Abstract
Humans acquire hantavirus infection by the inhalation of aerosolized excreta of infected rodent hosts. There is no treatment for hantavirus diseases at present. Therapeutic intervention during early stages of viral infection can improve the outcome of this zoonotic viral illness. The interaction between an evolutionary conserved sequence at the 5′ terminus of hantaviral genomic RNA and hantavirus nucleocapsid protein plays a critical role in the hantavirus replication cycle. This unique interaction is a novel target for therapeutic intervention of hantavirus disease. We developed a very sensitive, tractable, and cost-effective fluorescence-based assay to monitor the interaction between the nucleocapsid protein and the target RNA sequence. The assay was optimized for high-throughput screening of chemical libraries to identify molecules that interrupt this RNA–protein interaction. The assay was validated using a library of 6880 chemical compounds. This validation screen demonstrated the reproducibility and validity of required statistical criteria for high-throughput screening. The assay is ready to use for high-throughput screening of large chemical libraries to identify antihantaviral therapeutic molecules and can be amenable to similar targets in other viruses.
Keywords: hantavirus replication, nucleocapsid protein, high-throughput screen, Bunyaviridae family, negative-strand RNA viruses
Introduction
Hantaviruses are emerging negative-strand RNA viruses and members of the Bunyaviridae family. They are carried by rodents, and humans get infected by the inhalation of aerosolized excreta, such as saliva and urine droppings of infected rodent hosts. Hantavirus infections cause hemorrhagic fever with renal syndrome (HFRS) and hantavirus cardiopulmonary syndrome (HCPS) with mortality rates of 15% and 50%, respectively. Annually, 150,000–200,000 cases of hantavirus infections are reported worldwide, for which there is no treatment at present. The infected patients present nonspecific symptoms, such as fever, headache, and dizziness, to the clinic. Severely ill patients complain of shortness of breath, and most of them succumb due to complications in lung, renal, and heart function. The increased capillary leakage without physical damage to the vascular endothelium is the hallmark of hantavirus-associated diseases.1 HCPS is predominantly caused by sin nombre virus (SNV) and Andes virus (ANDV) in North and South America, respectively.
The trisegmented hantavirus genome composed of S-, M-, and L-segment RNA molecules encodes nucleocapsid protein (N protein), glycoprotein precursor (GPC), and RNA-dependent RNA polymerase (RdRp), respectively. The RdRp synthesizes viral mRNA by a unique cap-snatching mechanism during which it cleaves the host mRNA 10–14 nucleotides downstream of the 5′ cap and uses the cleaved capped RNA fragment as primer to initiate transcription. However, as a routine RNA turnover process, the host cell decapping machinery actively removes 5′ caps from cellular transcripts destined for degradation, posing a threat for hantavirus transcription initiation by controlling the availability of 5′ caps for cap snatching. We previously reported that N protein binds to the host mRNA 5′ caps and protects them from the attack of cellular decapping machinery.2 The nascent capped viral transcripts have to compete with the host cell mRNA for translation by the same translation apparatus. We previously reported that hantaviruses have evolved a unique N protein–mediated translation strategy that facilitates mRNA translation without the requirement of the eIF4F complex.3 We demonstrated that N protein–mediated translation strategy likely facilitates the translation of viral mRNA with the assistance of viral mRNA 5′ untranslated region (UTR). Thus, N protein plays a crucial role in the virus replication cycle at both the transcription and translational levels. In addition, N protein in association with the viral genome generates ribonucleocapsids that serve as templates for viral RdRp. The role of N protein in transcription and translation of viral mRNA is primarily mediated by a specific interaction between N protein and a highly conserved sequence at the 5′ UTR of the viral mRNA. This specific RNA–protein interaction is a novel target for therapeutic intervention of hantavirus disease. We developed a tractable fluorescence anisotropy– based assay to monitor the interaction between N protein and the viral UTR. The assay was validated for high-throughput screening (HTS) of chemical libraries to identify inhibitors that specifically interrupt N protein–UTR interaction. The validated assay was used to screen a large chemical library of more than 100,000 chemical compounds that resulted in the identification of a lead chemical scaffold that holds promise for the development of the first antihantaviral therapeutic.4 Here, we report the validated assay that is ready to use for screening the desired chemical libraries and can be amenable to similar targets in other viruses.
Materials and Methods
Expression and Purification of SNV N Protein
The C-terminally His-tagged SNV N protein was expressed and purified from bacteria using the plasmid pSNVN, as previously described.4 Briefly, BL21 cells were transformed with pSNVN and grown in 500 mL cultures. The bacterial cultures were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) when the optical density at 600 nm (OD600) reached 0.4. Cells were allowed to grow for 4 h at 37 °C post–IPTG induction. Cells were harvested by centrifugation at 3000 rpm for 30 min at room temperature. The cell pellets were resuspended in lysis buffer (20 mM HEPES [pH 8.0], 300 mM NaCl, 2 mM CHAPS, 8 M urea, 10 mM imidazole, and protease inhibitors) (Halt Protease Inhibitor Cocktail, Thermo Scientific, Waltham, MA). Cell lysate was centrifuged and clear supernatant was applied to Ni-NTA beads, previously equilibrated with lysis buffer. The beads were washed with lysis buffer containing increasing concentrations of 25, 50, and 100 mM imidazole. The bound protein was finally eluted with lysis buffer containing 250 mM imidazole. Purified protein was refolded by the gradual removal of urea using step dialysis in 20 mM HEPES (pH 8.0), 200 mM NaCl, 5% glycerol, and 1 mM DTT. The refolded protein was quantified and stored in 100 μL aliquots at −80 °C.
Fluorescence Polarization Anisotropy Assay Development
A 40-nucleotide-long fragment from the 5′ terminus of S-segment RNA UTR, harboring the N binding site at the 5′ terminus (5′-6FAM/GUAGUAGUAUGCUCCUUGAAAA GCAAUCAAGAAUUUACUU), was commercially synthesized. The RNA molecule was labeled with 6-carboxyfluorescien (6-FAM) at the 5′ end as previously reported.4 Interaction of the FAM-labeled RNA with purified N protein was carried out in 384-well plates for the development of the HTS assay. Each binding reaction was carried out in 40 μL of assay buffer (40 mM HEPES [pH 7.4], 80 mM NaCl, 20 mM KCl, and 1 mM DTT and RNAsin) containing the labeled RNA (3 nM) and N protein (100 nM). The reaction mixture was incubated for 90 min at 25 °C. The fluorescence polarization anisotropy (FAP) value was recorded for each sample using PerkinElmer Envision plate reader (Waltham, MA). The plate reader was calibrated for G values that generated a value of 27 mP for 1 nM fluorescein. The mP values were calculated using the equation mP = 1000* [(S − P*G)/(S + P*G)], where S = fluorescence signal parallel to excitation plane (S channel), P = fluorescence signal perpendicular to excitation plane (P channel), and G = instrument G factor. The assay was optimized for the time of incubation and DMSO sensitivity. Similar high-throughput assays have been previously developed for oncogenic mRNA binding protein IMP-1,5 Rift Valley fever virus nucleocapsid protein,6 and other protein nucleic acid interactions.7
Plate Uniformity Assay
We performed plate uniformity assays to study the signal variability between plates and between days the assays were set up, as previously reported.4 Briefly, the binding reactions in two 384-well plates were set up independently twice for 3 days. In both plates, each set of 128 wells contained binding reactions that would generate a maximum signal (max), a minimum signal (min), and a signal in the midrange (mid). The reaction mixtures containing N protein and RNA were incubated for 90 min, followed by the fluorescence polarization measurements. The means of the max, min, and mid signals, standard deviation (SD), and coefficient of variation (CV) were calculated for each plate. Both the signal windows (SW) and Z′ factor (Z′) were computed for each plate over a period of 3 days.
Compound Libraries
The following three compound collections were used to optimize the assay for screening the chemical libraries in high-throughput mode: (1) Micro-Source Spectrum (2320 compounds containing Food and Drug Administration (FDA)–approved drugs, bioactives, and natural products; www.msdiscovery.com), (2) Prestwick Chemical Library (1200 compounds; Prestwick Chemical, Washington, DC), and (3) the University of Kansas, Center of Excellence in Chemical Methodologies & Library Development (KU-CMLD; 3360 compounds with novel diverse scaffolds).
Results and Discussion
Targeting the N Protein–UTR Interaction for Therapeutic Intervention
Hantaviruses contain highly conserved noncoding sequences at the 5′ and 3′ termini of their genome. The sequences are partially complementary and undergo base pairing to form a panhandle hairpin structure (Fig. 1). We have previously reported that trimeric N protein specifically binds to the panhandle and the interaction is mediated by the terminal conserved nine nucleotides of the panhandle. After specific binding, the RNA helicase activity of N protein unwinds the panhandle and N protein remains associated with the unwound 5′ terminus (Fig. 1).8 We suggested that panhandle unwinding and association of N protein with the 5′ terminus creates an opportunity for viral RdRp to initiate transcription at the accessible 3′ terminus of the viral genome. This is consistent with the observations from other investigators that the panhandle functions as a promoter for the Bunyaviridae RdRp. In addition, we recently reported that N protein directly binds to the C-terminal domain of the RdRp. The N protein–RdRp interaction likely plays a role in the recruitment of RdRp at the promoter. However, other investigators have suggested that interaction of hantavirus N protein with the 5′ UTR is required for selective encapsidation of vRNA into viral nucloecapsids.9 The nucleocapsids are required for the packaging of the viral genome into new virions during the replication cycle. In addition, nucleocapsids are used as templates by the RdRp for the transcription and replication of the viral genome.
Figure 1.
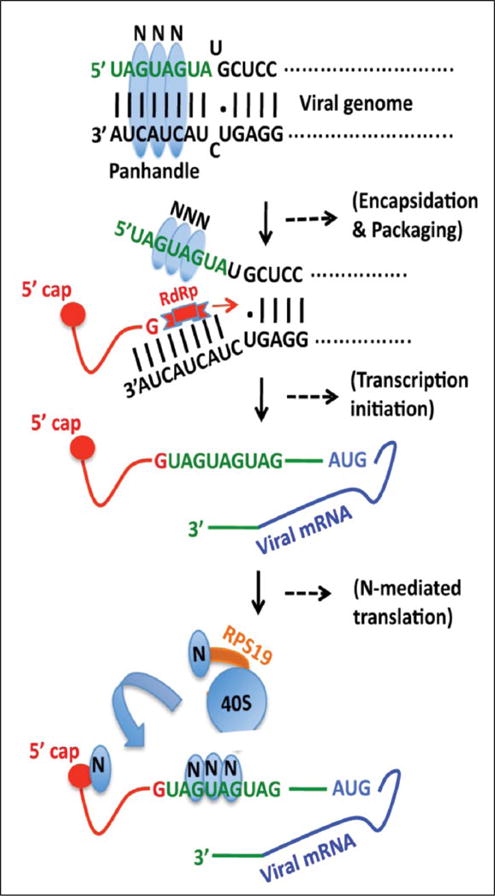
Schematic representation showing the role of N protein in the hantavirus replication cycle. The trimeric hantavirus nucleocapsid protein (blue) binds to the panhandle structure, formed by the base pairing of complementary nucleotides at the 5′ and 3′ termini of viral S-segment RNA. The nucleotides in green are highly conserved in both vRNA and mRNA. The RNA helix unwinding activity of N protein unwinds the panhandle and N protein remains bound to the unwound 5′ terminus, creating an opportunity for the RdRp (red) to initiate transcription at the 3′ end. The RdRp uses a capped RNA primer having a 5′ “G” residue (red line) to initiate transcription. The binding of N protein to the vRNA 5′ UTR has been reported to play a role in the selective encapsidation of vRNA into ribonucleocapsids. Hantaviral mRNAs are capped, contain 5′ and 3′ UTRs (green), and lack the 3′ poly (A) tail. The capped RNA primer remains attached to the 5′ terminus of the viral mRNA (red). Hantaviral mRNAs are preferentially translated by the N protein–mediated translation strategy, during which a monomeric and a trimeric N protein molecule bind to the mRNA 5′ cap and heptanucleotide region GUAGUAG of the viral mRNA 5′ UTR, respectively. Another N protein molecule binds to the ribosomal protein S19 (RPS19) (orange). N protein–associated 40S ribosomal subunits are efficiently loaded on viral mRNA 5′ UTR by an unknown mechanism.
The hantavirus mRNAs contain a viral encoded 5′ UTR of 35–52 nucleotides, which is further elongated due to the attachment of a caped RNA primer at the 5′ terminus during the cap-snatching mechanism of transcription initiation (Fig. 1). The viral encoded UTR contains a highly conserved triplet repeat sequence (UAGUAGUAG) at the 5′ terminus (Fig. 1). Hantaviruses have evolved a unique N protein–mediated translation mechanism that facilitates mRNA translation without the requirement of a canonical eIF4F cap binding complex. Our published results suggest that the N protein–mediated translation strategy likely favors the translation of viral transcripts in virus-infected cells to avoid the competition from host cell mRNAs for the same translation machinery. N protein binds to the 40S ribosomal subunit via the ribosomal protein S19 (RPS19). In addition, N protein also binds to both the viral mRNA 5′ cap and the heptanucleotide region GUAGUAG of the triplet repeat sequence at the viral mRNA 5′ UTR (Fig. 1). Our published data suggest that the specific binding of N protein to both the RPS19 and the heptanucleotide region aids the preferential engagement of N protein–associated ribosomes with the viral mRNA 5′ UTR by an unknown mechanism. Mutations in the heptanucleotide region abrogated the N protein–UTR interaction, which impacted the preferential translation of the downstream open reading frame (ORF). The triplet repeat sequence at the 5′ terminus of both vRNA and mRNA is highly conserved (Fig. 1). These observations have demonstrated that interaction of N protein with this region plays a critical role in the encapsidation of the viral genome, transcription initiation in conjunction with RdRp, and preferential translation of viral mRNA (Fig. 1). Thus, the interruption in this interaction will inhibit virus replication by the abrogation of viral transcription, encapsidation of vRNA, and translation of viral mRNA by the N protein– mediated translation strategy.
Development of FPA-Based Assay to Monitor the Interaction between N Protein and Viral UTR
A 40-nucleotide-long viral UTR sequence harboring the heptanucletide region at the 5′ terminus was commercially synthesized and labeled with a fluorescent moiety 6-FAM at the 5′ terminus, as mentioned in Materials and Methods. To confirm the N protein–UTR interaction, the fluorescent RNA was serially diluted from 800 to 1.5 nM. A fixed concentration of the fluorescent RNA was incubated with increasing input concentrations of bacterially expressed and purified N protein. The FAP (mp) of each sample was recorded and plotted versus input N protein concentration, as mentioned in Materials and Methods. The highest mp signal was observed at lower RNA concentration (Fig. 2A). A similar experiment with shorter RNA molecules of 20 and 30 nucleotides in length, harboring the heptanucleotide sequence, showed a poor mp signal (Fig. 2B). We observed that N protein binds to the fluorescent RNA (40 nucleotides) with a dissociation constant (Kd) of 21 ± 9 nM. The dissociation constant (Kd) represents the concentration of N protein at which mp signal was half saturated. To get insights about the kinetics of the N protein–UTR interaction, a fixed concentration of 40-nucleotide FAM-labeled RNA (3 or 6.25 nM) was incubated with increasing concentrations of N protein and mp signal was recorded at different time points. As shown in Figure 2C, the mp signal was saturated at 50 nM N protein and the formation of the N protein–UTR complex was very rapid. Since the compound libraries are maintained in 100% DMSO, its effect on the binding reactions was examined. The reactions containing either RNA alone or RNA along with N protein were incubated at 25 °C with increasing concentrations of DMSO, and the mp signal was recorded. This analysis revealed the assay was tolerant to DMSO at 10% (v/v) up to 4 h of incubation at 25 °C (Fig. 2D). To calculate the signal-to-background ratio, varying concentrations of three RNA molecules were incubated with increasing concentrations of N protein and mp signal was recorded for each sample (Fig. 3A). An optimal signal-to-background ratio of 3 was obtained with N protein (100 nM) and RNA (3 nM) (Fig. 3A). These concentrations were selected as optimal for performing the assay to study RNA–protein interactions in greater detail. An incubation period of 90 min at 25 °C was selected for the ease of high-throughput workflow.
Figure 2.
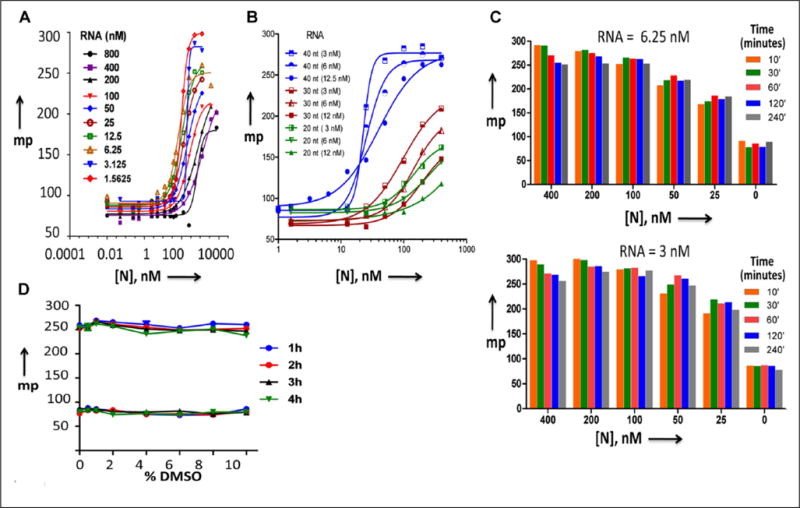
Binding of hantavirus N protein with the FAM-labeled RNA. (A,B) Dose–response curves for N protein–RNA interaction obtained by FPA assay, as mentioned in the text. Panel A shows the dose–response curves for 40-nucleotide-long RNA, and panel B shows the same curves, in which shorter RNAs (30 and 20 nucleotides) were also included. (C) Kinetics for the interaction of N protein with the FAM-labeled RNA. A fixed concentration of FAM-labeled RNA at a concentration of either 6.25 or 3 nM was incubated with increasing concentrations of N protein, shown in the upper and lower panels, respectively. The mixture of N protein and RNA was incubated for increasing time intervals before the mp signal was recorded and plotted versus the N protein concentration. (D) DMSO sensitivity of N protein–RNA interactions. Binding reactions containing RNA and N protein or RNA alone were incubated with the varying concentrations of DMSO for up to 4 h. The FPA readout was recorded at every hour to monitor the effect of DMSO on N protein–RNA interaction.
Figure 3.
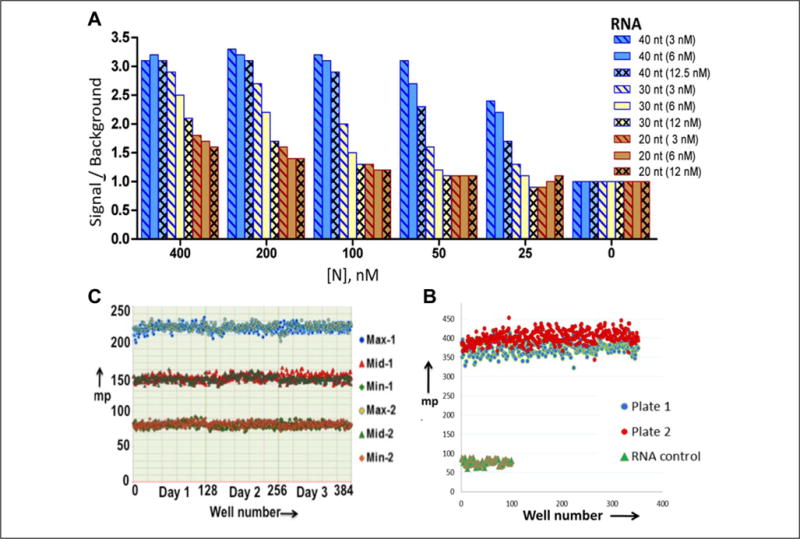
Characterization of assay robustness. (A) To calculate the signal-to-background ratios of N protein–RNA interaction, the FAM-labeled RNAs at a concentration of either 3, 6, or 12.5 nM were incubated with increasing concentrations of N protein, and mp signal was recorded. The signal-to-background ratio was calculated and plotted versus the N protein concentration. (B) RNA stability in HTS liquid handling. Both the RNA and N protein or RNA alone were dispensed using HTS liquid handling instrumentation into all wells of two 384-well plates. The FPA signal was recorded using Enspire. (C) To assess the plate uniformity and signal variability, two 384-well plates were run twice over 3 days to evaluate uniformity and separation of maximum (Max), minimum (Min), and midrange signals. Mean, SD, and CV of the mean were calculated for each Max, Mid, and Min signal. The overall Z′ value was found to be around 0.9, and signal-to-window ratios were >2. A scatter plot of Max, Mid, and Min signals shows the absence of drifts and edge effects over a 3-day assessment period.
Assay Robustness
The assay was tested for its robustness and reproducibility to enable its utility for the identification of small molecules that inhibit the N protein–UTR interaction. As the first step toward optimization for HTS, reagent stability was tested using HTS automation. Binding reactions containing RNA alone or RNA along with N protein were dispensed using WellMate into two 384-well plates, and the stability of the mp signal was determined. The signal readouts between RNA alone and RNA along with N protein remained well separated during the dispensing of reagents in all wells of two test 384-well plates. A well-defined signal separation was observed in each of the two 384-well plates tested (Fig. 3B). Plate uniformity assays were performed, as described in Materials and Methods. The FAP data from experiments performed on three independent days was plotted in the scattergram (Fig. 3C). This analysis revealed a signal window of >20, average Z′ value of 0.9, and CV of <20%. No edge effects or drifts in signal were detected.
Assay Validation
The assay was validated using a compound library of 6880 compounds. The library was screened at a final concentration of 30 μM containing 0.3% DMSO. The raw data collected was processed, and the parameters defining the quality and characteristics of a high-throughput screen were calculated. The average mp and Z′ values for all 22 plates are shown in Figure 4A, B. The Z′ scores of >0.5 are indicative of the suitability of the fluorescence polarization assays for HTS and reflect good separation between the median plate values and those of the positive and negative controls. The validation screen revealed an average Z′ score of 0.86 ± 0.029. The percent inhibition of N protein–UTR interaction was calculated for each compound and plotted as a scatter plot (Fig. 4C). Only 50 out of 6880 compounds inhibited N protein–UTR interaction ≥ plate median + 3 SD, giving an overall hit rate of 0.74%. Based on these statistically acceptable criteria, the assay is well suited for HTS. To confirm the activity of primary screen actives, an eight-concentration dose–response experiment was performed. A total of 192 compounds, including 50 primary compounds having activity ≥ plate median + 3 SD and remaining compounds having activity between plate median plus 2 SD and 3 SD, were cherry-picked from the library for reconfirmation. The compounds were tested in 40 μL reactions containing 100 nM N protein, 3 nM FAM-labeled 40-nucleotide RNA, and increasing concentrations of the compound of interest. The representative curves for several compounds are shown in Figure 4E. The analysis of dose–response curves revealed an overall reconfirmation rate of 85%. As shown in Figure 4D, the reconfirmation rate was higher for compounds whose initial activity was ≥ plate median + 3 SD. The validated assay was used to screen a large chemical library that led to the identification of several lead inhibitors.4
Figure 4.
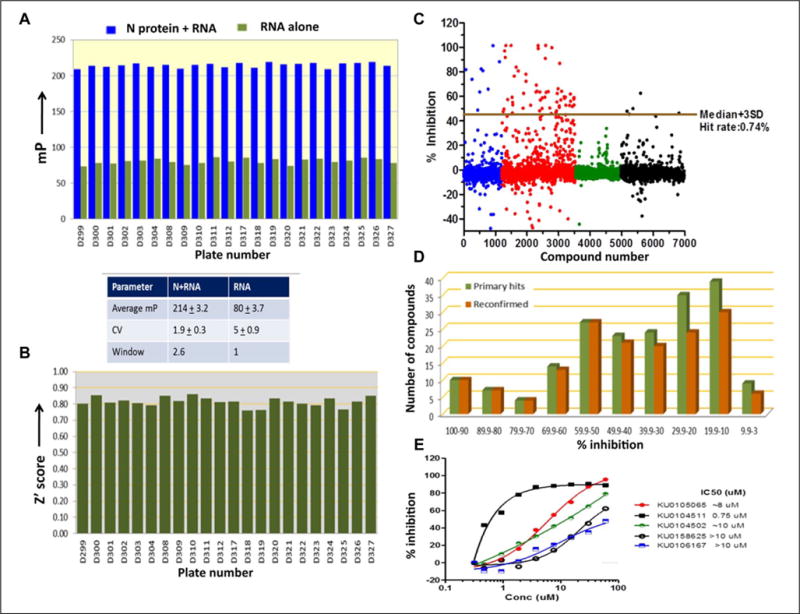
Characterization of assay robustness. (A) A library of 6880 compounds was screened at 30 μM using the optimized FPA assay. Distribution of mp values for wells containing N protein either alone or along with RNA. The average mp values and their SDs are reported in the table below panel A. (B) Plot of Z′ values across all 384-well plates tested in the validation screen. (C) Scatter plot of percent inhibition of N protein–UTR interaction by 6880 compounds. The primary screen actives were defined as compounds with activity greater than plate median + 3 SD. (D) Dose–response reconfirmation of primary screen actives. Actives from the primary screen were cherry-picked and tested for their inhibitory activity in the FPA assay. (E) The representative dose–response curves of some of the reconfirmed compounds.
Hantavirus infections are a growing public health concern due to the lack of treatment. Both HFRS and HCPS are acute infections affecting the cardiac, renal, pulmonary, central nervous, and hormonal systems. The recent HCPS outbreak in Yosemite National Park in California had a mortality rate of 30%, which has further stressed the need for the development of antihantaviral therapeutics. Viable treatment strategies attempted for hantavirus diseases include immunotherapy, vaccination, and antiviral therapeutics. At present, there are no published reports or clinical trials being conducted for immunotherapy, although several studies have indicated that passive introduction of neutralizing monoclonal antibodies (mAbs) or polyclonal sera against Hantaan virus (HTNV) can protect animal models from viral challenges. On the other hand, several attempts have been made to develop vaccines. In South Korea and China, rodent brain– and cell culture–derived inactivated vaccines are used locally.10 However, the danger associated with the mass production of virus under BSL-4 conditions has discouraged such vaccine trials in developed countries. Attempts have also been made to use recombinant proteins and virus-like particles for vaccine development with little success. However, DNA-based vaccines have been reported to provide protection against hantavirus diseases in animal models.11
To date, no antiviral drugs are available for the treatment of hantavirus diseases. However, ribavirin has been reported to provide protection against lethal HCPS in a Syrian hamster model exposed to ANDV.12 In addition, ribavirin was shown to be beneficial for HFRS patients when administered in the early disease course. A clinical study carried out between 1987 and 2005 on 38 individuals reported the reduced disease severity by ribavirin treatments. On the other hand, a clinical trial in HCPS patients reported that ribavirin did not show beneficial effects.13 Despite its ability to reduce disease severity, ribavirin has been shown to cause hemolytic anemia in patients that may worsen cardiac diseases and lead to fatal myocardial infractions. Due to significant manifestations of the heart and lungs during hantavirus infection, the efficacy of ribavirin is surely questionable for HFRS and HCPS. Apart from ribavirin, a riboside 1-β-D-ribofuranosyl-3-ethynyl-[1,2,4] triazole (ETAR) was tested in HTNV infections that showed significantly less efficiency than ribavirin.14
Due to early diagnosis, an early therapeutic intervention is possible and will significantly improve the outcome of hantavirus disease. As shown in Figure 1, the interaction of N protein with the highly conserved sequence in both viral genomic RNA and viral mRNA plays critical roles in the virus replication cycle. It facilitates the translation of viral mRNA to boost the expression of viral proteins in the host cell. This interaction facilitates transcription of viral mRNA in conjunction with viral RdRp. It also has a role in encapsidation and packaging of the viral genome. Thus, this unique RNA–protein interaction is a novel target for therapeutic intervention of hantavirus disease. We developed a highly sensitive fluorescence-based in vitro assay to monitor the interaction of N protein with the heptanucleotide sequence of viral UTR. The assay is cost-effective, requiring very little purified N protein and RNA. The assay has a good signal-to-background ratio and is tolerant of DMSO. The assay is robust and reproducible, having a very stable fluorescence readout. Plate uniformity assays revealed the statistical parameters (signal window of >20, average Z′ value of 0.9, and CV of <20%) that met the criteria for HTS. The assay was validated using a library of 6880 compounds. The validation screen further confirmed the reproducibility and validity of statistical parameters for HTS. The activity of primary screen actives of the validation screen was further confirmed by a dose–response experiment, which demonstrated an overall reconfirmation rate of 85%. The assay has met all the requirements for HTS and is ready to use for screening large chemical libraries to identify chemical inhibitors that interrupt N protein–UTR interaction and thereby inhibit hantavirus replication.
Acknowledgments
Anuradha Roy did all the experiments and provided all figures except Figure 1 used in this article. Mohammad Mir supervised the research and wrote the article. Both authors read and approved the article for publication.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH grants RO1 AI095236-01 and 1R21 AI097355-01.
Footnotes
Declaration of Conflicting Interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Voelkel NF. High-Altitude Pulmonary Edema. N Engl J Med. 2002;346:1606–1607. doi: 10.1056/NEJM200205233462102. [DOI] [PubMed] [Google Scholar]
- 2.Mir MA, Duran WA, Hjelle BL, et al. Storage of Cellular 5′ mRNA Caps in P Bodies for Viral Cap-Snatching. Proc Natl Acad Sci USA. 2008;105:19294–19299. doi: 10.1073/pnas.0807211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mir MA, Panganiban AT. A Protein That Replaces the Entire Cellular eIF4F Complex. EMBO J. 2008;27:3129–3139. doi: 10.1038/emboj.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salim NN, Ganaie SS, Roy A, et al. Targeting a Novel RNA-Protein Interaction for Therapeutic Intervention of Hantavirus Disease. J Biol Chem. 2016;229:24702–27714. doi: 10.1074/jbc.M116.750729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahapatra L, Mao C, Andruska N, et al. High-Throughput Fluorescence Anisotropy Screen for Inhibitors of the Oncogenic mRNA Binding Protein, IMP-1. J Biomol Screen. 2014;19:427–436. doi: 10.1177/1087057113499633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellenbecker M, Lanchy JM, Lodmell JS. Identification of Rift Valley Fever Virus Nucleocapsid Protein-RNA Binding Inhibitors Using a High-Throughput Screening Assay. J Biomol Screen. 2012;17:1062–1070. doi: 10.1177/1087057112448100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rye-McCurdy T, Rouzina I, Musier-Forsyth K. Fluorescence Anisotropy-Based Salt-Titration Approach to Characterize Protein-Nucleic Acid Interactions. Methods Mol Biol. 2015;1259:385–402. doi: 10.1007/978-1-4939-2214-7_23. [DOI] [PubMed] [Google Scholar]
- 8.Mir MA, Panganiban AT. The Bunyavirus Nucleocapsid Protein Is an RNA Chaperone: Possible Roles in Viral RNA Panhandle Formation and Genome Replication. RNA. 2006;12:272–282. doi: 10.1261/rna.2101906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne JC, Elliott RM. RNA Binding Properties of Bunyamwera Virus Nucleocapsid Protein and Selective Binding to an Element in the 5′ Terminus of the Negative-Sense S Segment. J Virol. 2000;74:9946–9952. doi: 10.1128/jvi.74.21.9946-9952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piyasirisilp S, Schmeckpeper BJ, Chandanayingyong D, et al. Association of HLA and T-Cell Receptor Gene Polymorphisms with Semple Rabies Vaccine-Induced Autoimmune Encephalomyelitis. Ann Neurol. 1999;45:595–600. doi: 10.1002/1531-8249(199905)45:5<595::aid-ana7>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 11.Maes P, Clement J, Van Ranst M. Recent Approaches in Hantavirus Vaccine Development. Expert Rev Vaccines. 2009;8:67–76. doi: 10.1586/14760584.8.1.67. [DOI] [PubMed] [Google Scholar]
- 12.Ogg M, Jonsson CB, Camp JV, et al. Ribavirin Protects Syrian Hamsters against Lethal Hantavirus Pulmonary Syndrome—After Intranasal Exposure to Andes Virus. Viruses. 2013;5:2704–2720. doi: 10.3390/v5112704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mertz GJ, Miedzinski L, Goade D, et al. Placebo-Controlled, Double-Blind Trial of Intravenous Ribavirin for the Treatment of Hantavirus Cardiopulmonary Syndrome in North America. Clin Infect Dis. 2004;39:1307–1313. doi: 10.1086/425007. [DOI] [PubMed] [Google Scholar]
- 14.Chung RT, Gale M, Jr, Polyak SJ, et al. Mechanisms of Action of Interferon and Ribavirin in Chronic Hepatitis C: Summary of a Workshop. Hepatology. 2008;47:306–320. doi: 10.1002/hep.22070. [DOI] [PMC free article] [PubMed] [Google Scholar]


