Abstract
BACKGROUND
The majority of cleft lip with or without cleft palate (CL/P) cases appear as an isolated, non-syndromic entity (NSCLP). With the advent of next generation sequencing, whole exome sequencing (WES) has been used to identify single nucleotide variants and insertion/deletions which cause or increase risk of NSCLP. However, to our knowledge, there are no published studies using WES in NSCLP to investigate copy number changes (CNCs), which are a major component of human genetic variation. Our study aimed to identify CNCs associated with NSCLP in a Honduran population using WES.
METHODS
WES was performed on two to four members of 27 multiplex Honduran families. CNCs were identified using two algorithms, CoNIFER and XHMM. Priority was given to CNCs that were identified in more than one patient and had variant frequencies of less than 5% in reference data sets.
RESULTS
WES completion was defined as >90% of the WES target at >8× coverage and >80% of the WES target at >20× coverage. 24 CNCs that met our inclusion criteria were identified by both CoNIFER and XHMM. These CNCs were confirmed using qPCR. Pedigree analysis produced three CNCs corresponding to ADH7, AHR, and CRYZ segregating with NSCLP. Two of the three CNCs implicate genes, AHR and ADH7, whose known biological functions could plausibly play a role in NSCLP.
CONCLUSIONS
WES can be used to detect candidate CNCs that may be involved in the pathophysiology of NSCLP.
Keywords: cleft lip, cleft palate, copy number changes, copy number variants, whole exome sequencing
Introduction
Cleft lip with or without palate (CL/P) is one of the most common birth defects worldwide, and is associated with major comorbidities, including feeding difficulties, speech delay, impaired hearing, dental problems, and psychiatric disease. CL/P occurs in approximately 1 in every 700 live births worldwide, but the prevalence may be as high as 1 in 500 in Asian and Amerindian populations (Dixon, 2011; Marazita, 2012). CL/P may occur as part of a syndrome, but the majority of cases (70%) appear as an isolated entity, nonsyndromic cleft lip with or without palate (NSCLP) (Calzolari, 2007).
It has long been thought that risk of developing NSCLP has a large genetic component (Dixon, 2011). However, identifying the specific genetic factors underlying risk for NSCLP has proven challenging for several reasons. For example, inheritance of NSCLP often departs from traditional Mendelian modes of inheritance and, in some families, penetrance appears to be incomplete (Beiraghi, 2007; Wyszynski, 2002). Additionally, assigning affectation status can be difficult because subclinical features, such as orbicularis oris defects, are part of the phenotypic spectrum of NSCLP (Neiswanger, 2007).
Prior targeted genetic studies have identified multiple candidate loci, including IRF6, TGFA, RARA, and TGFB3 (Lidral and Moreno, 2005). Genome-wide linkage scans and genome-wide association studies have revealed additional candidate genes and susceptibility loci (Beaty, 2010; Lidral and Moreno, 2005; Ludwig, 2012; Mangold, 2010; Prescott, 2000; Wyszynski, 2003). In addition, environmental factors – such as smoking (Honein, 2007; Little, 2004; Zeiger, 2005), alcohol use (Chevrier, 2005; Romitti, 2007; Shaw and Lammer, 1999), nutrition, teratogens, and viral infections – also influence disease risk (Dixon, 2011; Mossey, 2009; Murray, 2002). These environmental factors may interact with genetic variants to modulate the risk of developing orofacial clefts. Such gene-environment interactions have been demonstrated with maternal smoking and TGFA (Zeiger, 2005), maternal folic acid consumption and MTHFR (van Rooij, 2003), and maternal multivitamin use and NAT1 (Lammer, 2004).
The advent of next-generation sequencing has accelerated the discovery of new loci underlying Mendelian conditions and birth defects. Whole exome sequencing (WES) offers an efficient and powerful method for detecting pathogenic mutations, as exons comprise only 1% of the human genome but harbor 85% of disease-causing mutations (Choi, 2009). WES approaches have identified pathogenic single nucleotide variants (SNVs) and small insertions and deletions (indels) in NSCLP (Bureau, 2014; Jezewski, 2003; Vieira, 2005). However, fewer studies have investigated the role of copy number changes (CNCs) in NSCLP.
CNCs refer to portions of the genome present in variable number of copies between individuals or in comparison to a reference genome. CNCs have been defined at lengths of at least 50 base pairs (bp) to at least one kilobase (kb) (Feuk, 2006; Zarrei, 2015). In 2004, two genome-wide studies revealed that CNCs are prevalent in human genomes and are an important source of genetic and phenotypic diversity (Iafrate, 2004; Sebat, 2004). Since then, CNCs have been studied and implicated in a wide breadth of human diseases including autism, schizophrenia, obesity, type 1 diabetes, and various developmental disorders (Zarrei, 2015).
In the early years of CNC investigation, the predominant methods of CNC detection included fluorescent in situ hybridization, array comparative genomic hybridization, and single nucleotide polymorphism (SNP) arrays. These methods could detect variants several kb to megabases in size (Stankiewicz and Lupski, 2010). With such methods, prior studies of CNCs in NSCLP have identified candidate regions (such as 7p14.1) or candidate genes (such as SUMO1, BMP2, and CLPTM1L), some of which have been validated in animal studies (Klamt, 2016; Sahoo, 2011; Shi, 2009; Williams, 2012; Younkin, 2014). In recent years, algorithms for calling CNCs from WES data have been developed, allowing for identification of CNCs as small as 50 bp (Fromer, 2012; Krumm, 2012; Tan, 2014). To our knowledge, no prior studies have employed WES for this purpose in NSCLP.
Herein, we use WES to identify CNCs in a cohort of multiplex Honduran families with NSCLP. Studying multiplex families — those in which two or more persons have the same phenotype — increases the likelihood of finding alleles with larger effects that underlie NSCLP. Furthermore, it is advantageous to study the Honduran population because of their increased rate of clefting (given their Mesoamerican ancestry) and their relative genetic isolation due to limited influx of other ethnic populations (Dixon, 2011; Moreno-Estrada, 2013).
Materials and Methods
SAMPLES
Subjects were identified from patients at Hospital Escuela, a public hospital in Tegucigalpa, Honduras between 2001 and 2013. We recruited over 130 families with two or more members affected by NSCLP, although not all affected members were present to participate in this study. A medical history, family history, and physical exam were performed to characterize the type of cleft, exclude syndromic conditions, and construct pedigrees. Venous blood samples were obtained from both probands and available relatives. This study was approved by the Institutional Review Boards at Columbia University Medical Center (CUMC) and Hospital Escuela in Tegucigalpa, Honduras.
GENOTYPING AND WHOLE EXOME SEQUENCING
We selected 27 multiplex families with NSCLP for analysis, prioritizing families with DNA samples available for 2 or more affected individuals. The subjects included 52 affected individuals and 139 relatives. DNA was extracted from whole blood samples using the Qiagen Flexigene DNA kit (Qiagen, Valencia, CA). Two families were sequenced at Columbia University. The remaining samples were sent to the University of Washington Center for Mendelian Genomics (UW-CMG) in Seattle, Washington for sequencing. For sample quality control (QC), 191 subjects were genotyped using Illumina’s Human Core Exome BeadChip. Variants missing >5% of genotypes were excluded.
PLINK v1.90 was used to confirm pedigree relationships using Mendelian error checking (http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell, 2007). The BeadChip data were then used to estimate relationships using Kinship-based INference for Genome-wide association studies (http://people.virginia.edu/#wc9c/KING/) (Manichaikul, 2010). QC included verification of sample/pedigree relationships. When discrepancies were observed, we collected new blood samples for confirmation testing. In cases where correction was not possible, QC led to the exclusion of two complete families as well as select individuals from three other families (one individual each from two families and two siblings from another). All other samples from affected individuals underwent WES. In four families, an unaffected member was included for variant phasing, bringing the total number of samples to 59 that underwent WES.
Exome capture was performed using Perkin-Elmer Janus II in 96-well plate format and Roche/Nimblegen SeqCap EZ at UW-CMG, as has been previously described (http://uwcmg.org/#/instruction) (Aylward, 2016). Library concentration was determined using quantitative PCR. An Illumina HiSeq sequencer was used to massively parallel sequence samples and generate base calls. Unaligned BAM files were created using Picard Extract Illumina Barcodes and IlluminaBasecallsToSam and aligned to human reference GRCh37/hg19 using the Burrows-Wheeler Aligner v0.6.2 (Li and Durbin, 2009). QC measures were performed according to UW-CMG protocol (http://uwcmg.org/#/instruction) (Aylward, 2016). Briefly, WES completion was defined as >90% of the WES target at >8× coverage and >80% of the WES target at >20× coverage.
CNC CALLING
CNC variant calling was performed using a large set of reference WES data from the UW-CMG (N=6085) and the NHLBI GO Exome Sequencing Project (ESP; N=3635) to reduce noise and improve calling quality (Fromer, 2012; Krumm, 2012). Because there is no gold standard for CNC calling using WES data, we restricted analysis to events detected by both XHMM v1.0 (Fromer, 2012) and CoNIFER v.02.2 (Krumm, 2012) to decrease the number of false positives. The default software parameters were used for both tools. BED files from the two callsets were generated and Bedtools intersect was used to extract the intersecting calls (http://bedtools.readthedocs.org/en/latest/content/tools/intersect.html). Intersecting calls were defined as CNCs identified by both algorithms that shared the same genomic sections for 50% or more of their length.
CNC VALIDATION
Quantitative PCR (qPCR) was used to validate each CNC. Primers were designed using Primer3 (Rozen and Skaletsky, 2000) (Supplementary Table 1) and qPCR was performed on an Applied Biosystems 7500 Real Time PCR Instrument. Each sample was analyzed in triplicate, either in 25 µl or 50 µl reaction mixture, using SYBR Green I master mix (Roche Molecular Biochemicals), 200 nM of each primer, and 20 ng of genomic DNA. The default conditions supplied by the manufacturer (Applied Biosystems) were used for amplification. Data were normalized against the reference gene beta actin and relative gene expression was determined using the Livak method (Livak and Schmittgen, 2001). Each qPCR experiment was performed in triplicate for each suspected CNC carrier, along with three control samples selected at random from a cohort of 100 healthy Honduran pediatric patients undergoing minor surgical procedures and not affected by clefting or other known genetic diseases. Relative gene expression values of ≥1.4 and ≤0.6 were considered evidence of duplication and deletion, respectively (Supplementary Table 1). For gene expression values between 0.6 and 1.4, we ruled out the corresponding CNCs as WES calling errors.
In addition, the population frequencies of the CNCs shown to segregate with NSCLP were calculated. Allele frequencies for CNCs were estimated by identifying events of the same type (duplication vs. deletion) with a minimum overlap of 50% of the observed CNC using Bedtools (v2.25.0) within reference data sets. Reference data included the 1000 genomes project integrated structural variant map (Sudmant, 2015) and the Exome Aggregation Consortium (ExAC) release 0.3.1 data (Ruderfer, 2016).
Results
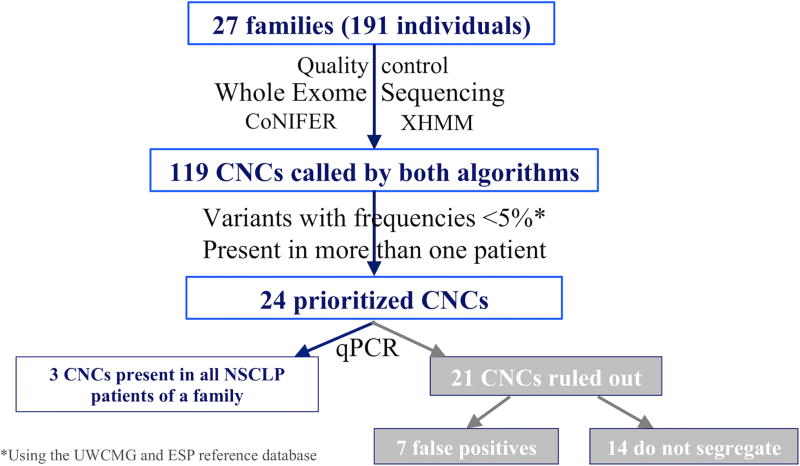
CNC events observed in the WES data from multiplex Honduran families with a history of NSCLP were prioritized for validation as outlined in Figure 1.
FIGURE 1.
Summary of CNC calling, prioritization, and confirmation.
XHMM identified 2545 CNCs while CoNIFER identified 1168 CNCs. After intersecting both call sets, 119 CNCs were identified by both XHMM and CoNIFER. To prioritize the CNCs most likely to be pathogenic, CNCs were filtered to focus on those identified in more than one patient and with variant frequencies <5% in the UW-CMG (N= 6085 individuals) and Exome Sequencing Project (ESP, N=3635 individuals) reference data sets. This narrowed the list of CNCs of interest from 119 to 24 CNCs. These 24 CNCs ranged in size from approximately 4 kb to 226 kb and the genes encompassed by the CNCs are listed in Table 1. After qPCR verification, seven CNCs were ruled out as CNC calling errors and 14 were excluded as they were not present in all patients affected by NSCLP within a pedigree (Figure 1).
TABLE 1.
Twenty-four CNCs identified by both XHMM and CoNIFER and present in more than one NSCLP patient.
| Gene | Event type |
Length (kb) |
Chromosome | Start/End coordinates* |
Families | qPCR testing |
Pedigree analysis** |
CoNIFER Scores*** |
XHMM Scores*** |
|---|---|---|---|---|---|---|---|---|---|
| ODF2L | Deletion | 4.79 | 1 | 86847923 – 86852712 | M28 | TRUE | Does not segregate | ||
| KIFAP3 | Deletion | 56.42, 60.35 | 1 | 169947224 – 170003639 169947224 – 170007574 | M45, M94 | FALSE | |||
| HOOK1 | Deletion | 2.14, 8.89 | 1 | 60305970 – 60314857 60312716 – 60314857 | M45 | FALSE | |||
| CRYZ | Deletion | 10.27 | 1 | 75180237 – 75190507 | M1 | TRUE | Segregates | −1.71 – −1.64 | −7.0 – −7.4 |
| WDPCP | Deletion | 50.1 | 2 | 63664553 – 63714656 | M55, M62 | FALSE | |||
| APLF | Deletion | 10.95 | 2 | 68729861 – 68740814 | M28, M62 | FALSE | |||
| FSIP2**** | Deletion | 8.82 | 2 | 186397377 – 186406199 | M55, M62 | TRUE | Does not segregate | ||
| ARPP21 | Deletion | 9.26 | 3 | 35723242 – 35732499 | M28, M62 | TRUE | Does not segregate | ||
| CACNA1D | Duplication | 226.17 | 3 | 53699685 – 53925858 | M71 | TRUE | Does not segregate | ||
| ADH7 | Duplication | 7.69 | 4 | 100334266 – 100341952 | M1 | TRUE | Segregates | 1.72 | 4.56 – 5.93 |
| ZNF608 | Duplication | 11.85 | 5 | 123973545 – 123985398 | M55 | TRUE | Does not segregate | ||
| WDR36 | Deletion | 6.86, 9.38, 20.56 | 5 | 110436286 – 110443138 110436286 – 110456839 110439483 – 110448852 | M28, M45, M46 | TRUE | Does not segregate | ||
| UFL1 | Deletion | 14.15, 14.56, 15.69, 19.27 | 6 | 96982117 – 97001381 96985248 – 96999808 96985248 – 96999401 96984118 – 96999808 | M17, M28, M55, M62 | FALSE | |||
| TRDN | Deletion | 31.02, 33.88 | 6 | 123539745 – 123573621 123542598 – 123573621 | M28, M62 | FALSE | |||
| NKAIN2 | Duplication | 165.09 | 6 | 124979330 – 125144422 | M46 | TRUE | Does not segregate | ||
| CACNA2D1 | Deletion | 4.74, 10.53 | 7 | 81593347 – 81603871 81596453 – 81601188 | M45, M62 | TRUE | Does not segregate | ||
| AHR | Deletion | 13.3, 23.6 | 7 | 17349559 – 17843203 17362123 – 17841282 17367382 – 17841282 | M45 | TRUE | Segregates | −0.735 – −0.517 | −2.76 |
| DDX18**** | Deletion | 133.09 | 9 | 118289457 – 118422544 | M28, M62 | TRUE | Does not segregate | ||
| DHTKD1 | Duplication | 36.35, 39.42, 51.86 | 10 | 12123469 – 12162889 12126537 – 12162889 12111031 – 12162889 | M16, M67 | TRUE | Does not segregate | ||
| FAM76B | Deletion | 8.4, 11.71 | 11 | 95504718 – 95513118 95504718 – 95516430 | M28, M45 | FALSE | |||
| PCNX | Duplication | 147.71 | 14 | 71428941 – 71576654 | M94 | TRUE | Does not segregate | ||
| VPS13C | Deletion | 32.2, 35.63 | 15 | 62304283 – 62336484 62300852 – 62336484 | M28, M45 | TRUE | Does not segregate | ||
| CGNL1 | Duplication | 23.9 | 15 | 57730196 – 57754092 | M53, M71 | TRUE | Does not segregate | ||
| C18orf54 | Deletion | 8.59, 11.53 | 18 | 57730196 – 57754092 | M28, M45 | TRUE | Does not segregate |
The CNC details and families in which they were called are listed.
Start and end coordinates from XHMM are listed.
Segregation of a CNC with NSCLP is defined as presence of the CNC in all family members affected by NSCLP within a family.
The CoNIFER score represents the median signal strength of the calls (median_svdzrpkm). The XHMM score shows the mean normalized read depth z-scores over the interval.
For CNC calls corresponding to intergenic regions, the gene for the closest exon is listed.
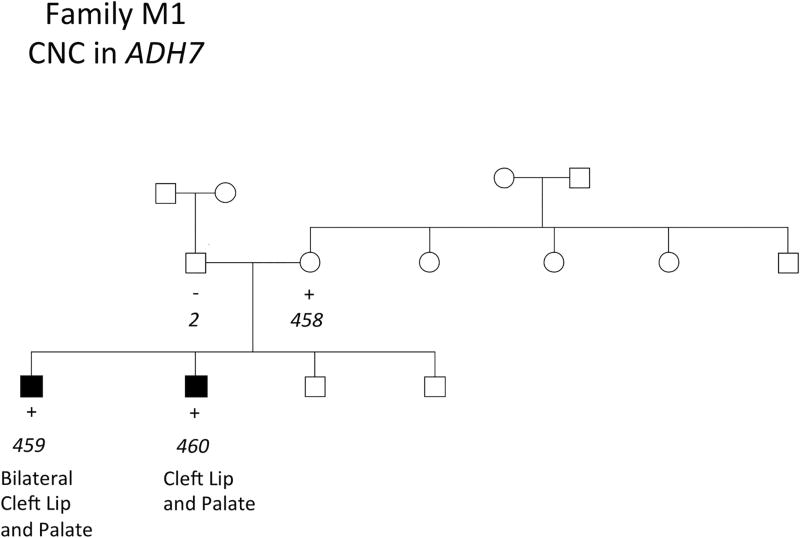
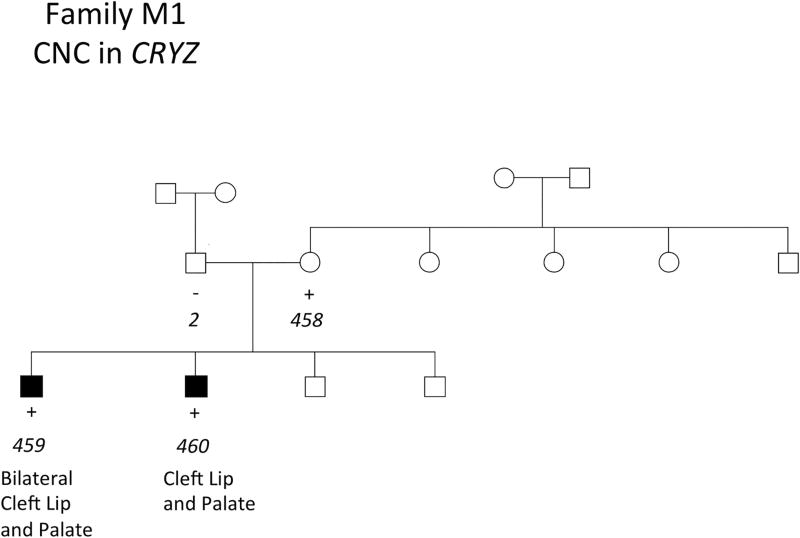
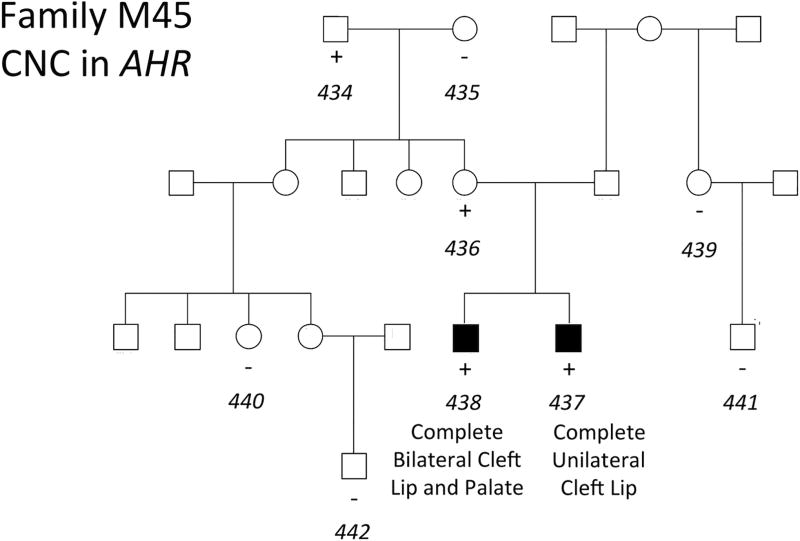
The three remaining CNCs were present in all members of a pedigree affected by NSCLP. Two of these CNCs were identified in a single family, observed in two affected brothers and their unaffected mother (Figures 2 and 3). These CNCs were a duplication event of 7.7 kb on chromosome 4 and a deletion event of 10.3 kb on chromosome 1 corresponding to ADH7 and CRYZ, respectively (Table 1). A CNC in AHR was identified in another family, observed in two affected brothers, their unaffected mother, and unaffected grandfather (Figure 4). This was a deletion event of 13.3 kb and 23.6 kb in the affected brothers. The genomic contexts and population frequencies of these three CNCs are shown in Figure 5 and Table 2, respectively. These CNCs appear to fit inside single genes and there are other events intersecting our variants in an online repository of CNCs with phenotypic information, DECIPHER (https://decipher.sanger.ac.uk) (Swaminathan, 2012). However, other than for the CNC associated with ADH7 (described in the Discussion section), none of the overlapping CNCs correspond to a phenotype of cleft lip or palate.
FIGURE 2.
Presence of CNCs in ADH7 in NSCLP patients in a Honduran family. Key: + = positive for a CNC in ADH7. DNA was not available for subjects without + or − denoted. Sample identification (ID) numbers are italicized and correspond to Supplementary Table 1.
FIGURE 3.
Presence of CNCs in CRYZ in NSCLP patients in family M1. Key: + = positive for a CNC in CRYZ. Sample ID numbers are italicized and correspond to Supplementary Table 1.
FIGURE 4.
Presence of CNCs in AHR with NSCLP in family M45. Key: + = positive for a CNC in AHR. Sample ID numbers are italicized and correspond to Supplementary Table 1.
FIGURE 5.

Genomic contexts of the CNCs identified in NSCLP patients. This figure was generated using UCSC Genome Browser. The genomic regions spanned by the CNCs corresponding to CRYZ (panel A), ADH7 (panel B), and AHR (panel C) are shown and represented by the black bar(s) labeled “Chromosome coordinates list.” The boundaries of each CNC are indicated by the red box on the chromosome ideogram and the encompassed RefSeq genes are shown. In addition, other CNVs that have been previously reported in DECIPHER are shown in the red (deletion/loss) or blue (duplication/gain) bars.
TABLE 2.
Population frequencies of CNCs in the genes CRYZ, ADH7, and AHR that segregated with NSCLP in a cohort of Honduran patients.
| 1000 Genomes Frequencies | ExAC frequencies | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Event | AFR | AMR | EAS | EUR | SAS | AFR | AMR | EAS | FIN | NFE | OTH | SAS |
| CRYZ | deletion | 0/1322 | 2/694 | 0/1008 | 0/1006 | 0/978 | 0/10406 | 1/11578 | 0/8654 | 0/6614 | 0/66740 | 0/908 | 0/16512 |
| ADH7 | duplication | 0/1322 | 0/694 | 0/1008 | 0/1006 | 0/978 | 6/10406 | 0/11578 | 0/8654 | 0/6614 | 1/66740 | 0/908 | 0/16512 |
| AHR | deletion | 0/1322 | 0/694 | 0/1008 | 0/1006 | 0/978 | 0/10406 | 0/11578 | 0/8654 | 0/6614 | 2/66740 | 0/908 | 0/16512 |
Allele frequencies are presented as the number of observations over the maximum number of chromosomes within a data set, as exact numbers of chromosomes observed at a given position were not provided in the ExAC data release. These values should then be considered minimum frequencies in the reference data set. Subpopulations include African (AFR), American (AMR), East Asian (EAS), European (EUR), Finnish (FIN), Non-Finnish European (NFE), other (OTH), and South Asian (SAS) populations.
Discussion
This study identifies candidate CNCs for NSCLP using WES technology. By utilizing this technique on a cohort of multiplex families affected by NSCLP, we identified CNCs of potential significance corresponding to the genes ADH7 (formerly referred to as ADH3), AHR, and CRYZ using our CNC filtering and prioritization criteria. Two of these genes, ADH7 and AHR, share involvement in biological pathways linked to environmental factors known to influence NSCLP.
A CNC in ADH7 was identified in two affected siblings as well as their unaffected mother. Interestingly, prior analysis of WES data in this cohort did not identify causal single nucleotide variants, insertions, or deletions that segregated with NSCLP in this family (M1) (Aylward, 2016). ADH7 encodes a member of the alcohol dehydrogenase family, class IV alcohol dehydrogenase, expressed ubiquitously during embryogenesis (Molotkov, 2002). Compared to other members of its class, it is less efficient in ethanol oxidation and most active as a retinol dehydrogenase (Satre, 1994). Thus, ADH7 may participate in the synthesis of retinoic acid, the active form of vitamin A. Retinoic acid plays an important role in cellular differentiation (Niederreither and Dolle, 2008) and is a well-established cause of cleft palate (Abbott and Birnbaum, 1990; Abbott, 1989b). Furthermore, prior studies of CL/P identified a significant association with a locus in its receptor, retinoic acid receptor (RARA) (Chenevix-Trench, 1992; Shaw, 1993). There are no published studies linking ADH7 to orofacial clefts, but there are additional cases with CNCs involving ADH7 linked to CL/P in DECIPHER (Swaminathan, 2012). A duplication event of 8.08 Mb was associated with a non-midline cleft lip in one patient (DECIPHER ID: 270855) and a deletion event of 5.81 Mb was associated with bilateral CL/P in the other patient (DECIPHER ID: 285906) (Figure 5). Both patients had other related disorders, suggesting these presentations of CL/P were syndromic. This CNC has also been identified in a Non-Finnish European population in ExAC at a frequency of <0.001% (Table 2).
CNCs in AHR were identified in two affected siblings as well as their unaffected mother and maternal grandfather, consistent with an autosomal dominant mode of inheritance with incomplete penetrance. AHR encodes the arylhydrocarbon receptor (AHR), which is expressed in the developing mouse palate and upregulated early in palatogenesis (Abbott, 1999). This receptor mediates the toxicities of aromatic hydrocarbons that may result in teratogenesis, cancers, and birth defects (Nebert, 2004; Whitlock, 1990). Hydrocarbons bind to AHR and activate downstream signaling pathways resulting in various toxicities (Landers and Bunce, 1991; Poland and Knutson, 1982). Of particular relevance to NSCLP, the aromatic hydrocarbon dioxin (2,3,7,8-tetrachlorodibenzo-p-dioxin) is a ligand of AHR and has been shown to induce cleft palate in pregnant mice (Abbott, 1989a; Pratt, 1984; Takagi, 2000). Interestingly, retinoic acid is involved in and necessary for the development of dioxin-induced cleft palate in mice by regulating AHR expression (Jacobs, 2011). There is considerable support for a role for AHR in cleft palate development in mice, but it may have less significance to cleft palate development in humans. In one study, AHR mRNA in human embryos was found at levels 350-fold less than in mouse embryos (Abbott, 1999). In addition, humans may be less sensitive to the pathogenic effects of dioxin than are mice (Moriguchi, 2003). Induction of cleft palate using dioxin required a much higher concentration of dioxin in human embryos than in mouse embryos (Abbott and Birnbaum, 1991). Nonetheless, CNCs in AHR warrant further investigation given their segregation with NSCLP in one of our multiplex families and the gene’s overall association with craniofacial development. Furthermore, SNPs in AHR’s cofactor, AHR nuclear translocator (ARNT), have been associated with nonsyndromic orofacial clefts in the Japanese population (Kayano, 2004). From our literature search, this CNC does not overlap with those that have been previously reported in association with NSCLP. This CNC has been identified in the African subpopulation in ExAC at a frequency of 0.06% and the Non-Finnish European subpopulation at frequency <0.01% (Table 2).
We also identified a CNC in CRYZ present in patients affected by NSCLP. The pedigree corresponding to CRYZ suggests an autosomal dominant inheritance pattern with incomplete penetrance in family M1 (Figure 3). It worth noting that members in this same family also had CNCs in ADH7. This finding of multiple candidate variants within the same patient and/or family has also been demonstrated by prior studies of SNVs or indels in our Honduran families (Aylward, 2016) and of CNCs in other cohorts (Simioni, 2015). These findings would be consistent with the polygenic nature of NSCLP and incomplete penetrance. However, there is less evidence supporting a major role for this gene in the pathogenesis of NSLCP. CRYZ encodes crystallin zeta, a quinone reductase expressed in the eye lens of vertebrates (Gonzalez, 1994). Its biologic function in humans is less well established, though a GWAS associated CRYZ with regulation of resistin, a hormone implicated in diabetes in cardiovascular disease (Qi, 2012). Our literature search did not find reports of an association of CRYZ with NSCLP, though this CNC has been identified in the American subpopulation in 1000 Genomes project and ExAC at frequencies of 0.29% and 0.09%, respectively (Table 2).
Conclusion
We have identified candidate CNCs that rank highly based on prioritization criteria in three separate genes. Two of these genes, ADH7 and AHR, play roles in craniofacial development. Interestingly, both ADH7 and AHR interact with environmental factors, retinoic acid and dioxin, respectively, both of which have well-established roles in clefting. Specifically, ADH7 participates in retinoic acid synthesis and retinoic acid regulates the expression of AHR. Our preliminary results suggest that these candidate genes identified via CNC analysis in exome sequencing data warrant further investigation as risk factors for NSCLP. Replication of these findings in a larger cohort of families with NSCLP or a different population is necessary to confirm or refute their role in the pathogenesis of NSCLP.
Supplementary Material
Acknowledgments
We thank the staff at Hospital Escuela, Tegucigalpa, Honduras and the Honduran Medical Institute for their assistance in identifying patients and obtaining samples. Sequencing was provided by the University of Washington Center for Mendelian Genomics (UW-CMG) and was funded by the National Human Genome Research Institute and the National Heart, Lung and Blood Institute grant 2UM1HG006493 to Drs. Debbie Nickerson, Michael Bamshad, and Suzanne Leal.
This study makes use of data generated by the DECIPHER Consortium. A full list of centres who contributed to the generation of the data is available from https://decipher.sanger.ac.uk/ and via email from decipher@sanger.ac.uk. Funding for the project was provided by the Wellcome Trust. Those who carried out the original analysis and collection of the Data bear no responsibility for the further analysis or interpretation of it by the Recipient or its Registered Users.
The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009-003], a parallel funding partnership between the Wellcome Trust and the Department of Health, and the Wellcome Trust Sanger Institute [grant number WT098051]. The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network.
Footnotes
Conflicts of interest and financial disclosures: None to disclose.
References
- Abbott BD, Birnbaum LS. Retinoic acid-induced alterations in the expression of growth factors in embryonic mouse palatal shelves. Teratology. 1990;42(6):597–610. doi: 10.1002/tera.1420420604. [DOI] [PubMed] [Google Scholar]
- Abbott BD, Birnbaum LS. TCDD exposure of human embryonic palatal shelves in organ culture alters the differentiation of medial epithelial cells. Teratology. 1991;43(2):119–132. doi: 10.1002/tera.1420430205. [DOI] [PubMed] [Google Scholar]
- Abbott BD, Diliberto JJ, Birnbaum LS. 2,3,7,8-Tetrachlorodibenzo-p-dioxin alters embryonic palatal medial epithelial cell differentiation in vitro. Toxicology and applied pharmacology. 1989a;100(1):119–131. doi: 10.1016/0041-008x(89)90096-3. [DOI] [PubMed] [Google Scholar]
- Abbott BD, Harris MW, Birnbaum LS. Etiology of retinoic acid-induced cleft palate varies with the embryonic stage. Teratology. 1989b;40(6):533–553. doi: 10.1002/tera.1420400602. [DOI] [PubMed] [Google Scholar]
- Abbott BD, Held GA, Wood CR, Buckalew AR, Brown JG, Schmid J. AhR, ARNT, and CYP1A1 mRNA quantitation in cultured human embryonic palates exposed to TCDD and comparison with mouse palate in vivo and in culture. Toxicological sciences : an official journal of the Society of Toxicology. 1999;47(1):62–75. doi: 10.1093/toxsci/47.1.62. [DOI] [PubMed] [Google Scholar]
- Aylward A, Cai Y, Lee A, Blue E, Rabinowitz D, Haddad J, Jr University of Washington Center for Mendelian G. Using Whole Exome Sequencing to Identify Candidate Genes With Rare Variants In Nonsyndromic Cleft Lip and Palate. Genetic epidemiology. 2016;40(5):432–441. doi: 10.1002/gepi.21972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, Liang KY, Wu T, Murray T, Fallin MD, Redett RA, Raymond G, Schwender H, Jin SC, Cooper ME, Dunnwald M, Mansilla MA, Leslie E, Bullard S, Lidral AC, Moreno LM, Menezes R, Vieira AR, Petrin A, Wilcox AJ, Lie RT, Jabs EW, Wu-Chou YH, Chen PK, Wang H, Ye X, Huang S, Yeow V, Chong SS, Jee SH, Shi B, Christensen K, Melbye M, Doheny KF, Pugh EW, Ling H, Castilla EE, Czeizel AE, Ma L, Field LL, Brody L, Pangilinan F, Mills JL, Molloy AM, Kirke PN, Scott JM, Arcos-Burgos M, Scott AF. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nature genetics. 2010;42(6):525–529. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beiraghi S, Nath SK, Gaines M, Mandhyan DD, Hutchings D, Ratnamala U, McElreavey K, Bartoloni L, Antonarakis GS, Antonarakis SE, Radhakrishna U. Autosomal dominant nonsyndromic cleft lip and palate: significant evidence of linkage at 18q21.1. American journal of human genetics. 2007;81(1):180–188. doi: 10.1086/518944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau A, Parker MM, Ruczinski I, Taub MA, Marazita ML, Murray JC, Mangold E, Noethen MM, Ludwig KU, Hetmanski JB, Bailey-Wilson JE, Cropp CD, Li Q, Szymczak S, Albacha-Hejazi H, Alqosayer K, Field LL, Wu-Chou YH, Doheny KF, Ling H, Scott AF, Beaty TH. Whole exome sequencing of distant relatives in multiplex families implicates rare variants in candidate genes for oral clefts. Genetics. 2014;197(3):1039–1044. doi: 10.1534/genetics.114.165225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzolari E, Pierini A, Astolfi G, Bianchi F, Neville AJ, Rivieri F. Associated anomalies in multi-malformed infants with cleft lip and palate: An epidemiologic study of nearly 6 million births in 23 EUROCAT registries. American journal of medical genetics Part A. 2007;143A(6):528–537. doi: 10.1002/ajmg.a.31447. [DOI] [PubMed] [Google Scholar]
- Chenevix-Trench G, Jones K, Green AC, Duffy DL, Martin NG. Cleft lip with or without cleft palate: associations with transforming growth factor alpha and retinoic acid receptor loci. American journal of human genetics. 1992;51(6):1377–1385. [PMC free article] [PubMed] [Google Scholar]
- Chevrier C, Perret C, Bahuau M, Nelva A, Herman C, Francannet C, Robert-Gnansia E, Cordier S. Interaction between the ADH1C polymorphism and maternal alcohol intake in the risk of nonsyndromic oral clefts: an evaluation of the contribution of child and maternal genotypes. Birth defects research Part A, Clinical and molecular teratology. 2005;73(2):114–122. doi: 10.1002/bdra.20103. [DOI] [PubMed] [Google Scholar]
- Choi M, Scholl UI, Ji W, Liu T, Tikhonova IR, Zumbo P, Nayir A, Bakkaloglu A, Ozen S, Sanjad S, Nelson-Williams C, Farhi A, Mane S, Lifton RP. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(45):19096–19101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon MJ, Marazita ML, Beaty TH, Murray JC. Cleft lip and palate: understanding genetic and environmental influences. Nature reviews Genetics. 2011;12(3):167–178. doi: 10.1038/nrg2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nature reviews Genetics. 2006;7(2):85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, Handsaker RE, McCarroll SA, O'Donovan MC, Owen MJ, Kirov G, Sullivan PF, Hultman CM, Sklar P, Purcell SM. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. American journal of human genetics. 2012;91(4):597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez P, Rao PV, Zigler JS., Jr Organization of the human zeta-crystallin/quinone reductase gene (CRYZ) Genomics. 1994;21(2):317–324. doi: 10.1006/geno.1994.1272. [DOI] [PubMed] [Google Scholar]
- Honein MA, Rasmussen SA, Reefhuis J, Romitti PA, Lammer EJ, Sun L, Correa A. Maternal smoking and environmental tobacco smoke exposure and the risk of orofacial clefts. Epidemiology. 2007;18(2):226–233. doi: 10.1097/01.ede.0000254430.61294.c0. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nature genetics. 2004;36(9):949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Jacobs H, Dennefeld C, Feret B, Viluksela M, Hakansson H, Mark M, Ghyselinck NB. Retinoic acid drives aryl hydrocarbon receptor expression and is instrumental to dioxin-induced toxicity during palate development. Environmental health perspectives. 2011;119(11):1590–1595. doi: 10.1289/ehp.1003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jezewski PA, Vieira AR, Nishimura C, Ludwig B, Johnson M, O'Brien SE, Daack-Hirsch S, Schultz RE, Weber A, Nepomucena B, Romitti PA, Christensen K, Orioli IM, Castilla EE, Machida J, Natsume N, Murray JC. Complete sequencing shows a role for MSX1 in non-syndromic cleft lip and palate. Journal of medical genetics. 2003;40(6):399–407. doi: 10.1136/jmg.40.6.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayano S, Suzuki Y, Kanno K, Aoki Y, Kure S, Yamada A, Matsubara Y. Significant association between nonsyndromic oral clefts and arylhydrocarbon receptor nuclear translocator (ARNT) American journal of medical genetics Part A. 2004;130A(1):40–44. doi: 10.1002/ajmg.a.30023. [DOI] [PubMed] [Google Scholar]
- Klamt J, Hofmann A, Bohmer AC, Hoebel AK, Golz L, Becker J, Zink AM, Draaken M, Hemprich A, Scheer M, Schmidt G, Martini M, Knapp M, Mangold E, Degenhardt F, Ludwig KU. Further Evidence for Deletions in 7p14.1 Contributing to Nonsyndromic Cleft Lip with or without Cleft Palate. Birth Defects Res A. 2016;106(9):767–772. doi: 10.1002/bdra.23539. [DOI] [PubMed] [Google Scholar]
- Krumm N, Sudmant PH, Ko A, O'Roak BJ, Malig M, Coe BP, Project NES. Quinlan AR, Nickerson DA, Eichler EE. Copy number variation detection and genotyping from exome sequence data. Genome research. 2012;22(8):1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer EJ, Shaw GM, Iovannisci DM, Finnell RH. Periconceptional multivitamin intake during early pregnancy, genetic variation of acetyl-N-transferase 1 (NAT1), and risk for orofacial clefts. Birth defects research Part A. Clinical and molecular teratology. 2004;70(11):846–852. doi: 10.1002/bdra.20081. [DOI] [PubMed] [Google Scholar]
- Landers JP, Bunce NJ. The Ah receptor and the mechanism of dioxin toxicity. The Biochemical journal. 1991;276(Pt 2):273–287. doi: 10.1042/bj2760273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidral AC, Moreno LM. Progress toward discerning the genetics of cleft lip. Current opinion in pediatrics. 2005;17(6):731–739. doi: 10.1097/01.mop.0000185138.65820.7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J, Cardy A, Munger RG. Tobacco smoking and oral clefts: a meta-analysis. Bulletin of the World Health Organization. 2004;82(3):213–218. [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Ludwig KU, Mangold E, Herms S, Nowak S, Reutter H, Paul A, Becker J, Herberz R, AlChawa T, Nasser E, Bohmer AC, Mattheisen M, Alblas MA, Barth S, Kluck N, Lauster C, Braumann B, Reich RH, Hemprich A, Potzsch S, Blaumeiser B, Daratsianos N, Kreusch T, Murray JC, Marazita ML, Ruczinski I, Scott AF, Beaty TH, Kramer FJ, Wienker TF, Steegers-Theunissen RP, Rubini M, Mossey PA, Hoffmann P, Lange C, Cichon S, Propping P, Knapp M, Nothen MM. Genome-wide meta-analyses of nonsyndromic cleft lip with or without cleft palate identify six new risk loci. Nature genetics. 2012;44(9):968–971. doi: 10.1038/ng.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangold E, Ludwig KU, Birnbaum S, Baluardo C, Ferrian M, Herms S, Reutter H, de Assis NA, Chawa TA, Mattheisen M, Steffens M, Barth S, Kluck N, Paul A, Becker J, Lauster C, Schmidt G, Braumann B, Scheer M, Reich RH, Hemprich A, Potzsch S, Blaumeiser B, Moebus S, Krawczak M, Schreiber S, Meitinger T, Wichmann HE, Steegers-Theunissen RP, Kramer FJ, Cichon S, Propping P, Wienker TF, Knapp M, Rubini M, Mossey PA, Hoffmann P, Nothen MM. Genome-wide association study identifies two susceptibility loci for nonsyndromic cleft lip with or without cleft palate. Nature genetics. 2010;42(1):24–26. doi: 10.1038/ng.506. [DOI] [PubMed] [Google Scholar]
- Manichaikul A, Mychaleckyj JC, Rich SS, Daly K, Sale M, Chen WM. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26(22):2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marazita ML. The evolution of human genetic studies of cleft lip and cleft palate. Annual review of genomics and human genetics. 2012;13:263–283. doi: 10.1146/annurev-genom-090711-163729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molotkov A, Fan X, Deltour L, Foglio MH, Martras S, Farres J, Pares X, Duester G. Stimulation of retinoic acid production and growth by ubiquitously expressed alcohol dehydrogenase Adh3. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(8):5337–5342. doi: 10.1073/pnas.082093299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, Gignoux CR, Ortiz-Tello PA, Martinez RJ, Hedges DJ, Morris RW, Eng C, Sandoval K, Acevedo-Acevedo S, Norman PJ, Layrisse Z, Parham P, Martinez-Cruzado JC, Burchard EG, Cuccaro ML, Martin ER, Bustamante CD. Reconstructing the population genetic history of the Caribbean. PLoS genetics. 2013;9(11):e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi T, Motohashi H, Hosoya T, Nakajima O, Takahashi S, Ohsako S, Aoki Y, Nishimura N, Tohyama C, Fujii-Kuriyama Y, Yamamoto M. Distinct response to dioxin in an arylhydrocarbon receptor (AHR)-humanized mouse. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(10):5652–5657. doi: 10.1073/pnas.1037886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossey PA, Little J, Munger RG, Dixon MJ, Shaw WC. Cleft lip and palate. Lancet. 2009;374(9703):1773–1785. doi: 10.1016/S0140-6736(09)60695-4. [DOI] [PubMed] [Google Scholar]
- Murray JC. Gene/environment causes of cleft lip and/or palate. Clinical genetics. 2002;61(4):248–256. doi: 10.1034/j.1399-0004.2002.610402.x. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. The Journal of biological chemistry. 2004;279(23):23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- Neiswanger K, Weinberg SM, Rogers CR, Brandon CA, Cooper ME, Bardi KM, Deleyiannis FW, Resick JM, Bowen A, Mooney MP, de Salamanca JE, Gonzalez B, Maher BS, Martin RA, Marazita ML. Orbicularis oris muscle defects as an expanded phenotypic feature in nonsyndromic cleft lip with or without cleft palate. American journal of medical genetics Part A. 2007;143A(11):1143–1149. doi: 10.1002/ajmg.a.31760. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nature reviews Genetics. 2008;9(7):541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Poland A, Knutson JC. 2,3,7,8-tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annual review of pharmacology and toxicology. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- Pratt RM, Dencker L, Diewert VM. 2,3,7,8-Tetrachlorodibenzo-p-dioxin-induced cleft palate in the mouse: evidence for alterations in palatal shelf fusion. Teratogenesis, carcinogenesis, and mutagenesis. 1984;4(5):427–436. doi: 10.1002/tcm.1770040505. [DOI] [PubMed] [Google Scholar]
- Prescott NJ, Lees MM, Winter RM, Malcolm S. Identification of susceptibility loci for nonsyndromic cleft lip with or without cleft palate in a two stage genome scan of affected sib-pairs. Human genetics. 2000;106(3):345–350. doi: 10.1007/s004390051048. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, Menzaghi C, Smith S, Liang L, de Rekeneire N, Garcia ME, Lohman KK, Miljkovic I, Strotmeyer ES, Cummings SR, Kanaya AM, Tylavsky FA, Satterfield S, Ding J, Rimm EB, Trischitta V, Hu FB, Liu Y, Qi L. Genome-wide association analysis identifies TYW3/CRYZ and NDST4 loci associated with circulating resistin levels. Human molecular genetics. 2012;21(21):4774–4780. doi: 10.1093/hmg/dds300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romitti PA, Sun L, Honein MA, Reefhuis J, Correa A, Rasmussen SA. Maternal periconceptional alcohol consumption and risk of orofacial clefts. American journal of epidemiology. 2007;166(7):775–785. doi: 10.1093/aje/kwm146. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods in molecular biology. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Ruderfer DM, Hamamsy T, Lek M, Karczewski KJ, Kavanagh D, Samocha KE, Exome Aggregation C. Daly MJ, MacArthur DG, Fromer M, Purcell SM. Patterns of genic intolerance of rare copy number variation in 59,898 human exomes. Nature genetics. 2016;48(10):1107–1111. doi: 10.1038/ng.3638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T, Theisen A, Sanchez-Lara PA, Marble M, Schweitzer DN, Torchia BS, Lamb AN, Bejjani BA, Shaffer LG, Lacassie Y. Microdeletion 20p12.3 involving BMP2 contributes to syndromic forms of cleft palate. American journal of medical genetics Part A. 2011;155A(7):1646–1653. doi: 10.1002/ajmg.a.34063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satre MA, Zgombic-Knight M, Duester G. The complete structure of human class IV alcohol dehydrogenase (retinol dehydrogenase) determined from the ADH7 gene. The Journal of biological chemistry. 1994;269(22):15606–15612. [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, Navin N, Lucito R, Healy J, Hicks J, Ye K, Reiner A, Gilliam TC, Trask B, Patterson N, Zetterberg A, Wigler M. Large-scale copy number polymorphism in the human genome. Science. 2004;305(5683):525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Shaw D, Ray A, Marazita M, Field L. Further evidence of a relationship between the retinoic acid receptor alpha locus and nonsyndromic cleft lip with or without cleft palate (CL +/− P) American journal of human genetics. 1993;53(5):1156–1157. [PMC free article] [PubMed] [Google Scholar]
- Shaw GM, Lammer EJ. Maternal periconceptional alcohol consumption and risk for orofacial clefts. The Journal of pediatrics. 1999;134(3):298–303. doi: 10.1016/s0022-3476(99)70453-1. [DOI] [PubMed] [Google Scholar]
- Shi M, Mostowska A, Jugessur A, Johnson MK, Mansilla MA, Christensen K, Lie RT, Wilcox AJ, Murray JC. Identification of microdeletions in candidate genes for cleft lip and/or palate. Birth defects research Part A, Clinical and molecular teratology. 2009;85(1):42–51. doi: 10.1002/bdra.20571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simioni M, Araujo TK, Monlleo IL, Maurer-Morelli CV, Gil-da-Silva-Lopes VL. Investigation of genetic factors underlying typical orofacial clefts: mutational screening and copy number variation. J Hum Genet. 2015;60(1):17–25. doi: 10.1038/jhg.2014.96. [DOI] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annual review of medicine. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Hsi-Yang Fritz M, Konkel MK, Malhotra A, Stutz AM, Shi X, Paolo Casale F, Chen J, Hormozdiari F, Dayama G, Chen K, Malig M, Chaisson MJ, Walter K, Meiers S, Kashin S, Garrison E, Auton A, Lam HY, Jasmine Mu X, Alkan C, Antaki D, Bae T, Cerveira E, Chines P, Chong Z, Clarke L, Dal E, Ding L, Emery S, Fan X, Gujral M, Kahveci F, Kidd JM, Kong Y, Lameijer EW, McCarthy S, Flicek P, Gibbs RA, Marth G, Mason CE, Menelaou A, Muzny DM, Nelson BJ, Noor A, Parrish NF, Pendleton M, Quitadamo A, Raeder B, Schadt EE, Romanovitch M, Schlattl A, Sebra R, Shabalin AA, Untergasser A, Walker JA, Wang M, Yu F, Zhang C, Zhang J, Zheng-Bradley X, Zhou W, Zichner T, Sebat J, Batzer MA, McCarroll SA, Genomes Project C. Mills RE, Gerstein MB, Bashir A, Stegle O, Devine SE, Lee C, Eichler EE, Korbel JO. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526(7571):75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan GJ, Bragin E, Chatzimichali EA, Corpas M, Bevan AP, Wright CF, Carter NP, Hurles ME, Firth HV. DECIPHER: web-based, community resource for clinical interpretation of rare variants in developmental disorders. Human molecular genetics. 2012;21(R1):R37–44. doi: 10.1093/hmg/dds362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi TN, Matsui KA, Yamashita K, Ohmori H, Yasuda M. Pathogenesis of cleft palate in mouse embryos exposed to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) Teratogenesis, carcinogenesis, and mutagenesis. 2000;20(2):73–86. doi: 10.1002/(sici)1520-6866(2000)20:2<73::aid-tcm3>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Tan R, Wang Y, Kleinstein SE, Liu Y, Zhu X, Guo H, Jiang Q, Allen AS, Zhu M. An evaluation of copy number variation detection tools from whole-exome sequencing data. Human mutation. 2014;35(7):899–907. doi: 10.1002/humu.22537. [DOI] [PubMed] [Google Scholar]
- van Rooij IA, Vermeij-Keers C, Kluijtmans LA, Ocke MC, Zielhuis GA, Goorhuis-Brouwer SM, van der Biezen JJ, Kuijpers-Jagtman AM, Steegers-Theunissen RP. Does the interaction between maternal folate intake and the methylenetetrahydrofolate reductase polymorphisms affect the risk of cleft lip with or without cleft palate? American journal of epidemiology. 2003;157(7):583–591. doi: 10.1093/aje/kwg005. [DOI] [PubMed] [Google Scholar]
- Vieira AR, Avila JR, Daack-Hirsch S, Dragan E, Felix TM, Rahimov F, Harrington J, Schultz RR, Watanabe Y, Johnson M, Fang J, O'Brien SE, Orioli IM, Castilla EE, Fitzpatrick DR, Jiang R, Marazita ML, Murray JC. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS genetics. 2005;1(6):e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock JP., Jr Genetic and molecular aspects of 2,3,7,8-tetrachlorodibenzo-p-dioxin action. Annual review of pharmacology and toxicology. 1990;30:251–277. doi: 10.1146/annurev.pa.30.040190.001343. [DOI] [PubMed] [Google Scholar]
- Williams ES, Uhas KA, Bunke BP, Garber KB, Martin CL. Cleft palate in a multigenerational family with a microdeletion of 20p12.3 involving BMP2. American journal of medical genetics Part A. 2012;158A(10):2616–2620. doi: 10.1002/ajmg.a.35594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski DF. Cleft Lip and Palate: From Origin to Treatment. USA: Oxford University Press; 2002. [Google Scholar]
- Wyszynski DF, Albacha-Hejazi H, Aldirani M, Hammod M, Shkair H, Karam A, Alashkar J, Holmes TN, Pugh EW, Doheny KF, McIntosh I, Beaty TH, Bailey-Wilson JE. A genome-wide scan for loci predisposing to non-syndromic cleft lip with or without cleft palate in two large Syrian families. American journal of medical genetics Part A. 2003;123A(2):140–147. doi: 10.1002/ajmg.a.20283. [DOI] [PubMed] [Google Scholar]
- Younkin SG, Scharpf RB, Schwender H, Parker MM, Scott AF, Marazita ML, Beaty TH, Ruczinski I. A genome-wide study of de novo deletions identifies a candidate locus for non-syndromic isolated cleft lip/palate risk. Bmc Genetics. 2014;15 doi: 10.1186/1471-2156-15-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nature reviews Genetics. 2015;16(3):172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- Zeiger JS, Beaty TH, Liang KY. Oral clefts, maternal smoking, and TGFA: a meta-analysis of gene-environment interaction. The Cleft palate-craniofacial journal : official publication of the American Cleft Palate-Craniofacial Association. 2005;42(1):58–63. doi: 10.1597/02-128.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.