WTG1 is involved in early chloroplast development and may function in RNA editing by enhancing the function of RNA editosomes in Arabidopsis
Keywords: Arabidopsis thaliana, chloroplast, early development, MORFs, plastid gene, PPR, RNA editing, TPR, virescent
Abstract
The chloroplast is essential for plant photosynthesis and production, but the regulatory mechanism of chloroplast development is still elusive. Here, a novel gene, WHITE TO GREEN1 (WTG1), was identified to have a function in chloroplast development and plastid gene expression by screening Arabidopsis leaf coloration mutants. WTG1 encodes a chloroplast-localized tetratricopeptide repeat protein that is expressed widely in Arabidopsis cells. Disruption of WTG1 suppresses plant growth, retards leaf greening and chloroplast development, and represses photosynthetic gene expression, but complemented expression of WTG1 restored a normal phenotype. Moreover, WTG1 protein is associated with the organelle RNA editing factors MORF8 and MORF9, and RNA editing of the plastid petL-5 and ndhG-50 transcripts was affected in wtg1 mutants. These results indicate that WTG1 affects both transcriptional and posttranscriptional regulation of plastid gene expression, and provide evidence for the involvement of a tetratricopeptide repeat protein in chloroplast RNA editing in Arabidopsis.
Introduction
Chloroplast biogenesis is crucial for higher plant growth and development, on which all life ultimately depends (Waters and Langdale, 2009). The process by which the proplastids develop into functional chloroplasts is rather complex, and numerous proteins have been reported to be involved. These proteins are either nuclear-encoded or plastid-encoded, and their correct assembly and proper function require coordination of the two organelles at the transcriptional, RNA processing, translational, and post-translational levels (Pogson and Albrecht, 2011). When this coordination is undermined, the plant may exhibit severely affected phenotypes. In Arabidopsis, the nuclear-encoded sigma (sig) factors mediate plastid gene transcription directly, and chloroplast biogenesis is significantly delayed in sig2 or sig6 null mutants (Ishizaki et al., 2005, Chi et al., 2010). The plastid transcriptionally active chromosome (pTAC) 3/10/12 genes, also encoded by nuclear genes, are required for plastid transcription, and loss of any of them results in lethality without exogenous carbon sources (Pfalz et al., 2006, Yagi et al., 2012, Pfalz and Pfannschmidt, 2013). The rice nuclear gene VIRESCENT2 encodes a guanylate kinase, and the Osvir2 mutation disrupts the chloroplast translation machinery, causing a chlorotic phenotype (Iba et al., 1991, Sugimoto et al., 2004, Sugimoto et al., 2007). In maize (Zea mays), the DNA- and RNA-binding protein ZmWHY2, which is encoded by a nuclear gene, is essential for chloroplast development, and the mutant allele causes a severe phenotype of albino seedlings lacking plastid ribosomes (Prikryl et al., 2008).
Among the factors present in the chloroplast that are encoded by nuclear genes, the most notable ones are the helical repeat proteins: the pentatricopeptide repeat (PPR) and tetratricopeptide repeat (TPR) proteins (Schmitz-Linneweber and Small, 2008, Stern et al., 2010, Shikanai and Fujii, 2013). The function of PPRs has been well characterized. Chloroplast-localized PPR proteins have been demonstrated to be involved in regulating plastid RNA editing (Lurin et al., 2004, Kotera et al., 2005, Tillich et al., 2005, Okuda et al., 2006, 2007, 2009, 2010, Chateigner-Boutin et al., 2008, Hammani et al., 2009, Yu et al., 2009, Bentolila et al., 2012, Hayes et al., 2013, Yagi et al., 2013, Kindgren et al., 2015, Wagoner et al., 2015, Yap et al., 2015), RNA splicing (Schmitz-Linneweber et al., 2006, Ichinose et al., 2012), RNA processing (Fisk et al., 1999, Meierhoff et al., 2003, Hattori et al., 2007), RNA stability (Yamazaki et al., 2004, Beick et al., 2008), and translation (Williams and Barkan, 2003, Tavares-Carreón et al., 2008).
RNA editing is a posttranscriptional process that converts specific cytidines (C) to uridines (U) in mitochondria and plastids (Covello and Gray, 1989, Hiesel et al., 1989, Hoch et al., 1991). Considerable evidence has shown that PPR proteins play crucial roles in RNA editing as the sequence-specific trans-factors that recognize editing sites (Okuda et al., 2006, Okuda and Shikanai, 2012). In addition, members of the RIP (RNA-editing factor interacting protein)/MORF (multiple organellar RNA editing factor) family, the ORRM (organelle RNA recognition motif-containing) family, and the RanBP2-type zinc finger protein family, as well as one tetrapyrrole biosynthetic enzyme, have also been identified as factors involved in editing Arabidopsis plastid RNA transcripts (Takenaka et al., 2012, Sun et al., 2013, 2015, Zhang et al., 2014, Shi et al., 2015, 2016a, 2016b). In Arabidopsis, the RIP/MORF family contains 10 members, including RIP1/MORF8, which targets both plastids and mitochondria, and RIP2/MORF2 and RIP9/MORF9, which target plastids (Takenaka et al., 2012). Defects in any of these factors affect the majority of RNA editing sites in plastids (Bentolila et al., 2012; Takenaka et al., 2012).
The TPR proteins, which are similar to PPR proteins, contain tandem repeats of 34 amino acids (one amino acid fewer than the number in a PPR motif) (Hirano et al., 1990, Sikorski et al., 1990). Repeated PPR and TPR motifs form a super-helix with a central groove that bonds a target molecule (Das et al., 1998, Delannoy et al., 2007); it has been suggested that PPR domains may bond preferably to RNAs (Delannoy et al., 2007), while TPR domains may bond preferably to host proteins (Das et al., 1998, D’Andrea and Regan, 2003). TPR proteins in the plastid always play an important role in chloroplast gene expression (Hu et al., 2014), protein turnover (Park et al., 2007), photosystem assembly and repair (Park et al., 2007, Heinnickel et al., 2016), chlorophyll biosynthesis, and thylakoid membrane biogenesis (Schottkowski et al., 2009). For example, the Chlamydomonas reinhardtii CGL71 is a TPR protein integral to chloroplast thylakoid membranes, and the cgl71 mutant cannot perform normal photosynthesis (Heinnickel et al., 2016). The orthologous protein of CGL71 in Arabidopsis, Pale Yellow Green7 (PYG7), is required for photosystem I accumulation, and deletion of Pyg7 results in alterations in leaf coloration and severely reduced growth rates (Stöckel et al., 2006). SLOW-GREENING1 (SG1), a chloroplast-localized TPR protein in Arabidopsis, has been demonstrated to be involved in regulating the expression of genes associated with photosynthesis, chlorophyll biosynthesis, and chloroplast development, while its mutant displays a slow-greening phenotype (Hu et al., 2014).
Here, a novel TPR protein was identified to affect chloroplast development in Arabidopsis and designated WHITE TO GREEN1 (WTG1). T-DNA insertion mutant plants (wtg1) have an albinic and dwarf phenotype, retarded chloroplast development, disturbed expression of chloroplast-related genes, and significantly lowered editing rates at two (petL-5 and ndhG-50) of 43 plastid RNA editing sites. WTG1 did not bind the cis-elements of petL-5 and ndhG-50 sites; instead, WTG1 interacted with plastid-localized MORFs. These findings suggest that WTG1 is required for chloroplast development and may function in RNA editing in Arabidopsis by enhancing the function of RNA editosomes.
Materials and methods
All primers are listed in Supplementary Table S1 at JXB online.
Plant material and culture
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild type in this work. The T-DNA insertion alleles in WTG1 (wtg1-1, wtg1-2, and wtg1-3) were obtained from the Arabidopsis Biological Resource Center (ABRC) stock center (SALK_071495, SALK_006120, and SALK_015164, respectively). All seeds were cultivated on agar plates containing 50% Murashige and Skoog medium (Murashige, 1962) and transplanted to soil in a greenhouse at 22 °C with a 16 h/8 h light (100 μmol m−2 s−1)/dark cycle.
Genetic analysis
Total genomic DNA was isolated as described previously (Edwards et al., 1991). Gene-specific primers, together with the Lba1 primer, were used to test the T-DNA insertion lines. Homozygous plants were identified and used for the following phenotypic analyses.
RNA extraction and RT-PCR
Total RNA was extracted from tissue samples using Trizol reagent (Invitrogen) according to the manufacturer’s instructions. To synthesize cDNA, total RNA was reverse-transcribed using the primeScript 1st Strand cDNA Synthesis Kit (TaKaRa). Primer sets L2/R2 and L2/LBa1 were used to determine the abundance of the WTG1 mRNA transcript in the wild-type, wild-type/wtg1 heterozygote, and wtg1 homozygote plants.
To investigate the expression pattern of WTG1, total RNA isolated from roots, stems, leaves, inflorescences, and siliques was subjected to RT-PCR using the L1/R1 primers. UBQ5 was amplified as a control using the UBQ5F/UBQ5R primers.
To detect the expression levels of chloroplast-related genes, quantitative real-time PCR (qRT-PCR) analysis was performed. qRT-PCR amplification was carried out in a LightCycler® 480 Real-Time PCR System. Primer sets are listed in Supplementary Table S1. Relative quantification of gene expression data was performed as described by Livak and Schmittgen (2001).
RNA-seq analysis
Total RNA was isolated from leaves of wild-type, wtg1, and complemented lines at the 18- and 50-day-old stages. mRNA enriched from total RNA was fragmented and reverse-transcribed using random hexamer primers. The library was then constructed and sequenced using an Illumina HiSeq 4000 (BIOPIC-Beijing). Clean reads were aligned to the A. thaliana genome (TAIR 10.0). Levels of gene expression were calculated using the RPKM (reads per kilobase transcript per million reads) method.
The significance of differentially expressed genes (DEGs) was determined through iSeq (http://iseq.cbi.pku.edu.cn) by fold change less than 0.33 or greater than 3. Gene ontology analysis was performed by DAVID 6.7 (Huang et al., 2009).
Plasmid construction
For genetic complementation and subcellular localization
The WTG1 coding sequence (CDS) without the stop codon was PCR-amplified from wild-type cDNA with 53080cds_L1/53080link_R1 primers as a WTG1-link fragment. green fluorescent protein (GFP) was amplified from the pGreen0029-DUO1-DIPS-GFP-NOS plasmid with linkGFP_L1/linkGFP_R1 primers to yield a link-GFP fragment. Using the WTG1-link and link-GFP fragments as a template, 53080cds_L1/linkGFP_R1 primers were used to generate a WTG1-GFP fragment, which was digested with BamHI and SalI and ligated into the BamHI-SalI site of pWM101 to yield a pWM101-35S-WTG1-GFP vector. A 35S-GFP construct was used as a negative control.
For protein expression
The full-length WTG1 coding region was amplified from genomic DNA (ecotype Col-0) using BamHI-53080-F/53080-EcoRI-R primers, after which the product was cut by BamHI and EcoRI and cloned into the pGEX-4T-1 vector (GE Healthcare) within the BamHI and EcoRI sites to yield a pGEX-WTG1 vector.
For yeast two-hybrid assay
For the yeast two-hybrid assay, the CDSs of WTG1, OTP82, MORF2, MORF8, MORF9, ORRM1, OZ1, and PPO1 were amplified from wild-type cDNA using gene-specific primer sets. Full-length WTG1 was ligated into pGBKT7 (BD) to generate a pBD-WTG1 plasmid, while full-length OTP82, MORF2, MORF8, MORF9, ORRM1, OZ1, and PPO1 were cloned into pGADT7 (AD) to yield pAD-OTP82, pAD-ORF2, pAD-MORF8, pAD-MORF9, pAD-ORRM1, pAD-OZ1, and pAD-PPO1 plasmids, respectively (see Supplementary Table S1 for primer sequences and cloning sites).
For bimolecular fluorescence complementation assay
For the bimolecular fluorescence complementation (BiFC) assay, MORF8, MORF9, and WTG1 were cloned into binary BiFC vectors pSPYNE173 and pSPYCE (M) to produce MORF8/9-eYNE and WTG1-eYCE plasmids (Waadt and Kudla, 2008), respectively (see Supplementary Table S1 for primer sequences and cloning sites).
Genetic complementation
The constructed pWM101-35S-WTG1-GFP vector was introduced into Agrobacterium tumefaciens strain GV3101 by electroporation and transformed into homozygous mutants (wtg1-1/wtg1-1) by floral dipping (Clough and Bent, 1998). The homozygous wtg1-1 mutants rescued by 35S-WTG1-GFP transgenic plants were confirmed by hygromycin selection and genotyping.
Detection of chlorophyll
Total chlorophyll was determined according to the method described by Lichtenthaler and Wellburn (1983). Extracts were obtained from 50 mg of fresh tissue from 18-, 35- and 50-day-old plants and homogenized in 100 ml of 80% acetone. Spectrophotometric quantification was carried out in a U-1800 spectrophotometer (Hitachi).
Transmission electron microscopy
We harvested 50-day-old leaves from wild-type plants, wtg1 homozygotes, and complemented lines (all three lines), as well as 18-day-old albino leaves and 35-day-old pale green leaves from wtg1 mutants. Samples (approximately 1 mm2) were cut with a new blade, fixed with 4% glutaraldehyde for 4 h at room temperature, and post-fixed in 2% osmium tetroxide for 1 h at room temperature. The samples were rinsed twice in 20 mM phosphate buffer (pH 7.0) and dehydrated in 30 min steps in a graded series of ethanol concentrations (10%, 30%, 50%, 70%, 85%, 95%, and two changes of 100%). We transferred the dehydrated samples to a gradient mixture of Spurr’s embedding medium and 100% ethanol (1:3, 1:1, and 3:1 respectively, v/v; 4 h in each concentration) at room temperature. Next, the samples were transferred into pure Spurr’s medium and incubated in a mixer for 24 h at room temperature. Finally, the samples were allowed to sit at 65 °C for 20 h to complete the embedding. Ultrathin sections of the samples were cut with a diamond knife on an ultramicrotome (Leica UC7) and collected on single-mesh copper grids. The sections were stained with 2% aqueous uranyl acetate and lead citrate before being viewed using an electron microscope (FEI Tecnai G2 20).
Subcellular localization
The constructed pWM101-35S-WTG1-GFP and pWM101-35S-GFP vectors were introduced separately into A. tumefaciens strain GV3101. Wild-type plants were stably transformed with the pWM101-35S-WTG1-GFP and pWM101-35S-GFP transformants via the floral dipping method (Clough and Bent, 1998). Transgenic plants were confirmed by hygromycin selection and genotyping, after which mesophyll cells were obtained. The GFP signal was observed using a DMI 6000 B microscope (Leica).
RNA editing analysis
Total RNA was extracted from plants at specific stages and reverse-transcribed as templates.
Bulk sequencing
Thirty-four distinct RNA editing sites were amplified and sequenced by specific primers (Cai et al., 2009). RNA editing rates were estimated by the relative heights of nucleotide peaks in the analyzed sequence.
Pyrosequencing
To verify the C (unedited) to U (edited) ratios at the petL-5 and ndhG-50 sites, we performed PCR with petLpyro-F/petLpyro-R (biotinylated) and ndhGLpyro-F/ndhGpyro-R (biotinylated) primers, respectively. PCR products were converted into single-stranded DNA templates using a PyroMark Q24 Vacuum Workstation (Qiagen) according to the manufacturer’s instructions. Pyrosequencing reactions were performed in a PyroMark Q24 Advanced System (Qiagen). C:U ratios were analyzed using PyroMark Q24 Advanced Software (Qiagen).
RNA electrophoretic mobility-shift assay
RNA electrophoretic mobility-shift assay (REMSA) was performed as described previously (Lin and Xu, 2012). Briefly, to prepare the template, equimolar oligonucleotides were mixed, heat-denatured, and annealed in Taq DNA polymerase buffer (NEB). Next, the template (0.5 μM final concentration) was used for in vitro RNA transcription by T7 RNA polymerase (NEB) following the manufacturer’s instructions. The templates were digested with RNase-free DNase I (TaKaRa), after which RNA probes were purified with the RNeasy Mini Kit (Qiagen) and then incubated with WTG1 protein at 30 °C for 30 min in a 20 μl system containing 20 mM Tris-acetate (pH 7.9), 50 mM potassium acetate, 10 mM magnesium, 2.5 mM dithiothreitol, 500 μg/ml BSA, and 40 units of RNase inhibitor. To prevent non-specific binding, 500 μg/ml BSA was added to the reaction system. RNA–protein complexes were resolved on a 1.5% agarose gel and detected by GelRed staining using the methods described by Lin and Xu (2012).
RNA immunoprecipitation
35S-WTG1-GFP transgenic seedlings (18 days old) were fixed with 1% formaldehyde. Chloroplast isolation and subsequent immunoprecipitation of specific RNA–protein complexes were performed as described previously (Ketcham et al., 1984, Terzi and Simpson, 2009). Anti-GFP (AbCam) and Dynabeads® ProteinG (Thermo Fisher) beads were used for immunoprecipitation. Next, RNA was isolated and reverse-transcribed. For real-time PCR, 1 μl of cDNA was loaded as the template. The negative control consisted of the sample without an antibody.
Yeast two-hybrid assay
The pGBKT7-WTG1 plasmid was cotransformed into yeast strain AH109 with pGADT7-OTP82, pGADT7-MORF2, pGADT7-MORF8, pGADT7-MORF9, pGADT7-ORRM1, pGADT7-OZ1, or pGADT7-PPO1. Yeast transformation, screening for positive clones, and subsequent assays were performed according to the manufacturer’s instructions (Clontech). Plasmids without WTG1 or other editing factors were used as negative controls.
Bimolecular fluorescence complementation assay
Agrobacterium tumefaciens strain GV3103 was co-transformed with MORF8-eYNE and WTG1-eYCE, or MORF9-eYNE and WTG1-eYCE. The MORF2-eYNE and WTG1-eYCE combination was used as the negative control, whereas the MORF2-eYNE and MORF9-eYCE combination was used as the positive control. Each combination was introduced into A. tumefaciens strain GV3103 by electroporation. The transformants were then used to infiltrate Nicotiana benthamiana leaves as described previously (Waadt and Kudla, 2008). After 48 h, the infiltrated leaves were subjected to confocal imaging analysis using an LSM 710 NLO laser scanning confocal microscope (Zeiss).
Accession numbers
Sequence data from this report can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under accession numbers At5g53080 (WTG1) and AP000423 (Arabidopsis plastid genome).
Results
WTG1 is a nuclear single-copy gene required for normal seedling growth and pigmentation
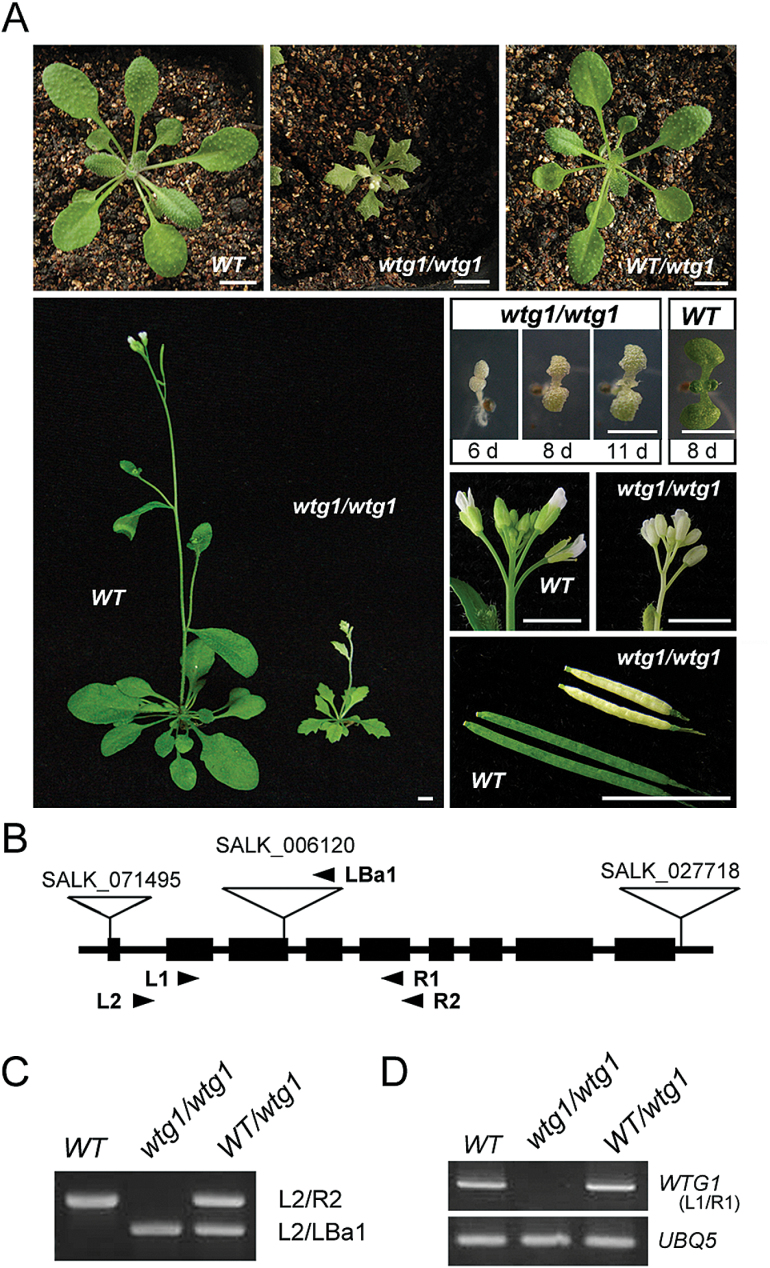
To study the mechanism of chloroplast development, we screened abnormal leaf coloration mutants from T-DNA insertion Arabidopsis lines (Alonso and Stepanova, 2003) and identified a mutant (SALK_006120) termed White-To-Green1-1 (wtg1-1). The homozygous mutants (wtg1-1/wtg1-1) germinated as dwarf and albino seedlings with serrated leaves, which, interestingly, turned green as they matured; these plants were fertile and had short siliques with a reduced seed set (Fig. 1A; Supplementary Fig. S1). Self-pollinated heterozygotes produced 607 offspring plants, among which the ratio of albino plants to green plants was 157:450 [χ2 (3:1)=0.241<χ20.85]. These results indicate that the defective leaf coloration phenotype of wtg1-1 mutants was regulated by a recessive allele and inherited by Mendelian genetic principles.
Fig. 1.
A WTG1 T-DNA insertional mutant exhibits a phenotype of dwarfism and albinism. (A) Phenotypes of wild-type (WT), wtg1 homozygotes, and heterozygotes. (B) Schematic representation of the WTG1 gene with exons shown as black rectangles and T-DNA insertions shown as triangles. The primers used for RT-PCR are indicated by arrowheads. (C) Identification of WT, wtg1 homozygotes, and wtg1/+ heterozygotes using insertion site analysis. (D) RT-PCR confirmation of wtg1 mutants. UBQ5 served as the loading control.
Specific PCR sequencing analysis indicated that the T-DNA was inserted at the end of the third exon, 804 bp downstream from the initiation codon of the At5g53080 locus (Fig. 1B, C). RT-PCR analysis showed that WTG1 transcripts were absent in wtg1-1 homologous mutants (Fig. 1D), indicating that transcription of WTG1 was disrupted by T-DNA insertion. In addition, two other T-DNA insertion lines related to WTG1, SALK_071495 (wtg1-2) and SALK_027718 (wtg1-3) (Fig. 1B), were obtained from the Arabidopsis Biological Resource Center and analyzed. The T-DNAs were inserted into the beginning of the first exon and the 3′-UTR region of the At5g53080 locus in the wtg1-2 and wtg1-3 alleles, respectively. However, expression of WTG1 was not disrupted in wtg1-2 and wtg1-3; therefore, we chose wtg1-1 for further analysis. To confirm that the abnormal phenotype of wtg1-1 homozygotes was caused by disruption of At5g53080, a genetic complementation assay was performed. The complete 1692 bp CDS (excluding the stop codon) of At5g53080 was transformed into homozygous wtg1 plants. In total, 42 T1 transgenic plants were identified; all transformants displayed normal morphology and were indistinguishable from wild-type plants (Fig. 2A). These results indicate that the WTG1 gene is At5g53080. Moreover, the 1692 bp coding region of WTG1 complemented the mutant phenotype.
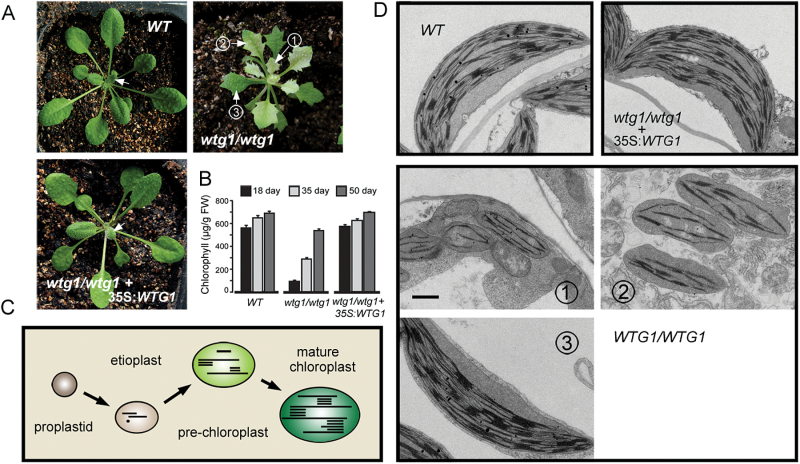
Fig. 2.
Loss of WTG1 postpones leaf greening and delays chloroplast development. (A) The wtg1 homozygotes show a delayed greening phenotype, and Pro35S:WTG1 restored the wild-type (WT) appearance. Arrows indicate the sites sampled for transmission electron microscopy. (B) Chlorophyll content in WT, wtg1, and complemented lines at different growth stages. Data are presented as mean±SD of triplicates. (C) The mode pattern of chloroplast biogenesis. (D) Transmission electron micrographs of chloroplasts from WT, wtg1, and complemented plants. Bar=1 μm.
WTG1 is required for chloroplast biogenesis
Chlorophyll content was measured as the leaves of wtg1 mutants transitioned from albino to pale green to green, representing three stages of development (Fig. 2A). Consistent with the phenotypes, the chlorophyll content of the leaves of wtg1 mutants increased as the leaves turned green, although they contained less chlorophyll than wild-type plants at each growth stage. The complemented lines possessed normal chlorophyll content (Fig. 2B).
Chloroplast biogenesis is a multistage process in which proplastids develop into fully differentiated and functional chloroplasts (Fig. 2C) (Rudowska et al., 2012, Jarvis and López-Juez, 2013). Chloroplast development was examined in wtg1 mutants using transmission electron microscopy. Albino leaves (stage 1) possessed etioplasts, whereas pale green leaves (stage 2) possessed pre-chloroplasts, and green leaves (stage 3) possessed mature chloroplasts (Fig. 2D). In contrast, mature chloroplasts were observed in the early leaves of wild-type and complemented plants (young leaves, as indicated by the arrows in Fig. 2A, were sampled) (Fig. 2D). These results indicate that chloroplast biogenesis was delayed in the wtg1 mutants. Therefore, we conclude that WTG1 is required for chloroplast biogenesis during the early stage of leaf development.
WTG1 is expressed ubiquitously in Arabidopsis and localized in chloroplasts
Expression data from Genevestigator(http://www.genevestigator.com) showed that WTG1 is widely expressed in Arabidopsis (Zimmermann et al., 2004). RT-PCR confirmed the ubiquitous expression pattern of WTG1 and showed that the greatest transcript abundance was in cauline leaves, whereas low transcript abundance was measured in siliques and roots (Supplementary Fig. S2A).
To clarify the subcellular localization of WTG1, we implemented a transgenic approach to induce wild-type Arabidopsis to express a WTG1-GFP fusion gene under the control of the 35S promoter. The fluorescent signal was localized within chloroplasts in homozygous transgenic plant cells (Supplementary Fig. S2B), indicating that WTG1 is a chloroplast protein.
WTG1 is required for plastid gene expression during early chloroplast development
Deficient chloroplast development is always associated with abnormal expression of plastid genes (Wang et al., 2016a, 2016b, Zhang et al., 2016). To analyze the effect of the wtg1 mutation on gene expression, we determined the transcription profiles of wild-type, wtg1, and complemented seedlings using RNA-seq (Supplementary Fig. S3). The results showed that, in 18-day old seedlings, 5013 genes exhibited differential expression (2463 up-regulated and 2550 down-regulated) between wtg1 and the wild type, indicating an abnormal expression pattern in wtg1 (Fig. 3A; Supplementary Data S1 and Supplementary Fig. S3). These transcriptional abnormalities were rescued by introduction of the WTG1 CDS into wtg1 mutants (Fig. 3A), suggesting that WTG1 plays a role in plastid gene expression during early chloroplast development.
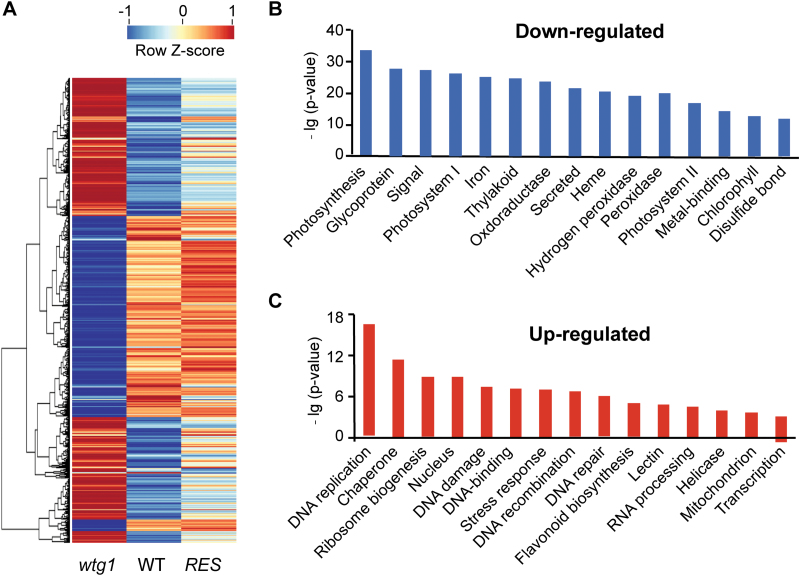
Fig. 3.
Transcriptional profiles in wild-type (WT), wtg1 and complemented/rescued (RES) seedlings obtained by RNA-seq. (A) Heatmap for differentially expressed genes (DEGs). (B) Top 15 GO terms for DEGs down-regulated in wtg1. (C) Top 15 GO terms for DEGs up-regulated in wtg1. Genes were classified by functional categories under the ontology category of biological process.
To determine whether these affected genes belonged to particular gene classes, we analyzed their gene ontology (GO) classifications in the biological process category (Fig. 3B, C). The GO enrichment results showed that the down-regulated genes were significantly enriched for terms related to photosynthesis (P=5.3e-34) and cell communications, e.g. ‘glycoprotein’ (P=6.6e-29) and ‘signal’ (P=3.8e-28), while the up-regulated DEGs were enriched for GO terms related to DNA processing and metabolism, including ‘DNA replication’ (P=3.7e-17), ‘DNA damage’ (P=4.1e-8), ‘DNA recombination’ (P=1.5e-7), and ‘DNA repair’ (P=4.5e-7).
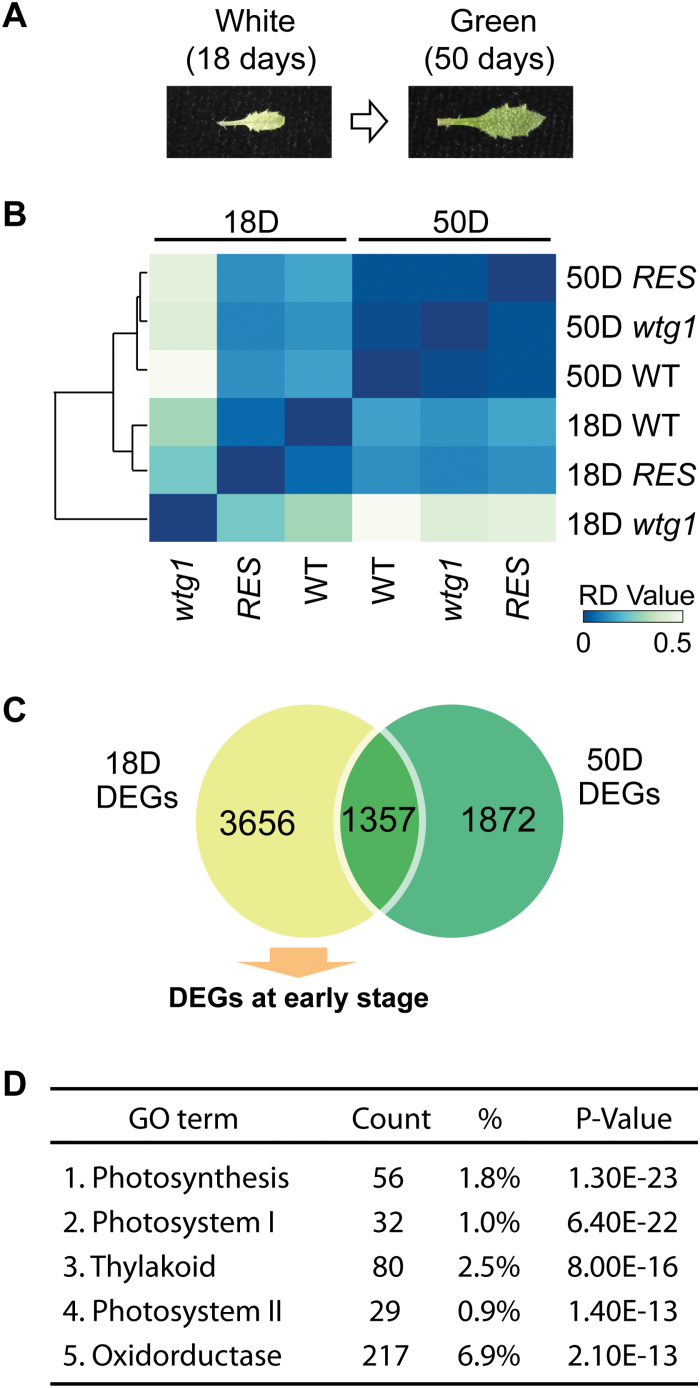
To determine whether the gene expression profiles changed as wtg1 leaves turned green, we compared the transcriptional levels in 50-day-old and 18-day-old leaves in wtg1, wild-type, and complemented seedlings by RNA-seq (Fig. 4A; Supplementary Fig. S3). Strikingly, the differences in gene expression profiles between 18-day-old wtg1 and wild-type leaves were the most significant, while the expression profiles tended to be consistent between the wild type and wtg1 in 50-day-old leaves (Fig. 4B), implying that WTG1 mainly functions during the early stage of chloroplast development. In 18-day-old wtg1 seedlings, there were 5013 DEGs compared with the wild-type seedlings (Fig. 4C), among which 1357 DEGs were overlapped with the DEGs in 50-day-old wtg1 plants. Because the chloroplasts in 50-day-old green wtg1 leaves were normal (Fig 2A, D), we hypothesized that the DEGs found only at the early stage were required for chloroplast development; thus, we performed GO enrichment analysis using the 3656 DEGs identified at the early stage (Fig. 4C, D). The results showed that the most enriched GO terms were all related to chloroplast functions, especially photosynthesis, which was consistent with our expectation (Fig. 4D).
Fig. 4.
Disruption of WTG1 causes a delayed greening phenotype and retarded developmental expression of photosystem genes. (A) wtg1 mutants show a delayed greening phenotype. (B) Clustered heatmap visualizing the similarity relationships among samples from 18-day-old and 50-day-old wtg1, wild-type (WT), and complemented/rescued (RES) plants. (C) Venn diagram showing the unique and shared relationships of differentially expressed genes (DEGs) in 18-day-old and 50-day-old plants. The DEGs that existed only in 18-day-old mutant were assigned as DEGs at the early stage. (D) List of the top five gene ontology (GO) terms for the DEGs at the early stage.
Expression of genes involved in photosynthesis is repressed in wtg1
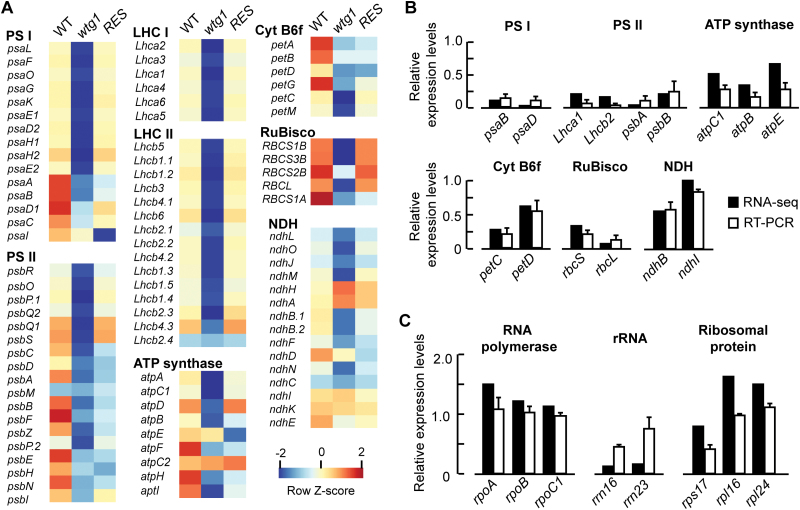
There are two classes of genes responsible for chloroplast biogenesis and metabolism: photosynthetic genes and non-photosynthetic housekeeping genes (Li and Chiu, 2010). To assess the impacts of WTG1 deficiency on the expression of these genes, we compared their transcriptional profiles among 18-day-old wtg1 mutants, wild-type, and complemented plants. As shown in Fig. 5A, many genes involved in photosynthesis were significantly repressed, including genes encoding proteins of photosystem I (PSI), photosystem II (PSII), light-harvesting complex, ATP synthase, and carbon fixation-related proteins; these results were partially verified using real-time PCR (Fig. 5B; Supplementary Fig. S4). Additionally, chlorophyll synthesis genes were down-regulated, consistent with the lower chlorophyll content in wtg1 mutants compared with wild-type plants (Fig. 2B; Supplementary Fig. S4). Furthermore, the expression levels of most of these affected genes were rescued in the complemented lines (Fig. 5A). For non-photosynthetic housekeeping genes, the effects of WTG1 mutation on gene expression varied. According to the RNA-seq and qRT-PCR results, the expression levels of plastid-encoded RNA polymerase genes (rpoA, rpoB, and rpoC1) and two ribosomal protein-encoding genes (rpl24 and rpl16) in the wtg1 mutants were almost the same as, or even higher than, those in the wild type. However, the relative expression levels of the ribosomal RNA genes rrn16 and rrn23 were significantly reduced in the mutant compared with the wild type. Expression of the ribosomal protein small subunit gene rps17 was also downregulated in the wtg1 mutants, with a transcript level of 41.61% of that in the wild type. (Fig. 5C). These results indicated that expression of the plastid genes was affected by the wtg1 mutation.
Fig. 5.
Effects of WTG1 deficiency on the expression of chloroplast-related genes. (A) Heatmap of the transcription profiles of photosynthesis-related genes in 18-day-old seedlings of wild-type (WT), wtg1 and complemented/rescued (RES) lines. Values were calculated as log2 ratio and colors are scaled per row, with red representing up-regulated genes and blue respresenting down-regulated genes. The heatmap was generated from http://iseq.cbi.pku.edu.cn. (B) qRT-PCR validation of the RNA-seq results. Fifteen genes were randomly selected to validate the changes in their expression levels obtained by RNA-seq (black bars) through qRT-PCR analysis (white bars). These genes belonged to different functional complexes involved in photosynthesis. (C) Expression levels of non-photosynthetic genes. Eight non-photosynthetic genes were selected to validate the changes in their expression levels obtained by RNA-seq (black bars) through qRT-PCR analysis (white bars). The expression relative to wild-type is set to 1; data are presented as mean±SD of triplicates in (B) and (C).
The WTG1 gene encodes a TPR protein
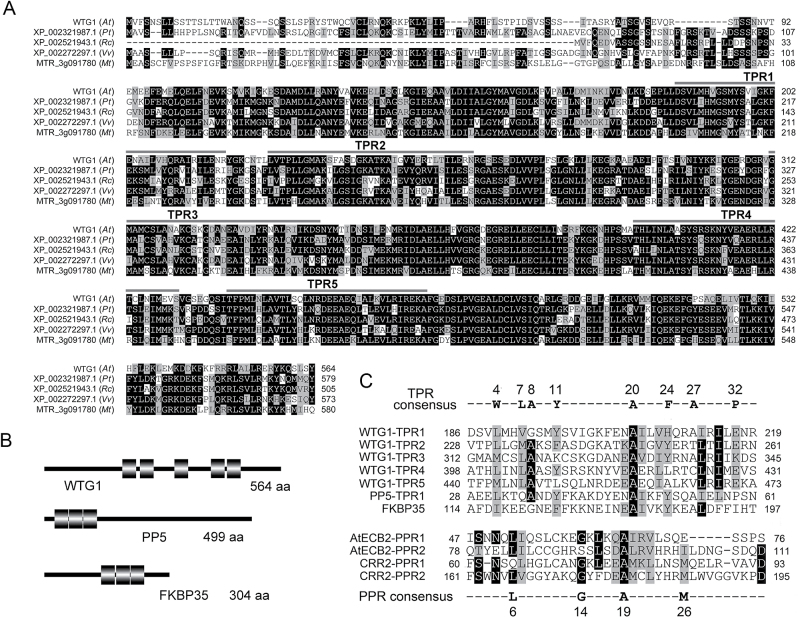
BLAST searches demonstrated that WTG1 is a single-copy nuclear gene that is conserved in flowering plants and encodes a putative polypeptide of 564 amino acids with a calculated molecular mass of 63.1 kDa (Fig. 6A). Domain analysis by PROSITE and SMART revealed that WTG1 possesses five TPR motifs (Fig. 6B), which meets the TPR protein definition criterion (i.e. containing 3–16 TPR motifs) (Blatch and Lässle, 1999).
Fig. 6.
WTG1 belongs to the TPR protein family. (A) Sequence alignment of WTG1 from Arabidopsis thaliana (At) and its homologs from Populus trichocarpa (Pt), Ricinus communis (Rc), Vitis vinifera (Vv), and Medicago truncatula (Mt). The sequences were aligned using ClustalW (Thompson et al., 1994) with the conserved residue shading mode. The TPR motifs are indicated at the top of each sequence. (B) Schematic diagrams of WTG1 and two other typical TPR proteins. Black rectangles represent TRP motifs. (C) Alignments and comparisons of TPR and PPR motifs. The upper sequence alignment consists of five TPR motifs in WTG1 and the first TPR motifs of two typical TPR proteins, PP5 and FKBP35. The reported TPR consensus is aligned at the top. The lower sequence alignment consists of two PPR motifs of the PPR proteins AtECB and CRR2. The reported PPR consensus is aligned at the bottom. The alignment was performed using ClustalW with the conserved residue shading mode.
The sequence of WTG1 was compared with those of well-characterized TPR proteins: human Ser/Thr phosphatase PP5 and plasmodium FKBP35 (Das et al., 1998, Alag et al., 2009). Using Bioedit software, we found that the five TPR coding sequences in WTG1 shared 60% similarity with the first TPR coding sequences of PP5 and FKBP35 (Fig. 6C). In particular, all five WTG1 TPR motifs contained the conserved residues typical of the TPR consensus sequence at positions 4 (W/L/F), 7 (L/I/M), 8 (G/A/S), 11 (Y/L/F), 20 (A/S/E), 24 (F/Y/L), 27 (A/S/L), and 32 (P/K/E) (Lamb et al., 1995, D’Andrea and Regan, 2003). The TPR motifs of WTG1 were then compared with the motif sequences of Arabidopsis PPR proteins AtECB2 and CRR2 by using the consensus sequence at positions 6 (L), 14 (G), 19 (A), and 26 (M) (Hashimoto et al., 2003, Yu et al., 2009) for comparisons (Fig. 6C), but revealed no similarities. These results indicate that WTG1 is a TPR protein rather than a PPR protein.
Disruption of WTG1 has an effect on RNA editing of petL and ndhG
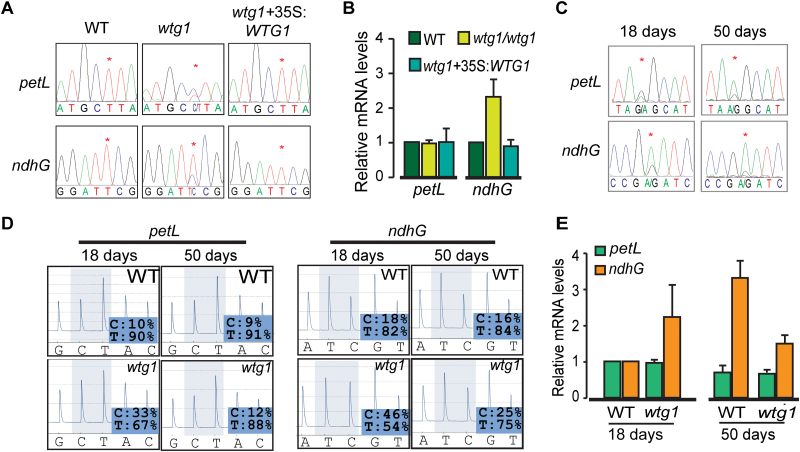
The albino phenotype with chloroplast biogenesis deficiency is reminiscent of some mutants with defective plastid RNA editing (Chateigner-Boutin et al., 2008, Yu et al., 2009, Zhou et al., 2009, Takenaka et al., 2012, Sun et al., 2015, Yap et al., 2015, Zhang et al., 2015). We therefore compared the states of chloroplast RNA editing in 18-day-old wild-type plants, wtg1 mutants, and the complemented transgenic line by bulk sequencing. Of the 34 plastid RNA editing sites reported previously (Tillich et al., 2005, Chateigner-Boutin and Small, 2007, Bentolila et al., 2012, Ruwe et al., 2013), 32 sites in the wtg1 mutants were edited in the same manner as those of the wild-type plants; however, the other two editing sites, in petL-5 and ndhG-50, exhibited remarkable differences (Fig. 7A; Supplementary Table S2). The petL-5 editing rate in the wtg1 mutants was approximately 40%, which was remarkably lower than that of the wild-type plants (nearly 100%). Similarly, the editing rate of the ndhG-50 site in the wtg1 mutants was approximately 60%, which was notably lower than that of the wild-type plants (nearly 100%). It is notable that the editing rates of both sites within the complemented transgenic line were identical to those of the wild-type plants (both nearly 100% edited). The transcript abundance of petL was unchanged in the wtg1 mutants, whereas that of ndhG-50 was increased for unknown reasons (Fig. 7B). These results indicate that WTG1 protein is required for full editing of petL-5 and ndhG-50 transcripts in early leaves.
Fig. 7.
WTG1 is required for RNA editing at petL-5 and ndhG-50 sites in plastids. (A) Loss of WTG1 disrupts editing of plastid petL-5 and ndhG-50 sites in 18-day-old seedlings as demonstrated by bulk sequencing. Asterisks indicate editable Cs. (B) Relative mRNA abundance of petL and ndhG in 18-day-old seedlings of wild-type (WT), wtg1, and wtg1 complemented with Pro35S:WTG1. Data are presented as mean±SD of triplicates. (C) Defects in editing of petL-5 and ndhG-50 sites are restored as leaves turn green. Asterisks indicate editable Cs. (D) Validation of petL-5 and ndhG-50 editing extent in WT and wtg1 mutant plants by pyrosequencing. (E) Relative mRNA abundance of petL and ndhG in WT and wtg1 plants at different growth stages. Data are presented as mean±SD of triplicates.
To detect whether these editing deficiencies could recover as albino leaves turned green, we sampled pale green (18-day-old) and green (50-day-old) leaves of wtg1 mutant plants and examined the extent of editing in RNA transcripts from these leaves. Bulk sequencing showed that the editing rates of petL-5 and ndhG-50 were mostly restored during leaf growth and greening (Fig. 7C). The editing rates of petL-5 and ndhG-50 were approximately 45% and 70%, respectively, in pale green leaves, but reached approximately 80% and 90%, respectively, in green leaves. Compared with bulk sequencing, pyrosequencing is a more sensitive and quantitative method of assessing RNA editing. Therefore, we reassessed the extent of RNA editing using pyrosequencing (Fig. 7D). The editing rate of petL-5 in pale green leaves was 67%, but this rate increased to 88% in green leaves; this editing rate was nearly equal to that of the wild-type plants (approximately 90% as determined with this method). At the ndhG-50 site in wtg1 mutants, the editing rate was 54% in pale green leaves and 75% in green leaves, while the editing rate of wild-type plants was determined to be approximately 83% using pyrosequencing. Moreover, we detected a decrease in ndhG and petL RNA abundance in the 50-day-old leaves compared with the 18-day-old leaves of wtg1 (Fig. 7E), implying that the increased editing rate in green leaves may be caused by the decrease of transcript abundance.
WTG1 may affect RNA editing through interactions with MORFs
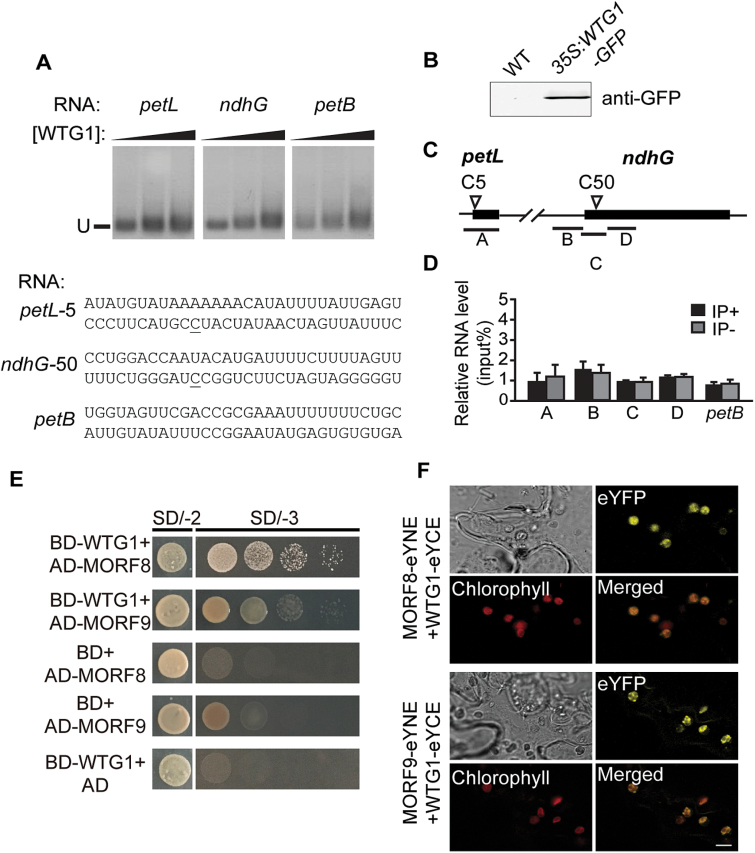
We presumed that WTG1 may participate in RNA editing through protein–protein interactions, as is typical for TPR proteins (D’Andrea and Regan, 2003), rather than by recognizing the cis-element 5′-adjacent to the target editable Cs, as is typical for PPR proteins (Chaudhuri and Maliga, 1996, Okuda et al., 2006, Hayes and Hanson, 2007). To assess this possibility, we first performed RNA binding assays with purified WTG1 protein. Gel mobility-shift assays were performed with WTG1 and synthetic RNAs of WTG1-related editing sites petL-5 and ndhG-50. A petB 5′-UTR lacking editing sites was used as the negative control (Sun et al., 2013). WTG1 exhibited no affinity for any of the three tested RNAs, showing that WTG1 does not bind to the two putative cis-elements of the petL-5 and ndhG-50 editing sites (Fig. 8A). To verify this result, RNA immunoprecipitation assays were performed (Fig. 8B–D). There was no pronounced enrichment of petL-5 and ndhG-50 in comparison with the petB 5′-UTR control in RNA samples isolated from 35S:WTG1-GFP transgenic plants, indicating that WTG1 does not specifically associate with these target sequences in the transcripts.
Fig. 8.
WTG1 has no association with petL and ndhG transcripts but interacts directly with MORF8 and MORF9. (A) REMSA showing no binding of WTG1 to the petL or ndhG transcript. The assays were performed with WTG1 protein at the indicated concentrations and the biotin-labeled RNAs shown below (edited site underlined). The petB sequence was not edited and served as a negative control. U, unbound RNA. (B) Validation of WTG1-GPF protein expression. Total protein was extracted from 18-day-old wild-type (WT) and 35S:WTG1-GFP transgenic plants, after which immunoblot analysis was performed with anti-GFP antibodies. (C) Diagram of petL and ndhG genes analyzed in the RIP assay. Regions analyzed by PCR are underlined. (D) RNA immunoprecipitation followed by a qRT-PCR assay using 35S:WTG1-GFP plants and anti-GFP antibodies. IP+, anti-GFP immunoprecipitation; IP-, mock immunoprecipitation. Data are presented as mean±SD of triplicates. (E) Yeast two-hybrid assay. AD, GAL4 activation domain; BD, GAL4 DNA binding domain. SD/-2 and SD/-3 indicate SD/-Trp-Leu and SD/-Trp-Leu-His dropout plates, respectively. Yeast colonies grown on SD/-3 plates indicate interaction between proteins. (F) Bimolecular fluorescence complementation assay showing the interactions between WTG1 and MORF8 or MORF9, which lead to the production of YFP fluorescence in chloroplasts. Chlorophyll autofluorescence is shown in red. Bar=5 μm.
Next, we employed a series of yeast two-hybrid assays to assess whether WTG1 interacts with editing factors at the petL-5 and ndhG-50 sites (Supplementary Table S3). These experiments were based on our presumption that WTG1 may participate in RNA editing through protein–protein interactions. WTG1 interacted with MORF8 and MORF9, but not with other factors known to be relevant to petL-5 and/or ndhG-50 editing, including OTP82, MORF2, ORRM1, OZ1, and PPO1 (Fig. 8E). Direct interaction between WTG1 and MORF8 and MORF9 was verified using a BiFC assay. Co-expression of the C-terminal YFP fusion of WTG1 (WTG1-eYCE) and the N-terminal YFP fusion of MORF8 (MORF8-eYNE) or MORF9 (MORF9-eYNE) reconstituted a functional YFP in chloroplasts (Fig. 8F; Supplementary Fig. S5). These results provide evidence that WTG1 may affect RNA editing through physical interactions with MORF proteins in chloroplasts.
Discussion
WTG1 is a TPR protein required for early chloroplast development
Disrupted chloroplast development usually results in abnormal leaf coloration, which severely impacts the biomass or survival of plants. Many mutants with delayed greening characteristics have been described. The dg1 mutant has very pale young leaves but greens gradually, eventually appearing similar to wild-type plants (Chi et al., 2008). In sg1 mutants, the newly formed leaves are initially albino but the wild-type phenotype is restored after 3 weeks (Hu et al., 2014). Mutation of the purine biosynthetic enzyme ATase2 of Arabidopsis results in chlorotic young leaves, which recover to be green upon maturity (Yang et al., 2015). In another recently identified mutant, dg238, the young leaves exhibit a chlorotic phenotype but this lessens as the plant develops (Wang et al., 2016a). In this study, we reported the characterization of a new mutant, wtg1, which also has a pronounced delay in greening and exhibits dwarf and serration phenotypes as well. Molecular cloning and complementation assays revealed that the phenotype of wtg1 was controlled by a recessive gene that encodes a TPR-containing protein.
The most interesting phenotype of wtg1 is the retarded greening of both its cotyledons and true leaves. The young leaves of wtg1 mutants were initially albino, and then gradually greened during development, indicating that WTG1 plays an important role in the early stages of chloroplast development. Like wtg1, all the above-mentioned mutants with delayed greening characteristics exhibit a severe chlorotic phenotype with defective chloroplasts only at the early stage of plant development; once fully grown, the chloroplasts become normal, making the plants photoautotrophic. There are two possible explanations for this phenomenon. One is that other proteins may show functional redundancy. That is, some homologous proteins or factors with similar functions may exist and partly compensate for the mutant proteins during later stages of development (Ishizaki et al., 2005, Chi et al., 2008). However, when we performed protein alignments we found no other proteins in Arabidopsis with homology to WTG1. The other possibility is that chloroplast development is an integrated outcome that depends on the balance of plastid protein accumulation and degradation. According to a previously hypothesized threshold model (Yu et al., 2005, 2008, Wang et al., 2016b), the products of plastid genes may accumulate at a faster rate than they are than degraded in the wtg1 mutant during plant growth; when the concentrations of certain proteins exceed a threshold, normal chloroplasts are produced, and the leaves turn green.
WTG1 plays an important role in the regulation of chloroplast gene expression
Chloroplast development requires the coordinated expression of genes encoded by both the nuclear and plastid genomes. Disruption of WTG1, a nuclear gene, dramatically reduced the expression levels of chloroplast-related genes (Fig. 5), indicating an important role for WTG1 in regulating chloroplast gene expression. In plastids, gene transcription depends on two types of RNA polymerases: the nuclear-encoded plastid RNA polymerase (NEP) and the plastid-encoded plastid RNA polymerase (PEP) (Maliga et al., 1988, Hajdukiewicz et al., 1997, Hedtke et al., 1997). NEP mainly transcribes non-photosynthetic housekeeping genes, while PEP specifically transcribes the photosynthetic genes. According to our RNA-seq results, the expression of most NEP- and PEP-transcribed genes was altered significantly in the wtg1 mutant (Supplementary Fig. S4), implying a distinct role of WTG1 in regulating gene expression compared with factors specifically involved in PEP transcription, such as the DG1, ATase2, and DG238 proteins (Chi et al., 2008, Yang et al., 2015, Wang et al., 2016a). In addition, we observed reduced expression of most nuclear-encoded chloroplast genes and chlorophyll biosynthesis genes (Fig. 5; Supplementary Fig. S4). It has been reported that the expression of a set of nuclear genes that encode chloroplast-localized proteins is controlled by signaling from the chloroplast via a process called retrograde signaling (Oelmuller, 1989, Surpin et al., 2002). We examined the expression levels of GUN1, GUN2 (Susek et al., 1993), EXECUTER1 and EXECUTER2 (Kim and Apel, 2013), which were proven to be involved in retrograde signaling, in wtg1 mutants. Although the expression level of EXECUTER2 was decreased, the transcript abundance of three other genes in wtg1 was equivalent to that in the wild type (Supplementary Fig. S4), suggesting that WTG1 is unlikely to be an upstream regulator in the retrograde signaling pathway. However, WTG1 may participate in the retrograde signaling process through protein–protein interactions or other unknown mechanisms. Further studies are required to clarify the mechanisms by which WTG1 regulates chloroplast development.
WTG1 may be involved in RNA editing of petL and ndhG transcripts
The extent of RNA editing can vary with RNA abundance. This can be observed in the correlation between transcript and editing levels. Reported examples include the mitochondrial gene nad3 (NADH-dehydrogenase subunit 3) in a Petunia hybrid (Lu and Hanson, 1992) and plastid genes in Nicotiana tabacum and Zea mays (Chateigner-Boutin and Hanson, 2002, Peeters and Hanson, 2002). However, the direct factor that affects RNA editing is the efficiency of the editosome, the functional editing protein complex composed of PPRs, MORFs, and other components. Experimental evidence has verified that when a component is deficient, the editing of one or several sites is impaired (Yu et al., 2005, Mach, 2009, Bentolila et al., 2012, Takenaka et al., 2012, Sun et al., 2013, 2015, Zhang et al., 2014, Shi et al., 2016b).
Our results showed that the expression profiles of plastid-encoded genes in wtg1 were largely altered compared with the wild type (Fig. 5). As described above, we cannot exclude the possible influence of RNA editing in wtg1 plants on the changes in RNA abundance. However, after careful analysis of our results, we believe that the influence of petL and ndhG RNA editing on RNA abundance in wtg1 is minor; that is, WTG1 is likely to participate in RNA editing. This is because, among the genes with greatly altered transcript abundances, such as accD, psbF, and ndhB (Fig. 5; Supplementary Fig. S4), only two sites in petL and ndhG showed a deficiency in RNA editing (Supplementary Table S2). In fact, the abundance of petL transcripts remained unchanged in wtg1 plants (Fig. 7B). More importantly, our results confirmed that WTG1 interacts with the known editing factors MORF8 and MORF9 (Fig. 8E, F). These results provide evidence that WTG1 affected RNA editing and suggest the involvement of WTG1 in the editing process. Given that WTG1 does not bind RNA (Fig. 8A–D), consistent with the notion that the TPR domain mainly mediates protein–protein interactions (Das et al., 1998, Blatch and Lässle, 1999, D’Andrea and Regan, 2003, Zeytuni and Zarivach, 2012), we propose that the involvement of WTG1 in RNA editing would be a procedure that stabilizes the editosome through interaction with MORFs. If this is true, WTG1 would add another level of complexity to the plant editosome. This would shed light on the mechanism of plant RNA editing because, to our knowledge, the involvement of a TPR protein in RNA editing has not been suggested before.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Loss of WTG1 reduces the yield trait of Arabidopsis.
Fig. S2. WTG1 is expressed ubiquitously in Arabidopsis and targeted to chloroplasts.
Fig. S3. RNA-seq analysis of wild-type and wtg1 leaves.
Fig. S4. Transcript analysis of chloroplast-associated genes in 18-day-old wtg1 by RNA-seq.
Fig. S5. Controls for the bimolecular fluorescence complementation assay.
Table S1. List of primers used in this study.
Table S2. Plastid RNA editing sites affected in the wtg1 mutant plants.
Table S3. Trans-factors involved in petL-5 and/or ndhG-50 editing.
Data S1. List of genes that are differentially expressed in wild-type and wtg1 plants.
Supplementary Material
Acknowledgements
We thank the TOLOBIO company for performing the REMSA experiments; Zhang Chao for providing the method to analyze RNA-seq data; and Professor Li Yi (Peking University) and Professor W. Sakamoto for providing the pWM101 and pGreen0029-DUO1-DIPS-GFP-NOS plasmids, respectively. This work was supported by grant 2013CB126905 from the National Basic Research Program of China (S) and grant 2014ZX0800938B from the Ministry of Agriculture of China for Transgenic Research (ZC).
References
- Alag R, Bharatham N, Dong A, Hills T, Harikishore A, Widjaja AA, Shochat SG, Hui R, Yoon HS. 2009. Crystallographic structure of the tetratricopeptide repeat domain of Plasmodium falciparum FKBP35 and its molecular interaction with Hsp90 C-terminal pentapeptide. Protein Science 18, 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN. 2003. T-DNA mutagenesis in Arabidopsis. Methods in Molecular Biology 236, 177–188. [DOI] [PubMed] [Google Scholar]
- Beick S, Schmitz-Linneweber C, Williams-Carrier R, Jensen B, Barkan A. 2008. The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Molecular and Cellular Biology 28, 5337–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Heller WP, Sun T, Babina AM, Friso G, van Wijk KJ, Hanson MR. 2012. RIP1, a member of an Arabidopsis protein family, interacts with the protein RARE1 and broadly affects RNA editing. Proceedings of the National Academy of Sciences, USA 109, E1453–E1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatch GL, Lässle M. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21, 932–939. [DOI] [PubMed] [Google Scholar]
- Cai W, Ji D, Peng L, Guo J, Ma J, Zou M, Lu C, Zhang L. 2009. LPA66 is required for editing psbF chloroplast transcripts in Arabidopsis. Plant Physiology 150, 1260–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Hanson MR. 2002. Cross-competition in transgenic chloroplasts expressing single editing sites reveals shared cis elements. Molecular and Cellular Biology 22, 8448–8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Ramos-Vega M, Guevara-García A et al. 2008. CLB19, a pentatricopeptide repeat protein required for editing of rpoA and clpP chloroplast transcripts. The Plant Journal 56, 590–602. [DOI] [PubMed] [Google Scholar]
- Chateigner-Boutin AL, Small I. 2007. A rapid high-throughput method for the detection and quantification of RNA editing based on high-resolution melting of amplicons. Nucleic Acids Research 35, e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri S, Maliga P. 1996. Sequences directing C to U editing of the plastid psbL mRNA are located within a 22 nucleotide segment spanning the editing site. The EMBO Journal 15, 5958–5964. [PMC free article] [PubMed] [Google Scholar]
- Chi W, Ma J, Zhang D, Guo J, Chen F, Lu C, Zhang L. 2008. The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiology 147, 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W, Mao J, Li Q, Ji D, Zou M, Lu C, Zhang L. 2010. Interaction of the pentatricopeptide-repeat protein DELAYED GREENING 1 with sigma factor SIG6 in the regulation of chloroplast gene expression in Arabidopsis cotyledons. The Plant Journal 64, 14–25. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Covello PS, Gray MW. 1989. RNA editing in plant mitochondria. Nature 341, 662–666. [DOI] [PubMed] [Google Scholar]
- D’Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends in Biochemical Sciences 28, 655–662. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PW, Barford D. 1998. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. The EMBO Journal 17, 1192–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delannoy E, Stanley WA, Bond CS, Small ID. 2007. Pentatricopeptide repeat (PPR) proteins as sequence-specificity factors in post-transcriptional processes in organelles. Biochemical Society Transactions 35, 1643–1647. [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C. 1991. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk DG, Walker MB, Barkan A. 1999. Molecular cloning of the maize gene crp1 reveals similarity between regulators of mitochondrial and chloroplast gene expression. The EMBO Journal 18, 2621–2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou JY, Miller LM, Hou G, Yu XH, Chen XY, Liu CJ. 2012. Acetylesterase-mediated deacetylation of pectin impairs cell elongation, pollen germination, and plant reproduction. The Plant Cell 24, 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz PT, Allison LA, Maliga P. 1997. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. The EMBO Journal 16, 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammani K, Okuda K, Tanz SK, Chateigner-Boutin AL, Shikanai T, Small I. 2009. A study of new Arabidopsis chloroplast RNA editing mutants reveals general features of editing factors and their target sites. The Plant Cell 21, 3686–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Endo T, Peltier G, Tasaka M, Shikanai T. 2003. A nucleus-encoded factor, CRR2, is essential for the expression of chloroplast ndhB in Arabidopsis. The Plant Journal 36, 541–549. [DOI] [PubMed] [Google Scholar]
- Hattori M, Miyake H, Sugita M. 2007. A pentatricopeptide repeat protein is required for RNA processing of clpP pre-mRNA in moss chloroplasts. Journal of Biological Chemistry 282, 10773–10782. [DOI] [PubMed] [Google Scholar]
- Hayes ML, Giang K, Berhane B, Mulligan RM. 2013. Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by C-terminal cytidine deaminase-like domains. Journal of Biological Chemistry 288, 36519–36529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes ML, Hanson MR. 2007. Identification of a sequence motif critical for editing of a tobacco chloroplast transcript. RNA 13, 281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A. 1997. Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science 277, 809–811. [DOI] [PubMed] [Google Scholar]
- Heinnickel M, Kim RG, Wittkopp TM, Yang W, Walters KA, Herbert SK, Grossman AR. 2016. Tetratricopeptide repeat protein protects photosystem I from oxidative disruption during assembly. Proceedings of the National Academy of Sciences, USA 113, 2774–2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiesel R, Wissinger B, Schuster W, Brennicke A. 1989. RNA editing in plant mitochondria. Science 246, 1632–1634. [DOI] [PubMed] [Google Scholar]
- Hirano T, Kinoshita N, Morikawa K, Yanagida M. 1990. Snap helix with knob and hole: essential repeats in S. pombe nuclear protein nuc2+. Cell 60, 319–328. [DOI] [PubMed] [Google Scholar]
- Hoch B, Maier RM, Appel K, Igloi GL, Kössel H. 1991. Editing of a chloroplast mRNA by creation of an initiation codon. Nature 353, 178–180. [DOI] [PubMed] [Google Scholar]
- Hu Z, Xu F, Guan L, Qian P, Liu Y, Zhang H, Huang Y, Hou S. 2014. The tetratricopeptide repeat-containing protein slow green1 is required for chloroplast development in Arabidopsis. Journal of Experimental Botany 65, 1111–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4, 44–57. [DOI] [PubMed] [Google Scholar]
- Iba K, Takamiya KI, Toh Y, Satoh H, Nishimura M. 1991. Formation of functionally active chloroplasts is determined at a limited stage of leaf development in virescent mutants of rice. Developmental Genetics 12, 342–348. [Google Scholar]
- Ichinose M, Tasaki E, Sugita C, Sugita M. 2012. A PPR-DYW protein is required for splicing of a group II intron of cox1 pre-mRNA in Physcomitrella patens. The Plant Journal 70, 271–278. [DOI] [PubMed] [Google Scholar]
- Ishizaki Y, Tsunoyama Y, Hatano K, Ando K, Kato K, Shinmyo A, Kobori M, Takeba G, Nakahira Y, Shiina T. 2005. A nuclear-encoded sigma factor, Arabidopsis SIG6, recognizes sigma-70 type chloroplast promoters and regulates early chloroplast development in cotyledons. The Plant Journal 42, 133–144. [DOI] [PubMed] [Google Scholar]
- Jarvis P, López-Juez E. 2013. Biogenesis and homeostasis of chloroplasts and other plastids. Nature Reviews. Molecular Cell Biology 14, 787–802. [DOI] [PubMed] [Google Scholar]
- Ketcham SR, Davenport JW, Warncke K, McCarty RE. 1984. Role of the gamma subunit of chloroplast coupling factor 1 in the light-dependent activation of photophosphorylation and ATPase activity by dithiothreitol. Journal of Biological Chemistry 259, 7286–7293. [PubMed] [Google Scholar]
- Kim C, Apel K. 2013. 1O2-mediated and EXECUTER-dependent retrograde plastid-to-nucleus signaling in norflurazon-treated seedlings of Arabidopsis thaliana. Molecular Plant 6, 1580–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Yap A, Bond CS, Small I. 2015. Predictable alteration of sequence recognition by RNA editing factors from Arabidopsis. The Plant Cell 27, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotera E, Tasaka M, Shikanai T. 2005. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433, 326–330. [DOI] [PubMed] [Google Scholar]
- Lamb JR, Tugendreich S, Hieter P. 1995. Tetratrico peptide repeat interactions: to TPR or not to TPR?Trends in Biochemical Sciences 20, 257–259. [DOI] [PubMed] [Google Scholar]
- Li HM, Chiu CC. 2010. Protein transport into chloroplasts. Annual Review of Plant Biology 61, 157–180. [DOI] [PubMed] [Google Scholar]
- Lichtenthaler W, Wellburn AR. 1983. Determination of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochemical Society Transactions 11, 591–592. [Google Scholar]
- Lin PC, Xu RM. 2012. Structure and assembly of the SF3a splicing factor complex of U2 snRNP. The EMBO Journal 31, 1579–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu B, Hanson MR. 1992. A single nuclear gene specifies the abundance and extent of RNA editing of a plant mitochondrial transcript. Nucleic Acids Research 20, 5699–5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S et al. 2004. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. The Plant Cell 16, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach J. 2009. Chloroplast RNA editing by pentatricopeptide repeat proteins. The Plant Cell 21, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga P, Svab Z, Harper EC, Jones JD. 1988. Improved expression of streptomycin resistance in plants due to a deletion in the streptomycin phosphotransferase coding sequence. Molecular and General Genetics 214, 456–459. [DOI] [PubMed] [Google Scholar]
- Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G. 2003. HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. The Plant Cell 15, 1480–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige S. 1962. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Oelmuller R. 1989. Photooxidative destruction of chloroplasts and its effect on nuclear gene-expression and extraplastidic enzyme levels. Photochemistry and Photobiology 49, 229–239. [Google Scholar]
- Okuda K, Chateigner-Boutin AL, Nakamura T, Delannoy E, Sugita M, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. 2009. Pentatricopeptide repeat proteins with the DYW motif have distinct molecular functions in RNA editing and RNA cleavage in Arabidopsis chloroplasts. The Plant Cell 21, 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Hammani K, Tanz SK, Peng L, Fukao Y, Myouga F, Motohashi R, Shinozaki K, Small I, Shikanai T. 2010. The pentatricopeptide repeat protein OTP82 is required for RNA editing of plastid ndhB and ndhG transcripts. The Plant Journal 61, 339–349. [DOI] [PubMed] [Google Scholar]
- Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T. 2007. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proceedings of the National Academy of Sciences, USA 104, 8178–8183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Nakamura T, Sugita M, Shimizu T, Shikanai T. 2006. A pentatricopeptide repeat protein is a site recognition factor in chloroplast RNA editing. Journal of Biological Chemistry 281, 37661–37667. [DOI] [PubMed] [Google Scholar]
- Okuda K, Shikanai T. 2012. A pentatricopeptide repeat protein acts as a site-specificity factor at multiple RNA editing sites with unrelated cis-acting elements in plastids. Nucleic Acids Research 40, 5052–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Khamai P, Garcia-Cerdan JG, Melis A. 2007. REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiology 143, 1547–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters NM, Hanson MR. 2002. Transcript abundance supercedes editing efficiency as a factor in developmental variation of chloroplast gene expression. RNA 8, 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmüller R. 2006. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. The Plant Cell 18, 176–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J, Pfannschmidt T. 2013. Essential nucleoid proteins in early chloroplast development. Trends in Plant Science 18, 186–194. [DOI] [PubMed] [Google Scholar]
- Pogson BJ, Albrecht V. 2011. Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiology 155, 1545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prikryl J, Watkins KP, Friso G, van Wijk KJ, Barkan A. 2008. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Research 36, 5152–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudowska L, Gieczewska K, Mazur R, Garstka M, Mostowska A. 2012. Chloroplast biogenesis—correlation between structure and function. Biochimica et Biophysica Acta 1817, 1380–1387. [DOI] [PubMed] [Google Scholar]
- Ruwe H, Castandet B, Schmitz-Linneweber C, Stern DB. 2013. Arabidopsis chloroplast quantitative editotype. FEBS Letters 587, 1429–1433. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Small I. 2008. Pentatricopeptide repeat proteins: a socket set for organelle gene expression. Trends in Plant Science 13, 663–670. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A. 2006. A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. The Plant Cell 18, 2650–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottkowski M, Ratke J, Oster U, Nowaczyk M, Nickelsen J. 2009. Pitt, a novel tetratricopeptide repeat protein involved in light-dependent chlorophyll biosynthesis and thylakoid membrane biogenesis in Synechocystis sp. PCC 6803. Molecular Plant 2, 1289–1297. [DOI] [PubMed] [Google Scholar]
- Shi X, Bentolila S, Hanson MR. 2016a. Organelle RNA recognition motif-containing (ORRM) proteins are plastid and mitochondrial editing factors in Arabidopsis. Plant Signaling & Behavior 11, e1167299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Germain A, Hanson MR, Bentolila S. 2016b. RNA recognition motif-containing protein ORRM4 broadly affects mitochondrial RNA editing and impacts plant development and flowering. Plant Physiology 170, 294–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X, Hanson MR, Bentolila S. 2015. Two RNA recognition motif-containing proteins are plant mitochondrial editing factors. Nucleic Acids Research 43, 3814–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai T, Fujii S. 2013. Function of PPR proteins in plastid gene expression. RNA Biology 10, 1446–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Boguski MS, Goebl M, Hieter P. 1990. A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60, 307–317. [DOI] [PubMed] [Google Scholar]
- Stern DB, Goldschmidt-Clermont M, Hanson MR. 2010. Chloroplast RNA metabolism. Annual Review of Plant Biology 61, 125–155. [DOI] [PubMed] [Google Scholar]
- Stöckel J, Bennewitz S, Hein P, Oelmüller R. 2006. The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiology 141, 870–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto H, Kusumi K, Noguchi K, Yano M, Yoshimura A, Iba K. 2007. The rice nuclear gene, VIRESCENT 2, is essential for chloroplast development and encodes a novel type of guanylate kinase targeted to plastids and mitochondria. The Plant Journal 52, 512–527. [DOI] [PubMed] [Google Scholar]
- Sugimoto H, Kusumi K, Tozawa Y, Yazaki J, Kishimoto N, Kikuchi S, Iba K. 2004. The virescent-2 mutation inhibits translation of plastid transcripts for the plastid genetic system at an early stage of chloroplast differentiation. Plant & Cell Physiology 45, 985–996. [DOI] [PubMed] [Google Scholar]
- Sun T, Germain A, Giloteaux L, Hammani K, Barkan A, Hanson MR, Bentolila S. 2013. An RNA recognition motif-containing protein is required for plastid RNA editing in Arabidopsis and maize. Proceedings of the National Academy of Sciences, USA 110, E1169–E1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Shi X, Friso G, Van Wijk K, Bentolila S, Hanson MR. 2015. A zinc finger motif-containing protein is essential for chloroplast RNA editing. PLoS Genetics 11, e1005028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M, Larkin RM, Chory J. 2002. Signal transduction between the chloroplast and the nucleus. The Plant Cell 14, S327–S338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. 1993. Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. [DOI] [PubMed] [Google Scholar]
- Takenaka M, Zehrmann A, Verbitskiy D, Kugelmann M, Hartel B, Brennicke A. 2012. Multiple organellar RNA editing factor (MORF) family proteins are required for RNA editing in mitochondria and plastids of plants. Proceedings of the National Academy of Sciences, USA 109, 5104–5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares-Carreón F, Camacho-Villasana Y, Zamudio-Ochoa A, Shingú-Vázquez M, Torres-Larios A, Pérez-Martínez X. 2008. The pentatricopeptide repeats present in Pet309 are necessary for translation but not for stability of the mitochondrial COX1 mRNA in yeast. Journal of Biological Chemistry 283, 1472–1479. [DOI] [PubMed] [Google Scholar]
- Terzi LC, Simpson GG. 2009. Arabidopsis RNA immunoprecipitation. The Plant Journal 59, 163–168. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Funk HT, Schmitz-Linneweber C, Poltnigg P, Sabater B, Martin M, Maier RM. 2005. Editing of plastid RNA in Arabidopsis thaliana ecotypes. The Plant Journal 43, 708–715. [DOI] [PubMed] [Google Scholar]
- Waadt R, Kudla J. 2008. In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protocols 2008, pdb.prot4995. [DOI] [PubMed] [Google Scholar]
- Wagoner JA, Sun T, Lin L, Hanson MR. 2015. Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. Journal of Biological Chemistry 290, 2957–2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Jiang L, Da Q, Liu J, Feng D, Wang J, Wang HB, Jin HL. 2016a. DELAYED GREENING 238, a nuclear-encoded chloroplast nucleoid protein, is involved in the regulation of early chloroplast development and plastid gene expression in Arabidopsis thaliana. Plant & Cell Physiology 57, 2586–2599. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang C, Zheng M et al. 2016b. WHITE PANICLE1, a Val-tRNA synthetase regulating chloroplast ribosome biogenesis in rice, is essential for early chloroplast development. Plant Physiology 170, 2110–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Langdale JA. 2009. The making of a chloroplast. The EMBO Journal 28, 2861–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PM, Barkan A. 2003. A chloroplast-localized PPR protein required for plastid ribosome accumulation. The Plant Journal 36, 675–686. [DOI] [PubMed] [Google Scholar]
- Yagi Y, Ishizaki Y, Nakahira Y, Tozawa Y, Shiina T. 2012. Eukaryotic-type plastid nucleoid protein pTAC3 is essential for transcription by the bacterial-type plastid RNA polymerase. Proceedings of the National Academy of Sciences, USA 109, 7541–7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi Y, Tachikawa M, Noguchi H, Satoh S, Obokata J, Nakamura T. 2013. Pentatricopeptide repeat proteins involved in plant organellar RNA editing. RNA Biology 10, 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Tasaka M, Shikanai T. 2004. PPR motifs of the nucleus-encoded factor, PGR3, function in the selective and distinct steps of chloroplast gene expression in Arabidopsis. The Plant Journal 38, 152–163. [DOI] [PubMed] [Google Scholar]
- Yang Z, Shang Z, Wang L, Lu Q, Wen X, Chi W, Zhang L, Lu C. 2015. Purine biosynthetic enzyme ATase2 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Photosynthesis Research 126, 285–300. [DOI] [PubMed] [Google Scholar]
- Yap A, Kindgren P, Colas des Francs-Small C, Kazama T, Tanz SK, Toriyama K, Small I. 2015. AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. The Plant Journal 81, 661–669. [DOI] [PubMed] [Google Scholar]
- Yu F, Liu X, Alsheikh M, Park S, Rodermel S. 2008. Mutations in SUPPRESSOR OF VARIEGATION1, a factor required for normal chloroplast translation, suppress var2-mediated leaf variegation in Arabidopsis. The Plant Cell 20, 1786–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Park S, Rodermel SR. 2005. Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiology 138, 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QB, Jiang Y, Chong K, Yang ZN. 2009. AtECB2, a pentatricopeptide repeat protein, is required for chloroplast transcript accD RNA editing and early chloroplast biogenesis in Arabidopsis thaliana. The Plant Journal 59, 1011–1023. [DOI] [PubMed] [Google Scholar]
- Zeytuni N, Zarivach R. 2012. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20, 397–405. [DOI] [PubMed] [Google Scholar]
- Zhang F, Tang W, Hedtke B, Zhong L, Liu L, Peng L, Lu C, Grimm B, Lin R. 2014. Tetrapyrrole biosynthetic enzyme protoporphyrinogen IX oxidase 1 is required for plastid RNA editing. Proceedings of the National Academy of Sciences, USA 111, 2023–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Wang C, Lin Q et al. 2015. A tetratricopeptide repeat domain-containing protein SSR1 located in mitochondria is involved in root development and auxin polar transport in Arabidopsis. The Plant Journal 83, 582–599. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Tan J, Shi Z et al. 2016. Albino Leaf1 that encodes the sole octotricopeptide repeat protein is responsible for chloroplast development. Plant Physiology 171, 1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Cheng Y, Yap A, Chateigner-Boutin AL, Delannoy E, Hammani K, Small I, Huang J. 2009. The Arabidopsis gene YS1 encoding a DYW protein is required for editing of rpoB transcripts and the rapid development of chloroplasts during early growth. The Plant Journal 58, 82–96. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.