Abstract
Prognostic biomarkers that stratify patients with cancer are needed. Recent studies from Asia have implicated SALL4, a stem cell marker, as useful in identifying aggressive cases of hepatocellular carcinoma (HCC), and over 50% of the cases tested had upregulation by microarray or dense immunoreactivity. Given the differences in predominant etiologic factors between the Asian and Western HCC, we sought to determine the prevalence of SALL4 immunoreactivity and its clinical relevance in Western HCC patients. We constructed tissue microarrays from 236 adult HCC. Two cores each of tumor and nontumor tissue were included for each case. SALL4 immunohistochemistry was scored in a semi-quantitative manner and the results correlated with recurrence-free and overall survival, in addition to standard demographics. Among the 236 cases, 165 (70.0%) were male. The median age was 59 years (range: 19–83 years). The majority (78.4%) of patients were Caucasian, followed by African American (15.7%), Asian (3.8%), Hispanic (1.7%), and Native American (0.4%). The majority of patients had hepatitis C (42.8%), followed by alcoholic liver disease and hepatitis B (both 8.9%), and nonalcoholic steatohepatitis (3.8%). SALL4 immunoreactivity was detected in a total of 3 cases (1.3%), and nonreactivity was validated on tissue sections from 73 cases. By univariate analysis, the SALL4-positive cases had significantly higher tumor grade (P = 0.0251), more frequent lymphovascular invasion (P = 0.0150), shorter recurrence-free survival (7.90 vs. 57.54 months; P = 0.0115) and overall survival (7.90 vs. 64.87 months; P = 0.0018). While SALL4 immunoreactivity in Western HCC is correlated with higher grade and poor prognosis, this is a rare event. Therefore, universal application of SALL4 as a biomarker for HCC should be performed with caution.
Keywords: stem cell, survival, recurrence, transplantation, immunostain, biomarker
Introduction
The worldwide incidence of hepatocellular carcinoma (HCC) is increasing. (1–6) The most common associations with hepatocellular carcinoma are chronic liver diseases due to viral hepatitis B and C and alcohol; nonalcoholic fatty liver with or without cirrhosis is also increasingly recognized in association with HCC. (2, 7–9) There is geographical variation in dominant etiology: while Western HCC patients usually develop tumor after hepatitis C, alcohol, and nonalcoholic fatty liver disease, Asian HCC patients more commonly are carriers of hepatitis B. (1, 2) Despite the advances in surveillance technology and treatment options, HCC remains a deadly disease. Standard therapies include surgery, transarterial chemoembolization, and radiofrequency ablation. (5, 10) More recently, several multikinase inhibitors (sorafenib, sunitinib, etc) have been shown to be effective in prolonging survival in late stage HCC. (11–13) These newer agents, however, while prolonging stable disease for several weeks, achieve only modest response. (10–12) Given the limited resource of donor livers and relative high cost in molecular targeted therapeutics, it is important to develop biomarkers that could aid in stratification of patients.
Any ideal prognostic biomarker should be based on disease pathobiology, with high sensitivity and specificity, and can be carried out by an assay that is easy to perform. Recent advances in prognostic biomarker development include gene and expression analysis. (8, 14–17) While there are exciting findings and the results can be linked directly to potential oncogenic pathways and hence, the possibility of selecting appropriate therapies, the cost and technical aspects may prevent widespread applications. Therefore, attention has been turned to immunohistochemical markers, most of which can be performed on formalin-fixed, paraffin-embedded tissue utilized in routine preparation in diagnostic evaluation.
Recently, translational work identified “stem cell features” as important indicators of poor prognosis in various cancer types, including HCC. (18–22) Subsequent validation by morphologic studies followed with the use of immunohistochemistry against keratin 19. (23, 24) Expressions of target proteins that are keys to the biology of cancer stem cell/progenitor cell components are thus promising immunohistochemical biomarkers. SALL4, an oncofetal protein, is expressed in fetal livers and various malignancies, including acute myeloid leukemia, lymphoma, yolk sac tumor, among others. (25–27) Two recent reports indicate that SALL4 may be important in carcinogenesis of HCC and implicates a more aggressive behavior. (28, 29) Importantly, in these reports, SALL4 immunoreactivity was seen in 56 – 85% of cases, and upregulation by microarray analysis was detected in 50% of cases. (28, 29) Interestingly, the patient cohorts analyzed in the studies to date were Asians, with a high prevalence of hepatitis B-associated HCC. We aimed, therefore, to examine the use of SALL4 as a prognostic marker in a large cohort of HCC in Western patients. We correlated the expression of SALL4 in 236 HCC cases with clinical outcomes.
Materials and Methods
Patients
All resected or transplanted pure, non-fibrolamellar HCC cases between 1990 and 2009 were identified by a computer database search of the Departments of Surgery and Pathology and Immunology, Washington University School of Medicine. The cases were reviewed, and only cases with sufficiently viable tissue material for assembling tissue microarray were included. Medical records were reviewed, and pertinent demographic data (age, gender, underlying diseases), pathology readouts (tumor size, tumor stage, degree of fibrosis, and lymphovascular invasion), and clinical data (previous therapies, recurrence-free survival, overall survival) extracted. The study was approved by the Institutional Review Board of Washington University School of Medicine.
Tissue Microarray (TMA)
The histology of each case was reviewed by a liver pathologist (E.M.B.) and representative tissue blocks were selected for TMA construction. For each case, two cores of 2 microns each were punched from both tumor and nontumor liver to construct TMA blocks. Nonhepatic tissue cores served as starting and ending cores for each block.
SALL4 immunohistochemistry
Immunohistochemistry of SALL4 was carried out as described previously. (27, 30) Briefly, antigen retrieval was performed using 1mM EDTA at pH 8.0. SALL4 immunohistochemistry was performed using a Benchmark XT autostainer (Ventana; Tucson, Arizona). SALL4 antibody was obtained from Sigma-Aldrich (Saint Louis, MO, USA; catalog number WH0057167-M03). The Ultraview universal DAB detection system was used for signal detection (Ventana). Appropriate positive and negative controls were included for each run of immunohistochemistry. SALL4 expression in tissue microarrays was scored by a liver pathologist (E.M.B.). Two patterns of nuclear reactivity were detected; one was granular and one was diffuse in which the nucleolus was completely obscured. Only the diffuse pattern of nuclear reactivity was considered positive for SALL4; patchy granular nuclear reactivity was noted but not scored as a positive. (29) A semi-quantitative score was used to classify SALL4 expression based on the percentage of tumor cells that showed the diffuse nuclear immunoreactive pattern for SALL4, as described previously: (29) 0: < 5%; 1: 5 – 30%; 2: 31 – 50%; 3: 51 – 80%; 4: >80%. Of note, the score 0 includes cases with focal granular nuclear staining pattern that was present in <5% of tumor cells. For cases with focal granular nuclear staining pattern seen on TMA, corresponding tumor blocks from which the TMAs were constructed were stained with SALL4 to exclude the possibility of undersampling. Fifty-three additional cases with nonreactivity on TMA were also randomly selected and the corresponding tumor blocks from which the TMAs were constructed were stained with SALL4.
Statistical analysis
For each parameter, a univariate analysis was performed. Unpaired t test or Chi-Square was used for comparison between two separate groups. Kaplan-Meier curves were created for recurrence-free survival and overall survival, and statistical analysis was performed using Log-rank test. Significance was defined as P < 0.05.
Results
Table 1 summarizes the demographics and additional clinical information of the HCC patients. Of the 236 patients whose tumors were included in the TMA analysis, 165 (70.0%) were male and 71 (30.0%) were female. The median age was 59 years old, and the mean age was 50 years old (range: 19–83 years old). The majority (n=185; 78.4%) of patients were Caucasian, followed by African American (n=37; 15.7%), Asian (n=9; 3.8%), Hispanic (n=4; 1.7%), and Native American (n=1; 0.4%). Twenty-one (8.9%) cases had hepatitis B, 101 (42.8%) had hepatitis C, 21 (8.9%) had alcoholic liver disease, 9 (3.8%) had nonalcoholic steatohepatitis, 2 (0.8%) had hemochromatosis, and 1 (0.4%) each had autoimmune hepatitis, primary sclerosing cholangitis, Budd-Chiari Syndrome, Allagille’s syndrome, or Byler’s Syndrome. Of note, among the patients, 15 (6.4%) had more than one etiology. One hundred twenty-seven patients (53.8%) received orthotopic liver transplantation, whereas 109 (46.2%) underwent partial hepatic resection for HCC.
Table 1.
Patient demographics and etiology.
| Percentage (%) | ||
|---|---|---|
| Total patient number | 236 | 100 |
| Male: Female | 165:71 | 70:30 |
| Median age (range) | 59 years old (19–83) | |
| Ethnicity | ||
| Caucasian | 185 | 78.4 |
| African American | 37 | 15.7 |
| Asian | 9 | 3.8 |
| Hispanic | 4 | 1.7 |
| Native American | 1 | 0.4 |
| Etiology of Liver Diseases | ||
| Hepatitis B | 15 | 6.4 |
| Hepatitis C | 87 | 36.9 |
| Alcohol | 10 | 4.2 |
| Nonalcoholic steatohepatitis | 9 | 3.8 |
| Hemochromatosis | 1 | 0.4 |
| Autoimmune hepatitis | 1 | 0.4 |
| Primary sclerosing cholangitis | 1 | 0.4 |
| Budd-Chiari syndrome | 1 | 0.4 |
| Allagille’s syndrome | 1 | 0.4 |
| Byler’s syndrome | 1 | 0.4 |
| Hepatitis B + Hepatitis C | 3 | 1.3 |
| Hepatitis B + Alcohol | 1 | 0.4 |
| Hepatitis C + Alcohol | 8 | 3.4 |
| Hepatitis C + Hemochromatosis | 1 | 0.4 |
| Hepatitis B + Hepatitis C + Alcohol | 2 | 0.8 |
| No Liver Disease | 94 | 39.8 |
Table 2 summarizes the clinical and pathologic features of the HCCs. The average tumor size was 4.70 cm (range, 0.3 – 24.0 cm). Child-Pugh score was documented in 205 patients, among which, 107 (52.2%) were stage A, 56 (27.3%) were stage B, and 42 (20.5%) were stage C. Barcelona Clinic Liver Cancer (BCLC) staging information was available in 214 patients: 79 (36.9%) were stage A, 34 (15.9%) were stage B, 58 (27.1%) were stage C, and 43 (20.1%) were stage D. Sixty-three patients (26.7%) had received prior therapy. The pathologic evaluation confirmed that 149 (63.6%) were cirrhotic. Seventy-seven (32.6%) of the cases had multiple lesions. Mean serum alpha fetoprotein level was 1604.7 ng/ml; 168 patients (71.2%) showed elevated serum alpha fetoprotein levels. Lymphovascular invasion was seen in 56 (23.7%). Among the 186 patients with available tumor grading information, 85 (45.7%) were well-differentiated, 83 (44.6%) were moderately-differentiated, and 18 (9.7%) were poorly-differentiated HCC.
Table 2.
Clinical and pathologic features of HCC and correlation with SALL4 immunoreactivity.
| Features | SALL4-negative (n=233) | SALL4-positive (n=3) | p value |
|---|---|---|---|
| Cirrhosis | 148/233 (63.5%) | 1/3 (33.3%) | 0.5565 |
| Child-Pugh stage (n=205) | 0.4249 | ||
| A | 105/203 (51.7%) | 2/2 (100%) | |
| B | 56/203 (27.6%) | ||
| C | 42/203 (20.7%) | ||
| BCLC stage (n=214) | 0.6032 (A+B vs. C+D) | ||
| A | 78/211 (37.0%) | 1/3 (33.3%) | |
| B | 34/211 (16.1%) | ||
| C | 56/211 (26.5%) | 2/3 (66.7%) | |
| D | 43/211 (20.4%) | ||
| Elevated serum AFP | 167/233 (71.7%) | 1/3 (33.3%) | 0.4116 |
| Average maximal tumor size (cm) | 4.67 | 6.40 | 0.3252 |
| Multinodularity | 76/233 (32.6%) | 1/3 (33.3%) | 0.9437 |
| Tumor grade (differentiation; n=186) | 0.0251 (well + moderately-diff. vs. poorly-diff.) | ||
| Well | 85/183 (46.4%) | ||
| Moderate | 82/183 (44.8%) | 1/3 (33.3%) | |
| Poor | 16/183 (8.7%) | 2/3 (66.7%) | |
| Lymphovascular invasion | 53/233 (22.7%) | 3/3 (100%) | 0.0150 |
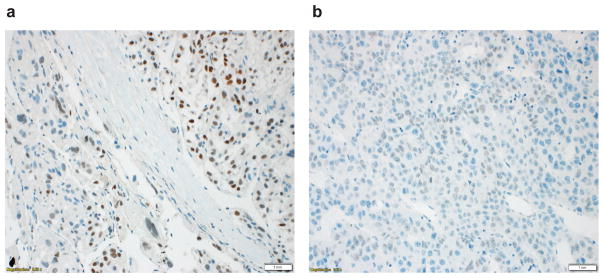
Using the quantification system for SALL4 described by Yong et. al. (29), among the 236 HCC specimens, only 3 (1.3%) showed score of ≥1. All three cases showed a diffuse nuclear staining pattern (Fig. 1a). Twenty additional cases showed focal (<5%) SALL4 immunoreactivity with a granular nuclear staining pattern (Fig. 1b). To exclude the possibility of inadequate sampling in these cases, we subsequently performed SALL4 immunohistochemistry on tissue blocks from which the TMAs were constructed. None of these cases showed ≥ 5% immunoreactivity or diffuse nuclear staining pattern on tissue sections (data not shown). As these cases had no differences in outcome from those without granular nuclear reactivity (data not shown), they were not further analyzed. We also additionally performed SALL4 immunohistochemistry on 53 HCC tumor blocks from the cases which showed nonimmunoreactivity on TMA. None of these cases showed nuclear staining on tissue sections. The 3 SALL4-positive HCCs (immunoreactivity score ≥ 1) are described in more detail in Table 3. Two were women; underlying diseases were HCV plus HBV (1); HCV (1) and in one case, there was no underlying liver disease. This patient had 2 recurrences of HCC. Only 1 case had elevated AFP (> 99,000 ng/ml). The tumor was multinodular and overall had measured 11 cm. The other two were smaller at 7 cm and 2.2 cm. All three cases had lymphovascular invasion. The tumor grades were moderately-differentiated in 1 and poorly-differentiated in 2. Univariate analysis showed that compared to SALL4-negative HCCs, the SALL4-positive tumors were more likely to be higher grade (P = 0.0251) and have lymphovascular invasion (P = 0.0150), whereas no significant differences were seen in correlations with age, gender, ethnicity, presence of cirrhosis, Child-Pugh and BCLC stage, tumor size, or multinodularity (Table 2).
Figure 1.
Different SALL4 staining patterns in HCC. (a) Diffuse nuclear SALL4 staining pattern was seen in 3 cases. These cases were defined as SALL4-positive. (b) Granular nuclear staining pattern was seen in 20 cases. Immunohistochemistry performed on the tumor blocks of these cases confirmed that these cases were SALL4-negative. Scale bar: 1 mm.
Table 3.
Detailed information on SALL4 immunoreactive HCC.
| Case | Age | Sex | Race | Proce dure |
HBV | HCV | Alcohol | NASH | Fibrosis stage |
BCLC stage |
Tumor grade (differentiati on) |
AFP (ng/ml) |
Multi- nodularity |
Size (cm) |
LVSI | SALL4 score |
RFS (mos) |
OS (mos) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1* | 52 | F | C | RES | 0 | 0 | 0 | 0 | 2 | C | poor | 0 | 0 | 7 | 1 | 2 | 11.51 | 17.18 |
| 2 | 59 | M | C | RES | 0 | 1 | 0 | 0 | 1 | A | moderate | 99950 | 1 | 11 | 1 | 3 | 7.10 | 7.10 |
| 3 | 56 | F | A | OLT | 1 | 1 | 0 | 0 | 4 | C | poor | 0 | 0 | 2.2 | 1 | 4 | 7.90 | 7.90 |
A: Asian; AFP: alpha fetoprotein; BCLC: Barcelona Clinical Liver Cancer; C: Caucasian; F: female; LVSI: lymphovascular invasion; M: male; NASH: nonalcoholic steatohepatitis; OLT: orthotopic liver transplantation; OS: overall survival (months); RES: resection; RFS: recurrence-free survival (months).
Case 1: had no underlying liver disease; was initially thought to be an adenoma.
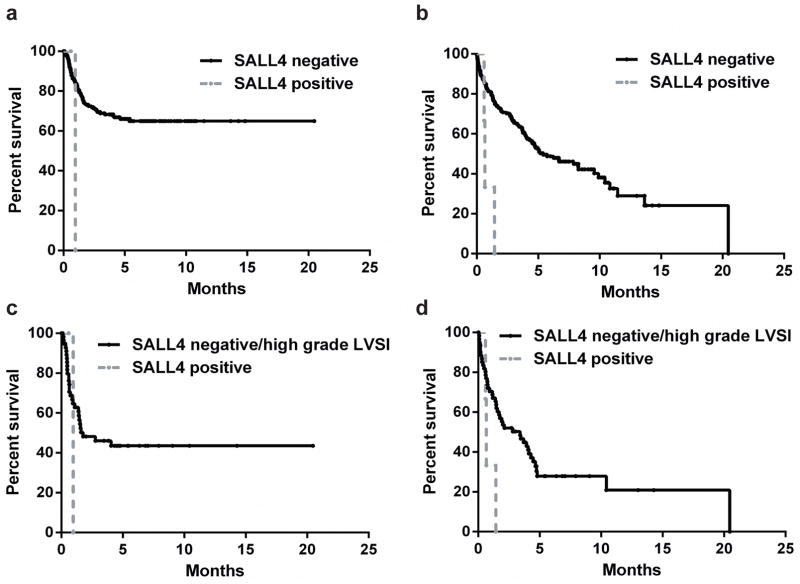
We next sought to determine whether the SALL4 immunoreactivity correlated with prognosis. There was no significant difference in recurrence-free survival and overall survival between cases that showed no SALL4 immunoreactivity and those that showed focal (<5%) granular nuclear reactivity (still considered score 0; data not shown). Therefore, for prognosis analysis, these cases were not separated. As shown in Fig. 2a and 2b, the three SALL4-positive cases had significantly shorter recurrence-free survival (7.90 vs. 57.54 months; P = 0.0115) and overall survival (7.90 vs. 64.87 months; P = 0.0018). We also performed a sub-analysis to compare the outcome between the 3 SALL4-positive cases and the SALL4-negative cases that were either high grade and/or had lymphovascular invasion (n = 64). As shown in Fig. 2c and 2d, there was no significant difference between the SALL4-negative/high tumor grade-lymphvascular invasion group and the SALL4-positive group in recurrence-free survival (1.775 vs. 0.959 months; P = 0.9750). However, there was significant difference between the SALL4-negative/high tumor grade-lymphovascular invasion group and the SALL4-positive group in overall survival (3.414 vs. 0.658 months; P = 0.0357). Due to the small sample size of the SALL4-positive group, multivariate analysis could not be performed.
Figure 2.
Clinical outcome of SALL4-positive and SALL4-negative HCCs. (a) Recurrence-free survival and (b) overall survival between SALL4-positive and SALL4-negative HCCs. Black line: SALL4-negative group (n=233); grey line: SALL4-positive group (n=3). SALL4-positive cases had significantly shorter recurrence-free survival (P = 0.0115) and overall survival (P = 0.0018). (c) Recurrence-free survival and (d) overall survival between SALL4-positive HCCs and SALL4-negative HCCs but with either high tumor grade or lymphovascular invasion. Black line: SALL4-negative/high grade lymphovascular invasion group (n=64); grey line: SALL4-positive group (n=3). The two groups had no significant difference in recurrence-free survival (P = 0.9750), but the SALL4-positive group had significantly shorter overall survival (P = 0.0357). LVSI: lymphovascular invasion.
Discussion
In this study, we examined the prevalence and clinical relevance of SALL4 immunoreactivity in a large Western HCC cohort. In contrast to recent reported findings in Asian HCC patients, SALL4 immunoreactivity (defined as ≥5% of tumor cells) (29) was seen only in 3 of 236 cases (1.3%) in our series. Among these 3 patients, 1 had hepatitis C, 1 had hepatitis B +C, and 1 had no underlying liver disease. The three SALL4-reactive tumors had moderately and poorly differentiated tumors and all had lymphovascular invasion. These cases also showed significantly shorter recurrence-free survival and overall survival compared to those with nonreactivity for SALL4.
Our results have several significant clinical implications. Recent in vitro and in vivo studies highlighted the role of SALL4 in hepatocarcinogenesis as well as its potential clinical relevance as a marker of aggressiveness. (28, 29) As a marker for stem cells, SALL4 expression is proposed to represent a stem cell phenotype in HCC, as shown in other cancer types. (31–35) These studies indicate that stem cell features, defined by nuclear SALL4 immunohistochemistry and/or mRNA expression, correlate with an aggressive course and poor prognosis. Importantly, in both HCC studies, SALL4 immunoreactivity was seen in 55.6% (n=171) and 85% (n=20) of cases, respectively, and overexpression by microarray analysis was detected in 50% of cases. (28, 29) This is in sharp contrast to our results. Interestingly, one recent study reported prevalence of SALL4 immunoreactivity that is closer to our data (0 of 60 cases examined). (36) In addition, a separate study examining 20 combined HCC-cholangiocarcinoma also found none of the cases reacted with SALL4 antibody. (37) It has been commented that technical aspects of antibody retrieval strongly affects SALL4 detection; (28) in our study, the methodology was similar to the protocol described by Yong et. al.. (29) Furthermore, our laboratory has previously validated the staining protocol. (27, 30, 38–41) Thus, it is unlikely that the methodology explains our differences.
The mechanisms of the discrepancy of results between ours and the study published by Yong et. al. may provide further insight into the clinical relevance of SALL4 as a prognostic marker in HCC management. It is possible that the discrepancy reflects the difference(s) in etiology of the HCC. Whereas the Asian cohorts in the previous studies are predominantly hepatitis B-related (> 50%), Western HCCs, as in our cohort, are enriched for hepatitis C and nonalcoholic steatohepatitis. (1, 5, 6) The prevalence of hepatitis B in our study was < 10%. The etiology of the HCC cases included in the study by Ushiku et. al. is unclear. (36) It has been proposed that the X protein encoded by hepatitis B virus (HBx), activates beta-catenin and epigenetically upregulates miR-181, impacts the expression of epithelial cell adhesion molecule, (42, 43) thereby promotes hepatocarcinogenesis through upregulating expression of multiple “stemness” markers. (42, 44, 45) Feng et. al. has shown that in mice and patients with chronic hepatitis B, the increased expression of interleukin-22 promotes proliferation of liver stem/progenitor cells, thus likely contributing to the formation of HCC. (46) Many other studies have supported the correlation between hepatitis B infection and HCC stem/progenitor cells. (47) While hepatitis C virus has been shown to induce liver cancer stem cells, (48, 49) the mechanisms are less understood, with Hedgehog and Toll-like receptor 4 signaling pathways being proposed as main events. (20, 50–52) Thus, it is possible that hepatitis B and hepatitis C viruses act through activating distinct signaling pathways, thereby resulting in different effects on the downstream expression profile of SALL4.
In summary, utilizing tissue microarrays constructed from a large HCC patient cohort, our study has shown that SALL4 expression in HCC patients should be interpreted with caution. While it is associated with higher tumor grade, lymphovascular invasion, and correlates with poor prognosis, the rarity of the prevalence (1.3%) limits its application for clinical management. Thus, it may be necessary to integrate additional biomarkers to provide a synthetic readout for prognosis prediction. Further validating studies from different ethnic and liver disease etiology groups are warranted. Mechanistic studies on how SALL4 expression is regulated in different hepatocarcinogenic pathways may also provide insights into the biology of HCC.
Supplementary Material
Acknowledgments
Financial support: this work was supported by the departmental fund by Department of Pathology, Washington University School of Medicine.
Abbreviations
- HCC
hepatocellular carcinoma
- IHC
immunohistochemistry
- TMA
tissue microarray
- BCLC
Barcelona Clinic Liver Cancer
Footnotes
Conflict of interest: none.
References
- 1.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 2.Bruix J, Boix L, Sala M, et al. Focus on hepatocellular carcinoma. Cancer Cell. 2004;5:215–219. doi: 10.1016/s1535-6108(04)00058-3. [DOI] [PubMed] [Google Scholar]
- 3.Llovet JM, Bruix J. Novel advancements in the management of hepatocellular carcinoma in 2008. J Hepatol. 2008;48(Suppl 1):S20–37. doi: 10.1016/j.jhep.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Llovet JM. Major achievements in hepatocellular carcinoma. Lancet. 2009;373:614–616. doi: 10.1016/S0140-6736(09)60381-0. [DOI] [PubMed] [Google Scholar]
- 5.Bruix J, Llovet JM. Two decades of advances in hepatocellular carcinoma research. Semin Liver Dis. 2010;30:1–2. doi: 10.1055/s-0030-1247219. [DOI] [PubMed] [Google Scholar]
- 6.Sherman M. Hepatocellular carcinoma: screening and staging. Clin Liver Dis. 2011;15:323–334. doi: 10.1016/j.cld.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Varela M, Sala M, Llovet JM, et al. Review article: natural history and prognostic prediction of patients with hepatocellular carcinoma. Aliment Pharmacol Ther. 2003;17(Suppl 2):98–102. doi: 10.1046/j.1365-2036.17.s2.11.x. [DOI] [PubMed] [Google Scholar]
- 8.Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 9.Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51:1820–1832. doi: 10.1002/hep.23594. [DOI] [PubMed] [Google Scholar]
- 10.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llovet JM, Bruix J. Molecular targeted therapies in hepatocellular carcinoma. Hepatology. 2008;48:1312–1327. doi: 10.1002/hep.22506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 13.Newell P, Toffanin S, Villanueva A, et al. Ras pathway activation in hepatocellular carcinoma and anti-tumoral effect of combined sorafenib and rapamycin in vivo. J Hepatol. 2009;51:725–733. doi: 10.1016/j.jhep.2009.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Llovet JM, Chen Y, Wurmbach E, et al. A molecular signature to discriminate dysplastic nodules from early hepatocellular carcinoma in HCV cirrhosis. Gastroenterology. 2006;131:1758–1767. doi: 10.1053/j.gastro.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Hoshida Y, Villanueva A, Kobayashi M, et al. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med. 2008;359:1995–2004. doi: 10.1056/NEJMoa0804525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tovar V, Alsinet C, Villanueva A, et al. IGF activation in a molecular subclass of hepatocellular carcinoma and pre-clinical efficacy of IGF-1R blockage. J Hepatol. 2010;52:550–559. doi: 10.1016/j.jhep.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villanueva A, Hoshida Y, Battiston C, et al. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology. 2011;140:1501–1512. e1502. doi: 10.1053/j.gastro.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliva J, French BA, Qing X, et al. The identification of stem cells in human liver diseases and hepatocellular carcinoma. Exp Mol Pathol. 2010;88:331–340. doi: 10.1016/j.yexmp.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yeh CT, Kuo CJ, Lai MW, et al. CD133-positive hepatocellular carcinoma in an area endemic for hepatitis B virus infection. BMC Cancer. 2009;9:324. doi: 10.1186/1471-2407-9-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Machida K, Chen CL, Liu JC, et al. Cancer stem cells generated by alcohol, diabetes, and hepatitis C virus. J Gastroenterol Hepatol. 2012;27(Suppl 2):19–22. doi: 10.1111/j.1440-1746.2011.07010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marquardt JU, Raggi C, Andersen JB, et al. Human hepatic cancer stem cells are characterized by common stemness traits and diverse oncogenic pathways. Hepatology. 2011;54:1031–1042. doi: 10.1002/hep.24454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JS, Heo J, Libbrecht L, et al. A novel prognostic subtype of human hepatocellular carcinoma derived from hepatic progenitor cells. Nat Med. 2006;12:410–416. doi: 10.1038/nm1377. [DOI] [PubMed] [Google Scholar]
- 23.Uenishi T, Kubo S, Yamamoto T, et al. Cytokeratin 19 expression in hepatocellular carcinoma predicts early postoperative recurrence. Cancer Sci. 2003;94:851–857. doi: 10.1111/j.1349-7006.2003.tb01366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang XR, Xu Y, Shi GM, et al. Cytokeratin 10 and cytokeratin 19: predictive markers for poor prognosis in hepatocellular carcinoma patients after curative resection. Clin Cancer Res. 2008;14:3850–3859. doi: 10.1158/1078-0432.CCR-07-4338. [DOI] [PubMed] [Google Scholar]
- 25.Ma Y, Cui W, Yang J, et al. SALL4, a novel oncogene, is constitutively expressed in human acute myeloid leukemia (AML) and induces AML in transgenic mice. Blood. 2006;108:2726–2735. doi: 10.1182/blood-2006-02-001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui W, Kong NR, Ma Y, et al. Differential expression of the novel oncogene, SALL4, in lymphoma, plasma cell myeloma, and acute lymphoblastic leukemia. Mod Pathol. 2006;19:1585–1592. doi: 10.1038/modpathol.3800694. [DOI] [PubMed] [Google Scholar]
- 27.Cao D, Li J, Guo CC, et al. SALL4 is a novel diagnostic marker for testicular germ cell tumors. Am J Surg Pathol. 2009;33:1065–1077. doi: 10.1097/PAS.0b013e3181a13eef. [DOI] [PubMed] [Google Scholar]
- 28.Oikawa T, Kamiya A, Zeniya M, et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–1483. doi: 10.1002/hep.26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yong KJ, Gao C, Lim JS, et al. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;368:2266–2276. doi: 10.1056/NEJMoa1300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao D, Guo S, Allan RW, et al. SALL4 is a novel sensitive and specific marker of ovarian primitive germ cell tumors and is particularly useful in distinguishing yolk sac tumor from clear cell carcinoma. Am J Surg Pathol. 2009;33:894–904. doi: 10.1097/PAS.0b013e318198177d. [DOI] [PubMed] [Google Scholar]
- 31.Fujii Y, Yoshihashi K, Suzuki H, et al. CDX1 confers intestinal phenotype on gastric epithelial cells via induction of stemness-associated reprogramming factors SALL4 and KLF5. Proc Natl Acad Sci U S A. 2012;109:20584–20589. doi: 10.1073/pnas.1208651109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Chai L, Liu F, et al. Bmi-1 is a target gene for SALL4 in hematopoietic and leukemic cells. Proc Natl Acad Sci U S A. 2007;104:10494–10499. doi: 10.1073/pnas.0704001104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Chai L, Gao C, et al. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood. 2008;112:805–813. doi: 10.1182/blood-2007-11-126326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobayashi D, Kuribayshi K, Tanaka M, et al. SALL4 is essential for cancer cell proliferation and is overexpressed at early clinical stages in breast cancer. Int J Oncol. 2011;38:933–939. doi: 10.3892/ijo.2011.929. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi D, Kuribayashi K, Tanaka M, et al. Overexpression of SALL4 in lung cancer and its importance in cell proliferation. Oncol Rep. 2011;26:965–970. doi: 10.3892/or.2011.1374. [DOI] [PubMed] [Google Scholar]
- 36.Ushiku T, Shinozaki A, Shibahara J, et al. SALL4 represents fetal gut differentiation of gastric cancer, and is diagnostically useful in distinguishing hepatoid gastric carcinoma from hepatocellular carcinoma. Am J Surg Pathol. 2010;34:533–540. doi: 10.1097/PAS.0b013e3181d1dcdd. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda H, Sato Y, Yoneda N, et al. alpha-Fetoprotein-producing gastric carcinoma and combined hepatocellular and cholangiocarcinoma show similar morphology but different histogenesis with respect to SALL4 expression. Hum Pathol. 2012;43:1955–1963. doi: 10.1016/j.humpath.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 38.Mei K, Liu A, Allan RW, et al. Diagnostic utility of SALL4 in primary germ cell tumors of the central nervous system: a study of 77 cases. Mod Pathol. 2009;22:1628–1636. doi: 10.1038/modpathol.2009.148. [DOI] [PubMed] [Google Scholar]
- 39.Wang B, Li L, Ni F, et al. Mutational analysis of SAL-Like 4 (SALL4) in Han Chinese women with premature ovarian failure. Mol Hum Reprod. 2009;15:557–562. doi: 10.1093/molehr/gap046. [DOI] [PubMed] [Google Scholar]
- 40.Wang F, Liu A, Peng Y, et al. Diagnostic utility of SALL4 in extragonadal yolk sac tumors: an immunohistochemical study of 59 cases with comparison to placental-like alkaline phosphatase, alpha-fetoprotein, and glypican-3. Am J Surg Pathol. 2009;33:1529–1539. doi: 10.1097/PAS.0b013e3181ad25d5. [DOI] [PubMed] [Google Scholar]
- 41.Cao D, Humphrey PA, Allan RW. SALL4 is a novel sensitive and specific marker for metastatic germ cell tumors, with particular utility in detection of metastatic yolk sac tumors. Cancer. 2009;115:2640–2651. doi: 10.1002/cncr.24308. [DOI] [PubMed] [Google Scholar]
- 42.Arzumanyan A, Friedman T, Ng IO, et al. Does the hepatitis B antigen HBx promote the appearance of liver cancer stem cells? Cancer Res. 2011;71:3701–3708. doi: 10.1158/0008-5472.CAN-10-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C, Yang W, Yan HX, et al. Hepatitis B virus X (HBx) induces tumorigenicity of hepatic progenitor cells in 3,5-diethoxycarbonyl-1,4-dihydrocollidine-treated HBx transgenic mice. Hepatology. 2012;55:108–120. doi: 10.1002/hep.24675. [DOI] [PubMed] [Google Scholar]
- 44.Huang J, Shen L, Lu Y, et al. Parallel induction of cell proliferation and inhibition of cell differentiation in hepatic progenitor cells by hepatitis B virus X gene. Int J Mol Med. 2012;30:842–848. doi: 10.3892/ijmm.2012.1060. [DOI] [PubMed] [Google Scholar]
- 45.Yoon SM, Gerasimidou D, Kuwahara R, et al. Epithelial cell adhesion molecule (EpCAM) marks hepatocytes newly derived from stem/progenitor cells in humans. Hepatology. 2011;53:964–973. doi: 10.1002/hep.24122. [DOI] [PubMed] [Google Scholar]
- 46.Feng D, Kong X, Weng H, et al. Interleukin-22 promotes proliferation of liver stem/progenitor cells in mice and patients with chronic hepatitis B virus infection. Gastroenterology. 2012;143:188–198. e187. doi: 10.1053/j.gastro.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun YL, Yin SY, Xie HY, et al. Stem-like cells in hepatitis B virus-associated cirrhotic livers and adjacent tissue to hepatocellular carcinomas possess the capacity of tumorigenicity. J Gastroenterol Hepatol. 2008;23:1280–1286. doi: 10.1111/j.1440-1746.2008.05342.x. [DOI] [PubMed] [Google Scholar]
- 48.Ali N, Allam H, May R, et al. Hepatitis C virus-induced cancer stem cell-like signatures in cell culture and murine tumor xenografts. J Virol. 2011;85:12292–12303. doi: 10.1128/JVI.05920-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziol M, Nault JC, Aout M, et al. Intermediate hepatobiliary cells predict an increased risk of hepatocarcinogenesis in patients with hepatitis C virus-related cirrhosis. Gastroenterology. 2010;139:335–343. e332. doi: 10.1053/j.gastro.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 50.de Pereira TA, Witek RP, Syn WK, et al. Viral factors induce Hedgehog pathway activation in humans with viral hepatitis, cirrhosis, and hepatocellular carcinoma. Lab Invest. 2010;90:1690–1703. doi: 10.1038/labinvest.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machida K, Tsukamoto H, Mkrtchyan H, et al. Toll-like receptor 4 mediates synergism between alcohol and HCV in hepatic oncogenesis involving stem cell marker Nanog. Proc Natl Acad Sci U S A. 2009;106:1548–1553. doi: 10.1073/pnas.0807390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Machida K. TLRs, Alcohol, HCV, and Tumorigenesis. Gastroenterol Res Pract. 2010;2010:518674. doi: 10.1155/2010/518674. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.