Abstract
Danshen, the dried root of Salvia miltiorrhiza Bunge (Lamiaceae), is one of the traditional Chinese medicines (TCMs) most commonly used for the treatment of cardiovascular and cerebrovascular diseases. However, little is known about the chemical and metabolic profiles of danshen in vitro or in vivo. In particular, more information is needed in relation to the 50% ethanol extracts usually used in danshen formulations such as Fufang Xueshuantong Capsules and Fufang Danshen tablets. High-performance liquid chromatography coupled with a linear ion trap-Orbitrap mass spectrometer (HPLC-LTQ-Orbitrap) provides a sensitive and accurate method for analyzing the composition of samples. This method was used to determine the in vitro and in vivo chemical and metabolic profiles of danshen. Sixty-nine components of danshen extract and 118 components of danshen in rat plasma, urine, feces, and bile were unambiguously or tentatively identified. These results not only revealed the material composition of danshen, but also provided a comprehensive research approach for the identification of multi-constituents in TCMs.
Keywords: Danshen, Chemical profile, Metabolic profile, HPLC-LTQ-Orbitrap
1. Introduction
Recently, high-performance liquid chromatography-mass spectrometry (HPLC-MS), especially for high-resolution mass spectrometry (HRMS), has become a powerful tool for detecting and identifying known and unknown metabolites of drugs owing to its high mass accuracy and high sensitivity (Liu et al., 2011; Wang et al., 2011; Liang et al., 2013). MS/MS data provide abundant information for elucidating the structure of compounds. Thus, this method provides an effective and powerful tool for the identification of compounds in complex matrices, such as traditional Chinese medicines (TCMs) and bio-samples. The linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap), an electrostatic Fourier-transform mass spectrometer, combines a high trapping capacity and MSn scanning function of the linear ion trap with accurate mass measurements to within 5 ppm (parts per million) and a resolving power of up to 100 000 (Cai et al., 2015; Zhang et al., 2015). Data-dependent MS/MS scanning can obtain more fragmentation information, improving the efficiency and accuracy of identification (Wang et al., 2016).
Danshen, the dried root of Chinese sage, Salvia miltiorrhiza Bunge (Lamiaceae), is one of the TCMs most commonly used in China and elsewhere, and is used either alone or in formulations. It has been widely used in the treatment of cardiovascular and cerebrovascular diseases, such as coronary artery disease (Ji et al., 2003), myocardial infarction (Sun et al., 2005), and stroke (Lam et al., 2003). It has also been used to treat other conditions, such as renal diseases (Kang et al., 2004) and diabetes (Belin et al., 2009). Many formulations containing danshen, for instance the Fufang Danshen Dripping Pill and Fufang Xueshuantong Capsule, are now frequently used in the clinical treatment of cardiovascular diseases and eye diseases (Duan et al., 2013; Yang et al., 2014). There are two principal bioactive components in danshen: water-soluble phenolic acids and liposoluble tanshinones. The phenolic acids include danshensu, rosmarinic acid, lithospermic acid, salvianolic acid A, salvianolic acid B, and other salvianolic acids. The tanshinones include tanshinone I, tanshinone IIA, tanshinone IIB, cryptotanshinone, 15,16-dihydrotanshinone I, and other tanshinones (Zhang et al., 2005; Wu et al., 2006).
Previous in vivo studies have focused mainly on the water decoction of danshen (Zhao et al., 2015) or its effective parts and components (Li et al., 2007; Sun et al., 2007). Danshen has often been used only as a component of ethanol extracts, especially in formulations, because of its complex composition and compatibility with other herbs. Also, there has been limited research on the excretion of danshen in feces and bile (Sun et al., 2007). Therefore, comprehensive and systematic studies are needed of the chemical and metabolic profiles of danshen in vitro and in vivo. In the present study, we analyzed the chemical profile of a 50% ethanol extract of danshen, as such extracts are often used in its formulation. The metabolic profile of danshen was determined in bio-samples from rats. An HPLC-LTQ-Orbitrap method coupled with an extracted ion chromatogram (EIC) data-processing technique was applied to elucidate the chemical and metabolic profiles. A total of 69 components of danshen extract and 118 components of danshen in rat plasma, urine, feces, and bile were unambiguously or tentatively identified. The present study provides a basis for research on the quality control and pharmacology of danshen, and establishes a comprehensive and reliable method for identification of multi-components of TCMs both in vitro and in vivo.
2. Materials and methods
2.1. Materials and reagents
Danshen crude drug was provided by the Guangdong Zhongsheng Pharmaceutical Co., Ltd. (Guangzhou, China) and was authenticated by Professor Jian-mei HUANG. Voucher specimens were deposited in the School of Chinese Materia Medica, Beijing University of Chinese Medicine, China.
Eleven reference standards, including caffeic acid, protocatechuic aldehyde, protocatechuic acid, danshensu, ferulic acid, isoferulic acid, rosmarinic acid, tanshinone I, dihydrotanshinone I, tanshinone IIA, and cryptotanshinone, were purchased from the Chengdu Must Bio-Technology Co., Ltd. (Chengdu, China). Three reference standards of tanshinol B, danshenxinkun B, and tanshinone IIB were purchased from the Shanghai Yuanye Bio-Technology Co., Ltd. (Shanghai, China). Three authentic standards, namely salvianolic acids A, B, and C, were obtained from the School of Chinese Materia Medica, Beijing University of Chinese Medicine, China.
HPLC-grade methanol and acetonitrile, and LC/MS-grade formic acid were purchased from Fisher Scientific (Fisher, Fair Lawn, NJ, USA).
2.2. Instrumentation and analytical conditions
Chromatographic analysis was performed using a Thermo Accela 600 HPLC system (Thermo Scientific, Bremen, Germany) equipped with a binary pump and an autosampler. Samples were separated on a Waters XBridge-C18 column (5 μm, 150 mm×4.6 mm) at room temperature. A gradient elution of solvent acetonitrile (A) and water containing 0.1% formic acid (B) was applied according to the following program: 0–10 min, 5%–20% A; 10–25 min, 20%–30% A; 25–35 min, 30%–70% A; 35–60 min, 70% A. The flow rate was set at 1.0 ml/min. Sample solution (10 μl) was injected into the HPLC-MS/MS system.
MS analysis was performed using an LTQ-Orbitrap mass spectrometer (Thermo Scientific, Bremen, Germany). The mass spectrometer was connected to the Accela HPLC system by an electrospray ionization (ESI) source and operated in both positive and negative ion modes. Compounds were detected by full scan mass analysis from m/z 100 to 1000 at a resolving power of 30 000 with data-dependent MSn (n=3) analysis. The optimized source parameters in positive (and negative) mode were as follows: capillary temperature, 350 °C; sheath gas flow, 30 arbitrary unit (arb); auxiliary gas flow, 10 arb; source voltage, 4.0 kV; capillary voltage, 35 V; tube lens voltage, 110 V. The isolation width was 2 Da, and the normalized collision energy (CE) was 35%.
2.3. Preparation of drugs
2.3.1 Preparation of danshen freeze-dried powder
Danshen freeze-dried powder was prepared by refluxing the extract twice with 50% (v/v) ethanol (100 g/700 ml for 3 h the first time, and 100 g/500 ml for 2 h the second time) after soaking in 50% ethanol for 30 min. Each decoction was mixed, filtered, vacuum-evaporated, and freeze-dried. The yield of powdered extract was about 42.3% (w/w).
2.3.2 Preparation of danshen extract
A total of 1.05 g danshen freeze-dried powder was accurately weighed and ultrasonicated with 30 ml of 50% ethanol for 30 min. The supernatants were filtered through a 0.22-μm membrane filter. The filtrates were collected and stored at 4 °C until HPLC-MS/MS analysis.
2.3.3 Preparation of danshen suspension
Danshen freeze-dried powder was accurately weighed and suspended in deionized water to obtain a final concentration of 1.5 g/ml (crude drug) for intragastric administration.
2.3.4 Preparation of standard solutions
Individual standard stock solutions of the seventeen standards were prepared by accurately weighing and then dissolving each standard in methanol, with concentrations ranging from 0.09 to 1.20 mg/ml. A working solution of each of the seventeen standards was obtained by diluting each stock solution with methanol to the desired concentration. Working solutions were stored at 4 °C before analysis.
2.4. Animals and drug administration
Twelve male Sprague-Dawley rats, weighing (250±20) g, were purchased from the Si Bei Fu Experimental Animal Science and Technology Co., Ltd. (Beijing, China). The rats were divided into two groups: a control group (n=3, one each for blank plasma, urine and feces, and bile) and a drug group (n=9, 3 for dosed plasma, 3 for dosed urine and feces, and 3 for dosed bile). The rats were housed in a controlled environment (12-h light/12-h dark cycle, at consistent temperature and humidity) for three days before the experiment. Danshen was administered orally to the drug group once a day at a dose of 1 ml/100 g body weight for three days. An equal dose of deionized water was administered by oral gavage to the rats of the control group.
Animal experiments were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals, and all experimental protocols were reviewed and approved by the Institutional animal Experimentation Committee of Beijing University of Chinese Medicine.
2.5. Biological sample collection
Before the last administration, the rats were deprived of food for 12 h. Blood samples (0.4 ml) were collected from the orbital vein and gathered into heparinized tubes at 0.25, 0.5, 0.75, 1, 1.5, 2, 4, 6, 8, and 12 h, respectively. All blood samples were then centrifuged at 3000g for 10 min to obtain plasma samples. Plasma samples from different rats and different time points in each group were then mixed in the same proportions to produce pooled plasma samples, which were stored at −80 °C until additional extraction and analysis.
The urine and feces of rats in each group were collected over a 24-h period starting immediately after the last administration. The urine and feces samples in each group were combined separately and stored at −80 °C until additional extraction and analysis.
Rats were fixed on a wooden plate and anesthetized with ethylurethanm following the last administration. An abdominal incision was made and the common bile duct was cannulated with PE10 tubing (inside diameter (ID)=0.28 mm, San Diego, CA USA) for collection of the bile samples. Bile samples from each group were collected for 24 h and combined and stored at −80 °C until additional extraction and analysis.
2.6. Biological sample pretreatment
An aliquot of 2 ml plasma for positive ion detection was suspended in 8 ml methanol. Another aliquot of 2 ml plasma for negative ion detection was suspended in 200 µl 10% (v/v) hydrochloric acid and 8 ml methanol, and then mixed by vortex for 3 min to precipitate protein, followed by centrifugation at 10 000g for 10 min. The supernatants were evaporated to dryness under nitrogen gas at room temperature, and the residues were dissolved in 200 µl 70% (v/v) methanol. After centrifugation at 12 000g for 10 min, 10 µl of the supernatant was injected into the HPLC-MS/MS system for analysis.
Urine sample (3 ml) was dissolved in 12 ml methanol, and then mixed by vortex for 3 min to precipitate protein, followed by centrifugation at 10 000g for 10 min. The supernatant was evaporated to dryness under nitrogen gas at room temperature, and the residue was dissolved in 600 µl 70% methanol. After centrifugation at 12 000g for 10 min, 10 µl of the supernatant was injected into the HPLC-MS/MS system for analysis.
Bile sample (3 ml) was dissolved in 12 ml methanol, and then mixed by vortex for 3 min to precipitate protein, followed by centrifugation at 10 000g for 10 min. The supernatant was evaporated to dryness under nitrogen gas at room temperature, and the residue was dissolved in 1.5 ml 70% methanol. After centrifugation at 12 000g for 10 min, 10 µl of the supernatant was injected into the HPLC-MS/MS system for analysis.
Feces were dried at 37 °C and grinded into powder. Feces sample (1.5 g) was extracted with 30 ml 70% methanol in an ultrasonic bath for 30 min, followed by filtration. Filtrate (2 ml) was evaporated to dryness under nitrogen gas at room temperature, and the residue was dissolved in 400 µl 70% methanol. After centrifugation at 12 000g for 10 min, a 10-µl aliquot of the supernatant was injected into the HPLC-MS/MS system for analysis.
2.7. Data processing
Thermo Xcalibur 2.1 workstation (Thermo Fisher Scientific, Bremen, Germany) was used for data acquisition and processing. Metworks (Thermo Scientific, Bremen, Germany) was used for data-filtering and identification of possible metabolites. The maximum mass error between the measured and calculated values was 5 ppm.
3. Results
3.1. Analysis of the chemical profile of danshen in vitro
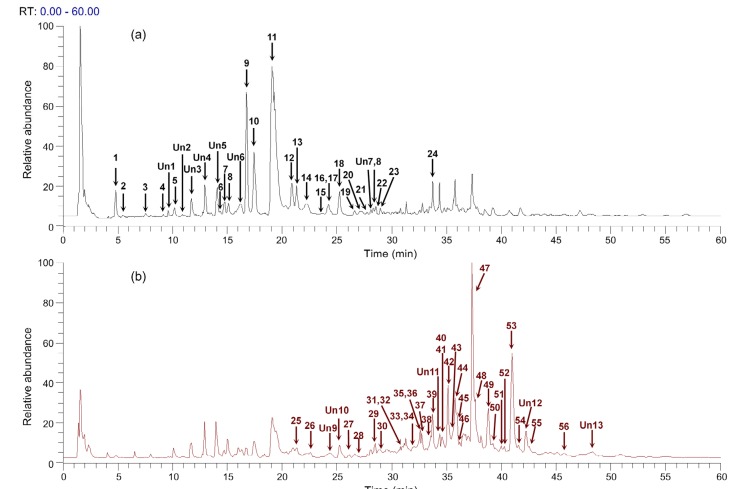
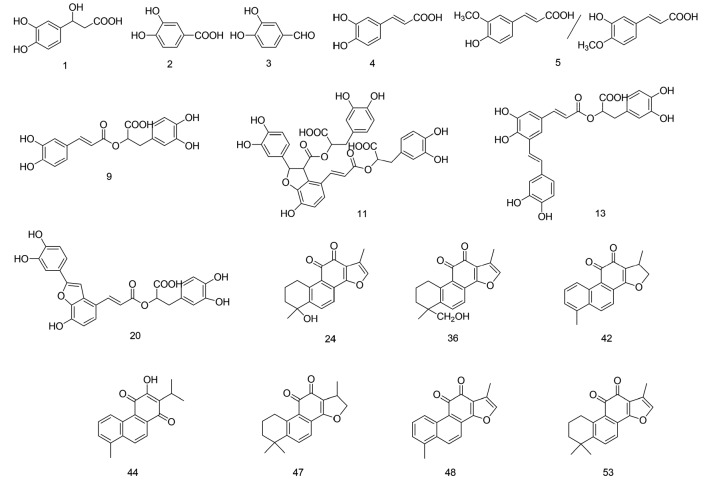
The results from total ion chromatography (TIC) of the danshen extract and the reference standards in positive mode and negative mode are shown in Fig. 1. Based on accurate mass measurements, MS/MS fragmentations, retention time, and reference data (Liu AH et al., 2007; Liu M et al., 2007; Su et al., 2015), a total of 69 components of danshen, including 23 phenolic acids, 33 tanshinones, and 13 unknown compounds, were identified. Their accurate mass measurements, retention time, and HPLC-MS/MS data are shown in Table 1. Among them, 16 compounds were unambiguously confirmed by comparison with reference standards. Their structures are shown in Fig. 2.
Fig. 1.
Total ion chromatography of danshen extract in negative (a) and positive (b) ion modes
Table 1.
HPLC-MS/MS data and identification of components of danshen
| No. | t R (min) | Ion | Theoretical mass (m/z) | Experimental mass (m/z) | Error (ppm) | Formula [M−H]−/[M+H]+ | MS/MS fragment | Identification | ||
| Phenolic acids | ||||||||||
| 1 | 4.83 | [M−H]− | 197.0444 | 197.0441 | −1.6 | C9H9O5 | MS2[197]: 179(100) MS3[179]: 135(100) | Danshensua | ||
| 2 | 5.70 | [M−H]− | 153.0182 | 153.0184 | 0.9 | C7H5O4 | Protocatechuic acida | |||
| 3 | 7.58 | [M−H]− | 137.0233 | 137.0236 | 1.7 | C7H5O3 | MS2[137]: 137(100) | Protocatechuic aldehydea | ||
| 4 | 9.16 | [M−H]− | 179.0339 | 179.0336 | −1.3 | C9H7O4 | MS2[179]: 135(100) | Caffeic acida | ||
| 5 | 10.08 | [M−H]− | 193.0495 | 193.0493 | −1.2 | C10H9O4 | MS2[193]: 178(100), 149(38), 134(81) MS3[178]: 134(100) | Ferulic acid/isoferulic acida | ||
| 6 | 14.42 | [M−H]− | 735.1556 | 735.1552 | −0.5 | C36H31O17 | MS2[735]: 537(100), 519(20) MS3[519]: 519(51), 357(73), 321(65), 297(100) | Hydrated salvianolic acid B | ||
| 7 | 14.76 | [M−H]− | 537.1028 | 537.1028 | 0.1 | C27H21O12 | MS2[537]: 339(100), 295(37) MS3[339]: 321(33), 295(100) | Salvianolic acid H/I | ||
| 8 | 15.02 | [M−H]− | 735.1556 | 735.1553 | −0.4 | C36H31O17 | MS2[735]: 537(100), 519(49) MS3[537]: 519(51), 339(20), 321(70), 297(100) | Hydrated salvianolic acid B | ||
| 9 | 16.74 | [M−H]− | 359.0761 | 359.0758 | −1.0 | C18H15O8 | MS2[359]: 197(24), 179(23), 161(100) MS3[161]: 161(38), 133(100) | Rosmarinic acida | ||
| 10 | 17.45 | [M−H]− | 493.1129 | 493.1129 | 0.0 | C26H21O10 | MS2[493]: 295(100) MS3[295]: 280(13), 277(65), 159(100) | Salvianolic acid A isomer | ||
| 11 | 19.07 | [M−H]− | 717.1450 | 717.1434 | −2.2 | C36H29O16 | MS2[717]: 519(100), 321(15) MS3[519]: 339(21), 321(100) | Salvianolic acid Ba | ||
| 12 | 20.89 | [M−H]− | 717.1450 | 717.1450 | 0.0 | C36H29O16 | MS2[717]: 519(100), 321(18) MS3[519]: 339(21), 321(100) | Salvianolic acid E | ||
| 13 | 21.32 | [M−H]− | 493.1129 | 493.1135 | 1.1 | C26H21O10 | MS2[493]: 295(100) MS3[295]: 280(14), 277(66), 159(100) | Salvianolic acid Aa | ||
| 14 | 22.26 | [M−H]− | 731.1607 | 731.1598 | −1.2 | C37H31O16 | MS2[731]: 533(100) MS3[533]: 353(51), 335(100) | Methyl salvianolic acid B | ||
| 15 | 23.68 | [M−H]− | 565.1341 | 565.1341 | 0.1 | C29H25O12 | MS2[565]: 519(87), 367(87), 339(15), 321(100) | Dimethyl lithospermate | ||
| 16 | 24.23 | [M−H]− | 491.0973 | 491.0982 | 1.9 | C26H19O10 | MS2[491]: 311(60), 293(100) MS3[293]: 276(32), 275(11), 265(100), 249(89), 247(13) | Isosalvianolic acid C | ||
| 17 | 24.44 | [M−H]− | 565.1341 | 565.1341 | 0.1 | C29H25O12 | MS2[565]: 367(76), 339(17), 321(100) | Dimethyl lithospermate | ||
| 18 | 25.23 | [M−H]− | 565.1341 | 565.1345 | 0.7 | C29H25O12 | MS2[565]: 519(88), 339(15), 321(100) MS3[321]: 293(31), 277(100), 249(76) | Dimethyl lithospermate | ||
| 19 | 26.65 | [M−H]− | 565.1341 | 565.1345 | 0.7 | C29H25O12 | MS2[565]: 367(100) | Dimethyl lithospermate | ||
| 20 | 27.27 | [M−H]− | 491.0973 | 491.0977 | 0.9 | C26H19O10 | MS2[491]: 311(22), 293(100) MS3[293]: 276(24), 275(18), 265(100), 249(21), 247(36) | Salvianolic acid Ca | ||
| 21 | 27.65 | [M−H]− | 745.1763 | 745.1760 | −0.5 | C38H33O16 | MS2[745]: 547(87), 519(100), 321(73) | Dimethyl salvianolic acid B | ||
| 22 | 28.62 | [M−H]− | 745.1763 | 745.1762 | −0.1 | C38H33O16 | MS2[745]: 547(87), 519(100), 321(74) | Dimethyl salvianolic acid B | ||
| 23 | 28.97 | [M−H]− | 313.0707 | 313.0709 | 0.8 | C17H13O6 | MS2[313]: 269(36), 161(100) MS3[161]: 161(29), 133(100) | Salvianolic acid F | ||
| Tanshinones | ||||||||||
| 24 | 33.73 | [M−H]− | 295.0965 | 295.0970 | 1.7 | C18H15O4 | MS2[295]: 277(16), 267(27), 265(100) MS3[265]: 265(100) | Tanshinol Ba | ||
| 25 | 21.24 | [M+H]+ | 313.1071 | 313.1056 | −4.8 | C18H17O5 | MS2[313]: 295(100), 267(22), 265(87) MS3[295]: 277(100), 267(40), 249(98) | Tanshindiol A/B/C | ||
| 26 | 22.57 | [M+H]+ | 313.1071 | 313.1056 | −4.6 | C18H17O5 | MS2[313]: 295(100), 267(7) MS3[295]: 267(100) | Tanshindiol A/B/C | ||
| 27 | 26.14 | [M+H]+ | 313.1071 | 313.1056 | −4.7 | C18H17O5 | MS2[313]: 295(100), 267(9) MS3[295]: 267(100) | Tanshindiol A/B/C | ||
| 28 | 27.01 | [M+H]+ | 293.0808 | 293.0797 | −3.9 | C18H13O4 | MS2[293]: 249(100) MS3[249]: 234(18), 221(24), 193(100), 178(39) | Hydroxyl tanshinone I | ||
| 29 | 28.49 | [M+H]+ | 313.1434 | 313.1422 | −3.9 | C19H21O4 | MS2[313]: 269(100) MS3[269]: 254(25), 251(16), 223(22), 199(100) | Hydroxyl cryptotanshinone | ||
| 30 | 29.28 | [M+H]+ | 283.0965 | 283.0954 | −3.7 | C17H15O4 | MS2[283]: 265(51), 255(27), 241(15), 237(100) MS3[237]: 222(11), 219(100), 209(87), 191(35), 181(25) | Dihydronortanshinone | ||
| 31 | 30.79 | [M+H]+ | 293.0808 | 293.0799 | −3.3 | C18H13O4 | MS2[293]: 249(100) MS3[249]: 234(18), 221(23), 193(100), 178(38) | Przewaquinone B | ||
| 32 | 30.97 | [M+H]+ | 295.0965 | 295.0953 | −4.1 | C18H15O4 | MS2[295]: 277(100), 267(44), 24(51) MS3[277]: 259(82), 249(100), 231(27) | Hydrated tanshinone I/trijuganone A | ||
| 33 | 32.08 | [M+H]+ | 295.0965 | 295.0951 | −4.8 | C18H15O4 | MS2[295]: 277(100), 249(11) MS3[277]: 249(100), 221(17) | Hydrated tanshinone I/trijuganone A | ||
| 34 | 32.26 | [M+H]+ | 313.1423 | 313.1423 | −3.5 | C19H21O4 | MS2[313]: 295(100), 267(73) MS3[295]: 277(100), 267(55), 249(52) | Hydroxyl cryptotanshinone | ||
| 35 | 32.44 | [M+H]+ | 301.1434 | 301.1424 | −3.6 | C18H21O4 | MS2[301]: 283(100) MS3[283]: 265(100), 255(26) | Salvianonol | ||
| 36 | 32.61 | [M+H]+ | 311.1278 | 311.1264 | −4.6 | C19H19O4 | MS2[311]: 293(100), 283(22), 267(80), 225(12) MS3[293]: 278(16), 275(100), 265(19), 251(80) | Tanshinone IIBa | ||
| 37 | 32.77 | [M+H]+ | 311.1278 | 311.1263 | −4.6 | C19H19O4 | MS2[311]: 293(14), 275(11), 267(100) MS3[267]: 252(100), 239(11), 225(63), 185(47) | Hydroxyl tanshinone IIA | ||
| 38 | 33.46 | [M+H]+ | 341.1384 | 341.1371 | −3.8 | C20H21O5 | MS2[341]: 281(100), 263(43) MS3[281]: 263(100), 235(19) | Methyl dihydrotanshinonate | ||
| 39 | 33.69 | [M+H]+ | 327.1214 | 327.1248 | −3.9 | C19H19O5 | MS2[327]: 309(100) MS3[309]: 265(100) | Hydroxyl tanshinone IIB | ||
| 40 | 34.56 | [M+H]+ | 281.1536 | 281.1524 | −4.1 | C19H21O2 | MS2[281]: 266(38), 263(50), 253(28), 239(100) MS3[239]: 224(22), 221(100), 193(54) | Dehydromiltirone | ||
| 41 | 34.64 | [M+H]+ | 293.1172 | 293.1159 | −4.4 | C19H17O3 | MS2[293]: 275(100), 265(11), 247(39) MS3[275]: 260(13), 247(100) | Dehydrotanshinone IIA | ||
| 42 | 35.19 | [M+H]+ | 279.1016 | 279.1004 | −4.3 | C18H15O3 | MS2[279]: 261(100), 233(5) MS3[261]: 233(100) | Dihydrotanshinone Ia | ||
| 43 | 35.71 | [M+H]+ | 315.1591 | 315.1576 | −4.7 | C19H23O4 | MS2[315]: 297(100) MS3[297]: 279(100), 251(57) | Neocryptotanshinone | ||
| 44 | 35.84 | [M+H]+ | 281.1172 | 281.1160 | −4.2 | C18H17O3 | MS2[281]: 263(100), 253(7), 235(71) MS3[263]: 248(16), 245(10), 235(100) | Danshenxinkun Ba | ||
| 45 | 35.98 | [M+H]+ | 339.1227 | 339.1219 | −2.3 | C20H19O5 | MS2[339]: 279(100) MS3[279]: 261(100) | Methyl tanshinonate | ||
| 46 | 36.24 | [M+H]+ | 295.1329 | 295.1319 | −3.3 | C19H19O3 | MS2[295]: 277(100), 267(15), 249(47) MS3[277]: 249(100) | Dehydrocryptotanshinone | ||
| 47 | 37.34 | [M+H]+ | 297.1485 | 297.1474 | −3.7 | C19H21O3 | MS2[297]: 279(100), 251(81) MS3[279]: 251(100) | Cryptotanshinonea | ||
| 48 | 37.62 | [M+H]+ | 277.0859 | 277.0850 | −3.5 | C18H13O3 | MS2[277]: 249(100), 231(13) | Tanshinone Ia | ||
| 49 | 38.84 | [M+H]+ | 279.1016 | 279.1003 | −4.7 | C18H15O3 | MS2[279]: 261(100), 233(6) MS3[261]: 233(100), 205(3) | Dihydrotanshinone I | ||
| 50 | 38.98 | [M+H]+ | 293.1172 | 293.1166 | −2.2 | C19H17O3 | MS2[293]: 275(100), 265(11), 247(38) MS3[275]: 260(13), 247(100) | Dehydrotanshinone IIA | ||
| 51 | 40.05 | [M+H]+ | 281.1536 | 281.1524 | −4.1 | C19H21O2 | MS2[281]: 266(17), 263(43), 253(82), 221(100) MS3[221]: 206(17), 193(100) | Dehydromiltirone | ||
| 52 | 40.28 | [M+H]+ | 293.1172 | 293.1159 | −4.5 | C19H17O3 | MS2[293]: 275(100), 265(11), 247(37) MS3[275]: 260(13), 247(100) | Dehydrotanshinone IIA | ||
| 53 | 41.17 | [M+H]+ | 295.1329 | 295.1317 | −4.0 | C19H19O3 | MS2[295]: 277(100), 249(14) MS3[277]: 249(100) | Tanshinone IIAa | ||
| 54 | 41.54 | [M+H]+ | 281.1536 | 281.1526 | −3.7 | C19H21O2 | MS2[281]: 266(18), 263(43), 253(93), 221(100) MS3[221]: 206(14), 193(100) | Dehydromiltirone | ||
| 55 | 42.34 | [M+H]+ | 283.1693 | 283.1682 | −3.6 | C19H23O2 | MS2[283]: 265(100), 241(47), 223(63) MS3[265]: 237(64), 223(100) | Miltirone | ||
| 56 | 45.75 | [M+H]+ | 557.1959 | 557.1942 | −3.1 | C36H29O6 | MS2[557]: 539(28), 529(100), 511(48) MS3[529]: 511(100), 501(81), 483(46) | Neoprzewaquinone A | ||
| Others | ||||||||||
| Un1 | 9.62 | [M−H]− | 509.2229 | 509.2230 | 0.2 | C22H37O13 | MS2[509]: 463(100) MS3[463]: 331(100), 161(27) | Unknown | ||
| Un2 | 10.91 | [M−H]− | 571.1082 | 571.1080 | −0.5 | C27H23O14 | MS2[571]: 527(21), 483(100), 439(73) | Unknown | ||
| Un3 | 11.73 | [M−H]− | 627.4044 | 627.4059 | 2.4 | C41H55O5 | MS2[627]: 610(14), 581(70), 564(100) | Unknown | ||
| Un4 | 12.96 | [M−H]− | 723.5042 | 723.5013 | −3.9 | C41H71O10 | MS2[723]: 678(100) MS3[678]: 659(100), 451(25), 338(25) | Unknown | ||
| Un5 | 14.07 | [M−H]− | 836.5856 | 836.5854 | −0.2 | C44H84O14 | MS2[836]: 791(100) MS3[791]: 773(100), 565(28) | Unknown | ||
| Un6 | 16.21 | [M−H]− | 717.1450 | 717.1452 | 0.3 | C36H29O16 | MS2[717]: 519(100), 321(16) MS3[519]: 339(22), 321(100) | Unknown | ||
| Un7 | 28.14 | [M−H]− | 341.1020 | 341.1023 | 0.9 | C19H17O6 | MS2[341]: 297(100), 253(14) MS3[297]: 253(100) | Unknown | ||
| Un8 | 28.33 | [M−H]− | 671.1395 | 671.1402 | 1.0 | C35H27O14 | MS2[671]: 473(100) MS3[473]: 429(100), 321(100) | Unknown | ||
| Un9 | 24.40 | [M+H]+ | 327.1227 | 327.1212 | −4.6 | C19H19O5 | MS2[327]: 283(100), 265(8) MS3[283]: 265(29), 254(100) | Unknown | ||
| Un10 | 25.27 | [M+H]+ | 369.0969 | 369.0955 | −3.7 | C20H17O7 | MS2[369]: 323(100), 295(77) MS3[323]: 295(100) | Unknown | ||
| Un11 | 34.38 | [M+H]+ | 297.1485 | 297.1471 | −4.7 | C19H21O3 | MS2[297]: 253(100) MS3[253]: 238(33), 211(100) | Unknown | ||
| Un12 | 42.22 | [M+H]+ | 283.1693 | 283.1677 | −5.4 | C19H23O2 | MS2[283]: 265(100), 241(47), 223(63) MS3[265]: 237(20), 223(100) | Unknown | ||
| Un13 | 50.83 | [M+H]+ | 587.2064 | 587.2059 | −0.9 | C37H31O7 | MS2[587]: 569(100), 541(24) MS3[569]: 551(100), 541(71) | Unknown | ||
Fig. 2.
Chemical structures of confirmed compounds in danshen extract
The numbering of compounds is consistent with that in Table 1
3.2. Analysis of the metabolic profile of danshen in vivo
For the identification of original components in bio-samples, the extract ion chromatograms (EICs) combined with the accurate mass measurements, MS/MS fragmentations, and retention time were compared with those of blank samples. For the identification of possible metabolites in bio-samples, firstly, all of the possible metabolic pathways of one component were input in Metworks; secondly, all of the possible metabolites proposed by the software were summarized in an Excel table; thirdly, the EICs, mass measurements, and MS/MS fragmentations of each metabolite were compared with those of blank samples.
As a result, 118 components were unambiguously or tentatively identified, including 38 original components and 80 transformative components (Table 2). Among these components, 7 phenolic acids and 28 tanshinones were identified in rat plasma; 17 phenolic acids and 46 tanshinones were tentatively identified in rat urine; 25 phenolic acids and 37 tanshinones were identified in rat feces; and 1 phenolic acid and 17 tanshinones were identified in rat bile.
Table 2.
Metabolites identified in bio-samples from rats after oral administration of danshen extract
| No. | t R (min) | Ion | Theoretical mass (m/z) | Experimental mass (m/z) | Formula [M−H]−/[M+H]+ | Error (ppm) | MS/MS fragment | Identification | Plasma | Urine | Feces | Bile |
| Phenolic acids | ||||||||||||
| 1 | 2.11 | [M−H]− | 277.0013 | 277.0020 | C9H9O8S | 2.8 | MS2[277]: 259(57), 215(35), 197(100) | Sulfate danshensu | × | × | √ | × |
| 2 | 4.26 | [M−H]− | 277.0013 | 277.0010 | C9H9O8S | −0.8 | MS2[277]: 215(40), 197(100) MS3[196]: 179(100) | Sulfate danshensu | × | √ | × | × |
| 3 | 4.77 | [M−H]− | 197.0444 | 197.0446 | C9H9O5 | 0.6 | MS2[197]: 179(100) MS3[179]: 135(100) | Danshensua,b | √ | √ | √ | × |
| 4 | 7.35 | [M−H]− | 211.0601 | 211.0603 | C10H11O5 | 0.9 | MS2[211]: 193(100), 165(23) MS3[193]: 149(29), 134(100) | Methyl danshensu | √ | √ | √ | √ |
| 5 | 7.47 | [M−H]− | 179.0339 | 179.0344 | C9H7O4 | 3.1 | MS2[179]: 135(100) MS3[135]: 135(100) | Demethyl ferulic acid | × | √ | × | × |
| 6 | 7.52 | [M−H]− | 137.0233 | 137.0236 | C7H5O3 | 1.7 | MS2[137]: 137(100) | Protocatechuic aldehydea,b | × | × | √ | × |
| 7 | 7.62 | [M−H]− | 181.0495 | 181.0502 | C9H9O4 | 3.7 | MS2[181]: 163(100) MS3[162]: 119(100) | Dihydro caffeic acid | × | √ | √ | × |
| 8 | 8.29 | [M−H]− | 179.0339 | 179.0341 | C9H7O4 | 1.2 | MS2[179]: 135(100) MS3[135]: 135(100) | Acetylated protocatechuic aldehyde | × | √ | × | × |
| 9 | 8.59 | [M−H]− | 165.0546 | 165.0550 | C9H9O3 | 2.4 | MS2[165]: 121(100) MS3[121]: 121(100) | Decarbonyl ferulic acid | × | √ | × | × |
| 10 | 9.38 | [M−H]− | 361.0918 | 361.0910 | C18H17O8 | −2.2 | MS2[361]: 317(39), 273(41), 239(100), 221(68) MS3[239]: 195(100), 151(19) | Dihydro rosmarinic acid | × | × | √ | × |
| 11 | 9.86 | [M−H]− | 193.0495 | 193.0502 | C10H9O4 | 3.5 | Ferulic acid/isoferulic acida,b | × | × | √ | × | |
| 12 | 12.03 | [M−H]− | 343.0812 | 343.0806 | C18H15O7 | −1.8 | MS2[343]: 299(100) MS3[299]: 255(100) | Hydroxyl and methyl salvianolic acid F | × | × | √ | × |
| 13 | 13.62 | [M−H]− | 535.1082 | 535.1072 | C24H23O14 | −1.9 | MS2[535]: 359(100) MS3[359]: 197(23), 179(20), 161(100) | Rosmarinic acid glucuronide conjugate | × | √ | × | × |
| 14 | 13.99 | [M−H]− | 539.1184 | 539.1169 | C27H23O12 | −2.7 | MS2[539]: 399(100), 297(67) MS3[297]: 219(100), 201(70) | Dihydro salvianolic acid H/I | × | √ | √ | × |
| 15 | 14.67 | [M−H]− | 537.1028 | 537.1025 | C27H21O12 | −0.4 | MS2[537]: 339(100), 295(38) | Salvianolic acid H/Ib | × | × | √ | × |
| 16 | 15.03 | [M−H]− | 735.1556 | 735.1539 | C36H31O17 | −2.3 | Hydrated salvianolic acid Bb | × | × | √ | × | |
| 17 | 15.17 | [M−H]− | 343.0812 | 343.0806 | C18H15O7 | −1.7 | MS2[343]: 255(100) MS3[255]: 237(91), 148(100) | Hydroxyl and methyl salvianolic acid F | × | × | √ | × |
| 18 | 16.55 | [M−H]− | 555.1133 | 555.1125 | C27H23O13 | −1.5 | MS2[555]: 375(100), 357(39) MS3[375]: 331(100), 269(60) | Hydrated salvianolic acid H/I | × | × | √ | × |
| 19 | 16.71 | [M−H]− | 359.0761 | 359.0761 | C18H15O8 | −0.2 | MS2[359]: 271(100) MS3[271]: 149(100), 135(46), 121(74) | Rosmarinic acida,b | √ | × | × | × |
| 20 | 17.01 | [M−H]− | 523.1235 | 523.1225 | C27H23O11 | −1.9 | MS2[523]: 505(53), 281(100) MS3[281]: 263(26), 174(100) | Demethyl and hydroxyl salvianolic acid A | × | × | √ | × |
| 21 | 17.14 | [M−H]− | 315.0863 | 315.0863 | C17H15O6 | 0.0 | MS2[315]: 297(18), 285(100), 267(12) | Dihydro salvianolic acid F | × | √ | × | × |
| 22 | 17.46 | [M−H]− | 493.1129 | 493.1130 | C26H21O10 | 0.1 | MS2[493]: 295(100) MS3[295]: 277(56), 159(100) | Iso salvianolic acid Ab | √ | √ | √ | × |
| 23 | 19.02 | [M−H]− | 717.1450 | 717.1439 | C36H29O16 | −1.5 | MS2[717]: 519(100), 321(17) MS3[519]: 339(22), 321(100) | Salvianolic acid Ba,b | × | √ | √ | × |
| 24 | 20.40 | [M−H]− | 763.1869 | 763.1854 | C38H35O17 | −1.9 | MS2[763]: 565(100), 520(57), 321(15) | Hydrated dimethyl salvianolic acid B | × | × | √ | × |
| 25 | 20.80 | [M−H]− | 717.1450 | 717.1441 | C36H29O16 | −1.3 | MS2[717]: 519(100), 321(19) MS3[519]: 339(23), 321(100) | Salvianolic acid Eb | × | √ | √ | × |
| 26 | 21.26 | [M−H]− | 493.1129 | 493.1129 | C26H21O10 | −0.1 | MS2[493]: 295(100) MS3[295]: 277(59), 159(100) | Salvianolic acid Aa,b | √ | √ | √ | × |
| 27 | 21.70 | [M−H]− | 551.1184 | 551.1177 | C28H23O12 | −1.3 | MS2[551]: 519(43), 371(100), 353(51), 339(73) | Methyl salvianolic acid H/I | × | × | √ | × |
| 28 | 22.20 | [M−H]− | 731.1607 | 731.1597 | C37H31O16 | −1.4 | MS2[731]: 533(100) MS3[533]: 353(49), 335(100) | Methyl salvianolic acid Bb | √ | √ | √ | × |
| 29 | 24.15 | [M−H]− | 491.0973 | 491.0971 | C26H19O10 | −0.3 | MS2[491]: 293(100) MS3[293]: 276(33), 264(100), 249(91) | Iso salvianolic acid Cb | × | × | √ | × |
| 30 | 24.35 | [M−H]− | 565.1341 | 565.1334 | C29H25O12 | −1.1 | MS2[565]: 519(84), 367(78), 321(100) | Dimethyl lithospermateb | × | × | √ | × |
| 31 | 25.16 | [M−H]− | 565.1341 | 565.1331 | C29H25O12 | −1.7 | MS2[565]: 519(87), 367(79), 321(100) | Dimethyl lithospermateb | √ | √ | √ | × |
| 32 | 26.60 | [M−H]− | 565.1341 | 565.1331 | C29H25O12 | −1.8 | Dimethyl lithospermateb | × | √ | × | × | |
| 33 | 27.21 | [M−H]− | 491.0973 | 491.0972 | C26H19O10 | −0.1 | MS2[491]: 293(100) | Salvianolic acid Ca,b | × | × | √ | × |
| Tanshinones | ||||||||||||
| 34 | 9.08 | [M+H]+ | 345.0969 | 345.0976 | C18H17O7 | 2.0 | MS2[345]: 327(100), 281(53) MS3[327]: 309(14), 281(100) | Demethyl and trihydroxyl tanshinone IIB | × | × | √ | × |
| 35 | 12.06 | [M+H]+ | 517.1704 | 517.1683 | C26H29O11 | −2.1 | Methyl dihydrotanshinonate glucuronide conjugate | × | √ | × | × | |
| 36 | 12.77 | [M+H]+ | 343.0812 | 343.0814 | C18H15O7 | 0.5 | MS2[343]: 325(92), 297(100) MS3[297]: 279(100), 255(56) | Demethyl and carboxylated tanshindiol A/B/C | × | × | √ | × |
| 37 | 13.06 | [M+H]+ | 329.1020 | 329.1023 | C18H17O6 | 1.1 | MS2[329]: 311(100), 265(58) MS3[311]: 283(25), 265(100) | Demethyl and two hydroxyl tanshinone IIB | × | × | √ | × |
| 38 | 13.37 | [M+H]+ | 620.1909 | 620.1917 | C28H34O11N3S | 1.3 | MS2[620]: 602(55), 584(32), 455(100) | Tanshindiol A/B/C glutathione conjugate | × | × | × | √ |
| 39 | 13.66 | [M+H]+ | 343.0812 | 343.0814 | C18H15O7 | 0.5 | MS2[343]: 325(95), 297(100) MS3[297]: 279(100), 255(60) | Demethyl and carboxylated tanshindiol A/B/C | × | × | √ | × |
| 40 | 14.04 | [M+H]+ | 471.1286 | 471.1286 | C24H23O10 | 0.1 | MS2[471]: 295(57), 277(23), 267(100) MS3[267]: 249(50), 237(100) | Hydroxyl and glucuronidated dihydrotanshinone I | × | √ | × | × |
| 41 | 14.25 | [M−H]− | 487.1235 | 487.1232 | C24H23O11 | −0.6 | MS2[487]: 311(100) MS3[311]: 283(30), 281(100) | Hydroxyl and glucuronidated tanshinol B | × | √ | × | × |
| 42 | 14.64 | [M+H]+ | 473.1442 | 473.1440 | C24H25O10 | −0.5 | MS2[473]: 297(80), 279(100), 251(39) MS3[279]: 261(100), 251(100) | Hydroxyl and glucuronidated danshenxinkun B | × | √ | × | × |
| 43 | 16.60 | [M+H]+ | 327.0863 | 327.0866 | C18H15O6 | 0.7 | MS2[327]: 309(100), 281(67), 265(12) MS3[309]: 291(13), 281(100) | Hydroxyl and dehydro tanshindiol A/B/C | × | × | √ | × |
| 44 | 18.89 | [M+H]+ | 345.1333 | 345.1338 | C19H21O6 | 1.5 | MS2[345]: 327(100), 309(23) MS3[327]: 309(100), 281(19) | Hydrated and hydroxyl tanshinone IIB | × | √ | × | × |
| 45 | 19.32 | [M+H]+ | 301.1434 | 301.1433 | C18H21O4 | −0.4 | MS2[301]: 283(100) | Demethyl neocryptotanshinone | √ | × | × | × |
| 46 | 19.43 | [M+H]+ | 301.1434 | 301.1439 | C18H21O4 | 1.6 | MS2[301]: 283(100) MS3[283]: 265(100), 255(26) | Demethyl and two hydroxyl miltirone | × | √ | × | × |
| 47 | 19.81 | [M−H]− | 313.0707 | 313.0708 | C17H13O6 | 0.4 | MS2[313]: 269(27), 252(100) | Demethyl and two hydroxyl tanshinol B | × | × | √ | × |
| 48 | 20.40 | [M+H]+ | 355.1176 | 355.1157 | C20H19O6 | −2.0 | MS2[355]: 337(100), 309(45) | Hydroxyl methyltanshinonate | × | √ | × | × |
| 49 | 20.47 | [M+H]+ | 489.1391 | 489.1397 | C24H25O11 | 1.1 | MS2[489]: 313(100), 295(57) | Tanshindiol A/B/C glucuronide conjugate | √ | × | × | × |
| 50 | 21.17 | [M+H]+ | 313.1071 | 313.1077 | C18H17O5 | 2.0 | MS2[313]: 295(100), 265(78) | Tanshindiol A/B/Cb | √ | × | √ | × |
| 51 | 21.54 | [M+H]+ | 339.1227 | 339.1209 | C20H19O5 | −1.8 | MS2[339]: 321(100) | Methyl tanshinonate | × | √ | × | × |
| 52 | 22.00 | [M+H]+ | 297.1121 | 297.1129 | C18H17O4 | 2.4 | MS2[297]: 279(100), 251(49), 237(36) | Hydrated dihydrotanshinone I | × | √ | × | × |
| 53 | 22.49 | [M+H]+ | 313.1071 | 313.1077 | C18H17O5 | 2.0 | MS2[313]: 295(100), 267(7) MS3[295]: 267(100) | Tanshindiol A/B/Cb | √ | √ | √ | × |
| 54 | 22.98 | [M+H]+ | 309.0758 | 309.0763 | C18H13O5 | 1.8 | MS2[309]: 291(18), 265(20), 235(100) MS3[235]: 207(18), 179(100) | Dihydroxyl tanshinone I | × | √ | × | × |
| 55 | 23.60 | [M+H]+ | 295.0601 | 295.0603 | C17H11O5 | 0.6 | MS2[295]: 267(100) MS3[267]: 239(100) | Demethyl and two hydroxyl tanshinone I | × | × | √ | × |
| 56 | 23.87 | [M+H]+ | 299.1278 | 299.1281 | C18H19O4 | 0.5 | MS2[299]: 281(42), 271(49), 253(100) | Demethyl and hydroxyl cryptotanshinone | × | √ | × | × |
| 57 | 23.89 | [M−H]− | 311.0914 | 311.0915 | C18H15O5 | 0.3 | MS2[311]: 283(32), 281(100) MS3[281]: 253(100) | Hydroxyl tanshinol B | × | √ | × | × |
| 58 | 24.85 | [M+H]+ | 299.1278 | 299.1284 | C18H19O4 | 1.1 | MS2[299]: 281(19), 271(44), 253(100) | Demethyl and hydroxyl cryptotanshinone | √ | √ | × | × |
| 59 | 25.78 | [M−H]− | 325.0707 | 325.0706 | C18H13O6 | −0.3 | MS2[325]: 297(31), 295(100), 268(30) MS3[294]: 267(100) | Demethyl and carboxylated tanshinol B | × | √ | × | × |
| 60 | 26.06 | [M+H]+ | 313.1071 | 313.1074 | C18H17O5 | 1.3 | MS2[313]: 295(100) | Tanshindiol A/B/Cb | √ | √ | √ | × |
| 61 | 26.21 | [M+H]+ | 345.1333 | 345.1341 | C19H21O6 | 0.5 | MS2[345]: 327(100) MS3[327]: 299(100), 281(13) | Demethyl and carboxylated neocryptotanshinone | × | √ | × | × |
| 62 | 28.01 | [M−H]− | 297.1121 | 297.1124 | C18H17O4 | 0.9 | MS2[297]: 253(100), 239(74), 221(26) MS3[253]: 238(100) | Dihydro tanshinol B | × | √ | × | × |
| 63 | 28.04 | [M+H]+ | 343.1176 | 343.1184 | C19H19O6 | 0.1 | MS2[343]: 325(100) | Dihydroxyl tanshinone IIB | × | √ | √ | × |
| 64 | 28.27 | [M+H]+ | 372.1806 | 372.1811 | C21H26O5N | 1.7 | MS2[372]: 354(100), 311(55), 283(60) MS3[354]: 326(100), 311(59) | Neocryptotanshinone glycine conjugate | × | √ | √ | × |
| 65 | 28.44 | [M+H]+ | 313.1434 | 313.1437 | C19H21O4 | −0.5 | MS2[313]: 269(100) MS3[269]: 254(25), 223(21), 199(100), 171(74) | Hydroxyl cryptotanshinoneb | √ | √ | √ | × |
| 66 | 28.70 | [M−H]− | 471.1286 | 471.1286 | C24H23O10 | 0.0 | MS2[471]: 295(100) | Tanshinol B glucuronide conjugate | × | √ | × | × |
| 67 | 28.73 | [M+H]+ | 331.1540 | 331.1545 | C19H23O5 | 0.7 | MS2[331]: 313(100), 295(38) | Hydroxyl and methyl salvianonol | √ | × | × | × |
| 68 | 28.80 | [M+H]+ | 459.1286 | 459.1287 | C23H23O10 | 1.5 | MS2[459]: 283(100) MS3[283]: 265(25), 237(100) | Dihydronortanshinone glucuronide conjugate | × | √ | × | × |
| 69 | 28.99 | [M−H]− | 471.1286 | 471.1286 | C24H23O10 | 0.1 | MS2[471]: 295(100) | Tanshinol B glucuronide conjugate | × | √ | × | × |
| 70 | 29.00 | [M+H]+ | 618.2116 | 618.2119 | C29H36O10N3S | −0.4 | MS2[618]: 471(44), 309(100) | Tanshinone IIB glutathione conjugate | × | × | × | √ |
| 71 | 29.01 | [M+H]+ | 327.1227 | 327.1231 | C19H19O5 | 1.6 | MS2[327]: 309(24), 299(22), 281(100), 263(57) MS3[281]: 263(100), 235(36) | Hydroxyl tanshinone IIB | × | √ | × | × |
| 72 | 29.41 | [M+H]+ | 309.1121 | 309.1121 | C19H17O4 | −2.0 | MS2[309]: 265(100) MS3[265]: 247(51), 223(100), 195(18) | Hydroxyl and dehydro tanshinone IIA | √ | × | × | × |
| 73 | 29.54 | [M+H]+ | 343.1176 | 343.1185 | C19H19O6 | 1.1 | MS2[343]: 325(100) | Dihydroxyl tanshinone IIB | × | √ | × | × |
| 74 | 30.06 | [M+H]+ | 313.1434 | 313.1438 | C19H21O4 | 1.1 | MS2[313]: 269(35), 251(100) MS3[251]: 223(100) | Hydroxyl cryptotanshinone | × | √ | × | × |
| 75 | 30.44 | [M+H]+ | 445.1857 | 445.1864 | C24H29O8 | −1.8 | MS2[445]: 269(100) MS3[269]: 254(100), 239(14) | Decarbonylated and glucuronidated cryptotanshinone | × | × | × | √ |
| 76 | 30.63 | [M+H]+ | 293.0808 | 293.0813 | C18H13O4 | 2.4 | MS2[293]: 275(22), 263(100), 249(23) MS3[263]: 235(100) | Hydroxyl tanshinone I | √ | √ | √ | × |
| 77 | 31.05 | [M+H]+ | 295.0965 | 295.0966 | C18H15O4 | 1.8 | MS2[295]: 251(100) MS3[251]: 223(97), 195(95), 169(100) | Hydrated tanshinone Ib | √ | √ | × | × |
| 78 | 31.08 | [M+H]+ | 297.1121 | 297.1124 | C18H17O4 | 0.6 | MS2[297]: 279(100), 261(36) MS3[279]: 261(100) | Demethyl and hydroxyl tanshinone IIA | √ | √ | √ | × |
| 79 | 31.30 | [M+H]+ | 285.1485 | 285.1489 | C18H21O3 | 1.0 | MS2[285]: 243(100), 229(6) MS3[243]: 228(71), 225(92), 1815(100) | Demethyl and hydroxyl miltirone | √ | √ | × | × |
| 80 | 31.32 | [M−H]− | 311.0914 | 311.0912 | C18H15O5 | −0.7 | MS2[311]: 267(100), 223(29) | Hydroxyl tanshinol B | × | √ | × | × |
| 81 | 31.36 | [M+H]+ | 473.1806 | 473.1806 | C25H29O9 | 2.3 | MS2[473]: 297(36), 269(100) MS3[269]: 241(100), 213(92), 199(87) | Cryptotanshinone glucuronide conjugate | × | × | √ | × |
| 82 | 31.61 | [M+H]+ | 339.1227 | 339.1210 | C20H19O5 | 1.2 | MS2[339]: 321(100), 295(21) | Methyl tanshinonate | × | √ | × | × |
| 83 | 32.13 | [M+H]+ | 459.2014 | 459.2012 | C25H31O8 | 2.4 | MS2[459]: 283(100) | Miltirone glucuronide conjugate | × | × | × | √ |
| 84 | 32.36 | [M+H]+ | 381.1333 | 381.1318 | C22H21O6 | 2.2 | MS2[381]: 363(60), 337(100) | Acetylated methyltanshinone | √ | × | × | × |
| 85 | 32.65 | [M+H]+ | 311.1278 | 311.1282 | C19H19O4 | 1.4 | MS2[311]: 293(40), 275(37), 267(100) MS3[267]: 252(100) | Tanshinone IIBa,b | √ | √ | √ | √ |
| 86 | 32.84 | [M+H]+ | 287.1642 | 287.1645 | C18H23O3 | 0.8 | MS2[287]: 269(100) MS3[269]: 251(58), 241(100), 213(86) | Decarbonylated neocryptotanshinone | √ | × | × | × |
| 87 | 33.16 | [M+H]+ | 299.1642 | 299.1646 | C19H23O3 | 1.4 | MS2[299]: 281(100), 255(68), 253(62) | Hydroxyl miltirone | × | √ | × | × |
| 88 | 33.24 | [M+H]+ | 267.1016 | 267.1022 | C17H15O3 | 0.3 | MS2[267]: 249(100), 221(22) | Decarbonylated hydrated tanshinone I | √ | × | × | × |
| 89 | 33.31 | [M+H]+ | 457.1493 | 457.1496 | C24H25O9 | 0.5 | MS2[457]: 281(100), 263(73), 261(41) MS3[281]: 263(100) | Danshenxinkun B glucuronide conjugate | × | × | × | √ |
| 90 | 33.59 | [M+H]+ | 341.1384 | 341.1388 | C20H21O5 | 1.2 | MS2[341]: 281(100), 263(42) MS3[281]: 263(100), 235(18) | Methyl dihydrotanshinonateb | × | × | √ | × |
| 91 | 33.67 | [M+H]+ | 279.1016 | 279.1021 | C18H15O3 | −0.2 | MS2[279]: 261(100) MS3[261]: 233(100), 205(13) | Dihydro tanshinone I | √ | × | × | √ |
| 92 | 33.69 | [M−H]− | 295.0965 | 295.0968 | C18H15O4 | 1.2 | MS2[295]: 277(17), 267(28), 265(100), 238(27) MS3[265]: 237(100) | Tanshinol Ba,b | √ | √ | √ | √ |
| 93 | 33.73 | [M+H]+ | 309.1121 | 309.1120 | C19H17O4 | 2.5 | MS2[309]: 265(100) MS3[265]: 247(55), 223(100) | Hydroxyl and dehydro tanshinone IIA | √ | √ | × | × |
| 94 | 34.29 | [M+H]+ | 297.1485 | 297.1489 | C19H21O3 | 0.3 | MS2[297]: 253(100) MS3[253]: 238(33), 225(24), 211(100), 209(16) | Hydroxyl dehydromiltirone | √ | × | × | × |
| 95 | 34.61 | [M+H]+ | 293.1172 | 293.1174 | C19H17O3 | 1.5 | MS2[293]: 275(100), 247(39) MS3[275]: 247(100) | Dehydrotanshinone IIAb | × | × | √ | × |
| 96 | 34.63 | [M+H]+ | 297.1485 | 297.1490 | C19H21O3 | 1.6 | MS2[297]: 279(100), 251(81) MS3[279]: 251(100), 237(67) | Dihydro tanshinone IIA | × | × | × | √ |
| 97 | 34.72 | [M+H]+ | 311.1278 | 311.1284 | C19H19O4 | 1.8 | MS2[311]: 283(100) MS3[283]: 265(100), 237(17) | Hydroxyl tanshinone IIA | √ | × | × | × |
| 98 | 35.14 | [M+H]+ | 279.1016 | 279.1020 | C18H15O3 | 1.4 | MS2[279]: 261(100), 233(5) MS3[261]: 233(100), 215(7), 205(14) | Dihydrotanshinone Ib | × | √ | √ | √ |
| 99 | 35.36 | [M+H]+ | 293.0808 | 293.0815 | C18H13O4 | 2.2 | MS2[293]: 249(100) MS3[249]: 221(26), 193(100), 178(52) | Hydroxyl hydrotanshinone I | × | × | × | √ |
| 100 | 35.69 | [M+H]+ | 315.1591 | 315.1594 | C19H23O4 | 1.0 | MS2[315]: 297(100) MS3[297]: 279(100), 268(13), 254(18), 251(58) | Hydrated cryptotanshinone | √ | × | × | √ |
| 101 | 35.83 | [M+H]+ | 281.1172 | 281.1178 | C18H17O3 | 2.2 | MS2[281]: 263(100), 235(72) MS3[263]: 235(100) | Danshenxinkun Ba b | × | √ | √ | × |
| 102 | 35.96 | [M+H]+ | 339.1227 | 339.1235 | C20H19O5 | 2.5 | MS2[339]: 279(100) MS3[279]: 261(100) | Methyl tanshinonateb | × | × | √ | √ |
| 103 | 36.22 | [M+H]+ | 295.1329 | 295.1331 | C19H19O3 | 0.9 | MS2[295]: 277(100), 249(40) MS3[277]: 262(53), 249(100), 235(38) | Dehydrocryptotanshinoneb | × | × | √ | × |
| 104 | 36.59 | [M+H]+ | 309.1121 | 309.1124 | C19H17O4 | 0.8 | MS2[309]: 265(100) MS3[265]: 247(52), 223(100) | Hydroxyl and methyl dihydrotanshinone I | × | × | × | √ |
| 105 | 36.67 | [M+H]+ | 301.1798 | 301.1802 | C19H25O3 | 1.4 | MS2[301]: 271(100) MS3[271]: 256(100) | Hydrated miltirone | × | × | √ | × |
| 106 | 36.96 | [M+H]+ | 313.1434 | 313.1439 | C19H21O4 | 1.5 | MS2[313]: 295(100), 277(31), 271(33), 267(25) MS3[295]: 277(100), 253(27), 249(23) | Hydroxyl cryptotanshinone | √ | × | × | × |
| 107 | 37.31 | [M+H]+ | 297.1485 | 297.1488 | C19H21O3 | 1.0 | MS2[297]: 279(100), 251(81) MS3[279]: 251(100), 237(73) | Cryptotanshinonea b | × | √ | √ | × |
| 108 | 37.58 | [M+H]+ | 277.0859 | 277.0863 | C18H13O3 | 1.4 | MS2[277]: 249(100), 231(13) MS3[249]: 234(18), 221(89), 193(100), 178(30) | Tanshinone Ia b | √ | √ | √ | × |
| 109 | 38.82 | [M+H]+ | 279.1016 | 279.1017 | C18H15O3 | 0.6 | MS2[279]: 261(100) MS3[261]: 233(100) | Dihydrotanshinone Ia b | × | √ | √ | √ |
| 110 | 38.97 | [M+H]+ | 293.1172 | 293.1178 | C19H17O3 | 2.1 | MS2[293]: 275(100), 247(40) MS3[275]: 247(100) | Dehydrotanshinone IIAb | √ | √ | √ | √ |
| 111 | 40.01 | [M+H]+ | 281.1536 | 281.1540 | C19H21O2 | 1.4 | Dehydromiltironeb | × | × | √ | × | |
| 112 | 40.23 | [M+H]+ | 293.1172 | 293.1175 | C19H17O3 | 1.0 | Dehydrotanshinone IIAb | √ | √ | √ | × | |
| 113 | 40.58 | [M−H]− | 277.0859 | 277.0865 | C18H13O3 | 2.1 | MS2[277]: 249(100), 221(53) | Dehydrated tanshinol B | × | × | √ | × |
| 114 | 41.15 | [M+H]+ | 295.1329 | 295.1331 | C19H19O3 | 0.9 | MS2[295]: 277(100), 249(14) MS3[277]: 262(29), 249(100) | Tanshinone IIAa b | √ | √ | √ | √ |
| 115 | 41.27 | [M−H]− | 277.0859 | 277.0863 | C18H13O3 | 1.4 | MS2[277]: 249(100), 221(60) | Dehydrated tanshinol B | × | × | √ | × |
| 116 | 42.32 | [M+H]+ | 283.1693 | 283.1694 | C19H23O2 | 0.4 | MS2[283]: 265(100), 241(47), 223(63) MS3[265]: 237(62), 223(100) | Miltironeb | × | √ | √ | × |
| 117 | 42.70 | [M+H]+ | 269.1536 | 269.1537 | C18H21O2 | 0.4 | MS2[269]: 254(100) MS3[254]: 239(100) | Demethyl miltirone | × | × | √ | × |
| 118 | 47.15 | [M+H]+ | 299.1642 | 299.1646 | C19H23O3 | 1.3 | MS2[299]: 281(100), 256(52), 253(49) | Dihydro cryptotanshinone | × | × | √ | × |
| Total | 35 | 63 | 62 | 18 | ||||||||
Confirmed by reference standards;
Original components in danshen extract.
"×":detected;"√":undetected
4. Discussion
To better identify the metabolites of danshen in vivo after oral administration, the original components of danshen were identified by HPLC-MS/MS in both negative and positive modes. According to the literature (Wei et al., 2007; Lv et al., 2010), the responses of phenolic acids are more sensitive to negative mode, while those of tanshinones are more sensitive to positive mode. In this study, the phenolic acids were detected in negative mode, and exhibited their parent ions as [M−H]−; the tanshinones were detected in positive mode, and exhibited their parent ions of [M+H]+ and/or [M+Na]+.
Although 10% hydrochloric acid was added to rat plasma to increase the recovery ratios of phenolic acids, few phenolic acids were detected. This may have been because of the low bioavailability and transformation of phenolic acids in vivo (Gao et al., 2009; Sun et al., 2013). Under our experimental conditions, very few phenolic acids and their metabolites were detected in rat bile, except methyl danshensu. Compared with plasma and bile, many more metabolites were detected and unambiguously identified in rat urine and feces. This suggests that urine and feces might be the major route for elimination of danshen after oral administration.
From the analysis of metabolites, we found that hydroxylation (36 out of 118), methylation/demethylation (35 out of 118), glucuronidation (14 out of 118), hydration/dehydration (8 out of 118), and hydrogenation/dehydrogenation (11 out of 118) might be the main metabolic pathways of danshen in vivo. The metabolic pathway for phenolic acids was mainly methylation/demethylation (11 out of 33), while tanshinones mostly showed hydroxylation (31 out of 85) and methylation/demethylation (19 out of 85). Hydrogenation, sulfation, acetylation, and glutathione conjugation were found to be the possible metabolic pathways of danshen. This research provided a comprehensive in vitro chemical profile and in vivo metabolic profile of danshen after oral administration, which could be useful in research on the quality control and pharmacology of danshen.
5. Conclusions
Using HPLC-MS/MS methods, our research provided the most comprehensive chemical and metabolic profiles of danshen. A total of 69 compounds in danshen extract and 118 metabolites were identified, including 35 in plasma, 63 in urine, 62 in feces, and 18 in bile. This analysis of chemical and metabolic components of danshen lays a foundation for further studies of the material composition of danshen, and provides a useful means for identification of multi-components of TCMs both in vitro and in vivo.
Footnotes
Project supported by the Ministry of Science and Technology of China (No. 2011ZX09201-201-22)
Compliance with ethics guidelines: Huan-huan PANG, Mei-fang JIANG, Qin-hui WANG, Xiao-ye WANG, Wei GAO, Zhi-hao TIAN, and Jian-mei HUANG declare that they have no conflict of interest.
All institutional and national guidelines for the care and use of laboratory animals were followed.
References
- 1.Belin DCE, Vessieres E, Guihot AL, et al. Type 2 diabetes severely impairs structural and functional adaptation of rat resistance arteries to chronic changes in blood flow. Cardiovasc Res. 2009;81(4):788–796. doi: 10.1093/cvr/cvn334. [DOI] [PubMed] [Google Scholar]
- 2.Cai W, Zhang JY, Dong LY, et al. Identification of the metabolites of Ixerin Z from Ixeris sonchifolia Hance in rats by HPLC-LTQ-Orbitrap mass spectrometry. J Pharm Biomed Anal. 2015;107:290–297. doi: 10.1016/j.jpba.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Duan H, Huang J, Li W, et al. Protective effects of Fufang Xueshuantong on diabetic retinopathy in rats. Evid Based Complement Alternat Med, 2013:408268. 2013 doi: 10.1155/2013/408268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao DY, Han LM, Zhang LH, et al. Bioavailability of salvianolic acid B and effect on blood viscosities after oral administration of salvianolic acids in beagle dogs. Arch Pharm Res. 2009;32(5):773–779. doi: 10.1007/s12272-009-1517-2. [DOI] [PubMed] [Google Scholar]
- 5.Ji X, Tan BK, Zhu YC, et al. Comparison of cardioprotective effects using ramipril and DanShen for the treatment of acute myocardial infarction in rats. Life Sci. 2003;73(11):1413–1426. doi: 10.1016/S0024-3205(03)00432-6. [DOI] [PubMed] [Google Scholar]
- 6.Kang DG, Oh H, Sohn EJ, et al. Lithospermic acid B isolated from Salvia miltiorrhiza ameliorates ischemia/reperfusion-induced renal injury in rats. Life Sci. 2004;75(15):1801–1816. doi: 10.1016/j.lfs.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 7.Lam BY, Lo AC, Sun X, et al. Neuroprotective effects of tanshinones in transient focal cerebral ischemia in mice. Phytomedicine. 2003;10(4):286. doi: 10.1078/094471103322004776. [DOI] [PubMed] [Google Scholar]
- 8.Li X, Yu C, Lu Y, et al. Pharmacokinetics, tissue distribution, metabolism, and excretion of depside salts from Salvia miltiorrhiza in rats. Drug Metab Dispos. 2007;35(2):234–239. doi: 10.1124/dmd.106.013045. [DOI] [PubMed] [Google Scholar]
- 9.Liang J, Xu F, Zhang Y, et al. The profiling and identification of the absorbed constituents and metabolites of Paeoniae Radix Rubra decoction in rat plasma and urine by the HPLC-DAD-ESI-IT-TOF-MSn technique: a novel strategy for the systematic screening and identification of absorbed constituents and metabolites from traditional Chinese medicines. J Pharmaceut Biomed. 2013;83:108–121. doi: 10.1016/j.jpba.2013.04.029. [DOI] [PubMed] [Google Scholar]
- 10.Liu AH, Guo H, Ye M, et al. Detection, characterization and identification of phenolic acids in Danshen using high-performance liquid chromatography with diode array detection and electrospray ionization mass spectrometry. J Chromatogr A. 2007;1161(1-2):170–182. doi: 10.1016/j.chroma.2007.05.081. [DOI] [PubMed] [Google Scholar]
- 11.Liu M, Li YG, Zhang F, et al. Chromatographic fingerprinting analysis of Danshen root (Salvia miltiorrhiza Radix et Rhizoma) and its preparations using high performance liquid chromatography with diode array detection and electrospray mass spectrometry (HPLC-DAD-ESI/MS) J Sep Sci. 2007;30(14):2256–2267. doi: 10.1002/jssc.200700149. [DOI] [PubMed] [Google Scholar]
- 12.Liu M, Zhao S, Wang Z, et al. Tentative identification of new metabolites of epimedin C by liquid chromatography-mass spectrometry. J Sep Sci. 2011;34(22):3200. doi: 10.1002/jssc.201100581. [DOI] [PubMed] [Google Scholar]
- 13.Lv Y, Zhang X, Liang X, et al. Characterization of the constituents in rat biological fluids after oral administration of Fufang Danshen tablets by ultra-performance liquid chromatography/quadrupole time-of-flight mass spectrometry. J Pharm Biomed Anal. 2010;52(1):155–159. doi: 10.1016/j.jpba.2009.12.013. [DOI] [PubMed] [Google Scholar]
- 14.Su CY, Ming QL, Rahman K, et al. Salvia miltiorrhiza: traditional medicinal uses, chemistry, and pharmacology. Chin J Nat Med. 2015;13(3):163–182. doi: 10.1016/S1875-5364(15)30002-9. [DOI] [PubMed] [Google Scholar]
- 15.Sun J, Huang SH, Tan BK, et al. Effects of purified herbal extract of Salvia miltiorrhiza on ischemic rat myocardium after acute myocardial infarction. Life Sci. 2005;76(24):2849–2860. doi: 10.1016/j.lfs.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 16.Sun J, Zhang L, Song J, et al. Pharmacokinetic study of salvianolic acid A in beagle dog after oral administration by a liquid chromatography-mass spectrometry method: a study on bioavailability and dose proportionality. J Ethnopharmacol. 2013;148(2):617–623. doi: 10.1016/j.jep.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Sun JH, Yang M, Wang XM, et al. Identification of tanshinones and their metabolites in rat bile after oral administration of TTE-50, a standardized extract of Salvia miltiorrhiza by HPLC-ESI-DAD-MSn . J Pharm Biomed Anal. 2007;44(2):564–574. doi: 10.1016/j.jpba.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Wang F, Zhang Q, Lu Z, et al. Identification of chemical constituents in traditional Chinese medicine formula using HPLC coupled with linear ion trap-Orbitrap MS from high doses of medicinal materials to equivalent doses of formula: study on Xiang-Sha-Liu-Jun-Zi-Jia-Jian granules. J Sep Sci. 2016;39(9):1619–1627. doi: 10.1002/jssc.201501223. [DOI] [PubMed] [Google Scholar]
- 19.Wang YH, Qiu C, Wang DW, et al. Identification of multiple constituents in the traditional Chinese medicine formula Sheng-Mai San and rat plasma after oral administration by HPLC-DAD-MS/MS. J Pharm Biomed Anal. 2011;54(5):1110–1127. doi: 10.1016/j.jpba.2010.11.034. [DOI] [PubMed] [Google Scholar]
- 20.Wei Y, Li P, Shu B, et al. Analysis of chemical and metabolic components in traditional Chinese medicinal combined prescription containing Radix Salvia miltiorrhiza and Radix Panax notoginseng by LC-ESI-MS methods. Biomed Chromatogr. 2007;21(8):797–809. doi: 10.1002/bmc.775. [DOI] [PubMed] [Google Scholar]
- 21.Wu YT, Chen YF, Hsieh YJ, et al. Bioavailability of salvianolic acid B in conscious and freely moving rats. Int J Pharm. 2006;326(1-2):25–31. doi: 10.1016/j.ijpharm.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 22.Yang R, Chang L, Guo BY, et al. Compound danshen dripping pill pretreatment to prevent contrast-induced nephropathy in patients with acute coronary syndrome undergoing percutaneous coronary intervention. Evid Based Complement Alternat Med, 2014:256268. 2014 doi: 10.1155/2014/256268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, He Y, Cui M, et al. Metabolic studies on the total phenolic acids from the roots of Salvia miltiorrhiza in rats. Biomed Chromatogr. 2005;19(1):51–59. doi: 10.1002/bmc.415. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Cai W, Zhou Y, et al. Profiling and identification of the metabolites of baicalin and study on their tissue distribution in rats by ultra-high-performance liquid chromatography with linear ion trap-Orbitrap mass spectrometer. J Chromatogr B Anal Technol Biomed Life Sci. 2015;985:91–102. doi: 10.1016/j.jchromb.2015.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Zhao X, Yang DH, Xu F, et al. The in vivo absorbed constituents and metabolites of Danshen decoction in rats identified by HPLC with electrospray ionization tandem ion trap and time-of-flight mass spectrometry. Biomed Chromatogr. 2015;29(2):285–304. doi: 10.1002/bmc.3275. [DOI] [PubMed] [Google Scholar]