Abstract
T-cell immunotherapy is a promising approach for cancer, infection, and autoimmune diseases. However, significant challenges hamper its therapeutic potential, including insufficient activation, delivery, and clonal expansion of T-cells into the tumor environment. To facilitate T-cell activation and differentiation in vitro, core–shell microparticles are developed for sustained delivery of cytokines. These particles are enriched by heparin to enable a steady release of interleukin-2 (IL-2), the major T-cell growth factor, over 10+ d. The controlled delivery of cytokines is used to steer lineage specification of cultured T-cells. This approach enables differentiation of T-cells into central memory and effector memory subsets. It is shown that the sustained release of stromal cell-derived factor 1α could accelerate T-cell migration. It is demonstrated that CD4+ T-cells could be induced to high concentrations of regulatory T-cells through controlled release of IL-2 and transforming growth factor beta. It is found that CD8+ T-cells that received IL-2 from microparticles are more likely to gain effector functions as compared with traditional administration of IL-2. Culture of T-cells within 3D scaffolds that contain IL-2-secreting microparticles enhances proliferation as compared with traditional, 2D approaches. This yield a new method to control the fate of T-cells and ultimately to new strategies for immune therapy.
Keywords: cytokine delivery, microfluidics, microparticles, T-cells
In this work, we developed biocompatible microparticles that delivered a variety of cytokines that influence the proliferation and differentiation of cultured, primary T-cells. Augmentation and engineering of immune responses have major applications in combating cancers, and enriching regulatory immune cells has potential roles in suppressing transplant rejection and in mitigating autoimmune and allergic diseases. There are a number of challenges to adoptive T-cell therapies,[1] especially in skewing bulk cultures of T-cells toward desired outcomes. Both cytotoxic (CD8+) T-cells, endowed with the ability to kill, and helper (CD4+) T-cells, endowed with the dedicated production of cytokines, are needed for clearing cancers and modulating aberrant responses. Controlled manipulation of these populations will be the key to successful immunotherapies.
A number of groups have developed polymeric carriers, such as microparticles, liposomes, or micelles for delivering biomolecules.[2] Such particles have be used to modulate the immune system by delivering antigens, vaccines, adjuvants, and drugs.[3] While the focus of prior work has mainly been on enhancing stability or solubility of the delivered molecules, little has been done on sustained release and its impact on therapeutic efficacy compared to burst release.[4] Here, we demonstrate enhancements over traditional approaches due to controlled release of cytokines to T-cells.
Tuning the effector functions of T-cells requires the presence of cytokines during activation. Certain cytokines, such as interleukin-2 (IL-2), enhance proliferation and affect the balance of effector versus memory cytotoxic T-cell differentiation.[5] The “strength” of IL-2 signals plays a crucial role in facilitating the acquisition of effector phenotypes in CD8+ T-cells.[5] However, few studies have been carried out to investigate the effect of engineered administration of this cytokine.[6] In undifferentiated CD4+ T-cells, IL-2 can act in synergy with transforming growth factor-beta (TGF-β) to promote the generation of induced regulatory T-cells (iT-reg).[7]
In this study, polymeric particulate systems loaded with cytokines were designed to exhibit tunable release profiles. Alginate and heparin, natural biopolymers, were chosen for our platform to encapsulate IL-2 due to their biocompatibility and facile gelation via calcium ions.[8] Traditional methods for the formation of polymeric microparticles such as microemulsions have been gradually replaced by microfluidic approaches[9] because of the better control over particle size and distribution (narrower polydispersity). Here, we employed a T-shaped droplet generator to form our polymeric microparticles at desired sizes. A protective shell of chitosan with desired thickness was applied to tune the rate of IL-2 release. To assess the potential for activation in a 3D environment and the effect of extended provision of cytokines, lymphocytes were seeded into bioengineered scaffolds imbued with cytokine release. Our data show that the tuning of release of cytokines can be used to regulate the suppressive and cytotoxic potentials of T-cells.
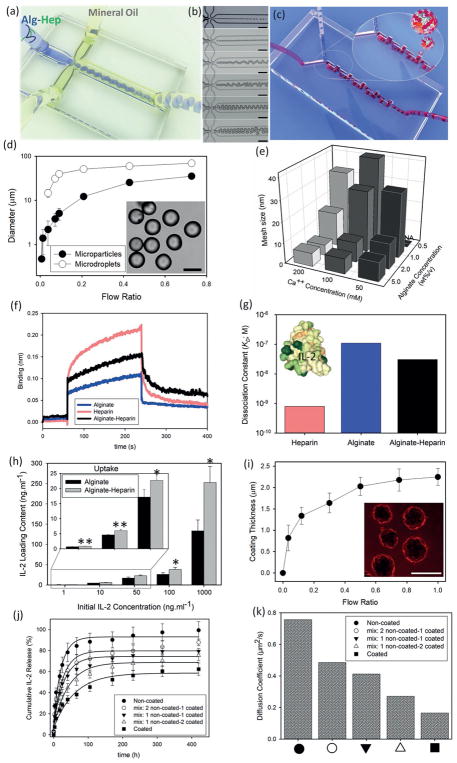
To form microparticles, we used a microfluidic device featuring a droplet generator (Figure 1a). Using a hydrophobic sheath fluid of mineral oil+ Span 8%, we generated alginate-heparin (Alg-Hep) microparticles comprising various concentrations of polymer from 0.5 to 5 wt%/v (as shown in Figure 1b, and a well-controlled micromixer (Figure 1c)). The flow ratios were tuned to make spherical Alg-Hep microdroplets of different diameters from 400 nm to 30 μm (Figure 1d). To achieve complete gelation of the resulting particles, they were stored in CaCl2 solution with different Ca++ concentrations (50–200 × 10−3M) for 40 min [Materials and Methods are available as Supporting Information]. Tuning the mesh size as an indicator for nanoscale porosity of the particles can affect the release rate of the entrapped proteins. To calculate the mesh sizes, we measured the swelling ratio of the microparticles. Calculations were done based on the Flory–Rehner method.[10] Estimated mesh size values of the Alg-Hep microparticles as a function of calcium ion and polymer concentrations are shown in Figure 1e. Higher cross-linking densities resulted in microparticles with smaller mesh sizes. These results show that a variety of particle and mesh sizes were achieved in our work.
Figure 1.
Microfluidic generation and coating of alginate-heparin (Alg-Hep) microparticles for controlled release of IL-2. a) Schematic representation of a microfluidic droplet generator device, that is applied to form (Alg-Hep) microparticles via sheath hydrophobic flows. b) Diameter of Alg-Hep microparticles (1 wt%/v) is controlled by changing the volumetric flow rate ratio of aqueous to oil phase (○). Equilibrium diameter of microparticles as characterized after gelation process at calcium ion bath (●). Inset: microscopy image of a typical batch of Alg-Hep particles formed at flow ratio of 0.45. c) Microscopic images of Alg-Hep microdroplets formed at different flow ratios resulting in different sizes from 400 nm to 30 μm. d) Schematic illustration of the serpentine micromixer used to coat Alg-Hep particles with chitosan layer. e) Quantification of mesh size analysis of microparticles as a function of calcium ion and Alg-Hep concentrations. f) Biolayer interferometry sensorgrams showing the binding kinetic of IL-2 to alginate, heparin, and Alg-Hep functionalized surfaces. g) Calculated dissociation constants (KD = koff/kon) based on the curve fittings for IL-2 bindings at various concentrations of 5–200 × 10−9M; Inset is the 3D representation of IL-2 (PDB ID: 1M47; colored based on hydrophobicity). h) IL-2 binding efficiency of alginate and Alg-Hep microparticles at various initial concentrations of IL-2 after 12 h of incubation. The presented data are expressed as average ± SD. The results were analyzed using unpaired t-tests. NA: We could not make particles from 0.5% alginate and 50 × 10−3M calcium. i) Microfluidic tuning of coating thickness via changing the flow ratio between Alg-Hep particles and chitosan polymers. Inset: Fluorescence images of RITC-labeled chitosan coated Alg-Hep particles. IL-2 release kinetic of microfluidic-synthesized microparticles prepared from different precursor formulations in PBS at 37 °C j) and calculated diffusion coefficients of studied microparticles (k).
To enhance cytokine binding, we modified the alginate biopolymer with heparin. To incorporate heparin into alginate structure, activated carboxylic acid groups on heparin chains were covalently reacted with diamine-modified alginate through an aqueous carbodiimide reaction. We examined the relationship between the initial amount of heparin and the final conjugated value, and found that our process saturated at 1.5 nmol of heparin per mg alginate (see Figure S1 in the Supporting Information). The intrinsic affinity of IL-2 to heparin has been reported before.[11] To measure the binding of IL-2 to alginate, heparin, and Alg-Hep functionalized surfaces, we employed real-time biolayer interferometry (Figure 1h). We found that Alg-Hep and IL-2 undergo a high affinity interaction (KD = 30.6 × 10−9M) compared to the unmodified alginate (110 × 10−9M) (Figure 1f). These results show that our heparin-modified particles improved the binding of IL-2 over alginate particles alone.
To assess the amount of IL-2 we could bind to our particles, they were incubated with different concentrations of IL-2 (1–1000 ng mL−1) for 12 h at 37 °C and after washing, microparticles were dissolved and the amount of released IL-2 was measured using enzyme-linked immunosorbent assay (ELISA). As shown in Figure 1g, the presence of heparin significantly enhanced the IL-2 loading content and efficiency (Figure 2, Supporting Information) of the microparticles. The IL-2 binding efficiency of alginate and Alg-Hep microparticles was also evaluated when designated amounts of IL-2 (1–100 ng mL−1) were mixed with alginate prior to microfluidic droplet formation and subsequent washing steps (see Figure 3 in the Supporting Information). To assess whether there was a size dependence to the loading, we tested the binding efficiency of Alg-Hep particles sized 1–35 μm. We found a modest relationship of IL-2 loading to the particle size using both loading methods (Figure 4, Supporting Information). These results show that large amounts of IL-2 can be integrated into our particles.
To control the timing of the release of IL-2 from the particles, we sought to coat the alginate-heparin particles with a layer (“shell”) of chitosan. We prepared a microfluidic device that controls residence time of particles and thus allows for increasing amounts of coating over time. In the second microfluidic device (serpentine micromixer; Figure 1d), mixing and laminar flows are the predominant mechanisms of mass transfer, where the coating process mainly occurs at the microscale and is controlled through manipulation of the residence time.[12] The serpentine micromixer was designed using a 3D printing and structure-removal approach[13] (see Figure S5 in the Supporting Information). By controlling the flow rate, and thus residence time, of particles in this device, we coated them with varying amounts of chitosan. To assess the thickness of chitosan, we coated with chitosan–rhodamine-B-isothiocyanate (RITC) with various residence times and measured the resulting thickness of the chitosan shell by fluorescence microscopy (Figure 1e). These results show that our microfluidic device could successfully coat alginate-heparin particles with chitosan.
To assess the effect of chitosan coating on the timing of release of IL-2, we measured the release of IL-2 from Alg-Hep microparticles in the presence or absence of chitosan layer sequentially over 18 d. We found that release of IL-2 from chitosan-coated microparticles was slower than from noncoated ones. By mixing coated and noncoated particles we could tune the release profile of IL-2 (Figure 1j). This tuning capability over the microscale release of IL-2 and its diffusion coefficient (Figure 1k) under different applied conditions were also investigated. To estimate the diffusion coefficient of IL-2 out of the particles, we examined the initial linear part of the plots (Figure S6, Supporting Information) and employed Fick’s law [Equation (1)][14]
| (1) |
where Mt/M∞ represents the fraction of released drug at time t, D is the diffusion coefficient of IL-2 molecules, and R is the radius of the particles (6.1 μm). The calculated diffusion coefficients are shown in Figure 1k. These results show that chitosan-coated particles have lower diffusion coefficients than noncoated ones, which is desirable for controlling the release of cytokines over time.
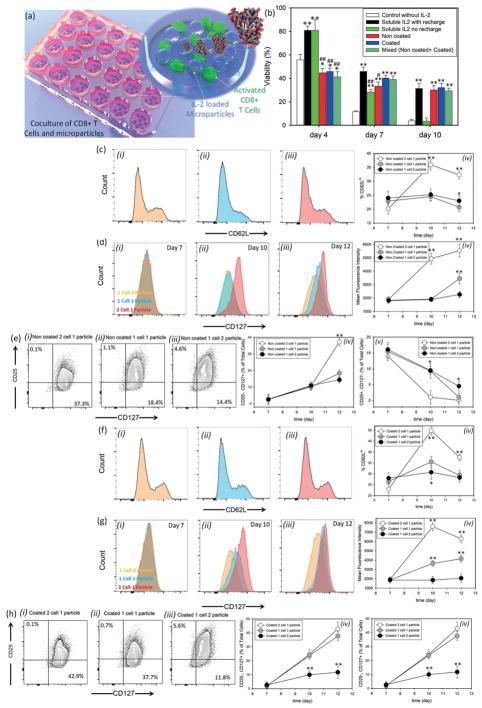
To test whether application of IL-2 could improve cytotoxic T-cell activation, we employed our particles in the activation of CD8+ T-cells. Different microparticles with similar loading efficiencies were synthesized and loaded with IL-2. Purified naïve CD8+ T-cells were activated with anti-CD3 and anti-CD28 as described in Experimental Section. After 2 d, cells were replated with different types/ratios of particles (Figure 2a). To first ensure that the particles were not toxic, we evaluated the viability of CD8+ T-cells after 12 and 48 h of coculture with non-coated and coated particles, and saw that even at very high ratios of particle (coated or noncoated) to cells (100:1) the cellular viability was more than 79% after 48 h of culture (Figure S7, Supporting Information). This confirmed the nontoxicity of the particles. IL-2 is critical to support T-cell survival and proliferation. Without addition of IL-2 in culture, cell viability decreased dramatically over a few days (Figure 2b, white bars). We compared the viability of T-cells in the presence of particles versus two conditions: first, refreshing media and IL-2 approximately every 2 d (Figure 2b, black bars); and second, supplied with IL-2 only at the beginning of experiment and then fresh media was changed every 2 d (Figure 2b, green bars). We found that CD8+ T-cell viability was comparable when employing IL-2 releasing particles (coated, uncoated, and mixed) as when frequently refreshing soluble IL-2 (Figure 2b). This result demonstrates that sustained release of IL-2 from our particles could sustain the viability of T-cells over 10 d.
Figure 2.
Role of interleukin-2 presentation on differentiation of cytotoxic T-cells. a) Schematic representation of CD8+ T-cells and microparticles coculture. b) Effects of IL-2 release on viability of CD8+ T-cells. All data are presented as average ± SD. The results were analyzed using unpaired t-tests. For all of the tests, the threshold was set to p < 0.05 for “statistically significant” and p < 0.01 was for “statistically very significant.” The thresholds *p < 0.05 and **p < 0.01 were obtained between each group and the control group without IL-2 treatment (negative control). On the other hand, #p < 0.05 and ##p < 0.01 are between each treatment and the cells treated by IL-2, with recharge (positive control). c) Kinetics of CD62L expression when purified naïve CD8+ T-cells were stained with antibodies against CD62L ex vivo and after priming for 2 d with anti-CD3 + anti-CD28 (day 2). Histograms are representative of day 12 marker expression on cells after treatment with one noncoated particle per two cells i), one particle per cell ii), and two particles per cell iii) as IL-2 releasing agents. iv) The MFI of CD62L expression during the treatment. d) Regulation of IL-7Ra (CD127) re-expression after priming in three different ratios of cell to particle at days 7 i), 10 ii), and 12 iii) and MFI of cells in these conditions over time iv). e) Flow cytometric analysis of IL-2Ra and IL-7Ra coexpression when treated with noncoated microparticles at particle: cell ratios of i) 1:2, ii) 1:1, and iii) 2:1. Percentage of cells that fall in the category of memory T-cells (CD 25-, CD127+) iv), or effector cells (CD25+, CD127−) v) in three different IL-2 concentrations over time. f) Kinetics of CD62L expression after incubation of activated CD8+ T-cells with coated microparticles stained with antibodies against CD62L ex vivo at particle: cell ratios of 1:2 i), 1:1 ii), and 2:1 iii). Histograms are representative of day 12 marker expression. iv) The MFI of CD62L staining is shown. g) The regulation of IL-7Ra (CD127) re-expression after priming in three different ratios of cells to coated particles at days 7 i), 10 ii), and 12 iii) and MFI of cells in these conditions over time iv). h) Analysis of IL-2Ra and IL-7Ra coexpression when treated with noncoated microparticles at three different ratios of i) 1:2, ii) 1:1, and iii) 2:1 to the cells. Percentage of cells that fall in the category of memory T-cells (CD 25−, CD127+) iv), or effector cells (CD25+, CD127−) v) in three different IL-2 concentrations over time is represented. In (c–h), the presented data are expressed as average ± SD. The results were analyzed using unpaired t-tests.
To distinguish the effect of IL-2 signal strength versus its rate of elaboration from the beads, we studied the differentiation of CD8+ T-cells upon fixing the microparticle type and adjusted the ratio of particles to cells. Activated CD8+ T-cells downregulate the IL-7 receptor alpha (CD127), upregulate the IL2 receptor alpha (CD25), and cleave the extracellular domain of CD62L. To this end, experiments were done under three different conditions where we had an equal number of cells and particles at the beginning of the experiment; the number of particles was twice the number of cells or vice versa. At different time points during a 12 d period, cells were harvested and stained against certain cell surface markers known to be associated with effector or memory cell phenotypes. Up/downregulation of CD62L, CD127, CD25, CD44, and CCR7 proteins on cell surface was monitored by flow cytometry over time, and we showed that the presence of microparticles did not interfere with staining for flow cytometry (Figure S8, Supporting Information). We found that after 7 d in culture with the particles, CD8+ T-cells in the wells with a 1:2 ratio of noncoated particles:cells (experiencing stronger IL-2 signals) showed the proportion of cells that were CD62Lhi was 32.3%, whereas the cells exposed to 1:1 or 1:2 ratios of cells:particles showed only 20.7% and 23.0% CD62Lhi, respectively (Figure 2c). Similarly, we saw the high ratio cells showed more downregulation of CD127 than the ones exposed to a lower ratio of particles (Figure 2d). In addition, we saw that the high ratio cells showed more upregulation of CD25 than the ones exposed to a lower ratio of particles (Figure 2e). These results show that higher exposure to IL-2 signal induced more effector-like phenotypes (CD62Llo, CD127lo, and CD25hi), whereas cells exposed to lower IL-2 became a central memory-like phenotype (CD62Lhi, CD127hi, and CD25lo).[15,16] It has also been shown that microparticles do not interfere with FACS staining (Figure S8, Supporting Information)
The same set of evaluations was performed on CD8+ cells cultured with different ratios of coated particles. These cells experienced the same overall dosage of IL-2 comparable to their noncoated compartments but the coating reduced the rate of IL-2 release. For the activation markers chosen, the general pattern of responses that the CD8 T-cells showed was similar (Figure 2f–h). Thus, the number of particles was the dominant factor in activation and differentiation. Our results suggest that the ratio of IL-2 secreting particles to cells drives a major impact in CD8+ T-cell differentiation.
Memory T-cells are classified according to heterogeneity of surface marker expression, effector functions, and homing tissue compatibility. Effector memory T cells (TEMs) provide immediate protection, while central memory T cells (TCMs) are responsible for the generation of additional effector cells.[17] Whether these two subsets behave differently in terms of cytolytic activity or which of them holds higher promise for further applications is unclear.[17,18] All activated T-cells and memory T-cells, even quiescent, show an upregulation and splicing change of CD44, which facilitates rapid extravasation into inflammatory sites through interaction with hyaluronan.[19] The regulation of the chemokine receptor CCR7 as an essential surface marker for migration of T-cells into peripheral lymph nodes is well established.[20] CCR7 expression is a key factor in distinguishing TCMs from TEMs.[17] Generally, TCMs are considered CCR7+CD44hi CD62Lhi cells, while TEMs are considered CCR7-CD44hi CD62Llow cells.
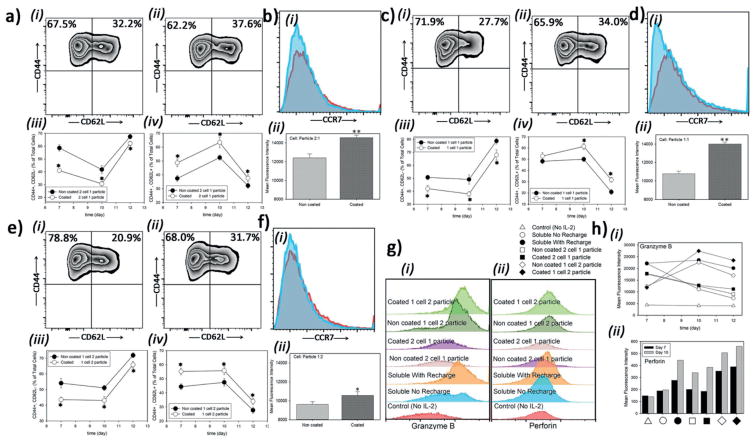
To assess whether the rate of administration of IL-2 impacts cell fate, we monitored the differentiation of CD8+ T-cells under conditions where the release profiles were different but the ratios of particle to cells was held constant. We found that after 10 d, CD8+ T-cells cultured with coated particles at 1:1 ratio (cell: particle) showed the proportion of cells that were CD62Lhi was 35.5%, whereas the cells cultured with noncoated particles showed 10% less CD62Lhi cells (Figure 2c–v,f–iv). In culture with coated particles at 1:1 cell: particle ratio, the CD8+ T-cells showed a CCR7 MFI of 14 000, whereas the cells exposed to noncoated particles showed an MFI of 10 800 (Figure 3d). All T-cells showed the CD44hi marker of memory cells; it would be expected that recently activated, effector T-cells downregulate CD44 by d 5 to 6 after activation in the absence of ongoing antigen signal.[21] These results show that coated particles favor the formation of a central memory phenotype. (Figure 3a–ii,iv,c–ii,iv,e–ii,iv), while culture with noncoated particles favored the formation of a more effector memory phenotype (Figure 3a–i,iii,c–i,iii,e–i, iii).
Figure 3.
Different profiles of IL-2 administration to CD8+ T-cells impacts differentiation of them to central or effector memory cells. a) Flow cytometric analysis of CD44 and CD62L coexpression at two cells per particle a,i–iv), one particle per cell c,i–iv), and two particles per cell e,i–iv). i) noncoated particles or ii) coated particles in each set (day 12). Percentage of effector cells (CD44+CD62L−) in each case iii) or memory (CD44+CD62L+) phenotype iv). Histograms of CCR7 expression of memory subset of cells,b),d), and f), when cells are treated with noncoated particles i) or coated ii) particles and their MFIs iii) at day 12. g,h) Flow cytometric analysis of granzyme B (i) and perforin (ii) expressions as a function of treatment modality and time. The presented data are expressed as average ± SD. The results were statistically analyzed using unpaired t-tests.
We attempted to further tune the release profile by mixing coated and noncoated particles (Figure 1j). We cultured CD8+ T-cells with a constant ratio of one particle per cell but employed coated, noncoated, and three different mixtures of un/coated particles (Figure S9, Supporting Information). Downregulation of CD127 was correlated with the rate of IL-2 release. This result adds to our conclusion that the rate of IL-2 exposure (as well as the dose) to activated T-cells can skew the outcome of their differentiation.
During CD8+ T-cell activation, these cells induce expression of proteins such as the pore forming perforin and the protease granzyme B that are critical for their cytotoxic function.[5] In activated cells, different expression levels of each protein have been correlated to development of effector or memory cytotoxic T lymphocytes (CTLs).[22] Here, we sought to understand if different kinetics of IL-2 administration affected the expression of perforin and granzyme B. Naïve CD8+ T-cells were cultured on anti-CD3 anti-CD28 for 3 d and then moved to a nonstimulatory well that contained IL-2 in the media. Naïve T-cells expressed a minimal amount of perforin and very little granzyme B (data not shown). Granzyme B expression was higher when IL-2 was “recharged” into the culture media every other day as compared to a single load of IL-2 at the start (Figure 3g). We found even higher granzyme expression in culture conditions where there was a high ratio of particles to cells. (Figure 3h–i). Perforin expression levels were higher with stronger and sustained IL-2 signals compared to low IL-2 or when IL-2 was not renewed (Figure 3hii). These results show that cytotoxic T-cell effect functions can be intensified over conventional approaches by providing IL-2 from microparticles.
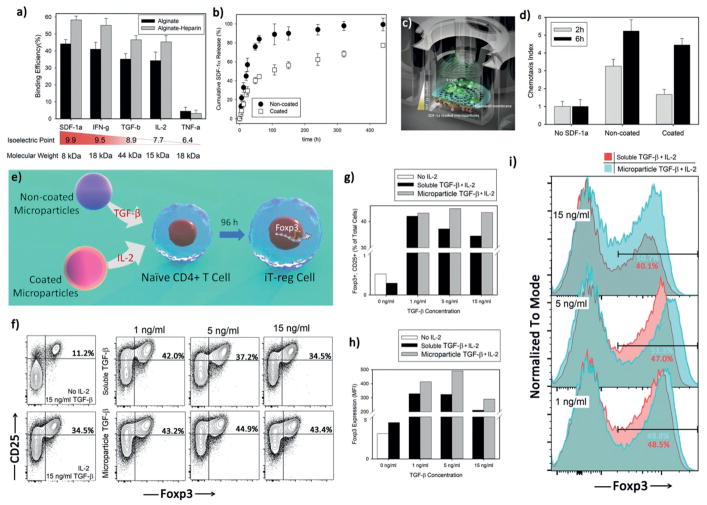
Our microparticle platform has a broad capability to deliver a multitude of cytokines and growth factors. The electrostatic charge of heparin should allow for a broad set of proteins to be captured based on their isoelectric point. For example, we expected to easily load into our microparticles positively charged proteins like stromal cell-derived factor 1 (SDF-1α; pI: 9.9), TGF-β (pI: 8.5), and interferon gamma protein (IFN-γ ; pI; 9.5). We performed loading experiments with these cytokines at 50 ng mL−1 and measured binding efficiency (Figure 4a). We found excellent loading, with improved loading in the heparin-coated particles as compared to alginate alone. As shown, this platform has limited functionality for absorbing proteins that have negative charge at neutral pH like tumor necrosis factor alpha (TNF-α; pI; 6.4).
Figure 4.
Engineered microparticles can deliver other signals beside IL-2. a) Binding capacity of alginate-heparin microparticles was evaluated after incubating them with various cytokines at constant concentration of 50 ng mL−1 for 12 h at 4 °C and under continuous, gentle shaking. b) Release profile of SDF-1α cytokine from noncoated Alg-Hep microparticles at 37 °C. SDF-1α releasing microparticles (in the bottom of the well) generate a sustained gradient to recruit local T-cells as schematically illustrated in (c). d) Quantification of relative T-cell’s migration (chemotactic index) for active T-cells after 2 h of coculture with noncoated Alg-Hep microparticles at different initial loadings. The presented data are expressed as average ± SD. e) Sustained release of TGF-β mediates development of induced regulatory T-cells (iT-reg cells). f–i) Flow cytometric analysis of iT-reg development was assessed and judged by Foxp3 and CD25 coexpression after coculture of naïve CD4+ T-cells with anti-CD3 and anti-CD28 for 4 d. The indicated range of TGF-β concentrations was applied either in a soluble format or via noncoated microparticles at the same time as activation signal. All the samples contained IL-2 releasing coated microparticles that delivered 20 U mL−1 IL-2 during treatment. Control experiments (f) left panels) were also performed to show the effect of IL-2 presence in the constant concentration of TGF-β (15 ng mL−1).
To study the impact on migration of naïve and activated T-cells, we employed alginate-heparin particles to encapsulate SDF-1α. Based on binding kinetics data (Figure S12, Supporting Information), heparin incorporation enhanced SDF-1α interaction with the alginate matrix. The sustained release of SDF-1α could also be controlled by a protective shell similar to the one proposed for IL-2 (Figure 4b) or it can be tuned based on the initial loading content of the cytokine (Figure S12c,d, Supporting Information). We measured migration of T-cells using a standard Transwell based technique where SDF-1α particles were placed below the Transwell and T-cells were placed above. T-cells were allowed to migrate in response to the eluted chemotactic gradient established across the Transwell. T-cells were counted on both sides of the Transwell and their chemotactic index calculated. We found that the particles significantly enhanced the chemotaxis of T-cells across the Transwell (Figure 4c,d). This result shows that chemokine-secreting particles can be used to direct the migration of T-cells.
We tested the ability of our microparticles to secrete TGF-β, a well-known growth factor that induces differentiation of naïve T-cells into iT-reg. T-regs express the transcription factor Foxp3 and play antigen-specific suppressive roles in a variety of autoimmune diseases including type 1 diabetes.[23] To evaluate the ability of our particles to generate iT-regs, we loaded Alg-Hep particles with TGF-β that released at three different concentrations (1, 5, and 15 ng mL−1). We cultured rigorously naïve CD4+ T-cells with anti-CD3 and anti-CD28 for 4 d in the wells containing the Alg-Hep particles loaded with TGF-β at different concentrations and compared them to the soluble (no particles) counterparts at identical concentrations. The expression of Foxp3 was measured by flow cytometry (Figure 4h–i). In all of the conditions, either when TGF-β was manually added in the beginning of experiment or delivered using the particles, coated particles were added to deliver IL-2 (20 U mL−1) to the cells. To show the importance of the role that IL-2 plays in making iT-regs, for soluble TGF-β at the concentration of 15 ng mL−1, we did the measurements in both the absence and presence of IL-2 (Figure 4f left panels). Interestingly, in the presence of IL-2, Foxp3 expression was ≈threefold greater compared to TGF-β only conditions with no addition of IL-2. In all concentrations of TGF-β as provided by the particles, Foxp3 expression was high (50%+). On the other hand, we found that at very high soluble concentrations of TGF-β, Foxp3 expression plateaued and was even slightly suppressed (Figure 4g). Furthermore, the MFI of Foxp3 directly relates to the suppressive capacity of the regulatory T-cells.[24] We found that the MFI of Foxp3 was highest when using particles to deliver TGF-β (Figure 4h). These results show that induced regulatory T-cells can be potently generated by the use of microparticles that secrete TGF-β and IL-2.
It has been reported that the inflammatory cytokines interferon gamma (IFN-γ) and tumor necrosis factor-α (TNF-α) can inhibit differentiation and cause apoptosis of stem cells.[25] Here, we tested whether the sustained release of IFN-γ from microparticles could affect the fate of human bone marrow mesenchymal stem cells. We measured apoptosis using flow cytometry after 7 d of coculture with stem cells and particles. We found that 30% of stem cells showed evidence of apoptosis when cultured with IFN-γ releasing coated/noncoated microparticles after 1 week. In comparison, particles lacking IFN-γ showed only 5% apoptosis (Figure S13, Supporting Information). These results show that particles can release IFN-γ that influences stem cell survival.
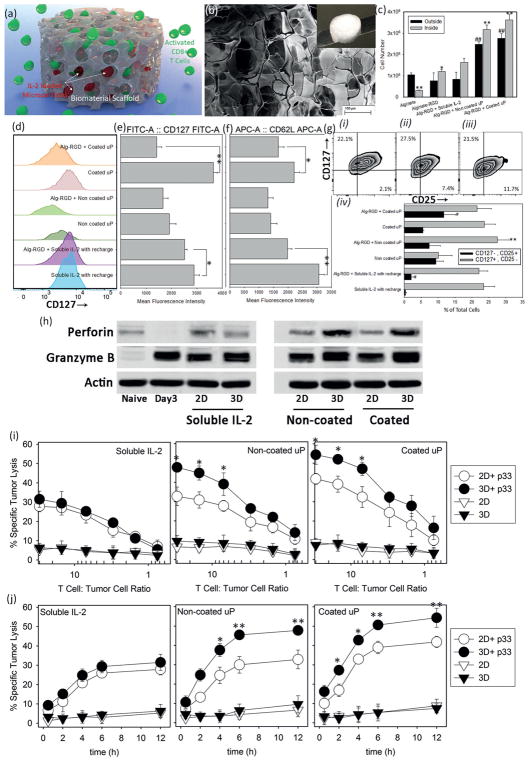
We sought to investigate the combination of secreted cytokines and 3D culture versus 2D culture in driving T-cell proliferation and differentiation (Figure 5a). We prepared a series of alginate-based scaffolds in the presence or absence of IL-2-carrying microparticles. These 3D environments can mimic the in vivo context encountered by activated lymphocytes and give a better idea of the effects of spatiotemporal variables on cellular fate.[26] Scanning electron microscope (SEM) images of alginate-RGD scaffolds demonstrated their highly porous microstructure with an average pore size of 65 ± 4 μm (Figure 5b), which is ample space for mobility of T-cells. To turn the scaffold into a more favorable material for cell trafficking, RGD modification was performed (see Experimental Section). IL-2 was delivered to the cells either via soluble administration or through embedded particles. To assess proliferation, the number of T-cells inside and outside the scaffold was measured after 10 d of culture (Figure 5c). To test if RGD modification alone made the scaffold more favorable for cells to propagate, we measured the number of T-cells proliferating in alginate scaffolds versus alginate + RGD. We found a fourfold increase in the number of cells within the RGD-bearing scaffold. To assess the impact of stimulatory signals on proliferation, IL-2 was either recharged manually at the concentration of 100 U mL−1 (about 7 ng mL−1) or IL-2 was released from coated and noncoated particles. We loaded the particles such that the total amount of IL-2 released from the one-particle-per-cell condition was the same as the soluble 100 U mL−1 condition. Our results show a significant increase in both the total number of cells and the number of cells in/outside the scaffold in particle-enriched scaffolds compared to the manual administration of soluble IL-2. These findings prove that particulate delivery of IL-2 is superior to soluble IL-2 in 3D structures. For a better clarification of the effect of 3D culture, the same surface markers, CD62L, CD127, and CD25 of each set of cells were compared to their 2D culture compartments (Figure 5d–g) alongside their mean fluorescence intensities (MFIs). We tested if release of IL-2 from the plain alginate scaffold, without particles, could provide a similar effect as the IL-2 particles. We found that in absence of particles it is not possible to provide sustain IL-2 release (Figure S14, Supporting Information), and treatment of the T-cells in such a scaffold failed to improve the number of memory or effector T-cells over time (data not shown). The differences between the released amounts were statistically very significant (p < 0.01). Here, we also evaluated the effect of 3D culture on expression of perforin and granzyme B after 10 d of culturing in alginate-RGD scaffolds. We found that both molecules were significantly induced in 3D culture and to a higher degree when particles were used to provide IL-2 (Figure 5h). Taken together, these data show the advantage of culturing CD8 T-cells in a supportive 3D microenvironment coupled with IL-2 secretion from microparticles.
Figure 5.
Performance boost from encapsulation in scaffold. a) Schematic representation of scaffold approach to deliver pre-activated T-cells and IL-2 loaded microparticles. b) Electron microscopy image of the prepared alginate-RGD scaffold. Scale bar: 100 μm; Inset: Macroscopic image of a freeze-dried scaffold. c) Number of cells inside and outside the scaffolds 10 d after incorporation of T-cells into the scaffold and in absence or presence of IL-2 (soluble administration or released by microparticles). The presented data are expressed as average ± SD. The results were statistically analyzed using unpaired t-tests. *p < 0.05 and **p < 0.01 are between numbers of cells inside each scaffold and the control alginate-RGD scaffold with soluble IL-2 (positive control). #p < 0.05 and ##p < 0.01 are between number of cells outside each scaffold and the control alginate-RGD scaffold with soluble IL-2 (positive control). d) Regulation of IL-7Ra (CD127) re-expression after 10 d of treatment in presence or absence of scaffold and MFI of cells in these conditions e). f) The quantification (MFI) of CD62L expression after 10 d of treatment. *p < 0.05 and **p < 0.01 are between the samples treated with and without using the alginate-RGD scaffold. g) Flow cytometric analysis of co-expression of IL-2Ra and IL-7Ra for scaffold-based treatment with soluble IL-2 i), noncoated ii), and coated iii) microparticles at constant ratio of one particle per cell. Quantitative evaluation of the percentage of cells that fall in the category of memory T-cells (CD 25−, CD127 +) versus effector cells (CD25+, CD127−) after 10 d of treatment in different conditions iv). **p < 0.01 is between the samples treated with and without using the alginate-RGD scaffold; CD25+, CD127− group. #p < 0.05 is between the samples treated with and without using the alginate-RGD scaffold; CD 25−, CD127+ group at day 10. h) CD8+ T-cells were MACS-purified, FACS-sorted, and cocultured with soluble IL-2 or IL-2 loaded microparticles in 2D or 3D cultures for 10 d. Western blot analysis show levels of perforin and granzyme B. β-actin was used as loading control. i) Chromium (51Cr) release assays comparing antigen (p33)-specific (circles) and nonspecific (triangle) cytotoxicity of treated T-cells with different methods in 2D (empty circles) versus 3D (filled circles) cultures after 12 h in the indicated T-cells to tumor cells ratios. j) Kinetics of cytotoxic activity of p33-specific T-cells after treatment with soluble IL-2, noncoated, and coated IL-2-loaded microparticles in 2D (empty circles) and in 3D alginate-RGD scaffold (filled circles). Cytotoxic activity of spleen-derived, in vitro treated, CD8+ T-cells was analyzed using chromium assay. Specific tumor lysis was determined at the indicated time points (30 min, 2, 4, 6, and 12 h) of incubation. Cytotoxic activity was examined at four different ratios (30:1, 7:1, 3:1, and 0.75:1) of treated T-cells to tumor cells. The data are presented as average ± SD of five independent samples. The presented data are expressed as average ± SD. The results are analyzed using unpaired t-tests. In (h) and (j), p33-treated and nontreated T-cells are significantly different (p < 0.001).
To further assess the functionality of CD8+ T-cells activated in a 3D environment, we tested the in vitro cytotoxicity of T-cells using the standard chromium (51Cr) release assays. The cells were first stimulated with p33 antigen to test the antigen-specific cytotoxicity at different ratios of T-cells to tumor cells. All treated cells showed specific lysis of p33-coated target tumor cells (p < 0001). CD8+ T-cells cultured initially within a 3D scaffold and in presence of IL-2 secreting microparticles exhibited stronger cytolytic potential compared to the T-cells that were cocultured with microparticles in 2D. We also evaluated the kinetics of gaining cytotoxic activity of spleen-derived p33-specific T-cells after treatment with soluble IL-2, noncoated, and coated IL-2-loaded microparticles in 2D or 3D-alginate-RGD scaffolds (Figure 5j). T-cells treated with coated particles (i.e., that had slower IL-2 release), both in 2D and 3D, demonstrated faster cytotoxicity compared to those cultured with noncoated particles or that received soluble administration of IL-2 (Figure 5j, Figures S15–17, Supporting Information). This effect was enhanced in the 3D environment as compared with 2D. These results show that 3D culture of cytotoxic T-cells in conjunction with IL-2-bearing microparticles produces a potent system for antitumor activity.
In this paper, cytokines released from microparticles were shown to skew the activation, differentiation, proliferation, and effector responses of T-cells. This new capability is easily adaptable to influence the behaviors of other immune cells, including NK cells and B cells. The scaffold demonstrated here could be engineered to release chemoattractants such as SDF-1α to enhance recruitment of T-cells into a tumor site or could release TGF-β to promote suppression of tissue specific autoimmunity.
Supplementary Material
Acknowledgments
F.S.M. and M.M.H.-S. contributed equally to this work. Interlukin-2 used for this study was provided by the BRB Preclinical Repository of the National Cancer Institute, Frederick, MD, USA. The authors acknowledge Dr. Nassir Mokarram from Georgia Tech for technical assistance.
Footnotes
The ORCID identification number(s) for the author(s) of this article can be found under https://doi.org/10.1002/adma.201703178.
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Fatemeh S. Majedi, Department of Bioengineering, University of California, 420 Westwood Plaza, 5121 Engineering V, Los Angeles, CA 90095-1600, USA
Dr. Mohammad Mahdi Hasani-Sadrabadi, Department of Chemistry and Biochemistry, University of California, 607 Charles E. Young Drive South, Los Angeles, CA 90095-1569, USA. Weintraub Center for Reconstructive Biotechnology, Division of Advanced Prosthodontics, School of Dentistry, University of California, Los Angeles, CA 90095-1668, USA. Parker H. Petit Institute for Bioengineering and Bioscience, G.W. Woodruff School of Mechanical Engineering, Georgia Institute of Technology, Atlanta, GA 30332-0405, USA
Dr. Yoko Kidani, Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles, CA 90095-1489, USA
Dr. Timothy J. Thauland, Division of Immunology, Allergy, and Rheumatology, Department of Pediatrics, University of California, Los Angeles, CA 90095, USA
Prof. Alireza Moshaverinia, Weintraub Center for Reconstructive Biotechnology, Division of Advanced Prosthodontics, School of Dentistry, University of California, Los Angeles, CA 90095-1668, USA
Prof. Manish J. Butte, Division of Immunology, Allergy, and Rheumatology, Department of Pediatrics, University of California, Los Angeles, CA 90095, USA. Department of Molecular & Medical Pharmacology, University of California, Los Angeles, CA 90095-1735, USA. The Molecular Biology Institute and Jonsson Comprehensive Cancer Center, University of California, Los Angeles, CA 90095-1781, USA. California NanoSystems Institute, University of California, 570 Westwood Plaza, Los Angeles, CA 90095-7227, USA
Prof. Steven J. Bensinger, Department of Microbiology, Immunology, and Molecular Genetics, University of California, Los Angeles, CA 90095-1489, USA. Department of Molecular & Medical Pharmacology, University of California, Los Angeles, CA 90095-1735, USA. The Molecular Biology Institute and Jonsson Comprehensive Cancer Center, University of California, Los Angeles, CA 90095-1781, USA
Prof. Louis-S. Bouchard, Department of Bioengineering, University of California, 420 Westwood Plaza, 5121 Engineering V, Los Angeles, CA 90095-1600, USA. Department of Chemistry and Biochemistry, University of California, 607 Charles E. Young Drive South, Los Angeles, CA 90095-1569, USA. The Molecular Biology Institute and Jonsson Comprehensive Cancer Center, University of California, Los Angeles, CA 90095-1781, USA. California NanoSystems Institute, University of California, 570 Westwood Plaza, Los Angeles, CA 90095-7227, USA
References
- 1.a) Klebanoff CA, Gattinoni L, Restifo NP. Immunol Rev. 2006;211:214. doi: 10.1111/j.0105-2896.2006.00391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) June CH, Riddell SR, Schumacher TN. Sci Transl Med. 2015;7:280ps7. doi: 10.1126/scitranslmed.aaa3643. [DOI] [PubMed] [Google Scholar]
- 2.a) Yoo JW, Irvine DJ, Discher DE, Mitragotri S. Nat Rev Drug Discovery. 2011;10:521. doi: 10.1038/nrd3499. [DOI] [PubMed] [Google Scholar]; b) Langer R. Science. 1990;249:1527. doi: 10.1126/science.2218494. [DOI] [PubMed] [Google Scholar]
- 3.Moon JJ, Huang B, Irvine DJ. Adv Mater. 2012;24:3724. doi: 10.1002/adma.201200446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Li J, Mooney DJ. Nat Rev Mat. 2016;1:16071. doi: 10.1038/natrevmats.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang Y, Kohane DS. Nat Rev Mat. 2017;2:17020. [Google Scholar]; c) Tibbitt MW, Dahlman JE, Langer R. J Am Chem Soc. 2016;138:704. doi: 10.1021/jacs.5b09974. [DOI] [PubMed] [Google Scholar]
- 5.Pipkin ME, Sacks JA, Cruz-Guilloty F, Lichtenheld MG, Bevan MJ, Rao A. Immunity. 2010;32:79. doi: 10.1016/j.immuni.2009.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.a) Steenblock ER, Fadel T, Labowsky M, Pober JS, Fahmy TM. J Biol Chem. 2011;286:34883. doi: 10.1074/jbc.M111.276329. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Özbaş-Turan S, Akbuga J, Aral C. J Pharm Sci. 2002;91:1245. doi: 10.1002/jps.10122. [DOI] [PubMed] [Google Scholar]; c) Jhunjhunwala S, Balmert SC, Raimondi G, Dons E, Nichols EE, Thomson AW, Little SR. J Controlled Release. 2012;159:78. doi: 10.1016/j.jconrel.2012.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Jin W, Hardegen N, Lei K-j, Li L, Marinos N, McGrady G, Wahl SM. J Exp Med. 2003;198:1875. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu L, Mooney DJ. Nat Rev Cancer. 2016;16:56. doi: 10.1038/nrc.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.a) Zhao X, Liu S, Yildirimer L, Zhao H, Ding R, Wang H, Cui W, Weitz D. Adv Funct Mater. 2016;26:2809. [Google Scholar]; b) Bazban-Shotorbani S, Dashtimoghadam E, Karkhaneh A, Hasani-Sadrabadi MM, Jacob KI. Langmuir. 2016;32:4996. doi: 10.1021/acs.langmuir.5b04645. [DOI] [PubMed] [Google Scholar]
- 10.Sperling LH. Introduction to Physical Polymer Science. John Wiley & Sons; Hoboken, NJ: 2005. [Google Scholar]
- 11.a) Hasan M, Najjam S, Gordon MY, Gibbs RV, Rider CC. J Immunol. 1999;162:1064. [PubMed] [Google Scholar]; b) Najjam S, Mulloy B, Theze J, Gordon M, Gibbs R, Rider CC. Glycobiology. 1998;8:509. doi: 10.1093/glycob/8.5.509. [DOI] [PubMed] [Google Scholar]; c) Najjam S, Gibbs RV, Gordon MY, Rider CC. Cytokine. 1997;9:1013. doi: 10.1006/cyto.1997.0246. [DOI] [PubMed] [Google Scholar]
- 12.Hasani-Sadrabadi MM, Taranejoo S, Dashtimoghadam E, Bahlakeh G, Majedi FS, VanDersarl JJ, Janmaleki M, Sharifi F, Bertsch A, Hourigan K. Adv Mater. 2016;28:4134. doi: 10.1002/adma.201502697. [DOI] [PubMed] [Google Scholar]
- 13.Saggiomo V, Velders AH. Adv Sci. 2015;2:1500125. doi: 10.1002/advs.201500125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majedi FS, Hasani-Sadrabadi MM, VanDersarl JJ, Mokarram N, Hojjati-Emami S, Dashtimoghadam E, Bonakdar S, Shokrgozar MA, Bertsch A, Renaud P. Adv Funct Mater. 2014;24:432. [Google Scholar]
- 15.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Nat Immunol. 2003;4:1191. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima H, Liu XW, Wynshaw-Boris A, Rosenthal LA, Imada K, Finbloom DS, Hennighausen L, Leonard WJ. Immunity. 1997;7:691. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- 17.Sallusto F, Lenig D, Förster R, Lipp M, Lanzavecchia A. Nature. 1999;401:708. [PubMed] [Google Scholar]
- 18.a) Wherry EJ, Teichgräber V, Becker TC, Masopust D, Kaech SM, Antia R, Von Andrian UH, Ahmed R. Nat Immunol. 2003;4:225. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]; b) Masopust D, Vezys V, Marzo AL, Lefrançois L. Science. 2001;291:2413. [PubMed] [Google Scholar]
- 19.Kuipers HF, Rieck M, Gurevich I, Nagy N, Butte MJ, Negrin RS, Wight TN, Steinman L, Bollyky PL. Proc Natl Acad Sci USA. 2016;113:1339. doi: 10.1073/pnas.1525086113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baars PA, Sierro S, Arens R, Tesselaar K, Hooibrink B, Klenerman P, van Lier RA. Eur J Immunol. 2005;35:3131. doi: 10.1002/eji.200425770. [DOI] [PubMed] [Google Scholar]
- 21.Kaech SM, Ahmed R. Nat Immunol. 2001;2:415. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.a) Bannard O, Kraman M, Fearon DT. Science. 2009;323:505. doi: 10.1126/science.1166831. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Harrington LE, Janowski KM, Oliver JR, Zajac AJ, Weaver CT. Nature. 2008;452:356. doi: 10.1038/nature06672. [DOI] [PubMed] [Google Scholar]; c) Opferman JT, Ober BT, Ashton-Rickardt PG. Science. 1999;283:1745. doi: 10.1126/science.283.5408.1745. [DOI] [PubMed] [Google Scholar]
- 23.Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. J Exp Med. 2009;206:3015. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wan YY, Flavell RA. Nature. 2007;445:766. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 25.a) Liu Y, Wang L, Kikuiri T, Akiyama K, Chen C, Xu X, Yang R, Chen W, Wang S, Shi S. Nat Med. 2011;17:1594. doi: 10.1038/nm.2542. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Akiyama K, Chen C, Wang D, Xu X, Qu C, Yamaza T, Cai T, Chen W, Sun L, Shi S. Cell Stem Cell. 2012;10:544. doi: 10.1016/j.stem.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ansari S, Chen C, Hasani-Sadrabadi MM, Yu B, Zadeh HH, Wu BM, Moshaverinia A. Acta Biomater. 2017;60:181. doi: 10.1016/j.actbio.2017.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stephan SB, Taber AM, Jileaeva I, Pegues EP, Sentman CL, Stephan MT. Nat Biotechnol. 2015;33:97. doi: 10.1038/nbt.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.