Abstract
Fritillaria spp. constitute important traditional Chinese medicinal plants. Xinjiang is one of two diversity hotspots in China in which eight Fritillaria species occur, two of which are endemic to the region. Furthermore, the phylogenetic relationships of Xinjiang Fritillaria species (including F. yuminensis) within the genus are unclear. In the present study, we sequenced the chloroplast (cp) genomes of seven Fritillaria species in Xinjiang using the Illumina HiSeq platform, with the aim of assessing the global structural patterns of the seven cp genomes and identifying highly variable cp DNA sequences. These were compared to previously sequenced Fritillaria cp genomes. Phylogenetic analysis was then used to evaluate the relationships of the Xinjiang species and assess the evolution of an undivided stigma. The seven cp genomes ranged from 151,764 to 152,112 bp, presenting a traditional quadripartite structure. The gene order and gene content of the seven cp genomes were identical. A comparison of the 13 cp genomes indicated that the structure is highly conserved. Ten highly divergent regions were identified that could be valuable in phylogenetic and population genetic studies. The phylogenetic relationships of the 13 Fritillaria species inferred from the protein-coding genes, large single-copy, small single-copy, and inverted repeat regions were identical and highly resolved. The phylogenetic relationships of the species corresponded with their geographic distribution patterns, in that the north group (consisting of eight species from Xinjiang and Heilongjiang in North China) and the south group (including six species from South China) were basically divided at 40°N. Species with an undivided stigma were not monophyletic, suggesting that this trait might have evolved several times in the genus.
Introduction
The genus Fritillaria L. (Liliaceae) consists of approximately 140 species and is widely distributed in Europe (mostly in the Mediterranean region), Central Asia, China, Japan, and North America [1]. Twenty-four species occur in China, of which 15 are endemic. They are distributed throughout most provinces in China, among which Sichuan and Xinjiang constitute two diversity hotspots. Seven species occur in Xinjiang, and F. tortifolia X.Z.Duan & X.J.Zheng and F. yuminensis X.Z.Duan are endemic to this region. Two further species, F. tachengensis X.Z.Duan & X.J.Zheng (endemic) and F. ferganensis Losinsk, recorded in Flora Xinjiangensis, were reduced to the synonyms of F. yuminensis and F. walujewii Regel, respectively, in the Flora of China (FOC, http://foc.eflora.cn/).
The morphological traits of Fritillaria species, particularly the Fritillaria cirrhosa D.Don complex (referring to F. cirrhosa and closely-related species in morphology), which are widely distributed in southwest China [2], are complex due to the high variability of several characters, including leaf width; leaf curling; petals tessellated or not, and bract number. However, the mechanism of the variation is not clear and the current classification of some species is only temporary. More comprehensive studies into the morphological variation in the genus are required to facilitate a precise and reasonable species classification [2]. Furthermore, the species occurring in Xinjiang also exhibit significant morphological variation due to the diversity of microclimates (mountains, swamps, saline conditions, and other habitats). Currently, 16 variants are recorded in Flora Xinjiangensis, though they are treated as synonyms of the corresponding accepted species names in the FOC and The Plant List (www.theplantlist.org). Certain character variations of some individuals are prominent and beyond the characteristic range of the genus, such as 8–12 petals, 4–8 stamens, and a 3–5-lobed stigma. Moreover, the stigma of most Fritillaria species is 3-lobed, but in a few species, i.e., F. yuminensis and F. karelinii (Fisch. ex D.Don) Baker, it is undivided. It has been proposed that an undivided stigma is a primitive characteristic [3], but physiological and molecular evidence is required to test this hypothesis and to assess the evolution of this trait within the genus.
The bulbs of some Fritillaria species, including F. thunbergii Miq., F. cirrhosa, F. walujewii, and F. pallidiflora Schrenk, have long been used in traditional Chinese medicine [4]. As a result, long-term excessive harvesting has led to substantial declines in the size of wild Fritillaria populations. At present, all of the eight species in Xinjiang have been classified as vulnerable according to the list of rare endangered endemic higher plants of Xinjiang [5], which has attracted scientific interest. The genetic diversity of some species in the genus was previously assessed, and corresponding conservation areas were proposed [6, 7]; however, some species with very narrow distributions and greater extinction threat require evaluation. A scientific approach to conservation requires an accurate understanding of the population genetic diversity and structure. The diversity estimated by different markers, such as plastid DNA, genomic inter-simple sequence repeats (ISSRs), and single nucleotide polymorphisms (SNPs), can be used to comprehensively inform conservation strategies.
The classification of the genus was previously revised where it was subdivided into eight subgenera, including Davidii, Liliorhiza, Japonica, Fritillaria, Rhinopetalum, Petilium, Theresia, and Korolkowia [8]. A later phylogenetic analysis of 37 Fritillaria species using matK, trnK intron, rp116 intron, and nrDNA ITS [1] supported this subgeneric classification [8]. Khourang et al. investigated the phylogenetic position of nine species in Iran using the ITS and trnL-F regions [9], and showed that members of the subgenera Fritillaria and Rhinopetalum formed one clade. However, a phylogenetic study of 92 species using matK, rbcL, and rpl16 [10] indicated that, in contrast to the results of [1, 9], Fritillaria appeared to be polyphyletic. Additionally, the monophyly of seven out of the eight newly classified subgenera by Rix [8] (F. subgenus davidii, Liliorhiza, Japonica, Rhinopetalum, Petilium, Theresia, and Korolkowia) was well supported. The largest subgenus (F. subgenus Fritillaria) formed two strongly supported clades, with one clade comprising taxa that occur mainly in Europe, the Middle East, and North Africa, and the other clade comprising taxa occurring in China and Central Asia [10]. However, the relationships of some of these species were not well resolved, particularly F. thunbergii Miq. and F. cirrhosa. The phylogenetic position of the Xinjiang-endemic species F. yuminensis remains unclear.
The chloroplast (cp) genome in angiosperms is highly conserved, with a quadripartite structure consisting of a large single copy (LSC) region, a small single copy (SSC) region, and two copies of a larger inverted repeat (IR). The gene orders in these regions are also similar; however, structural rearrangements and gene losses can be found in some lineages [11, 12]. Plastid sequences have been widely used for deciphering phylogenetic relationships and in DNA barcoding to identify plant species [13]. However, DNA barcoding for species identification and phylogenetic analysis is hampered by weak resolution in some plants [14–16]. Complete cp genomes have therefore emerged as a means of improving the resolution of phylogenies that have varied among, or been unresolved in, earlier single- and multi-gene studies [17–20]. With the rapid development of next-generation sequencing techniques, it is now more convenient and relatively inexpensive to obtain cp genome sequences and extend gene-based phylogenetics to phylogenomics.
To date, a total of six Fritillaria cp genomes have been sequenced and are available on GenBank. Park et al. reported the cp genomes of F. ussuriensis and F. cirrhosa and performed a comparative analysis with four Fritillaria cp genomes available on GenBank, the outcome of which has provided a basic understanding of the cp genome characteristics of the genus [21]. In the present study, we sequenced the cp genomes of seven Fritillaria species from Xinjiang using the Illumina HiSeq platform. The aims of this study were to (1) analyze the global structural patterns of the seven cp genomes and compare them with the six cp genomes available on GenBank; (2) discover highly divergent DNA markers that can be used for population genetics; and (3) evaluate the phylogenetic relationships of the Xinjiang species, particularly the position of F. yuminensis, and assess the evolution of an undivided stigma in the genus.
Materials and methods
Plant materials
Fresh leaves of seven Fritillaria species were collected from Tacheng and Yili Prefecture of Xinjiang Uygur Autonomous Region, China. The geographic origin and coordinates of sampling locations were listed in S1 Table. The sample collection was approved by the Forestry Bureau of Tacheng Prefecture and Yili Prefecture. For each species, two to five individuals were sampled. Voucher specimens were deposited at the Xinjiang Institute of Ecology and Geography, Chinese Academy of Sciences (S1 Table).
Genome sequencing
Total DNA was extracted from approximately 100 mg of fresh leaves using the CTAB method following Yao et al. [22]. Illumina paired-end libraries were constructed and sequenced by the Illumina HiSeq X-Ten platform (Illumina Inc., USA) at the Germplasm Bank of Wild Species in Southwest China, Kunming Institution of Botany, Chinese Academy of Sciences. Each individual of each species was sequenced independently. In total, 22 individuals of seven species were sequenced. Because the cp genome sequences of repeat individuals of each species were almost identical, therefore, we reported only one genome of each species.
Genome assembly and annotation
The raw reads were trimmed and assembled into contigs using SPAdes [23]. Contigs representing the cp genome were obtained after a BLAST search using the cp genome sequence of F. cirrhosa (GenBank No. KY646167) as a reference sequence. The resulting contigs were assembled after being aligned to the reference genome using Geneious 4.8 [24] and annotated using the Dual Organellar GenoMe Annotator (DOGMA) database [25]. The cp genome map was generated using OGDRAW (http://ogdraw.mpimp-golm.mpg.de/) [26]. The raw sequencing data were deposited in GenBank SRA database (SAMN08348372–SAMN08348378, https://submit.ncbi.nlm.nih.gov/subs/sra). The annotated seven cp genomes were deposited in GenBank (accession number MG200070, MG211818-MG211823).
Genome comparison
A comparative plot consisting of full alignments of the cp genomes with annotations was produced by mVISTA using F. cirrhosa as the reference. The sequences were aligned using MEGA 6 [27] and then manually adjusted using BioEdit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Subsequently, a sliding window analysis was conducted to evaluate the nucleotide diversity (Pi) of the cp genome using DnaSP 5.1 [28]. The step size and window length was set to 200 bp and 600 bp, respectively. The number of variable sites and the Pi across the complete cp genomes, LSC, SSC, and IR regions were calculated using DnaSP 5.1. The p-distance among species was calculated in MEGA 6 to evaluate the divergence of Fritillaria species.
Phylogenetic analyses
Sequences of the 13 Fritillaria species and three Lilium species were aligned using MEGA 6. Phylogenies were constructed by maximum likelihood (ML) and Bayesian Inference (BI) analyses using the protein-coding genes (PCGs), LSC, SSC, and IR regions. ML analyses were conducted in MEGA 6, while BI analyses were conducted using BEAST 1.7 [29]. GTR+G+I and GTR+G were selected as the best substitution models for the ML and BI analyses according to the Akaike information criterion (AIC) [30] and Bayesian information criterion (BIC) [31], respectively, and were estimated using MrModeltest 2.3 [32]. For ML, initial tree(s) for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach. The tree was drawn to scale, with branch lengths measured in the number of substitutions per site. All alignment positions containing more than 5% gaps were eliminated. For BI, two independent Markov Chain Monte Carlo chains were conducted simultaneously for 2 × 107 generations and sampled every 1,000 generations. Each run was assessed using Tracer 1.6 [33] to evaluate whether a sufficient effective sample size (ESS) had been reached. The two runs were considered as converged when the ESS of all relevant parameters was above 200. A consensus maximum clade credibility (MCC) tree was generated from the 75% post-burn-in trees using TreeAnnotator 1.7.
Results
Genome sequencing, assembly, and genome features
Illumina sequencing generated 3.5 to 7.1 Gb of raw reads and 164,351 to 697,002 paired-end reads for the seven Fritillaria species. After the de novo assembly, eight to 19 contigs covering the whole chloroplast genome were used to generate a complete cp genome (S1 Table). Using reference-guided assembly, seven Fritillaria cp genomes were obtained, with coverage of 162× to 688× for each species.
The full-length cp genomes of the seven species ranged from 151,764 in F. meleagroides Patrin ex Schult. & Schult.f. to 152,112 bp in F. karelinii (Table 1). The cp genome presented a typical quadripartite structure including one LSC region (81,533–81,879 bp), one SSC region (17,277–17,526 bp), and a pair of IR regions (52,654–52,778 bp; 26,327–26,389 each).
Table 1. The chloroplast genomic characteristics of 13 Fritillaria species.
| Species | Genome (bp) | LSC (bp) | SSC (bp) | IRs (bp) | PCG | tRNA | rRNA | GC (%) | GenBank accession No. |
|---|---|---|---|---|---|---|---|---|---|
| F. pallidiflora Schrenk | 152,078 | 81,787 | 17,513 | 26,389 | 78 | 30 | 4 | 37 | MG211822 |
| F. tortifolia X.Z.Duan & X.J.Zheng | 152,005 | 81,778 | 17,509 | 26,359 | 78 | 30 | 4 | 37 | MG211819 |
| F. walujewii Regel | 151,920 | 81,743 | 17,523 | 26,327 | 78 | 30 | 4 | 36.9 | MG211820 |
| F. verticillata Willd. | 151,959 | 81,730 | 17,509 | 26,360 | 78 | 30 | 4 | 36.9 | MG211823 |
| F. karelinii (Fisch. ex D.Don) Baker | 152,112 | 81,879 | 17,473 | 26,381 | 78 | 30 | 4 | 36.9 | MG211818 |
| F. meleagroides Patrin ex Schult. & Schult.f. | 151,764 | 81,833 | 17,277 | 26,327 | 78 | 30 | 4 | 36.9 | MG211821 |
| F. yuminensis X.Z.Duan | 151,813 | 81,533 | 17,526 | 26,377 | 78 | 30 | 4 | 36.9 | MG200070 |
| F. ussuriensis Maxim. | 151,524 | 81,732 | 17,114 | 26,339 | 78 | 30 | 4 | 36.95 | KY646166 |
| F. cirrhosa D.Don | 151,083 | 81,390 | 17,537 | 26,078 | 78 | 30 | 4 | 36.96 | KY646167 |
| F. hupenesis P.K.Hsiao & K.C.Hsia | 152,145 | 81,898 | 17,553 | 26,347 | 77 | 30 | 4 | 37 | NC024736 |
| F. taipaiensis P.Y.Li | 151,693 | 81,390 | 17,550 | 26,352 | 78 | 30 | 4 | 37 | NC023247 |
| F. unibracteata P.K.Hsiao & K.C.Hsia | 151,009 | 81,290 | 17,541 | 26,089 | 78 | 30 | 4 | 37 | KF769142 |
| F. thunbergii Miq. | 152,155 | 81,890 | 17,565 | 26,350 | 78 | 30 | 4 | 37 | KY646165 |
The gene content and order were identical in the seven species. A total of 114 distinct genes were annotated, including 78 PCGs, 30 tRNA genes, four rRNA genes, infA (translation initiation factor gene), and hypothetical ORF ycf15 (S2 Table). Eighteen genes were duplicated in the cp genome, including eight tRNAs (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC, trnH-GUG), four rRNAs (rrn16, rrn23, rrn4.5, rrn5), and six PCGs (ndhB, rpl2, rpl23, rps12, ycf2, rps7). Gene rps12 was trans-spliced because the 5′end was located in the LSC region and the 3′ end in the IR region. Gene ycf1 in the junction region between SSC and IRb was the only pseudogene found due to the incomplete duplication of the normal copy in the junction region (Fig 1). There were 18 intron-containing genes, among which two genes (ycf3 and clpP) had two introns each, while the other 16 had one intron, including 10 PCGs (atpF, rpoC1, rpl2, ndhB, ndhA, petB, petD, rpl16, rps16, rps12) and six tRNA genes (trnA-UGC, trnG-GCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) (Fig 1).
Fig 1. Gene map of the Fritillaria cp genome.
Genes belonging to different functional groups are color-coded. The dashed area in the inner circle indicates the GC content. Cp genome size ranges are provided for the seven Xinjiang Fritillaria species.
The Pi of the seven species was 0.00648. SSC had the highest Pi value, while IR had the lowest value (Table 2). The mean p-distance among the seven species was 0.00558, ranging from 0.003 to 0.01. The distance between F. karelinii /F. meleagroides and the other five species was larger than that between the five species (S3 Table), indicating that F. karelinii and F. meleagroides were most divergent.
Table 2. Variable site analyses in Fritillaria chloroplast genomes.
| Among 13 Fritillaria species | Among seven Xinjiang species | Among other six species | |||||
|---|---|---|---|---|---|---|---|
| Region | Total sites | Variable sites | Pi | Variable sites | Pi | Variable sites | Pi |
| LSC | 84,114 | 2,227 | 0.00737 | 1,734 | 0.00782 | 1,368 | 0.00638 |
| SSC | 17,854 | 672 | 0.01044 | 518 | 0.0112 | 346 | 0.00774 |
| IR | 26,550 | 152 | 0.00148 | 126 | 0.00169 | 59 | 0.00094 |
| Complete cp genome | 154,837 | 3,199 | 0.00557 | 2,498 | 0.00684 | 1,625 | 0.00419 |
Genome sequence divergence
We compared the Pi of the LSC, SSC, and IR regions of the cp genome. In total, 3,199 variable sites were found (Pi = 0.00557), indicating moderate genetic divergence of the Fritillaria cp genomes. The IR regions exhibited the lowest Pi (0.00148), while SSC had the highest Pi (0.01044) (Table 2). The p-distances among the Fritillaria species ranged from 0.0001 to 0.01, and F. karelinii, F. meleagroides, and F. ussuriensis exhibited the greatest sequence divergence. The Pi of the Xinjiang species (0.00648) was higher than that of the species from the other regions (0.00419), as the two highly divergent species F. karelinii and F. meleagroides are from Xinjiang.
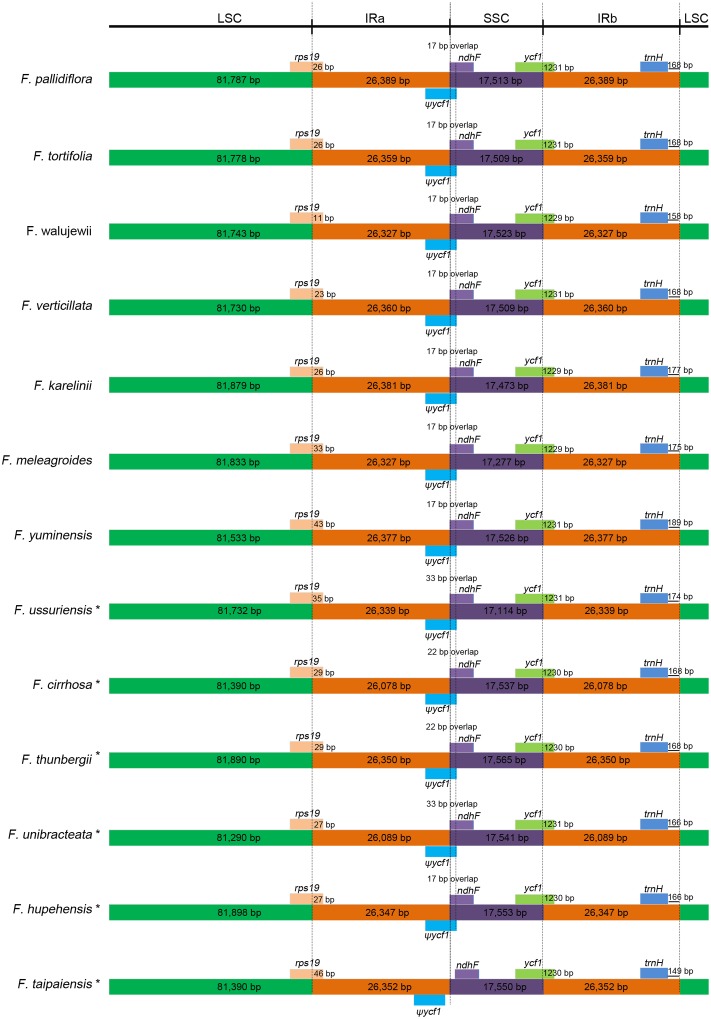
Junction characteristics
The junction of the LSC, SSC, and IR regions of the seven species are shown in Fig 2. The rps19 gene located in the LSC was extended into the IRa by 11–43 bp. The border between IRa/SSC and SSC/IRb extended into the ycf1 genes. Overlaps of 17 bp were found between the ycf1 pseudogene and the ndhF gene. The trnH genes were all located in the IR region, 158–189 bp from the IRb/LSC boundary.
Fig 2. Comparison of LSC, SSC, and IR border regions among the 13 Fritillaria cp genomes.
Colored boxes for genes represent the gene position. ψ: pseudogenes. *: these six species were redrawn according to Park et al. [21].
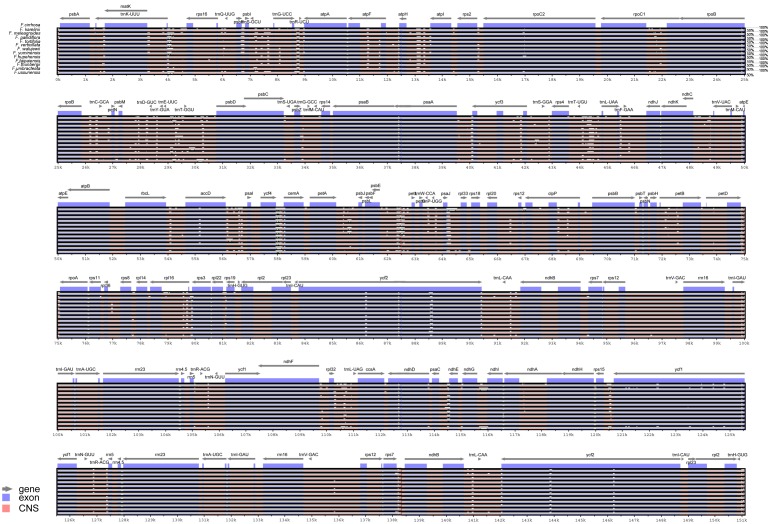
Genome-wide comparative analyses
We aligned the 13 Fritillaria cp genomes using mVISTA, and found that the gene order and clusters were very similar in all the species (Fig 3). Using sliding window analysis, we identified the 10 most divergent regions that could be utilized as potential molecular markers for population genetic and phylogenetic studies in Fritillaria. These regions included matK-rps16, trnS-trnG, atpH-atpI, trnC-petN, trnE-trnT-psbT, trnT-trnL-trnF, rps12-psbB, rpl32-trnL in IGS, and the petB intron and ycf1 in the coding region (Fig 4). Additionally, psbB-psbH, petD-rpoA, ycf4-cemA, and ycf2 also constitute potential candidates.
Fig 3. Comparison of 13 Fritillaria cp genomes with F. cirrhosa as the reference.
LSC: long single copy region; SSC: short single copy region; IRa and IRb: inverted regions. Gray arrow: gene and translation direction; blue block: exon of the gene; red block: conserved non-coding sequences (CNS). Sequence identities are labeled at the right side and range between 50%-100%.
Fig 4. Sliding window analysis of the entire chloroplast genome of 13 Fritillaria species.
Phylogenetic analyses
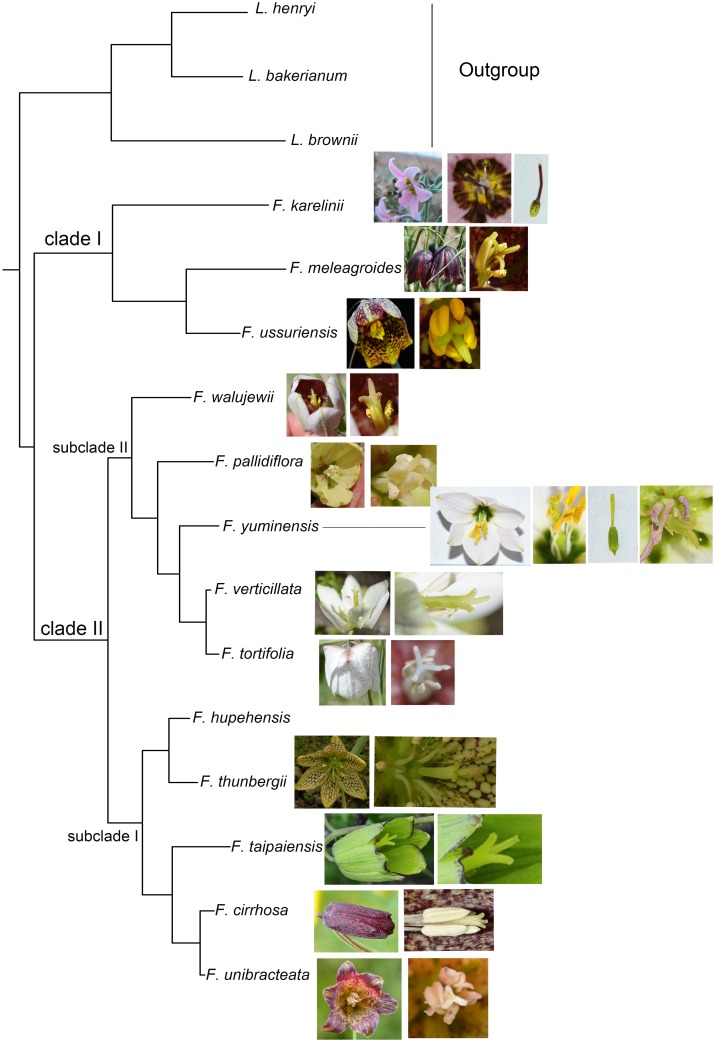
The phylogenetic analyses were conducted with the PCGs, LSC, SSC, and IR regions using ML and BI inference methods. All the analyses revealed congruent tree topologies, and all branches were highly supported (S1 Fig). Two clades were identified among the 13 Fritillaria species. Clade I contained F. ussuriensis, F. meleagroides, and F. karelinii. The sister clade (clade II) comprised the remaining 10 species, in which two subclades were strongly supported. Subclade I contained species from South China (F. cirrhosa, F. unibracteata, F. taipaiensis, F. hupehensis, and F. thunbergii). Subclade II included five Xinjiang species, namely F. tortifolia, F. verticillata, F. yuminensis, F. pallidiflora, and F. walujewii. The seven Xinjiang species did not form a monophyletic group, as F. meleagroides and F. karelinii were separated from the other five species. Additionally, F. yuminensis had a close phylogenetic relationship with F. tortifolia and F. verticillata.
Discussion
In this study, seven new cp genomes of Fritillaria were sequenced and ranged in size from 151,764 to 152,112 bp. The reported Fritillaria cp genome size in this study is consistent with previously sequenced Fritillaria cp genomes, and is also within the cp genome size range of angiosperms. The gene content and gene order were the same in the seven Xinjiang species, containing 78 PCGs, 30 tRNA genes, four rRNA genes, and the infA and ycf15 genes. Compared with the other six species, the tRNA and rRNA genes were identical, but the PCGs differed, ranging from 77 to 78 due to the absence of the clpP gene in the cp genome of F. hupehensis. The hypothetical gene ycf68 was present in the cp genome of F. unibracteata, while ycf15 was absent from F. taipaiensis, F. thunbergii, and F. ussuriensis. The absence or presence of some genes in a particular species of a genus has also been observed in Ipomoea [34]. The functions of ycf15 and ycf68 are ambiguous in various land plants; for instance, ycf15 in Ipomoea purpurea and Ageratina adenophora encode a complete RF15 protein, but the former has no ycf68, while the latter has one incomplete ycf68 open reading frame. In Musa acuminata, these two genes were determined as non-functional due to the presence of several stop codons in the gene sequence [34].
The boundaries between IR and LSC or SSC were identical, except in F. taipaiensis. The LSC/IRa boundary of Fritillaria is located in the rps19 gene, and a small section of the 5′end of rps19 is in the IRb region, which is similar to Ilex [22] and Brassicaceae species [35–38]. In contrast, rps19 does not extend into the IR in Lupinus luteus [39] and Millettia pinnata [40], while in others, such as Phaseolus vulgaris [41] and Oryza [42], the whole gene is contained inside the IR. ψycf1 spans the SSC/IRa boundary and overlaps with the ndhF gene in most of the Fritillaria species. However, these are separated and located at each side of the boundary in the F. taipaiensis cp genome, which has also been observed in Petroselinum crispum (HM596073), Tiedemania filiformis (HM596071), and Panax ginseng (AY582139). The SSC/IRb boundary is inside the ycf1 gene, which is consistent with many plants, including those from Asteraceae [43], Ilex [22], Lilium [44], and Ananas [45]. Conversely, in Cryptochloa strictiflora [46] and Ipomoea batata [34], the junction falls into the ndhF gene due to the loss of the ycf1 gene. The trnH gene is duplicated in the IRs in Fritillaria, as observed in Lilium [44], whereas trnH is a single cope gene located in the LSC of other species, such as Ipomoea batata [34], Datura stramonium [47], and Citrus aurantiifolia [48].
The genomic structure and gene order of the Fritillaria cp genomes are highly conserved, and no rearrangement has occurred. The IRs of the Fritillaria species were about 26 kb, which is within the size range of most angiosperm cp genomes (20–28 kb). The IR usually varies between 200 and 300 nucleotides in seed plants. However, the extreme expansion of the IRs has been observed in Oenothera (54 kb) [49], Fabaceae (50 kb) [50], and Pelargonium×hortorum (75 kb) [51]. In contrast, the loss or near loss of the IR has also been also detected in Erodium and Sarcocaulon [52]. These significant contractions and expansions of the IR contribute towards genome size variation.
Several variable cp DNA markers have been used in phylogenetic studies of Fritillaria, for instance rbcL, matK, and atpB. Some divergent intergenic spacers, i.e., trnH-psbA, rpl32-trnL, psbB-psbH, and trnS-trnG, are more informative and suitable in lower taxonomic ranks [53]. Upon comparison of the 13 cp genomes, the 10 most divergent regions were identified, and included matK-rps16, trnS-trnG, atpH-atpI, trnC-petN, trnE-trnT-psbT, trnT-trnL-trnF, rps12-psbB, and rpl32-trnL in IGS, and the petB intron and ycf1 in the coding region. The Pi of these regions ranged from 0.015 to 0.022. Additionally, psbB-psbH, petD-rpoA, ycf4-cemA, and ycf2 also constitute potential candidates, which corroborates previous studies [44]. These highly divergent regions (also called hotspots) in the cp genome are useful for further phylogenetic and population genetics studies. However, in contrast to Park et al. [21], we found the petB intron to be highly divergent. Furthermore, the clpP intron was also found to be highly variable, as reported in Acacia ligulata [54]. Gene ycf1 is considered as the most promising plastid DNA barcode of land plants [55].
Universal DNA barcoding is widely used in the identification of plant species, but has several limitations [14–16]. The complete cp genome, as a super DNA barcode, has been successfully used in numerous phylogenetic studies of seed plants [56, 57] and in resolving species relationships at lower taxonomic levels [58]. Park et al. conducted a phylogenetic study of six Fritillaria species based on the cp genome and concluded that plastome phylogenies are suitable for uncovering relationships among Fritillaria species, and obtain good support with high bootstrap values [21]. We constructed phylogenetic trees of 13 Fritillaria species using the PCG, LSC, SSC, and IR datasets. The phylogenetic relationships within the genus were identical and strongly supported in all of the phylogenies. In this study, Fritillaria appears to be a monophyletic group, which differs from the results of Day et al. [10] and may be attributed to our smaller sampling size. However, the positions of F. cirrhosa and F. thunbergii are far more highly resolved in our study (S1 Fig).
With some exceptions, our phylogenies are largely consistent with Day et al. [10] and support the polyphyletic classification of F. subgenus Fritillaria by Rix [8]. Two species of F. subgenus Fritillaria (F. meleagroides and F. ussuriensis) clustered together and are sister to F. karelinii of F. subgenus Rhinopetalum in clade I (Figs 5 and 6), which is similar to the results of Khourang et al. [9], and may be attributed to the small sample size. The other 10 species of F. subgenus Fritillaria formed a strongly supported clade (clade II), and two subclades were resolved in clade II. The five species from outside Xinjiang formed a strongly supported subclade (subclade I), which was sister to subclade II containing the other five Xinjiang species. This indicated that the Xinjiang species had a close genetic affinity. Interestingly, we found that the eight species in clade I and subclade II of clade II originate from Xinjiang and Heilongjiang in North China (named the “north group”), and the five species in subclade I of clade II originate from South China (named the “south group”). An alternative explanation of the phylogenetic pattern is that the southern taxa diverged from the northern taxa and become distinct due to limited seed flow or genetic contact.
Fig 5. Map indicating the distribution of Fritillaria in China.
The distribution area of each species is drawn according to the records in the FOC and Flora Xinjiangensis. Photographs of the species are also provided.
Fig 6. The evolutionary progression of stigmatic traits within the genus.
The seven Xinjiang species did not form a monophyletic group, as F. karelinii and F. meleagroides were highly divergent from the other five species, and, interestingly, also differ in their morphology and habitat. Specifically, F. meleagroides occurs in a variety of habitats, including hilly slopes, shallow waters in mountainous areas, saline areas, and shallow swamps, while F. karelinii can usually be found in the plains of Artemisia desert habitats (desert habitat dominated by some drought tolerant Artemisia species) or low gravel hills. This species has a style that is longer than the stamens, and the stigma is scarcely lobed and slightly inflated at the top (Figs 5 and 6).
Endemic species are often limited to specific geographic areas, and in many instances have evolved vicariantly [59]. Previous studies have demonstrated that specific limiting factors in an environment can significantly influence the geographic distribution patterns of species, including physical factors (i.e., temperature, light, moisture, aridity) and biotic factors (i.e., competition, predation, food availability). These factors usually influence the survival and propagation ability of plants. For instance, Corynephorus canescens is widely distributed in mid and south Europe, and its northern distribution limit in Europe coincides with the 15°C isotherm in July, as its germination and flowering are affected by low temperature [60]. The winter distribution and abundance patterns of several avian species are directly linked to their physiological limits, with the northern range limit being associated with the −4°C isotherm of the average minimum January temperature [61]. As high solar radiation and temperature are most favorable for the C4 photosynthetic pathway, C4 grass abundance patterns in North America are separated at 40°N, where the C4 grass abundance is above 50% north of 40°N and below 50% south of 40°N [62].
Interestingly, we also discovered that the northern and southern groups were largely separated at 40°N (Fig 5). However, the determining factor(s) influencing the distribution of Fritillaria species are not investigated in the present study. However, we hypothesize that soil moisture is an important environmental constraint influencing the growth of Fritillaria and other spring ephemeral plants. A semi-arid or desert climate prevails in Xinjiang and the precipitation is very low. Adequate water supply is only available from snow melting during March to June. From late June, the climate turns dry and hot, and is not suitable for growth. They have therefore adapted to a complete growth cycle ahead of the hot summer. Conversely, in south China, such as Sichuan, Hubei, and Zhejiang, precipitation is greater in summer, and thus some species have much longer growth cycles and can thrive from August to October (i.e., F. cirrhosa) (S4 Table).
The stigma in the majority of Fritillaria species is 3-lobed; however, in a few species, i.e., F. yuminensis and F. karelinii, the stigma is undivided (Fig 6). We surveyed 48 species in FOC and Flora of USSR, and found that only four species possess a scarcely lobed stigma. It was proposed that the trait of an undivided stigma might be a primitive characteristic [3]. Our results do not support this hypothesis. The phylogenies demonstrate that F. karelinii diverges early, while F. yuminensis does not, and F. karelinii is closely related to F. tortifolia and F. verticillata. Moreover, in comparison to the phylogeny of Day et al. [10], F. karelinii is not resolved as a basal species. Therefore, at this stage, we cannot infer a definite evolutionary trend for this trait. More cp genomes need to be sequenced to gain a comprehensive and accurate assessment of the evolutionary progression of the stigma. Furthermore, as F. yuminensis and F. karelinii do not form a monophyletic clade, this suggests that this trait might have evolved independently several times in the genus.
Additionally, wild Fritillaria populations have been dramatically reduced due to excessive harvesting in recent decades. During our field investigation, we noted that F. meleagroides and F. karelinii were rare in the wild. The endemic species F. yuminensis is now endangered and can only be found in remote areas that are uninhabited by humans and livestock. Small populations of the other endemic species F. tortifolia can only be found in remote areas and natural reserves. Although all seven species in Xinjiang are listed in the class I protection plant list of Xinjiang, conservation action is urgently required. Population diversity is an important index in the formulation of a scientific conservation strategy. The newly sequenced cp genomes of these seven Fritillaria species would be useful for the development of SSR markers, and together with the identified divergent regions DNA regions, could be used to comprehensively assess the genetic diversity of wild populations in order to inform the protection of these valuable medicinal resources.
As there are more than 140 species in the genus, the currently sequenced species only represent a very limited sample. However, we provide evidence that the cp genome can increase the resolution of phylogenetic relationships within the genus. More cp genomes are required to clarify the taxonomic and phylogenetic relationships of Fritillaria species at lower taxonomic levels, and can be used to estimate the population genetic diversity in order to formulate effective protection strategies.
Supporting information
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(A) PCGs, (B) LSC region, (C) SSC region, and (D) IR region. Values above the branches represent ML bootstrap values/BI posterior probability values. Outgroup: Lilium brownii KY748296; L. bakerianum KY748301; L. henryi KY748302.
(TIF)
Acknowledgments
We thank Zhou Yao, Xu Yechun, Liu Jun, Hua Guojun, Liu Zhaolong, Zhu Xinxin, and Duan Liaochuan for providing pictures of some Fritillaria species, and the authorization of PPBC for their publication. We also thank Yin Gang for providing the map of China. We would like to thank LetPub for providing linguistic assistance during the preparation of this manuscript.
Data Availability
The raw sequencing data were deposited in GenBank SRA database (SAMN08348372–SAMN08348378, https://submit.ncbi.nlm.nih.gov/subs/sra). The annotated seven cp genomes were deposited in GenBank (accession number MG200070, MG211818-MG211823).
Funding Statement
This work is funded by the Natural Science Foundation of China (31500309, 31560131), and a Grant of the Large-scale Scientific Facilities of the Chinese Academy of Sciences (No. 2017-LSF-GBOWS-02).
References
- 1.Rønsted N, Law S, Thornton H, Fay MF, Chase MW. Molecular phylogenetic evidence for the monophyly of Fritillaria and Lilium Liliaceae; Liliales) and the infrageneric classification of Fritillaria. Mol Phylogenet Evol. 2005; 35: 509–527. [DOI] [PubMed] [Google Scholar]
- 2.Luo YB, Chen SC. A revision of Fritillaria L. (Liliaceae) in the Hengduan Mountains and adjacent regions, China (1)- a study of Fritillaria cirrhosa D. Don and its related species. Acta Phytotaxon Sin. 1996; 34: 304–312.(In Chinese) [Google Scholar]
- 3.Duan XZ. Fritillaria yuminensis X.Z. Duan. Act Phyt of Sin. 1981;19(2): 257–258. (In Chinese). [Google Scholar]
- 4.Pharmacopoeia of the People’s Republic of China vol. 1: Chinese Pharmacopoeia Commission. China Science Medical Press, Beijing; 2010. [Google Scholar]
- 5.Yin LK, Tan LX, Wang B. Rare endanged endemic higher plants in Xinjiang of China. Science Press: Urumqi, China; 2006. [Google Scholar]
- 6.Su ZH, Pan BR, Sanderson SC, Jiang XL, Zhang ML. Conservation genetics and geographic patterns of genetic variation of the endangered officinal herb Fritillaria pallidiflora. Nord J Bot. 2015; 33: 506–512. [Google Scholar]
- 7.Su ZH, Pan BR, Sanderson SC, Shi XJ, Jiang XL. Conservation genetics and geographic patterns of genetic variation of the vulnerable officinal herb Fritillaria walujewii (Liliaceae). Aust J Bot. 2015; 63: 467–476. [Google Scholar]
- 8.Rix EM. Fritillaria: A revised classification together with an updated list of species. Publication of the Fritillaria Group of the Alpine Garden Society, UK; 2001. [Google Scholar]
- 9.Khourang M, Babaei A, Sefidkon F, Naghavi MR, Asgari D, Potter D. Phylogenetic relationship in Fritillaria spp. of Iran inferred from ribosomal ITS and chloroplast trnL-trnF sequence data. Biochem Syst Ecol. 2014;57: 451–457. [Google Scholar]
- 10.Day PD, Berger M, Hill L, Fay MF, Leitch AR, Leitch IJ, et al. Evolutionary relationships in the medicinally important genus Fritillaria L. (Liliaceae). Mol Phylogenet Evol. 2014; 80: 11–19. doi: 10.1016/j.ympev.2014.07.024 [DOI] [PubMed] [Google Scholar]
- 11.Wicke S, Schneeweiss GM, dePamphilis CW, Muller KF, Quandt D. The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol Biol. 2011; 76: 273–297. doi: 10.1007/s11103-011-9762-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo XY, Liu JQ, Hao GQ, Zhang L, Mao KS, Wang XJ, et al. Plastome phylogeny and early diversification of Brassicaceae. BMC Genomics. 2017;18(1):176 doi: 10.1186/s12864-017-3555-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. P Natl Acad Sci USA. 2005;102: 8369–8374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Feng Y, Wang XY, Liu B, Lv GH. Failure of DNA barcoding in discriminating Calligonum species. Nord J Bot. 2014; 32: 511–517. [Google Scholar]
- 15.Percy DM, Argus GW, Cronk QC, Fazekas AJ, Kesanakurti PR, Burgess KS, et al. Understanding the spectacular failure of DNA barcoding in willows (Salix): does this result from a trans-specific selective sweep? Mol Ecol. 2014; 23: 4737–4756. doi: 10.1111/mec.12837 [DOI] [PubMed] [Google Scholar]
- 16.Burke SV, Wysocki WP, Zuloaga FO, Craine JM, Pires JC, Edger PP, et al. Evolutionary relationships in Panicoid grasses based on plastome phylogenomics (Panicoideae; Poaceae). BMC Plant Biol. 2016; 16(1): 140 doi: 10.1186/s12870-016-0823-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu JM, Zhang N, Du XY, Wen J, Li DZ. Chloroplast phylogenomics resolves key relationships in ferns. J Syst Evol. 2015; 53: 448–457. [Google Scholar]
- 18.Saarela JM, Wysocki WP, Barrett CF, Soreng RJ, Davis JI, Clark LG, et al. Plastid phylogenomics of the cool-season grass subfamily: clarification of relationships among early-diverging tribes. Aob Plants. 2015; 7: plv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng PK, Lin SM, Lim PE, Liu LC, Chen CM, Pai TW. Complete chloroplast genome of Gracilaria firma (Gracilariaceae, Rhodophyta), with discussion on the use of chloroplast phylogenomics in the subclass Rhodymeniophycidae. BMC Genomics. 2017; 18(1): 40 doi: 10.1186/s12864-016-3453-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wei R, Yan YH, Harris AJ, Kang JS, Shen H, Xiang QP, et al. Plastid phylogenomics resolve deep relationships among eupolypod II ferns with rapid radiation and rate heterogeneity. Genome Biol Evol. 2017;9: 1646–1657. doi: 10.1093/gbe/evx107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park I, Kim WJ, Yeo SM, Choi G, Kang YM, Piao R, et al. The complete chloroplast genome sequences of Fritillaria ussuriensis Maxim. and Fritillaria cirrhosa D. Don, and comparative analysis with other Fritillaria species. Molecules. 2017; 22: 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao X, Tan YH, Liu YY, Song Y, Yang JB, Corlett RT. Chloroplast genome structure in Ilex (Aquifoliaceae). Sci Rep. 2016; 6: 28559 doi: 10.1038/srep28559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012; 19: 455–477. doi: 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012; 28: 1647–1649. doi: 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004; 20: 3252–3255. doi: 10.1093/bioinformatics/bth352 [DOI] [PubMed] [Google Scholar]
- 26.Lohse M, Drechsel O, Kahlau S, Bock R. OrganellarGenomeDRAW-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013; 41: W575–W581. doi: 10.1093/nar/gkt289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Bio Evol. 2013; 30(12): 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009; 25: 1451–1452. doi: 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- 29.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012; 29: 1969–1973. doi: 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akaike H. 1973. Information theory and an extension of the maximum likelihood principle. In: B.N. Petrov, F. Csaki (eds.) 2nd International symposium on information theory. Akademiai Kiado, Budapest. 1973; pp 267–281.
- 31.Schwarz G. Estimating the dimension of a model. Ann Stat. 1978; 6: 461–464. [Google Scholar]
- 32.Nylander JAA. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. 2004.
- 33.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.6. 2014. http://tree.bio.ed.ac.uk/software/tracer/
- 34.Yan L, Lai XJ, Li XD, Wei CH, Tan XM, Zhang YZ. Analyses of the complete genome and gene expression of chloroplast of sweet potato (Ipomoea batata). Plos One. 2015; 10(4): e0124083 doi: 10.1371/journal.pone.0124083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohta Y, Hirata Y, Motegi T, Hattori G, Noguchi T. Analysis of chloroplast genome of two cytoplasmic male sterile lines derived from interspecific chimera and intergeneric somatic hybrid in Brassicaceae. Breeding Sci. 2006; 56: 1–5. [Google Scholar]
- 36.Hu QJ, Hu H, Guo XY, Ma YZ, Liu JQ, Ma T. Characterization of the complete chloroplast genome of two sister species of Pugionium (Brassicaceae). Conserv Genet Resour. 2016; 8: 243–245. [Google Scholar]
- 37.Shang HY, Li YS, Guo XY, Wang XJ. Characterization of the complete chloroplast genome of two sister species of salt cress (Brassicaceae). Conserv Genet Resour. 2017; 9: 237–239. [Google Scholar]
- 38.Zhou T, Yang YC, Hu YH, Zhang X, Bai GQ, Zhao GF. Characterization of the complete chloroplast genome sequence of Lepidium meyenii (Brassicaceae). Conserv Genet Resour. 2017; 9: 405–408. [Google Scholar]
- 39.Martin GE, Rousseau-Gueutin M, Cordonnier S, Lima O, Michon-Coudouel S, Naquin D, et al. The first complete chloroplast genome of the Genistoid legume Lupinus luteus: evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Ann Bot. 2014; 113: 1197–1210. doi: 10.1093/aob/mcu050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kazakoff SH, Imelfort M, Edwards D, Koehorst J, Biswas B, Batley J, et al. Capturing the biofuel wellhead and powerhouse: the chloroplast and mitochondrial genomes of the leguminous feedstock tree Pongamia pinnata. Plos One. 2012; 7(12): e51687 doi: 10.1371/journal.pone.0051687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo XW, Castillo-Ramirez S, Gonzalez V, Bustos P, Fernandez-Vazquez JL, Santamaria RI., et al. Rapid evolutionary change of common bean (Phaseolus vulgaris L) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics. 2007; 8(1): 228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wambugu PW, Brozynska M, Furtado A, Waters DL, Henry RJ. Relationships of wild and domesticated rices (Oryza AA genome species) based upon whole chloroplast genome sequences. Sci Rep. 2015; 5: 13957 doi: 10.1038/srep13957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curci PL, de Paola D, Danzi D, Vendramin GG, Sonnante G. Complete chloroplast genome of the multifunctional crop globe artichoke and comparison with other Asteraceae. Plos One. 2015; 10(3): e0120589 doi: 10.1371/journal.pone.0120589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du YP, Bi Y, Yang FP, Zhang MF, Chen XQ, Xue J, et al. Complete chloroplast genome sequences of Lilium: insights into evolutionary dynamics and phylogenetic analyses. Sci Rep. 2017; 7(1): 5751 doi: 10.1038/s41598-017-06210-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nashima K, Terakami S, Nishitani C, Kunihisa M, Shoda M, Takeuchi M, et al. Complete chloroplast genome sequence of pineapple (Ananas comosus). Tree Genet Genomes. 2015; 11(3): 60. [Google Scholar]
- 46.Burke SV, Grennan CP, Duvall MR. Plastome sequences of two New World bamboos—Arundinaria gigantea and Cryptochloa strictiflora (Poaceae)—extend phylogenomic understanding of Bambusoideae. Am J Bot. 2012; 99 (12): 1951–1961. doi: 10.3732/ajb.1200365 [DOI] [PubMed] [Google Scholar]
- 47.Yang Y, Dang YY, Li Q, Lu JJ, Li XW, Wang YT. Complete chloroplast genome sequence of Poisonous and medicinal plant Datura stramonium: organizations and implications for genetic engineering. Plos One. 2014; 9(11): e110656 doi: 10.1371/journal.pone.0110656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Su HJ, Hogenhout SA, Al-Sadi AM, Kuo CH. Complete chloroplast genome sequence of omani lime (Citrus aurantiifolia) and comparative analysis within the Rosids. Plos One. 2014; 9(11): e113049 doi: 10.1371/journal.pone.0113049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hupfer H, Swiatek M, Hornung S, Herrmann RG, Maier RM, Chiu WL, et al. Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Mol Gen Genet. 2000; 263: 1071–1071. [DOI] [PubMed] [Google Scholar]
- 50.Doyle JJ, Doyle JL, Ballenger JA, Palmer JD. The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol Phylogenet Evol. 1996; 5: 429–438. doi: 10.1006/mpev.1996.0038 [DOI] [PubMed] [Google Scholar]
- 51.Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006; 23: 2175–2190. doi: 10.1093/molbev/msl089 [DOI] [PubMed] [Google Scholar]
- 52.Price RA, Calie PJ, Downie SR, Logsdon JM, Palmer JD. Chloroplast DNA variation in the Geraniaceae: a preliminary report. In Proceedings of the International Geraniaceae Symposium, editor P. Vorster (Stellenbosch(RSA)), 1990; 235–244.
- 53.Downie SR, Jansen RK. A Comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst Bot. 2015; 40: 336–351. [Google Scholar]
- 54.Williams AV, Boykin LM, Howell KA, Nevill PG, Small I. The complete sequence of the Acacia ligulata chloroplast genome reveals a highly divergent clpP1 gene. Plos One. 2015; 10(5): e0125768 doi: 10.1371/journal.pone.0125768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong WP, Xu C, Li CH, Sun JH, Zuo YJ, Shi S, et al. ycf1, the most promising plastid DNA barcode of land plants. Sci Rep. 2015; 5: 8348 doi: 10.1038/srep08348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jansen RK, Cai Z, Raubeson LA, Daniell H, Depamphilis CW, Leebens-Mack J, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. P Natl Acad Sci USA. 2007; 104: 19369–19374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams AV, Miller JT, Small I, Nevill PG, Boykin LM. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia. Mol Phylogenet Evol. 2016; 96: 1–8. doi: 10.1016/j.ympev.2015.11.021 [DOI] [PubMed] [Google Scholar]
- 58.Li XW, Yang Y, Henry RJ, Rossetto M, Wang YT, Chen SL. Plant DNA barcoding: from gene to genome. Biol Rev. 2015; 90: 157–166. doi: 10.1111/brv.12104 [DOI] [PubMed] [Google Scholar]
- 59.Thorne K. Endemic species. Springer: Netherlands: 2016. [Google Scholar]
- 60.Marshall JK. Factors limiting the survival of Corynephorus canescens (L.) Beauv. in Great Britain at the northern edge of its distribution. Oikos. 1968; 19: 206–216. [Google Scholar]
- 61.Root T. Energy constraints on Avian distributions and abundances. Ecology. 1988; 69: 330–339. [Google Scholar]
- 62.Teeri JA, Stowe LG. Climatic patterns and the distribution of C4 grasses in North America. Oecologia. 1976; 23: 1–12. doi: 10.1007/BF00351210 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(A) PCGs, (B) LSC region, (C) SSC region, and (D) IR region. Values above the branches represent ML bootstrap values/BI posterior probability values. Outgroup: Lilium brownii KY748296; L. bakerianum KY748301; L. henryi KY748302.
(TIF)
Data Availability Statement
The raw sequencing data were deposited in GenBank SRA database (SAMN08348372–SAMN08348378, https://submit.ncbi.nlm.nih.gov/subs/sra). The annotated seven cp genomes were deposited in GenBank (accession number MG200070, MG211818-MG211823).