Using a genome-wide association mapping approach, we identified a new role for the GRAS transcription factor RAD1 in supporting infection by an oomycete root pathogen. Previously, RAD1 was exclusively implicated in symbiosis signalling.
Keywords: Arbuscular mycorrhiza, genome-wide association mapping, host susceptibility, Medicago truncatula, MtSymSCL3, Phytophthora palmivora, RAD1, root colonization, symbiosis, oomycete
Abstract
The roots of most land plants are colonized by symbiotic arbuscular mycorrhiza (AM) fungi. To facilitate this symbiosis, plant genomes encode a set of genes required for microbial perception and accommodation. However, the extent to which infection by filamentous root pathogens also relies on some of these genes remains an open question. Here, we used genome-wide association mapping to identify genes contributing to colonization of Medicago truncatula roots by the pathogenic oomycete Phytophthora palmivora. Single-nucleotide polymorphism (SNP) markers most significantly associated with plant colonization response were identified upstream of RAD1, which encodes a GRAS transcription regulator first negatively implicated in root nodule symbiosis and recently identified as a positive regulator of AM symbiosis. RAD1 transcript levels are up-regulated both in response to AM fungus and, to a lower extent, in infected tissues by P. palmivora where its expression is restricted to root cortex cells proximal to pathogen hyphae. Reverse genetics showed that reduction of RAD1 transcript levels as well as a rad1 mutant are impaired in their full colonization by AM fungi as well as by P. palmivora. Thus, the importance of RAD1 extends beyond symbiotic interactions, suggesting a general involvement in M. truncatula microbe-induced root development and interactions with unrelated beneficial and detrimental filamentous microbes.
Introduction
Host compatibility genes facilitate the colonization of plants by pathogenic microbes (van Schie and Takken, 2014). Knowing such host genes and the processes in which they function provides inroads for efforts to establish disease resistance. For example, the knock-out mutant of MLO in barley has been successfully deployed to provide resistance against powdery mildew (Acevedo-Garcia et al., 2014). Plant host compatibility genes also function in beneficial root endosymbiosis, such as with arbuscular mycorrhiza (AM) fungi (Gutjahr and Parniske, 2013) and nitrogen-fixing rhizobia (Oldroyd et al., 2011; Popp and Ott, 2011). For instance, commercial wheat varieties often possess a Reduced height (Rht) dominant allele of DELLA suppressing gibberellic acid (GA) signalling, which was recently and unexpectedly found to favour mycorrhiza (Floss et al., 2013). Therefore, breeding for higher yield and reduced height can directly impact AM symbiosis, but may also increase susceptibility to other biotrophic pathogens (Saville et al., 2012). Compatibility genes often control processes essential to general plant biology and physiology, thereby explaining the conservation of those facilitating entry of pathogenic microbial intruders.
Compatibility loci may be a rewarding target to reduce pathogen susceptibility or to support symbiotic associations, but their identification often is challenging. First, genetic detection of susceptibility traits can be masked in pathosystems that are dominated by effective disease resistance responses (Jones and Dangl, 2006; Dangl et al., 2013). Secondly, adapted pathogens may not be fully reliant on basic compatibility genes, since they may specifically reprogram the host to achieve their accommodation in planta (Mukhtar et al., 2011). Finally, potential trade-offs between inactivation of host susceptibility genes and implementation of symbiosis-promoting alleles need to be examined (Rey and Schornack, 2013; Evangelisti et al., 2014).
To avoid some of these pitfalls, biotrophic pathogens with broad host range can be used in non-co-evolving pathosystems to uncover susceptibility genes, followed by informed transfer into crops and assessment of crop-relevant pathogens with similar lifestyles (Rey and Schornack, 2013). The oomycete Phytophthora palmivora represents a suitable microbe to pursue this strategy, since it infects a number of tropical cash crops but can also colonize Medicago truncatula, a model plant for nitrogen-fixing symbiosis and AM symbiosis studies (Evangelisti et al., 2014; Rey et al., 2015). Phytophthora palmivora infections start with mobile flagellated zoospores which encyst once settled on targeted root tissues, preferably near the root tip. A germ tube emerges which develops an appressorium on the root epidermis (Wang et al, 2012). Entry into the inner root tissues occurs through or between epidermal cells or through wounds. In the root cortex, P. palmivora grows largely intercellularly but projects short, specialized hyphae termed haustoria into individual living cortex cells to suppress immunity. This biotrophic stage is followed by necrotrophy when P. palmivora kills host tissues through the release of cell wall-degrading enzymes. At this stage, P. palmivora can enter the plant vasculature and systemically infect the shoot. Sporangiophores carrying sporangia are formed in the dying tissues, and sporangia serve as propagules or can release several mobile zoospores, thereby completing the asexual life cycle with 2–3 d.
The wealth of genomic (Young et al., 2011), genetic (Pislariu et al., 2012; Urbański et al., 2012), and transcriptomic tools (Benedito et al., 2008; Verdier et al., 2013) available in M. truncatula can considerably accelerate molecular characterization of the colonization principles involved in association with P. palmivora and possible commonalities with symbiotic processes. Forward genetics of M. truncatula identified the dual contribution of a glycerol phosphate acyl transferase gene termed RAM2 (Reduced Arbuscular Mycorrhization 2) in the colonization by AM fungi and the oomycete pathogens P. palmivora (Wang et al., 2012) and Aphanomyces euteiches (Gobbato et al., 2013). Complementarily, reverse genetics approaches based on surveying mutants defective in symbiosis for their interaction with pathogens revealed that symbiosis-related LysM-RLK receptors, transcription factors, and hormone signalling genes may have broader functions in the associations with microbes (Laffont et al., 2015; Moreau et al., 2013; Rey et al., 2013, 2015, 2016).
As an alternative to a forward-genetics approach, the Medicago HapMap Project (http://www.medicagohapmap.org/) has performed genome resequencing of a core collection of inbred natural accessions of M. truncatula to detect single nucleotide polymorphisms (SNPs) at very high resolution, thereby permitting genome-wide association mapping (GWAM) (Atwell et al., 2010; Branca et al., 2011; Stanton-Geddes et al., 2013; Curtin et al., 2017). HapMap accessions have already been used to map the genetic bases of flowering time and traits related to nitrogen fixation (Stanton-Geddes et al., 2013), resistance to osmotic stress (Kang et al., 2015), resistance to the root legume pathogen Aphanomyces euteiches (Bonhomme et al., 2014), as well as of the nutritional value of seeds (Le Signor et al., 2017).
Here, we used GWAM on Medicago HapMap accessions to identify new genetic components underlying the response of M. truncatula to infection with P. palmivora. We measured variation in seedling length upon infection with the oomycete. GWAM revealed a single genomic locus containing two SNPs significantly associated with P. palmivora-dependent seedling length, in the promoter region of MtSymSCL3, a GRAS transcription factor encoding a gene previously shown to be involved in regulation of root nodule nitrogen-fixing symbiosis (Kim and Nam, 2013). More recently, this locus was termed Required for Arbuscule Development 1 (RAD1) and was found to be involved in endomycorrhizal symbiosis of M. truncatula and Lotus japonicus (Park et al., 2015; Xue et al., 2015). RAD1 transcript levels were induced during colonization by P. palmivora and AM fungi. Transcript knock-down in composite plants and transposon insertion-mediated knock-out in seedlings independently confirmed that RAD1 is required for P. palmivora infection and AM symbiosis. Thus, we demonstrated that RAD1 is a host susceptibility factor contributing to the colonization of roots by unrelated filamentous symbionts and pathogens.
Materials and methods
Medicago truncatula HapMap collection and in vitro cultivation
Medicago truncatula germplasm seeds in the collection of the Medicago HapMap project (http://www.medicagohapmap.org/) were kindly provided by Dr Jean-Marie Prospéri (INRA-Montpellier). A total of 172 lines from the core collection CC192, including the A17 reference line, were phenotyped for P. palmivora resistance, whilst 190 lines were used in the uninfected condition. Each accession was phenotyped in three independent repeats with P. palmivora and two independent experiments for the uninfected condition. To set up the phenotyping assays, the seeds were gently scratched on sandpaper and left in water for 2 h before being placed on inverted agar plates for 48 h at 20 °C to allow germination. Germinated seedlings were then placed on 1% water–agarose for growth and inoculation with P. palmivora. Data for all tested seedlings and mean value for each accession are available in Supplementary dataset S1A–D at JXB online.
Medicago truncatula mutant plants
The M. truncatula R108 rad1 transposon insertion mutant NF-9554 (BC1-F4) was obtained from M. Harrison (Boyce Thompson Institute, Ithaca, NY, USA) and has previously been reported (Park et al., 2015).
Medicago truncatula composite plants
To generate composite plants with transgenic roots, A17 seeds were scarified using concentrated sulphuric acid, rinsed three times in water, sterilized with 3% bleach, and rinsed again before soaking for 2 h in sterile water. Seeds were then plated on 2% agar and placed in the dark at 4 °C for 16 h to synchronize their germination, and subsequently transferred to 20 °C in the dark for 24 h before transformation. Agrobacterium rhizogenes ARqua1 strain AR1193 (kindly provided by Allan Downie) was electroporated with binary vectors and used to generate composite plants comprising a transgenic hairy root system with non-transformed shoots and leaves (Boisson-Dernier et al., 2001). Transformed plants at 3 or 4 weeks old were selected based on the red fluorescence conferred by the T-DNA insertion, and non-transformed roots were excised prior to further experiments (see ‘Design of constructs‘).
P. palmivora inoculation assays
The strains and cultivation methods used in the current study were previously described by Rey et al. (2015). Phytophthora palmivora isolates were cultivated on V8 vegetable juice agar plates from which zoospores were released in cold water. The spore concentration was adjusted to 105 spores ml–1. For seedling inoculation assays, 10 µl droplets of P. palmivora AJ-td or Lili-td carrying a pTOR::TdTomato vector (accession nos P6390 and P16830) were placed at the root tip of M. truncatula seedlings plated on 0.8% agarose dissolved in ultrapure water. For inoculation of composite plants, transgenic roots were flooded with fluorescently labelled or Lili-YKDEL (accession no. P16830) zoospores carrying a pTOR::CALYKDEL construct (kindly provided by Howard Judelson).
Arbuscular mycorrhizal assays
Arbuscular mycorrhizal assays were conducted in sand substrate as control, which was supplemented with a proprietary preparation of AM fungi provided by Agrauxine in a concentration of 100 propagule units per plant (http://www.agrauxine.com/; France) to assess establishment of AM symbiosis (1:10 w/w). Ten germinated seedlings or composite plants were transferred to pots containing mycorrhizal substrate. Plants were grown for 6 weeks in glasshouse conditions (day temperature 26 °C, night 19 °C, 65% humidity, and 16 h of 165 W m–2 light) and watered every 2 d with Long-Ashton nutrient solution (Balzergue et al., 2011). The fertilizer phosphate concentration was 0.37 g NaH2PO4×H2O per litre. At the end of the assay, seedlings were suspended in water to separate them and remove sand. For composite plants, newly formed untransformed roots were removed prior to scoring of the extent of mycorrhizal colonization based on absence of a DsRed fluorescent signal in their tissues (see ‘Design of constructs’). Assessment of mycorrhizal colonization was performed on each plant both microscopically (Balzergue et al., 2011) and by quantification (see ‘Gene expression assays’) of M. truncatula AM markers MtBCP1 and MtPT4 (Lauressergues et al., 2012).
Phenotype modelling and association mapping
SNP data were available for 160 lines used in the P. palmivora dataset and 175 lines used in the non-infected dataset. We calculated adjusted means of root length for each M. truncatula accession by fitting a mixed linear model in which accessions had fixed effects and the biological repeat and boxes were considered as random effects. GWAM was performed on these adjusted means with the compressed mixed linear model approach and a Q+K model accounting for population structure and kinship among M. truncatula accessions in the core collection (Yu et al., 2006; Kang et al., 2010; Zhang et al., 2010), implemented in the software TASSEL (Bradbury et al., 2007), with parameters described in Bonhomme et al. (2014). A set of 5 329 189 SNPs with minor allele frequency (MAF) ≥0.05 with maximum 10% missing counts was used for GWAM. To identify significant associations, we applied a genome-wide 5% significance threshold with Bonferroni correction for the number of linkage disequilibrium (LD) blocks in the genome, leading to a significance threshold of 10–6 for the P-value, following Bonhomme et al. (2014).
Gene expression assays
RNA extraction was performed with 100 mg of pooled plant material for each biological replicate (RNeasy Plant Minikit, QIAGEN), reverse transcription of first-strand cDNA (Transcriptor First Strand cDNA Synthesis Kit, Roche), and quantitative PCR (qPCR) analyses were performed and analysed as described in Rey et al. (2015). In brief, M. truncatula stable transcripts MtH3l, MtUBQ, and MtTefa were used to standardize expression of M. truncatula RAD1 and defence genes of interest (Mtr.9569.1.S1_at, Medtr4g083710, and Medtr1g075340) in seedlings and hairy roots (Nars et al., 2013). In addition, quantitative reverse transcription–PCR (RT–qPCR) was also use to examine accumulation of P. palmivora biomass by measuring expression of the oomycete housekeeping genes Elongation factor 1α (Ef1α) and ribosomal protein S3a (WS21), which were identified by homology to Phytophthora parasitica transcripts (Yan and Liou, 2006). Expression of the in planta induced P. palmivora effector gene REX3 (Evangelisti et al., 2017) was normalized to WS21 to track its biotrophy-stage induction. The 2−ΔCp and 2−ΔΔCp methods were used to display gene expression levels (Livak and Schmittgen, 2001). Primers used in this study are listed in Supplementary Table S1.
Design of constructs
The 3′-untranslated region (UTR) of RAD1 (Medtr4g104020) was identified in the M. truncatula v 4.0 genome release and supported by RNAseq coverage (http://jcvi.org/medicago/). A 177 bp fragment of the 3'UTR was amplified from M. truncatula A17 cDNA using Phusion DNA polymerase (New England Biolab Inc., UK) with primers listed in Supplementary Table S1. This sequence was unique in the Medicago genome and did not return any blast hit other than the query. Amplicons were immediately ligated into the pENTR/D-TOPO Cloning Kit (Invitrogen) used as an entry Gateway vector, and transformations were carried out into TOP10 chemically competent Escherichia coli (Invitrogen). Minipreps were carried out using a QIAprep Spin Miniprep Kit (QIAGEN). Then recombination was performed into a pK7GWIWG2_II-Red Root vector (VIB, Ghent University, Belgium) using Gateway LR Clonase II Enzyme Mix (Invitrogen) to produce hpRAD1 binary constructs. A control recombination with the pENTR-GUS clone provided with the LR kit was performed, and the resulting vector was used as a control in the experiment and designated hpuidA.
Microscopy
Imaging of P. palmivora colonization in infected root sections was performed on excised root tissues mounted in water and covered by coverslips. Using a Leica TCS SP8 confocal microscope equipped with HyD detectors, the P. palmivora AJ-td fluorescence was visualized with the 561 nm laser for excitation and the HyD detector detecting fluorescence in a 570–600 nm window.
Quantification of P. palmivora colonization in R108 (and the corresponding rad1 mutant background) was performed on confocal images of infected seedlings at 24 h after inoculation. Confocal images were obtained at the same developmental area of each seedling radicle (2–3 mm from the radicle tip). After selecting the saturation channel, background subtraction was performed. Then, the area of pathogen was selected by creating a binary image, with black pixels representing the P. palmivora signal distribution and white pixels representing non-colonized tissue. The area of black pixels in the binary images was then quantified using ImageJ/Fiji to determine the extent of pathogen spread.
For the imaging of transgenic hairy roots, we used the P. palmivora Lili-YKDEL strain to avoid interference with the DsRed fluorescence expressed by transgenic roots. Hence, the yellow fluorescent protein (YFP) signal was detected using 514 nm excitation and 520–550 nm emission, and DsRed was detected using 561 nm excitation and 570–600 nm emission. Images represent stacks of 20–25 slices of 1 μm in maximum-intensity projections merged with inverted transillumination images to outline cells. Line averaging of 4 was applied to reduce the noise-to-signal ratio. Pictures were processed with ImageJ software v1.46 including Fiji plug-ins. Complementary light microscopy imaging was carried out using a Leica M165 FC fluorescence stereomicroscope equipped with a DFC310FX camera. The DSR filter (10447412) was used to detect the tdTomato produced by P. palmivora AJ-td. The same settings were used to screen for monomeric DsRed-expressing hairy roots of M. truncatula composite plants. For mycorrhiza assays, the percentage mycorrhization was established using the grid intersect method as described in Lauressergues et al. (2012). Root samples were boiled in 10% KOH for 7 min, washed three times in water, and ink-stained at 95 °C for 3 min (Scheaffer ink, 5% solution) before being rinsed in 0.5% acetic acid and then mounted between slides and coverslips. SEM was performed in variable pressure mode using Zeiss EVO HD15.RNA in situ hybridizations of root sections.
Probe synthesis and histochemical detection of transcripts
To generate gene-specific probes, cDNA fragments were amplified using primers 5'-CACCATGTCACCTGCACTTTATGCTAGTACC-3' and 5'-GCATTTCCAGCAAGAAA CTGCAACAATAGG-3' for RAD1, and 5'-ATGGTGAGCAAGGGCGAG-3' and 5'-CTTGTACAGC TCGTCCAT-3' for GFP (green fluorescent protein). The fragments were ligated into the pGEM-T Easy vector (Promega) and verified by sequencing. The constructs were then used as templates for in vitro transcription using the DIG RNA Labeling Kit (Roche). Medicago truncatula A17 roots were fixed in FAA (formaldehyde, acetic acid, ethanol), embedded in wax, and cut into 8 μm sections. The sections were processed according to http://www.its.caltech.edu/~plantlab/protocols/insitu.pdf and hybridized with RAD1 or GFP probes. After incubation with anti-digoxigenin antibody (Roche), the signals were detected by NBT/BCIP colour reaction (Roche).
Accession numbers
Sequence data referred to in this article can be found in the EMBL/GenBank databases under the following accession numbers: MtSymSCL3/RAD1 (MTR_4g104020), MtTefa (MTR_6g021800), MtUBQ (MTR_3g091400), MtBCP1 (MTR_7g086190), MtPT4 (MTR_1g028600), Thaumatin-PR5 (Mtr.9569.1.S1_at), Anionic-Peroxidase (MTR_4g083710), and Protease-Inhibitor (MTR_1g075340).
Results
Genome-wide association mapping of infected seedling length identifies RAD1
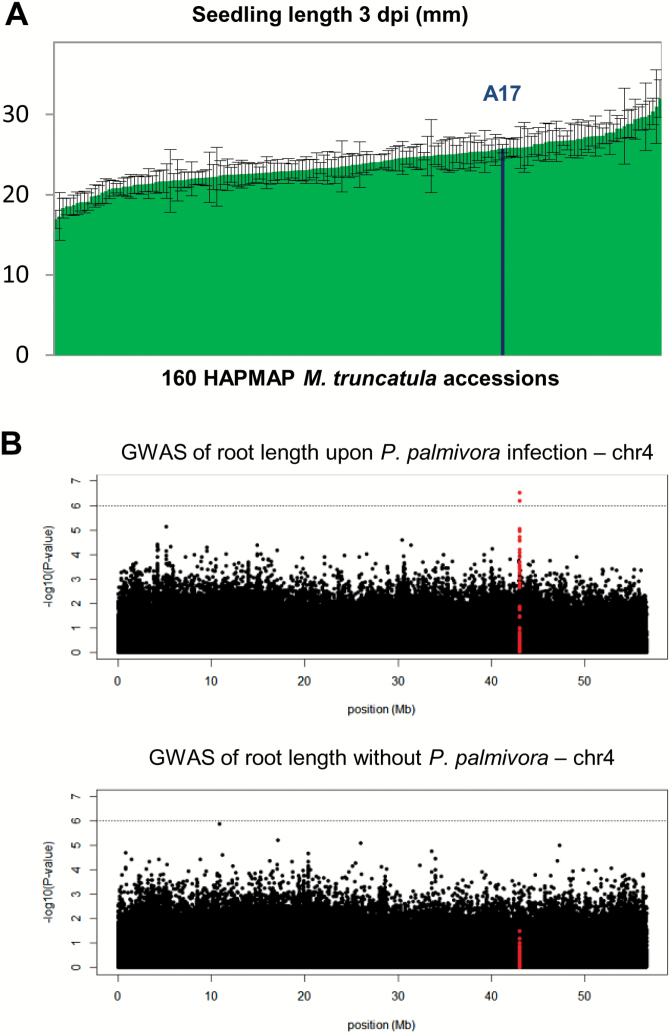
To assess the diversity of M. truncatula responses to root colonization by P. palmivora, we surveyed the collection of natural M. truncatula accessions sequenced by the Medicago HapMap project. In total, germinated seedlings of 172 lines, among which 160 had SNP data available (on average 13 seedlings per line), were infected with P. palmivora AJ-td zoospores (Supplementary dataset S1A, B). We noticed a striking variation in seedling length of the infected HapMap lines ranging from 16 mm to 32 mm at 3 days post-infection (dpi) (Fig. 1A), when first root rot symptoms can be observed, due to the shift between biotrophic and necrotrophic stages in the pathogen’s life cycle (Rey et al., 2015) (Supplementary Fig. S1).
Fig. 1.
Variation of length of seedlings infected with P. palmivora across the HapMap collection of M. truncatula accessions revealed a contribution of Medtr4g104020. (A) Variation in seedling length across all tested accessions at 3 days post-infection (dpi) with P. palmivora AJ-td. (B) Genome-wide association mapping identified a unique genomic region significantly associated (P<10–6) with seedling length upon infection of M. truncatula with P. palmivora. Manhattan plot of SNP positions on chromosome 4 and their significance values [–log10(P)] shows two candidate SNPs positioned upstream of Medtr4g104020 (SNPs in red cover 5 kb upstream plus Medtr4g104020 gene model). (This figure is available in colour at JXB online.)
To investigate the genetic components contributing to this variation in seedling length, a GWAM was carried out using genotypic data at 5 329 189 SNPs on 160 lines spanning the range of M. truncatula diversity. These SNPs, characterized by MAF ≥0.05 and maximum 10% missing counts, were analysed in a mixed linear model (Q and K matrix) to reduce false positives generated by population structure and kinship among accessions (for further details, see the Materials and methods).
Narrow-sense heritability was estimated at 0.35 for seedling length upon infection in the M. truncatula collection, and we detected a single locus in the last third of chromosome 4 (Fig. 1B) with two significant SNPs (P≤10–6) located at positions –4251 and –2467 upstream of the predicted ATG codon of RAD1, a GRAS (GAI, RGA, SCR) transcription factor-encoding gene (Medtr4g104020) (Table 1; Supplementary Fig. S2; Supplementary dataset S1E). LD analysis of the region encompassing the two SNPs showed that Medtr4g104020 is the only gene within an LD block of ~20.6 kb (Supplementary Fig. S2), suggesting that it is a strong candidate for the observed seedling length variation upon infection. Conversely, a similar GWAM on seedling length of 175 uninfected accessions (Supplementary dataset S1C, D) did not detect any significant SNPs associated with seedling length in this region (Fig. 1B). In addition, no significant SNPs reaching the –log10–6 threshold could be detected in a genome-wide analysis of uninfected seedling length (Supplementary Fig. S3). This suggests that natural variation linked to RAD1 is specifically implicated in infected seedling responses and that RAD1 is the major regulator of seedling length variation in the conditions of our screening.
Table 1.
Candidate SNPs (Mt4 genome version) associated with extent of symptoms and length of seedlings upon P. palmivora inoculation
| Chromosome | Position | P-value | Reference allele | SNP | Gene region | Gene model | Annotation |
|---|---|---|---|---|---|---|---|
| 4 | 43024916 | 6.5 × 10−7 | C | T | Non-coding | Medtr4g104020 | MtSymSCL3/RAD1 |
| 4 | 43026700 | 3.03 × 10−7 | T | C | Non-coding | Medtr4g104020 | MtSymSCL3/RAD1 |
RAD1 is expressed locally during P. palmivora root cortex colonization
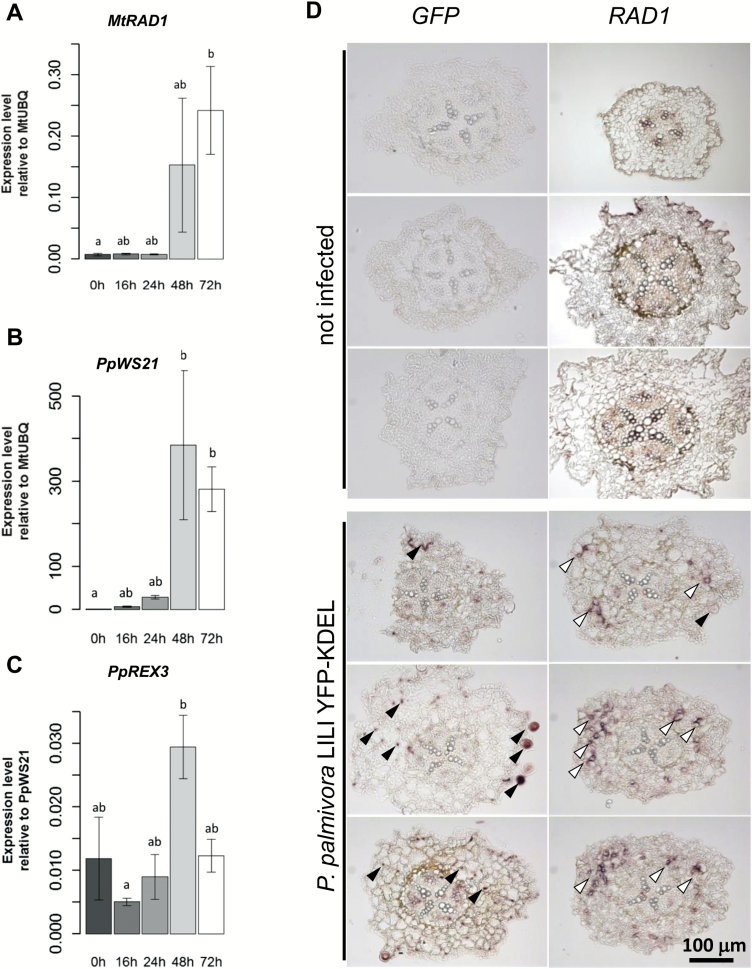
We assessed RAD1 expression levels during a time course of infection with P. palmivora LILI-YKDEL, which constitutively expresses an endoplasmic reticulum-targeted YFP, allowing for in vivo imaging of infection structures. RAD1 expression levels increased 48 h post-inoculation (hpi; Fig. 2A) when the PpWS21 housekeeping gene expression started to accumulate in host tissues (Fig. 2B), during the biotrophic infection stage where P. palmivora colonized the cortex (Supplementary Fig. S1). Furthermore, RAD1 expression induction coincided with peak expression levels of the RXLR effector gene REX3 (Evangelisti et al., 2017) (Fig. 2C) and the occurrence of haustoria (Rey et al., 2015), both indicators for the biotrophic stage of infection. We carried out in situ hybridization of M. truncatula A17 seedlings under mock conditions or upon biotrophic colonization by P. palmivora. Wax-embedded seedlings were cut and hybridized with a GFP probe detecting YFP-expressing hyphae of P. palmivora LILI-YKDEL in the root cortex as well as sporangia formed outside the root (Fig. 2D). When we hybridized similar sections from the same samples with a full-length RAD1 probe encompassing the ORF, we found localized hybridization signals resembling the pattern of P. palmivora colonization, whereas the majority of the root tissues did not show any hybridization (Fig. 2D). No strong RAD1 probe signal was observed in uninfected seedlings. From these data, we conclude that RAD1 expression is induced during biotrophic colonization by P. palmivora and restricted to a few cells.
Fig. 2.
Expression of MtRAD1 is locally induced during cortex infection with P. palmivora. (A–C) Expression levels of P. palmivora WS21 (a), M. truncatula RAD1 (b), and the P. palmivora REX3 effector candidate gene (c) at different hours post-infection (four replicates). Expression levels are calculated relative to MtUBQ (a, b) and relative to PpWS21 (c). Bars and errors bars indicate means ±SD of n=4. Comparisons made using Kruskal–Wallis and Nemenyi’s test of multiple comparisons for independent samples (Tukey). Means with different group letters are significantly different (P<0.05) for (a) and (c). and significantly different (P<0.01) for (b). (D) In situ hybridization using GFP or RAD1 probes on uninfected and infected M. truncatula A17 seedling root sections. GFP probes label expression of YFP-KDEL inside hyphae and sporangia of P. palmivora (black arrows). The RAD1 probe labels localized expression within cortex cells (white arrows).
Knock-down of RAD1 impairs colonization by arbuscular mycorrhiza fungi
To assess a requirement for RAD1 in microbial colonization, we devised a hairpin silencing construct (hpRAD1) specifically targeting only the single-copy M. truncatula A17 RAD1 gene. Nucleotide BLAST (basic local alignment sequence tool) searches against the M. truncatula genome Mt4.0v1 did not identify any other sequences with significant similarity, making off-target effects highly unlikely.
To confirm functional knock-down of RAD1, we transformed roots of M. truncatula A17 with either hpRAD1 or an hpuidA control construct using Agrobacterium rhizogenes-mediated transformation (Boisson-Dernier et al., 2001) and tested for a reduction in AM fungus colonization which was recently reported (Park et al., 2015). A constitutively co-expressed DsRed encoded by the same T-DNA served as a red fluorescent cellular marker to identify transgenic roots. We did not observe significant macroscopic alterations of root system development upon transformation (Supplementary Fig. S4).
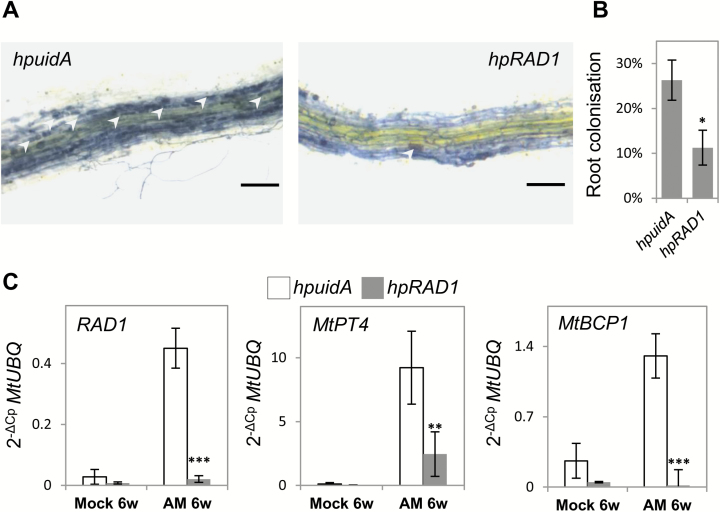
We grew composite plants carrying hpRAD1 or hpuidA for 6 weeks in a sand substrate with or without added AM fungi inoculum. After 6 weeks, transformed roots of each composite plant were subjected to ink staining to detect intraradical fungal structures. RNA extraction was carried out to validate the efficiency of RAD1 silencing and to quantify transcriptional induction of M. truncatula AM symbiosis marker genes. Using the grid intersect method (Balzergue et al., 2011), we detected 26% mycorrhizal colonization in hpuidA control roots versus 11% in hpRAD1 (Fig. 3A, B). Visual inspection of mycorrhizal structures showed a lower amount of arbuscules in hpRAD1 (Fig. 3A) at the time of harvest. RT–qPCR analysis validated a 17-fold transcriptional induction of RAD1 in AM fungus-colonized roots carrying the hpuidA control construct, and this transcriptional induction was abolished in hpRAD1 roots (Fig. 3C). To assess the amount of arbuscules formed in hpuidA and hpRAD1 roots, we quantified arbuscule-specific gene expression of the phosphate transporter MtPT4 and the blue copper-binding protein MtBCP1 (Lauressergues et al., 2012). While these genes are respectively up-regulated 76 and 5 times in hpuidA-expressing mycorrhized roots, we detected significantly lower transcript levels in hpRAD1-expressing mycorrhized roots (Fig. 3C). Thus, our RT–qPCR and visual assessment of the extent of mycorrhizal colonization in hpRAD1 roots confirmed a role for RAD1 in AM fungus colonization and also the functionality of our hairpin knock-down construct.
Fig. 3.
Expression of hpRAD1 reduces RAD1 transcript levels as well as the degree of mycorrhization by mixed arbuscular mycorrhiza (AM). (A) Ink staining of mycorrhizal structures in hpuidA and hpRAD1 hairy roots (scale bars=200 µm); white arrowheads indicate mycorrhizal arbuscules; brightness has been enhanced in both images to increase visibility of arbuscule-filled cells. (B) Quantification of the overall degree of AM fungal colonization within root systems expressing hairpin silencing constructs targeting uidA or RAD1. (C) Transcript levels of RAD1 and mycorrhizal symbiosis markers MtPT4 and MtBCP1 in roots expressing hpuidA and hpRAD1 constructs and grown in control conditions (n=3) or in AM fungi mixed inoculum (n=4). Each sample consists of five composites plants comprising at least four transformed roots, Student’s t-test was applied between constructs in each condition to compare standardized gene expression using MtUBQ as housekeeping gene and the 2−ΔCp method (**P<0.01, ***P<0.001). Error bars show the SE.
RAD1 is required for full P. palmivora colonization
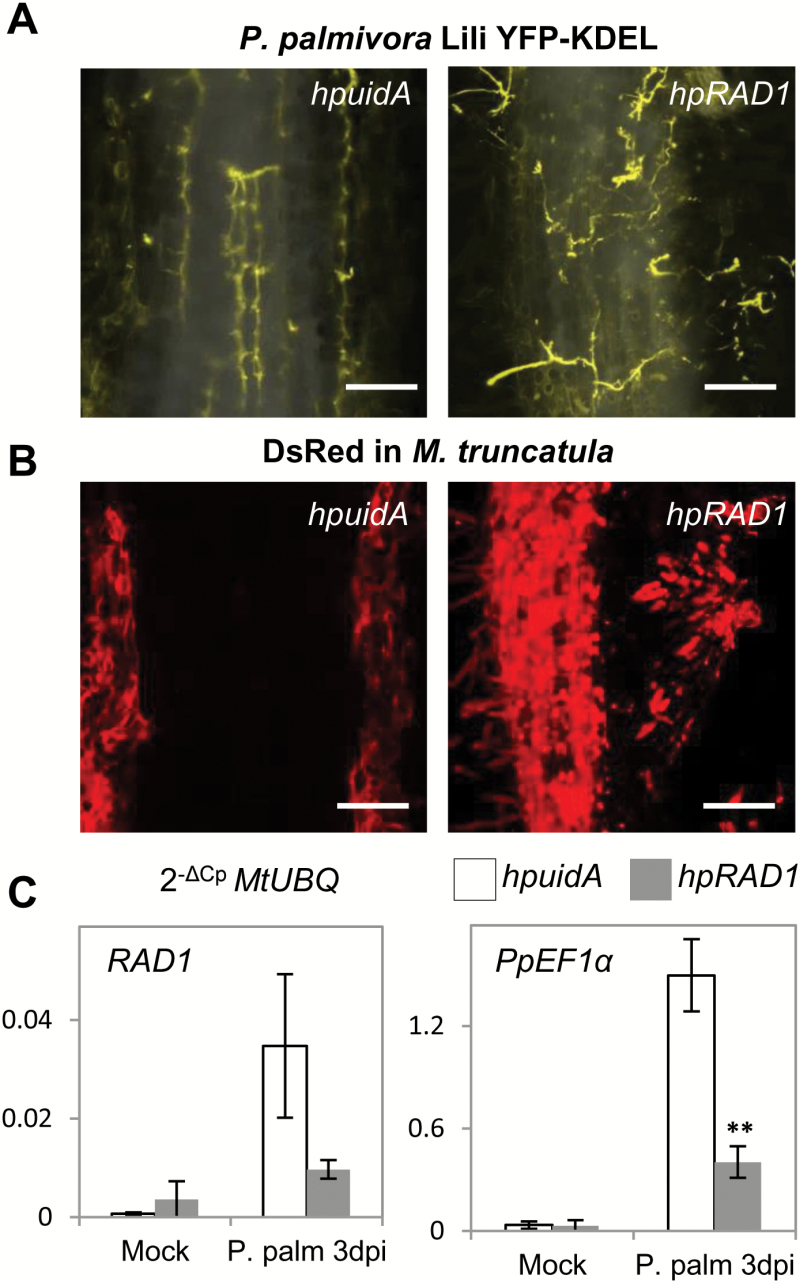
To decipher the relevance of RAD1 in the interaction with P. palmivora, we infected 6-week-old M. truncatula A17 roots expressing hpuidA or hpRAD1 with P. palmivora Lili-YKDEL (Chaparro-Garcia et al., 2011; Rey et al., 2015) spores and carried out visual, microscopic, and molecular analysis of the colonization. At 3 dpi, oomycete hyphae were spreading in cortical tissues along the hpuidA roots (Fig. 4A). Colonization was associated with a decrease in red fluorescence from DsRed-co-expressing cells harbouring the hairpin uidA constructs, indicating the start of disintegration of infected tissues (Fig. 4B). In contrast, hyphae were detected mostly at the surface of hpRAD1 roots, and the underlying tissues remained red fluorescent and viable (Fig. 4A, B). We quantified the expression of RAD1 in these roots and observed 47 times higher steady-state transcript levels in hpuidA roots infected with P. palmivora at 3 dpi compared with uninfected roots (P=0.09; Fig. 4C). By comparison, RAD1 was very weakly induced in hpRAD1-expressing roots (P=0.3), reaching only 27% of the expression level detected in colonized hpuidA roots (P=0.09; Fig. 4C). Therefore, we concluded that the hpRAD1 construct successfully restricted the P. palmivora-triggered up-regulation of RAD1 transcript levels, preventing a full transcriptional response. To quantify P. palmivora in these roots, we then measured the expression of P. palmivora Ef1α as a biomass marker and observed 75% lower levels in hpRAD1 roots versus hpuidA (P=0.008; Fig. 4C). Hence, full RAD1 transcript induction is required for optimal establishment of P. palmivora colonization. Activation of genes implicated in defence was not significantly higher in plants expressing hpRAD1, suggesting that the observed reduction in pathogen biomass in these lines is not attributable to a stronger defence response (Supplementary Fig. S5).
Fig. 4.
Medicago truncatula A17 roots expressing hpRAD1 silencing constructs are impaired in colonization by P. palmivora Lili-YKDEL. (A) Overlay of maximum projections of inverted transmitted light from rhizodermis and P. palmivora Lili-YKDEL yellow fluorescence (scale bars=200 µm). (B) Maximum projection of red fluorescence from expression of nucleocytoplasmic DsRED (scale bars=200 µm). (C) Transcript levels of RAD1 and PpEF1α in roots expressing hpuidA and hpRAD1 constructs grown in control conditions (white bars, n=3) or upon infection with P. palmivora Lili-YKDEL (grey bars, n=4). Student’s t-test was applied to compare gene expression between constructs in each condition using MtUBQ as housekeeping gene and the 2−ΔCp method (**P<0.01). Error bars show the SE.
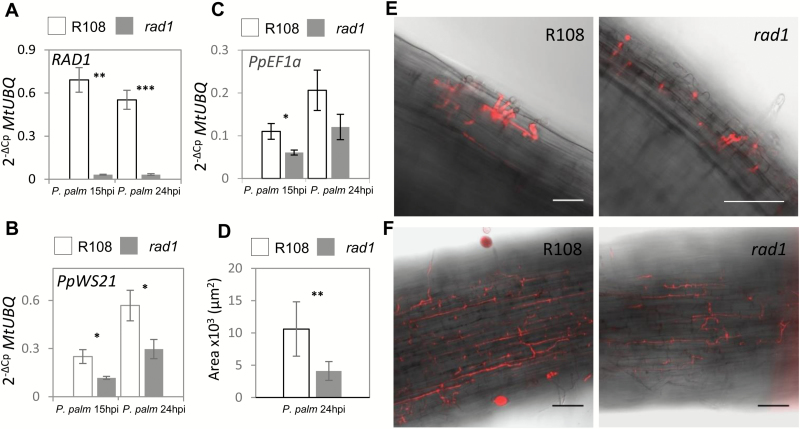
We furthermore assessed P. palmivora accumulation in infected M. truncatula rad1 mutant seedlings (Park et al., 2015) and their corresponding M. truncatula R108 wild-type genotype. Transposon-insertion rad1 lines displayed strongly reduced rad1 transcript levels compared with the wild type (Fig. 5A). Quantification of P. palmivora WS21 or EF1α transcript levels was carried out at 15 hpi and 24 hpi as the infection progresses much faster in M. truncatula R108 compared with A17 (Huisman et al., 2015). We found a significant reduction in P. palmivora biomass in rad1 mutants compared with the wild type at both time points (Fig. 5B, C). At 24 hpi, we observed significantly larger amounts of P. palmivora hyphae in infected wild-type R108 roots as compared with rad1 roots (Fig. 5D, F). At an earlier time point of 5 hpi when P. palmivora had just crossed the epidermal barrier, there was no clear difference between the mutant and wild type (Fig. 5E). This suggests that rad1 insertion lines are impaired in root cortex colonization, matching the observed cortex-based expression of RAD1 (Fig. 2). In summary, RAD1 contributes to P. palmivora colonization processes in the M. truncatula accessions A17 and R108.
Fig. 5.
The M. truncatula rad1 mutant is impaired in colonization by P. palmivora. (A) Transcript levels of RAD1 in R108 (white bars) and rad1 (grey bars) upon infection with P. palmivora Lili-Td. (B) Transcript levels of PpEF1α in R108 (white bars) and rad1 (grey bars) upon infection with P. palmivora Lili-Td. (C) Transcript levels of PpWS21 in R108 (white bars) and rad1 (grey bars) upon infection with P. palmivora Lili-Td. For each data point, n=4 biological replicates were analysed. Student’s t-test was applied to compare gene expression between lines in each condition using MtUBQ as housekeeping gene and the 2−ΔCp method. (D) P. palmivora progress at 24 hpi in seedling roots of R108 (n=13) and rad1 (n=15) measured as surface area after binary conversion (Fiji) of confocal images. (E) Confocal microscopy of M. truncatula R108 and the rad1 mutant 5 hpi with P. palmivora Lili-Td. (F) Confocal microscopy of M. truncatula R108 and the rad1 mutant 24 hpi with P. palmivora Lili-Td.(*P<0.05; **P<0.01; ***P<0.001). Error bars show the SD. Scale bars=30 µm.
Discussion
We used GWAM to identify genes contributing to seedling length upon root colonization of M. truncatula by the broad host range hemibiotrophic oomycete pathogen P. palmivora. Medicago truncatula is a model system for root–microbe interactions with a strong emphasis on symbiotic associations formed with AM fungi and nitrogen-fixing bacteria (Boisson-Dernier et al., 2001). In contrast to the AM symbiosis-incapable GWAM workhorse Arabidopsis (Atwell et al., 2010), M. truncatula enables cross-checking the role of compatibility genes identified for either AM symbiosis or pathogen colonization without the need to identify orthologous genes of the other species (Evangelisti et al., 2014; Rey and Schornack, 2013).
Here, we identified two SNPs upstream of RAD1 as significantly correlated with the length of P. palmivora-infected seedlings, but not with seedling length in uninfected conditions. Notably, RAD1 was not identified as a candidate in a GWAM study of another Medicago root-infecting oomycete, Aphanomyces euteiches (Bonhomme et al., 2014). This may suggest that RAD1 contemporary evolution contributes differentially to different root infections, and further experiments are required to test this hypothesis. Attempts to use GWAM in the detection of genetic loci under natural variation in AM have been unsuccessful (Dreher et al., 2017); therefore, the use of a biotrophic pathogen such as P. palmivora to identify common genetic traits underlying root colonization may provide inroads to identifying novel symbiosis-relevant genes.
Mutants in RAD1 have reduced, but not abolished AM fungal colonization (Park et al., 2015). Furthermore, hairpin silencing of RAD1, then named MtSymSCL3, resulted in higher nodule numbers (Kim and Nam, 2013), implicating RAD1 in two different symbiotic plant–microbe interactions and lending further support to its dual role in root development and plant–microbe interactions. In contrast to the pathogen-inducible RAD1, the constitutively expressed L. japonicus GRAS transcription factor gene NSP2 contributes to seedling root length in uninfected scenarios (Murakami et al., 2013). RAD1 transcript accumulation during P. palmivora infection was spatially restricted and similar in distribution to the root cortex-colonizing biotrophic hyphae. Furthermore, the highest RAD1 expression levels during infection coincided with those of the RXLR effector gene REX3. REX3 and other RXLR effector genes have been demonstrated previously to have peak expression levels during biotrophy (Haas et al., 2009; Evangelisti et al., 2017) and can therefore be used as markers for this developmental stage of infection. Thus, it is likely that RAD1 expression is limited to the root cortex cell in the vicinity of biotrophic P. palmivora hyphae, similar to the observed expression pattern of LjRAD1 promoter–GUS (β-glucuronidase) reporters in arbuscule-containing tissues (Park et al., 2015; Xue et al., 2015).
To study the requirement for RAD1 to achieve a full P. palmivora root infection, we employed both a silencing approach in the previously established M. truncatula A17–P. palmivora pathosystem (Rey et al., 2015) and a recently described transposon-insertion line in seedlings of the accession R108 background. Knock-down of RAD1 transcripts in M. truncatula A17 significantly reduced P. palmivora infection as well as AM fungal colonization and arbuscule formation, and the rad1 mutant has previously been characterized as being impaired in AM fungal symbiosis (Park et al., 2015). Importantly, we demonstrate a role for RAD1 in both infections of fully developed root systems of composite plants and seedling root infections. Therefore, RAD1 is required for full colonization by a filamentous pathogen as well as the unrelated beneficial AM fungi. RAD1 is unlikely to function as a negative regulator of defence since we did not observe higher defence gene expression in roots expressing hpRAD1 constructs which displayed reduced pathogen biomass (Supplementary Fig. S5).
The evolutionary history of RAD1 predicts a conservation for AM fungal symbiosis (Bravo et al., 2017). Our results demonstrate that plant genes which were hypothesized to be specifically involved in AM formation can also be exploited by pathogenic filamentous microbes, as previously demonstrated (Wang et al., 2012). It will be interesting to test further genes whose presence is evolutionarily linked with AM symbiosis for their role in plant–pathogen interactions to unravel the extent of overlap between both types of interaction outcomes.
RAD1 has previously also been implicated in the formation of intracellular arbuscules by AM fungi in L. japonicus roots (Park et al., 2015; Xue et al., 2015). We did not observe haustorium formation in hpRAD1-expressing hairy roots, but this is likely to be attributable to an overall lack of intraradical colonization. Thus, a potential role for RAD1 in haustorium formation remains to be addressed.
GRAS proteins are transcription regulators and form homo- and heterodimers to fulfil their function (Hirsch et al., 2009; Gobbato et al., 2013). RAD1 has been shown to interact with the symbiosis-associated GRAS proteins NSP1, NSP2 (Nod Signaling Pathway 1,2), RAM1 (Required for Arbuscular Mycorrhiza 1), and the GA signalling component DELLA (Park et al., 2015; Xue et al., 2015; Floss et al., 2016). Therefore, this RAD1 associated network of GRAS proteins can integrate microbe-derived signals such as lipo-chitooligosaccharides or short chitooligosaccharides (Oldroyd, 2013) as well as endogenous hormonal cues (Floss et al., 2016; Pimprikar et al., 2016). RAD1-containing GRAS complexes shape the root system, and symbiotic and immune responses, thereby controlling the outcome of interactions with beneficial and detrimental microbes, and it is therefore not unexpected that our GWAM of infected seedling lengths picked up RAD1 as a candidate gene. Nonetheless, the array of downstream targets of RAD1 in the control of seedling elongation upon P. palmivora colonization, and susceptibility to oomycete and AM colonization remains largely unknown. In mycorrhizal symbiosis, the RAM2 compatibility gene involved in lipid metabolism is under the control of RAM1 (Gobbato et al., 2013; Bravo et al., 2017). In contrast, RAM2 controls surface entry by P. palmivora in a RAM1-independent manner (Rey et al., 2015). It would therefore be relevant to characterize interactors of RAD1 such as TF80 and TF124 (Park et al., 2015) for their contribution in seedling elongation and oomycete compatibility to understand further whether and how RAD1 recruits different partners in order to direct such diverse signalling inputs.
In conclusion, we demonstrate that root interactions with a pathogen, the oomycete P. palmivora, involve a Medicago gene previously implicated specifically in root symbiosis with bacterial and filamentous microbes. Furthermore, our root length association study points to a pathogen-inducible modulation of seedling root length via RAD1. Our findings broaden the view of common principles of plant–microbe interactions and provide new options to breed for quantitative resistance against diseases caused by oomycete pathogens.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Time course of P. palmivora AJ-td infection in M. truncatula A17.
Fig. S2. Linkage disequilibrium (LD) analysis of the region encompassing RAD1.
Fig. S3. Genome-wide display of negative logarithms of the association P-value for all single nucleotide polymorphisms (SNPs).
Fig. S4. Root expression of hpRAD1 does not affect overall development of composite plants.
Fig. S5. Medicago truncatula A17 roots expressing hpRAD1 silencing constructs are not impaired in the defence response triggered by P. palmivora Lili-YKDEL.
Dataset S1. Total numbers and root length of each individual P. palmivora-infected and uninfected seedling for each accession in all repetitions and alleles present in each accession for the two SNPs significantly associated with mean root length
Table S1. List of primers used in this study, their applications, and genomic identifier of target sequences
Acknowledgements
We would like to thank Jean Marie Prospéri and Pascal Joly for providing seeds of the Medicago HAPMAP project. We are grateful to Howard Judelson and Allan Downie for provision of vectors and pathogen strains, to Maria Harrison for providing the rad1 transposon insertion line, to Edouard Evangelisti and Lara Busby for support on seedling length measurements and data analysis, and to Guillaume Chesneau for technical support. We are also grateful to three anonymous reviewers for their constructive comments. S.S. acknowledges funding by the Gatsby Charitable Foundation (GAT3273/GLD), Royal Society (UF110073), and European Research Council (ERC-2014-STG, H2020, 637537). This work was also supported by the French laboratory of Excellence project (LABEX) ‘TULIP’ (ANR-10-LABX-41).
References
- Acevedo-Garcia J, Kusch S, Panstruga R. 2014. Magical mystery tour: MLO proteins in plant immunity and beyond. New Phytologist 204, 273–281. [DOI] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ et al. 2010. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465, 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzergue C, Puech-Pagès V, Bécard G, Rochange SF. 2011. The regulation of arbuscular mycorrhizal symbiosis by phosphate in pea involves early and systemic signalling events. Journal of Experimental Botany 62, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedito VA, Torres-Jerez I, Murray JD et al. 2008. A gene expression atlas of the model legume Medicago truncatula. The Plant Journal 55, 504–513. [DOI] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Bécard G, Rosenberg C, Barker DG. 2001. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Molecular Plant-Microbe Interactions 14, 695–700. [DOI] [PubMed] [Google Scholar]
- Bonhomme M, André O, Badis Y et al. 2014. High-density genome-wide association mapping implicates an F-box encoding gene in Medicago truncatula resistance to Aphanomyces euteiches. New Phytologist 201, 1328–1342. [DOI] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. 2007. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23, 2633–2635. [DOI] [PubMed] [Google Scholar]
- Branca A, Paape TD, Zhou P, Briskine R, Farmer AD, Mudge J, Bharti AK, Woodward JE, May GD, Gentzbittel L. 2011. Whole-genome nucleotide diversity, recombination, and linkage disequilibrium in the model legume Medicago truncatula. Proceedings of the National Academy of Sciences, USA 108, E864–E870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, Brands M, Wewer V, Dörmann P, Harrison MJ. 2017. Arbuscular mycorrhiza-specific enzymes FatM and RAM2 fine-tune lipid biosynthesis to promote development of arbuscular mycorrhiza. New Phytologist 214, 1631–1645. [DOI] [PubMed] [Google Scholar]
- Bravo A, York T, Pumplin N, Mueller LA, Harrison MJ. 2016. Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nature Plants 2, 15208. [DOI] [PubMed] [Google Scholar]
- Chaparro-Garcia A, Wilkinson RC, Gimenez-Ibanez S, Findlay K, Coffey MD, Zipfel C, Rathjen JP, Kamoun S, Schornack S. 2011. The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS One 6, e16608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin SJ, Tiffin P, Guhlin J et al. 2017. Validating genome-wide association candidates controlling quantitative variation in nodulation. Plant Physiology 173, 921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ. 2013. Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher D, Yadav H, Zander S, Hause B. 2017. Is there genetic variation in mycorrhization of Medicago truncatula?PeerJ 5, e3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti E, Gogleva A, Hainaux T, Doumane M, Tulin F, Quan C, Yunusov T, Floch K, Schornack S. 2017. Time-resolved dual transcriptomics reveal early induced Nicotiana benthamiana root genes and conserved infection-promoting Phytophthora palmivora effectors. BMC Biology 15, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelisti E, Rey T, Schornack S. 2014. Cross-interference of plant development and plant–microbe interactions. Current Opinion in Plant Biology 20, 118–126. [DOI] [PubMed] [Google Scholar]
- Floss DS, Lévesque-Tremblay V, Park HJ, Harrison MJ. 2016. DELLA proteins regulate expression of a subset of AM symbiosis-induced genes in Medicago truncatula. Plant Signaling and Behavior 11, e1162369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss DS, Levy JG, Lévesque-Tremblay V, Pumplin N, Harrison MJ. 2013. DELLA proteins regulate arbuscule formation in arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Sciences, USA 110, E5025–E5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobbato E, Wang E, Higgins G, Bano SA, Henry C, Schultze M, Oldroyd GE. 2013. RAM1 and RAM2 function and expression during arbuscular mycorrhizal symbiosis and Aphanomyces euteiches colonization. Plant Signaling and Behavior 8, e26049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr C, Parniske M. 2013. Cell and developmental biology of arbuscular mycorrhiza symbiosis. Annual Review of Cell and Developmental Biology 29, 593–617. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Kamoun S, Zody MC et al. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461, 393–398. [DOI] [PubMed] [Google Scholar]
- Hirsch S, Kim J, Muñoz A,Heckmann AB, Downie JA, Oldroyd GE. 2009. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. The Plant Cell 21, 545–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman R, Bouwmeester K, Brattinga M, Govers F, Bisseling T, Limpens E. 2015. Haustorium formation in Medicago truncatula roots infected by Phytophthora palmivora does not involve the common endosymbiotic program shared by arbuscular mycorrhizal fungi and rhizobia. Molecular Plant-Microbe Interactions 28, 1271–1280. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. 2010. Variance component model to account for sample structure in genome-wide association studies. Nature Genetics 42, 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Sakiroglu M, Krom N, Stanton-Geddes J, Wang M, Lee YC, Young ND, Udvardi M. 2015. Genome-wide association of drought-related and biomass traits with HapMap SNPs in Medicago truncatula. Plant, Cell and Environment 38, 1997–2011. [DOI] [PubMed] [Google Scholar]
- Kim G-B, Nam Y-W. 2013. A novel GRAS protein gene MtSymSCL1 plays a role in regulating nodule number in Medicago truncatula. Plant Growth Regulation 71, 77–92. [Google Scholar]
- Laffont C, Rey T, André O, Novero M, Kazmierczak T, Debellé F, Bonfante P, Jacquet C, Frugier F. 2015. The CRE1 cytokinin pathway is differentially recruited depending on Medicago truncatula root environments and negatively regulates resistance to a pathogen. PLoS One 10, e0116819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauressergues D, Delaux PM, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier JP. 2012. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. The Plant Journal 72, 512–522. [DOI] [PubMed] [Google Scholar]
- Le Signor C, Aimé D, Bordat A et al. 2017. Genome-wide association studies with proteomics data reveal genes important for synthesis, transport and packaging of globulins in legume seeds. New Phytologist 214, 1597–1613. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Moreau S, Fromentin J, Vailleau F, Vernié T, Huguet S, Balzergue S, Frugier F, Gamas P, Jardinaud MF. 2014. The symbiotic transcription factor MtEFD and cytokinins are positively acting in the Medicago truncatula and Ralstonia solanacearum pathogenic interaction. New Phytologist 201, 1343–1357. [DOI] [PubMed] [Google Scholar]
- Mukhtar MS, Carvunis AR, Dreze M et al. 2011. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 333, 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y, Yokoyama H, Fukui R, Kawaguchi M. 2013. Down-regulation of NSP2 expression in developmentally young regions of Lotus japonicus roots in response to rhizobial inoculation. Plant and Cell Physiology 54, 518–527. [DOI] [PubMed] [Google Scholar]
- Nars A, Rey T, Lafitte C et al. 2013. An experimental system to study responses of Medicago truncatula roots to chitin oligomers of high degree of polymerization and other microbial elicitors. Plant Cell Reports 32, 489–502. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE. 2013. Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nature Reviews. Microbiology 11, 252–263. [DOI] [PubMed] [Google Scholar]
- Oldroyd GE, Murray JD, Poole PS, Downie JA. 2011. The rules of engagement in the legume–rhizobial symbiosis. Annual Review of Genetics 45, 119–144. [DOI] [PubMed] [Google Scholar]
- Park HJ, Floss DS, Levesque-Tremblay V, Bravo A, Harrison MJ. 2015. Hyphal branching during arbuscule development requires reduced arbuscular mycorrhiza1. Plant Physiology 169, 2774–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimprikar P, Carbonnel S, Paries M et al. 2016. A CCaMK–CYCLOPS–DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Current Biology 26, 987–998. [DOI] [PubMed] [Google Scholar]
- Pislariu CI, Murray JD, Wen J et al. 2012. A Medicago truncatula tobacco retrotransposon insertion mutant collection with defects in nodule development and symbiotic nitrogen fixation. Plant Physiology 159, 1686–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp C, Ott T. 2011. Regulation of signal transduction and bacterial infection during root nodule symbiosis. Current Opinion in Plant Biology 14, 458–467. [DOI] [PubMed] [Google Scholar]
- Rey T, Chatterjee A, Buttay M, Toulotte J, Schornack S. 2015. Medicago truncatula symbiosis mutants affected in the interaction with a biotrophic root pathogen. New Phytologist 206, 497–500. [DOI] [PubMed] [Google Scholar]
- Rey T, Laporte P, Bonhomme M, Jardinaud MF, Huguet S, Balzergue S, Dumas B, Niebel A, Jacquet C. 2016. MtNF-YA1, a central transcriptional regulator of symbiotic nodule development, is also a determinant of Medicago truncatula susceptibility toward a root pathogen. Frontiers in Plant Science 7, 1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey T, Nars A, Bonhomme M et al. 2013. NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytologist 198, 875–886. [DOI] [PubMed] [Google Scholar]
- Rey T, Schornack S. 2013. Interactions of beneficial and detrimental root-colonizing filamentous microbes with plant hosts. Genome Biology 14, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville RJ, Gosman N, Burt CJ, Makepeace J, Steed A, Corbitt M, Chandler E, Brown JK, Boulton MI, Nicholson P. 2012. The ‘Green Revolution’ dwarfing genes play a role in disease resistance in Triticum aestivum and Hordeum vulgare. Journal of Experimental Botany 63, 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton-Geddes J, Paape T, Epstein B et al. 2013. Candidate genes and genetic architecture of symbiotic and agronomic traits revealed by whole-genome, sequence-based association genetics in Medicago truncatula. PLoS One 8, e65688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbański DF, Małolepszy A, Stougaard J, Andersen SU. 2012. Genome-wide LORE1 retrotransposon mutagenesis and high-throughput insertion detection in Lotus japonicus. The Plant Journal 69, 731–741. [DOI] [PubMed] [Google Scholar]
- van Schie CC, Takken FL. 2014. Susceptibility genes 101: how to be a good host. Annual Review of Phytopathology 52, 551–581. [DOI] [PubMed] [Google Scholar]
- Verdier J, Torres-Jerez I, Wang M, Andriankaja A, Allen SN, He J, Tang Y, Murray JD, Udvardi MK. 2013. Establishment of the Lotus japonicus gene expression atlas (LjGEA) and its use to explore legume seed maturation. The Plant Journal 74, 351–362. [DOI] [PubMed] [Google Scholar]
- Wang E, Schornack S, Marsh JF, Gobbato E, Schwessinger B, Eastmond P, Schultze M, Kamoun S, Oldroyd GE. 2012. A common signaling process that promotes mycorrhizal and oomycete colonization of plants. Current Biology 22, 2242–2246. [DOI] [PubMed] [Google Scholar]
- Xue L, Cui H, Buer B, Vijayakumar V, Delaux PM, Junkermann S, Bucher M. 2015. Network of GRAS transcription factors involved in the control of arbuscule development in Lotus japonicus. Plant Physiology 167, 854–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan HZ, Liou RF. 2006. Selection of internal control genes for real-time quantitative RT-PCR assays in the oomycete plant pathogen Phytophthora parasitica. Fungal Genetics and Biology 43, 430–438. [DOI] [PubMed] [Google Scholar]
- Young ND, Debellé F, Oldroyd GE et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH et al. 2006. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nature Genetics 38, 203–208. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ et al. 2010. Mixed linear model approach adapted for genome-wide association studies. Nature Genetics 42, 355–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.