skeletons. Earth Sci Rev. 115:249–261. [Google Scholar]
- Wrange AL, Valero J, Harkestad LS, Strand Ø, Lindegarth S, Christensen HT, Dolmer P, Kristensen PS, Mortensen S. 2010. Massive settlements of the Pacific oyster, Crassostrea gigas, in Scandinavia. Biol Invasions 12:1145–1152. [Google Scholar]
- Wright PE, Dyson HJ. 1999. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 293:321–331. [DOI] [PubMed] [Google Scholar]
- Yarra T, Gharbi K, Blaxter M, Peck LS, Clark MS. 2016. Characterization of the mantle transcriptome in bivalves: Pecten maximus, Mytilus edulis and Crassostrea gigas. Mar Genomics 27:9–15. [DOI] [PubMed] [Google Scholar]
- Yeretssian G, Labbé K, Saleh M. 2008. Molecular regulation of inflammation and cell death. Cytokine 43:380–390. [DOI] [PubMed] [Google Scholar]
- Yonezawa M, Sakuda S, Yoshimura E, Suzuki M. 2016. Molecular cloning and functional analysis of chitinases in the fresh water snail, Lymnaea stagnalis. J Struct Biol. doi: 10.1016/j.jsb.2016.02.021. [DOI] [PubMed] [Google Scholar]
- Zhang C, Xie L, Huang J, Chen L, Zhang R. 2006. A novel putative tyrosinase involved in periostracum formation from the pearl oyster (Pinctada fucata). Biochem Biophys Res Commun. 342:632–639. [DOI] [PubMed] [Google Scholar]
- Zhang G, Fang X, Guo X, Li L, Luo R, Xu F, Yang P, Zhang L, Wang X, Qi H, et al. 2012. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490:49–54. [DOI] [PubMed] [Google Scholar]
- Zhang H, Song L, Li C, Zhao J, Wang H, Qiu L, Ni D, Zhang Y. 2008. A novel C1q-domain-containing protein from Zhikong scallop Chlamys farreri with lipopolysaccharide binding activity. Fish Shellfish Immunol. 25:281–289. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.