Abstract
Ca2+-selective transient receptor potential vanilloid subfamily member 6 (TRPV6) channels play a critical role in calcium uptake in epithelial tissues1–4. Altered TRPV6 expression is associated with a variety of human diseases5, including cancers6. TRPV6 channels are constitutively active1,7,8 and their open probability depends on the lipidic composition of the membrane, increasing significantly in the presence of phosphatidylinositol 4,5-bisphosphate (PIP2)7,9. We previously solved crystal structures of detergent-solubilized rat TRPV6 in the closed state10,11. Corroborating previous electrophysiological studies3, these structures demonstrated that the Ca2+ selectivity of TRPV6 arises from a ring of aspartate side chains in the selectivity filter that tightly binds Ca2+. However, it has remained unknown how TRPV6 channels open and close their pores for ion permeation. Here we present cryo-EM structures of human TRPV6 in the open and closed states. The channel selectivity filter adopts similar conformations in both states, consistent with its explicit role in ion permeation. The iris-like channel opening is accompanied by an α-to-π helical transition in the pore-lining S6 helices at an alanine hinge just below the selectivity filter. As a result of this transition, the S6 helices bend and rotate, exposing different residues to the ion channel pore in the open and closed states. This novel gating mechanism, which defines the constitutive activity of TRPV6, is unique for tetrameric ion channels and provides new structural insights for understanding their diverse roles in physiology and disease.
We expressed the full-length human TRPV6 (hTRPV6) channel in HEK 293 cells, where it exhibited typical Ca2+ permeability12 (Fig. 1a–b) and current-voltage relationships13–17 (Extended Data Fig. 1a). To structurally characterize hTRPV6, we purified it separately in nanodiscs and amphipols (Methods) and solved the corresponding structures using cryo-EM (Extended Data Figs. 2–3 and Extended Data Table 1) to 3.6 Å and 4.0 Å, respectively. While the reconstructions in nanodiscs and amphipols were nearly identical, the structure solved in nanodiscs had better overall resolution and will be our primary descriptor of hTRPV6. Two-dimensional class averages showed diverse orientations and easily discernable secondary structure features (Fig. 1c). The resulting 3D reconstruction (Fig. 1d–e) showed higher resolution features for the core of the molecule when compared to its periphery (Extended Data Fig. 2c) and was of sufficient quality (Extended Data Fig. 4) to build each subunit (residues 28 to 638) of the hTRPV6 homotetramer de novo.
Figure 1. Function and cryo-EM of hTRPV6.

a–b, Functional characterization of hTRPV6 using ratiometric fluorescence measurements. a, Fluorescence curves recorded from HEK 293 cells expressing hTRPV6 in response to the application of Ca2+ (arrow) at different concentrations. These experiments were repeated independently three times with similar results. b, Ca2+ dose-response curve for the maximal value of fluorescence fitted with the logistic equation. Calculated IC50 is the mean ± SEM (n = 3). c, Two-dimensional class averages of hTRPV6 particles, showing diverse orientations. d–e, hTRPV6 3.6 Å cryo-EM reconstruction, with density shown at 0.035 threshold level (UCSF Chimera) representing hTRPV6 subunits colored green, cyan, pink and yellow, lipid in purple and ions in red.
The structure of hTRPV6 (Fig. 2a–b) has the same overall architecture as rat TRPV6 (rTRPV6)10. While no discernible lipid densities were previously observed in the crystal structures of rTRPV610,11, the hTRPV6 cryo-EM reconstruction reveals 16 (4 per subunit) well-resolved non-protein densities that are intercalated in subunit interfaces and likely represent lipids (Fig. 2c). Similarly positioned densities in the structure of TRPV118 were modeled with phosphatidylinositol (PI), phosphatidylcholine (PC), and phosphatidylethanolamine (PE) lipids. Of the four putative lipid densities in hTRPV6, the fourth density has a clear head-and-two-tails appearance. Fitting different lipid molecules into density 4 (Extended Data Fig. 5a–c) suggests that the chemical environment around the lipid head group, including the negatively charged aspartate D525 and polar Y349, Y509, Q513 and Y524, supports binding of PE or PC rather than PIP2. Densities 1–3 have sausage-like appearances and might represent a wider variety of lipid-like molecules, including cholesterol or cholesterol hemisuccinate (CHS) (Extended Data Fig. 5d–e). In physiological conditions, some of these sites can bind PIP27,9. For example, positively charged R470 and K484 as well as polar T479, Q483 and Q596 around density 2 create a permissive chemical environment for the head group of PIP2. However, the poor fit of PIP2 into density 2 (Extended Data Fig. 5f) suggests that in our cryo-EM structure it represents a different molecule.
Figure 2. Structure of hTRPV6.

a–b, Side (a) and top (b) views of hTRPV6 tetramer, with each subunit (A–D) shown in different color. Putative lipid densities at 3.5σ and ion densities at 4σ are illustrated by purple and red mesh, respectively. c, Expanded view of the four (1–4) putative lipid densities per hTRPV6 subunit. d–e, Expanded views of the putative ion densities at 4σ at the (d) selectivity filter and (e) S6 helices bundle crossing.
Previously, we crystallized rTRPV6 in the closed state, where the M577 side chains formed a hydrophobic “seal” on the cytoplasmic side of the S6 helices10,11. Conversely, interatomic distances within the pore confirmed that the hTRPV6 channel pore is open (Fig. 3). The pore surface is lined by the side chains of D542, T539, N572, I575, D580 and W583, as well as the backbone carbonyl oxygens of I540, I541 and G579. The narrowest part of the upper pore, the selectivity filter, is formed by the D542 side chains, one from each subunit, which project towards the center of the pore (Fig. 3a,c,f). We propose that D542 in hTRPV6, similar to D541 in rTRPV6, plays an important role in Ca2+ permeation by directly coordinating dehydrated Ca2+ ions11. The narrowest part of the hTRPV6 lower pore (9.6-Å interatomic distance) is defined by the side chains of I575 at the S6 bundle crossing (Fig. 3a,c,d). This part of the pore is comparable in size to the pore of open TRPV1 (9.3-Å interatomic distance, measured between side chains of I679)18 and is wide-open for conductance of hydrated Na+ or Ca2+ ions.
Figure 3. Open and closed ion channel pore.

a–b, Ion conduction pathway (green) in (a) open hTRPV6 and (b) closed hTRPV6-R470E, with residues lining the selectivity filter and around the gate shown as sticks. Only two of four subunits are shown, with the front and back subunits removed for clarity. c, Pore radius calculated using HOLE29 for hTRPV6 (orange) and hTRPV6-R470E (blue). d–e, Intracellular view of the S6 bundle crossing in (d) hTRPV6 and (e) hTRPV6-R470E. f, Superposition of the selectivity filter regions in hTRPV6 (orange) and hTRPV6-R470E (blue), viewed extracellularly. g, Superposition of the P-loop and S6 in hTRPV6 (orange) and hTRPV6-R470E (blue), viewed parallel to the membrane. The straight line shows the pore axis, red arrow indicates the position of the gating hinge alanine A566 and black arrows illustrate ~100° rotation and ~11° bending away from the pore axis of the C-terminal part of S6.
Along the axis of the hTRPV6 pore, there is a strong density ~3.9 Å away from the side chains of D542 (Fig. 2d) likely representing a Ca2+ ion bound to a site homologous to the main Ca2+ binding site (Site 1) at D541 in the pore of rTRPV610,11. An additional strong density along the hTRPV6 pore is observed at the bundle crossing of the S6 helices, in the vicinity of D580 (~8.0 Å apart) and W583 (~6.6 Å apart), suggesting that these residues may play an important role in ion permeation (Fig. 2e). Indeed, W583 is conserved in TRPV6 and TRPV5 channels and is involved in regulation of calcium uptake, as evidenced by mutation W583A in TRPV5, which induces cell death due to increased calcium influx19. The density at the S6 helices bundle crossing, which was not observed in the pore of the closed-state rTRPV610,11, most likely represents another permeant ion bound in the open pore of hTRPV6.
In order to determine the structure of hTRPV6 in the closed state, we decided to shift the open-closed state gating equilibrium towards the closed state by interfering with channel activation. Because the open probability of TRPV6 is strongly dependent on membrane lipids7,9, disturbing lipid binding by mutagenesis might result in channel closure. TRPV1, for example, is activated through an intramembrane vanilloid binding site (Fig. 4a), which accommodates agonists, such as resiniferatoxin (RTX) and capsaicin, and antagonists, like capsazepine (CPZ)18. In the absence of ligands, this site is occupied by the lipid PI favoring the closed pore conformation18. The TRPV1 vanilloid binding site coincides with hTRPV6 lipid density 2, which may represent the binding site for natural lipid agonists (Fig. 4b).
Figure 4. Activation-related lipid binding pocket.
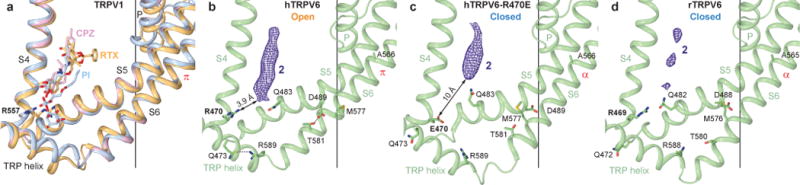
a, Superposition of the agonist binding site in TRPV1 structures in the PI-bound closed state (blue, PDB ID: 5IRZ), antagonist CPZ-bound closed state (pink, PDB ID: 5IS0) and agonist RTX-bound open state (orange, PDB ID: 5IRX). b–d, Putative activating lipid binding site in (b) open hTRPV6, (c) closed hTRPV6-R470E and (d) closed rTRPV6, with densities filtered to the same resolution (4.24 Å) and shown at 5.3σ as purple mesh. Residues involved in gating are shown as sticks. Dashed lines in b indicate bonds between the residues.
To test whether this site is critical for channel activation, we mutated R470 to glutamate (R470E). An analogous mutation had been previously shown to eliminate capsaicin-evoked currents in TRPV120. hTRPV6-R470E channels were functional (Extended Data Fig. 1b) but showed ~10 times slower calcium uptake compared to wild type channels (Extended Data Fig. 1e–f), consistent with their less frequent openings. Additionally, 2-APB, a TRPV6 inhibitor that acts through the membrane, displayed increased affinity to and decreased maximum inhibition of hTRPV6-R470E compared to wild type channels (Extended Data Fig. 1i–k), consistent with the R470E mutation altering TRPV6 regulation by lipids.
We solved the hTRPV6-R470E structure in amphipols by cryo-EM to 4.2 Å resolution (Extended Data Fig. 6). Consistent with the idea that the site 2 density represents an activating lipid, its size in hTRPV6-R470E became smaller (Fig. 4c) than in hTRPV6 (Fig. 4b). Confirming different physical occupancy of site 2, the side chain of Q483 in hTRPV6-R470E has an altered conformation that would cause clashing with the lipid density in wild type hTRPV6 (Fig. 4b–c). Supporting the role of Q483 in lipid recognition, an hTRPV6-Q483A mutant, while being functional (Extended Data Fig. 1c), demonstrated a ~5 times slower calcium uptake compared to wild type channels (Extended Data Fig. 1e,g). More importantly, the ion channel in hTRPV6-R470E appears to be closed (Fig. 3b,c,e). Indeed, the size of the pore at the S6 bundle crossing becomes comparable to the narrowest point of the selectivity filter. While the latter is formed by the side chains of D542, which directly coordinate calcium ions for selective permeation, the S6 bundle crossing is formed by the side chains of L574 and M578, which create a hydrophobic seal impermeable to ions and water, and therefore represent the channel gate.
The closed-state structure of hTRPV6-R470E is nearly identical to the closed-state crystal structure of rat TRPV6 (rTRPV6)10 and their superposition yields a root mean square deviation (RMSD) of 0.917 Å. To verify that the rTRPV6 crystal structure represents the physiologically-relevant conformation, we solved the structure of rTRPV6 by cryo-EM to 3.9 Å using a lipid nanodisc preparation similar to that used for hTRPV6 (Extended Data Fig. 7). Strikingly, the cryo-EM structure of rTRPV6 is nearly identical (RMSD = 0.781 Å) to the crystal structure of rTRPV6 (Extended Data Fig. 8a–c). Since the hTRPV6-R470E structure is nearly identical to both the rTRPV6 cryo-EM structure (Extended Data Fig. 8d–e, RMSD = 0.932 Å) and the rTRPV6 crystal structure, we contend that it represents the true closed state of hTRPV6. Consistently, a much weaker density at site 2 in the cryo-EM structure of rTRPV6 (Fig. 4d) suggests either lower occupancy or greater mobility of the putative bound lipid. Because rTRPV6 and hTRPV6 were purified in similar conditions, have 89% overall sequence identity, and have identical amino acid compositions of their site 2 lipid binding pockets, it remains unclear why one channel was closed and the other open. For example, some lipids within the membranes of the protein-expressing HEK cells may be important for opening of hTRPV6 but not rTRPV6. Different conformations of rTRPV6 and hTRPV6 might also reflect the ease with which these constitutively active channels rapidly transition between gating states, and that very subtle changes can push this equilibrium towards one state or the other. Such subtle changes, for instance, can originate from different interactions of membrane-mimicking environment (amphipols or nanodiscs) with S1–S3. These helices contain the largest number of membrane lipid-facing residues (69%), of which only 80% are identical between rTRPV6 and hTRPV6.
To understand the structural changes that occur during TRPV6 opening, we compared our hTRPV6-R470E and hTRPV6 structures. The principal changes occur in the pore-lining helix S6 and originate at A566, a residue highly conserved in TRPV5 and TRPV6 (Extended Data Fig. 9k) and located right below the selectivity filter (Fig. 3g). Confirming its important role in gating, substituting A566 with threonine, the homologous residue conserved in TRPV1–4, greatly reduces the TRPV6 current amplitude (Extended Data Fig. 1d) and slows down calcium uptake by ~30 times (Extended Data Fig. 1e,h). Upon opening, S6, which has an α-helical conformation in the closed state, undergoes a local transition to a π-helix. Interestingly, such a transition was earlier hypothesized based on a comparison between TRPV1 and TRPV2 structures21. Concurrently, the lower part of S6, which forms the gate in the closed state, rotates by ~100° and bends away from the pore axis by ~11° (Fig. 3g). Importantly, these rearrangements not only widen the pore for permeant ions but also change the set of residues that face the pore axis (e.g., N572 and I575 in the open state compared to L574 and M578 in the closed state). Alanine A566, therefore, acts as a hinge to allow TRPV6 gating at the S6 bundle crossing without changing the conformation of the selectivity filter (Fig. 3f). Correspondingly, the selectivity filter appears to play a crucial role in TRPV6 channel ion permeation rather than gating. Gating-related conformational changes induced by the α-to-π helical transition in S6 seem to only involve the intracellular portions of S5 and S6, the S4–S5 linker and the TRP helix. Indeed, superposition of the corresponding regions (residues 469–500 plus 566–608) in hTRPV6 and hTRPV6-R470E gives an RMSD of 1.74 Å, while the rest of the molecules superpose with a much lower RMSD of 0.218 Å.
Within the regions involved in gating, pore opening in hTRPV6 is accompanied by the formation of two electrostatic bonds per subunit (Fig. 4b). A salt bridge forms between Q473 in the S4–S5 elbow and R589 in the TRP helix, and a hydrogen bond forms between D489 in the S5 helix and T581 in the S6 helix. Neither interaction is present in the closed-state structures of hTRPV6-R470E or rTRPV6 and the formation of the hydrogen bond (D489-T581 in hTRPV6-R460E or D488-T580 in rTRPV6) is prevented by the side chain of M577 or M576, respectively (Fig. 4c–d, Extended Data Fig. 8f–g). The importance of the D489–T581 interaction for hTRPV6 opening is supported by the previous observation that a mutation equivalent to T581A reduces the excessive constitutive activity of an hTRPV6-G516S mutant22. We speculate that formation of the electrostatic bonds compensates the energetic cost of the unfavorable α-to-π helical transition in S6 during channel opening. This structural solution therefore maintains the relative stabilities and similar energy levels of both gating states and supports the constitutive activity of TRPV6. Accordingly, the open and closed conformations of TRPV6 remain in a state of readily tunable equilibrium that can be shifted towards either state by different stimuli, including lipids7,9.
Our structures of hTRPV6 reveal a gating mechanism novel for tetrameric ion channels (Fig. 5, Supplementary Video 1). Despite other representatives of the TRP channel family have a local α-to-π helical transition in the middle of S618,23–25, they lack the alanine gating hinge (Extended Data Fig. 9). As a result, S6 maintains its secondary structure throughout the entire TRPV1 gating cycle, the same residues are facing the pore in the closed and open states, and pore widening is observed at both the S6 bundle crossing and the selectivity filter18. On the other hand, K+ channels do have a gating hinge in their pore-forming inner helices26,27. However, this hinge is formed by a glycine located one residue C-terminally compared to the gating hinge alanine in TRPV6 and permits bending of the inner helices by ~30° without an α-to-π transition. The glycine hinge, like the alanine hinge in TRPV6, allows gating of K+ channels to occur at the inner helices bundle crossing without changing the selectivity filter. However, unlike TRPV6, the glycine gating hinge in K+ channels does not introduce a ~100° rotation of the lower parts of the pore-forming helices and correspondingly does not change the residues lining the pore gate region. An alanine gating hinge is present in the pore-forming helices of ionotropic glutamate receptor (iGluR) family tetrameric ion channels28. However, this alanine gating hinge is located at the ion channel gate region. Correspondingly, bending the pore-forming helices at the iGluR alanine gating hinge directly alters the diameter of the pore in close proximity to the gate without an α-to-π transition. The alanine gating hinge in TRPV5/6 channels is therefore a unique structural element that is likely associated with their exclusive physiological role as calcium uptake channels.
Figure 5. TRPV6 channel gating mechanism.

Cartoons represent the structural changes associated with TRPV6 channel gating. Transition from the closed to open state, stabilized by the formation of salt bridges (dashed lines), leads to permeation of ions (green spheres) and is accompanied by a local α-to-π helical transition in S6 that maintains the selectivity filter conformation, while the lower part of S6 bends by ~11° and rotates by ~100°. These movements result in a different set of residues (blue versus pink symbols) lining the pore in the vicinity of the channel gate.
Methods
Construct
The full-length human TRPV6 (residues 1–725) and rat TRPV6 (rTRPV6, residues 1–727) were each introduced into a pEG BacMam vector30, with the C-terminal thrombin cleavage site (LVPRG) followed by the streptavidin affinity tag (WSHPQFEK). The R470E mutation in hTRPV6 was introduced by conventional mutagenesis. The rTRPV6 construct previously used for crystallographic studies, rTRPV6*10, was also introduced into a pEG BacMam vector but with the eGFP inserted between the thrombin site and the streptavidin tag. Compared to wild type rTRPV6, the rTRPV6* construct is C-terminally truncated by 59 residues and contains three point mutations in the ankyrin repeat domain, I62Y, L92N and M96Q.
Expression and purification
All constructs were expressed and purified similarly to TRPV6cryst.11 Bacmids and baculoviruses were made using a standard method30. The P2 baculovirus, produced in Sf9 cells (Thermo Fisher Scientific, mycoplasma test negative, GIBCO #12659017), was added to HEK 293S cells lacking N-acetyl-glucosaminyltransferase I (GnTI−) and grown in suspension (mycoplasma test negative, ATCC #CRL-3022) in Freestyle 293 media (GIBCO-Life Technologies #12338-018) supplemented with 2% FBS at 37°C and 5% CO2. 8–12 hours post-transduction, 10 mM sodium butyrate was added to enhance protein expression and the temperature was reduced to 30°C. At 48–72 hours post-transduction, cells were harvested by low-speed centrifugation in a Sorvall Evolution RC Centrifuge (Thermo Scientific) at 5471 g for 15 minutes, washed in phosphate buffer saline (PBS) pH 8.0, and pelleted in an Eppendorf Centrifuge 5810 at 3,202 g for 10 minutes. The cell pellet was resuspended and subjected to sonication with a Misonix sonicator (12 × 15 s, power level 8) in a buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 8.0), 1 mM βME (beta-mercaptoethanol) and protease inhibitors (0.8 μM aprotinin, 2 μg/ml leupeptin, 2 μM pepstatin A and 1 mM phenylmethysulfonyl fluoride) – 50 ml was used per 800 ml of HEK 293 cell culture. Subsequently, the lysate was clarified after centrifugation using a Sorvall RC-5C Plus centrifuge at 9,900 g for 15 minutes, and the membranes were collected by ultracentrifugation in a Beckman Coulter ultracentrifuge equipped with Beckman Coulter Type 45 Ti Rotor rotor at 186,000 g for one hour. The membranes were then mechanically homogenized, and solubilized for 1–2 hours in 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 1% DDM (n-Dodecyl-β-D-Maltopyranoside), 0.1% Cholesteryl hemisuccinate (CHS), and 1 mM βME. Insoluble material was removed by ultracentrifugation for 40 minutes in a Beckman Coulter Type 45 Ti Rotor at 186,000 g and the supernatant was added to streptavidin-linked resin and rotated for 10–14 hours at 4°C. Next, the resin was washed with 10 column volumes of wash buffer containing 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 1 mM βME, 0.1% DDM, and 0.01% CHS. The bound protein was eluted in wash buffer to which 2.5 mM D-desthiobiotin was added. All constructs were purified by size exclusion chromatography using a Superose 6 column equilibrated in 150 mM NaCl, 20 mM Tris-HCl pH 8.0, 1mM βME, 0.1% DDM, and 0.01% CHS. Tris(2-carboxyethyl)phosphine (TCEP; 10 mM) was added to the peak fractions which were pooled and concentrated for channel reconstitution in nanodisc or amphipols. The rTRPV6 and rTRPV6* constructs were expressed and purified similarly to the hTRPV6 constructs but without the addition of CHS to any buffer. Additionally, after elution from the streptavidin-linked resin, the rTRPV6* fusion protein was concentrated to ~1.0 mg/ml and subjected to thrombin digestion in mass ration of 1:100 (thrombin: protein) for one hour at 22°C rocking, before size exclusion chromatography. Prior to reconstitution in nanodiscs or amphipols, the concentration of each construct was adjusted to approximately 1.2 mg/ml.
Reconstitution of TRPV6 protein into nanodiscs and amphipols
Both hTRPV6 and rTRPV6 were incorporated into conventional MSP2N2 lipid nanodisc as described previously18. Briefly, soybean polar lipid extract (Avanti #541602) was solubilized in buffer containing 20 mM Tris pH 8.0, 150 mM NaCl, 2 mM TCEP, and 15 mM DDM to create a 10-mM stock. Purified sample was mixed with the soybean polar lipid extract stock (~7.6 mg/ml) and MSP2N2 (~5.3 mg/ml) in a molar ratio of approximately 1:3:166 for both hTRPV6 (monomer:MSP2N2:lipid) and rTRPV6 (monomer:MSP2N2:lipid) and rocked at room temperature for one hour. Subsequently, 10 mg of Bio-beads SM2 (Bio-rad) prewet in buffer (20 mM Tris pH 8.0, 150 mM NaCl, 1mM BME) was added to 0.5 ml of mixture and the mixture was rotated at 4°C. After one hour, an additional 10 mg of Bio-beads SM2 was added and the resulting mixture was rotated at 4°C for ~ 20 hours. The Bio-beads SM2 were removed by pipetting and TRPV6 reconstituted in nanodiscs was isolated from empty nanodiscs by size exclusion chromatography using a Superose 6 column equilibrated in 150 mM NaCl, 20 mM Tris-HCl pH 8.0, and 1 mM βME.
cNW11 circularized nanodiscs were prepared as described previously31. Purified rTRPV6* was incorporated into cNW11 (2.0 mg/ml) circularized nanodiscs using the procedure described above for the MS2N2 nanodiscs but with a molar ratio of 1:10:267 (rTRPV6* monomer:cNW11:lipid).
For reconstitution in A8–35 amphipols (Anatrace #A835), we adapted the previously described procedure32. hTRPV6 or hTRPV6-R470E were each mixed with amphipols in a 1:3 mass ratio (protein:amphipols) and incubated for three hours with rotation at 4°C. After three hours, 7–8 mg per 0.5 ml of mixture of Bio-beads SM2 prewet in buffer containing 20 mM Tris pH 8.0, 150 mM NaCl, 1mM BME was added to the protein-amphipols mixture to facilitate the reconstitution of TRPV6 into amphipols. The mixture was rotated for ~20 hours at 4 °C and the amphipols-solubilized TRPV6 was purified as described above.
Cryo-EM sample preparation and data collection
Au/Au grids were prepared as described in the literature33. Briefly, grids were prepared by first coating C-flat (Protochips, Inc., Morrisville, NC) CF-1.2/1.3-2Au 200 mesh holey carbon grids with ~50 nm of gold using an Edwards Auto 306 evaporator. Subsequently, an Ar/O2 plasma treatment (6 minutes, 50 watts, 35.0 sccm Ar, 11.5 sccm O2) was used to remove the carbon with a Gatan Solarus (model 950) Advanced Plasma Cleaning System. The grids were again plasma treated (H2/O2, 20s, 10 watts, 6.4 sccm H2, 27.5 sccm O2) prior to sample application in order to make their surfaces hydrophilic. A Vitrobot Mark IV (FEI, Hillsboro, OR, USA) was used to plunge-freeze the grids after the application of 3 μl protein solution with 100% humidity at 5 °C, a blot time of 2 or 3 seconds, blot force set to 3, and a wait time of 20 seconds. A concentration of 0.5 mg/ml was used for the nanodiscs-solubilized protein and 0.3 mg/ml for the amphipols-solubilized protein.
The hTRPV6-nanodiscs data were collected on a Tecnai F30 Polara (Cs 2.26 mm) at 300 kV equipped with a Gatan K2 Summit electron detection (DED) camera (Gatan, Pleasanton, CA, USA) using Leginon34. 1733 micrographs were collected with a super-resolution pixel size of 0.98 Å across a defocus range of −1.5 μm to −3.5 μm. The total dose, ~67 e− Å−2, was attained by using a dose rate of ~8.0 e− pixel−1 s−1 across 40 frames for 8 seconds total exposure time. 1538 hTRPV6-amphipols micrographs and 1301 rTRPV6* micrographs were collected as described above. 2167 rTRPV6 micrographs were collected as described above but in counting mode with a pixel size of 0.98 Å. The hTRPV6-R470E data were collected on a Cs-corrected Titan Krios (FEI) equipped with a post-column GIF Quantum energy filter at 300 kV. 3540 micrographs were collected in counting mode with a pixel size of 1.10 Å across a defocus range of −1.5 μm to −3.5 μm. The total dose, ~67 e− Å−2, was attained by using a dose rate of ~8.0 e− pixel−1 s−1 across 50 frames for 10 seconds total exposure time.
Image processing
Data were collected using the Gatan K2 Summit electron detection (DED) camera (Gatan, Pleasanton, CA, USA) in super-resolution mode and binned 2 × 2. Frame alignment was done using MotionCor235. CTF correction, using CTFFIND436 for the hTRPV6-nanodiscs data set and gCTF37 for all other data sets, was performed on non-dose-weighted micrographs while subsequent data processing was done on dose-weighted micrographs. All other data processing, unless stated otherwise, was performed using Relion 2.038. For each data set, 1000–2000 particles were manually selected to generate 2D classes for use in auto-picking.
In processing the hTRPV6-nanodiscs data set, seven 2D classes were used for automatically picking 509,569 particles from the 1733 collected micrographs. The particle images were binned to a pixel size of 1.96 Å/pixel and screened by 2D classification to remove aberrantly picked particles. The remaining 508,019 particles were subjected to 3D classification into 10 classes with no symmetry imposed. A density map was generated in chimera from the crystal structure of rTRPV6 (PDB ID: 5IWK), low-pass filtered to 40 Å, and used as an initial reference. Five classes, comprising 313,369 particle images, exhibited structural features of a quality that warranted further processing. Of the five, one showed structural features of higher detail and comprised 71,582 particles images. The particle images composing this class were extracted without binning (0.98 Å/pixel), refined with C4 symmetry using the same reference (unbinned) as the prior round of classification, low-pass filtered to 40 Å, and post-processed. The resulting map was then used as a reference for the second round of 3D classification in which the particles composing the best five aforementioned classes were extracted with binning (1.98 Å/pixel) and split into ten classes with C4 symmetry imposed. Two new classes, comprising a total of 67,034 particles, exhibited structural features of a quality that warranted further processing. These 67,034 particle images were extracted without binning (0.98 Å/pixel), refined with C4 symmetry using the same reference (unbinned) as the prior round of classification low-pass filtered to 40 Å, and post-processed. The resulting map was used for the final round of 3D classification in which the two best classes from the prior round of 3D classification were without binning, and with C4 symmetry imposed, classified into 10. The four best classes, comprising 46,124 particle images, were refined together and post-processed to generate the final 3.6 Å map. This relatively small number of particles compared to the initial pool of picked up 509,569 particles indicates that the majority of picked up particles represent either artifacts or contaminants or TRPV6 molecules in alternative conformations including different gating states or unnatural conformations produced by artificial environment of the cryo-EM grid.
Each data set was processed using a workflow similar to that described above and the reported resolutions were estimated using the Fourier shell correlation (FSC=0.143) criterion on masking-effect-corrected FSC curves calculated between two independent half-maps39,40. The local resolutions were estimated with unfiltered half-maps using ResMap and EM density maps were visualized using UCSF Chimera42.
The cryo-EM data collected for full-length rTRPV6 yielded a low-resolution (6.4 Å) reconstruction that was sufficient to conclude that it represents the closed-state conformation similar to rTRPV6*. Since it is lacking high-resolution details, it is not described in the main text or the Extended Data sections.
Model building
To build the open- and closed-state models of TRPV6 in COOT43, we used the rTRPV6* crystal structure10 as a guide. The resulting models were refined against un-filtered half-maps in real space with constraints using PHENIX44. The refined models were tested for overfitting (Extended Data Figs. 2f, 3f, 6f, 7f) by shifting their coordinates 0.5 Å with shake in PHENIX and building their corresponding densities in Chimera42 from the shaken models. FSC was calculated between the densities from the shaken models, the half maps used in PHENIX refinement (work), the second half-maps (free) and the unfiltered sum maps, using EMAN245. The local resolutions in the transmembrane regions of our hTRPV6-nanodiscs and hTRPV6-R470E maps reached 2.5 Å as estimated by ResMap41. These high resolutions allowed us to unambiguously define the conformation of S6 in the open and closed state as well as the existence of the π-helix in extracellular half of S6 in the open state. Structures were visualized and figures were prepared in Pymol46.
Fura 2-AM measurements
Wild type hTRPV6 or TRPV6-R470E fused to C-terminal strep tag was expressed in HEK cells. 50–60 hours after transduction, cells were harvested by centrifugation at 600 g for 5 minutes. The cells were resuspended in pre-warmed modified HEPES buffered saline (HBS) (118 mM NaCl, 4.8 mM KCl, 1 mM MgCl2, 5 mM D-glucose, 10 mM HEPES pH 7.4) containing 5 μg/mL of Fura2-AM (Life Technologies) and incubated at 37°C for 45 minutes. The loaded cells were then centrifuged for 5 minutes at 600 g, and resuspended in prewarmed, modified HBS and incubated again at 37°C for 25–35 minutes in the dark. The cells were subsequently pelleted and washed twice, then resuspended in modified HBS for experiments. The cells were kept on ice in the dark for a maximum of ~2 hours before fluorescence measurements, which were conducted using a spectrofluorometer QuantaMaster™ 40 (Photon Technology International) at room temperature in a quartz cuvette under constant stirring. Intracellular Ca2+ was measured by taking the ratio of two excitation wavelengths (340 and 380 nm) at one emission wavelength (510 nm). The excitation wavelength was switched at 1-s intervals.
Electrophysiology
HEK293 cells (ATCC #CRL-1573) grown on glass cover slips in 35 mm dishes and were transduced with the same P2 virus as was used for large-scale protein production. Recordings were made at room temperature, 36 to 72 hours post-transduction. Currents from whole cells, typically held at a 0 or −60 mV membrane potential, were recorded using Axopatch 200B amplifier (Molecular Devices, LLC), filtered at 5 kHz and digitized at 10 kHz using low-noise data acquisition system Digidata 1440A and pCLAMP software (Molecular Devices, LLC). The external solution contained (in mM): 140 NaCl, 6 CsCl, 1 MgCl2, 10 HEPES pH 7.4 and 10 glucose. To evoke monovalent currents, 1 mM EGTA was added to the external solution. The internal solution contained (in mM): 100 CsAsp, 20 CsF, 10 EGTA, 3 MgCl2, 4 NaATP and 20 HEPES pH 7.2. TRPV6 currents were recorded in response to 50-ms voltage ramps from −120 mV to 120 mV (see Extended Data Fig. 1). Data analysis was performed using the computer program Origin 9.1.0 (OriginLab Corp.).
Data Availability
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMDB-7120 (hTRPV6-nanodiscs), EMDB-7121 (hTRPV6-amphipols), EMDB-7122 (hTRPV6-R470E) and EMDB-7123 (rTRPV6*). Model coordinates have been deposited in the Protein Data Bank (PDB) under accession numbers 6BO8 (hTRPV6-nanodiscs), 6BO9 (hTRPV6-amphipols), 6BOA (hTRPV6-R470E) and 6BOB (rTRPV6*). All other data are available from the corresponding author upon request.
Extended Data
Extended Data Figure 1. Functional characterization of wild type and mutant hTRPV6 channels.

a–d, Whole-cell patch-clamp recordings from HEK 293 cells expressing (a) wild type hTRPV6, (b) hTRPV6-R470E, (c) hTRPV6-Q483A and (d) hTRPV6-A566T. Shown are leak-subtracted currents in response to voltage ramp protocols illustrated above the recordings. Although the shape of the currents for wild type and mutant hTRPV6 channels was similar, their amplitudes were different. The average current amplitudes at −60 mV membrane potential (mean ± SEM) were 3171 ± 767 pA (n = 11) for wild type hTRPV6, 918 ± 267 pA (n = 9) for hTRPV6-R470E, 2239 ± 398 pA (n = 7) for hTRPV6-Q483A and 145 ± 52 pA (n = 5) for hTRPV6-A566T. e–h, Kinetics of calcium uptake using Fura-2 AM ratiometric fluorescence measurements. Shown are representative fluorescence curves for (e) wild type hTRPV6, (f) hTRPV6-R470E, (g) hTRPV6-Q483A and (h) hTRPV6-A566T in response to application of 2 mM Ca2+ (arrow). Exponential fits are shown in red, with the time constants indicated. Over five measurements, the time constants (mean ± SEM) were 4.2 ± 0.5 s for hTRPV6, 47 ± 13 s for hTRPV6-R470E, 18.9 ± 0.8 s for hTRPV6-Q483A and 121 ± 12 s for hTRPV6-A566T. At n = 5 and P = 0.05, the time constant values for wild type and mutant channels were statistically different (two-sided t-test). i–j, Fluorescence curves for (i) wild type hTRPV6 and (j) hTRPV6-R470E in response to application of 2 mM Ca2+ after pre-incubation of cells in different concentrations of 2-APB. These experiments were repeated independently three times with similar results. k, Dose-response curves for 2-APB inhibition calculated for wild type hTRPV6 (black) and hTRPV6-R470E (red) (n = 3 for all measurements). The changes in the fluorescence intensity ratio at 340 and 380 nm (F340/F380) evoked by addition of 2 mM Ca2+ after pre-incubation with various concentrations of 2-APB were normalized to the maximal change in F340/F380 after addition of 2 mM Ca2+ in the absence of 2-APB. Straight lines through the data points are fits with the logistic equation, with the mean ± SEM values of half-maximum inhibitory concentration (IC50), 274 ± 27 μM and 85 ± 5 μM, and the maximal inhibition, 72.6 ± 2.7 % and 50.3 ± 1.1 %, for hTRPV6 and hTRPV6-R470E, respectively. The leftward shift of the 2-APB dose-response curve of hTRPV6-R470E, when compared to the dose-response curve of wild type hTRPV6, indicates an increased affinity of the channel for 2-APB. This is likely the result of the R470E mutation reducing the affinity of the channel for an activating lipid ligand. On the other hand, the reduced maximum inhibition of hTRPV6-R470E at high concentrations of 2-APB, when compared to that of wild type hTRPV6, indicates a reduced efficacy of 2-APB that could be a result of the R470E mutation disrupting the mechanism by which 2-APB binding is allosterically coupled to channel gating.
Extended Data Figure 2. Overview of single-particle cryo-EM for hTRPV6 in nanodiscs.

a, Example cryo-EM micrograph for hTRPV6 in nanodiscs. b, Orientations of particles that contribute to the final 3.6 Å reconstruction. Longer red rods represent orientations that comprise more particles. c, Local resolution mapped on density at 0.013 threshold level (UCSF Chimera) calculated using Resmap and two unfiltered half maps, with the highest resolution observed for the channel core. d, FSC curve calculated between half-maps. e, Cross-validation FSC curves for the refined model versus unfiltered half maps (only half map1 was used for PHENIX refinement) and the unfiltered summed map.
Extended Data Figure 3. Overview of single-particle cryo-EM for hTRPV6 in amphipols and comparison to the reconstruction in nanodiscs.

a, Example cryo-EM micrograph for hTRPV6 in amphipols. b, Reference-free two-dimensional class averages of hTRPV6 in amphipols illustrating different particle orientations. c, Local resolution mapped on density at 0.01 threshold level (UCSF Chimera) calculated using Resmap and two unfiltered half-maps, with the highest resolution observed for the channel core. d, Orientations of particles that contribute to the final 4.0 Å reconstruction. Longer red rods represent orientations that comprise more particles. e, FSC curve calculated between half-maps. f, Cross-validation FSC curves for the refined model versus unfiltered half maps (only half map1 was used for PHENIX refinement) and the unfiltered summed map. g–h, Comparison of putative lipid densities for hTRPV6 in (g) amphipols and (h) nanodiscs, filtered to the same (4.0 Å) resolution and shown at 3.5σ as purple mesh.
Extended Data Figure 4. Cryo-EM density for hTRPV6 in nanodiscs.

a, Cryo-EM density at 4σ for a single hTRPV6 subunit, with the protein shown in ribbon and colored according to domains. b–g, Fragments of the hTRPV6 transmembrane domain with the corresponding cryo-EM density.
Extended Data Figure 5. Fitting lipids into cryo-EM density.

a–c, Molecules of (a) phosphatidylethanolamine (PE), (b) phosphatidylcholine (PC) and (c) phosphatidylinositol 4,5-bis phosphate (PIP2) fitted into the site 4 lipid density shown at 3.5σ as purple mesh. d–f, Molecules of (d) cholesterol, (e) cholesterol hemisuccinate (CHS) and (f) PIP2 fitted into the site 2 putative activating lipid density shown at 5.3σ.
Extended Data Figure 6. Overview of single-particle cryo-EM for hTRPV6-R470E in amphipols.

a, Example cryo-EM micrograph for hTRPV6-R470E in amphipols. b, Reference-free two-dimensional class averages of hTRPV6-R470E in amphipols illustrating different particle orientations. c, Local resolution mapped on density at 0.017 threshold level (UCSF Chimera) calculated using Resmap and two unfiltered half-maps, with the highest resolution observed for the channel core. d, Orientations of particles that contribute to the final 4.2 Å reconstruction. Longer red rods represent orientations that comprise more particles. e, FSC curve calculated between half-maps. f, Cross-validation FSC curves for the refined model versus unfiltered half maps (only half map1 was used for PHENIX refinement) and the unfiltered summed map.
Extended Data Figure 7. Overview of single-particle cryo-EM for rTRPV6 in CNW11 nanodiscs.

a, Example cryo-EM micrograph for rTRPV6 in CNW11 nanodiscs. b, Reference-free two-dimensional class averages of rTRPV6 in CNW11 nanodiscs illustrating different particle orientations. c, Local resolution mapped on density at 0.011 threshold level (UCSF Chimera) calculated using Resmap and two unfiltered half maps, with the highest resolution observed for the channel core. d, Orientations of particles that contribute to the final 3.9 Å reconstruction. Longer red rods represent orientations that comprise more particles. e, FSC curve calculated between half-maps. f, Cross-validation FSC curves for the refined model versus unfiltered half-maps (only half-map1 was used for PHENIX refinement) and the unfiltered summed map.
Extended Data Figure 8. Comparison of cryo-EM and crystal structures of rTRPV6, cryo-EM structures of hTRPV6-R470E and rTRPV6 and regions in hTRPV6 and hTRPV6-R470E encompassing D489 and T581.

a–c, Superimposed are (a) the transmembrane domain of a single subunit, and (b–c) the pore-forming region viewed (b) parallel to the membrane or (c) intracellularly for the cryo-EM (green) and crystal (orange) structures of rTRPV6. Only two of four rTRPV6 subunits are shown in b, with the front and back subunits omitted for clarity. Residues lining the selectivity filter and gate are shown as sticks. d–e, Superposition of the P-loop and S6 in cryo-EM structures of hTRPV6-R470E (blue) and rTRPV6 (green), viewed (d) parallel to the membrane, and (e) intracellularly. In d, only two of four subunits are shown, with the front and back subunits removed for clarity. The residues lining the pore are shown as sticks. f–g, Regions in (f) hTRPV6 and (g) hTRPV6-R470E encompassing D489 and T581. The closest distance between D489 and T581 is indicated by dashed lines. Note, M485 and M577 either surround the potentially interacting D489 and T581 (f, hTRPV6) or reside between these residues (g, hTRPV6-R470E), apparently preventing their interaction. Blue mesh shows cryo-EM density at 4σ.
Extended Data Figure 9. Structural superposition and sequence alignment of the pore domain in tetrameric ion channels.

a–i, Pairwise superposition of the pore domain in hTRPV6 with (a) rat TRPV118 (PDB ID: 5IRX; RMSD = 2.065 Å), (b) rabbit TRPV221 (PDB ID: 5AN8; RMSD = 3.757 Å), (c) rat TRPV224 (PDB ID: 5HI9; RMSD = 4.399 Å), (d) human TRPA123 (PDB ID: 3J9P; RMSD = 1.429 Å), (e) human PKD225 (PDB ID: 5T4D; RMSD = 2.676 Å), (f) KcsA from Streptomyces lividans47 (PDB ID: 1BL8; RMSD = 2.708 Å), (g) MthK from M. thermautotrophicum48 (PDB ID: 1LNQ; RMSD = 2.947 Å), (h) rat Shaker49 (PDB ID: 2A79; RMSD = 2.487 Å) and (i) rat GluA2 AMPA-subtype iGluR28 (PDB ID: 5WEO; RMSD = 2.044 Å). j, Sequence alignment for the pore region of human TRPV3–6, TRPA1 and PKD2, rat TRPV1, 2, 6, Shaker and GluA2, rabbit TRPV2 and bacterial K+ channels KcsA and MthK. The selectivity filter residues in K+ channels and gating hinge residues in S6 (M3 in GluA2) are colored red. k, Aligned sequence logos for TRPV channels in S6, generated by WebLogo50 from 1200 TRPV1–6 sequences. The red rectangle and arrow indicate the position of the alanine gating hinge in TRPV6. Note, the relatively small side chain residues threonine or alanine at the next to the gating hinge alanine position in TRPV5 and TRPV6, instead of bulky hydrophobic phenylalanine or tyrosine in TRPV1–4, might be critical for the α-to-π helical transition in S6 during channel opening.
Extended Data Table 1. Cryo-EM data collection, refinement and validation statistics.
| hTRPV6-nanodiscs (EMDB-7120) (PDB 6B08) |
hTRPV6-amphipols (EMDB-7121) (PDB 6B09) |
hTRPV6-R470E (EMDB-7122) (PDB 6BOA) |
rTRPV6* (EMDB-7123) (PDB 6BOB) |
|
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 39,000× | 39,000 | 105,000× | 39,000× |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 67 | 67 | 67 | 67 |
| Defocus range (μm) | 1.5 to −3.5 | −1.5 to −3.5 | −1.5 to −3.5 | −1.5 to −3.5 |
| Pixel size (Å) | 0.98 | 0.98 | 1.10 | 0.98 |
| Symmetry imposed | C4 | C4 | C4 | C4 |
| Initial particle images (no.) | 509,569 | 306,784 | 1,243,159 | 248,836 |
| Final particle images (no.) | 46,124 | 65,259 | 59,298 | 20,808 |
| Map resolution (Å) | 3.56 | 4.00 | 4.24 | 3.92 |
| FSC threshold | ||||
| Map resolution range (Å) | 2.5 to 6.0 | 2.5 to 6.0 | 2.5 to 6.0 | 2.5 to 6.0 |
| Refinement | ||||
| Initial model used (PDB code) | 5IWK | This study | This study | This study |
| Model resolution (Å) | 3.56 | 4.00 | 4.24 | 3.92 |
| FSC threshold | ||||
| Model resolution range (Å) | 2.5 to 6.0 | 2.5 to 6.0 | 2.5 to 6.0 | 2.5 to 6.0 |
| Map sharpening B factor (Å2) | −165 | −206 | −239 | −173 |
| Model composition | ||||
| Non-hydrogen atoms | 19,048 | 19,048 | 19,040 | 19,340 |
| Protein residues | 611 | 611 | 611 | 611 |
| Ligands | N/A | N/A | N/A | N/A |
| B factors (Å2) | ||||
| Protein | 182.9 | 242.8 | 122.0 | 184.2 |
| Ligand | N/A | N/A | N/A | N/A |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.0079 | 0.0071 | 0.0101 | 0.0072 |
| Bond angles (°) | 1.33 | 1.3 | 1.46 | 1.37 |
| Validation | ||||
| MolProbity score | 1.97 | 2.06 | 2.22 | 1.98 |
| Clashscore | 7 | 8 | 5 | 7 |
| Poor rotamers (%) | 0.6 | 0.8 | 1.3 | 2.7 |
| Ramachandran plot | ||||
| Favored (%) | 88.96 | 87.95 | 88.75 | 88.55 |
| Allowed (%) | 11.04 | 12.05 | 11.25 | 11.15 |
| Disallowed (%) | 0.00 | 0.00 | 0.00 | 0.30 |
Supplementary Material
A morph between closed and open states of human TRPV6 represented by the hTRPV6-R470E and wild type hTRPV6 structures, respectively. Shown is the pore domain viewed intracellularly and illustrating the iris-like pore opening or viewed parallel to the membrane, with the front and back subunits disappearing for clarity, and illustrating the α-to-π helical transition in S6 during channel opening. Residues around the gate and critical for permeation aspartates D542 are shown in stick representation.
Acknowledgments
We thank Tibor Rohacs for advice on electrophysiological recordings, Joachim Frank for comments on the manuscript, H. Kao for computational support and members of the E.C. Greene lab for assistance with their fluorimeter. E.C.T is supported by the NIH (F31 NS093838). A.I.S. is supported by the NIH (R01 CA206573, R01 NS083660), the Amgen Young Investigator and Irma T. Hirschl Career Scientist Awards. Data were collected at the Columbia University Medical Center cryo-EM facility and at the Simons Electron Microscopy Center and National Resource for Automated Molecular Microscopy located at the New York Structural Biology Center, supported by grants from the Simons Foundation (349247), NYSTAR, and the NIH (GM103310).
Footnotes
Supplementary Information is linked to the online version of the paper.
Author Contributions: L.L.M., A.K.S. and A.I.S. designed the project, built models, analyzed data. L.L.M., A.K.S. carried out Fura-2 experiments, cryo-EM data collection and processing. L.L.M., A.K.S., K.S., M.V.Y. developed expression and purification protocols. L.L.M., A.K.S., K.S. designed constructs, prepared protein samples. M.V.Y. carried out electrophysiology experiments. E.C.T advised on cryo-EM workflow. R.A.G and E.C.T assisted with microscope operation. A.I.S. supervised the project. L.L.M., A.K.S., K.S., M.V.Y., E.C.T, A.I.S. wrote the manuscript.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Peng JB, et al. Molecular cloning and characterization of a channel-like transporter mediating intestinal calcium absorption. J Biol Chem. 1999;274:22739–22746. doi: 10.1074/jbc.274.32.22739. [DOI] [PubMed] [Google Scholar]
- 2.Yue L, Peng JB, Hediger MA, Clapham DE. CaT1 manifests the pore properties of the calcium-release-activated calcium channel. Nature. 2001;410:705–709. doi: 10.1038/35070596. [DOI] [PubMed] [Google Scholar]
- 3.Owsianik G, Talavera K, Voets T, Nilius B. Permeation and selectivity of TRP channels. Annu Rev Physiol. 2006;68:685–717. doi: 10.1146/annurev.physiol.68.040204.101406. [DOI] [PubMed] [Google Scholar]
- 4.Woudenberg-Vrenken TE, et al. Functional TRPV6 channels are crucial for transepithelial Ca2+ absorption. Am J Physiol Gastrointest Liver Physiol. 2012;303:G879–885. doi: 10.1152/ajpgi.00089.2012. [DOI] [PubMed] [Google Scholar]
- 5.Fecher-Trost C, Weissgerber P, Wissenbach U. TRPV6 channels. Handb Exp Pharmacol. 2014;222:359–384. doi: 10.1007/978-3-642-54215-2_14. [DOI] [PubMed] [Google Scholar]
- 6.Lehen’kyi V, Raphael M, Prevarskaya N. The role of the TRPV6 channel in cancer. J Physiol. 2012;590:1369–1376. doi: 10.1113/jphysiol.2011.225862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zakharian E, Cao C, Rohacs T. Intracellular ATP supports TRPV6 activity via lipid kinases and the generation of PtdIns(4,5) P(2) FASEB J. 2011;25:3915–3928. doi: 10.1096/fj.11-184630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.den Dekker E, Hoenderop JG, Nilius B, Bindels RJ. The epithelial calcium channels, TRPV5 & TRPV6: from identification towards regulation. Cell Calcium. 2003;33:497–507. doi: 10.1016/s0143-4160(03)00065-4. [DOI] [PubMed] [Google Scholar]
- 9.Velisetty P, et al. A molecular determinant of phosphoinositide affinity in mammalian TRPV channels. Scientific Reports. 2016;6:27652. doi: 10.1038/srep27652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singh AK, Saotome K, Sobolevsky AI. Swapping of transmembrane domains in the epithelial calcium channel TRPV6. Sci Rep. 2017;7:10669. doi: 10.1038/s41598-017-10993-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saotome K, Singh AK, Yelshanskaya MV, Sobolevsky AI. Crystal structure of the epithelial calcium channel TRPV6. Nature. 2016;534:506–511. doi: 10.1038/nature17975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodding M, Flockerzi V. Ca2+ dependence of the Ca2+-selective TRPV6 channel. J Biol Chem. 2004;279:36546–36552. doi: 10.1074/jbc.M404679200. [DOI] [PubMed] [Google Scholar]
- 13.Hoenderop JG, Nilius B, Bindels RJ. Calcium absorption across epithelia. Physiol Rev. 2005;85:373–422. doi: 10.1152/physrev.00003.2004. [DOI] [PubMed] [Google Scholar]
- 14.Hoenderop JG, et al. Function and expression of the epithelial Ca(2+) channel family: comparison of mammalian ECaC1 and 2. J Physiol. 2001;537:747–761. doi: 10.1111/j.1469-7793.2001.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovacs G, et al. Inhibition of the human epithelial calcium channel TRPV6 by 2-aminoethoxydiphenyl borate (2-APB) Cell Calcium. 2012;52:468–480. doi: 10.1016/j.ceca.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 16.Fecher-Trost C, et al. The in vivo TRPV6 protein starts at a non-AUG triplet, decoded as methionine, upstream of canonical initiation at AUG. J Biol Chem. 2013;288:16629–16644. doi: 10.1074/jbc.M113.469726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voets T, Janssens A, Prenen J, Droogmans G, Nilius B. Mg2+-dependent gating and strong inward rectification of the cation channel TRPV6. J Gen Physiol. 2003;121:245–260. doi: 10.1085/jgp.20028752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao Y, Cao EH, Julius D, Cheng YF. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Wijst J, et al. A Gate Hinge Controls the Epithelial Calcium Channel TRPV5. Sci Rep. 2017;7:45489. doi: 10.1038/srep45489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boukalova S, Marsakova L, Teisinger J, Vlachova V. Conserved residues within the putative S4-S5 region serve distinct functions among thermosensitive vanilloid transient receptor potential (TRPV) channels. J Biol Chem. 2010;285:41455–41462. doi: 10.1074/jbc.M110.145466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zubcevic L, et al. Cryo-electron microscopy structure of the TRPV2 ion channel. Nature Structural & Molecular Biology. 2016;23:180–186. doi: 10.1038/nsmb.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofmann L, Wang H, Beck A, Wissenbach U, Flockerzi V. A conserved gating element in TRPV6 channels. Cell Calcium. 2017;63:24–28. doi: 10.1016/j.ceca.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Paulsen CE, Armache JP, Gao Y, Cheng Y, Julius D. Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huynh KW, et al. Structure of the full-length TRPV2 channel by cryo-EM. Nature Communications. 2016;7:11130. doi: 10.1038/ncomms11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen PS, et al. The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs. Cell. 2016;167:763–773. doi: 10.1016/j.cell.2016.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang Y, et al. The open pore conformation of potassium channels. Nature. 2002;417:523–526. doi: 10.1038/417523a. [DOI] [PubMed] [Google Scholar]
- 27.Jiang Y, et al. X-ray structure of a voltage-dependent K+ channel. Nature. 2003;423:33–41. doi: 10.1038/nature01580. [DOI] [PubMed] [Google Scholar]
- 28.Twomey EC, Yelshanskaya MV, Grassucci RA, Frank J, Sobolevsky AI. Channel opening and gating mechanism in AMPA-subtype glutamate receptors. Nature. 2017;549:60–65. doi: 10.1038/nature23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MS. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 30.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nasr ML, et al. Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods. 2017;14:49–52. doi: 10.1038/nmeth.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Russo CJ, Passmore LA. Electron microscopy: Ultrastable gold substrates for electron cryomicroscopy. Science. 2014;346:1377–1380. doi: 10.1126/science.1259530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimanius D, Forsberg BO, Scheres SHW, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. Elife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9:853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen S, et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy. 2013;135:24–35. doi: 10.1016/j.ultramic.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 46.The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA, USA: p. 2002. [Google Scholar]
- 47.Doyle DA, et al. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 48.Jiang Y, et al. Crystal structure and mechanism of a calcium-gated potassium channel. Nature. 2002;417:515–522. doi: 10.1038/417515a. [DOI] [PubMed] [Google Scholar]
- 49.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 50.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A morph between closed and open states of human TRPV6 represented by the hTRPV6-R470E and wild type hTRPV6 structures, respectively. Shown is the pore domain viewed intracellularly and illustrating the iris-like pore opening or viewed parallel to the membrane, with the front and back subunits disappearing for clarity, and illustrating the α-to-π helical transition in S6 during channel opening. Residues around the gate and critical for permeation aspartates D542 are shown in stick representation.
Data Availability Statement
Cryo-EM density maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMDB-7120 (hTRPV6-nanodiscs), EMDB-7121 (hTRPV6-amphipols), EMDB-7122 (hTRPV6-R470E) and EMDB-7123 (rTRPV6*). Model coordinates have been deposited in the Protein Data Bank (PDB) under accession numbers 6BO8 (hTRPV6-nanodiscs), 6BO9 (hTRPV6-amphipols), 6BOA (hTRPV6-R470E) and 6BOB (rTRPV6*). All other data are available from the corresponding author upon request.


