Phosphate deficiency-responsive lncRNAs in Medicago truncatula are involved in the regulation of Pi deficiency signaling network and Pi transport.
Keywords: High-throughput sequencing, long non-coding RNAs (lncRNAs), phosphate deficiency, Medicago truncatula, phosphate acquisition, legume plants
Abstract
Emerging evidence indicates that long non-coding RNAs (lncRNAs) play important roles in the regulation of many biological processes. Inhibition of plant growth due to deficiency in soil inorganic phosphate (Pi) occurs widely across natural and agricultural ecosystems; however, we know little about the function of plant lncRNAs in response to Pi deficiency. To address this issue, we first identified 10 785 lncRNAs in the legume model species Medicago truncatula by sequencing eight strand-specific libraries. Out of these lncRNAs, 358 and 224 were responsive to Pi deficiency in the leaves and roots, respectively. We further predicted and classified the putative targets of those lncRNAs and the results revealed that they may be involved in the processes of signal transduction, energy synthesis, detoxification, and Pi transport. Finally, we functionally characterized three Phosphate Deficiency-Induced LncRNAs (PDILs) using their corresponding Tnt1 mutants. The results showed that PDIL1 suppressed degradation of MtPHO2, which encodes a ubiquitin-conjugating E2 enzyme regulated by miR399, while PDIL2 and PDIL3 directly regulated Pi transport at the transcriptional level. These findings demonstrate that PDILs can regulate Pi-deficiency signaling and Pi transport, highlighting the involvement of lncRNAs in the regulation of responses of plants to Pi deficiency.
Introduction
Improvements in high-throughput sequencing technology have revealed that over 90% of the genome can generate a large number of non-coding RNAs (ncRNAs) (Chekanova et al., 2007; Kapranov et al., 2007). These ncRNAs are categorized into small RNAs and long non-coding RNAs (lncRNAs) according to their length (Brosnan and Voinnet, 2009; Kim et al., 2011). LncRNAs have a length of more than 200 nucleotides and relatively less protein-coding capacity, and constitute the biggest class of ncRNAs (Rinn and Chang, 2012; Chekanova, 2015). The majority of lncRNAs can be transcribed by RNA Polymerase II, and they are generally expressed in a tissue-specific manner (Wilusz et al., 2009). According to their locations relative to protein-coding genes in the genome, lncRNAs can be further grouped into sense, antisense, bidirectional, intronic, and intergenic lncRNAs (Ponting et al., 2009).
The function of lncRNAs has long been ignored, and they have often been regarded as transcriptional ‘noise’ due to their low expression and low protein-coding potential. However, recent studies have provided convincing evidence to support regulatory roles of lncRNAs in numerous biological processes in plants (Chekanova, 2015; Liu et al., 2015). Transcriptional regulation of lncRNAs by regulating their targets is the most common way to modulate biological processes, and the lncRNAs can work either in close proximity (cis-acting) or at a distance (trans-acting) by sequence complementarity with RNA and DNA (Kornienko et al., 2013). The regulatory mechanisms of lncRNAs include transcriptional interference, chromatin remodeling, promoter inactivation, transcription factor activation, epigenetic silencing, as well as repression (Ponting et al., 2009).
The functions of plant lncRNAs are being revealed. For instance, Swiezewski et al. (2009) reported that lncRNAs are involved in the regulation of flowering by altering the expression of FLOWERING LOCUS C (FLC) in Arabidopsis. Two lncRNAs that were transcribed from the antisense strand of FLC were designed as COOLAIR, and were found to be able to silence the expression of FLC (Swiezewski et al., 2009). An intronic lncRNA, COLDAIR, can induce the epigenetic repression of FLC (Heo and Sung, 2011). Moreover, APOLO and ASCO have been reported to regulate root development in Arabidopsis (Ariel et al., 2014). PINOID is a key regulator of polar auxin transport, and its expression can be up-regulated by the change in chromatin formation induced by the expression of the lncRNA APOLO (Ariel et al., 2014). Nuclear Speckles RNA-binding proteins (NSRs) modulate the alternative splicing of a subset of genes involved in lateral root initiation, and this effect weakens when NSRs bind to ASCO (Bardou et al., 2014). The lncRNA HID1 mediates photomorphogenesis in Arabidopsis induced by red light (Wang et al., 2014). The low expression of the lncRNA LDMAR was found to lead to male sterility under long-day conditions (Ding et al., 2012), and Enod40 has been shown to be associated with nodulation in legume plants (Campalans et al., 2004). Under phosphate-deficient conditions, miR399 up-regulates the expression of Pi transport genes by cleaving PHOSPHATE2 (PHO2), while IPS1, At4, and Mt4 suppress the effect of miR399 by acting as mimics of PHO2 (Burleigh and Harrison, 1997; Shin et al., 2006; Franco-Zorrilla et al., 2007). A cis-natural antisense lncRNA, cis-NATPHO1;2, positively regulates the expression of a protein that is critical for phosphate homeostasis in rice (Jabnoune et al., 2013). Despite the progress made in these various studies, the functions and regulatory networks of lncRNAs in plants are less well known compared to those in mammals.
Advancements in high-throughput sequencing technology have allowed the accurate identification of lncRNAs in plants at the whole-genome level. LncRNAs have been identified in several model plant species, including Arabidopsis (Ben Amor et al., 2009; Liu et al., 2012; Zhu et al., 2014), rice (Liu et al., 2013; Xu et al., 2016), maize (Li et al., 2014), and poplar (Shuai et al., 2014; Tian et al., 2016). In contrast, little information is available regarding lncRNAs in the model legume Medicago truncatula. Medicago truncatula is an annual species that is widely used to study the functional genomics of legume plants because of its small diploid genome and amenability to transformation (Young et al., 2011). Legume plants account for one-third of primary crop production worldwide, and often suffer from abiotic stresses (Benedito et al., 2008). Emerging evidence indicates that lncRNAs play regulatory roles in responses to abiotic stress in plants (Liu et al., 2012; Shuai et al., 2014; Zhu et al., 2014). Given the the great numbers and poorly known functions of lncRNAs compared to protein-coding genes, the functional characterization of lncRNAs has the potential to shed important light on their roles and could provide an effective tool to enhance yields of legume crops under unfavorable growth conditions (Liu and Zhu, 2014).
In our previous studies, we identified a number of osmotic- and salt stress-responsive lncRNAs in M. truncatula. By predicting the targets of lncRNAs, they were found to be implicated in numerous biological processes such as signal transduction and detoxification under abiotic stress (Wang et al., 2015). Phosphorus (P) is one of the essential mineral nutrients for plant growth and development. Despite high total P content in soils, the amounts of inorganic phosphate (Pi) that are available to plants in many natural and agricultural ecosystems are often low, thus limiting plant growth and production (Hinsinger et al., 2011; Nestler and Wissuwa, 2016). To cope with Pi deficiency in soils, plants have evolved numerous strategies at morphological, physiological, and molecular levels (Ding et al., 2016; Nath and Tuteja, 2016; Dong et al., 2017; Li et al., 2017). Many protein-coding genes and microRNAs that are involved in sensing and responding to Pi deficiency have been identified (Plaxton and Tran, 2011). In contrast, few studies have specifically investigated the roles of lncRNAs in the regulation of physiological processes in response to Pi deficiency (Shin et al., 2006; Franco-Zorrilla et al., 2007; Jabnoune et al., 2013; Xu et al., 2016). In the present study, we identified a comprehensive set of Pi deficiency-responsive lncRNAs from M. truncatula by sequencing eight paired-end libraries, and functionally characterized three lncRNAs in response to Pi deficiency using the relevant mutants of M. truncatula.
Materials and methods
Plant material and stress treatments
Medicago truncatula Jemalong A17 is the model of legume plants, and its genome has been sequenced (Young et al., 2011). Medicago Tnt1 insertion mutants were generated based on M. truncatula R108 because of its relatively easy transformation (Tadege et al., 2008). This mutant resource has been used in many studies of Medicago. Seeds of M. truncatula Jemalong A17, R108, and Tnt1 insertion mutants were treated with concentrated sulfuric acid for 8 min, and then thoroughly rinsed with distilled water. After being chilled at 4 °C for 2 d, seeds were sown on 0.8% agar and germinated at 25 °C. When the radicals were about 2 cm long, the plants were transferred into plastic buckets filled with aerated nutrient solution under controlled conditions (26 °C day/22 °C night, and 14-h photoperiod). The composition of the full-strength nutrient solution was: 2.5 mM KNO3, 0.5 mM KH2PO4, 0.25 mM CaCl2, 1 mM MgSO4, 100 µM Fe-Na-EDTA, 30 µM H3BO3, 5 µM MnSO4, 1 µM ZnSO4, 1 µM CuSO4, and 0.7 µM Na2MoO4, at pH 6.0.
Three-week-old seedlings of Jemalong A17 were transferred from the full-strength nutrient solution into a solution with reduced KH2PO4 concentration (1 µM) for either 12, 24, or 48 h. After exposure to the Pi-deficient media, samples were collected at the same time to discount any circadian effects. Leaf and root samples from 10 individual plants grown in control (CK, Pi-sufficient) and Pi-deficient (PD) media were harvested and dried at 80 °C and dry weights were determined once constant weight had been achieved. Three-week-old seedlings of R108 (the wild-type, WT) and Tnt1 insertion mutants were treated using the same regime for 24 or 72 h. These materials were used to perform quantitative real-time PCR (qRT-PCR) and to determine P concentration, respectively.
Construction of cDNA libraries and high-throughput sequencing
Seedlings of M. truncatula Jemalong A17 were used to construct libraries. Total RNA was extracted from leaves and roots exposed to Pi-sufficient and Pi-deficient media for 24 h using TRIzol (Invitrogen) according to the manufacturer’s protocols. Ribosome RNAs of RNA samples were removed using a Ribo-Zero™ Magnetic Kit (Epicentre). The strand-specific sequencing libraries were constructed following previously described protocols (Borodina et al., 2011). Paired-end sequencing (2 × 100 bp) was performed on an Illumina Hiseq2000 sequencer. Two biological repeats were used in the construction of libraries.
Read mapping and transcriptome assembly
The resulting directional paired-end reads were quality-checked with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and adapter contaminations and the low-quality tags in the raw data were discarded. Ribosome RNA data were also removed from the remaining data by alignment. Thereafter, the clean reads from the eight cDNA libraries were merged and mapped to the M. truncatula genome sequence (Mt4.0) using the spliced read aligner TopHat (Trapnell et al., 2009). To construct transcriptomes, the mapped reads were assembled de novo using Cufflinks (Trapnell et al., 2010). All the transcripts were required to have exons greater than 1 and longer than 200 bp in length.
Identification of lncRNAs
The assembled transcripts were annotated using the Cuffcompare program from the Cufflinks package (Trapnell et al., 2010). The known protein-coding transcripts were identified according to the annotation of M. truncatula genome sequences (Mt4.0). The remaining unknown transcripts were used to screen putative lncRNAs. Transcripts less than 200 nt in length and with less than three reads were first excluded. Then, the coding potentials of the remaining transcripts were calculated using the Coding Potential Calculator (CPC) and Coding-Non-Coding Index (CNCI) (Kong et al., 2007; Sun et al., 2013). A transcript with a CPC value less than –1 and a CNCI value lower than 0 was taken as being a non-coding one.
Analysis of differential expression of lncRNAs
Expression levels of all transcripts, including those of putative lncRNAs and mRNAs, were quantified as fragments per kilobase of exon per million fragments mapped (FPKM) using the Cuffdiff program from the Cufflinks package (Trapnell et al., 2010). Differential gene expression was determined by a P-value less than 0.05.
Prediction of putative cis- and trans-targets of lncRNAs
The transcription of lncRNAs has been implicated in the regulation of the expression of genes in close genomic proximity (cis-acting regulation) and in the targeting of distant genes (trans-acting regulation) via multiple mechanisms (Ponting et al., 2009; Kornienko et al., 2013). Many studies have demonstrated that one important function of lncRNAs is to regulate the expression of neighboring protein-coding genes via epigenetic modification and/or transcriptional co-activation/repression (Rinn et al., 2007; Yu et al., 2008; Mercer et al., 2009). Therefore, the analyses were performed of genomic co-locations (<10 kb) of the lncRNAs and mRNAs according to previously described methods (Liao et al., 2011). In addition, the formation of near-complementary lncRNA–target duplexes is also an important way to regulate the expression of their trans-targets (Chekanova et al., 2007; Ponting et al., 2009). The trans-targets of lncRNAs were predicted by the complementarity of lncRNAs and their targets with expression markedly different under Pi-deficient conditions using RIsearch (Wenzel et al., 2012). Finally, those putative cis- and trans-targets of lncRNAs were analysed using gene ontology (GO) (Ashburner et al., 2000), and GO terms were considered to be enriched when the P value was less than 0.05 using Blast2GO (Conesa et al., 2005).
GFP analyses
Expression vectors of 35S:PDIL2, 35S:PDIL3, and 35S:Medtr1g074930:GFP were constructed for transient transformation of Nicotiana benthamiana following the protocols described by Franco-Zorrilla et al. (2007). Either PDIL2 or PDIL3 was co-transformed into leaves of N. benthamiana with Medtr1g074930:GFP by agroinfiltration. In this assay, the intensity of GFP (green fluorescent protein) indicates the expression level of Medtr1g074930. Images were taken under identical conditions using a fluorescence microscope (Nikon Eclipse Ti). At least five images were used to analyse the relative intensity of GFP using the ImageJ software.
Quantitative real-time PCR
Total RNA was isolated using RNAiso Plus reagent (TaKaRa) and treated with RNase-free DNase I (Promega). About 0.5 μg RNA was reverse-transcribed into first-strand cDNA with a PrimeScript® RT reagent Kit (TaKaRa). Quantitative real-time PCR (qRT-PCR) was performed using an ABI Stepone Plus Instrument. Gene-specific primers and internal control primers are listed in Supplementary Table S1 at JXB online.
All qRT-PCR reactions were performed in triplicates for each cDNA sample with an annealing temperature of 57 °C and a total of 40 cycles of amplification. The relative expression level was calculated by the comparative CT method (Livak and Schmittgen, 2001).
Identification and confirmation of homozygotic mutants
Generation of the M. truncatula Tnt1 insertional mutants was described previously (Tadege et al., 2008). Mutants were identified by aligning lncRNA sequences with the M. truncatula mutant database (http://medicago-mutant.noble.org/mutant/blast/blast.php).
Genomic DNA from mutants was isolated using a Plant Genomic DNA Kit (Tiangen). Homozygotes were identified using two sets of PCR primers as follows: for pdil1-1 (PDIL1-F+TNT1-R and PDIL1-F+PDIL1-R), for pdil1-2 (PDIL1-F+TNT1-F and PDIL1-F+PDIL1-R), for pdil2 (PDIL2-F+TNT1-F and PDIL2-F+PDIL2-R), and for pdil3 (PDIL3-F+TNT1-R and PDIL3-F+PDIL3-R). Primers are listed in Supplementary Table S1. If the first reaction was positive and the second reaction was negative, the mutant was a homozygote. The expression levels of PDILs in WT and mutants were monitored by RT-PCR. Seeds of homozygotes were harvested and used in this study.
Determination of P concentration in leaves and roots
Leaf and root samples exposed to Pi-sufficient and Pi-deficient media were harvested and dried at 80 °C to constant weight. A mixture of 50 mg of dry material, 5 ml of nitric acid, and 2 ml of hydrogen peroxide was placed in digestion tubes, and then samples were digested using a microwave system (MARS, CEM). After diluting and filtering, P concentrations were measured using an ICP-AES (Thermo).
Statistical analysis
Data were analysed using SPSS statistics 17.0. One-way ANOVA analysis with Duncan tests was performed for multiple comparisons, and t-tests were performed to test for significant differences between the two groups of data in this study.
Accession numbers
RNA-seq data are available in the Sequence Read Archive database (https://www.ncbi.nlm.nih.gov/sra) under accession numbers SRR1523070 and SRR3938213 for CK-L, SRR1523071 and SRR3938216 for CK-R, SRR1536246 and SRR3938252 for PD-L, and SRR1536247 and SRR3938256 for PD-R.
Results
Plant P concentrations
In this study, 3-week-old seedlings of M. truncatula previously grown in Pi-sufficient medium (0.5 mM Pi) were exposed to Pi-deficient (1 µM Pi) medium for 0, 12, 24 or 48 h. We sampled both leaves and roots for determination of biomass and P concentrations. As shown in Table 1, exposure to Pi-deficient medium for 24 h led to significant reductions in dry biomass of roots, and in P concentrations in leaves and roots. In contrast, foliar dry biomass was relatively constant after treatment with Pi deficiency for 24 h (Table 1). These results suggested that the plants exposed to P-deficient medium for 24 h were at an early stage of stress response, and that plants may actively mobilize genes and regulatory networks to cope with Pi deficiency. Therefore, we sampled roots and leaves at this point to construct cDNA libraries in order to identify Pi deficiency-responsive lncRNAs.
Table 1.
Time-dependent changes in plant growth and P concentrations in leaves and roots of M. truncatula Jemalong A17 seedlings in response to Pi deficiency
| Treatment time (h) | Dried biomass (mg plant−1) | P concentration (mg g−1 DW) | P content (mg plant−1) |
|---|---|---|---|
| Leaves | |||
| 0 | 35.700 ± 1.159a | 10.645 ± 0.486a | 0.380 ± 0.017a |
| 12 | 33.180 ± 0.961a | 10.502 ± 0.328a | 0.348 ± 0.011a |
| 24 | 32.980 ± 0.873a | 8.806 ± 0.191b | 0.290 ± 0.006b |
| 48 | 29.080 ± 1.161b | 7.642 ± 0.188c | 0.222 ± 0.005c |
| Roots | |||
| 0 | 16.360 ± 0.923a | 7.645 ± 0.487a | 0.125 ± 0.008a |
| 12 | 15.570 ± 0.522a | 7.253 ± 0.241a | 0.113 ± 0.004a |
| 24 | 13.530 ± 0.614b | 5.873 ± 0.080b | 0.080 ± 0.001b |
| 48 | 12.270 ± 0.693b | 4.909 ± 0.213c | 0.060 ± 0.003c |
Data are the means ±SE. For dried biomass, there were 10 replicates. For P concentration and content, there were three replicates. Different letters indicate significant differences among treatments (P<0.05).
RNA-seq of eight cDNA libraries
Leaf and root samples of M. truncatula Jemalong A17 exposed to Pi-deficient and Pi-sufficient media for 24 h were used to construct cDNA libraries and were sequenced by an Illumina-Solexa sequencer. Two biological repeats were used to construct the libraries. High-throughput RNA-sequencing (RNA-seq) of the eight libraries led to generation of 741 571 940 clean reads and 111.24 G clean bases (Table 2). A quality score (Q) for each base in the reads was calculated by a phred-like algorithm using FastQC (Ewing and Green, 1998), and the results showed that the data were highly credible (Supplementary Fig. S1).
Table 2.
Statistical data of the RNA-seq reads for the eight libraries constructed from leaves and roots of M. truncatula Jemalong A17 exposed to control (CK, Pi-sufficient) and Pi-deficient (PD) media
| CK | PD | |||
|---|---|---|---|---|
| Leaves | Roots | Leaves | Roots | |
| Raw reads | 95 999 176 + 77 703 184 | 93 999 446 + 92 619 970 | 164 698 304 + 91 928 280 | 89 588 642 + 78 257 344 |
| Raw bases | 14.40 G + 11.66 G | 14.10 G + 13.89 G | 24.70 G + 13.79 G | 13.44 G + 11.74 G |
| Clean reads | 87 897 824 + 75 538 710 | 87 087 836 + 90 435 180 | 153 503 868 + 89 656 196 | 81 349 080 + 76 103 276 |
| Clean bases | 13.18 G + 11.33 G | 13.06 G + 13.57 G | 23.03 G + 13.45 G | 12.20 G + 11.42 G |
| Unique lncRNAs | 7381 | 9454 | 7213 | 9614 |
Identification and characterization of lncRNAs
After the clean reads were mapped, assembled, and annotated according to the M. truncatula genome sequence (Mt4.0) using the TopHat and Cufflinks packages (Trapnell et al., 2009, 2010), the known protein-coding RNAs were first identified. The remaining reads were then filtered by length and coding potential, such that transcripts smaller than 200 bp were excluded, and the transcripts with CPC more than –1 and CNCI greater than 0 were removed. The remaining transcripts were considered as putative lncRNAs.
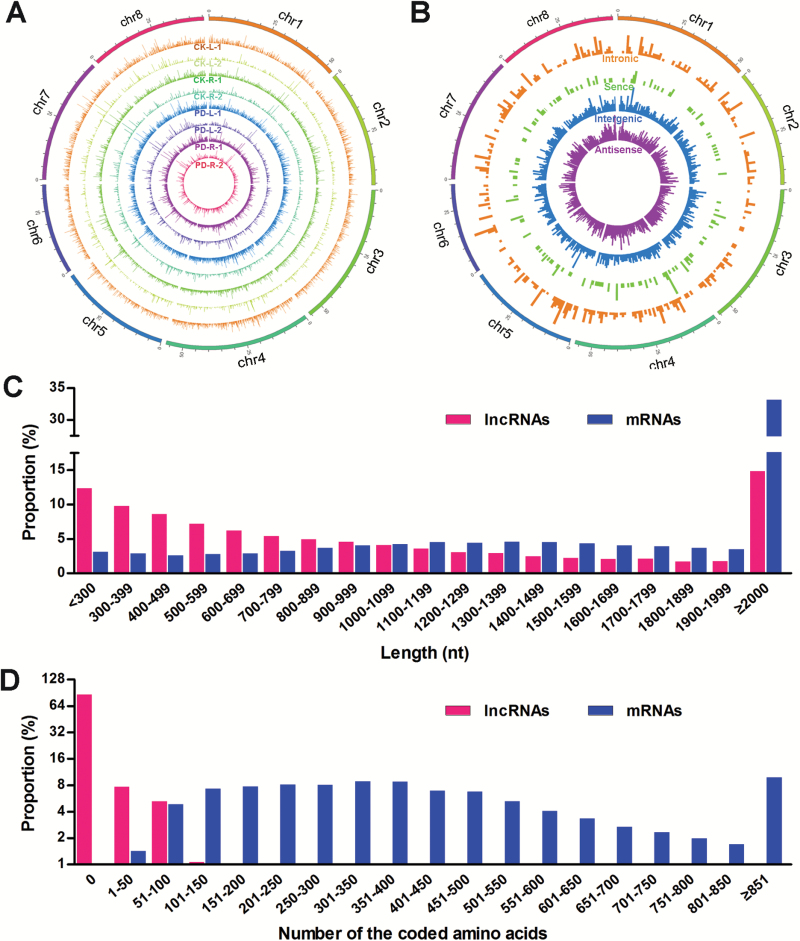
The number of unique lncRNAs identified in the leaves and roots of M. truncatula Jemalong A17 exposed to Pi-deficient and Pi-sufficient media for 24 h is shown in Table 2. In total, we obtained 10 785 unique lncRNAs, and their loci are shown in Supplementary Table S2. Using the Circos program (Krzywinski et al., 2009), the expressional distribution of lncRNAs from the eight libraries were drawn along eight chromosomes, and they exhibited no obvious preference for locations (Fig. 1A). In terms of the loci in the genome, 4811 intergenic, 599 intronic, 161 sense, and 5214 antisense lncRNAs were distinguished (Fig. 1B). Compared with protein-coding genes, the length of lncRNAs was much shorter (Fig. 1C). Among the lncRNAs identified in this study, 85.7% cannot code for proteins, and the remaining lncRNAs only coded for proteins with short chains of amino acids (Fig. 1D).
Fig. 1.
Characteristics of M. truncatula lncRNAs. (A) The expression level of lncRNAs (log10 FPKM) along the eight M. truncatula chromosomes. The eight concentric rings correspond to different samples: two control (Pi-sufficient) samples of leaves (CK-L), two control samples of roots (CK-R), two Pi-deficient samples of leaves (PD-L), and two Pi-deficient samples of roots (PD-R). (B) Distribution of different types of lncRNAs. The intronic, sense, intergenic, and antisense lncRNAs are represented by the different concentric rings from outer to inner, according to the loci of lncRNAs in the genome. (C) Length distribution of lncRNAs and mRNAs. (D) Length distribution of amino acids coded by lncRNAs and mRNAs.
Response of lncRNAs to Pi deficiency
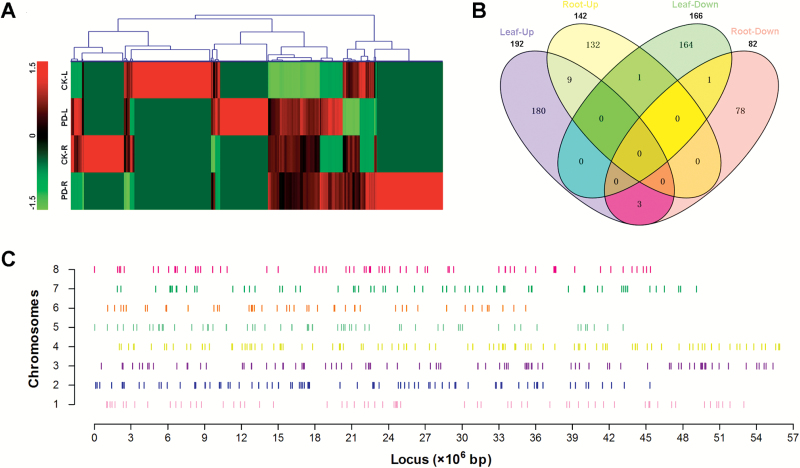
To identify the Pi deficiency-responsive lncRNAs, we calculated and compared the FPKM values in the four treatments (eight libraries) (Fig. 2A). Transcript levels of 358 lncRNAs in the leaves and 224 lncRNAs in the roots were altered by Pi deficiency (Supplementary Table S3), and they were designated as Phosphate Deficiency-Induced LncRNAs (PDILs). LncRNAs with expression up- or down-regulated by Pi deficiency in both leaves and roots are shown in the Venn diagram in Fig. 2B. We further grouped the Pi deficiency-responsive lncRNAs into those that were common or specific. For example, nine lncRNAs in both leaves and roots were up-regulated in response to Pi deficiency, while one lncRNA was down-regulated in both tissues. The Pi deficiency-responsive lncRNAs were found to be located across all the chromosomes, with the most abundant lncRNAs on Chromosome 4 (Fig. 2C).
Fig. 2.
Heatmap (A), Venn diagram (B), and locus (C) of Pi-deficiency lncRNAs. The gene tree in (A) was drawn by the method of Hierarchical Clustering, and the Z-values of log2 FPKM were used in this analysis. CK, control (Pi-sufficient); PD, Pi-deficient; L, leaves; R, roots. In (C) the short vertical lines indicate the loci of Pi-deficiency lncRNAs in eight chromosomes.
Functional analysis of Pi deficiency-responsive lncRNAs
Previous studies demonstrated that the genes encoding lncRNAs were preferentially located next to protein-coding genes, and that they regulated the expression of the cis- or trans-targets by forming near-complementary lncRNA–target duplexes (Rinn et al., 2007; Yu et al., 2008; Mercer et al., 2009; Kornienko et al., 2013). To examine the potential roles of Pi deficiency-responsive lncRNAs, we analysed the GO terms of putative targets of the lncRNAs.
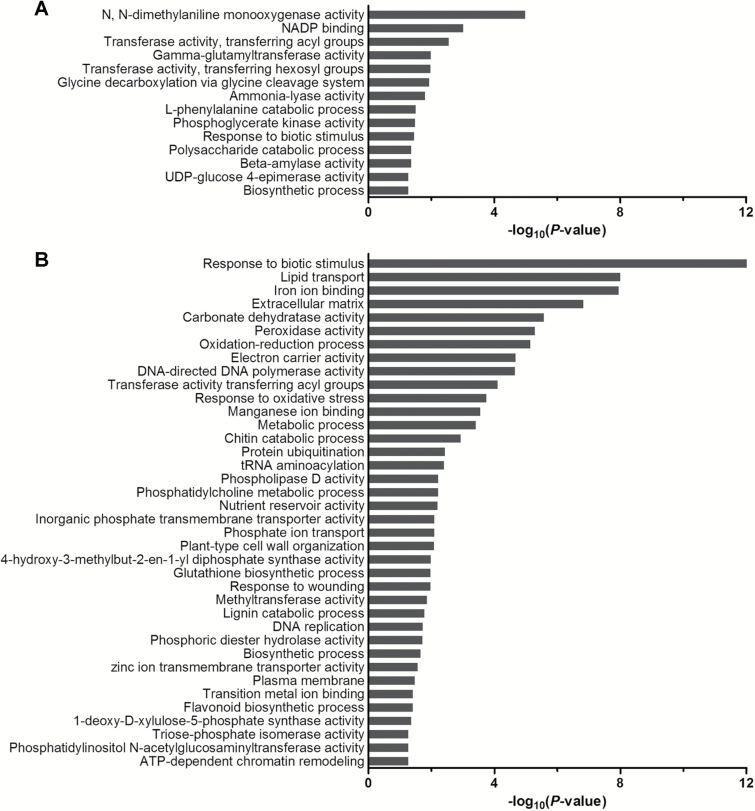
A significant enrichment of putative targets in 14 and 38 GO terms was detected in leaf and root samples, respectively, under the Pi-deficient conditions (Fig. 3, Supplementary Tables S4, S5). The higher number of enriched GO terms in the roots may suggest that they are more sensitive to Pi deficiency than the leaves. The findings also suggest that the Pi deficiency-responsive lncRNAs may regulate the expression of genes involved in many biological processes, including those of signal transduction, energy synthesis, and detoxification. Moreover, some lncRNAs may directly regulate the transport of phosphate (GO: 0006817, Phosphate transport).
Fig. 3.
GO enhancements of putative targets of lncRNAs in leaves (A) and roots (B) of M. truncatula under Pi deficiency. The reliability is calculated by –log10(P-value). More detailed information is given in Supplementary Tables S4 and S5.
Functional characterization of PDIL1, PDIL2, and PDIL3
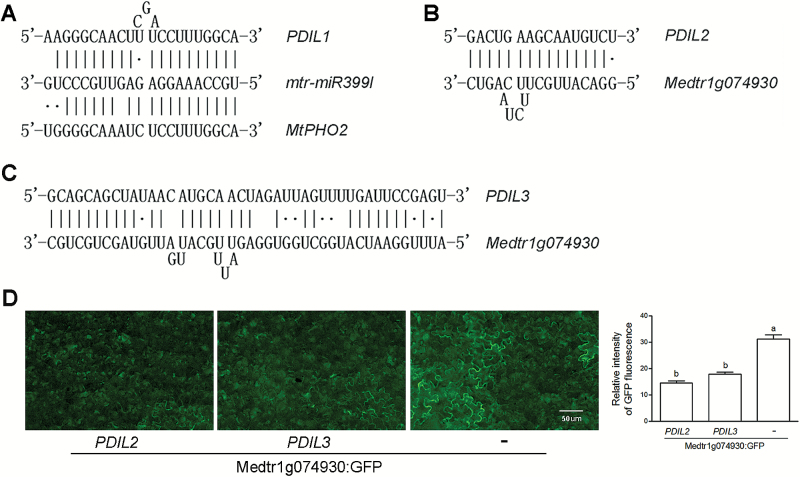
We identified that PDIL1 was an Mt4-like lncRNA by sequence alignment (Supplementary Fig. S2A). Moreover, both PDIL1 and MtPHO2 (Medtr2g013650) are predicted to be targeted by mtr-miR399l in M. truncatula (Fig. 4A). Therefore, PDIL1 can competitively inhibit MtPHO2 degradation as a target mimic of miR399. In addition, PDIL2 and PDIL3 may directly repress the expression of the Pi transporter gene Medtr1g074930 by complementary binding (Fig. 4B–D, Supplementary Figs S2B and S3).
Fig. 4.
Regulation of targets by PDILs. (A) Sequence complementary positions of mtr-miR399l, PDIL1, and MtPHO2. (B) Sequence complementary positions of PDIL2 and its target Medtr1g074930. (C) Sequence complementary positions of PDIL3 and its target Medtr1g074930. Perfect base pairing is depicted with a vertical line, while G–U wobble pairing is marked with a point. Alignments of full sequences between PDIL1 and Mt4, and between PDIL2, PDIL3, and Medtr1g074930 are shown in Supplementary Fig. S2. (D) Transient expression assays in N. benthamiana. Expression of Medtr1g074930:GFP was monitored by fluorescence microscopy. Control agroinfiltration using a strain with an empty vector is indicated by ‘–’.
By means of expression in N. benthamiana, Franco-Zorrilla et al. (2007) reported that Mt4-like lncRNA was involved in the regulation of a Pi transporter gene. We used the same method to test whether PDIL2 and PDIL3 play a role in the regulation of the Pi transporter gene Medtr1g074930. As shown in Fig. 4D, expression of PDIL2 and PDIL3 in N. benthamiana led to a reduction in the relative intensity of Medtr1g074930:GFP.
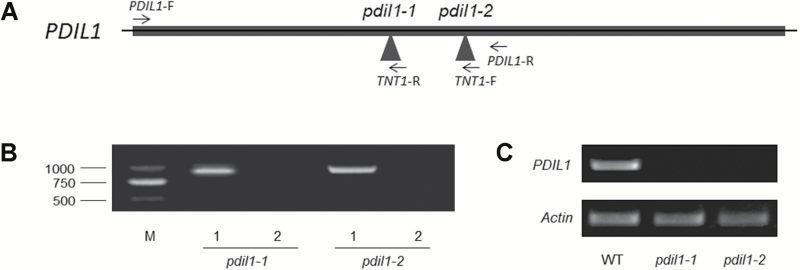
To further study the function of the Pi deficiency-responsive lncRNAs, mutants of these lncRNAs were identified from the M. truncatula mutant database (https://medicago-mutant.noble.org/mutant/). NF19212 (pdil1-1) and NF8919 (pdil1-2) were identified as two mutants for PDIL1 (Fig. 5A), and NF7430 (pdil2) and NF21369 (pdil3) were identified as mutants for PDIL2 and PDIL3, respectively (Supplementary Fig. S4A). Homozygotes of these mutants were validated using PCR (Fig. 5B, Supplementary Fig. S4B). We did not detect the expression of these lncRNAs in the mutants by RT-PCR (Fig. 5C, Supplementary Fig. S4C).
Fig. 5.
Identification and confirmation of pdil1 mutants. (A) The insertional positions of Tnt1 in pdil1 mutations. Triangles indicate insertional positions of Tnt1. The locations of primers used for identification of homozygotes are labelled. (B) Identification of homozygotic pdil1 mutants. Homozygotes were identified using two sets of PCR primers as follows: for pdil1-1, PDIL1-F+TNT1-R in lane 1 and PDIL1-F+PDIL1-R in lane 2; for pdil1-2, PDIL1-F+TNT1-F in lane 1 and PDIL1-F+PDIL1-R in lane 2. (C) The expression levels of PDIL1 in pdil1 mutants. Primers are listed in Supplementary Table S1.
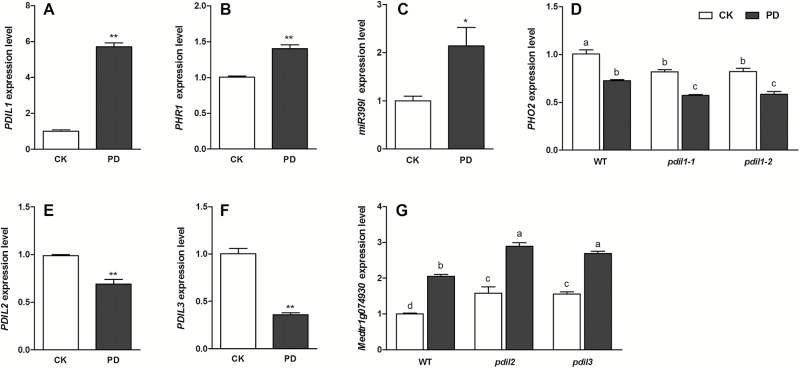
Under Pi-deficient conditions, the expression of a MYB transcription factor gene MtPHR1 (Phosphate Starvation Response 1) was up-regulated in roots, and mtr-miR399l was induced by Pi deficiency (Bari et al., 2006;). MtPHO2 was suppressed as the target of miR399, and it can minimize degradation of Pi transporters in the post-translational processes (Huang et al., 2013). At the same time, PDIL1 was positively regulated to suppress the effect of miR399, thus avoiding the superfluous accumulation of P. Expression-levels of PDIL1, PHR1, and miR3991 (Fig. 6A–C) in WT plants were up-regulated by Pi deficiency. In contrast, expression of PHO2 in WT plants was down-regulated by Pi deficiency (Fig. 6D). Moreover, the mutation of PDIL1 led to a lower expression of MtPHO2 under both Pi-sufficient and Pi-deficient conditions (Fig. 6D). The expression of PDIL2 and PDIL3 (Fig. 6E, F) in WT plants was down-regulated upon exposure to Pi-deficient medium, while expression of a Pi transporter gene Medtr1g074930 was up-regulated by the Pi deficiency (Fig. 6G). The expression-levels of Medtr1g074930 in pdil2 and pdil3 mutants were greater than in WT plants under both Pi-sufficient and Pi-deficient conditions (Fig. 6G).
Fig. 6.
The expression levels of PDIL1 (A), PHR1 (B), miR399l (C), PHO2 (D), PDIL2 (E), PDIL3 (F), and Medtr1g074930 (G) in roots of wild-type (WT) M. truncatula under Pi-sufficient (control, CK) or Pi-deficient (PD) conditions. The expression levels of PHO2 and Medtr1g074930 were detected in pdil mutants. Data with ‘*’ or different letters indicate significant differences (P<0.05) between treatments and control.
The mutants did not show any visible phenotypic differences relative to their WT counterparts, and exhibited similar biomass to WT plants after exposure to Pi-deficient medium for 3 d. In the Pi-sufficient medium, no significant differences in P concentrations of leaves and roots were detected between pdil1 mutants and WT plants (Table 3). P concentrations in the roots of pdil1 mutants and WT plants were comparable after exposure to Pi-deficient medium. However, P concentrations in leaves of pdil1 mutants were significantly higher than in those of WT plants under Pi-deficient conditions (Table 3). Similar to pdil1, pdil2 and pdil3 mutants also exhibited greater capability to acquire P, as evidenced by higher P concentrations in their shoots than in WT plants.
Table 3.
Effects of Pi deficiency on P concentrations in leaves and roots of the wild-type (WT) and different mutants for plants exposed to control (CK, Pi-sufficient) and Pi-deficient (PD) media
| Leaf P concentration (mg g−1 DW) | Root P concentration (mg g−1 DW) | |||
|---|---|---|---|---|
| CK | PD | CK | PD | |
| WT | 10.265 ± 0.283 | 7.353 ± 0.154 | 7.123 ± 0.385 | 3.972 ± 0.183 |
| pdil1-1 | 10.499 ± 0.519 | 8.638 ± 0.161* | 7.268 ± 0.249 | 4.181 ± 0.254 |
| pdil1-2 | 10.293 ± 0.400 | 8.758 ± 0.424* | 7.376 ± 0.139 | 4.051 ± 0.131 |
| pdil2 | 12.630 ± 0.447* | 8.788 ± 0.535* | 8.885 ± 0.592* | 4.286 ± 0.178 |
| pdil3 | 12.360 ± 0.930* | 9.568 ± 0.503** | 8.931 ± 0.853* | 4.326 ± 0.244 |
Data are the means ±SE (n=3). *P<0.05, **P<0.01: significant differences between treatments and control.
Discussion
Despite the greater quantity of lncRNAs than of protein-coding genes in the genomes, little is known about the functions of lncRNAs in plants. Functional characterization of lncRNAs and deciphering their regulatory mechanisms is crucial to advance our knowledge. Identification of lncRNAs at the whole-genome level has been conducted in several plant species by high-throughput sequencing (Ben Amor et al., 2009; Liu et al., 2012, 2013; Li et al., 2014; Shuai et al., 2014; Zhu et al., 2014; Tian et al., 2016; Xu et al., 2016); however, the methods used in some of the studies have not been comprehensive. For example, lncRNAs without poly(A) were not included and only the intergenic lncRNAs were identified (Shuai et al., 2014). In addition, most of the libraries used for sequencing of lncRNAs were without biological repeats, and the functions of lncRNAs have often been predicted by bioinformatics approaches without experimental data (Zhang et al., 2014).
In the present study, we identified all the sense, antisense, bidirectional, intronic, and intergenic lncRNAs, and included the lncRNAs both with and without poly(A) (Fig. 1B). Moreover, we detected low-expressional and tissue-specific lncRNAs using large amounts of data from leaf and root samples. More importantly, two biological repeats of libraries and highly credible data (Supplementary Fig. S1) ensured that our transcriptomic analysis is highly reproducible and reliable, and this was validated by qRT-PCR (Fig. 6A, E, F). We further characterized the functions of three Pi deficiency-responsive lncRNAs using their Tnt1 mutants. To the best of our knowledge, our results identified, for the first time, the most comprehensive Pi deficiency-responsive lncRNAs at the whole-genome level in the higher plants.
Phosphorus is a key component of many macromolecules and ATP in plant cells, and plays important roles in enzymatic reactions and signal transduction (Chiou and Lin, 2011; Dai et al., 2016). Despite high amounts of total P in soils, the inorganic phosphate (Pi) that can be directly acquired by plants is low, and globally approximately 70% of cultivated land suffers from Pi deficiency (Plaxton and Tran, 2011; Li et al., 2016). To cope with deficiency, plants have evolved numerous strategies to acquire Pi from soils. The involvements of SIZ1, PHR1, miR399, and PHO2 in the regulation of Pi acquisition have been well established in plants (Bari et al., 2006; Chiou and Lin, 2011). For instance, the expression of miR399 is positively regulated by the transcription factor PHR1 that is induced by Pi deficiency. PHO2, a target of miR399, encodes a ubiquitin-conjugating E2 enzyme that can degrade Pi transporters, such that cleavage of PHO2 confers greater acquisition of Pi by roots under Pi-deficient conditions (Bari et al., 2006; Huang et al., 2013; Park et al., 2014). There have been reports implying that lncRNAs participate in signal transduction of Pi in plants. For example, Pi starvation markedly induced expression of Mt4, and At4 and IPS1 were identified as two Mt4-like lncRNAs in Arabidopsis (Bazin and Bailey-Serres, 2015). At4 and IPS1 share a conserved motif, showing partial complementarity with miR399. Thus, they can competitively bind to miR399 as target mimics to protect PHO2 transcripts from degradation by miR399 (Shin et al., 2006; Franco-Zorrilla et al., 2007). The functions of At4 and IPS1 have been characterized by overexpression lines and mutants (Shin et al., 2006; Franco-Zorrilla et al., 2007), but the function of Mt4 in the regulation of Pi homeostasis has not been validated experimentally using genetic material in M. truncatula.
Here, we identified a Pi deficiency-responsive lncRNA, PDIL1, that is a close paralog of Mt4. To characterize the function of PDIL1, we obtained two mutants from the M. truncatula mutant database. The mutation of PDIL1 potentiated the negative regulation of miR399 to PHO2, leading to a lower expression of PHO2 in the pdil1 and pdil2 mutants than that of WT plants in Pi-sufficient medium (Fig. 6D). Moreover, the PHO2 transcripts were reduced by enhanced expression of miR399 in both WT and mutants under Pi-deficient conditions, while a lower expression level of PHO2 was observed in roots of mutants than WT plants (Fig. 6D). PHO2 encodes a ubiquitin-conjugating enzyme, UBC24, which mediates degradation of high-affinity Pi transporters. It is conceivable that the activity of Pi transporters in pdil1 is greater than that in the WT plants, thus conferring the pdil1 mutant greater ability to acquire Pi. Our results showed that P concentrations in roots of pdil1 were comparable to those in WT plants. However, mutation of pdil1 rendered a greater accumulation of P in shoots under Pi-deficient conditions (Table 3). In Arabidopsis, overexpression of At4 and IPS1 resulted in a decrease in shoot P concentration in Pi-sufficient medium, and mutation of At4 increased P concentrations in shoots under Pi-deficient conditions (Shin et al., 2006; Franco-Zorrilla et al., 2007). In the present study, we found that mutation of PDIL led to an increase in P concentrations in leaves (Table 3). By contrast, no significant differences in P concentrations in roots between the WT and the pdil1 mutants were observed (Table 3). This observation may suggest that P taken up by roots is translocated preferentially into shoots to participate in important biological processes such as photophosphorylation. In Pi-sufficient medium, we did not observe the greater accumulation of P in the pdli1 mutants. This suggests that PDIL1 may not directly regulate the Pi transporters; instead, it may indirectly control the Pi transport by PHO2 via unknown mechanisms, thus allowing the plant to maintain a constant P concentration under Pi-sufficient conditions. In addition, we found that Pi deficiency down-regulated expression of PDIL2 and PDIL3 (Fig. 6E, F). We further demonstrated that expression of PDIL2 and PDIL3 in N. benthamiana suppressed expression of the Pi transporter gene Medtr1g074930 (Fig. 4B–D). Mutations of the two lncRNAs resulted in a greater ability to acquire P by the mutants. In Pi-sufficient medium, accumulation of P in both leaves and roots of pdil2 and pdil3 was greater than that of the WT. However, P contents in the leaves of the pdil2 and pdil3 mutants under Pi-deficient conditions were higher than those in WT plants (Table 3). A similar explanation to that of the pdil1 mutant may be used to account for the difference. Functional elucidation of these lncRNAs highlights their important roles in the regulation of Pi homeostasis in plants.
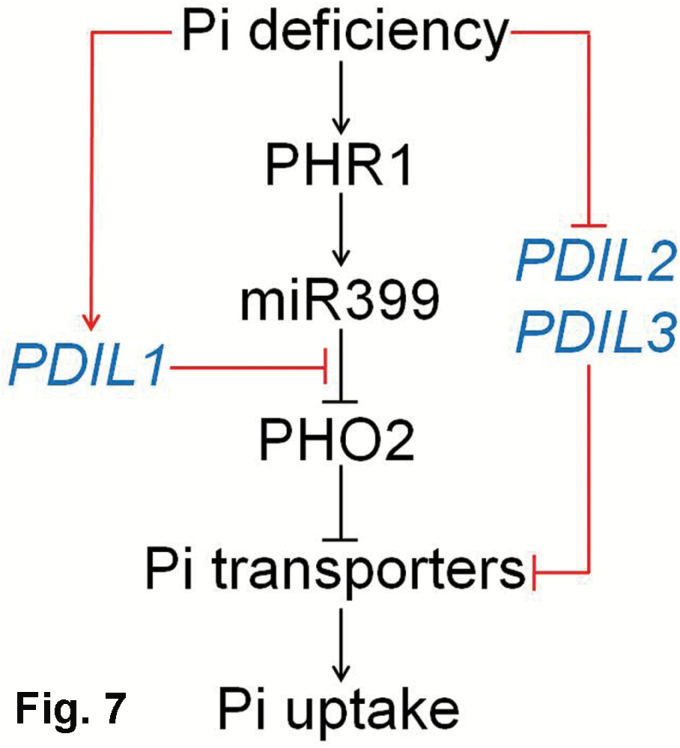
Based on our results, we propose a model for the involvement of lncRNAs in the regulation of a Pi-deficient signaling pathway (Fig. 7). In this model, the PHR1–miR399–PHO2 pathway has been established to play a central role in the regulation of P acquisition. Upon exposure to Pi-deficient medium, the MYB transcription factor genes of PHR1 and miR399 are up-regulated consecutively. The expression of PHO2, a target of miR399, is suppressed by miR399 under Pi- deficient conditions. PHO2 is involved in the degradation of Pi transporters by the ubiquitination pathway, thus leading to an increase in P acquisition by up-regulating the activity of Pi transporters. The two molecular pathways converge to form Pi-dependent signaling cascades. It is predicated that PDIL2 and PDIL3 negatively regulate the expression of the Pi transporter gene Medtr1g074930. The down-regulation of PDIL2 and PDIL3 evoked by Pi deficiency up-regulates the transcript of Medtr1g074930. Both changes in the PHR1–miR399–PHO2 and PDIL2/PDIL3 pathways can enhance P acquisition under Pi-deficient conditions. In contrast, the Mt4-like lncRNAs, including PDIL1, are induced by Pi deficiency, and they negatively regulate P acquisition. PDIL1 shares a conserved motif with PHO2 that can be identified by miR399 and PDIL1. Therefore, PDIL1 can competitively bind to miR399 as a target mimic to inhibit PHO2, leading to a negative regulation of P acquisition. The two directional efforts form a regulatory loop to maintain P homeostasis, thus allowing plants to perform vari- ous physiological processes at an appropriate Pi concentration.
Fig. 7.
A model for Pi signaling involving PHR1, miR399, PHO2, and three Pi deficiency-responsive lncRNAs (PDIL1, PDIL2, and PDIL3) in M. truncatula. The red lines represent the pathways identified in the present study. Arrows denote positive effects, whereas lines ending with a short bar indicate negative effects.
In conclusion, we obtained 111.24 G clean sequence data from sequencing eight paired-end libraries, and identified 10 785 lncRNAs from the legume model plant M. truncatula. By GO enrichment of the targets of Pi deficiency-responsive lncRNAs, we showed that these lncRNAs were involved in the regulation of signal transduction, energy synthesis, detoxification, and phosphate transport. We further demonstrated that the lncRNAs PDIL1-3 were involved in the regulation of the Pi-deficiency network and Pi transport. These results provide valuable information for our understanding of the functions of lncRNAs in response to Pi deficiency.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Sequences of primers used in this study.
Table S2. All putative lncRNAs identified in this study.
Table S3. Information relating to Pi deficiency-responsive lncRNAs.
Table S4. GO enhancements of the putative targets of lncRNAs in leaves under P deficiency.
Table S5. GO enhancements of the putative targets of lncRNAs in roots under P deficiency.
Fig. S1. Quality score values of RNA-seq from eight samples.
Fig. S2. Alignments of full sequences between PDIL1 and Mt4, and between PDIL2, PDIL3, and Medtr1g074930.
Fig. S3. Alignments of protein sequences between Medtr1g074930 and other phosphate transporters of Arabidopsis and rice.
Fig. S4. Identification and confirmation of pdil2 and pdil3 mutants.
Supplementary Material
Acknowledgements
This work was supported by the National Science Foundation of China [31300231, 31370300, and 31671270] and the Chinese Academy of Sciences [KFJ-STS-ZDTP-004 ]. Generation of M. truncatula mutants was supported by grants from the National Science Foundation, USA [DBI 0703285 and IOS 1127155] and in part by the Noble Research Institute. We greatly appreciate the constructive suggestions made on a previous version of the manuscript by three anonymous reviewers and by the editor. We thank Dr Linda Zhu of LC Biotech for help with the bioinformatics analysis.
References
- Ariel F, Jegu T, Latrasse D, Romero-Barrios N, Christ A, Benhamed M, Crespi M. 2014. Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Molecular Cell 55, 383–396. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA et al. . 2000. Gene ontology: tool for the unification of biology. Nature Genetics 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardou F, Ariel F, Simpson CG, Romero-Barrios N, Laporte P, Balzergue S, Brown JW, Crespi M. 2014. Long noncoding RNA modulates alternative splicing regulators in Arabidopsis.Developmental Cell 30, 166–176. [DOI] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. 2006. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiology 141, 988–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Bailey-Serres J. 2015. Emerging roles of long non-coding RNA in root developmental plasticity and regulation of phosphate homeostasis. Frontiers in Plant Science 6, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben Amor B, Wirth S, Merchan F et al. . 2009. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Research 19, 57–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD et al. . 2008. A gene expression atlas of the model legume Medicago truncatula. The Plant Journal 55, 504–513. [DOI] [PubMed] [Google Scholar]
- Borodina T, Adjaye J, Sultan M. 2011. A strand-specific library preparation protocol for RNA sequencing. Methods in Enzymology 500, 79–98. [DOI] [PubMed] [Google Scholar]
- Brosnan CA, Voinnet O. 2009. The long and the short of noncoding RNAs. Current Opinion in Cell Biology 21, 416–425. [DOI] [PubMed] [Google Scholar]
- Burleigh SH, Harrison MJ. 1997. A novel gene whose expression in Medicago truncatula roots is suppressed in response to colonization by vesicular-arbuscular mycorrhizal (VAM) fungi and to phosphate nutrition. Plant Molecular Biology 34, 199–208. [DOI] [PubMed] [Google Scholar]
- Campalans A, Kondorosi A, Crespi M. 2004. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. The Plant Cell 16, 1047–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA. 2015. Long non-coding RNAs and their functions in plants. Current Opinion in Plant Biology 27, 207–216. [DOI] [PubMed] [Google Scholar]
- Chekanova JA, Gregory BD, Reverdatto SV et al. . 2007. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell 131, 1340–1353. [DOI] [PubMed] [Google Scholar]
- Chiou TJ, Lin SI. 2011. Signaling network in sensing phosphate availability in plants. Annual Review of Plant Biology 62, 185–206. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. [DOI] [PubMed] [Google Scholar]
- Dai X, Wang Y, Zhang WH. 2016. OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. Journal of Experimental Botany 67, 947–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JH, Lu Q, Ouyang YD, Mao HL, Zhang PB, Yao JL, Xu CG, Li XH, Xiao JH, Zhang QF. 2012. A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proceedings of the National Academy of Sciences, USA 109, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding XD, Zhang SR, Wang RP, Li SY, Liao XR. 2016. AM fungi and rhizobium regulate nodule growth, phosphorous (P) uptake, and soluble sugar concentration of soybeans experiencing P deficiency. Journal of Plant Nutrition 39, 1915–1925. [Google Scholar]
- Dong J, Piñeros MA, Li X, Yang H, Liu Y, Murphy AS, Kochian LV, Liu D. 2017. An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Molecular Plant 10, 244–259. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Research 8, 186–194. [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. 2007. Target mimicry provides a new mechanism for regulation of microRNA activity. Nature Genetics 39, 1033–1037. [DOI] [PubMed] [Google Scholar]
- Heo JB, Sung S. 2011. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. [DOI] [PubMed] [Google Scholar]
- Hinsinger P, Betencourt E, Bernard L, Brauman A, Plassard C, Shen J, Tang X, Zhang F. 2011. P for two, sharing a scarce resource: soil phosphorus acquisition in the rhizosphere of intercropped species. Plant Physiology 156, 1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TK, Han CL, Lin SI et al. . 2013. Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. The Plant Cell 25, 4044–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabnoune M, Secco D, Lecampion C, Robaglia C, Shu Q, Poirier Y. 2013. A rice cis-natural antisense RNA acts as a translational enhancer for its cognate mRNA and contributes to phosphate homeostasis and plant fitness. The Plant Cell 25, 4166–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S et al. . 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316, 1484–1488. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Zheng B, Yu Y, Won SY, Mo B, Chen X. 2011. The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. The EMBO Journal 30, 814–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Research 35, W345–W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. 2013. Gene regulation by the act of long non-coding RNA transcription. BMC Biology 11, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Research 19, 1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li C, Zhang H, Liao H, Wang X. 2017. The purple acid phosphatase GmPAP21 enhances internal phosphorus utilization and possibly plays a role in symbiosis with rhizobia in soybean. Physiologia Plantarum 159, 215–227. [DOI] [PubMed] [Google Scholar]
- Li L, Eichten SR, Shimizu R et al. . 2014. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biology 15, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wu H, Fan H, Zhao T, Ling HQ. 2016. Characterization of the AtSPX3 promoter elucidates its complex regulation in response to phosphorus deficiency. Plant & Cell Physiology 57, 1767–1778. [DOI] [PubMed] [Google Scholar]
- Liao Q, Liu C, Yuan X et al. . 2011. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Research 39, 3864–3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH. 2012. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. The Plant Cell 24, 4333–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang H, Chua NH. 2015. Long noncoding RNA transcriptome of plants. Plant Biotechnology Journal 13, 319–328. [DOI] [PubMed] [Google Scholar]
- Liu R, Zhu JK. 2014. Non-coding RNAs as potent tools for crop improvement. National Science Review 1, 186–189. [Google Scholar]
- Liu TT, Zhu D, Chen W, Deng W, He H, He G, Bai B, Qi Y, Chen R, Deng XW. 2013. A global identification and analysis of small nucleolar RNAs and possible intermediate-sized non-coding RNAs in Oryza sativa. Molecular Plant 6, 830–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. 2009. Long non-coding RNAs: insights into functions. Nature Reviews Genetics 10, 155–159. [DOI] [PubMed] [Google Scholar]
- Nath M, Tuteja N. 2016. NPKS uptake, sensing, and signaling and miRNAs in plant nutrient stress. Protoplasma 253, 767–786. [DOI] [PubMed] [Google Scholar]
- Nestler J, Wissuwa M. 2016. Superior root hair formation confers root efficiency in some, but not all, rice genotypes upon P deficiency. Frontiers in Plant Science 7, 1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BS, Seo JS, Chua NH. 2014. NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. The Plant Cell 26, 454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaxton WC, Tran HT. 2011. Metabolic adaptations of phosphate-starved plants. Plant Physiology 156, 1006–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W. 2009. Evolution and functions of long noncoding RNAs. Cell 136, 629–641. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annual Review of Biochemistry 81, 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK et al. . 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin HS, Chen R, Harrison MJ. 2006. Loss of At4 function impacts phosphate distribution between the roots and the shoots during phosphate starvation. The Plant Journal 45, 712–726. [DOI] [PubMed] [Google Scholar]
- Shuai P, Liang D, Tang S, Zhang Z, Ye CY, Su Y, Xia X, Yin W. 2014. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. Journal of Experimental Botany 65, 4975–4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Luo H, Bu D, Zhao G, Yu K, Zhang C, Liu Y, Chen R, Zhao Y. 2013. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Research 41, e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462, 799–802. [DOI] [PubMed] [Google Scholar]
- Tadege M, Wen J, He J et al. . 2008. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. The Plant Journal 54, 335–347. [DOI] [PubMed] [Google Scholar]
- Tian J, Song Y, Du Q, Yang X, Ci D, Chen J, Xie J, Li B, Zhang D. 2016. Population genomic analysis of gibberellin-responsive long non-coding RNAs in Populus. Journal of Experimental Botany 67, 2467–2482. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28, 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TZ, Liu M, Zhao MG, Chen R, Zhang WH. 2015. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biology 15, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Fan XD, Lin F, He GM, Terzaghi W, Zhu DM, Deng XW. 2014. Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proceedings of the National Academy of Sciences, USA 111, 10359–10364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel A, Akbasli E, Gorodkin J. 2012. RIsearch: fast RNA–RNA interaction search using a simplified nearest-neighbor energy model. Bioinformatics 28, 2738–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes & Development 23, 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XW, Zhou XH, Wang RR, Peng WL, An Y, Chen LL. 2016. Functional analysis of long intergenic non-coding RNAs in phosphate-starved rice using competing endogenous RNA network. Scientific Reports 6, 20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE et al. . 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. 2008. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451, 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Han Z, Guo Q, Liu Y, Zheng Y, Wu F, Jin W. 2014. Identification of maize long non-coding RNAs responsive to drought stress. PLoS ONE 9, e98958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Stephen S, Taylor J, Helliwell CA, Wang MB. 2014. Long noncoding RNAs responsive to Fusarium oxysporum infection in Arabidopsis thaliana. New Phytologist 201, 574–584. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.