The plant-derived membrane surrounding the powdery mildew intracellular feeding structure, called the extrahaustorial membrane, accumulates the ER- and ER/cis-Golgi-associated Sar1 and Rab1 GTPases and stains with dyes for the ER.
Keywords: Barley organelle markers, endoplasmic reticulum (ER), extrahaustorial membrane (EHM), powdery mildew, Rab GTPase, Sar1 GTPase, secretion
Abstract
Many filamentous plant pathogens place specialized feeding structures, called haustoria, inside living host cells. As haustoria grow, they are believed to manipulate plant cells to generate a specialized, still enigmatic extrahaustorial membrane (EHM) around them. Here, we focused on revealing properties of the EHM. With the help of membrane-specific dyes and transient expression of membrane-associated proteins fused to fluorescent tags, we studied the nature of the EHM generated by barley leaf epidermal cells around powdery mildew haustoria. Observations suggesting that endoplasmic reticulum (ER) membrane-specific dyes labelled the EHM led us to find that Sar1 and RabD2a GTPases bind this membrane. These proteins are usually associated with the ER and the ER/cis-Golgi membrane, respectively. In contrast, transmembrane and luminal ER and Golgi markers failed to label the EHM, suggesting that it is not a continuum of the ER. Furthermore, GDP-locked Sar1 and a nucleotide-free RabD2a, which block ER to Golgi exit, did not hamper haustorium formation. These results indicated that the EHM shares features with the plant ER membrane, but that the EHM membrane is not dependent on conventional secretion. This raises the prospect that an unconventional secretory pathway from the ER may provide this membrane’s material. Understanding these processes will assist future approaches to providing resistance by preventing EHM generation.
Introduction
Many microbes of major importance are extremely intimate with higher organisms as they live entirely or partially within the boundaries of their host’s cells, where they take advantage of a continuous nutrient supply. This is also true for several clades of filamentous fungi and oomycetes that interact with plants, either pathogenically or symbiotically. These microbes include the beneficial mycorrhizal fungi, the oomycete pathogens causing late blight, downy mildew and white rust, and the rust and powdery mildew fungi. In host plant cells, specialized fungal structures, called arbuscules or haustoria, are formed. These are directly connected to hyphae outside the cells. As haustoria and arbuscules grow, the plant cells generate a membrane around them, which allows the invader and the host to have an intimate coexistence for an extended period of time. These periarbuscular and extrahaustorial membranes (EHMs) separate the microbe from the host cytosol and mediate nutrient absorption and transfer of virulence factors into the host cytosol (Szabo & Bushnell, 2001; O’Connell & Panstruga, 2006). Despite their obvious importance, the nature and source of these membranes remain unclear.
In plant cells, three major membrane trafficking pathways can be defined. These are the secretory pathway, the endocytic pathway and the vacuolar pathway (Kanazawa and Ueda, 2017). When Ustilago maydis attacks maize cells, the membranes surrounding the pathogen’s intracellular hyphae are labelled by plasma membrane (PM) markers (Djamei & Kahmann, 2012). This suggests that the PM is invaginated and/or that the endocytic pathway contributes to this membrane. Also, a number of PM-resident proteins and the marker for endocytosed membranes, FM4-64, co-localize with the EHM around haustoria of the oomycetes Hyaloperonospora arabidopsidis and Phytophthera infestans (Lu et al., 2012). Furthermore, the EHM in P. infestans-invaded Nicotiana benthamiana cells is labeled by the multivesicular body/tonoplast-localized GTPase NbRabG3c (Bozkurt et al., 2015). This suggests endocytic delivery via multivesicular bodies (MVBs) is involved in the formation of the EHM when plants are infected by these two oomycete pathogens.
Several PM markers failed to accumulate on the Arabidopsis EHM formed around Golovinomyces cichoracearum powdery mildew haustoria (Koh et al., 2005). This indicates that either the EHM formation in response to G. cichoracearum does not involve endocytosis from the PM or that a filtering process excludes PM-targeted proteins from the EHM. The EHMs in cells invaded by powdery mildew as well as by rust fungal species are separated from the host PM by a neck band (Manners & Gay, 1977; Szabo & Bushnell, 2001; Larous et al., 2008), which likely limits mixing of proteins and lipids between the membranes. A number of observations for the EHM around powdery mildew haustoria indicate that a conventional membrane trafficking pathway does not generate this membrane. Firstly, the EHM in the Arabidopsis–G. cichoracearum interaction is targeted by the resistance protein RPW8.2 in a vesicle-associated membrane protein (VAMP) 721/VAMP722-dependent process (Wang et al., 2009; Kim et al., 2014; Kuhn et al., 2016). These vesicle N-ethylmaleimide sensitive factor attachment protein receptors (v-SNAREs) are suggested to be involved in secretion by mediating vesicle fusions at the PM (Kwon et al., 2008; Kuhn et al., 2016), and therefore the data suggest a secretory pathway to the EHM. Secondly, application of the fungal toxin brefeldin A (BFA) affects neither RPW8.2 delivery to the EHM (Berkey et al., 2017) nor haustorial success rate (Nielsen et al., 2012). The latter indirectly shows that EHM formation is insensitive to BFA. BFA blocks recycling of PM material to the site of attack via the trans-Golgi network (TGN) and MVBs (Nielsen et al., 2012; Nielsen and Thordal-Christensen, 2013). Therefore, these BFA studies do not conflict with the existence of a secretory pathway to the EHM. Thirdly, Berkey et al. (2017) showed that FM4-64 does not reach the EHM following endocytosis. In plants, the TGN acts as an early endosome and a major sorting station for secretion, recycling, and trafficking to the vacuole via MVBs, and all these compartments readily stain with FM4-64 (Viotti et al., 2010; Richter et al., 2014). Therefore, the TGN is not the likely source of membrane delivery to the EHM, suggesting that neither secretion nor conventional vacuolar trafficking is involved in its formation. This conflicts with the fourth observation, namely that Rab5 GTPases associate with the EHM around powdery mildew haustoria in both the Arabidopsis–G. orontii and in the barley–Blumeria graminis f.sp. hordei (Bgh) interactions (Inada et al., 2016). Plant Rab5 GTPases are suggested to be recruited to the TGN and follow the MVBs during their maturation (Singh et al., 2014). Combined, these observations suggest an unconventional membrane trafficking route to the EHM.
While these observations do not allow conclusions to be drawn as to the likely membrane contributor to EHM formation, the close physical association of the endoplasmic reticulum (ER) tubules with the EHM (Harder et al., 1978; Leckie et al., 1995; Koh et al., 2005; Micali et al., 2011; Zhang et al., 2013) could suggest vesicle exchange from the ER membrane to the EHM. This is supported by data showing the ER-membrane dye DiOC6 labelling the EHM of powdery mildew haustoria in pea (Leckie et al., 1995). In fact, we show here that the EHM surrounding the haustoria of Bgh has ER-like properties. Our data suggest that it can be stained by ER-membrane dyes, and we find that two small GTPases, Sar1 and RabD2a, which are essential for COPII- and COPI-mediated vesicle trafficking between the ER and Golgi, are specifically associated with it. However, the EHM forms despite inhibition of conventional ER–Golgi trafficking after expression of dominant-negative versions of Sar1a and RabD2a. This leads us to suggest that the EHM obtains its ER-like properties from an unconventional trafficking pathway transporting membrane material from the ER.
Materials and methods
Plant and fungal growth
For this study, barley (Hordeum vulgare) cv. Golden Promise seedlings, grown at 20 °C with 16 h light (150 μE s−1 m−2) and 8 h dark, were used. Bgh (isolate DH14) was propagated by weekly inoculum transfer onto 1-week-old barley seedlings.
Particle bombardment
The first leaves of 7-day-old seedlings were transiently transformed by particle bombardment as described by Douchkov et al. (2005). For marker localization studies in powdery mildew-infected cells, the leaves were inoculated 2 h after bombardment. The leaves were examined by microscopy from 2 d after inoculation (dai).
Constructs and primers
The coding regions of HvSar1a, HvSar1b and HvSar1c, HvRabD2a and HvRabG3b, Genbank accessions AK252291.1, AK252372.1, AK250294.1, AK355333.1 and AK368361.1, respectively, were amplified using Gateway® compatible primers. The coding regions of HvERD2 and HvSec12, Genbank accessions AK250768.1 and AK356828.1, respectively, were amplified using two sets of cDNA specific primers. However, since AK356828.1 is a partial sequence, we designed the Sec12 forward primer (see Supplementary Table S1 at JXB online) based on the barley Sec12 genomic sequence and positioned it upstream from the start codon. See Supplementary Figure S1 for alignments of the encoded proteins to the closest Arabidopsis homologues. Subsequently, on the purified PCR product, a second PCR using Gateway® compatible primers was performed. The coding sequences were amplified from barley Golden Promise cDNA. With a Gateway® BP reaction (Invitrogen) the fragments were subsequently cloned into the pDONR201 donor vector. The dominant-negative mutations HvSar1a-T34N and HvRabD2a-N121I were introduced using overlapping primers encoding the planned mutation using the Quickchange XL mutagenesis kit (Stratagene). Primer sequences can be found in Table S1. After validation of the donor constructs by sequencing, the inserts were transferred into the destination vector with a Gateway® LR reaction (Invitrogen). The pUbi-mYFP-Gateway, pUbi-mCherry-Gateway, pUbi-Gateway-mYFP and pUbi-Gateway-mCherry destination vectors used in this study are described in Kwaaitaal et al. (2010), and the 35S-driven overexpression destination vectors p2FGW7 and p2WFHB-Gateway-GFP are described in Karimi et al. (2002) and Böhlenius et al. (2010), respectively. Table 1 lists the marker constructs used in this work.
Table 1.
Markers used in this study
| Marker/construct | Labels | Function | Reference |
|---|---|---|---|
| DiOC6 | ER membrane | — | — |
| ER-Tracker Blue-White | ER membrane | — | — |
| Hexyl rhodamine B | ER membrane | — | — |
| SP–YFP–HDEL | ER lumen | — | Irons et al. (2003) |
| SP–mCherry–HDEL | ER lumen | — | Nelson et al. (2007) |
| HvMLO–mCherry | Plasma membrane | Powdery mildew susceptibility | Kwaaitaal et al. (2010) |
| HvERD2–mCherry | ER membrane | HDEL receptor | Kwaaitaal et al. (2010) |
| HvLTP–mCherry | Plasma membrane | Lipid transfer protein | Kwaaitaal et al. (2010) |
| ST–YFP | Golgi | 52-amino acid signal anchor of sialyltransferase (from rat) | Brandizzi et al. (2002) |
| GFP–HvRabG3b | Tonoplast/cytosol | GTPase, membrane fusion | This work |
| HvSar1a–mCherry | ER membrane/cytosol | GTPase, vesicle budding | This work |
| HvSar1a–mYFP | ER membrane/cytosol | GTPase, vesicle budding | This work |
| HvSar1b–mCherry | ER membrane/cytosol | GTPase, vesicle budding | This work |
| HvSar1c–mYFP | ER membrane/cytosol | GTPase, vesicle budding | This work |
| HvSec12–mCherry | ER membrane/cytosol | GEF for Sar1 activation | This work |
| GFP and mYFP–HvRabD2a | ER/cis-Golgi/cytosol | GTPase, membrane fusion | This work |
| mCherry–HvRabD2a | ER/cis-Golgi/cytosol | GTPase, membrane fusion | This work |
| HvSar1aT34N–mYFP (GDP-locked) | ER membrane/cytosol | Inhibition of vesicle budding | This work |
| HvSar1aT34N–mCherry (GDP-locked) | ER membrane/cytosol | Inhibition of vesicle budding | This work |
| Golgi–mCherry | Golgi | α-1,2-Mannosidase I (soybean) | Nelson et al. (2007) |
| CFP–YFP | Cytosol | — | Bethke et al. (2009) |
| HvRabD2aN121I (nucleotide-free) | ER/cis-Golgi/cytosol | Inhibition of membrane fusion | This work |
| HvARA6–GFP | trans-Golgi network/MVB/cytosol | GTPase, MVB maturation | Inada et al. (2016) |
Confocal microscopy
Barley leaves with transformed epidermal cells were vacuum infiltrated with 0.01% Tween20 and subsequently mounted in the same buffer under a coverslip for confocal microscopy. Leica SP5-X, SP5-II and SP8 confocal laser scanning microscopes mounted with ×63 water immersion lenses with a numerical aperture of 1.2 were used. For detection and localization of the fluorophores, green fluorescent protein (GFP) was excited at 488 nm and detected between 500 and 540 nm, monomeric yellow fluorescent protein (mYFP) was excited at 514 nm and detected between 528 nm and 560 nm, while mCherry was excited at 543 nm (Leica SP5-II) or 587 nm using the super continuum white laser (Leica SP5-X) and detected between 600 and 640 nm. To limit signal bleed-through between channels, the measurements of each fluorophore were performed in independent tracks exciting only one fluorophore at a time.
Staining with the ER-specific dyes
The dyes, ER-Trackertm Blue-White, DiOC6 and hexyl rhodamine B (all Invitrogen; Table 1), were vacuum infiltrated into 1 cm barley leaf pieces in 0.01% Tween20 at final concentrations of 5, 10, and 1.6 μM, respectively. After incubation for 30 min, excess dye was removed by a short wash in 0.01% Tween20 and afterwards mounted in the same buffer for microscopy. ER-Tracker Blue-White was excited with a UV laser at 405 nm and the fluorescence was detected between 420 and 540 nm. DiOC6 was excited at 488 nm and the fluorescence was detected between 495 and 520 nm. Hexyl rhodamine B was excited at 543 nm and the fluorescence was detected between 580 and 640 nm.
Quantification of powdery mildew fungal infection efficiency
To determine the role of HvSar1a in powdery mildew haustorial establishment, barley leaf epidermal cells were transformed, using particle bombardment, with a p35S–mCherry construct as transformation reporter and equimolar amounts of pUbi–YFP (control), pUbi–HvSar1a or pUbi–HvSar1aT34N (GDP-bound). To determine the role of HvRabD2a, transformation was made with a β-glucuronidase (GUS) reporter construct (pUbiGUS) mixed in equimolar amounts with p2WF7HB (empty vector control) or p35S–HvRabD2aN121I (nucleotide-free). One day after transformation, the leaves were inoculated with powdery mildew spores at a density of 200 spores per square millimeter. Two days later, the total number of transformed cells was determined either by the mCherry signal using a standard epifluorescence microscope, or by GUS staining using a standard light microscope. At the same time the number of transformed cells containing a haustorium was scored. For additional details, see Böhlenius et al. (2010). The haustorial index was calculated as the fraction of the transformed cell that hosted a haustorium relative to the same fraction in the control treatment. Three independent biological replicates were performed with two technical replicates in each experiment.
Fluorescence redistribution after photobleaching
Fluorescence redistribution after photobleaching (FRAP) measurements were performed on epidermal cells 2 d after transformation with fluorescent constructs and inoculation with Bgh. For each FRAP measurement, an area of 4 × 4 μm positioned at a haustorial finger was selected. The fluorescent signal in this image area was initially recorded by scanning five times. Thereafter, the area was bleached with high excitation laser power after which the time-lapse acquisition was continued for 60 s to be able to follow and quantify fluorescence recovery in the bleached area. The recovery curves were evaluated and fit using the LAS AF lite software package (Leica Microsystems CMS GmbH) and Igor Pro 6 with the K_FRAPcalc v9i procedure developed by Kota Miura (EMBL, https://cmci-embl.gitlab.io/docs/analysis/frapcalc/, last accessed 15 November 2017). The values were normalized using Phair’s double normalization and fit with a model taking two diffusing species into account (Phair et al., 2004). An average diffusion coefficient was calculated taking the relative fractions of the two diffusing species into account.
Image processing and analysis
Raw image data was exported from the LAS AF lite software package and imported for processing in ImageJ (versions 1.45–1.48, NIH, USA). Using the ‘Window/Level’ function, the whole image was adjusted to use the complete grey scale range without or with a limited amount of saturated areas in the image. This adjustment allowed evaluation of the co-localization of signals independent of the overall fluorophore-dependent signal intensity. Subsequently, the images were blurred, using the ‘Gaussian Blur’ filter with a sigma radius of 1 to reduce background noise, and merged. False colors were assigned to the channels to evaluate co-localization. Maximum intensity projections of a series of consecutive z-stacks were performed using the Zprojection function in ImageJ. The extended depth of field projection shown in Fig. 1A was generated using the ‘Extended depth of field’ plug-in described in Forster et al. (2004) (http://bigwww.epfl.ch/demo/edf/index.html, last accessed 15 November 2017).
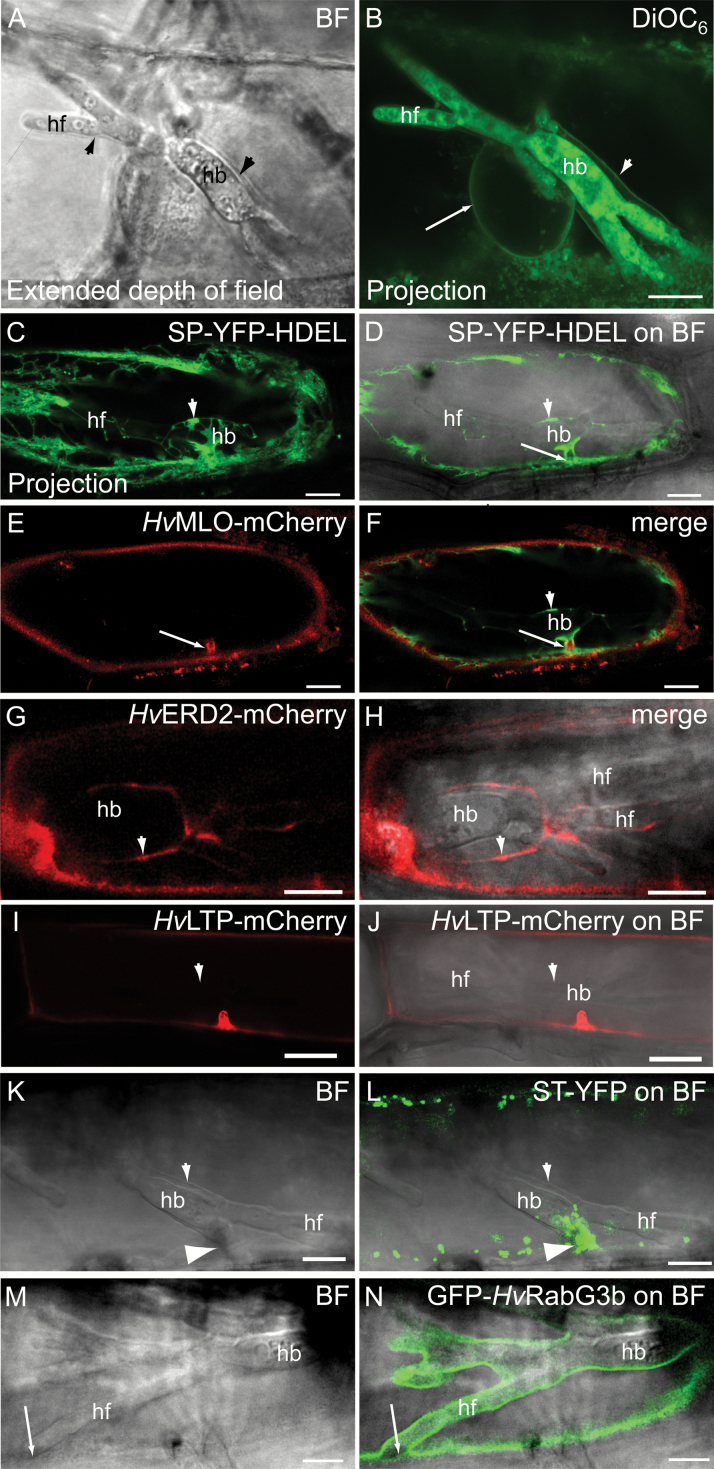
Fig. 1.
The extrahaustorial membrane, ER-membrane binding dyes and close association with the early secretory pathway. All micrographs show barley epidermal cells containing Bgh haustoria at 2 dai. (A, B) Cell stained with 10 μM DiOC6. (A) Bright field (BF) image showing the haustorial body, haustorial fingers and the EHM (arrowheads). (B) Maximum intensity projection of the DiOC6 fluorescence. Note labelling of the nuclear membrane ER membrane around the nucleus (arrow) and membrane around the haustorium (arrowhead). A single section of this projection can be seen in Supplementary Fig. S2D. (C–F) Cell co-expressing the PM protein HvMLO–mCherry and the ER marker SP–YFP–HDEL. Arrow: haustorial neck; arrowhead: ER tubules and cisternae around the haustorial body. (C) Maximum intensity projection of the SP–YFP–HDEL signal. This micrograph visualizes the ER tubules around the haustorial body. (G, H) Cell expressing the HvERD2–mCherry marker. Arrowhead: ER at haustorial body. More images of this cell can be seen in Supplementary Fig. S4D–H. (I, J) Cell expressing the PM protein HvLTP–mCherry. Arrowhead: EHM. (K, L) Cell expressing the ST–YFP Golgi marker. Note the accumulation of Golgi bodies at the haustorial neck (arrowhead). (M, N) Cell expressing the tonoplast protein GFP–HvRabG3b. Note that GFP–HvRabG3b stays closely associated with the haustorium, but retracts with the tonoplast at the tips of the haustorial fingers when these are close to the host cell circumference (arrow). This latter observation has been made at least seven times, including those shown in Figs 2 and 3 below. hb: haustorial bodies; hf: haustorial fingers. Scale bars: 10 μm.
Results
Membrane dyes suggest the EHM surrounding Bgh haustoria shares features with the ER membrane
In leaf epidermal cells of the barley host, the Bgh fungus forms an elaborate haustorium with a central body and finger-like protrusions, all surrounded by the EHM (Fig. 1A). Previous findings suggests that the host cell ER resides closely to the EHM (Leckie et al., 1995; Koh et al., 2005; Micali et al., 2011; Zhang et al., 2013). By transiently transforming single barley leaf epidermal cells with a luminal ER marker construct, encoding YFP with an N-terminal signal peptide and a C-terminal ER retention signal (SP–YFP–HDEL), we were able to confirm these findings (Fig. 1C–F). YFP-labeled ER tubules and cisternae localized to the fungal entry site and around the haustorial body, but notably less so around the haustorial fingers. Next, we cloned the barley homologue of ER RETENTION DEFICIENT 2 (HvERD2) and genetically fused it to mCherry. ERD2 is the transmembrane receptor for recycling of proteins containing the ER retention signal H/KDEL. ERD2 shuttles between ER and Golgi via COPII- and COPI-coated vesicles, and it generally marks ER exit sites (ERESs) and Golgi (Hanton et al., 2007, 2008). HvERD2 showed a similar pattern in relation to haustoria as the luminal ER marker, by being patchy around the haustorial body (arrowhead) and showing less signal around the haustorial fingers (Fig. 1G, H).
To study the ER–EHM association further, we used three dyes that all label the ER membrane: hexyl rhodamine B (Grabski et al., 1993; Zheng et al., 2005; Kankanala et al., 2007; Fitzgibbon et al., 2010; Zhao et al., 2010; Haslam et al., 2012), ER-Tracker Blue-White DPX (Ashtamker et al., 2007) and DiOC6 (Terasaki and Reese, 1992; Grabski et al., 1993; Zheng et al., 2004; Martens et al., 2006; Fitzgibbon et al., 2010). As expected, these dyes labelled a membrane around the nucleus, in agreement with their binding to the ER membrane (Fig. 1B and Supplementary Fig. S2A), but also intracellular fungal haustorial membranes (Fig. 1B and Supplementary Fig. S2B–D). All three dyes stained a continuous membrane proximal to the haustorium, which surrounded both the haustorial body and fingers. As the three dyes have the ER membrane as common target, our observations gave the first indication that the EHM may share features with this host membrane. Leckie et al. (1995) already showed data suggesting DiOC6 stains the EHM. However, it is challenging to separate the EHM from the tonoplast, as these are very close along most of the haustorium, and yet by focusing on cells in which the nucleus is close to the haustorium, Inada et al (2016) succeeded in discerning the EHM. A closer look at the DiOC6 signal in a single section of the z-stack in Fig. 1B (see Supplementary Fig. S2D) showed that DiOC6 stained the membrane between the haustorium and nucleus (diamond in Supplementary Fig. S2D). Furthermore, neither DiOC6 nor the two other dyes is reported to stain the tonoplast. If DiOC6 would stain the tonoplast, we would have expected to see this by a lumen of cytosol where the nucleus and the haustorium meet in Fig. 1B and Supplementary Fig. S2D. Therefore, the continuous DiOC6 stain around the haustorium is suggested to be at the EHM, and in fact we believe that the diamond in Supplementary Fig. S2D marks both the nuclear membrane and the EHM. However, confocal microscopy cannot reveal whether there in fact are two membranes at this site.
Major plant organelle markers are not found on the EHM
We then investigated in detail how other abundant membranes were positioned in the cell in relation to the haustorium. For this purpose we introduced several well-described cellular markers for the PM, Golgi, and tonoplast. The PM markers, HvMLO–mCherry and HvLTP–mCherry (Kwaaitaal et al., 2010), remained in the PM and did not localize to the EHM (Fig. 1E, F, I, J). However, they were deposited at the haustorial neck, and here HvMLO–mCherry was surrounded by the plant ER (Fig. 1E, F, I, J). The Golgi was visualized using a fusion of the 52-amino-acid signal anchor of a rat sialyltransferase (ST) to YFP (Brandizzi et al., 2002), which accumulated at the fungal entry site (Fig. 1K, L) as previously observed in Arabidopsis (Koh et al., 2005). As for the PM markers, the ST–YFP signal did not extend to the EHM. To visualize the tonoplast, we cloned the barley Ras-related small GTP-binding protein most closely related to AtRabG3b (see Supplementary Figs S1 and S3A), which is located at the tonoplast in Arabidopsis (Rutherford and Moore, 2002; Vernoud et al., 2003; Carter et al., 2004). A fluorescent fusion, GFP–HvRabG3b, of this Rab GTPase also localized to the tonoplast in barley (Supplementary Fig. S3B–D). In invaded cells, GFP–HvRabG3b surrounded the haustorium (Fig. 1M, N). However, since the tonoplast and the EHM are very close (Inada et al., 2016), confocal microscopy has difficulty in revealing directly to which membrane this marker was associated (see more details below). Therefore, we searched for sites where the haustorial fingers approached the cellular circumference. Here, we detected that GFP–HvRabG3b retracted from the EHM and instead followed the vacuole. Only a weak signal, presumed to derive from the soluble fraction of this fluorescent GTPase, remained at the tip of the haustorial finger (Fig. 1M, N). This situation has been observed in at least seven cases, including this one and the two cases below. In conclusion, none of the shown markers for the tonoplast, PM, and Golgi localized to the EHM.
Members of the HvSar1 family of small GTPases localize to the EHM
We have observations that the EHM can be stained by the ER membrane dyes. However, the ER integral membrane protein marker ERD2 did not label the EHM, nor did the ER luminal marker label the extrahaustorial matrix. This suggested that the EHM may have ER-like properties without being directly connected to the host cell ER. Therefore, we turned to monitor proteins that become associated with the ER membrane from the cytosolic side, such as members of the Sar1 family. These small GTPases bind to the ER membrane by means of an amphipathic domain (Lee et al., 2005), and they are involved in anterograde transport from the ER, where they control COPII-coated vesicle formation from ERESs (Memon, 2004). Barley has three Sar1 isoforms (Böhlenius et al., 2010; Supplementary Fig. S1), which we cloned to make C-terminal fusions to either monomeric (m)YFP or mCherry. Co-localization with the luminal ER marker SP–mCherry–HDEL and a Golgi marker, consisting of the cytoplasmic domain and the transmembrane domain of soybean α-1,2-mannosidase I fused to mCherry (Nelson et al., 2007), revealed that HvSar1a–mYFP localized to the cytosol, ER-tubules and bright fluorescent structures associated with the ER, representing ERESs (Fig. 2A–C and Supplementary Fig. S4A–C).
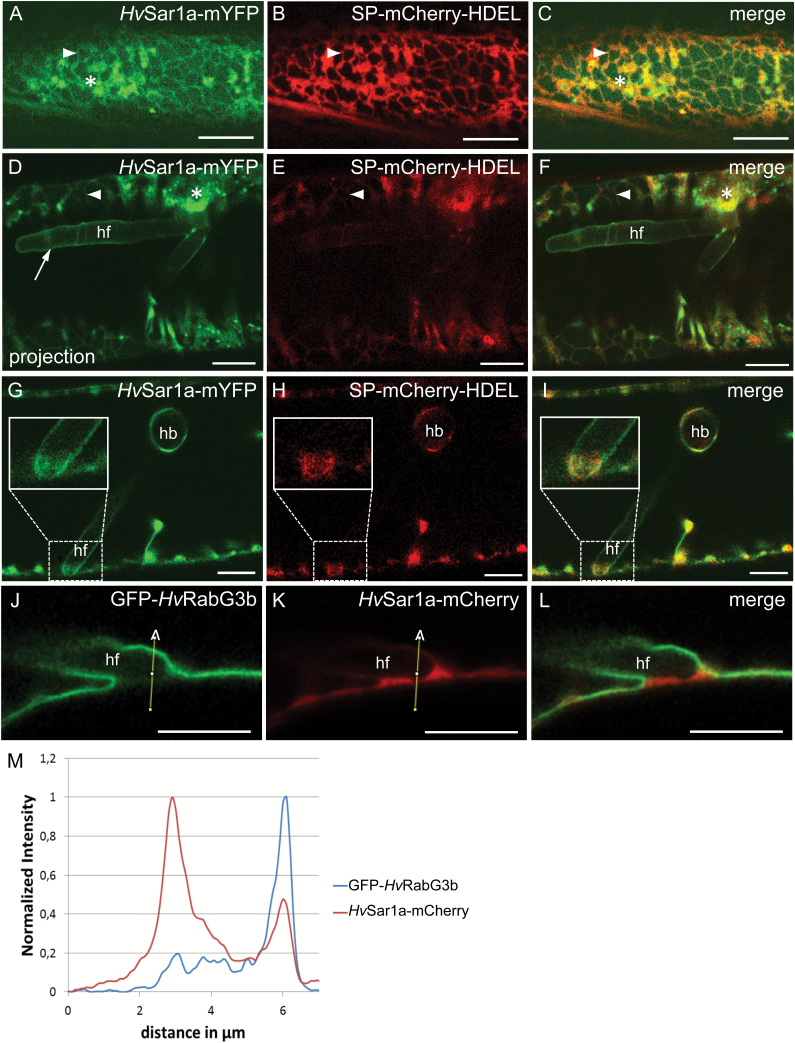
Fig. 2.
The small GTPase HvSar1a localized to the EHM. (A–C) Barley epidermal cell co-expressing HvSar1a–mYFP and the ER luminal marker SP–mCherry–HDEL. (D–L) Barley epidermal cells containing Bgh haustoria at 2 dai. (D–I) Cell at two focal planes, (D–F) and (G–I), co-expressing HvSar1a–mYFP and the ER marker SP–mCherry–HDEL. HvSar1a–mYFP accumulated at ERESs at the fungal entry site, and at EHM (arrow). Note that HvSar1a–mYFP remained associated with the EHM at the tip of haustorial fingers near the host cell circumference (insets in G–I). (J–L) Cell co-expressing GFP–HvRabG3b and HvSar1a–mCherry. (M) Signal quantification of GFP–HvRabG3b and HvSar1a–mCherry along the indicated path in (J, K). hb: haustorial bodies; hf: haustorial fingers; asterisk: ERES; arrowhead: ER tubules. Scale bars: 10 μm.
Next, we localized HvSar1 in barley cells containing a haustorium 2 d after inoculation with Bgh. Large numbers of HvSar1a–mYFP-labeled ERESs accumulated at the fungal entry site (Fig. 2D), suggesting active polarized COPII-mediated secretion at this location. In contrast to the ER luminal and transmembrane markers (Zhang et al., 2013) (Fig. 1C, D, F–H), HvSar1a–mYFP exhibited a continuous signal around fungal haustoria (Fig. 2D–I and Supplementary Fig. S4F–H). The signal intensity of HvSar1a–mYFP on the EHM is comparable to that on the non-ERES parts of the ER and markedly weaker than that of ERESs (Fig. 2D). HvSar1b and HvSar1c exhibited a similar localization to the EHM in Bgh-invaded cells as HvSar1a when fused to mCherry or mYFP (see Supplementary Fig. S4I–N). At sites where the haustorial fingers approached the cellular circumference, HvSar1a–mYFP remained associated with the haustorium (Fig. 2G–I). At this site, where the tonoplast, the PM and the EHM met, a cytosolic space occurred in which ER structures also accumulated (Fig. 2H). Yet, a continuous HvSar1a–mYFP signal, which could be distinguished from an ER signal, was present around the tip of the haustorial finger (Fig. 2G–I).
These observations suggested the ER-associated HvSar1a–mYFP, and not the tonoplast-associated GFP–HvRabG3b (Fig. 1N), labels the EHM. This was further tested by co-localizing GFP–HvRabG3b and HvSar1a–mCherry. Again GFP–HvRabG3b followed the tonoplast where a haustorial finger was near the PM (Fig. 2J, L). A minor signal, quantified in the intensity plot, between the haustorium and PM is considered to represent the cytosolic portion of this GTPase (Fig. 2J, M), similar to what was seen in Fig. 1N. Also here, the HvSar1 signal was observed around the haustorial finger. Yet, whether the HvSar1–mCherry signal in the cytosolic space at the fingertip was soluble, associated with the EHM, with the ER, or all three, could not be distinguished (Fig. 2K, M), unlike in Fig. 2G–I. We could furthermore observe that GFP–HvRabG3b, but not HvSar1a–mCherry, was excluded from sites where the cell nucleus and haustoria are in close contact (see Supplementary Fig. S5A–E), and that HvSar1a–mCherry is closer to the haustorium than GFP–HvRabG3b at specific sites (Supplementary Fig. S5F–I). Collectively, these observations indicate that the tonoplast marker, GFP–HvRabG3b, did not label the EHM, while they suggested that the Sar1 markers did.
We subsequently cloned the barley version of Sec12 (see Supplementary Fig. S1) and genetically fused it to mCherry. Sec12 is the ER membrane-localized guanine nucleotide exchange factor (GEF) activating Sar1 (daSilva et al., 2004). As expected, HvSec12–mCherry co-localized with HvSar1a–mYFP at the ER tubules (Supplementary Fig. S6A–C). Furthermore, the ER marker HvERD2–mCherry partially co-localized with HvSar1a–mYFP at ERESs (Supplementary Fig. S6D–F). These observations confirm an authentic localization of HvSar1a to ER and ERESs. Sar1, Sec12 and ERD2 all act in COPII vesicle formation (Hanton et al., 2007). As opposed to the Sar1 GTPases, it is noteworthy that HvERD2–mCherry did not label the EHM above background level (Fig. 1G, H and Supplementary Fig. S4D–H). The localization of HvSec12–mCherry was less conclusive as we did detect a weak signal following the EHM (Supplementary Fig. S6G–I). Combined, this shows that while the localization of HvSar1a to the EHM suggests that this membrane indeed has ER-like properties, the absence of HvERD2 on the EHM indicates that the ER and the EHM do not form a continuous entity, in agreement with the absence of ER luminal markers. Moreover, lack of HvERD2 at the EHM would suggest that neither COPI- nor COPII-mediated trafficking is directed to or from the EHM.
The small GTPase HvRabD2a localizes to the EHM
The mammalian Rab1 GTPase associates with tethering proteins essential for trafficking of both COPI- and COPII-coated vesicles and, by this, regulates both antero- and retrograde trafficking between the ER and Golgi (García et al., 2011). Likewise, members of the homologous RabD1 and RabD2 clades in Arabidopsis are essential for protein transport from the ER to the Golgi, and localize at the Golgi and the TGN (Batoko et al., 2000; Pinheiro et al., 2009). Rab GTPases in general bind to membranes via lipidation (Barr, 2013). We isolated the closest barley homologue of AtRabD2a (see Supplementary Figs S1 and S3A) and generated N-terminal fusions with GFP and mYFP. When co-expressed with the soluble ER marker SP–mCherry–HDEL, a spot-like distribution was observed of mYFP–HvRabD2a in association with the ER (Fig. 3A–C). Co-localization analysis with a Golgi marker and the ERES/cis-Golgi marker HvERD2–mCherry confirmed that HvRabD2a mainly co-localized with the ERES/Golgi (Supplementary Fig. S7A–F), as previously observed by Pinheiro et al. (2009). The remaining spots labelled by GFP–HvRabD2a that did not co-localize with the Golgi marker likely label the TGN, similarly to its AtRabD2a homologue in Arabidopsis (Pinheiro et al., 2009). GFP–HvRabD2a and HvSar1a–mCherry completely co-localized at the ER and ERESs (Supplementary Fig. S7G–I).
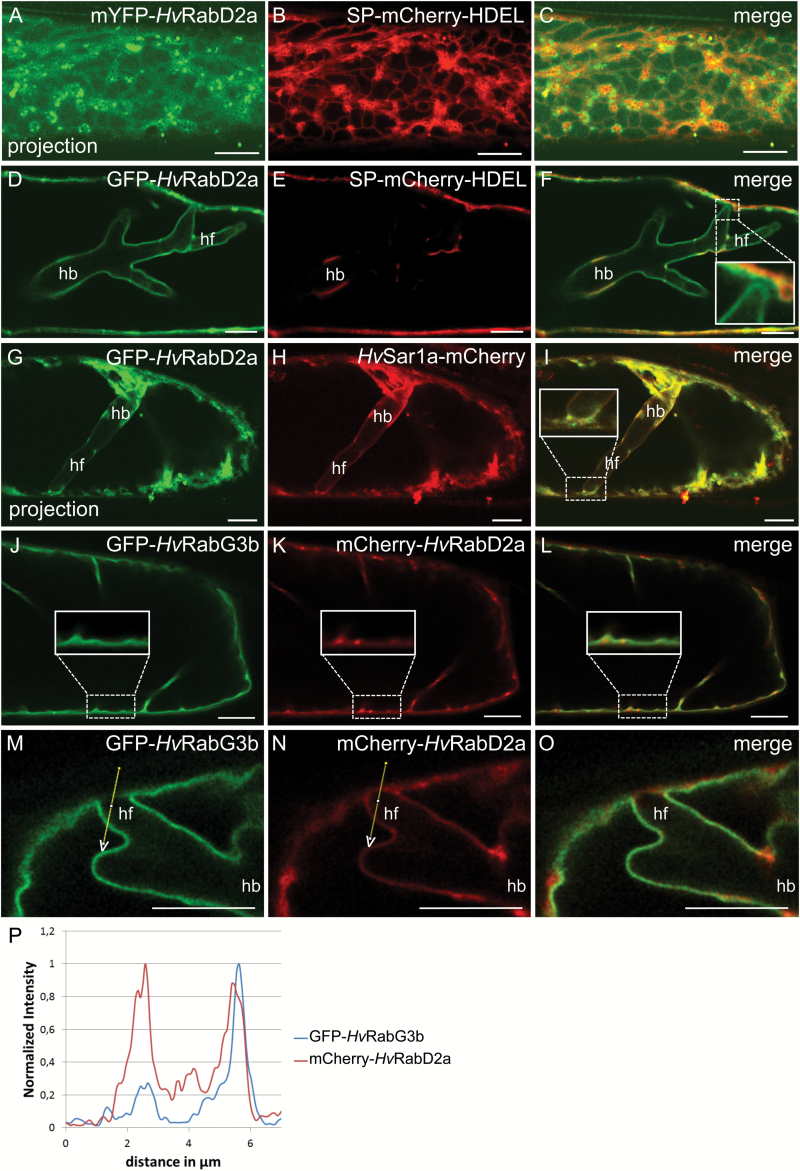
Fig. 3.
The small GTPase HvRabD2a localized to the EHM. Non-invaded (A–C, J–L) and Bgh haustorium-containing (D–I, M–O) barley epidermal cells at 2 dai. (A–F) Barley epidermal cells co-expressing mYFP–HvRabD2a (A, C) or GFP–HvRabD2a (D, F) and the ER luminal marker SP–mCherry–HDEL. Note that GFP–HvRabD2a remains associated with the EHM at the tip of haustorial fingers near the host cell circumference (insert in F). (G–I) Cell co-expressing GFP–HvRabD2a and HvSar1a–mCherry. (J–O) Barley cells co-expressing GFP–HvRabG3b and mCherry–HvRabD2a. Note that mCherry–HvRabD2a, but not GFP–HvRabG3b, appears associated with the EHM at the tip of haustorial fingers near the host cell circumference. (P) Signal quantification of GFP–HvRabG3b and mCherry–HvRabD2a along the indicated path in (M, N). Images in (A–C) and (G–I) are maximum intensity projection of a series of z-sections. hb: haustorial bodies; hf: haustorial fingers. Scale bars: 10 μm.
In cells invaded by Bgh, the GFP–HvRabD2a signal, unlike the ER and Golgi markers, and like the HvSar1 fluorescent protein fusions, localized around the haustorium (Fig. 3D–I). As suggested for HvSar1 (Fig. 2G–I), at sites where haustorial fingers approached the cellular circumference (Fig. 3F, inset), the GFP–HvRabD2a signal remained associated with the haustorium. Here, the GFP–HvRabD2a signal at the haustorial fingertip was clearly separated from other signals nearby, indicating that this GTPase is associated with the EHM. Next, in cells in which GFP–HvRabD2a and HvSar1a–mCherry were co-expressed, these two fluorescently tagged GTPases showed highly overlapping signals around the haustorium and elsewhere in the cell (Fig. 3G–I). Also in this case the signals appeared to follow the EHM around the haustorial fingertip near the cell PM to the extent that this could be distinguished from signals from other structures, potentially the ER (Fig. 3I, inset). As above, when mCherry–HvRabD2a was co-expressed with the tonoplast marker, GFP–HvRabG3b, it was confirmed that these two Rab GTPases associate with separate membranes in control cells (Fig. 3J–L). Here, it was clear that both had a significant cytosolic fraction. In cells harboring a Bgh haustorium, this distinct membrane labelling was confirmed, and like in the case of HvSar1a (Fig. 2J–M), the signal of HvRabD2a between the haustorium and the PM might originate from EHM, ER and cytosolic fractions (Fig. 3M–P).
HvRabD2a, wild-type and GDP-bound HvSar1a have a low diffusion rate at the EHM
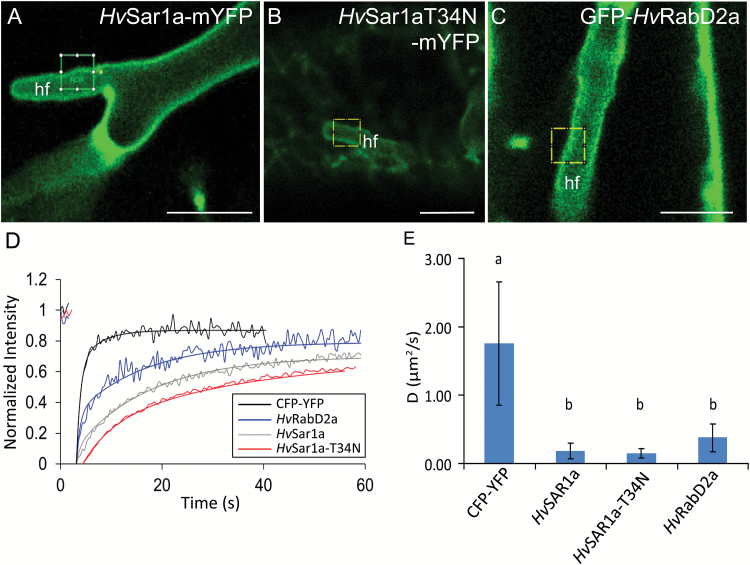
The finding that the signal intensity of HvSar1a at the EHM is comparable to the one at non-ERES ER domains might suggest that HvSar1a binds to the EHM in its GDP-bound state. To verify this observation, we generated an inactive, dominant-negative (DN) GDP-locked mutant, HvSar1aT34N–mYFP, as described by Ward et al. (2001). This DN version of HvSar1a localized as expected to ER tubules, like wild-type HvSar1a, but it did not accumulate at ERESs. It furthermore prevented the Golgi α-1,2-mannosidase marker (Nelson et al., 2007) from trafficking from the ER to the Golgi (see Supplementary Fig. S8A–C), as previously shown by Ward et al. (2001). This was unlike when the Golgi marker was co-expressed with wild-type Sar1a (Supplementary Fig. S4A–C). This documented the dominant-negative functionality of HvSar1aT34N–mYFP. Our prediction of GTP-independent binding of Sar1a to the EHM was confirmed using HvSar1aT34N–mYFP (Supplementary Fig. S8D).
We have clearly shown above that, as expected, fluorescent fusion versions of HvSar1a and HvRabD2a are not associated with the tonoplast. This indicates that the continuous signals around the haustoria from these two GTPases can be from soluble fractions trapped between the EHM and the tonoplast or from a fraction that physically interacts with the EHM, as suggested by the haustorial fingertip observations above. To resolve which of these two fractions provided the signals, we performed FRAP measurements using a soluble cyan fluorescent protein (CFP)–YFP fusion as control. This protein has a similar size as the fluorescent small GTPase fusion proteins. The measured fluorescence recovery rates at the haustorial fingers (Fig. 4A–C) and extrapolated average diffusion coefficients for HvSar1a–mYFP, HvSar1aT34N–mYFP and GFP–HvRabD2a were significantly lower than for the cytosolic CFP–YFP (Fig. 4D, E). These recordings were made at areas of the haustoria where no ER was present (Fig. 4A–C). ER was otherwise clearly discernable as bands of more intense signal (Figs 2D–F and 3D–F, and Supplementary Figs S4D–N and S6G–I). This shows that a large proportion of the three versions of the GTPases were temporarily or constitutively membrane-bound at the EHM, and not just freely diffusing in the cytosol.
Fig. 4.
Slow diffusion of HvSar1a, GDP-locked HvSar1a and HvRabD2a at the EHM. FRAP measurements of HvSar1a–mYFP, HvSar1aT34N–mYFP (GDP-locked), GFP–HvRabD2a, and CFP–YFP expressed in barley epidermal cells containing Bgh haustoria at 2 dai. (A–C) Example micrographs for HvSar1a–mYFP, HvSar1aT34N–mYFP, and GFP–HvRabD2a, respectively, with the bleached areas marked. Scale bars: 10 μm. (D) Examples of average recovery curves. To allow comparison of the recovery curves, the fluorescence intensity before the bleach was normalized to 1 and at the bleached point to 0. (E) Average diffusion coefficients (D) of CFP–YFP and the fluorescent fusion constructs of HvSar1a–mYFP, HvSar1aT34N–mYFP, and GFP–HvRabD2a. D values were calculated from 11 or more recovery curves originating from three or more biological replicates. hf: haustorial fingers. Error bars show SD. Significance of differences was evaluated using a one-way ANOVA with Tukey’s HSD post hoc analysis. The groups a and b differ significantly from one another (P<0.01).
HvSar1- and HvRabD2a-mediated transport appears not essential for EHM formation
Our data indicate that the association of HvSar1a to the EHM is not dependent on its activation state, suggesting that EHM binding may not have any functional implications, but may be indicative of the physicochemical properties of the membrane. Yet, we followed up on these findings by investigating the potential role of HvSar1a in mediating membrane transport important for EHM formation by using the GDP-locked version of this GTPase. Expressing this DN GTPase in barley cells did not appear to have any effect on the establishment of Bgh haustoria (see Supplementary Fig. S8E), in turn suggesting that EHM formation is not affected and does not require COPII vesicle formation. This observation further suggests that the EHM is generated despite the collapse of the ER and Golgi, as seen when overexpressing HvSar1aT34N–mYFP (Supplementary Fig. S8A–C). This indirectly suggests that Golgi-formed COPI retrograde vesicles are also not required for EHM formation. To confirm this hypothesis, we made use of the EHM-localized HvRabD2a and analysed whether this GTPase is required for EHM formation. Pinheiro et al. (2009) previously demonstrated that the nucleotide-free version of Arabidopsis RabD2a (RabD2aN121I) caused an otherwise secreted protein to be retained in the ER. Expression of HvRabD2aN121I in barley epidermal cells seems not to have an effect on Bgh haustorial establishment and thus cannot be linked to EHM formation (Supplementary Fig. S8F).
Discussion
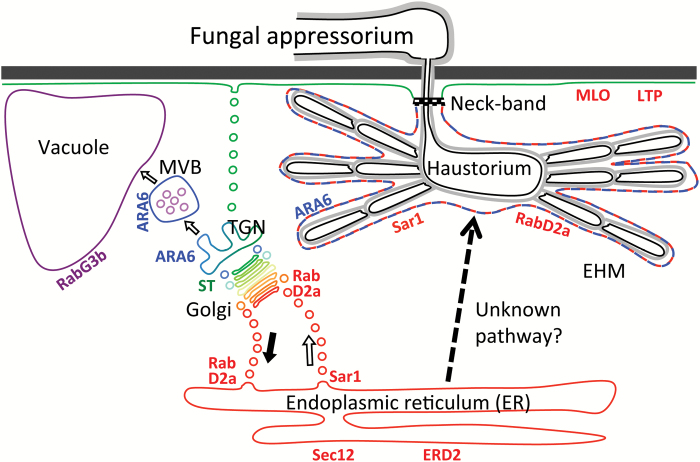
Summarized, our data suggested that the EHM stains with ER membrane-binding dyes, which led us to find that it binds the ER-associated proteins HvSar1 and HvRabD2a. In contrast, ER and Golgi integral membrane and luminal markers failed to accumulate at the EHM or in the extrahaustorial matrix, which suggested that the EHM is directly connected neither to the ER nor to the Golgi. Inada et al. (2016) recently demonstrated that the MVB Rab5 GTPase ARA6 binds to the powdery mildew EHM in Arabidopsis and barley. Our results confirm that barley ARA6 is localized around the Bgh haustorium (see Supplementary Fig. S9A–C). These observations, together with our suggestion of the EHM having ER properties, points to the EHM being a unique and complex biological membrane, not previously described. A model summarizing our marker localizations is provided in Fig. 5.
Fig. 5.
Model localizing protein markers used in this work and suggested pathway for membrane trafficking to the EHM.
COPII-coated vesicle formation at the ER is activated by the Sar1 GEF Sec12, which removes GDP and allows binding of GTP to Sar1 (Futai et al., 2004). GTP binding results in a conformational change of the protein, which exposes an N-terminal amphipathic helix that binds to the ER membrane (Lee et al., 2005). The GDP-locked form of Sar1a still binds ER-enriched microsomes, although with lower affinity than the GTP-locked form. However, it does not sequester the COPII components Sec23/24p and Sec13/31p (Matsuoka et al., 1998). Our images and diffusion measurements suggest that both the wild-type and GDP-locked form of HvSar1a–mYFP are associated with the EHM. The signal intensity of HvSar1a–mYFP at the EHM was similar to its intensity at ER tubules and much lower than at ERESs (Fig. 2A–F). These observations combined suggest that Sar1a binds to the EHM with the low, but significant, affinity of the GDP-bound stage. Therefore, its binding is likely due to ER-like properties of the EHM and not related to HvSar1a activity.
The membrane properties that allow staining with DiOC6, hexyl rhodamine B and ER-Tracker Blue-White in plants are poorly described. This is also the case for Rab GTPases (Barr, 2013). Active Sar1a preferentially binds membranes consisting of neutral unsaturated lipids (Matsuoka et al., 1998) and Sar1 GTPase activity is directed to liquid-disordered lipid phases (Long et al., 2010). However, what determines targeting of the GDP-locked HvSar1a to ER membranes is unknown. Therefore, no suggestions as to the biochemistry of the EHM can be made based on our observations. Yet, our results suggest it is most likely that the ER-like properties of the EHM somehow originate from ER-derived vesicles. Interestingly, the EHM around haustoria of the oomycete P. infestans is labelled by a close relative of HvRabG3b (Bozkurt et al., 2015), which we found to be excluded from EHM around powdery mildew haustoria. Even though we cannot rule out that a Rab closely related to HvRabG3b would be able to label the EHM, our data indicate that distinct cellular mechanisms are involved in EHM formation associated with these different pathogens.
One hypothesis to explain the ER-like properties of the EHM is that they are derived from COPI or COPII vesicles. However, our results do not support this hypothesis. The binding of Sar1 to the EHM is independent of GTP, and blocking COPII-coated vesicle budding at the ER by expression of GDP-locked Sar1 did not affect the establishment of haustoria (see Supplementary Fig. S8E). Furthermore, expression of GDP-locked Sar1 most likely indirectly interferes with retrograde COPI vesicle budding from the cis-Golgi, since interfering with COPII-coated vesicle formation leads to extensive Golgi stack fragmentation and a gradual loss of Golgi compartments (Osterrieder et al., 2010), as reflected by the retention of the Golgi marker in the ER (Supplementary Fig. S8A–C). Failure of nucleotide-free RabD2aN121I to affect the establishment of haustoria (Supplementary Fig. S8F) appears to support the hypothesis that COPI and COPII vesicles do not provide membrane material for the EHM, since expression of RabD2a with the same mutation in Arabidopsis interfered with ER–Golgi trafficking (Pinheiro et al., 2009). In addition, interference with the ER–Golgi traffic is likely to affect the entire conventional secretory pathway (Takeuchi et al., 2000; Hanton et al., 2008), indicating that membrane needed for the formation of the EHM is not provided by later steps in this pathway. Instead, we suggest that the ER-like properties of the EHM may originate from an unconventional secretory pathway originating from the ER as in the formation of peroxisomes (Hoepfner et al., 2005; Kaur et al., 2009), autophagosomes (Hamasaki et al., 2013; Lamb et al., 2013) or compartments for unconventional protein secretion (Malhotra, 2013). Alternatively, membrane material in the EHM may be derived from direct ER–tonoplast traffic, as suggested by Viotti et al. (2013) (see Fig. 5). Such a route might explain why the EHM has both ER properties and late endosome properties, as indicated by the ARA6 labeling (Inada et al., 2016; Supplementary Fig. S9).
Nonetheless, host cell membrane trafficking through the Golgi is important for penetration resistance against Bgh. This was demonstrated in barley by Ostertag et al. (2013). When they used RNA interference to silence components of the conserved oligomeric Golgi complex COG1 and COG3, the COPIγ coat protein, as well as HvRabD2a, or overexpressed the VTI1-like SNARE protein, the penetration success of Bgh was increased. Moreover, they showed that HvRabD2a co-localizes with a Golgi marker and like HvSar1a–mYFP (Fig. 2D) accumulates below the powdery mildew attack site (Ostertag et al., 2013).
The presence of a novel host membrane with ER-like and MVB-like properties in plant cells harboring fungal haustoria suggests that the pathogen hijacks the host membrane system for its own benefit. Similarly, the human pathogen Legionella pneumophilla uses effectors secreted into the host cytosol to redirect ER-derived vesicles to the Legionella-containing vacuole (LCV), in which the bacterium replicates. One of the secreted effectors, DrrA, acts as a Rab1 GEF, thereby controlling accumulation of Rab1 on the LCV membrane (Murata et al., 2006). We speculate that Bgh introduces effectors into the host cell to control vesicle traffic in a similar way and thereby facilitates the formation of this specialized EHM. The Bgh genome encodes hundreds of candidates for effector proteins (Spanu et al., 2010; Pedersen et al., 2012) and future efforts will include attempts to identify effectors controlling EHM generation. Understanding the vesicle trafficking pathway leading to EHM formation and how it may be regulated by effectors will be essential in future plant genome editing approaches aimed at preventing EHM formation and thereby preventing powdery mildew infection.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Alignments of barley proteins used in this study to their closest relatives in Arabidopsis.
Fig. S2. ER membrane dyes and the extrahaustorial membrane.
Fig. S3. Barley Rab GTPases.
Fig. S4. Three Sar1 GTPases, but not the HDEL receptor ERD2, labeled the extrahaustorial membrane.
Fig. S5. The tonoplast marker HvRabG3b did not label the EHM.
Fig. S6. Sar1a, ERD2, the Sar1 GEF Sec12, and the EHM.
Fig. S7. RabD2a co-localized with Golgi marker, ERD2 and Sar1a.
Fig. S8. Arresting ER–Golgi traffic did not affect haustorial formation.
Fig. S9. ARA6 labels EHM.
Table S1. Primer sequences.
Acknowledgements
We thank the Centre for Advanced Bioimaging (CAB) at the University of Copenhagen for use of their facilities. Furthermore, we thank Prof. Paul Schulze-Lefert and Prof. Ralph Panstruga for sharing constructs. We are very grateful as well to Prof. Panstruga as well for use of the Leica SP8 confocal microscope and lab facilities for part of the localization studies. We thank Prof. Chris Hawes and Dr Yumiko Sakuragi for making the ST–YFP construct available to us. We thank Dr Noriko Inada and Prof. Takashi Ueda for providing the ARA6–GFP construct. In addition, we thank Dr Kota Miura for making the FRAPcalc plugin available. We thank Prof. Andreas Nebenführ for the SP–mCherry–HDEL and Golgi–mCherry constructs. We thank Prof. Ralf Reski for generating the CFP–YFP marker. We thank The Danish Strategic Research Council, The Danish Council for Independent Research/Technology and Production Sciences, and the Villum Foundation, Denmark (VKR023502) for funding.
Glossary
Abbreviations:
- BFA
brefeldin A
- Bgh
Blumeria graminis f.sp. hordei
- EHM
extrahaustorial membrane
- ER
endoplasmic reticulum
- FRAP
fluorescence redistribution after photobleaching
- MVB
multivesicular body
- PM
plasma membrane
- TGN
trans-Golgi network.
References
- Ashtamker C, Kiss V, Sagi M, Davydov O, Fluhr R. 2007. Diverse subcellular locations of cryptogein-induced reactive oxygen species production in tobacco Bright Yellow-2 cells. Plant Physiology 143, 1817–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA. 2013. Rab GTPases and membrane identity: Causal or inconsequential?The Journal of Cell Biology 202, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H, Zheng HQ, Hawes C, Moore I. 2000. A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. The Plant Cell 12, 2201–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkey R, Zhang Y, Ma X, King H, Zhang Q, Wang W, Xiao S. 2017. Homologues of the RPW8 resistance protein are localized to the extrahaustorial membrane that is likely synthesized de novo. Plant Physiology 173, 600–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G, Unthan T, Uhrig JF, Pöschl Y, Gust AA, Scheel D, Lee J. 2009. Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proceedings of the National Academy of Sciences, USA 106, 8067–8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius H, Mørch SM, Godfrey D, Nielsen ME, Thordal-Christensen H. 2010. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. The Plant Cell 22, 3831–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt TO, Belhaj K, Dagdas YF, Chaparro-Garcia A, Wu CH, Cano LM, Kamoun S. 2015. Rerouting of plant late endocytic trafficking towards a pathogen interface. Traffic 16, 204–226. [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Snapp EL, Roberts AG, Lippincott-Schwartz J, Hawes C. 2002. Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: evidence from selective photobleaching. The Plant Cell 14, 1293–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV. 2004. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. The Plant Cell 16, 3285–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva LL, Snapp EL, Lippincott-Schwartz J, Hawes C, Brandizzi F, Luis LP. 2004. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. The Plant Cell 16, 1753–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djamei A, Kahmann R. 2012. Ustilago maydis: dissecting the molecular interface between pathogen and plant. PLoS Pathogens 8, e1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchkov D, Nowara D, Zierold U, Schweizer P. 2005. A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Molecular Plant-Microbe Interactions 18, 755–761. [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J, Bell K, King E, Oparka K. 2010. Super-resolution imaging of plasmodesmata using three-dimensional structured illumination microscopy. Plant Physiology 153, 1453–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster B, Van De Ville D, Berent J, Sage D, Unser M. 2004. Complex wavelets for extended depth-of-field: a new method for the fusion of multichannel microscopy images. Microscopy Research and Technique 65, 33–42. [DOI] [PubMed] [Google Scholar]
- Futai E, Hamamoto S, Orci L, Schekman R. 2004. GTP/GDP exchange by Sec12p enables COPII vesicle bud formation on synthetic liposomes. The EMBO Journal 23, 4146–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García IA, Martinez HE, Alvarez C. 2011. Rab1b regulates COPI and COPII dynamics in mammalian cells. Cellular Logistics 1, 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabski S, De Feijter AW, Schindler M. 1993. Endoplasmic reticulum forms a dynamic continuum for lipid diffusion between contiguous soybean root cells. The Plant Cell 5, 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamasaki M, Furuta N, Matsuda A et al. . 2013. Autophagosomes form at ER–mitochondria contact sites. Nature 495, 389–393. [DOI] [PubMed] [Google Scholar]
- Hanton SL, Chatre L, Matheson LA, Rossi M, Held MA, Brandizzi F. 2008. Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Molecular Biology 67, 283–294. [DOI] [PubMed] [Google Scholar]
- Hanton SL, Chatre L, Renna L, Matheson LA, Brandizzi F. 2007. De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. Plant Physiology 143, 1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harder DE, Rohringer R, Samborski DJ, Kim WK, Chong J. 1978. Electron-microscopy of susceptible and resistant near-isogenic (sr6-Sr6) lines of wheat infected by Puccinia graminis tritici. I. The host-pathogen interface in the compatible (sr6-P6) interaction. The Canadian Journal of Botany 56, 2955–2966. [Google Scholar]
- Haslam TM, Mañas-Fernández A, Zhao L, Kunst L. 2012. Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiology 160, 1164–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. 2005. Contribution of the endoplasmic reticulum to peroxisome formation. Cell 122, 85–95. [DOI] [PubMed] [Google Scholar]
- Inada N, Betsuyaku S, Shimada TL et al. . 2016. Modulation of plant RAB GTPase-mediated membrane trafficking pathway at the interface between plants and obligate biotrophic pathogens. Plant & Cell Physiology 57, 1854–1864. [DOI] [PubMed] [Google Scholar]
- Irons SL, Evans DE, Brandizzi F. 2003. The first 238 amino acids of the human lamin B receptor are targeted to the nuclear envelope in plants. Journal of Experimental Botany 54, 943–950. [DOI] [PubMed] [Google Scholar]
- Kankanala P, Czymmek K, Valent B. 2007. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. The Plant Cell 19, 706–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T, Ueda T. 2017. Exocytic trafficking pathways in plants: why and how they are redirected. New Phytologist 215, 952–957. [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kaur N, Reumann S, Hu J. 2009. Peroxisome biogenesis and function. The Arabidopsis Book 7, e0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, O’Connell R, Maekawa-Yoshikawa M, Uemura T, Neumann U, Schulze-Lefert P. 2014. The powdery mildew resistance protein RPW8.2 is carried on VAMP721/722 vesicles to the extrahaustorial membrane of haustorial complexes. The Plant Journal 79, 835–847. [DOI] [PubMed] [Google Scholar]
- Koh S, André A, Edwards H, Ehrhardt D, Somerville S. 2005. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. The Plant Journal 44, 516–529. [DOI] [PubMed] [Google Scholar]
- Kuhn H, Kwaaitaal M, Kusch S, Acevedo-Garcia J, Wu H, Panstruga R. 2016. Biotrophy at its best: Novel findings and unsolved mysteries of the Arabidopsis-powdery mildew pathosystem. The Arabidopsis Book 14, e0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwaaitaal M, Keinath NF, Pajonk S, Biskup C, Panstruga R. 2010. Combined bimolecular fluorescence complementation and Forster resonance energy transfer reveals ternary SNARE complex formation in living plant cells. Plant Physiology 152, 1135–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S et al. . 2008. Co-option of a default secretory pathway for plant immune responses. Nature 451, 835–840. [DOI] [PubMed] [Google Scholar]
- Lamb CA, Yoshimori T, Tooze SA. 2013. The autophagosome: origins unknown, biogenesis complex. Nature Reviews. Molecular Cell Biology 14, 759–774. [DOI] [PubMed] [Google Scholar]
- Larous L, Kameli A, Lösel DM. 2008. Ultrastructural observations on Puccinia menthae infections. The Journal of Plant Pathology 90, 185–190. [Google Scholar]
- Leckie CP, Callow JA, Green JR. 1995. Reorganization of the endoplasmic reticulum in pea leaf epidermal cells infected by the powdery mildew fungus Erysiphe pisi. New Phytologist 131, 211–221. [Google Scholar]
- Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. 2005. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 122, 605–617. [DOI] [PubMed] [Google Scholar]
- Long KR, Yamamoto Y, Baker AL, Watkins SC, Coyne CB, Conway JF, Aridor M. 2010. Sar1 assembly regulates membrane constriction and ER export. The Journal of Cell Biology 190, 115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YJ, Schornack S, Spallek T, Geldner N, Chory J, Schellmann S, Schumacher K, Kamoun S, Robatzek S. 2012. Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cellular Microbiology 14, 682–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra V. 2013. Unconventional protein secretion: an evolving mechanism. The EMBO Journal 32, 1660–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manners JM, Gay JL. 1977. The morphology of haustorial complexes isolated from apple, barley, beet and vine infected with powdery mildews. Physiological Plant Pathology 11, 261–266. [Google Scholar]
- Martens HJ, Roberts AG, Oparka KJ, Schulz A. 2006. Quantification of plasmodesmatal endoplasmic reticulum coupling between sieve elements and companion cells using fluorescence redistribution after photobleaching. Plant Physiology 142, 471–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. 1998. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell 93, 263–275. [DOI] [PubMed] [Google Scholar]
- Memon AR. 2004. The role of ADP-ribosylation factor and SAR1 in vesicular trafficking in plants. Biochimica et Biophysica Acta 1664, 9–30. [DOI] [PubMed] [Google Scholar]
- Micali CO, Neumann U, Grunewald D, Panstruga R, O’Connell R. 2011. Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cellular Microbiology 13, 210–226. [DOI] [PubMed] [Google Scholar]
- Murata T, Delprato A, Ingmundson A, Toomre DK, Lambright DG, Roy CR. 2006. The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nature Cell Biology 8, 971–977. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenführ A. 2007. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal 51, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Nielsen ME, Feechan A, Böhlenius H, Ueda T, Thordal-Christensen H. 2012. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proceedings of the National Academy of Sciences, USA 109, 11443–11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen ME, Thordal-Christensen H. 2013. Transcytosis shuts the door for an unwanted guest. Trends in Plant Science 18, 611–616. [DOI] [PubMed] [Google Scholar]
- O’Connell RJ, Panstruga R. 2006. Tête à tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytologist 171, 699–718. [DOI] [PubMed] [Google Scholar]
- Osterrieder A, Hummel E, Carvalho CM, Hawes C. 2010. Golgi membrane dynamics after induction of a dominant-negative mutant Sar1 GTPase in tobacco. Journal of Experimental Botany 61, 405–422. [DOI] [PubMed] [Google Scholar]
- Ostertag M, Stammler J, Douchkov D, Eichmann R, Hückelhoven R. 2013. The conserved oligomeric Golgi complex is involved in penetration resistance of barley to the barley powdery mildew fungus. Molecular Plant Pathology 14, 230–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen C, Ver Loren van Themaat E, McGuffin LJ et al. . 2012. Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phair RD, Gorski SA, Misteli T. 2004. Measurement of dynamic protein binding to chromatin in vivo, using photobleaching microscopy. Methods in Enzymology 375, 393–414. [DOI] [PubMed] [Google Scholar]
- Pinheiro H, Samalova M, Geldner N, Chory J, Martinez A, Moore I. 2009. Genetic evidence that the higher plant Rab-D1 and Rab-D2 GTPases exhibit distinct but overlapping interactions in the early secretory pathway. Journal of Cell Science 122, 3749–3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Kientz M, Brumm S et al. . 2014. Delivery of endocytosed proteins to the cell-division plane requires change of pathway from recycling to secretion. eLife 3, e02131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S, Moore I. 2002. The Arabidopsis Rab GTPase family: another enigma variation. Current Opinion in Plant Biolology 5, 518–528. [DOI] [PubMed] [Google Scholar]
- Singh MK, Krüger F, Beckmann H et al. . 2014. Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Current Biology 24, 1383–1389. [DOI] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J et al. . 2010. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330, 1543–1546. [DOI] [PubMed] [Google Scholar]
- Szabo LJ, Bushnell WR. 2001. Hidden robbers the role of fungal haustoria in parasitism of plants. Proceedings of the National Academy of Sciences, USA 98, 7654–7655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi M, Ueda T, Sato K, Abe H, Nagata T, Nakano A. 2000. A dominant negative mutant of Sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. The Plant Journal 23, 517–525. [DOI] [PubMed] [Google Scholar]
- Terasaki M, Reese TS. 1992. Characterization of endoplasmic reticulum by co-localization of BiP and dicarbocyanine dyes. Journal of Cell Science 101, 315–322. [DOI] [PubMed] [Google Scholar]
- Vernoud V, Horton AC, Yang Z, Nielsen E. 2003. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiology 131, 1191–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C, Bubeck J, Stierhof YD et al. . 2010. Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. The Plant Cell 22, 1344–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti C, Krüger F, Krebs M et al. . 2013. The endoplasmic reticulum is the main membrane source for biogenesis of the lytic vacuole in Arabidopsis. The Plant Cell 25, 3434–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wen Y, Berkey R, Xiao S. 2009. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal haustorium renders broad-spectrum resistance to powdery mildew. The Plant Cell 21, 2898–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. 2001. Maintenance of Golgi structure and function depends on the integrity of ER export. The Journal of Cell Biology 155, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WJ, Hanisch S, Kwaaitaal M, Pedersen C, Thordal-Christensen H. 2013. A component of the Sec61 ER protein transporting pore is required for plant susceptibility to powdery mildew. Frontiers in Plant Science 4, 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Katavic V, Li F, Haughn GW, Kunst L. 2010. Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1 (LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis. The Plant Journal 64, 1048–1058. [DOI] [PubMed] [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I. 2004. A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. The Plant Journal 37, 398–414. [DOI] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L. 2005. Disruptions of the Arabidopsis Enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. The Plant Cell 17, 1467–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.