Abstract
Aims/Hypothesis
One of the hallmarks of diabetes is impaired endothelial function. Previous studies showed that HDL can exert protective effects on endothelium stimulating NO production and protecting from inflammation and suggested that HDL in obese people with diabetes and dyslipidemia may have lower endothelial protective function. We aimed to investigate whether type 2 diabetes impairs HDL endothelium protective functions in people with otherwise normal lipid profile.
Methods
In a case-control study (n = 41 per group) nested in the Cooper Center Longitudinal Study we tested the ability of HDL to protect endothelium by stimulating endothelial nitric oxide synthase activity and suppressing NFκB-mediated inflammatory response in endothelial cells. In parallel we measured HDL protein composition, sphinogosine-1-phosphate and P-selectin.
Results
Despite similar levels of plasma HDL-C the HDL in individuals with type 2 diabetes lost almost 40% of its ability to stimulate eNOS activity (P<0.001) and 20% of its ability to suppress TNFα-dependent NFκB-mediated inflammatory response in endothelial cells (P<0.001) compared to non-T2D controls despite similar BMI and lipid profile (HDL-C, LDL-C, TC, TG). Significantly, the ability of HDL to stimulate eNOS activity was negatively associated with plasma levels of P-selectin, an established marker of endothelial dysfunction (r = −0.32, P<0.001). Furthermore, sphingosine-1-phosphate (S1P) levels were decreased in diabetic plasma (P = 0.017) and correlated with HDL-mediated eNOS activation.
Conclusions/Interpretations
Collectively, our data suggest that HDL in individuals with type 2 diabetes loses its ability to maintain proper endothelial function independent of HDL-C, perhaps due to loss of S1P, and may contribute to development of diabetic complications.
Introduction
Although dyslipidemia characterized by high triglycerides and low levels of HDL-cholesterol (HDL-C) is a hallmark of diabetes, mounting evidence supports the idea that HDL function rather than HDL-C concentration is the relevant measure of HDL protective properties. Three studies have demonstrated that HDL cholesterol efflux capacity (CEC), the ability to accept cholesterol from lipid-loaded macrophages, is impaired in people with cardiovascular disease and is inversely related to risk of CVD.[1–3] In addition to the reverse cholesterol transport from peripheral tissues, several other biological functions of HDL have been identified by which HDL exerts protective effects on endothelial function: namely its ability to reduce inflammation, apoptosis, and thrombosis.[4–6] In healthy individuals, HDL is anti-inflammatory; however, in chronic illnesses characterized by systemic inflammation, such as diabetes, HDL may become “dysfunctional” and actually promote inflammatory responses.[7, 8]
Studies of HDL endothelial protective functions suggest that endothelial and vascular effects of HDL are heterogeneous and may be impaired in many disease states such coronary artery disease, diabetes, and chronic kidney studies.[4, 9, 10] In particular HDL may protect endothelium through its ability to stimulate nitric oxide production by endothelial cells.[4, 11] Ex vivo studies of endothelial cells incubated with isolated HDL from healthy volunteers demonstrate that HDL activates endothelial nitric oxide synthase (eNOS) and stimulates NO production.[12] Furthermore, infusion of reconstituted HDL in patients with type 2 diabetes (T2D) restores endothelial function as measured by forearm blood flow responses.[13] The ability to stimulate eNOS activity and NO production, however, may not be preserved in HDL from obese people with T2D and metabolic syndrome (low HDL, high triglycerides, and large waist circumference).[14]
HDL activates eNOS in cultured endothelial cells in vitro, however, the specific molecular mechanism of the activation is not clear. The sphigosine-1-phosphate (S1P), a bioactive phospholipid primarily carried in plasma on HDL, can directly interact with endothelial S1P receptors, activating Akt and eNOS.[15] In agreement with this mechanism, HDL had no effect upon eNOS activation and NO production in mice lacking S1P receptors.[15] Other studies suggest that apoA-I, the main HDL protein, may be able to activate eNOS through its interaction with SR-BI on endothelial cells, and mice lacking SR-BI fail to stimulate eNOS and produce NO in response to HDL stimulation.[12] Furthermore, we have demonstrated previously that isolated HDL from humans increases eNOS activity and reduces endothelial inflammation in primary endothelial cells.[8] Finally, in recent studies, cholesterol efflux to HDL from endothelial cells via the ATP-binding transporter ABCG1 has also been shown to maintain endothelial function in mice fed a high cholesterol diet (a model known to induce endothelial dysfunction)[16] and in humans with coronary endothelial dysfunction.[17] Collectively, these studies suggest that HDL may play an important role in maintaining proper endothelial function, and that HDL dysfunction may contribute to endothelial dysfunction.
In this study, we aimed to determine whether T2D, in people with otherwise apparently normal lipid profiles, may impair HDL protective function of endothelial cells as assessed by ability of HDL to stimulate eNOS Ser1177 phosphorylation and/or inhibit TNF-α dependent NF-κB activation, and thus contribute to endothelial dysfunction associated with T2D. Our results show that HDL in people with T2D loses its endothelium protective properties independent of HDL-C levels and the loss of ability to stimulate eNOS activation is associated with decreased levels of S1P and increased circulating P-selectin, a marker of endothelial dysfunction. Moreover, the loss of HDL function was independent of the level of glycemic control.
Materials and methods
Study population
We obtained plasma samples from community dwelling participants from the Cooper Center Longitudinal Study (CCLS). The CCLS is an observational database of patient visits to the Cooper Clinic in Dallas, Texas, a preventive medicine practice established in 1970 which began collecting blood samples from participants in 1999. CCLS participants are generally healthy and are either self-referred or referred by their employers for preventive health examination which include a standardized medical examination, anthropometric measurements, fasting laboratory studies, and a maximal treadmill exercise test. Participants provided informed consent for the use of their data for research purposes. The CCLS database is maintained by The Cooper Institute, a nonprofit independent research institute and privacy precautions were maintained through institute policy. The data collection and informed consent processes were reviewed and approved by the IRB at The Cooper Institute.
The sub-sample of the CCLS utilized for this study was seen after 2008 during which 11,838 unique individuals had at least one visit with stored plasma acquisition in addition to the preventive medical examination. Key clinical variables included height and weight measured using a standard clinical stadiometer and scale to calculate body mass index and seated resting blood pressure measured with a calibrated sphygmomanometer. Laboratory studies obtained after a 12-hour fast included blood glucose, hemoglobin A1c, and cholesterol profile. Plasma samples were stored in 1.5 milliliter vials at -70°C and were not previously thawed.
The CCLS database was queried for all patients with available plasma and a diagnosis of type 2 diabetes, as defined by: self-report, blood glucose over 126 mg/dL, use of diabetic medication, or hemoglobin A1c of greater that 6.5% and 349 subjects with T2D were identified. Preliminary data suggested effect size approximately 0.73. We calculated that with n = 31 per group we will achieve 80% power to observe significant difference between people with diabetes and controls at significance level alpha = 0.05. We increased likelihood of significant findings by increasing power to 90% (n = 41) and randomly selected 41 patients with type 2 diabetes and 41 non-diabetic controls matched by age, sex, and BMI.
Cell-based assays
Primary human microvascular endothelial cells (HMEC) and primary bovine aortic endothelial cells (BAEC) (Invitrogen-Cascade Biological, Carlsbad, CA) were used for the studies. HMEC were used for the p-65 NFκB assay, and the BAEC were used in the eNOS stimulation assays as they proved superior in cell batch-to-batch reproducibility. In preliminary experiments of eNOS activation assay we used bovine aortic endothelial cells (BAEC) and in human aortic endothelial cells (HAEC). The control HDL isolated from apparently healthy people robustly stimulated eNOS Ser1177 phosphorylation compared to untreated cells (1.4x fold induction with 50 μg/mL HDL and maximal 1.6 and 1.7x induction with 100 nM insulin in HAEC and BAEC cell lines, respectively). Because BAEC cells responded to HDL in identical fashion as HAEC cells and provided more reproducible response to positive control, we used BAEC cells for all our eNOS experiments. Similarly, in preliminary experiments of HDL anti-inflammatory assay the HMEC cells provided similar response, but superior robustness to HAEC cells and therefore we used HMEC cells for our studies. Both cell types were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (Hyclone Laboratories, Logan, UT) and 12 μg/mL of bovine brain extract (Clonetics, Walkersville, MD), L-glutamine (2 mM), sodium pyruvate (1 mM) and nonessential amino acids in the presence of penicillin (100 units/mL) and maintained at 37°C in 5% CO2. For the assays HMEC and BAEC were plated in 24-well plates at 37°C in 5% CO2. For the anti-inflammatory assay the HMEC cells were then incubated with or without 50 μg/mL HDL in HMEC cell culture media (0.1% FBS) for 16h. The HMEC cells were washed extensively with PBS, and then incubated with an inflammatory stimulus (i.e. 5 ng/mL TNFα for 4 h) and cell lysates were harvested in the presence of protease and phosphatase inhibitors for analysis of phosphorylation of p65 (a marker of NFκB activation). For the eNOS activation assay the BAEC cells were treated with 50 μg/mL HDL for 30 min [18] and cell lysates were harvested in the presence of protease and phosphatase inhibitors. The cell lysates were assayed for phospho-Ser1179 eNOS (equivalent to human Ser1177) by Western blot analysis.
For the analysis the clinical samples were randomized and laboratory staff performing the analysis was blinded to the sample identity. Assay was performed in 24-well plates with all samples analyzed in parallel in a single batch as described above. For Western blot analysis the SDS-PAGE electrophoresis was performed using a 4–12% gradient gels, transferred to membranes and stained with specific antibodies (see below). The blots were scanned and the densitometry measurements from each gel were first normalized to a pooled normal HDL which was included in a random position on every gel to correct for gel-to-gel variability. Subsequently, for each sample, the signal of phosphorylated protein was normalized to total eNOS and p65, respectively. To calculate % activation the corrected and normalized data was then normalized to the values obtained from control samples incubated with vehicle alone (or TNF-alpha treated for the anti-inflammatory activity), and percent change relative to the control treatment condition was calculated.
Anti-phospho-p65 (#3033), anti-phospho-eNOS (Ser1177; #9570) (cross-reactive to bovine Ser1179 phospho-eNOS), anti-total p65 and eNOS were from Cell Signaling (Beverly, MA). Insulin levels were determined by ELISA kit (Crystal Chem, Inc. Downers Grove Il).
P-selectin
Plasma levels of P-selectin were measured by ELISA (R&D Systems, MN) according to manufacturer protocol.
HDL isolation
HDL was isolated from EDTA-anticoagulated plasma, using sequential ultracentrifugation (d = 1.063–1.21 mg/mL) essentially as described previously[19] in table-top ultracentrifuge Beckman Optima XL with TL100.1 rotor (Beckman, CA). Aliquots of HDL were immediately frozen and stored at -80 C prior to analysis as it was previously shown not to impact HDL protective effects on endothelial cells.[20]
Protein digestion
HDL (10 μg protein) was solubilized with 0.1% RapiGest (Waters, MA) in 200 mM ammonium bicarbonate, spiked with 1 μg of 15N-apoA-I as internal standard,[21] reduced with dithiothreitol, alkylated with iodoacetamide, and digested with two additions of trypsin (1:20, w/w HDL protein; Promega, WI) initially for 3 h at 37°C, and after second addition overnight. After acidic hydrolysis of RapiGest, samples were dried, and stored at −20°C until analysis. Samples were reconstituted in 5% acetonitrile, 0.1% formic acid to 0.25 μg/μL.[22]
Selection of the peptides and transitions for SRM analysis
At least two peptides for each protein were chosen for SRM analysis based on spectral count in shotgun experiments, observed frequency in the PeptideAtlas mass spectral database, and on screening experiments that evaluate the correlation of peptides from the same protein with one another across clinical study populations.[21] Peptides containing methionine residues were excluded from consideration. The transitions monitored for each peptide were selected based on signal intensity from a screening experiment. List of peptides and acquisition conditions is provided in S1 Table (isoforms of SAA, SAA1 and SAA2, were not distinguished and are reported together as SAA1/2).
Selected-reaction monitoring analysis
Tryptic digests of HDL were chromatographed using a nanoAquity UPLC (Waters, MA) with a C18 trapping column (XBridge BEH C18 100 Å, 5 μm, 0.1 x 40 mm, Waters, MA) (trapping flow rate 4 μL/min), a capillary XBridge BEH C18 analytical column (XBridge BEH C18 100Å, 3.5μm, 100x0.075 mm, Waters, MA), and a 30 min linear gradient of acetonitrile, 0.1% formic acid (7–35%) in 0.1% formic acid in water at a flow rate of 0.6 μL/min. The NanoAquity UPLC was connected to a Thermo TSQ Vantage triple-quadrupole mass spectrometer with electrospray ionization. The instrument was operated in SRM mode with 10 ms dwell time and the peptides were monitored collision energies optimized to maximize the signal. Peak areas were integrated using Skyline software.[23]
Measurement of S1P
Sphingosine-1-phosphate was extracted from 10 μL of plasma after dilution with 50μL with 30mM citric Acid/40mM sodium phosphate buffer pH 4.0 and spiking with 5uL internal standard (S1P-C17 base, Avanti LM-6002) to a concentration of 0.23μM.[24] After thorough mixing the sample was extracted with 275 μL of 1-butanol (Fisher A399), spun down and 220 μL were collected and dried down. The dried down samples were reconstituted for LCMS analysis in 125 μL of 50% methanol, 1% formic acid in 5mM ammonium formate. Samples were spun down and 10μL injected onto LCMS consisting of Shimadzu Prominence LC system and 4000 QTrap (ABSciex) in Turbo Ion Spray Mode. Separation was accomplished on a Luna 3μm C18 100Å 50x2.0mm column (Phenomenex) with 1.5 minute gradient from LC buffer A (50% methanol, 1% formic acid in 5mM ammonium formate) to LC buffer B (89% methanol, 10% 2−propanol, 1% formic acid in 5mM ammonium formate) and S1P was quantified together with the internal standard (S1P-C17) using multiple-reaction-monitoring. Calibration was accomplished using a single point calibrator repeatedly assayed throughout the analysis.
Statistical analysis
All HDL functional assays were performed in duplicate and the mean was computed and used for analysis. Triglyceride concentrations were log-transformed. Proteomics peptide measurements were normalized to 15N-apoA-I peptide VQPYLDDFQK and response of two peptides for each protein was averaged, log2 transformed and standardized prior to further analysis. Four samples failed the LCMS analysis and therefore were not used in the data analysis. Outliers in the data were identified using Median Absolute Deviation (MAD) approach[25] as data points exceeding 3*MAD from a median of a given group and were eliminated from corresponding analysis. Group comparisons were performed using Student’s t-test (non-parametric Mann-Whitney U test for proteomics) and associations between continuous variables were established by using linear regression and Pearson’s correlation coefficient, association with diabetes status were established using logistic regression. Multivariate logistic regression was used to investigate associations between diabetes status and HDL function controlling for potential confounders. Statistical significance was determined for P < 0.05 for all tests. Statistical analyses were performed using R software (version. 3.1).
Results
All subjects in the present study were moderately overweight (BMI>25, mean BMI 29.3±4.6 kg/m2). Consistent with their diabetes diagnosis, the subjects in the diabetic group had modest but significantly higher blood glucose and glycated hemoglobin A1c levels (Table 1). There were no differences between control and diabetic groups in any of the plasma lipids measures including HDL-C, LDL-C and triglycerides, as well as in blood pressure, or systemic measure of inflammation (hsCRP). Clinical and demographic characteristics of the study population are presented in the Table 1.
Table 1. Clinical characteristics of the study population.
| Control subjects (n = 41) |
T2D subjects (n = 41) |
P | |
|---|---|---|---|
| Age (yr) | 59.1 ± 7.4 | 60.1 ± 7.3 | 0.52 |
| Sex (F/M) | 7/34 | 7/34 | |
| Body mass index, kg/m2 | 29.1 ± 4.5 | 29.5 ± 4.7 | 0.71 |
| Fasting glucose, mg/dL | 96.5 ± 9.7 | 132 ± 30 | <0.001 |
| Hemoglobin A1C, % (mmol/mol) | 5.6 ± 0.4 (38 ± 4.4) |
6.8 ± 0.9 (51.0 ± 9.8) |
<0.001 |
| high sensitivity CRP, ng/mL | 2.6 ± 6.7 | 4.5 ± 5.6 | 0.16 |
| HDL cholesterol, mg/dL | 54 ± 15.8 | 53.7 ± 15 | 0.95 |
| LDL cholesterol, mg/dL | 94.8 ± 34.9 | 88.9 ± 38.9 | 0.47 |
| Total cholesterol, mg/dL | 172 ± 40.6 | 166.6 ± 45.2 | 0.57 |
| Triglycerides, mg/dL | 116.2 ± 49.4 | 119.8 ± 46.2 | 0.74 |
| Mean arterial pressure, mm Hg | 93 ± 8.1 | 92.9 ± 13.1 | 0.97 |
Values are presented as mean ± SD or number of subjects
HDL in type 2 diabetes loses anti-inflammatory properties
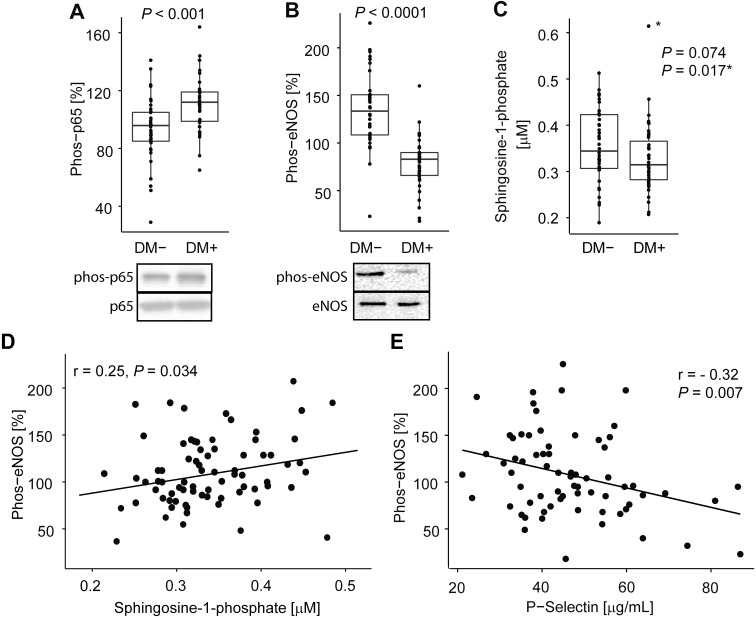
HDL has been shown to exert anti-inflammatory effects on endothelial cells suppressing activation of NFκB in human endothelial cells,[8] however, this function may be impaired in people with T2D. As a marker of NFκB activation we measured TNFα-dependent phosphorylation of p65, a key event in the NFκB pathway which is necessary for NFκB translocation and cellular inflammatory response,[26, 27] after pretreatment with HDL. When incubated with human endothelial cells, the HDL isolated from the non-diabetic controls suppressed the TNFα dependent NFκB activation in endothelial cells (mean = 93.2±22.4%; n = 41) (Fig 1A). In contrast, HDL from age and sex matched diabetic patients was unable to suppress the NFκB activation in these cells and even displayed pro-inflammatory effects (mean = 111.0±19.0%, n = 41).
Fig 1. Protective effects of HDL on endothelial cells are impaired in diabetes, correlate with S1P and negatively associate with in vivo measure of endothelial dysfunction.
(A) The ability of HDL to suppress NFκB activation was measured as phosphorylation of p65 in HMEC after 16 h incubation with HDL (50 μg/mL) followed by 4 h stimulation with TNFα (n = 41 per group). (B) The ability of HDL to stimulate eNOS Ser1179 phosphorylation was measured in BAEC after 30 min incubation with HDL (50 μg/mL) (n = 38 non-diabetic, n = 41 diabetic subjects). (Data is expressed relative to cells not treated with HDL). (C) Sphigosine-1-phosphate concentration measured by LC-MS is reduced in patients with diabetes and (D) correlates positively with HDL ability to stimulate eNOS phosphorylation (n = 40 non-diabetic, n = 39 diabetic subjects; *excluded outlier and P-value after exclusion). (E) The ability of HDL to stimulate eNOS is inversely correlated negatively with level of P-selectin in plasma, an in vivo measure of endothelial dysfunction (Pearson correlation coefficient; n = 81 after an outlier exclusion).
HDL in diabetes loses ability to stimulate eNOS activation
Diabetes is associated with endothelial dysfunction and HDL can protect endothelium by stimulating production of nitric oxide through activation of eNOS. We therefore measured the ability of HDL to stimulate phosphorylation of eNOS Ser1177, a marker of activation of endothelial nitric oxide synthase and NO production in endothelial cells.[28] Compared to the HDL from non-diabetic subjects, the HDL from the diabetic subjects lost its ability to stimulate Ser1179 phosphorylation and eNOS activation eliciting 40% less activity (P<0.0001) (Fig 1B). Notably, the response to the HDL from people with T2D was less than that of an untreated control, suggesting a suppressive rather than stimulatory effect.
Endothelial functions of HDL are independent of dyslipidemia and glycemic control
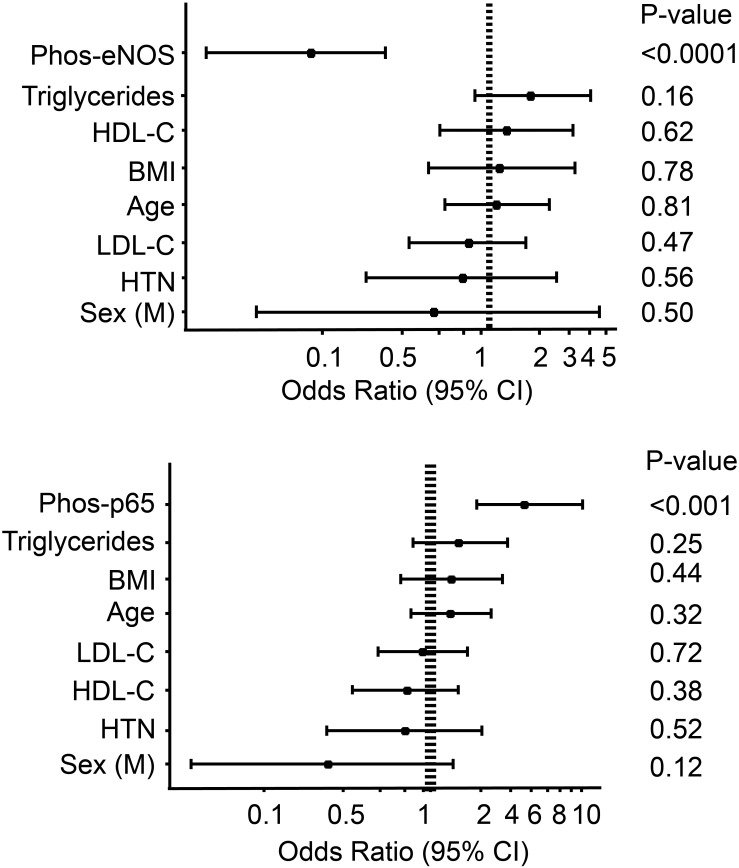
To further investigate factors, which may be associated with the impaired HDL functionality in diabetic patients, we employed multivariate logistic regression models. Both HDL anti-inflammatory activity (as measured by ability to suppress p65 phosphorylation) and stimulation of eNOS activity were independent of age, sex, and HDL-C (OR = 2.8, 95%CI 1.6–5.5, P = 0.001, and OR = 0.08, 95%CI 0.02–0.2, P<0.001, Table 2). The endothelium protective functions of HDL remained significantly associated with diabetes status even after adjusting for fasting glucose or glycated HbA1c (Table 2). In models with selected cardiovascular factors age, LDL-C, HDL-C, triglycerides or BMI only the HDL endothelium protective functions were strongly associated with the diabetes status (Fig 2). Moreover, linear regression analysis of HDL activity versus fasting glucose or glycated HbA1c, while revealing strong negative correlation of phos-eNOS (r = −0.32, P = 0.01, r = −0.35, P = 0.01 for glucose and HbA1c, respectively) explained only 12% of the HDL activity variance. The correlation with HDL anti-inflammatory properties was even weaker (r = 0.2, P = N.S., r = 0.25, P = 0.01 for fasting glucose and glycated HbA1c, respectively).
Table 2. Multivariate logistic regression analysis of diabetic status as a function of HDL activity adjusted for various potential confounders and measures of diabetes.
| Model | Tested variable | OR | 95% CI | P | |
|---|---|---|---|---|---|
| Univariate Models | Glucose | Glucose | 1.16 | 1.1–1.24 | <0.001 |
| HbA1c | HbA1c | 51.61 | 11.45–424.9 | <0.001 | |
| phos-p65# | phos-p65 | 2.78 | 1.59–5.46 | 0.001 | |
| phos-eNOS$ | phos-eNOS | 0.07 | 0.02–0.2 | <0.001 | |
| Multivariate models—age, sex and HDL-C | phos-p65+age+sex | phos-p65 | 2.76 | 1.57–5.45 | 0.001 |
| phos-p65+age+sex+HDL-C | phos-p65 | 2.78 | 1.59–5.48 | 0.001 | |
| phos-eNOS+age+sex | phos-eNOS | 0.07 | 0.02–0.21 | <0.001 | |
| phos-eNOS+age+sex+HDL-C | phos-eNOS | 0.07 | 0.02–0.21 | <0.001 | |
| Multivariate models—age, sex and glucose or HbA1c | phos-p65+age+sex+glucose | phos-p65 | 7.31 | 2.45–36.97 | 0.003 |
| phos-p65+age+sex+HbA1c | phos-p65 | 3.10 | 1.26–9.25 | 0.02 | |
| phos-eNOS+age+sex+glucose | phos-eNOS | 0.03 | 0–0.19 | 0.002 | |
| phos-eNOS+age+sex+HbA1c | phos-eNOS | 0.06 | 0.01–0.24 | 0.001 |
Odds ratio of diabetic status for 1-SD of HDL activity, or for 1 unit of fasting glucose or HbA1c
#—phos-p65—HDL anti-inflammatory activity
$—phos-eNOS—HDL eNOS stimulating activity
Fig 2. Odds ratios for type 2 diabetes according to HDL endothelial protective functions and selected dyslipidemia and common risk factors.
Multivariate logistic regression models included the listed dyslipidemia and other cardiovascular risk factors and odds ratios for continuous variables are presented per 1-SD increase for (A) HDL ability to stimulate eNOS activation, and (B) for the suppression of NFκB activation.
S1P levels are decreased in diabetes and associate with HDL function
Previous studies suggested that HDL effects on endothelial cells may be mediated at least in part through sphingosine-1-phosphate (S1P), for which HDL is the major carrier in plasma (over 70% of plasma levels of S1P are associated with HDL).[15] We therefore measured plasma levels of S1P. The S1P levels were 10% lower in diabetes compared to controls (P = 0.074, after excluding the single outlier in diabetic group, P = 0.017) (Fig 1C). Moreover, S1P levels were strongly associated with the ability of HDL to stimulate eNOS activity (r = 0.22, P = 0.055, r = 0.25, P = 0.034 after excluding a single outlier) (Fig 1D). Lack of association of the outlier S1P level with any other measures suggest that the high S1P value is likely due to assay technical issues. No association of S1P with HDL anti-inflammatory activity was observed.
The eNOS stimulation by HDL is negatively associated with endothelial dysfunction
Because our data implicate that diabetes impairs the ability of HDL to protect the endothelium, we investigated whether HDL function is associated with endothelial dysfunction in vivo. As an in vivo measure of endothelial dysfunction we quantified circulating levels of P-selectin, an established marker of endothelial dysfunction.[29] P-selectin was moderately elevated in diabetic patients (11% increase compared to non-diabetic patients, P = 0.14; 15% increase, P = 0.04 after excluding a single non-diabetic subject with extremely high P-selectin levels). Moreover, the ability of HDL to stimulate eNOS was negatively associated with the plasma P-selectin concentration (r = −0.32, P = 0.007) (Fig 1E).
HDL protein composition in type 2 diabetes
To investigate if HDL protein composition was associated with the loss of the protective properties of HDL, we employed the quantitative targeted proteomics approach that we have developed previously.[21] We quantified 36 major HDL associated proteins (Table 3) using 15N-isotope labeled apolipoprotein A-I for internal standardization and compared the relative abundance of these proteins between diabetic and non-diabetic subjects. While several proteins were significantly different between diabetic subjects and non-diabetic controls in univariate analysis, apoC-III (P = 0.03), SAA1/2 (SAA1 and SAA2 measured together) (P = 0.0032) increased and apoA-IV (P = 0.034) decreased, after controlling for multiple comparisons, none of the protein differences remained significant. Moreover, only SAA1/2 showed modest inverse correlation with HDL eNOS stimulating activity suggesting that it may in part contribute to the impairment of this HDL function in T2D (r = −0.26, P = 0.026). Collectively, our proteomics data indicate that HDL protein composition does not significantly contribute to HDL functional impairment in people with T2D.
Table 3. Protein quantified in the targeted proteomics analysis.
| Protein | Protein Name | Protein | Protein Name |
|---|---|---|---|
| ALB | Serum albumin | CETP | Cholesteryl ester transfer protein |
| APOA1 | Apolipoprotein A-I | CLU | Clusterin(Apolipoprotein J) |
| APOA2 | Apolipoprotein A-II | HPR | Haptoglobin-related protein |
| APOA4 | Apolipoprotein A-IV | HPX | Hemopexin (Beta-1B-glycoprotein) |
| APOB | Apolipoprotein B-100 | LCAT | Lecithin-cholesterol acyltransferase |
| APOC1 | Apolipoprotein C-I | LPA | Apolipoprotein(a) |
| APOC2 | Apolipoprotein C-II | PCYOX1 | Prenylcysteine oxidase 1 |
| APOC3 | Apolipoprotein C-III | PLTP | Phospholipid transfer protein |
| APOC4 | Apolipoprotein C-IV | PON1 | Serum paraoxonase/arylesterase 1 |
| APOD | Apolipoprotein D | PON3 | Serum paraoxonase/lactonase 3 |
| APOE | Apolipoprotein E | RBP4 | Retinol-binding protein 4 |
| APOF | Apolipoprotein F | SAA1_2 | Serum Amyloid A-1/Serum Amyloid-2 proteins |
| APOH | Beta-2-glycoprotein 1 | SAA4 | Serum amyloid A-4 protein |
| APOL1 | Apolipoprotein L1 | SERPINA1 | Alpha-1-antitrypsin |
| APOM | Apolipoprotein M | SERPINA4 | Kallistatin (Kallikrein inhibitor) |
| C3 | Complement C3 | VDBP | Vitamin D Binding Protein |
| C4A | Complement C4-A | VTN | Vitronectin |
Discussion
In the present study of community dwelling patients with T2D and normal lipid profiles, we found that diabetes significantly impaired HDL endothelial protective functions, as measured by ability of HDL to stimulate eNOS and attenuate NF-κB activation in response to TNFα, despite lack of differences, compared to non-diabetic subjects, in traditional clinical measures associated with metabolic syndrome and cardiovascular risk (HDL-C, TG, LDL-C, BMI, hypertension or CRP). Moreover, our results strongly suggest that in vivo endothelial dysfunction may be associated with the observed HDL dysfunction and that this HDL dysfunction is not associated with level of glycemic control. Collectively, our data indicate that the ability of HDL to maintain endothelial health is impaired in T2D and may contribute the endothelial dysfunction associated with T2D.
Multiple studies showed that diabetes is associated with endothelial dysfunction and that endothelial dysfunction may be associated with insulin resistance and precede diabetes onset[29–31] and HDL has been shown to stimulate eNOS/NO mediated vasodilation[14, 16] as well as suppress the endothelial inflammatory response to TNFα[32] and decrease endothelial cell exocytosis.[33] Elevation of HDL by infusion of reconstituted HDL increased endothelial function as measured by increased blood flow in response to acetylcholine,[13] improved flow-mediated vasodilation in type 2 diabetic patients[34] and the improved anti-inflammatory function of HDL was strongly correlated with elevation of apoA-I and HDL-C concentration.[35]
In a recent study the ability of HDL to protect the endothelium was impaired in people with T2D and metabolic syndrome (markedly low HDL, 33 vs. 53 mg/dL in controls; higher central obesity waist circumference 116 vs 91 cm, BMI 33 vs 27 kg/m2; and elevated triglycerides 225 vs 168 mg/dL),[14] raising the question whether the observed phenomena is merely reflection of low HDL-C levels. HDL inflammatory index (ability to protect from oxidized-LDL induced inflammation) was also reduced in obese, hypertensive subjects with previous CVD compared to healthy controls.[36] In contrast, our results clearly demonstrate that the HDL dysfunction associated with diabetes is independent of HDL-C levels or any other indicators of metabolic syndrome or other co-morbidities (Fig 2). Several studies suggest that the impairment of the HDL function may be associated with glycation of apoA-I.[37] In the current study, the HDL eNOS activity was only very modestly correlated with fasting glucose or glycated HbA1c and independent of it in multivariate regression models (Table 2), suggesting that the ability of the HDL to stimulate eNOS activity is also independent of glycemic control.
The ability of HDL to directly activate eNOS in primary aortic endothelial cells may be mediated by several mechanisms. Multiple lines of evidence suggest sphingosine-1-phosphate (S1P), primarily carried in plasma in HDL,[38] is one of the key effectors of HDL protective endothelial functions.[39–41] S1P can directly interact with endothelial S1P receptors (S1PR1, S1PR3), activating Akt and eNOS.[15] In the current study we found decreased levels of S1P in diabetic patients and a strong negative correlation between plasma S1P levels and ability of HDL to activate eNOS. In agreement with our results, a recent study in mouse models of diabetes showed decrease of plasma S1P levels in streptozotocin-induced diabetic mice.[42] Notably, S1P content of HDL has been associated with the presence of apoM.[43] However, we found that apoM levels were not different between T2D patients and controls suggesting that while apoM may control a large portion of HDL S1P content, it is not associated with decrease of S1P in diabetes and ultimately for HDL functionality. Indeed, a recent study showed that HDL may be replenished with S1P, even in the absence of apoM, completely restoring its eNOS stimulating activity.[44]
In addition to actions of HDL-carried S1P, apoA-I has also been reported to activate eNOS through its interaction with SR-BI on endothelial cells.[45] However, others have found that despite being the ligand for SR-BI, lipid free apoA-I failed to activate eNOS[46] suggesting that other HDL components may be important or are required to mediate the apoA-I effect and allow it to interact with SR-BI. While our proteomics analysis did not detect a significant difference in HDL-associated apoA-I levels between T2D and controls, other investigators have suggested that hyperglycemia results in nonenzymatic glycation of apoA-I[37] and that glycation of apoA-I, and advanced glycation end-products (AGEs) in general, may contribute to reduction of HDL ability to activate eNOS.[47] Although the T2D patients had elevated glucose and glycated HbA1c, the concentrations we observed only slightly correlated with the ability of HDL to activate eNOS. Multivariate regression suggested that the inability of HDL from people with T2D to activate eNOS is independent of glucose or hemoglobin A1c concentrations, indicating that glycation may contribute only partially to the diabetes associated HDL dysfunction.
The heterogeneity of the vascular effects of HDL may be attributed to HDL-associated proteome and lipid content as well as post-translational protein modification. The total pool of HDL within an individual is composed of numerous HDL subpopulations with distinct protein and lipid composition. Although proteomic studies have reported >90 HDL associated proteins, only a small fraction of these has been consistently found associated with HDL. We performed quantitative analysis of 35 proteins consistently associated with HDL including proteins with relative abundance as low as 0.1% of the abundance of apoA-I, and did not find any significant differences between T2D and control patients. These results suggest that differences in other, low abundance proteins not captured in our analysis and/or post translational modification of the measured proteins may be responsible for functional differences. HDL also carries a rich ensemble of phospholipids in addition to S1P, which may mediate HDL function and have been associated with insulin resistance and diabetes.[48] It is thus possible that other lipids then S1P may contribute to the HDL dysfunction in diabetes.
In future studies, it will be important to determine whether specific post-translational modifications and lipid components of HDL mediate the impairment of HDL endothelial protective function in diabetes and whether and how therapies targeting HDL affect the HDL ability to protect endothelium.
Strengths of our study include the unique patient population, the similar baseline characteristics of control and diabetic subjects, and the use of novel assays for quantifying HDL endothelial protective functions. Limitations include the lack direct measurement of in vivo endothelial function, S1P levels in HDL and data on HDL protein glycation or other post-translation modifications.
Conclusions
In summary, our study provided strong evidence suggesting that type 2 diabetes impairs HDL-mediated eNOS activation and HDL-mediated attenuation of NFκB signaling in endothelial cells and that this HDL dysfunction may be associated with in vivo endothelial dysfunction. This HDL dysfunction appears to be independent of plasma lipids (HDL-C, TG, LDL-C) or other risk factors associated with endothelial dysfunction (i.e. BMI or central obesity). These findings suggest that loss of HDL endothelial protective functions may contribute to increased risk of CVD in diabetes. Our findings also suggest that these HDL functions might be target for development new therapeutics for CVD.
Supporting information
(PDF)
Acknowledgments
The authors thank Kenneth H. Cooper, MD, MPH, for establishing the CCLS, the Cooper Clinic for collecting the data, and The Cooper Institute for maintaining the data and biobank. We acknowledge the support for the proteomics analysis by the University of Washington's Proteomics Resource (UWPR95794).
Data Availability
All relevant biochemical data are included in the paper. Proteomics data are available from figshare (DOI: 10.6084/m9.figshare.5890555), as well as the PanoramaWeb public repository for targeted proteomics results (https://panoramaweb.org/labkey/VaisarHDL_T2D.url).
Funding Statement
This study was supported by National Institute of Health (http://www.nih.gov/) Grants DK106703 (to FK), DK035816 and HL111375 (to ANH), American Heart Association (https://professional.heart.org/) 14GRNT18410022 (to TV), the Kenneth H. Cooper Endowed Professorship in Preventive Cardiology (FK). HDL isolation and proteomics analysis were supported by NIH grants HL092969 and DK017047. The proteomics work was supported in part by the University of Washington's Proteomics Resource (UWPR95794). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Khera AV, Cuchel M, de la Llera-Moya M, Rodrigues A, Burke MF, Jafri K, et al. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. N Engl J Med. 2011;364(2):127–35. doi: 10.1056/NEJMoa1001689 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohatgi A, Khera A, Berry JD, Givens EG, Ayers CR, Wedin KE, et al. HDL cholesterol efflux capacity and incident cardiovascular events. N Engl J Med. 2014;371(25):2383–93. doi: 10.1056/NEJMoa1409065 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saleheen D, Scott R, Javad S, Zhao W, Rodrigues A, Picataggi A, et al. Association of HDL cholesterol efflux capacity with incident coronary heart disease events: a prospective case-control study. Lancet Diabetes Endocrinol. 2015;3(7):507–13. doi: 10.1016/S2213-8587(15)00126-6 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riwanto M, Landmesser U. High density lipoproteins and endothelial functions: mechanistic insights and alterations in cardiovascular disease. J Lipid Res. 2013;54(12):3227–43. doi: 10.1194/jlr.R037762 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rye KA, Barter PJ. Cardioprotective functions of HDLs. J Lipid Res. 2014;55(2):168–79. doi: 10.1194/jlr.R039297 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mineo C, Shaul PW. Novel biological functions of high-density lipoprotein cholesterol. Circulation research. 2012;111(8):1079–90. doi: 10.1161/CIRCRESAHA.111.258673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kratzer A, Giral H, Landmesser U. High-density lipoproteins as modulators of endothelial cell functions: alterations in patients with coronary artery disease. Cardiovasc Res. 2014;103(3):350–61. doi: 10.1093/cvr/cvu139 . [DOI] [PubMed] [Google Scholar]
- 8.Cheng AM, Handa P, Tateya S, Schwartz J, Tang C, Mitra P, et al. Apolipoprotein A-I attenuates palmitate-mediated NF-kappaB activation by reducing Toll-like receptor-4 recruitment into lipid rafts. PLoS One. 2012;7(3):e33917 doi: 10.1371/journal.pone.0033917 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Santos-Gallego CG, Rosenson RS. Role of HDL in those with diabetes. Curr Cardiol Rep. 2014;16(8):512 doi: 10.1007/s11886-014-0512-5 . [DOI] [PubMed] [Google Scholar]
- 10.Lowenstein CJ, Cameron SJ. High-density lipoprotein metabolism and endothelial function. Curr Opin Endocrinol Diabetes Obes. 2010;17(2):166–70. doi: 10.1097/MED.0b013e32833727ee . [DOI] [PubMed] [Google Scholar]
- 11.Mineo C, Shaul PW. Regulation of signal transduction by HDL. Journal of lipid research. 2013;54(9):2315–24. doi: 10.1194/jlr.R039479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuhanna IS, Zhu Y, Cox BE, Hahner LD, Osborne-Lawrence S, Lu P, et al. High-density lipoprotein binding to scavenger receptor-BI activates endothelial nitric oxide synthase. Nat Med. 2001;7(7):853–7. doi: 10.1038/89986 . [DOI] [PubMed] [Google Scholar]
- 13.Spieker LE, Sudano I, Hurlimann D, Lerch PG, Lang MG, Binggeli C, et al. High-density lipoprotein restores endothelial function in hypercholesterolemic men. Circulation. 2002;105(12):1399–402. doi: 10.1161/01.CIR.0000013424.28206.8F . [DOI] [PubMed] [Google Scholar]
- 14.Sorrentino SA, Besler C, Rohrer L, Meyer M, Heinrich K, Bahlmann FH, et al. Endothelial-vasoprotective effects of high-density lipoprotein are impaired in patients with type 2 diabetes mellitus but are improved after extended-release niacin therapy. Circulation. 2010;121(1):110–22. doi: 10.1161/CIRCULATIONAHA.108.836346 . [DOI] [PubMed] [Google Scholar]
- 15.Nofer JR, van der Giet M, Tolle M, Wolinska I, von Wnuck Lipinski K, Baba HA, et al. HDL induces NO-dependent vasorelaxation via the lysophospholipid receptor S1P3. J Clin Invest. 2004;113(4):569–81. doi: 10.1172/JCI18004 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terasaka N, Yu S, Yvan-Charvet L, Wang N, Mzhavia N, Langlois R, et al. ABCG1 and HDL protect against endothelial dysfunction in mice fed a high-cholesterol diet. J Clin Invest. 2008;118(11):3701–13. doi: 10.1172/JCI35470 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Monette JS, Hutchins PM, Ronsein GE, Wimberger J, Irwin AD, Tang C, et al. Patients with coronary endothelial dysfunction have impaired cholesterol efflux capacity and reduced HDL particle concentration. Circ Res. 2016;119(1):83–90. doi: 10.1161/CIRCRESAHA.116.308357 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mineo C, Yuhanna IS, Quon MJ, Shaul PW. High density lipoprotein-induced endothelial nitric-oxide synthase activation is mediated by Akt and MAP kinases. The Journal of biological chemistry. 2003;278(11):9142–9. doi: 10.1074/jbc.M211394200 [DOI] [PubMed] [Google Scholar]
- 19.Vaisar T, Pennathur S, Green PS, Gharib SA, Hoofnagle AN, Cheung MC, et al. Shotgun proteomics implicates protease inhibition and complement activation in the antiinflammatory properties of HDL. J Clin Invest. 2007;117(3):746–56. doi: 10.1172/JCI26206 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denimal D, Monier S, Brindisi MC, Petit JM, Bouillet B, Nguyen A, et al. Impairment of the Ability of HDL From Patients With Metabolic Syndrome but Without Diabetes Mellitus to Activate eNOS: Correction by S1P Enrichment. Arterioscler Thromb Vasc Biol. 2017;37(5):804–11. doi: 10.1161/ATVBAHA.117.309287 . [DOI] [PubMed] [Google Scholar]
- 21.Hoofnagle AN, Becker JO, Oda MN, Cavigiolio G, Mayer P, Vaisar T. Multiple-reaction monitoring-mass spectrometric assays can accurately measure the relative protein abundance in complex mixtures. Clin Chem. 2012;58(4):777–81. doi: 10.1373/clinchem.2011.173856 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheung MC, Vaisar T, Han X, Heinecke JW, Albers JJ. Phospholipid transfer protein in human plasma associates with proteins linked to immunity and inflammation. Biochemistry. 2010;49(34):7314–22. doi: 10.1021/bi100359f . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26(7):966–8. doi: 10.1093/bioinformatics/btq054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ieronimakis N, Pantoja M, Hays AL, Dosey TL, Qi J, Fischer KA, et al. Increased sphingosine-1-phosphate improves muscle regeneration in acutely injured mdx mice. Skelet Muscle. 2013;3(1):20 doi: 10.1186/2044-5040-3-20 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leys C, Ley C, Klein O, Bernard P, Licata L. Detecting outliers: Do not use standard deviation around the mean, use absolute deviation around the median. Journal of Experimental Social Psychology. 2013;49(4):764–6. doi: 10.1016/j.jesp.2013.03.013 [Google Scholar]
- 26.Napetschnig J, Wu H. Molecular basis of NF-kappaB signaling. Annu Rev Biophys. 2013;42:443–68. Epub 2013/03/19. doi: 10.1146/annurev-biophys-083012-130338 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kempe S, Kestler H, Lasar A, Wirth T. NF-kappaB controls the global pro-inflammatory response in endothelial cells: evidence for the regulation of a pro-atherogenic program. Nucleic Acids Res. 2005;33(16):5308–19. Epub 2005/09/24. doi: 10.1093/nar/gki836 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fulton D, Gratton JP, McCabe TJ, Fontana J, Fujio Y, Walsh K, et al. Regulation of endothelium-derived nitric oxide production by the protein kinase Akt. Nature. 1999;399(6736):597–601. doi: 10.1038/21218 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meigs JB, Hu FB, Rifai N, Manson JE. Biomarkers of endothelial dysfunction and risk of type 2 diabetes mellitus. JAMA. 2004;291(16):1978–86. doi: 10.1001/jama.291.16.1978 . [DOI] [PubMed] [Google Scholar]
- 30.Chadderdon SM, Belcik JT, Bader L, Kirigiti MA, Peters DM, Kievit P, et al. Proinflammatory endothelial activation detected by molecular imaging in obese nonhuman primates coincides with onset of insulin resistance and progressively increases with duration of insulin resistance. Circulation. 2014;129(4):471–8. doi: 10.1161/CIRCULATIONAHA.113.003645 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gordon SM, Davidson WS, Urbina EM, Dolan LM, Heink A, Zang H, et al. The effects of type 2 diabetes on lipoprotein composition and arterial stiffness in male youth. Diabetes. 2013;62(8):2958–67. doi: 10.2337/db12-1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cockerill GW, Rye KA, Gamble JR, Vadas MA, Barter PJ. High-density lipoproteins inhibit cytokine-induced expression of endothelial cell adhesion molecules. Arterioscler Thromb Vasc Biol. 1995;15(11):1987–94. . [DOI] [PubMed] [Google Scholar]
- 33.Cameron SJ, Morrell CN, Bao C, Swaim AF, Rodriguez A, Lowenstein CJ. A Novel Anti-Inflammatory Effect for High Density Lipoprotein. PLoS One. 2015;10(12):e0144372 doi: 10.1371/journal.pone.0144372 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieuwdorp M, Vergeer M, Bisoendial RJ, op 't Roodt J, Levels H, Birjmohun RS, et al. Reconstituted HDL infusion restores endothelial function in patients with type 2 diabetes mellitus. Diabetologia. 2008;51(6):1081–4. doi: 10.1007/s00125-008-0975-2 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patel S, Drew BG, Nakhla S, Duffy SJ, Murphy AJ, Barter PJ, et al. Reconstituted high-density lipoprotein increases plasma high-density lipoprotein anti-inflammatory properties and cholesterol efflux capacity in patients with type 2 diabetes. J Am Coll Cardiol. 2009;53(11):962–71. doi: 10.1016/j.jacc.2008.12.008 . [DOI] [PubMed] [Google Scholar]
- 36.Morgantini C, Natali A, Boldrini B, Imaizumi S, Navab M, Fogelman AM, et al. Anti-inflammatory and antioxidant properties of HDLs are impaired in type 2 diabetes. Diabetes. 2011;60(10):2617–23. doi: 10.2337/db11-0378 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pu LJ, Lu L, Zhang RY, Du R, Shen Y, Zhang Q, et al. Glycation of apoprotein A-I is associated with coronary artery plaque progression in type 2 diabetic patients. Diabetes Care. 2013;36(5):1312–20. doi: 10.2337/dc12-1411 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hammad SM, Al Gadban MM, Semler AJ, Klein RL. Sphingosine 1-phosphate distribution in human plasma: associations with lipid profiles. J Lipids. 2012;2012:180705 doi: 10.1155/2012/180705 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lucke S, Levkau B. Endothelial functions of sphingosine-1-phosphate. Cell Physiol Biochem. 2010;26(1):87–96. doi: 10.1159/000315109 . [DOI] [PubMed] [Google Scholar]
- 40.Sattler K, Levkau B. Sphingosine-1-phosphate as a mediator of high-density lipoprotein effects in cardiovascular protection. Cardiovasc Res. 2009;82(2):201–11. doi: 10.1093/cvr/cvp070 . [DOI] [PubMed] [Google Scholar]
- 41.Argraves KM, Gazzolo PJ, Groh EM, Wilkerson BA, Matsuura BS, Twal WO, et al. High density lipoprotein-associated sphingosine 1-phosphate promotes endothelial barrier function. J Biol Chem. 2008;283(36):25074–81. doi: 10.1074/jbc.M801214200 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nojiri T, Kurano M, Tokuhara Y, Ohkubo S, Hara M, Ikeda H, et al. Modulation of sphingosine-1-phosphate and apolipoprotein M levels in the plasma, liver and kidneys in streptozotocin-induced diabetic mice. J Diabetes Investig. 2014;5(6):639–48. doi: 10.1111/jdi.12232 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christoffersen C, Obinata H, Kumaraswamy SB, Galvani S, Ahnstrom J, Sevvana M, et al. Endothelium-protective sphingosine-1-phosphate provided by HDL-associated apolipoprotein M. Proc Natl Acad Sci U S A. 2011;108(23):9613–8. doi: 10.1073/pnas.1103187108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sattler K, Graler M, Keul P, Weske S, Reimann CM, Jindrova H, et al. Defects of High-Density Lipoproteins in Coronary Artery Disease Caused by Low Sphingosine-1-Phosphate Content: Correction by Sphingosine-1-Phosphate-Loading. J Am Coll Cardiol. 2015;66(13):1470–85. doi: 10.1016/j.jacc.2015.07.057 . [DOI] [PubMed] [Google Scholar]
- 45.Drew BG, Fidge NH, Gallon-Beaumier G, Kemp BE, Kingwell BA. High-density lipoprotein and apolipoprotein AI increase endothelial NO synthase activity by protein association and multisite phosphorylation. Proc Natl Acad Sci U S A. 2004;101(18):6999–7004. doi: 10.1073/pnas.0306266101 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Beer MC, Durbin DM, Cai L, Jonas A, de Beer FC, van der Westhuyzen DR. Apolipoprotein A-I conformation markedly influences HDL interaction with scavenger receptor BI. J Lipid Res. 2001;42(2):309–13. . [PubMed] [Google Scholar]
- 47.Goldin A, Beckman JA, Schmidt AM, Creager MA. Advanced glycation end products: sparking the development of diabetic vascular injury. Circulation. 2006;114(6):597–605. doi: 10.1161/CIRCULATIONAHA.106.621854 . [DOI] [PubMed] [Google Scholar]
- 48.Hoofnagle AN, Vaisar T, Mitra P, Chait A. HDL lipids and insulin resistance. Curr Diab Rep. 2010;10(1):78–86. doi: 10.1007/s11892-009-0085-7 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Data Availability Statement
All relevant biochemical data are included in the paper. Proteomics data are available from figshare (DOI: 10.6084/m9.figshare.5890555), as well as the PanoramaWeb public repository for targeted proteomics results (https://panoramaweb.org/labkey/VaisarHDL_T2D.url).