Abstract
Objective
To define the relationships between molecular measures of viral persistence in blood (i.e., plasma viremia, cellular HIV-1 DNA, and mRNA) and expressed or inducible virus from resting CD4+T-cells of individuals on suppressive ART.
Design
We compared molecular measurements of HIV-1 in plasma and in uncultured peripheral blood mononuclear cells (PBMC) to the levels of virions produced by either unstimulated or phorbol myristate acetate and ionomycin (PMA/iono)-stimulated PBMC or resting CD4+T-cells from 21 donors on suppressive ART.
Results
We found that unstimulated virion release from cultured resting CD4+T-cells was positively correlated with the levels of plasma viremia in vivo (Spearman rho=0.67, p=0.0017). We also found that levels of both cellular HIV-1 DNA and unspliced HIV-1 mRNA per million uncultured PBMC were positively correlated with the levels of inducible virion release from both PMA/iono-stimulated PBMC (total HIV-1 DNA: rho=0.64, p=0.0017; unspliced HIV-1 RNA: rho=0.77, p<0.001) and PMA/iono-stimulated resting CD4+T-cells (total HIV-1 DNA: rho=0.75, p<0.001; unspliced HIV-1 RNA: rho=0.75, p<0.001).
Conclusions
These results show for the first time that there are strong associations between in vivo measures of HIV-1 persistence and ex vivo measures of spontaneous and inducible virus production from cultured PBMC and resting CD4+T-cells. Findings from this study provide insight into the biology of HIV-1 persistence and suggest methods to guide the evaluation of clinical strategies to reduce the size of the viral reservoir.
Keywords: HIV persistence, HIV reservoir, biomarkers of HIV persistence, persistent HIV viremia, suppressive antiretroviral therapy
Introduction
Antiretroviral therapy (ART) suppresses HIV-1 viral replication, which leads to rapid decreases in plasma viremia and concomitant increases in CD4+T-cell counts [1–4]. Quantifying the persistence of HIV-1 following the initiation of ART has been the subject of much investigation. Most studies have focused on evaluating molecular markers of HIV-1 persistence such as low-level plasma viremia, cellular proviral DNA, or cellular HIV-1 RNA. These studies have demonstrated that levels of viremia fall 4–5 orders of magnitude in the years after beginning ART, but that low levels of HIV-1 RNA (ca. 1 copy/ml) can remain detectable in plasma for 12 or more years after the start of therapy [5–8].
Other studies have focused on evaluating levels of total cellular HIV-1 DNA following the initiation of ART. Previous work has shown an approximately 10- to 15-fold decay of HIV-1 DNA over the first year [9–11]. Gandhi et al. also found that HIV-1 DNA levels continued to decline after 4 years on ART, with a half-life of approximately 13 years[11]. Cellular HIV-1 DNA persists to varying degrees in all subsets of CD4+T-cells, though predominantly in central memory CD4+T-cells [12, 13]. Levels of cellular HIV-1 DNA are lower in those who initiated ART during the primary infection stage, compared with chronic infection [9]; those who initiated ART at the earliest stage of infection (Fiebig I) have lower HIV-1 DNA levels than those who started later (Fiebig II–IV) [14]. Although multiple forms of HIV-1 DNA can be detected during suppressive ART, integrated DNA accumulates prior to the initiation of ART, and tends to dominate the pool of total HIV-1 DNA after long-term suppressive ART [15]. The large majority (>95%) of HIV-1 DNA in individuals on effective ART is defective due to insertions, deletions, or APOBEC-induced hypermutation [16].
Cellular HIV-1 RNA was initially used as a biomarker of progression to AIDS in untreated HIV-1 infection [17–20]. In contrast to the 10- to 15-fold decay in HIV-1 DNA that has been observed during the first 4 years of therapy, cellular HIV-1 RNA has recently been found to decay approximately 525-fold over the first 4 years of ART[11]. Early initiation of ART also leads to significantly lower levels of cellular unspliced HIV-1 RNA compared with patients who begin therapy during chronic infection[21].
In addition to molecular markers of HIV-1 persistence, a reservoir of cells carrying intact (replication-competent) and inducible proviruses persists despite ART at a frequency of approximately 1 cell per million resting CD4+T-cells[22–27], although higher frequencies have recently been reported[28]. The relationships between the HIV-1 latent reservoir and molecular measures of viral persistence have not been thoroughly elucidated. Most studies to date have evaluated the correlations between viral outgrowth assays (QVOA) and molecular measures. Viral outgrowth from resting CD4+T-cells has been reported to correlate with levels of integrated [29, 30] or total HIV-1 DNA in PBMC [30], the ratio of unspliced cellular HIV-1 RNA to DNA in gut-associated CD4+T-cells [29], and the levels of plasma viremia by the single-copy assay (SCA) [31], but correlations have been inconsistent across studies. A broader assessment of molecular measures that includes both spontaneous and inducible virus production has yet to be performed. Ideally, simple molecular measures would be strongly predictive of the latent HIV-1 reservoir.
Toward this end, we compared molecular measures of cellular HIV-1 nucleic acids to each other and to levels of virion release from cultured PBMC and resting CD4+T-cells isolated from donors on suppressive ART. We hypothesized that the frequency of infected cells, and their transcriptional activity in PBMC, would be correlated with both unstimulated and inducible virion release from resting CD4+T-cells.
Materials and methods
Participant cohort and study samples
Selection criteria for donors in this cross-sectional study were HIV-positive individuals ≥18 years old on suppressive ART with plasma HIV-1 RNA <50 copies per milliliter for ≥1 year. Consecutive donors were enrolled by Quest Clinical Research (San Francisco, CA). All donors provided written informed consent for their participation. This study was approved by the Western Institutional Review Board and the Institutional Review Board at the University of Pittsburgh. PBMC and plasma were obtained by leukapheresis and plasmapheresis, respectively.
Isolation of PBMC and resting CD4+T-cells
PBMC were isolated from leukapheresis products by Ficoll-Hypaque density gradient centrifugation. Resting CD4+T-cells (i.e., CD3+CD4+CD25−CD69−HLA−DR−) were isolated by negative selection from PBMC using a custom kit from StemCell Technologies™ containing a cocktail of antibodies targeting CD14, CD16, CD19, CD20, CD25, CD36, CD56, CD69, CD66b, CD123, HLA-DR, and glycophorin A. This negative selection routinely achieved >95% of cells with a resting CD4+T-cell phenotype by flow cytometric analysis [32]. PBMC were cryopreserved in 90% fetal bovine serum and 10% DMSO and stored in liquid nitrogen for downstream assessment of cellular HIV-1 DNA and RNA. Resting CD4+T-cells were stored at −80°C as cell pellets. Plasma was stored at −80°C for downstream assessment of low-level viremia.
Cell culture conditions
For all ex vivo measures of virus release, freshly isolated PBMC and resting CD4+T-cells were cultured in both unstimulated (i.e., medium only) and stimulated (i.e., stimulation for 7 days with 50 ng/ml phorbol myristate acetate [PMA] and 500 ng/ml ionomycin [iono]) conditions to assess virus release. PBMC were cultured at 15×106 cells per well, and the culture supernatant was harvested after 7 days of culture. Resting CD4+T-cells were cultured at 5×106 cells per well, and the supernatant was harvested after 6 days of culture. All cells were cultured in deep-well 96-well plates in 1.1 milliliters of RPMI 1640 without phenol red indicator, and contained 10% (volume/volume) fetal bovine serum and 0.6% (v/v) penicillin/streptomycin. To prevent viral propagation, 100 nM efavirenz and 100 nM elvitegravir were included in the culture medium. Supernatants were stored at −80°C until assessment of virus release by qRT-PCR.
Assessment of virion release in culture supernatants
To assess unstimulated and stimulated virus release, levels of HIV-1 RNA were measured in harvested culture supernatants by qRT-PCR using the Roche COBAS AmpliPrep/TaqMan v2.0 (CAP/CTM) platform [32, 33]. Culture supernatants assessed using CAP/CTM have previously been shown to be free of contaminating HIV-1 DNA[34].
Quantification of low-level viremia
Low-level viremia was quantified using a large-volume adaptation of the SCA targeting the integrase region of the HIV-1 genome as previously described[35]. Briefly, nucleic acid was isolated from large volumes (18–40 ml) of plasma in parallel replicate extractions of ~4 ml. Nucleic acid was isolated by guanidinium hydrochloride/proteinase K incubation followed by guanidinium thiocyanate/glycogen incubation and precipitation of nucleic acid in 100% isopropanol. After washing in 70% ethanol, the nucleic acid was eluted in 5 mM Tris, DTT and RNasin, and cDNA was reverse transcribed, followed by qRT-PCR as previously described[35].
Quantification of cellular HIV-1 RNA and DNA
Total cellular HIV-1 DNA and unspliced cellular HIV-1 RNA were quantified as previously described[36]. Briefly, total nucleic acid was isolated from PBMC or resting CD4+T-cells, and the final extract was split into two aliquots. One aliquot was frozen at −80°C for downstream assessment of total HIV-1 DNA and CCR5 to control for the total input cell number. The second aliquot was DNase treated to remove all DNA for the quantification of cellular unspliced HIV-1 RNA, and levels of the human gene IPO8 were quantified to serve as an internal control for the recovery of cellular RNA. The primers and probe used to quantify cellular HIV-1 DNA and RNA were identical to those used for large-volume SCA.
Statistical analysis
All figures (excluding the correlogram) were generated using Prism 6 for Mac OS X. The correlogram was generated in R[37] using the ggplot2 [38] and gplots[39] packages. Spearman’s correlation was used to test the significance of all correlations, and the results were corrected for multiple comparisons by setting the false discovery rate at 5%. The limit of detection for large-volume SCA depended on the input volume of plasma assayed: the limit of quantification of the assays for cellular HIV-1 DNA and RNA was one copy of nucleic acid each, and the limit of quantification of the Roche CAP/CTM was 20 copies per milliliter of culture supernatant. Values below the limit of detection of these assays were interpolated as 50% of the limit of detection, and interpolated values were used in all analyses. All statistical analyses were two-sided, and α<0.05 was considered significant.
Results
Study participant characteristics
Table 1 shows characteristics of the donors (n = 21). The median age of the participants was 54 years old (interquartile range (IQR): 52–58 years). All participants had CD4+T-cell counts above 500 cells/mm3 (median 725 cells/mm3; IQR: 607–796 cells/mm3); in percentages, a median of 34% of lymphocytes were CD4+T-cells (IQR: 27–40%). Median nadir CD4+T-cell count was 187 (IQR: 92–277) cells/mm3. At the time of the study, all participants were on suppressive ART regimens with HIV-1 RNA in plasma <50 copies per milliliter for ≥1 (median: 12; IQR: 6–16) years. Median low-level plasma viremia was 1.1 (IQR of 0.05–2.9) HIV-1 RNA copies per milliliter in the 19 participants for which sufficient plasma was available for large-volume iSCA. The median level of total cellular HIV-1 DNA was 283 (IQR: 160–504) copies per 106 PBMC, and the median level of cellular unspliced HIV-1 RNA was 37 (IQR: 13–74) copies per 106 PBMC. The median level of total cellular HIV-1 DNA in resting CD4+T-cell was 1159 (IQR: 547–2378) copies per 106 cells. Results for molecular measures of viral persistence and culture-based measures of virion production are included in supplementary Table 1. Purified resting CD4+T-cells were stored as cell pellets at −80°C, which is not adequate for RNA preservation, thus cellular HIV-1 RNA could not be accurately quantified in resting CD4+T-cells.
Table 1.
Characteristics of study donors.
| Participant ID | Age | Nadir CD4+T-cell count (cells/mm3) | Duration of virologic suppression (years) | CD4+ T-cell count (cells/mm3) | Percent CD4+ T-cells in lymphocytes |
|---|---|---|---|---|---|
| 1 | 69 | 210 | 6.00 | 742 | 41.2 |
| 2 | 54 | 253 | 19 | 653 | 36.3 |
| 3 | 57 | 300 | 2 | 587 | 36.7 |
| 4 | 55 | NA | NA | 762 | 23.8 |
| 5 | 51 | 0 | 8 | 692 | 17.3 |
| 6 | 54 | 425 | 3 | 607 | 26.4 |
| 7 | 61 | 325 | 16 | 1022 | 29.2 |
| 8 | 58 | NA | NA | 632 | 23.4 |
| 9 | 50 | NA | NA | 1345 | 26.9 |
| 10 | 54 | 187 | 12 | 590 | 26.8 |
| 11 | 52 | 67 | 14 | 422 | 28.1 |
| 12 | 53 | 150 | 7 | 958 | 38.3 |
| 13 | 52 | 96 | 1 | 832 | 39.6 |
| 14 | 64 | NA | NA | 725 | 48.3 |
| 15 | 64 | 7 | 15 | 538 | 33.6 |
| 16 | 39 | 325 | 6 | 556 | 39.7 |
| 17 | 56 | NA | NA | 882 | 42 |
| 18 | 54 | 120 | 15 | 699 | 36.8 |
| 19 | 57 | 190 | 19 | 780 | 39 |
| 20 | 47 | NA | NA | 796 | 36.2 |
| 21 | 62 | 88 | 24 | 783 | 43.5 |
| Median (Interquartile range) | 54 (52–58) | 187 (92–277) | 12 (6–16) | 725 (607–796) | 34 (27–40) |
NA, not available
Overall assessment of relationships between variables
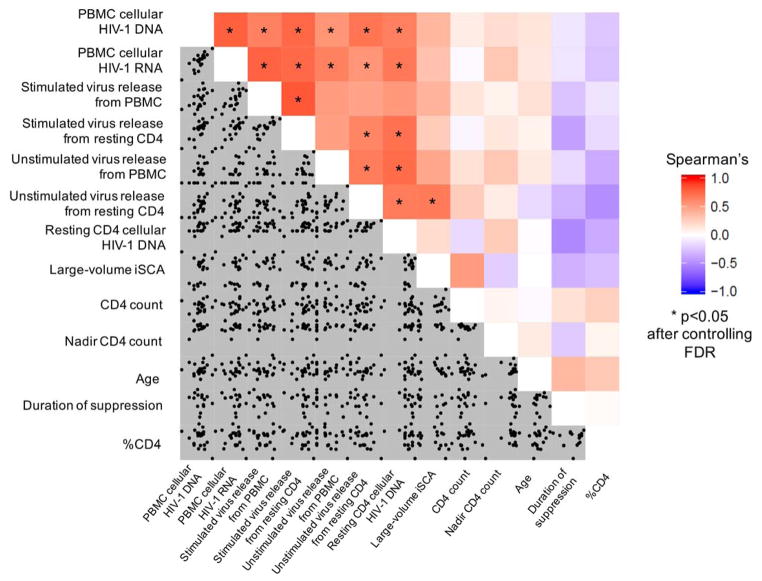
Figure 1 shows the relationships among 13 different continuous variables using Spearman’s correlations for 78 comparisons. We constructed a correlogram to display all the data and relationships simultaneously (data included in the correlogram are also shown in Table 1 and in supplementary Table 1). The bottom half of the correlogram shows scatter plots of the individual variables; the top half is a heat map quantifying the strength of the correlations between variables. Red on the heat map indicates a strong positive correlation, blue represents a strong negative correlation, and white indicates that no correlation was detected. Asterisks highlight the statistically significant relationships between variables at a false discovery rate (FDR) of 5%. In total, 18 of the 78 comparisons were statistically significant. Some variables showed a high degree of interrelatedness, including the frequency of infected cells in PBMC (i.e., total cellular HIV-1 DNA), levels of transcriptional activity in PBMC (i.e., cellular unspliced HIV-1 RNA), spontaneous virus release from resting CD4+T-cells, and inducible virus release from resting CD4+T-cells.
Fig. 1. Correlogram showing the interrelationships between the variables studied.
Each of the 13 variables studied is listed on the x- and y-axes. The bottom left-hand portion of the correlogram shows scatter plots between the individual variables, and the top right-hand portion of the plots shows a heat map of correlations between variables. The key on the right-hand side shows the colors corresponding to strong positive correlations (red), strong negative correlations (blue), and neutral correlations (white). Asterisks indicate comparisons that were statistically significant by Spearman’s correlation after correcting for multiple comparisons by maintaining a false discovery rate of 5%. In total, 18 of the 78 comparisons were statistically significant. Highly interrelated variables included total cellular HIV-1 DNA, cellular unspliced HIV-1 RNA, and virion release from stimulated resting CD4+T-cells.
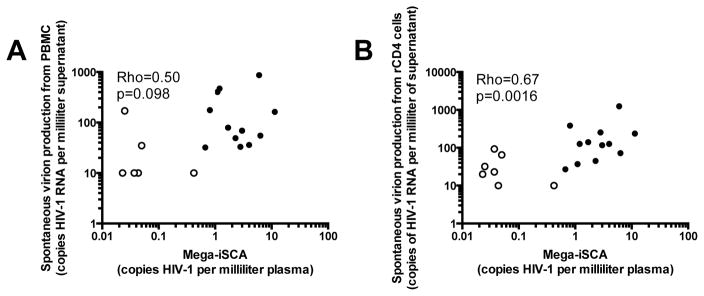
Spontaneous virion release ex vivo correlates with low-level viremia in vivo
We compared low-level viremia to levels of unstimulated virus release from cultured PBMC or resting CD4+T-cells isolated from peripheral blood (Figure 2). The median level of unstimulated virus release from PBMC was 36 (IQR: <20 to 164) HIV-1 RNA copies per milliliter of culture supernatant, whereas the median level of spontaneous virion release from resting CD4+T-cells was 72 (IQR: 27–126) copies per milliliter of culture supernatant. Surprisingly, we found that the in vivo levels of viremia were significantly correlated at the FDR of 5% with spontaneously released virions from resting CD4+T-cells (rho=0.67, p=0.0017), and were nearly significantly correlated with spontaneously released virions from cultured PBMC (rho=0.50, p=0.098) at this FDR level (without correcting for multiple comparisons by FDR, this correlation was statistically significant with rho=0.50, p=0.030). These results suggest that the spontaneous release of virions from cells obtained from blood may reflect overall virion release and clearance in vivo.
Fig. 2. Unstimulated virus release from cultured PBMC and resting CD4+T-cells correlates with persistent viremia on ART.
(a) Unstimulated virus release from 15×106 PBMC following 5 days of culture correlated with levels of plasma viremia (HIV-1 RNA) measured by large-volume iSCA (rho=0.50, p=0.098). (b) Similarly, unstimulated virus release from 5×106 resting CD4+T-cells after 7 days of culture correlated with the levels of plasma viremia by large-volume iSCA (rho = 0.67, p=0.0016). In both (a) and (b), open symbols denote samples that had undetectable HIV-1 RNA by large-volume iSCA (interpolated at 50% of the limit of detection based on plasma volume assayed) or CAP/CTM qRT-PCR of culture supernatant (interpolated at 50% of the limit of detection, or 10 copies per milliliter of culture supernatant assayed).
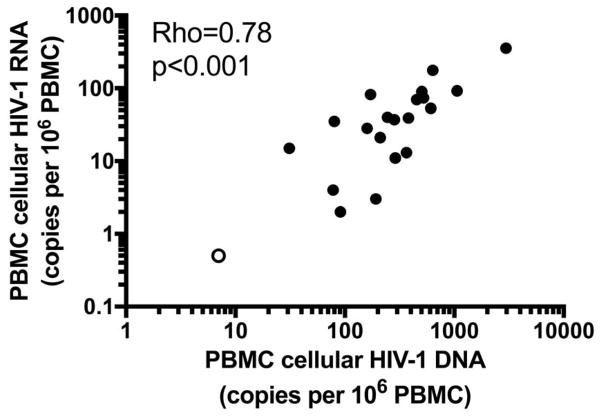
Cellular HIV-1 DNA and unspliced HIV-1 RNA are strongly correlated
We next investigated whether the frequency of infected cells in PBMC was related to their transcriptional activity (Figure 3). Consistent with recently published findings[36], we found a strong positive correlation (rho=0.78, p<0.001) between the levels of total HIV-1 DNA in PBMC and levels of unspliced cellular HIV-1 RNA. These data indicate that a higher frequency of infected cells is associated with higher levels of HIV-1 transcriptional activity.
Fig. 3. The frequency of infected cells (cellular HIV-1 DNA) is correlated with HIV-1 transcriptional activity in PBMC (cellular HIV-1 RNA).
Levels of total cellular HIV-1 DNA were significantly correlated with basal levels of unspliced HIV-1 RNA in PBMC that were cryopreserved after processing the leukapheresis product. The open symbol denotes a sample with undetectable HIV-1 RNA, interpolated at 50% of the limit of detection, or 0.5 copies of HIV-1 RNA per million PBMC.
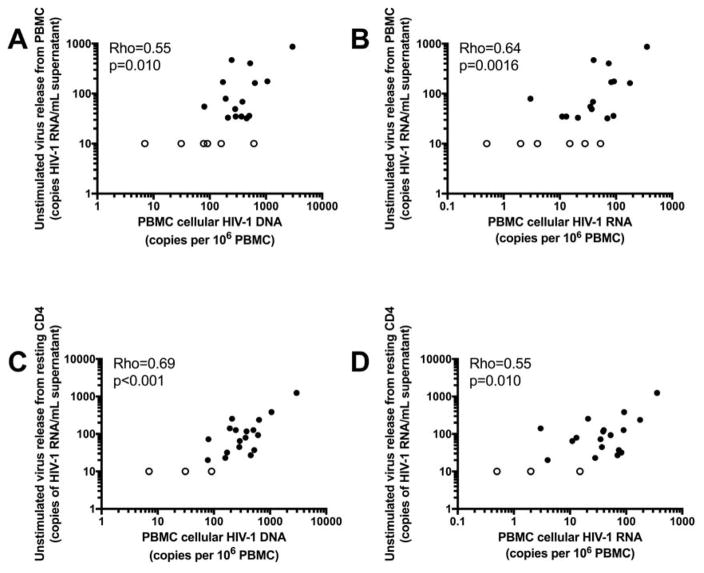
Unstimulated and stimulated virus release are correlated with cellular HIV-1 DNA and RNA
We next sought to assess whether levels of unstimulated virus release from PBMC and resting CD4+T-cells were related to the frequency of infection and transcriptional activity of infected cells in PBMC (Figure 4). In the 21 study participants evaluated, we found that levels of both total cellular HIV-1 DNA per million PBMC and cellular unspliced HIV-1 RNA per million PBMC were related to unstimulated virus release from PBMC (total HIV-1 DNA: rho=0.55, p=0.010; unspliced HIV-1 RNA: rho=0.64, p=0.0016). Levels of cellular HIV-1 DNA and RNA in PBMC were also correlated with unstimulated virus release from resting CD4+T-cells (total HIV-1 DNA: rho=0.69, p<0.001; unspliced HIV-1 RNA rho=0.55, p=0.010). These findings indicate that a higher frequency and a higher level of transcriptional activity of infected cells are associated with higher levels of spontaneous virus release. The frequency of infected resting CD4+T-cells was also significantly associated with spontaneous virion release from PBMC (rho=0.73, p<0.001) and from resting CD4+T-cells (rho=0.66, p=0.0019) (Figure 1).
Fig. 4. Spontaneous virion release is correlated with the frequency of infected cells and their transcriptional activity in PBMC.
(a) The frequency of infected cells (cellular HIV-1 DNA in PBMC) is correlated with the spontaneous release of virions from unstimulated cultured PBMC (rho=0.55, p=0.010). (b) The level of unspliced cellular HIV-1 RNA transcription in PBMC is correlated with spontaneous virion release from PBMC (rho=0.64, p=0.0016). (c) The frequency of infected cells in PBMC is correlated with spontaneous virion release from resting CD4+T-cells (rho=0.69, p<0.001). (d) The level of unspliced cellular HIV-1 RNA transcription in PBMC is correlated with spontaneous virion release from resting CD4+ T cells (rho=0.55, p=0.010). In (a), (b), (c), and (d), open circles represent samples that were undetectable by CAP/CTM qRT-PCR for spontaneous virion release, or one sample that had undetectable levels of cellular unspliced HIV-1 RNA. In all cases of non-detection, values were interpolated as 50% of the limit of detection of the assay (i.e., 10 copies per milliliter of supernatant for CAP/CTM and 0.5 copies of unspliced cellular HIV-1 RNA per million PBMC).
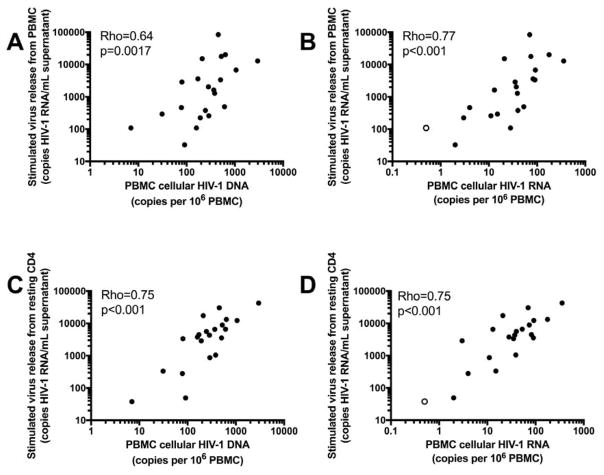
We next evaluated whether the frequency of infected cells and transcriptional activity in PBMC were related to induced virus release from PBMC and resting CD4+T-cells stimulated with PMA/iono (Figure 5). The median level of HIV-1 RNA released for stimulated PBMC was 1633 (IQR: 292–6729) copies of HIV-1 RNA per milliliter of culture supernatant, whereas resting CD4+T-cells released a median of 4329 (IQR: 1042–8948) copies of HIV-1 RNA per milliliter of culture supernatant following stimulation. This analysis showed that the frequency of infected cells is related to the level of inducible virus release from both PBMC (rho=0.64, p=0.0017) and resting CD4+T-cells (rho=0.75, p<0.001). Levels of viral transcription per million PBMC were also positively and significantly correlated with stimulated virus release from PBMC (rho=0.77, p<0.001) and resting CD4+T-cells (rho=0.75 p<0.001). The frequency of infection of resting CD4+T-cells was also correlated with inducible virion release from PBMC (rho=0.53, p=0.020) and resting CD4+T-cells (rho=0.71, p<0.001) (Figure 1).
Fig. 5. The frequency of infected cells and their transcriptional activity in PBMC are correlated with the level of inducible virion release from PBMC and resting CD4+T-cells.
(a) The frequency of infected cells in PBMC is correlated with the level of inducible virion release from cultured PBMC treated with PMA/iono for 5 days (rho=0.64, p=0.0017). (b) The level of cellular unspliced HIV-1 RNA transcription is correlated with the level of inducible virion release from cultured PBMC (rho=0.77, p<0.001). (c) The frequency of infected cells in PBMC was correlated with the level of inducible virion release from cultured resting CD4+T-cells treated with PMA/iono for 7 days (rho=0.75, p<0.001). (d) The level of cellular unspliced HIV-1 RNA in PBMC is correlated with the level of inducible virion release from resting CD4+T-cells stimulated with PMA/iono (rho=0.75, p<0.001). In (b) and (d), one sample (shown as an open circle) had undetectable levels of cellular unspliced HIV-1 RNA. The value of this sample was interpolated as 50% of the limit of detection of the assay, or 0.5 copies per million PBMC.
Discussion
To date there have been no published studies that include an integrated assessment of molecular measures of viral persistence in vivo and both spontaneous and stimulated virus release ex vivo. We therefore evaluated in vivo measures of persistence and ex vivo measures of virus release in donors on suppressive ART. We found that levels of total cellular HIV-1 DNA and cellular unspliced HIV-1 RNA in PBMC were strongly associated with each other, and with unstimulated and stimulated virus release from PBMC and resting CD4+T-cells. We also discovered that unstimulated virus release ex vivo from resting CD4+T-cells was correlated with the level of persistent plasma viremia in vivo, and that there was a positive association (albeit not statistically significant with a FDR of 5%) between spontaneous release of virions from PBMC and persistent plasma viremia.
Longitudinal single genome sequencing of plasma viral HIV-1 RNA from before the initiation of ART and after years of suppressive ART has revealed no evidence of continued viral evolution [40], demonstrating that ART is largely able to stop viral replication. However, low-level viremia remains detectable on long-term suppressive therapy at a level of approximately 1 copy per milliliter of plasma after up to 7 years of viral suppression [7], regardless of the ART regimen used [41]. The source of this low-level viremia is contentious, and data related to a quantitative relationship between low-level viremia and the size of the viral reservoir are sparse and conflicting [29, 31]. In the current study, we found that levels of unstimulated virus release from resting CD4+T-cells cultured ex vivo are significantly correlated with levels of persistent viremia in vivo when using large volumes of plasma to accurately quantify viremia. This connection between unstimulated virus release and low-level viremia establishes a link between an in vivo measure of active virus release, and an ex vivo measure of spontaneous release of virions from resting CD4+T-cells. Although statistical significance was not reached (after correcting for multiple comparisons) between persistent virema and spontaneous virion release from PBMC, there was a trend towards a positive association. This raises the intriguing possibility that spontaneous virion release from PBMC may be reflective of the balance between virion production and clearance in vivo. Future studies will need to evaluate the role effector cells (such as CD8+T-cells and natural killer cells) present in bulk PBMC play in clearance or inhibition of virion producing cells ex vivo, and the ways in which this is reflective of in vivo clearance of virus producing cells.
We also assessed whether the frequency of infected cells in PBMC was related to the level of viral transcription of unspliced cellular HIV-1 RNA. We found that the frequency of infected cells in PBMC was strongly related to the level of viral transcription in PBMC, which confirms a recently published finding from our group [36]. The frequency of infected, resting CD4+T-cells was also related to the level of total cellular HIV-1 DNA and RNA per million PBMC (Figure 1). These data show that there is proportionality between the frequency of infection and the level of viral transcription. Single-cell analysis will be needed to ascertain whether the source of cellular HIV-1 RNA is many cells transcribing small amounts of HIV-1 RNA, or a few cells transcribing high levels of HIV-1 RNA.
In the current study, we used total virus release from PMA/iono-stimulated PBMC and resting CD4+T-cells as the culture-based measure of latent HIV-1. The gold standard for assessing the size of the latent reservoir is the QVOA, but this assay has substantial barriers to widespread use, including the need for large numbers of resting CD4+T-cells, specialized culture medium, irradiated feeder cells from HIV-negative blood donors, target cells for the propagation of virus, and up to 28 days of co-culture [42]. Recent work has somewhat simplified the QVOA method, and has suggested that QVOA wells containing virions with properly poly-adenylated virion-associated HIV-1 RNA become positive for p24 after extended culture [43]. Nevertheless, because of the complexity of QVOA, we quantified total, inducible virus release by RT-PCR quantification of HIV-1 RNA in culture supernatants as a simpler approach to measuring latent HIV-1 that is sensitive for virus-producing cells and has a large dynamic range (3 log10). Because the current study did not measure inducible, replication-competent virus production, future work should evaluate the relationship between measures of persistence and virion production described here, and infectious virus outgrowth as measured by the more time- and resource-intensive QVOA.
We found that the frequency and transcriptional activity of infected PBMC are both significantly correlated with inducible virus release from both resting CD4+T-cells and PBMC. These findings are striking, given our recent work showing that only 1.5% of proviruses detectable by qPCR in resting CD4+T-cells could be reactivated to produce virions with one round of stimulation [32]. Although it is well established that many of the proviruses present in resting CD4+T-cells are defective [44], our results suggest that there is a degree of proportionality between the number of infected cells and the amount of virus produced. Similarly, the transcription of HIV-1 RNA in infected cells is correlated with inducible virus release, which demonstrates a proportional relationship between transcription and virus release.
The finding that levels of HIV-1 RNA transcription in PBMC are correlated with virus release following stimulation is consistent with the report that rebound viremia can sometimes be genetically linked to cells transcribing viral RNA prior to the cessation of ART [45]. Work by Rothenberger et al. has also shown that there are many sites of viral expression in lymph nodes across the body during suppressive ART [46], which is in line with the idea that rebound virus may originate from cells that are already transcribing HIV-1 RNA. Alternatively, rebound virus could also come from latently infected cells undergoing stochastic reactivation. Modeling work by Hill et al. [47] has similarly suggested that actively transcribing cells are likely responsible for the rapid viral rebound following the cessation of ART in most people with HIV-1, whereas latently infected cells that have become reactivated are likely responsible for the late viral rebound observed in the Boston patients [48, 49] and the Mississippi baby [50–52].
Our findings are also consistent with recently published in vivo assessments of reservoir size in treatment interruption studies, in which Li et al. found that levels of unspliced cellular HIV-1 RNA in PBMC are associated with time to viral rebound [53]. Similarly, Williams et al. found that levels of total HIV-1 DNA in PBMC were associated with time to viral rebound [54].
Combining our results with the two in vivo studies described above of the time to viral rebound following ART interruption suggests that simple measures of the frequency of HIV-1 infected cells and their transcriptional activity by q(RT)-PCR in unfractionated and uncultured PBMC can be used to estimate spontaneous and inducible virus production in persons with HIV-1 on suppressive ART. Ultimately, prospective studies of intensively monitored ART cessation and time to viral rebound will be required to validate whether simple measures of viral persistence in the periphery can accurately predict the size of the viral reservoir, and whether the relationships between molecular viral persistence and culture-based measures of virus production are maintained following interventions designed to reduce the size of the reservoir.
Supplementary Material
Acknowledgments
Funding sources: This project was supported by grants from the National Institute of Health (1R21AI113102-01) and Gilead Sciences. A.R.C. received support from the Pitt AIDS Research Training Grant (T32 AI065380-08).
We thank the study participants for donating samples for this study.
Footnotes
Conflicts of interest: A.T., A.I., J.K., and J.P.M. are employees of Gilead Sciences. D.D.S. is a former employee of Gilead Sciences, and a current employee of Agenovir. S.S.W. is an employee of Quest Clinical Research. J.W.M. is a consultant for Gilead Sciences and owns shares of Co-crystal Pharma, Inc. All other authors declare no relevant conflicts of interest.
References
- 1.Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, et al. Treatment with Indinavir, Zidovudine, and Lamivudine in Adults with Human Immunodeficiency Virus Infection and Prior Antiretroviral Therapy. New England Journal of Medicine. 1997;337(11):734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 2.Hammer SM, Squires KE, Hughes MD, Grimes JM, Demeter LM, Currier JS, et al. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N Engl J Med. 1997;337(11):725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 3.Gallant JE, DeJesus E, Arribas JR, Pozniak AL, Gazzard B, Campo RE, et al. Tenofovir DF, Emtricitabine, and Efavirenz vs. Zidovudine, Lamivudine, and Efavirenz for HIV. New England Journal of Medicine. 2006;354(3):251–260. doi: 10.1056/NEJMoa051871. [DOI] [PubMed] [Google Scholar]
- 4.Walmsley SL, Antela A, Clumeck N, Duiculescu D, Eberhard A, Gutiérrez F, et al. Dolutegravir plus Abacavir–Lamivudine for the Treatment of HIV-1 Infection. New England Journal of Medicine. 2013;369(19):1807–1818. doi: 10.1056/NEJMoa1215541. [DOI] [PubMed] [Google Scholar]
- 5.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271(5255):1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 6.Perelson AS, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, et al. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387(6629):188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 7.Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2008;105(10):3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riddler SA, Aga E, Bosch RJ, Bastow B, Bedison M, Vagratian D, et al. Continued Slow Decay of the Residual Plasma Viremia Level in HIV-1-Infected Adults Receiving Long-term Antiretroviral Therapy. J Infect Dis. 2016;213(4):556–560. doi: 10.1093/infdis/jiv433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray JM, McBride K, Boesecke C, Bailey M, Amin J, Suzuki K, et al. Integrated HIV DNA accumulates prior to treatment while episomal HIV DNA records ongoing transmission afterwards. Aids. 2012;26(5):543–550. doi: 10.1097/QAD.0b013e328350fb3c. [DOI] [PubMed] [Google Scholar]
- 10.Besson GJ, Lalama CM, Bosch RJ, Gandhi RT, Bedison MA, Aga E, et al. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis. 2014;59(9):1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gandhi RT, McMahon DK, Bosch RJ, Lalama CM, Cyktor JC, Macatangay BJ, et al. Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog. 2017;13(4):e1006285. doi: 10.1371/journal.ppat.1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, et al. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog. 2013;9(3):e1003211. doi: 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ananworanich J, Sacdalan CP, Pinyakorn S, Chomont N, de Souza M, Luekasemsuk T, et al. Virological and immunological characteristics of HIV-infected individuals at the earliest stage of infection. J Virus Erad. 2016;2:43–48. doi: 10.1016/S2055-6640(20)30688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Gunthard HF, et al. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis. 2008;197(3):411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- 16.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, et al. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med. 2016;22(9):1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furtado MR, Callaway DS, Phair JP, Kunstman KJ, Stanton JL, Macken CA, et al. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med. 1999;340(21):1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- 18.Furtado MR, Kingsley LA, Wolinsky SM. Changes in the viral mRNA expression pattern correlate with a rapid rate of CD4+ T-cell number decline in human immunodeficiency virus type 1-infected individuals. Journal of Virology. 1995;69(4):2092–2100. doi: 10.1128/jvi.69.4.2092-2100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saksela K, Stevens C, Rubinstein P, Baltimore D. Human immunodeficiency virus type 1 mRNA expression in peripheral blood cells predicts disease progression independently of the numbers of CD4+ lymphocytes. Proc Natl Acad Sci U S A. 1994;91(3):1104–1108. doi: 10.1073/pnas.91.3.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saksela K, Stevens CE, Rubinstein P, Taylor PE, Baltimore D. HIV-1 Messenger RNA in Peripheral Blood Mononuclear Cells as an Early Marker of Risk for Progression to AIDS. Annals of Internal Medicine. 1995;123(9):641–648. doi: 10.7326/0003-4819-123-9-199511010-00001. [DOI] [PubMed] [Google Scholar]
- 21.Schmid A, Gianella S, von Wyl V, Metzner KJ, Scherrer AU, Niederöst B, et al. Profound Depletion of HIV-1 Transcription in Patients Initiating Antiretroviral Therapy during Acute Infection. PLoS ONE. 2010;5(10):e13310. doi: 10.1371/journal.pone.0013310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chun TW, Stuyver L, Mizell SB, Ehler LA, Mican JA, Baseler M, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A. 1997;94(24):13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 24.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278(5341):1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 25.Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, et al. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nat Med. 2003;9(6):727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- 26.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, et al. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278(5341):1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 27.Crooks AM, Bateson R, Cope AB, Dahl NP, Griggs MK, Kuruc JD, et al. Precise Quantitation of the Latent HIV-1 Reservoir: Implications for Eradication Strategies. J Infect Dis. 2015;212(9):1361–1365. doi: 10.1093/infdis/jiv218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanyal A, Mailliard RB, Rinaldo CR, Ratner D, Ding M, Chen Y, et al. Novel assay reveals a large, inducible, replication-competent HIV-1 reservoir in resting CD4+ T cells. Nat Med. 2017;23(7):885–889. doi: 10.1038/nm.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9(2):e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, Vandekerckhove L. Integrated and Total HIV-1 DNA Predict Ex Vivo Viral Outgrowth. PLoS Pathog. 2016;12(3):e1005472. doi: 10.1371/journal.ppat.1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gandhi RT, Bosch RJ, Aga E, Bedison MA, Bastow B, Schmitz JL, et al. Residual plasma viraemia and infectious HIV-1 recovery from resting memory CD4 cells in patients on antiretroviral therapy: results from ACTG A5173. Antivir Ther. 2013;18(4):607–613. doi: 10.3851/IMP2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cillo AR, Sobolewski MD, Bosch RJ, Fyne E, Piatak M, Jr, Coffin JM, et al. Quantification of HIV-1 latency reversal in resting CD4+ T cells from patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A. 2014;111(19):7078–7083. doi: 10.1073/pnas.1402873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bui JK, Mellors JW, Cillo AR. HIV-1 Virion Production from Single Inducible Proviruses following T-Cell Activation Ex Vivo. J Virol. 2015;90(3):1673–1676. doi: 10.1128/JVI.02520-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei DG, Chiang V, Fyne E, Balakrishnan M, Barnes T, Graupe M, et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014;10(4):e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cillo AR, Vagratian D, Bedison MA, Anderson EM, Kearney MF, Fyne E, et al. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. J Clin Microbiol. 2014;52(11):3944–3951. doi: 10.1128/JCM.02060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong F, Aga E, Cillo A, Yates AL, Besson G, Fyne E, et al. Novel assays to measure total cell-associated HIV-1 DNA and RNA. J Clin Microbiol. 2016 doi: 10.1128/JCM.02904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Team RC. R, A Language and Environment for Statistical Computing; 2013. http://www.r-project.org/ [Google Scholar]
- 38.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; New York: 2009. [Google Scholar]
- 39.Warner GR, Bolker B, Bonebakker L, Gentleman R, Huber W, Liaw A, et al. gplots: Various R Programming Tools for Plotting Data. 2016. [Google Scholar]
- 40.Kearney MF, Spindler J, Shao W, Yu S, Anderson EM, O’Shea A, et al. Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 2014;10(3):e1004010. doi: 10.1371/journal.ppat.1004010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 2007;3(4):e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siliciano JD, Siliciano RF. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 43.Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, et al. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9(5):e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, et al. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155(3):540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kearney MF, Wiegand A, Shao W, Coffin JM, Mellors JW, Lederman M, et al. Origin of Rebound Plasma HIV Includes Cells with Identical Proviruses That Are Transcriptionally Active before Stopping of Antiretroviral Therapy. J Virol. 2015;90(3):1369–1376. doi: 10.1128/JVI.02139-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothenberger MK, Keele BF, Wietgrefe SW, Fletcher CV, Beilman GJ, Chipman JG, et al. Large number of rebounding/founder HIV variants emerge from multifocal infection in lymphatic tissues after treatment interruption. Proc Natl Acad Sci U S A. 2015;112(10):E1126–1134. doi: 10.1073/pnas.1414926112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill AL, Rosenbloom DI, Fu F, Nowak MA, Siliciano RF. Predicting the outcomes of treatment to eradicate the latent reservoir for HIV-1. Proc Natl Acad Sci U S A. 2014;111(37):13475–13480. doi: 10.1073/pnas.1406663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Henrich TJ, Hanhauser E, Marty FM, Sirignano MN, Keating S, Lee TH, et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med. 2014;161(5):319–327. doi: 10.7326/M14-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henrich TJ, Hu Z, Li JZ, Sciaranghella G, Busch MP, Keating SM, et al. Long-Term Reduction in Peripheral Blood HIV Type 1 Reservoirs Following Reduced-Intensity Conditioning Allogeneic Stem Cell Transplantation. The Journal of Infectious Diseases. 2013;207(11):1694–1702. doi: 10.1093/infdis/jit086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Luzuriaga K, Gay H, Ziemniak C, Sanborn KB, Somasundaran M, Rainwater-Lovett K, et al. Viremic relapse after HIV-1 remission in a perinatally infected child. N Engl J Med. 2015;372(8):786–788. doi: 10.1056/NEJMc1413931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Persaud D, Luzuriaga K. Absence of HIV-1 after treatment cessation in an infant. N Engl J Med. 2014;370(7):678. doi: 10.1056/NEJMc1315498. [DOI] [PubMed] [Google Scholar]
- 52.Persaud D, Gay H, Ziemniak C, Chen YH, Piatak M, Jr, Chun TW, et al. Absence of detectable HIV-1 viremia after treatment cessation in an infant. N Engl J Med. 2013;369(19):1828–1835. doi: 10.1056/NEJMoa1302976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, et al. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. AIDS. 2016;30(3):343–353. doi: 10.1097/QAD.0000000000000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams JP, Hurst J, Stohr W, Robinson N, Brown H, Fisher M, et al. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife. 2014;3:e03821. doi: 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.