DOFT and its interacting partner DOFTIP1 promote flowering in the orchid Dendrobium Chao Praya Smile, and changes in the expression of DOFT affect pseudobulb formation and flower development.
Keywords: Arabidopsis thaliana, DOFT, DOFTIP1, flowering time, orchid, reproductive development
Abstract
FLOWERING LOCUS T (FT) in Arabidopsis encodes the florigen that moves from leaves to the shoot apical meristem to induce flowering, and this is partly mediated by FT-INTERACTING PROTEIN 1 (FTIP1). Although FT orthologs have been identified in some flowering plants, their endogenous roles in Orchidaceae, which is one of the largest families of flowering plants, are still largely unknown. In this study, we show that DOFT and DOFTIP1, the orchid orthologs of FT and FTIP1, respectively, play important roles in promoting flowering in the orchid Dendrobium Chao Praya Smile. Expression of DOFT and DOFTIP1 increases in whole plantlets during the transition from vegetative to reproductive development. Both transcripts are present in significant levels in reproductive organs, including inflorescence apices, stems, floral buds, and open flowers. Through successful generation of transgenic orchids, we have revealed that overexpression or down-regulation of DOFT accelerates or delays flowering, respectively, while alteration of DOFT expression also greatly affects pseudobulb formation and flower development. In common with their counterparts in Arabidopsis and rice, DOFTIP1 interacts with DOFT and affects flowering time in orchids. Our results suggest that while DOFT and DOFTIP1 play evolutionarily conserved roles in promoting flowering, DOFT may have evolved with hitherto unknown functions pertaining to the regulation of storage organs and flower development in the Orchidaceae family.
Introduction
The floral transition from vegetative to reproductive development is the most dramatic phase change in the life cycle of a flowering plant, and is tightly controlled by various flowering pathways in response to developmental and environmental signals. In the model plant Arabidopsis thaliana, convergence of the flowering signals from various pathways mediates the transcriptional regulation of several floral pathway integrators, including FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), to precisely regulate the timing of the floral transition (Blázquez and Weigel, 2000; Lee et al., 2000; Samach et al., 2000; Li et al., 2008). FT encodes a phosphatidylethanolamine binding protein and acts as a key flowering regulator that relays the photoperiod signal to activate floral meristem identity genes in Arabidopsis (Kardailsky et al., 1999; Corbesier et al., 2007). Overexpression of FT results in extremely early flowering, while ft mutants exhibit a late-flowering phenotype under long-day conditions (LDs) (Koornneef et al., 1998). A major transcriptional regulator, CONSTANS (CO), in the photoperiod pathway activates FT mRNA expression in the vascular tissues of leaves under LDs (Samach et al., 2000; An et al., 2004; Wigge et al., 2005).
Several studies have suggested that the FT protein acts as a mobile florigen signal moving from the leaves to the shoot apical meristem (SAM) to induce flowering (Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007). In Arabidopsis, a multiple C2 domain and transmembrane-region protein (MCTP), FT-INTERACTING PROTEIN 1 (FTIP1), and a heavy-metal-associated (HMA) domain-containing protein, SODIUM POTASSIUM ROOT DEFECTIVE 1 (NaKR1), directly interact with FT and sequentially participate in the mediation of long-distance movement of FT from source leaves to the sink SAM in response to LDs (Liu et al., 2012; Zhu et al., 2016). FTIP1 is associated with FT in companion cells of the phloem and is specifically required for FT transport from companion cells to sieve elements through plasmodesmata (Liu et al., 2012). NaKR1 is activated by CO under LDs, and regulates long-distance movement of FT from sieve elements in leaves to those below the SAM (Zhu et al., 2016). In the SAM, it has been suggested that FT interacts with FD, a bZIP transcription factor, to directly activate the expression of SOC1 and a floral meristem identity gene, APETALA1 (AP1), thus initiating flower development (Abe et al., 2005; Wigge et al., 2005). Despite the progress made in understanding the flowering mechanisms involving FT and its interacting partners in Arabidopsis, the biological functions of their orthologs in orchids largely remain elusive.
Orchids belong to the family Orchidaceae, which is one of the largest families of angiosperms. As a group of important ornamental plants with great diversity and specialized floral morphology, this family is recognized as an economically important commodity in the international floriculture industry. They contribute to a large share of global floriculture trade as cut flowers and potted plants, partly because of their attractive flower morphology and long shelf life (Da Silva, 2013). In addition to their high economic value, orchids provide unique genetic material for the study of the mechanisms of plant reproductive development, such as flower development, pigment formation, and flower senescence, owing to their distinctive and colorful flower morphology as well as their specialized reproductive strategies (Da Silva et al., 2014; Gutiérrez, 2010; Yu and Goh, 2001). A major obstacle for breeding and for the use of orchids in economic and academic applications is the prolonged vegetative phase, which makes it time-consuming to select desirable reproductive traits through traditional breeding methods (Da Silva et al., 2014; Yu and Goh, 2001). Thus, it is important to understand the molecular mechanisms involved in the floral transition in orchids, so that the knowledge gained can be applied to classical breeding or to targeted manipulation of orchid varieties with desirable flowering traits.
In order to facilitate molecular studies of orchid development, we have previously developed a reproducible gene transformation coupled with an in vitro tissue culture system for the orchid Dendrobium Chao Praya Smile using L-methionine sulfoximine (MSO), as an agent for the selection of transgenic plants with the bialaphos resistance (bar) gene as a selectable marker (Yu and Goh, 2000a; Yu et al., 2001; Chai et al., 2007; Ding et al., 2013). In this study, we isolated the orchid orthologs of FT and FTIP1, namely DOFT and DOFTIP1, respectively, from Dendrobium Chao Praya Smile, and characterized their biological functions in orchids utilizing the established gene transformation system. Expression analysis demonstrated that transcripts of both DOFT and DOFTIP1 increased in orchid plantlets during the transition from vegetative to reproductive development. Expression of DOFT and DOFTIP1 rescued the late-flowering phenotype of their corresponding Arabidopsis mutants, ft-10 and ftip1-1, respectively. Through the creation of transgenic orchids, we further determined that alteration of DOFT expression in Dendrobium Chao Praya Smile not only significantly affected flowering time, but also influenced pseudobulb formation and flower development. In contrast, while DOFTIP1 interacted with DOFT, DOFTIP1 only affected flowering time in orchids. Therefore, our results suggest that although both DOFT and DOFTIP1 are involved in promoting flowering, DOFT could have evolved to exert novel functions in regulating the development of pseudobulbs and floral organs in orchids.
Materials and methods
Plant material and growth conditions
Dendrobium Chao Praya Smile, a hybrid of Dendrobium Pinky and Dendrobium Kiyomi Beauty, were grown under long days (16 h light/8 h dark). Under our in vitro orchid culture system for Dendrobium Chao Praya Smile, calli that developed from seeds served as the starting material and were cultured at 24 °C under a 16-h photoperiod of 35 µmol m–2 s–1 from daylight fluorescent lamps as previously described (Ding et al., 2013). Arabidopsis thaliana ecotype Columbia-0 (Col-0) plants were grown under long days at 23 ± 2 °C. The mutants used, ft-10 and ftip1-1, are in the Col-0 background.
Plant transformation
Agrobacterium tumefaciens-mediated transformation of Arabidopsis plants in the Col-0 background was carried out by a floral dipping method (Clough and Bent, 1998). All transgenic Arabidopsis plants generated were selected by Basta on soil. Genetic transformation of Dendrobium Chao Praya Smile was performed using particle bombardment or A. tumefaciens-mediated transformation coupled with the modified MSO selection system as previously reported (Chai et al., 2007; Ding et al., 2013).
Cloning of DOFT and DOFTIP1 from Dendrobium Chao Praya Smile
Total RNA was isolated from leaves of Dendrobium Chao Praya Smile using the RNeasy® Plant Mini Kit (Qiagen). Specific FT-like and FTIP1-like cDNA fragments were amplified with corresponding pairs of degenerate primers. The resulting cDNA fragments were cloned into the pGEM-T Easy vector (Promega) and sequenced. To further obtain the full-length sequences of the cDNAs, 3′-RACE and 5′-RACE were performed with gene-specific primers using the SMARTTM RACE cDNA Amplification Kit (BD Biosciences Clontech). The primers used for gene cloning are listed in Supplementary Table S1 at JXB online.
Sequence analysis
Alignment of deduced amino acid sequences was carried out using the software MEGA 6.0 and BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html). The protein sequences of FT and FTIP1 orthologs aligned in this study were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/). The phylogenetic tree was constructed with the Neighbor-joining algorithm using MEGA 6.0.
Plasmid construction
To construct 35S:DOFT and 35S:DOFTIP1, the cDNAs of DOFT and DOFTIP1 were amplified and ligated into pGreen 0229-35S containing 2 × 35S promoters. To create DOFT and DOFTIP1 RNAi constructs, the amplified antisense- and sense-specific fragments for DOFT and DOFTIP1 were ligated to the pGreen 0229 vector flanking a GUS fragment. To construct AmiR-doft and AmiR-doftip1, AmiRs were designed using the software on the WMD3 website (http://wmd3.weigelworld.org). Based on the DOFT and DOFTIP1 sequences, sets of four primers for each gene were generated and used for the PCR amplification according to the published protocol (Schwab et al., 2006). The resulting PCR fragments were cloned into the pGreen 0229-35S vector (Yu et al., 2004).
Southern blot analysis
Total DNA was isolated from leaves of Dendrobium orchids using the CTAB method (Porebski et al., 1997). A 20-μg aliquot of genomic DNA was digested with different restriction enzymes, resolved on a 0.8% (w/v) agarose gel, and blotted onto a nylon membrane. The blot was hybridized overnight with the specific digoxigenin-labeled DNA, then washed and detected as previously described (Yu and Goh, 2000b).
Expression analysis
Total RNA from either orchids or Arabidopsis was extracted using the FavorPrep Plant Total RNA mini-Kit (Favorgen) and reverse-transcribed using the M-MLV Reverse Transcriptase (Promega) according to the manufacturer’s instructions. Quantitative real-time PCR was performed on three biological replicates using the CFX384 Real-Time PCR Detection System (Bio-Rad) with the Maxima SYBR Green/ROX qPCR Master Mix (Fermentas). The orchid polyubiqutin gene DOUbi and the Arabidopsis TUB2 gene were used as the normalization controls for expression analysis. Gene expression levels were calculated as previously described (Liu et al., 2007). Primers used for quantitative real-time PCR are listed in Supplementary Table S1.
In situ hybridization
Non-radioactive in situ hybridization was performed as previously described (Yu and Goh, 2000b). Gene-specific regions of DOFT and DOFTIP1 were amplified and cloned into the pGEM-T Easy vector (Promega) to produce the templates for in vitro transcription using the DIG RNA Labeling Kit (Roche Molecular Biochemicals).
Yeast two-hybrid assay
To construct the vectors for yeast two-hybrid assays, the coding sequence of DOFT was amplified and cloned into pGADT7, while the full-length and N-terminal regions of DOFTIP1 were amplified and cloned into pGBKT7 (Clontech). The yeast two-hybrid assay was performed using the Yeastmaker Yeast Transformation System 2 (Clontech). The transformed cells were selected on SD–His/–Trp/–Leu medium supplemented with 3 mM 3-amino-1, 2, 4-triazole. β-galactosidase assays were performed according to the Yeast Protocols Handbook (Clontech).
Glutathione S-transferase (GST) pull-down assay
The coding sequence of DOFT was cloned into the pGEX-4T-1 vector (Pharmacia) and transformed into E. coli Rosetta competent cells. Protein expression was induced by isopropyl-β-D-thiogalactoside at 16 °C. The soluble GST fusion proteins were extracted and immobilized on glutathione sepharose beads (Amersham Biosciences) for subsequent pull-down assays. The DOFTIP1 N-terminal fragment containing the three C2 domains (N550) was cloned into the pGADT7 vector (Clontech). The resulting plasmid was added to the TNT T7 Quick Coupled Transcription/Translation System (Promega) to synthesize DOFTIP1 (N550)–HA (hemagglutinin) protein. The resulting fusion protein was then incubated with the immobilized GST and GST-FT fusion proteins. Proteins retained on the beads were resolved by SDS-PAGE and detected with anti-HA antibody (Santa Cruz Biotechnology).
Bimolecular fluorescence complementation (BiFC) analysis
The full-length coding regions of DOFT and DOFTIP1 were cloned into primary pSAT1 vectors. The resulting cassettes containing the fusion proteins driven by the constitutive promoters were cloned into a pGreen binary vector, pHY105, and transformed into Agrobacterium. These were in turn co-infiltrated into tobacco (Nicotiana benthamiana) leaves and observed through a confocal microscope.
Co-immunoprecipitation experiment
Total proteins were isolated from transiently transformed tobacco leaves with a Plant Total Protein Extraction Kit (Sigma-Aldrich). The protein extracts were then immunoprecipitated using an anti-myc antibody (Santa Cruz Biotechnology). The total protein extracts as inputs and the immunoprecipitated proteins were resolved by SDS-PAGE and detected by an anti-HA antibody (Santa Cruz Biotechnology).
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: DOFT, MF063061; DOFTIP1, MF063062; DOAP1, KY471451; DOSOC1, KC121576.
Results
Isolation of DOFT and DOFTIP1 from Dendrobium Chao Praya Smile
To isolate putative FT and FTIP1 orthologs from Dendrobium Chao Praya Smile, we designed degenerate primers from the conserved regions of FT-like and FTIP1-like genes, and amplified the cDNA fragments of around 400 bp and 750 bp, respectively. After confirming the sequence similarity between these fragments and the other orthologs of DOFT and DOFTIP1, we used the rapid amplification of cDNA ends (RACE) method to obtain the full-length cDNAs corresponding to these two fragments, designated DOFT and DOFTIP1 (GenBank accession No. MF063061 and MF063062, respectively).
DOFT encodes a protein with 204 amino acid residues and an estimated molecular weight of 23 kDa. Multiple protein sequence alignment showed that DOFT exhibited 73, 79, and 94% identity to Arabidopsis FT, rice Hd3a, and another orchid FT ortholog OnFT from Oncidium Gower Ramsey, respectively (see Supplementary Fig. S1A). In particular, we found that Tyr85 and Gln140, which are the most critical residues for distinguishing FT and its closest homolog TERMINAL FLOWER1 (TFL1) in Arabidopsis (Hanzawa et al., 2005; Ahn et al., 2006), are present in the corresponding positions of Tyr111 and Gln166 of DOFT (Supplementary Fig. S1A). To determine the evolutionary relationship between DOFT and other FT orthologs, we constructed a phylogenetic tree that showed that DOFT was clustered in the FT clade and was closely related to OnFT as expected (Supplementary Fig. S1B). These results indicate that DOFT is the putative FT ortholog from Dendrobium Chao Praya Smile.
DOFTIP1 encodes a protein with 815 amino acids and an estimated molecular weight of 92 kDa. Like other MCTPs in plants (Liu et al., 2012, 2013), the deduced amino acid sequence of DOFTIP1 contains three C2 domains and one PRT_C domain, which was predicted to be a membrane-targeted domain according to the topology analysis (see Supplementary Fig. S2A, B). Multiple sequence alignment revealed that DOFTIP1 shared 76% sequence identity with FTIP1, and even higher identity with FTIP1 orthologs in other monocots, such as OsFTIP1 in rice (83% identity; see Supplementary Fig. S2C) (Song et al., 2017). Construction of a phylogenetic tree based on DOFTIP1 and other FTIP1-like proteins showed that DOFTIP1 was closely related to rice OsFTIP1 (Supplementary Fig. S2D). These sequence analyses also suggested that DOFTIP1 is the putative ortholog of FTIP1 in Dendrobium Chao Praya Smile.
To investigate the genomic organization of DOFT and DOFTIP1 in Dendrobium orchids, we performed Southern blot analysis of genomic DNA digested with several enzymes using digoxigenin-labeled probes synthesized from the gene-specific regions of DOFT and DOFTIP1. DNA blot analysis revealed a single strong band for either DOFT or DOFTIP1 (see Supplementary Fig. S2E), indicating that DOFT or DOFTIP1 is present as a single-copy or low-copy number gene in the genome of Dendrobium Chao Praya Smile.
Spatial and temporal expression patterns of DOFT and DOFTIP1
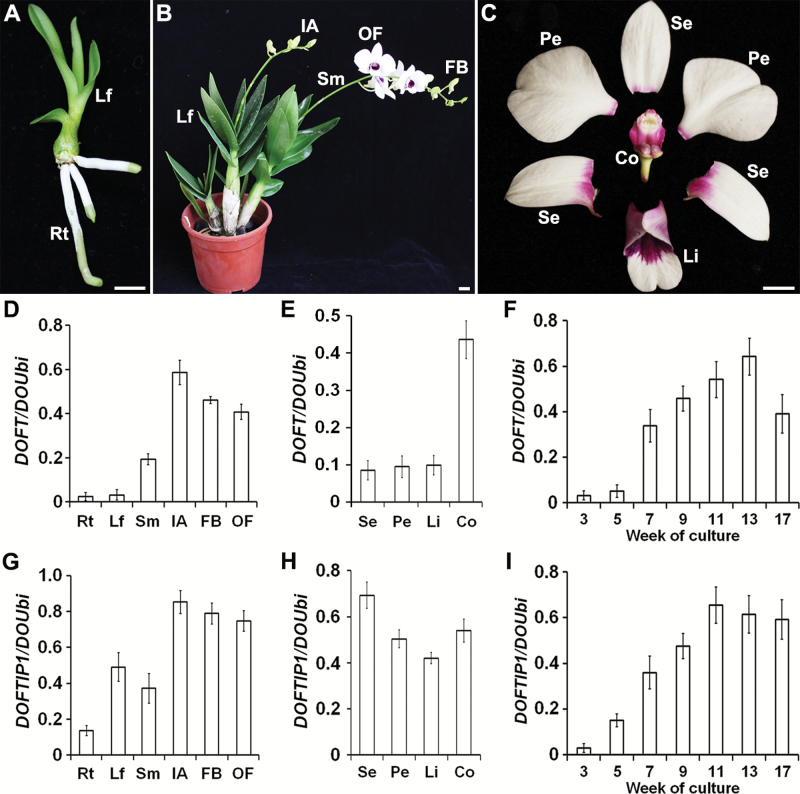
To characterize the functions of DOFT and DOFTIP1, we first carried out quantitative real-time PCR to examine their spatial expression patterns in various orchid tissues harvested at normal greenhouse growth conditions (Fig. 1A–C and Supplementary Fig. S3A). The transcripts of both genes were detectable at low levels in vegetative tissues, such as roots, leaves, and stems, but were present at obviously higher levels in reproductive organs, such as inflorescence apices, floral buds, and open flowers (Fig. 1A, B, D, G). In the different floral organs (Fig. 1C), DOFT was highly expressed in the column (gynostemium, a fused structure of stigmas, styles, and stamens) (Fig. 1E), whereas DOFTIP1 expression was high in all flower organs (Fig. 1H). In flowers at different development stages (see Supplementary Fig. S3A), expression levels of DOFT and DOFTIP1 gradually decreased from young floral buds to open flowers (Supplementary Fig. S3B, C). These expression data imply that DOFT and DOFTIP1 might be associated with reproductive organ development in Dendrobium Chao Praya Smile.
Fig. 1.
Quantitative analysis of DOFT and DOFTIP1 expression in Dendrobium Chao Praya Smile. (A) A vegetative seedling. Rt, root; Lf, leaf. (B) A flowering plant with inflorescences bearing flowers at various stages. Lf, leaf; Sm, stem; IA, inflorescence apex; OF, open flower; FB, floral bud. (C) An open flower consisting of three sepals (Se), two petals (Pe), one lip (Li), and the reproductive organ column (Co). Scale bars in (A–C) =1 cm. (D–F) Quantitative analysis of DOFT expression in various tissues (D), different floral organs (E), and orchid plants at various developmental stages (F). Error bars indicate SD. (G–I) Quantitative analysis of DOFTIP1 expression in various tissues (G), different floral organs (H), and orchid plants at different developmental stages (I). Error bars indicate SD. Expression levels in (D– I) were determined by quantitative real-time PCR analyses of three independently collected samples. The levels of gene expression were normalized to the expression of the orchid polyubiquitin gene (DOUbi).
We further examined the temporal expression of DOFT and DOFTIP1 in whole plants at various developmental stages under two growth conditions. In our established in vitro orchid culture system that allows rapid development of Dendrobium orchids from the vegetative to reproductive phase within about 3 months (Yu and Goh, 2000a; Ding et al., 2013), thin sections of protocorms that serve as starting material generate protocorm-like bodies (PLBs) about 0.5 cm in length within 4 weeks, which further develop into vegetative plantlets in the subsequent 4 weeks. Most of these plantlets enter the floral transitional stage following another 4 weeks of culture, after which they produce visible inflorescences and flowers. We found that DOFT expression was low in 3-week-old PLBs and 5-week-old vegetative plantlets, but it dramatically increased in 7-week-old plantlets before the floral transitional stage (Fig. 1F). Its expression further increased in 9- and 11-week-old plantlets during the floral transitional stage, and reached a maximum level in 13-week-old plantlets that were undergoing reproductive development. DOFTIP1 exhibited a temporal expression pattern similar to DOFT, but an obvious increase in its expression level was observed earlier in 5-week-old vegetative plantlets (Fig. 1I). In orchids grown under normal greenhouse conditions (see Supplementary Fig. S4A), the floral transition typically occurs 1 year after the starting material, which are young plantlets about 3 cm in height, are cultured in our greenhouse. We found that expression of both DOFT and DOFTIP1 was low in 2- and 8-month-old vegetative plantlets, but dramatically increased in 15-month-old plantlets at the floral transitional stage, and remained high in plantlets at the subsequent reproductive stage (Supplementary Fig. S4B, C). These temporal patterns in the orchids grown under normal greenhouse conditions were consistent with those displayed in the orchids grown under the in vitro culture condition, suggesting that up-regulation of DOFT and DOFTIP1 might be required for the floral transition in Dendrobium.
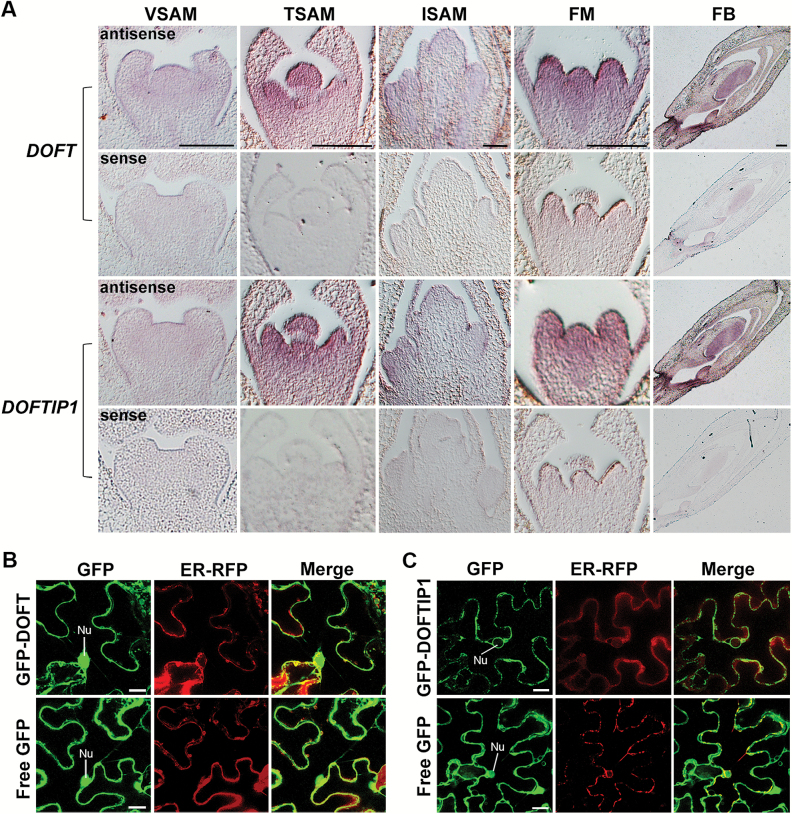
As both DOFT and DOFTIP1 were highly expressed in inflorescence apices and flowers (Fig. 1D, G), we then performed in situ hybridization using specific antisense DOFT and DOFTIP1 RNA probes to investigate their detailed localization in shoot apices and flowers harvested under the in vitro culture system. DOFT and DOFTIP1 expression was barely detectable in 6-week-old vegetative SAMs, but greatly increased in 12-week-old transitional SAMs (Fig. 2A). At the inflorescence and flower development stage, DOFT and DOFTIP1 transcripts were distributed throughout the inflorescence SAM and the flanking floral primordia, floral meristems, and floral buds with developing floral organs (Fig. 2A). These observations, together with the quantitative expression data (Fig. 1), provide evidence that DOFT and DOFTIP1 expression could be involved in the regulation of the floral transition and flower development in orchids.
Fig. 2.
mRNA and protein localization of DOFT and DOFTIP1. (A) In situ localization of DOFT and DOFTIP1 transcripts in Dendrobium Chao Praya Smile. Sections were hybridized with the gene-specific antisense and sense RNA probes of DOFT and DOFTIP1. VSAM, vegetative shoot apical meristem (6 weeks old); TSAM, transitional shoot apical meristem (12 weeks old); ISAM, inflorescence shoot apical meristem (16 weeks old); FM, floral meristem; FB, floral bud. Scale bars =200 μm. (B, C) Subcellular localization of GFP-DOFT (B) and GFP-DOFTIP1 (C) with free GFP in N. benthamiana leaf epidermal cells. Nu, nucleus. GFP, GFP fluorescence; ER-RFP, RFP fluorescence of an ER marker; Merge, merged images of GFP and RFP. Scale bars =20 μm.
To explore whether the expression of DOFT and DOFTIP1 was influenced by photoperiod, their mRNA levels in leaves of 16-month-old orchids (grown under greenhouse conditions) at the floral transitional stage were analysed at 4-h intervals over a 24-h period under LDs and short-days (SDs). DOFT was expressed at stable levels under SDs, but exhibited a diurnal oscillation under LDs with a peak at Zeitgeber time (ZT)-8 (8 h after the beginning of the light period) (see Supplementary Fig. S5A). In contrast, DOFTIP1 expression did not show an obvious circadian rhythm under either LDs or SDs (Supplementary Fig. S5B). These results suggest that, like their counterparts in Arabidopsis, day length affects the expression of DOFT but not DOFTIP1 in orchids.
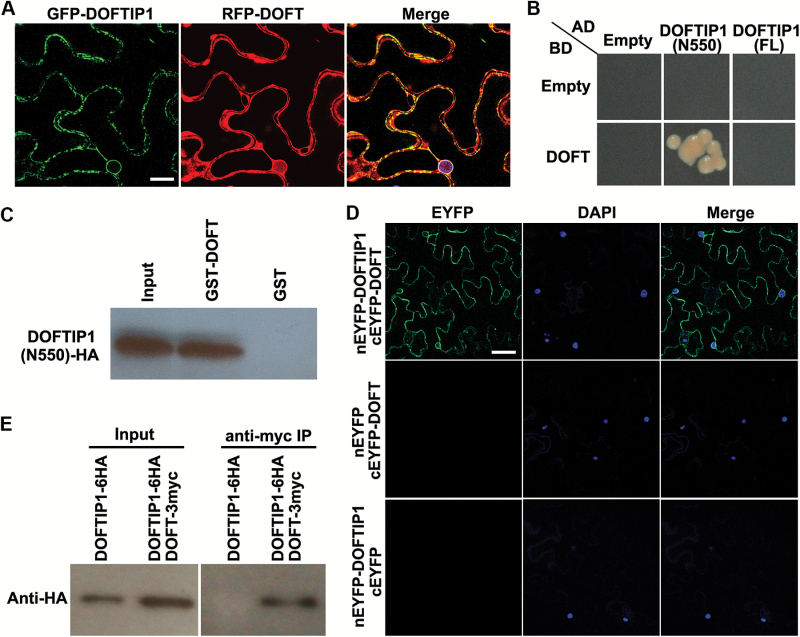
To assess the subcellular localization of DOFT and DOFTIP1, the coding sequences of DOFT and DOFTIP1 were fused with the green fluorescent protein (GFP) driven by the cauliflower mosaic virus (CaMV) 35S promoter. We transiently co-expressed 35S:GFP-DOFT or 35S:GFP-DOFTIP1 with an endoplasmic reticulum (ER) marker (ER-RFP) in N. benthamiana leaf epidermal cells, and found that GFP-DOFT and GFP-DOFTIP were similarly co-localized with ER-RFP, whereas only the GFP-DOFT signal appeared in the nucleus (Fig. 2B).
DOFT promotes flowering in Arabidopsis
To explore the biological role of DOFT, we created transgenic Arabidopsis plants in which DOFT was driven by the 35S promoter. Among 29 independent 35S:DOFT T1 transgenic plants created at the T1 generation, all lines flowered earlier than wild-type plants, with an average of 4.1 rosette leaves under LDs (see Supplementary Fig. S6A, B). We further overexpressed DOFT in Arabidopsis ft-10 loss-of-function mutants to test whether it could complement the loss of FT. A total of 22 independent ft-10 35S:DOFT transgenic plants were obtained at the T1 generation, and all of them flowered even earlier than wild-type plants, demonstrating a complete rescue of the late-flowering phenotype of ft-10 (Supplementary Fig. S6A, B). Semi-quantitative PCR analysis showed that DOFT expression was higher in both 35S:DOFT and 35S:DOFT ft-10 transgenic lines with stronger early-flowering phenotypes than in those with weak phenotypes (Supplementary Fig. S6C), indicating that DOFT might have a dosage-dependent effect on promoting flowering in Arabidopsis.
Previous studies in Arabidopsis have demonstrated that FT is expressed in the phloem, and that its protein movement to the SAM through the phloem system contributes to floral induction (Takada and Goto, 2003; Corbesier et al., 2007; Jaeger and Wigge, 2007; Mathieu et al., 2007). To examine whether DOFT plays a similar role in the phloem, we expressed the DOFT coding sequence under the phloem-specific SUCROSE TRANSPORTER 2 (SUC2) promoter in both wild-type and ft-10 backgrounds. Like 35S:DOFT, most of the SUC2:DOFT transgenic lines in both backgrounds displayed earlier flowering than the wild-type and ft-10 plants (see Supplementary Fig. S7A, B), suggesting that DOFT acts in a manner similar to FT in the Arabidopsis phloem.
DOFT promotes flowering in Dendrobium Chao Praya Smile
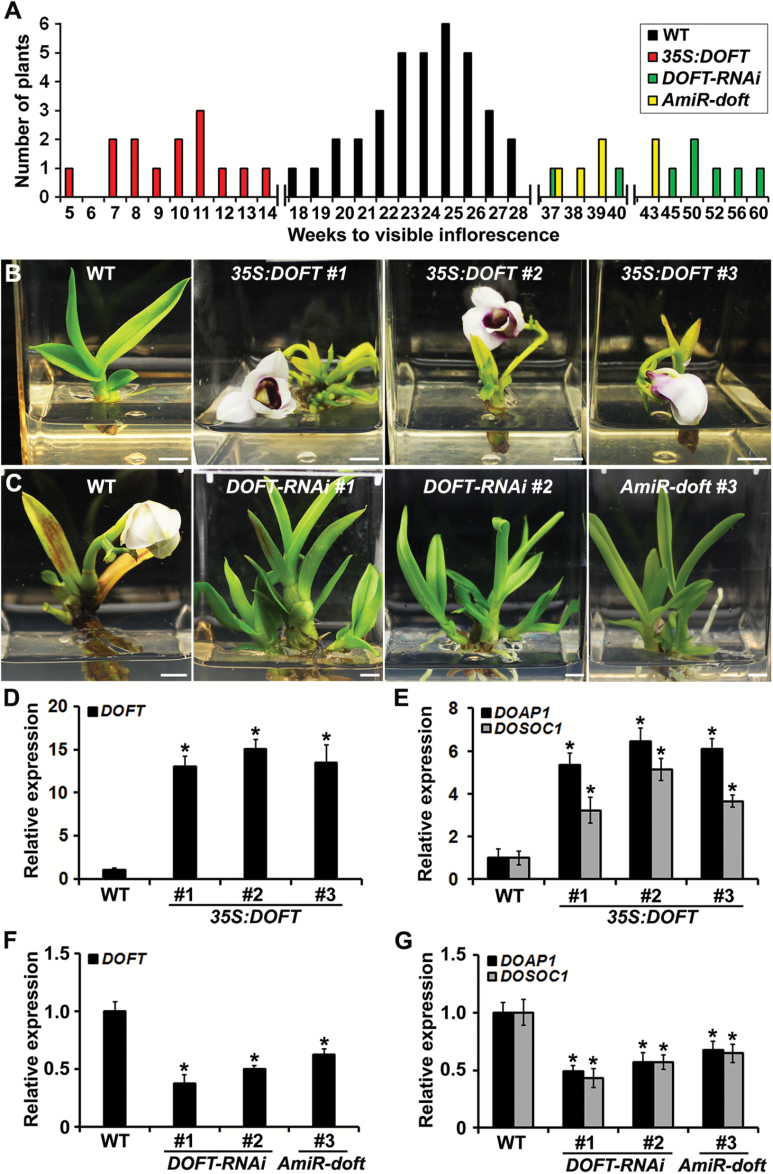
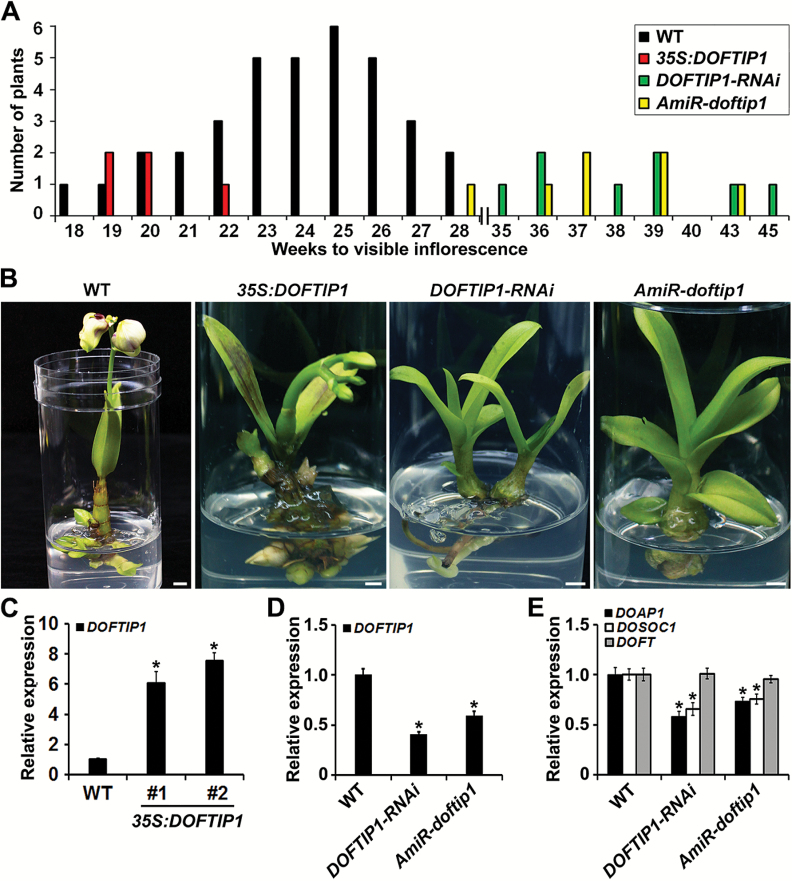
To understand the endogenous function of DOFT in Dendrobium orchids, we generated overexpression and knockdown transgenic orchids using particle bombardment or Agrobacterium-mediated transformation through an integrated gene transformation coupled with an in vitro tissue culture system (Yu and Goh, 2000a; Yu et al., 2001; Chai et al., 2007; Ding et al., 2013). The presence of the transgenes in the putative transgenic orchids was examined by PCR genotyping and Southern blot analysis (see Supplementary Fig. S8). We created 14 independent 35S:DOFT transgenic lines (Supplementary Fig. S8A, D), all of which generated the first visible inflorescence stalks at 5–14 weeks of culture (Fig. 3A, B). This was much earlier than the flowering time exhibited by wild-type orchids, which did not generate inflorescences until at least 18 weeks of culture (Fig. 3A). As expected, DOFT expression was much higher in leaves of the representative 35S:DOFT lines than in the wild-type (Fig. 3B, D). In Arabidopsis, FT induces flowering through up-regulating other downstream flowering regulators, such as SOC1 and AP1, in the shoot apex (Lee et al., 2000; Abe et al., 2005; Wigge et al., 2005). Similarly, we found that the expressions of DOSOC1 and DOAP1 (Ding et al., 2013; Sawettalake et al., 2017), the orchid orthologs of SOC1 and AP1 in Dendrobium Chao Praya Smile, were significantly higher in shoot apices of 35S:DOFT than in the wild-type (Fig. 3E).
Fig. 3.
DOFT promotes flowering in Dendrobium Chao Praya Smile. (A) Comparison of flowering time of wild-type and various transgenic orchids. The orchid flowering time is represented by the number of weeks of culture until the first inflorescence stalk was visible. (B) Overexpression of DOFT (35S:DOFT) in Dendrobium Chao Praya Smile results in earlier flowering than wild-type plants at 10 weeks of in vitro culture. Scale bars = 1 cm. (C) Knock-down of DOFT by RNA interference (DOFT-RNAi) or artificial microRNA interference (AmiR-doft) in Dendrobium Chao Praya Smile causes later flowering than wild-type plants at 25 weeks of in vitro culture. Scale bars = 1 cm. (D, E) Quantitative analysis of the expression of DOFT (D), and DOAP1 and DOSOC1 (E) in wild-type and representative 35S:DOFT transgenic orchids shown in (B) by real time RT-PCR. Error bars indicate SD. (F, G) Quantitative analysis of the expression of DOFT (F), and DOAP1 and DOSOC1 (G) in wild-type and representative DOFT knock-down transgenic orchids shown in (C) by real time RT-PCR. Error bars indicate SD. Total RNA extracted from leaves (D, F) or shoot apices (E, G) of orchid plants at 5 weeks of culture was used for expression analyses. The results in (D–G) were normalized against the expression levels of DOUbi, and the gene expression level in the wild-type plants was set as 1. Asterisks indicate significant differences in gene expression levels in various transgenic plants compared with those in corresponding wild-type plants (two-tailed paired Student’s t-test, P<0.05).
We also created DOFT knockdown transgenic plants by RNA interference (RNAi) and artificial microRNA (AmiR) interference. Among 10 DOFT-RNAi and 6 AmiR-doft independent transgenic lines created (see Supplementary Fig. S8B–D), 14 lines generated the first visible inflorescence stalks at 37–60 weeks of culture (Fig. 3A, C), while the other two lines never generated any visible inflorescence stalks under our culture conditions. Thus, these DOFT knockdown transgenic plants displayed much later flowering time than wild-type orchids, which flowered at the latest at 28 weeks of culture (Fig. 3A). The expression of DOFT, DOSOC1, and DOAP1 was consistently down-regulated in the representative knockdown lines compared to the wild-type (Fig. 3F, G). These observations suggest that DOFT plays a conserved role as FT in promoting orchid flowering, partially through engaging similar downstream regulators as revealed in Arabidopsis.
DOFT affects other developmental processes in Dendrobium Chao Praya Smile
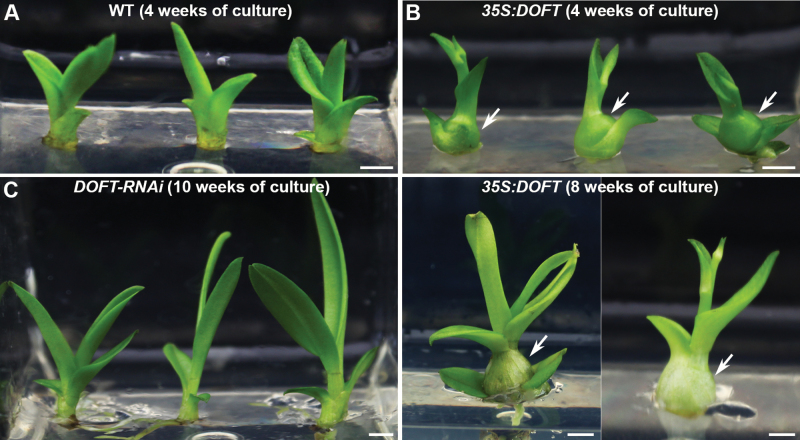
A marked morphological feature of Dendrobium, like many other epiphytic orchids, is the presence of a pseudobulb, which is a storage organ for supplying water, minerals, and carbohydrates (Zimmerman, 1990; Yong and Hew, 1995; Ng and Hew, 2000; He et al., 2011). It has been suggested that pseudobulb photosynthesis recycles respiratory carbon, and that carbohydrate reserves in pseudobulbs contribute to the development of new shoots and inflorescences in the orchids Catasetum viridiflavum and Oncidium Goldiana (Zimmerman, 1990; Yong and Hew, 1995; Ng and Hew, 2000; Blanchard and Runkle, 2008; Wang et al., 2008). Consistent with these previous findings, we found that pseudobulb formation usually occurred at 14 weeks under our in vitro tissue culture system for Dendrobium Chao Praya Smile at the beginning of the inflorescence and flower developmental stage (Yu and Goh, 2000a; Ding et al., 2013), indicating that pseudobulb formation is required for reproductive development. In agreement with the early-flowering phenotype, half of the 14 35S:DOFT transgenic orchids produced visible pseudobulbs early at 4–6 weeks of culture, while the other half showed visible pseudobulbs at 8–12 weeks of culture (Fig. 4A, B). In contrast, pseudobulb formation in DOFT-RNAi and AmiR-doft transgenic lines was much delayed, either not occurring until 32 weeks of culture at the earliest or never occurring at all (Fig. 4C). These observations suggest that DOFT promotes both pseudobulb formation and flowering in Dendrobium Chao Praya Smile.
Fig. 4.
DOFT affects pseudobulb formation in Dendrobium Chao Praya Smile. (A) Pseudobulb formation does not occur in wild-type plants at 4 weeks of culture. (B) 35S:DOFT transgenic orchid plants show obvious pseudobulb formation at 4 weeks (upper panel) and 8 weeks (lower panel) of culture. Arrows indicate pseudobulb formation. (C) Pseudobulb formation does not occur in DOFT-RNAi knock-down transgenic lines at 10 weeks of culture. Scale bars = 0.5 cm.
We further investigated the effects of DOFT on flower phenotypes under the in vitro tissue culture system, and found that changes in DOFT expression in both overexpression (35S:DOFT) and knockdown (DOFT-RNAi and AmiR-doft) transgenic orchids caused a higher percentage of abnormal or incomplete inflorescences and floral organs than in the wild-type (Fig. 5 and Supplementary Table S2). These results, together with high expression of DOFT in reproductive organs, including inflorescence apices, floral buds, and open flowers (Fig. 1D), strongly suggest that in addition to control of flowering time, DOFT is also required for inflorescence and floral development in Dendrobium orchids.
Fig. 5.
Alteration of DOFT expression compromises normal inflorescence and flower development in Dendrobium Chao Praya Smile. (A) A wild type orchid line. (B) A 35S:DOFT transgenic orchid line generates an aborted inflorescence apex, shown enlarged in the inset. (C–I) Various DOFT transgenic orchid lines, including 35S:DOFT (C–F), DOFT-RNAi (G, H), and AmiR-doft (I), generate flowers with either incomplete or abnormal floral organs. Scale bars: (A–F) = 0.5 cm, (G–I) = 1 cm.
DOFTIP1 accelerates flowering in Arabidopsis and Dendrobium Chao Praya Smile
In Arabidopsis, FTIP1 interacts with FT in companion cells and mediates its transport from companion cells to sieve elements through plasmodesmata (Liu et al., 2012). Notably, either loss or overexpression of FTIP1 causes a late-flowering phenotype, as the former compromises FT transport from the companion cells to sieve elements, whereas the latter deregulates FT protein transport out of the phloem system. To understand the biological function of DOFTIP1, we also created 35S:DOFTIP1 transgenic Arabidopsis plants in both wild-type and ftip1-1 backgrounds. Similar to 35S:FTIP1 (Liu et al., 2012), 17 out of 25 35S:DOFTIP1 transgenic lines flowered later than wild-type plants under LDs (see Supplementary Fig. S9A–C). In contrast, overexpression of FTIP1 in ftip1-1 evidently rescued the late-flowering phenotype of ftip1-1, and 13 out 14 ftip1-1 35S:DOFTIP1 lines displayed a comparable flowering time to the wild-type plants (Supplementary Fig. S9A–C). Thus, DOFTIP1 is able to play a similar role to FTIP1 in promoting flowering in Arabidopsis, but its expression in excessive amounts in wild-type plants could similarly deregulate FT transport, thus resulting in late flowering.
We further created DOFTIP1 overexpression and knockdown transgenic orchids to investigate the endogenous function of DOFTIP1. After PCR genotyping and Southern blot analysis (Supplementary Fig. S10), we obtained five, eight, and seven independent lines for 35S:DOFTIP1, DOFTIP1-RNAi, and AmiR-doftip1, respectively. All the 35S:DOFTIP1 lines exhibited a comparable flowering time to the wild-type orchids, although DOFTIP1 was overexpressed in the leaves of these transgenic lines (Fig. 6A–C). In contrast, most of the DOFTIP1-RNAi and AmiR-doftip1 lines generated the first visible inflorescence stalks at 35–45 weeks of culture, which was much later than the flowering time of the wild-type (Fig. 6A, B). DOFTIP1 expression was consistently down-regulated in leaves of the representative DOFTIP1-RNAi and AmiR-doftip1 lines (Fig. 6D). Furthermore, we found that while down-regulation of DOFTIP1 did not affect DOFT transcript levels, DOSOC1 and DOAP1 were significantly decreased in shoot apices of the DOFTIP1-RNAi and AmiR-doftip1 lines compared to their expression in wild-type orchids (Fig. 6E). These results suggest that DOFTIP1 promotes orchid flowering, but does not directly affect DOFT expression levels.
Fig. 6.
Down-regulation of DOFTIP1 delays flowering in Dendrobium Chao Praya Smile. (A) Comparison of flowering time of wild-type and various transgenic orchids. (B) Knock-down of DOFTIP1 by RNA interference (DOFTIP1-RNAi) or artificial microRNA interference (AmiR-doftip1) causes later flowering than wild-type and 35S:DOFTIP1 orchid plants at 25 weeks of in vitro culture. Scale bars = 1 cm. (C, D) Quantitative analysis of DOFTIP1 expression in leaves of representative 35S:DOFTIP1 (C) and DOFTIP1 knock-down (D) transgenic orchids by real time RT-PCR. Error bars indicate SD. (E) Quantitative analysis of the expression of DOFT, DOAP1, and DOSOC1 in 35S:DOFTIP1 and DOFTIP1 knock-down transgenic orchids by real time RT-PCR. Total RNA extracted from shoot apices of orchid plants at 10 weeks of culture was used for expression analysis. Error bars indicate SD. The results in (C–E) were normalized against the expression levels of DOUbi, and the expression level of each gene in the wild-type plants was set as 1. Asterisks indicate significant differences in gene expression levels in various transgenic plants compared with those in corresponding wild-type plants (two-tailed paired Student’s t-test, P<0.05).
DOFTIP1 interacts with DOFT
Our findings on the similar tissue expression patterns of DOFT and DOFTIP1 and their same roles in promoting flowering in both Arabidopsis and orchids prompted us to investigate whether they interact with each other, like their counterparts in Arabidopsis (Liu et al., 2012). To this end, we examined the protein interaction between DOFTIP1 and DOFT through a few approaches. First, we co-expressed 35S:RFP-DOFT and 35S:GFP-DOFTIP1 in N. benthamiana leaf epidermal cells, and found both RFP-DOFT and GFP-DOFTIP1 were co-localized to ER connected to the nuclear envelope (Figs 2B, C and 7A). Second, yeast two-hybrid assays further revealed that the truncated DOFTIP1 protein devoid of the PRT_C domain, DOFTIP1 (N550), interacted with DOFT, whereas no interaction was detected between the full-length DOFTIP1 and DOFT (Fig. 7B). Quantification of the yeast two-hybrid interaction by β-galactosidase assays confirmed the strong interaction between DOFTIP1 (N550) and DOFT (see Supplementary Fig. S11A). As the PRT_C domain could be a membrane-targeted domain (Supplementary Fig. S2A, B), the full-length DOFTIP1 protein might not be in the membrane-bound state in yeast cells. This may cause an appropriate folding of DOFTIP1 to prevent its interaction with DOFT. Third, a GST pull-down assay also demonstrated that HA-labeled DOFTIP1 (N550) bound to GST-DOFT, but not GST, induced in E. coli cells (Fig. 7C and Supplementary Fig. S11B). To test the in planta interaction between DOFTIP1 and DOFT, we further performed BiFC assays, and also found an enhanced YFP (EYFP) signal in N. benthamiana leaf epidermal cells except the nuclei (Fig. 7D). To verify this in planta interaction, we transiently transformed 35S:DOFTIP1-6HA and 35S:DOFT-3myc into tobacco leaves. Co-immunoprecipitation analysis of the protein extracts revealed that DOFTIP1-6HA interacted with DOFT-3myc only in the tobacco leaves transformed with both 35S:DOFTIP1-6HA and 35S:DOFT-3myc (Fig. 7E), substantiating the interaction between DOFTIP1 and DOFT in plant cells.
Fig. 7.
DOFTIP1 interacts with DOFT. (A) Co-localization of GFP-DOFTIP1 and RFP-DOFT in N. benthamiana leaf epidermal cells. Merge indicates merged images of GFP and RFP. Scale bar = 20 μm. (B) Yeast two-hybrid assay of the interaction between DOFT and the full-length DOFTIP1 (FL) and its N-terminus (amino acids 1–550; N550). Transformed yeast cells were grown on SD–His/–Trp/–Leu medium supplemented with 3 mM 3-amino-1,2,4-triazole. Empty refers to AD- or BD-containing vector only. (C) In vitro pull-down assay of the interaction between DOFT and DOFTIP1 (N550). ‘Input’ indicates 5% of HA-labeled DOFTIP1 (N550) subjected to pull-down by GST and GST-DOFT. (D) BiFC analysis of the interaction between DOFTIP1 and DOFT. DAPI, fluorescence of 4’,6-diamino-2-phenylindol; EYFP, fluorescence of enhanced yellow fluorescent protein; Merge, merged images of DAPI and EYFP. Scale bar = 10 μm. (E) In vivo interaction between DOFTIP1 and DOFT shown by co-immunoprecipitation. 35S:DOFTIP1-6HA and 35S:DOFT-3myc were transiently transformed into tobacco leaves, and total protein extracts were immunoprecipitated using an anti-myc antibody. The input and co-immunoprecipitated protein were detected using an anti-HA antibody.
Discussion
The proteins encoded by FT in Arabidopsis and its orthologs in several plant species have been identified as part of the mobile florigenic signals and play an important role in promoting flowering (Corbesier et al., 2007; Jaeger and Wigge, 2007; Lin et al., 2007; Mathieu et al., 2007; Tamaki et al., 2007). Although FT-like genes have also been isolated in a few orchid varieties (Hou and Yang, 2009; Huang et al., 2012; Li et al., 2012), these studies were unable to address their endogenous functions because of a lack of transgenic material. Thus, the biological functions of FT orthologs in the Orchidaceae, which is one of the largest families of flowering plants, are to date still largely unknown. In this study, we have characterized the functions of the orchid orthologs of FT and FTIP1, namely DOFT and DOFTIP1, respectively, from Dendrobium Chao Praya Smile, using an established gene transformation coupled with an in vitro tissue culture system. Our findings suggest that while both DOFT and DOFTIP1 play conserved roles in promoting flowering, DOFT has evolved with hitherto unknown functions in regulating the development of pseudobulbs and floral organs in orchids.
Our results provide several pieces of evidence suggesting that DOFT is the FT ortholog in Dendrobium Chao Praya Smile. First, DOFT shares high sequence similarity with FT and it orthologs in other plant species, including the signature residues Tyr85 and Gln140 of FT in the corresponding positions (see Supplementary Fig. S1A). Phylogenetic analysis based on the full-length amino acid sequences also demonstrated that DOFT is clustered with another orchid FT ortholog, OnFT, from Oncidium Gower Ramsey, in the FT clade (Supplementary Fig. 1B). Second, DOFT expression gradually increased during the floral transition in Dendrobium orchids under both in vitro culture and normal greenhouse conditions (Fig. 1F and Supplementary Fig. S4). This is similar to the FT expression pattern in Arabidopsis (Kardailsky et al., 1999; Kobayashi et al., 1999). Third, overexpression of DOFT caused early flowering and completely rescued the late-flowering phenotype of ft-10 in Arabidopsis (Supplementary Fig. S6). In addition, expression of DOFT driven by the phloem-specific SUC2 promoter in Arabidopsis exhibited a promotive effect on flowering similar to overexpression of DOFT (Supplementary Fig. S7). These results suggest that DOFT plays the same role as FT in promoting flowering in the Arabidopsis phloem. Lastly, overexpression or down-regulation of DOFT accelerated or delayed flowering in Dendrobium Chao Praya Smile (Fig. 3), demonstrating that DOFT is indeed required for orchid flowering. Notably, DOFT expression was positively correlated with the expression of DOSOC1 and DOAP1, the orthologs of SOC1 and AP1, respectively, in Dendrobium Chao Praya Smile (Ding et al., 2013; Sawettalake et al., 2017). This is consistent with the effect of FT in up-regulating SOC1 and AP1 in Arabidopsis (Lee et al., 2000; Abe et al., 2005; Wigge et al., 2005), implying that DOFT and FT may similarly control comparable downstream genes to promote flowering in Dendrobium Chao Praya Smile and Arabidopsis, respectively.
In Arabidopsis and rice, FTIP1 and its rice counterpart OsFTIP1 mediate transport of FT and its rice ortholog, RICE FLOWERING LOCUS T 1 (RFT1), respectively, from companion cells to sieve elements in the phloem, thus affecting their movement from leaves to the shoot apex (Liu et al., 2012; Song et al., 2017). In Dendrobium Chao Praya Smile, the orchid ortholog of FTIP1, DOFTIP1, exhibited several characteristics similar to FTIP1. In addition to sequence similarity between DOFTIP1 and FTIP1 or OsFTIP1, expression of DOFTIP1 rescued the late-flowering phenotype of its corresponding Arabidopsis mutant ftip1-1 (see Supplementary Fig. S9), while down-regulation of DOFTIP1 delayed flowering in Dendrobium orchids (Fig. 6), indicating that DOFTIP1 plays a similar role to FTIP1 in promoting flowering. Both RFP-DOFT and GFP-DOFTIP1 were co-localized to the ER in plant cells (Fig. 7A). Furthermore, DOSOC1 and DOAP1 were similarly down-regulated in shoot apices of both DOFT and DOFTIP1 knockdown orchids, whereas down-regulation of DOFTIP1 did not affect DOFT expression (Figs 3G and 6E), implying that DOFTIP1 may interact with DOFT at the protein level, thus affecting their common downstream genes. Indeed, several approaches confirmed the interaction between DOFTIP1 and DOFT in vitro and in planta (Fig. 7B–D). These observations all support the view that DOFTIP1 is the orchid counterpart of FTIP1 and interacts with DOFT to affect flowering time in Dendrobium Chao Praya Smile.
While DOFT plays an evolutionarily conserved role in promoting flowering in orchids, our findings have also revealed two interesting and hitherto unknown functions of FT-like genes in orchids. Firstly, DOFT affected the generation of pseudobulbs, which are a type of storage organ that contribute to reproductive development in many epiphytic orchids (Zimmerman, 1990; Yong and Hew, 1995; Ng and Hew, 2000; He et al., 2011). High expression of DOFT promoted pseudobulb formation, which was, however, delayed when DOFT was down-regulated (Fig. 4). As pseudobulb formation evidently appears after the floral transition in Dendrobium Chao Praya Smile (Yu and Goh, 2000a; Ding et al., 2013), it is highly possible that DOFT promotes flowering partly through positively regulating the formation of pseudobulbs so that these storage organs can provide sufficient quantities of the water, minerals, and carbohydrates that are essential for inflorescence and flower development in orchids. The effects of DOFT on pseudobulb formation and flowering in orchids is reminiscent of the similar role of FT-like genes in controlling flowering and tuberization in potato (Navarro et al., 2011), indicating that FT-like genes may play broader roles in regulating other developmental processes required for the floral transition.
Secondly, DOFT also contributes to inflorescence and flower development in Dendrobium Chao Praya Smile. Alteration in DOFT expression levels in 35S:DOFT, DOFT-RNAi, and AmiR-doft transgenic orchids resulted in more abnormal or incomplete inflorescences and floral organs than in wild-type plants (Fig. 5 and Supplementary Table S2), suggesting that an appropriate control of DOFT expression in inflorescences and developing floral organs is important for normal reproductive development in orchids. We consistently found that DOFT was expressed at high levels in reproductive organs, such as inflorescence apices, floral buds, and open flowers (Fig. 1D). Notably, among all the floral organs, DOFT was highly expressed in the column, which is a fused structure of reproductive organs, including stigmas, styles, and stamens (Fig. 1E). In agreement with this result, incomplete flowers without the column were often observed in DOFT knockdown lines, such as AmiR-doft (Fig. 5I), indicating that DOFT expression in the column is essential for reproductive organ development in orchids. In contrast to DOFT, changes in DOFTIP1 expression did not greatly affect inflorescence and flower development compared to the wild-type, suggesting that DOFTIP1 has a specific role in regulating flowering time rather than inflorescence and flower development in orchids.
To date, a number of FT orthologs have been isolated from a wide range of plant species. Besides the functional diversity of DOFT, as discussed above, some other FT orthologs have also been shown to have unique functions in various plant developmental processes; for example, PtFT1 and PtFT2 participate in SD-induced growth cessation and bud set in Populus (Böhlenius et al., 2006; Hsu et al., 2006), while SFT regulates fruit set, termination of sympodial meristems, and leaf architecture in tomato (Lifschitz et al., 2006). These observations demonstrate that FT-like genes not only encode important mobile flowering signals, but also participate in other diverse developmental processes that need to be further elucidated.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Sequence analysis of DOFT.
Fig. S2. Sequence analysis of DOFTIP1.
Fig. S3. Expression of DOFT and DOFTIP1 in flowers at different developmental stages.
Fig. S4. Quantitative analysis of temporal expression of DOFT and DOFTIP1 in Dendrobium Chao Praya Smile grown under normal greenhouse conditions.
Fig. S5. Quantitative analysis of DOFT and DOFTIP1 expression levels in leaves of Dendrobium Chao Praya Smile within a 24-h cycle under long and short days.
Fig. S6. Overexpression of DOFT promotes flowering in Arabidopsis.
Fig. S7. Expression of SUC2:DOFT promotes flowering in Arabidopsis.
Fig. S8. Molecular identification of DOFT transgenic orchids.
Fig. S9. Overexpression of DOFTIP1 rescues the late flowering phenotype of Arabidopsis ftip1-1.
Fig. S10. Molecular identification of DOFTIP1 transgenic orchids.
Fig. S11. Evidence that DOFTIP1 interacts with DOFT.
Table S1. List of primers used in this study.
Table S2. Comparison of flower development in wild-type and transgenic Dendrobium Chao Praya Smile after in vitro culture under our growth conditions.
Supplementary Material
Acknowledgements
We thank members of the Hao Yu laboratory for their critical reading of earlier versions of this article.
References
- Abe M, Kobayashi Y, Yamamoto S et al. 2005. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science Signaling 309, 1052–1056. [DOI] [PubMed] [Google Scholar]
- Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D. 2006. A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. The EMBO Journal 25, 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An H, Roussot C, Suarez-Lopez P et al. 2004. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131, 3615–3626. [DOI] [PubMed] [Google Scholar]
- Blanchard MG, Runkle ES. 2008. Temperature and pseudobulb size influence flowering of Odontioda orchids. HortScience 43, 1404–1409. [Google Scholar]
- Blázquez MA, Weigel D. 2000. Integration of floral inductive signals in Arabidopsis. Nature 404, 889–892. [DOI] [PubMed] [Google Scholar]
- Böhlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O. 2006. CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312, 1040–1043. [DOI] [PubMed] [Google Scholar]
- Chai D, Lee SM, Ng JH, Yu H. 2007. L-methionine sulfoximine as a novel selection agent for genetic transformation of orchids. Journal of Biotechnology 131, 466–472. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang SH et al. 2007. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316, 1030–1033. [DOI] [PubMed] [Google Scholar]
- Da Silva JT. 2013. Orchids: advances in tissue culture, genetics, phytochemistry and transgenic biotechnology. Floriculture and Ornamental Biotechnology 7, 1–52. [Google Scholar]
- Da Silva JAT, Aceto S, Liu W, Yu H, Kanno A. 2014. Genetic control of flower development, color and senescence of Dendrobium orchids. Scientia Horticulturae 175, 74–86. [Google Scholar]
- Ding L, Wang Y, Yu H. 2013. Overexpression of DOSOC1, an ortholog of Arabidopsis SOC1, promotes flowering in the orchid Dendrobium Chao Parya Smile. Plant & Cell Physiology 54, 595–608. [DOI] [PubMed] [Google Scholar]
- Gutiérrez RMP. 2010. Orchids: A review of uses in traditional medicine, its phytochemistry and pharmacology. Journal of Medicinal Plants Research 4, 592–638. [Google Scholar]
- Hanzawa Y, Money T, Bradley D. 2005. A single amino acid converts a repressor to an activator of flowering. Proceedings of the National Academy of Sciences, USA 102, 7748–7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Tan BH, Qin L. 2011. Source-to-sink relationship between green leaves and green pseudobulbs of C3 orchid in regulation of photosynthesis. Photosynthetica 49, 209–218. [Google Scholar]
- Hou CJ, Yang CH. 2009. Functional analysis of FT and TFL1 orthologs from orchid (Oncidium Gower Ramsey) that regulate the vegetative to reproductive transition. Plant & Cell Physiology 50, 1544–1557. [DOI] [PubMed] [Google Scholar]
- Hsu CY, Liu Y, Luthe DS, Yuceer C. 2006. Poplar FT2 shortens the juvenile phase and promotes seasonal flowering. The Plant Cell 18, 1846–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Fang Z, Zeng S, Zhang J, Wu K, Chen Z, Teixeira da Silva JA, Duan J. 2012. Molecular cloning and functional analysis of three FLOWERING LOCUS T (FT) homologous genes from Chinese Cymbidium. International Journal of Molecular Sciences 13, 11385–11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger KE, Wigge PA. 2007. FT protein acts as a long-range signal in Arabidopsis. Current Biology 17, 1050–1054. [DOI] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D. 1999. Activation tagging of the floral inducer FT. Science 286, 1962–1965. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. 1999. A pair of related genes with antagonistic roles in mediating flowering signals. Science 286, 1960–1962. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Peeters AJ. 1998. Genetic interactions among late-flowering mutants of Arabidopsis. Genetics 148, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. 2000. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes & Development 14, 2366–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Liu C, Shen L et al. 2008. A repressor complex governs the integration of flowering signals in Arabidopsis. Developmental Cell 15, 110–120. [DOI] [PubMed] [Google Scholar]
- Li R, Wang A, Sun S, Liang S, Wang X, Ye Q, Li H. 2012. Functional characterization of FT and MFT ortholog genes in orchid (Dendrobium nobile Lindl) that regulate the vegetative to reproductive transition in Arabidopsis. Plant Cell, Tissue and Organ Culture 111, 143–151. [Google Scholar]
- Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y. 2006. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proceedings of the National Academy of Sciences, USA 103, 6398–6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M-K, Belanger H, Lee Y-J et al. 2007. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. The Plant Cell 19, 1488–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, Yu H. 2007. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134, 1901–1910. [DOI] [PubMed] [Google Scholar]
- Liu L, Liu C, Hou X, Xi W, Shen L, Tao Z, Wang Y, Yu H. 2012. FTIP1 is an essential regulator required for florigen transport. PLoS Biology 10, e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhu Y, Shen L, Yu H. 2013. Emerging insights into florigen transport. Current Opinion in Plant Biology 16, 607–613. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Warthmann N, Küttner F, Schmid M. 2007. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Current Biology 17, 1055–1060. [DOI] [PubMed] [Google Scholar]
- Navarro C, Abelenda JA, Cruz-Oró E, Cuéllar CA, Tamaki S, Silva J, Shimamoto K, Prat S. 2011. Control of flowering and storage organ formation in potato by FLOWERING LOCUS T. Nature 478, 119–122. [DOI] [PubMed] [Google Scholar]
- Ng CKY, Hew CS. 2000. Orchid pseudobulbs – ‘false’ bulbs with a genuine importance in orchid growth and survival!Scientia Horticulturae 83, 165–172. [Google Scholar]
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Molecular Biology Reporter 15, 8–15. [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. 2000. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288, 1613–1616. [DOI] [PubMed] [Google Scholar]
- Sawettalake N, Bunnag S, Wang Y, Shen L, Yu H. 2017. DOAP1 promotes flowering in the orchid Dendrobium Chao Praya Smile. Frontiers in Plant Science 8, 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. 2006. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell 18, 1121–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Chen Y, Liu L et al. 2017. OsFTIP1-mediated regulation of florigen transport in rice is negatively regulated by the ubiquitin-like domain kinase OsUbDKγ4. The Plant Cell 29, 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada S, Goto K. 2003. TERMINAL FLOWER2, an Arabidopsis homolog of HETEROCHROMATIN PROTEIN1, counteracts the activation of FLOWERING LOCUS T by CONSTANS in the vascular tissues of leaves to regulate flowering time. The Plant Cell 15, 2856–2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. 2007. Hd3a protein is a mobile flowering signal in rice. Science 316, 1033–1036. [DOI] [PubMed] [Google Scholar]
- Wang CY, Chiou CY, Wang HL, Krishnamurthy R, Venkatagiri S, Tan J, Yeh KW. 2008. Carbohydrate mobilization and gene regulatory profile in the pseudobulb of Oncidium orchid during the flowering process. Planta 227, 1063–1077. [DOI] [PubMed] [Google Scholar]
- Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M, Lohmann JU, Weigel D. 2005. Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309, 1056–1059. [DOI] [PubMed] [Google Scholar]
- Yong J, Hew C. 1995. The importance of photoassimilate contribution from the current shoot and connected back shoots to inflorescence size in the thin-leaved sympodial orchid Oncidium Goldiana. International Journal of Plant Sciences 156, 450–459. [Google Scholar]
- Yu H, Goh CJ. 2000a. Differential gene expression during floral transition in an orchid hybrid Dendrobium Madame Thong-In. Plant Cell Reports 19, 926–931. [DOI] [PubMed] [Google Scholar]
- Yu H, Goh CJ. 2000b. Identification and characterization of three orchid MADS-box genes of the AP1/AGL9 subfamily during floral transition. Plant Physiology 123, 1325–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Goh CJ. 2001. Molecular genetics of reproductive biology in orchids. Plant Physiology 127, 1390–1393. [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Wellmer F, Meyerowitz EM. 2004. Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nature Genetics 36, 157–161. [DOI] [PubMed] [Google Scholar]
- Yu H, Yang S, Goh C. 2001. Agrobacterium-mediated transformation of a Dendrobium orchid with the class 1 knox gene DOH1. Plant Cell Reports 20, 301–305. [Google Scholar]
- Zhu Y, Liu L, Shen L, Yu H. 2016. NaKR1 regulates long-distance movement of FLOWERING LOCUS T in Arabidopsis. Nature Plants 2, 16075. [DOI] [PubMed] [Google Scholar]
- Zimmerman JK. 1990. Role of pseudobulbs in growth and flowering of Catasetum viridiflavum (Orchidaceae). American Journal Botany 77, 533–542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.