Abstract
Genomic analyses have identified only a handful of robust risk loci for major depressive disorder (MDD). In addition to the published genome-wide significant genes, it is believed that there are undiscovered ‘treasures’ underlying the current MDD genome-wide association studies (GWASs) and gene expression data sets, and digging into these data will allow better understanding of the illness and development of new therapeutic approaches. For this purpose, we performed a meta-analytic study combining three MDD GWAS data sets (23andMe, CONVERGE, and PGC), and then conducted independent replications of significant loci in two additional samples. The genome-wide significant variants then underwent explorative analyses on MDD-related phenotypes, cognitive function alterations, and gene expression in brains. In the discovery meta-analysis, a previously unidentified single-nucleotide polymorphism (SNP) rs9540720 in the PCDH9 gene was genome-wide significantly associated with MDD (p=1.69 × 10–8 in a total of 89 610 cases and 246 603 controls), and the association was further strengthened when additional replication samples were included (p=1.20 × 10–8 in a total of 136 115 cases and 355 275 controls). The risk SNP was also associated with multiple MDD-related phenotypes and cognitive function impairment in diverse samples. Intriguingly, the risk allele of rs9540720 predicted lower PCDH9 expression, consistent with the diagnostic analysis results that PCDH9 mRNA expression levels in the brain and peripheral blood tissues were reduced in MDD patients compared with healthy controls. These convergent lines of evidence suggest that PCDH9 is likely a novel risk gene for MDD. Our study highlights the necessity and importance of excavating the public data sets to explore risk genes for MDD, and this approach is also applicable to other complex diseases.
Introduction
Major depressive disorder (MDD) is a severe and complex mental disorder with a lifetime prevalence of ~15% (Hasin et al, 2005), and was estimated to be the second leading cause of disability in the near future (Kessler and Bromet, 2013) by the World Health Organization. The overall heritability of MDD is 31–42% (Sullivan et al, 2000), with certain subsets being more heritable (eg, recurrent early-onset MDD) (Levinson, 2006). Although this trait of MDD suggests potential opportunity to understand the disease via genetic analyses, such modest heritability has greatly complicated the search for risk or protective genetic loci. This is reflected by the fact that previous candidate gene studies (Bosker et al, 2011) and early genome-wide association studies (GWASs) (Kohli et al, 2011; Lewis et al, 2010; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al, 2013; Muglia et al, 2010; Rietschel et al, 2010; Shi et al, 2011; Wray et al, 2012) were unable to identify large numbers of robust MDD genetic variants. Until recently, two landmark MDD GWAS studies named 23andMe (European descent) (Hyde et al, 2016) and CONVERGE (Han Chinese population) (Converge consortium, 2015) were published, and have made promising contributions to the understanding of MDD. 23andMe GWAS used the largest sample size so far, and CONVERGE GWAS focused on female subjects with recurrent MDD to reduce phenotypic heterogeneity. These two GWASs successfully discovered 17 (15 by 23andMe and 2 by CONVERGE consortium) genetic loci yielding genome-wide significance (detailed information about the genome-wide significant risk variants is listed in Supplementary Table S1).
Despite the recent achievements, GWAS approaches in MDD so far still seem to be less successful than in other complex diseases such as schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014). This is likely because of the aforementioned higher prevalence but lower heritability of MDD compared with other adult psychiatric disorders (such as schizophrenia), and more cases are thus required to detect the same number of single-nucleotide polymorphism (SNP) associations (Levinson et al, 2014). Therefore, future meta-analyses of diverse genome-wide data with increasing sample sizes are in critical need. Indeed, meta-analysis has become essential in genetic analyses of complex human diseases, and many recent studies in which meta-analyses combine dozens of GWAS data sets have shed light on the genetic architectures of complex diseases such as schizophrenia (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014), Crohn’s disease (Franke et al, 2010), and type 2 diabetes mellitus (Voight et al, 2010). In addition to genome-wide meta-analysis of SNPs, convergent functional genomics approaches have also aided in identification of potential susceptibility genes and molecular mechanisms for psychiatric disorders including schizophrenia (Ayalew et al, 2012; Luo et al, 2014) and bipolar disorder (Le-Niculescu et al, 2007; Ogden et al, 2004). Integration of genome-wide meta-analysis of SNPs and convergent functional genomics data is believed to be an efficient strategy in dissecting the genetic and biological basis of complex illnesses.
To date, GWASs of MDD by 23andMe (Hyde et al, 2016), CONVERGE (Converge consortium, 2015), and PGC (Psychiatric Genomics Consortium) (Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al, 2013) consortia have (fully or partially) released their genome-wide results. In addition, many genome-wide gene expression analyses in brain tissues have also shared their data (Colantuoni et al, 2011; GTEx Consortium, 2013; Ramasamy et al, 2014), and analyses on subjects with MDD are included (Kim and Webster, 2009, 2010). We believe that systemic utilization of these public resources allows identification of novel MDD risk genes, and will provide valuable information that is beneficial for other psychiatric studies. Besides, MDD patients often exhibit certain related phenotypes and impaired cognitive abilities compared with healthy individuals (Souery et al, 2007; Taylor Tavares et al, 2007), and accumulating data indicate that genetic loci associated with MDD are also related to these phenotypes in humans (Demirkan et al, 2011; Schuhmacher et al, 2013; Vrijsen et al, 2015). So far, researchers have released the data and resources of several GWASs on MDD-related phenotypes such as neuroticism and depressive symptoms (Okbay et al, 2016a), preschool internalizing problems (Benke et al, 2014), anxiety symptoms (Otowa et al, 2016), as well as cognitive functions (Benyamin et al, 2014; Okbay et al, 2016b), providing valuable tools for further analyses on these phenotypes to strengthen the association between the risk SNPs and MDD and also to reveal potential biological mechanisms. In the current study, we conducted integrative analyses using these data to examine the genetic risk of MDD from convergent perspectives (Le-Niculescu et al, 2010; Niculescu, 2005; Ogden et al, 2004). We discovered a novel SNP rs9540720 in the PCDH9 gene conferring genome-wide significant risk of MDD, and this SNP was also associated with multiple MDD-related phenotypes and cognitive function alterations. Intriguingly, the risk allele of rs9540720 was associated with reduced expression of PCDH9, consistent with the significant downregulation of this gene in the brain and peripheral blood tissue of MDD patients compared with healthy controls. Collectively, our study supports the potential roles of PCDH9 in MDD susceptibility, and illustrates an example of comprehensive utilization of public resources to uncover the genetic risk factors of MDD.
Materials and methods
All the protocols and methods used in this study were approved by the institutional review board of the Kunming Institute of Zoology, Chinese Academy of Sciences.
MDD Discovery GWAS Data Set
23andMe GWAS sample
The 23andMe GWAS sample included 75 607 cases with self-declared depression (SDD) and 231 747 controls (Hyde et al, 2016). A previous study showed that SDD and clinically assessed MDD were highly correlated (r=1.00, SE=0.20) on common-variant-associated genetic effect (Zeng et al, 2016). We therefore consider SDD as an alternative phenotype for identifying common risk variants associated with MDD. The participating cohort was collected from the customer base of the consumer genetics company 23andMe (Eriksson et al, 2010; Hyde et al, 2016; Tung et al, 2011). Participants provided informed consents and the protocol was approved by an external AAHRPP-accredited institutional review board, Ethical and Independent Review Services. Detailed information of the samples, genotyping methods, and statistical analyses can be found in the original GWAS report (Hyde et al, 2016).
CONVERGE GWAS sample
The CONVERGE GWAS consisted of 5303 cases with MDD and 5337 controls (Converge consortium, 2015). Cases were diagnosed using the Composite International Diagnostic Interview (CIDI) (WHO lifetime version 2.1; Chinese version) following DSM-IV criteria. Preexisting records of bipolar disorder, psychosis or mental retardation, as well as drugs or alcohol abuse history before the first depressive episode were applied as excluding criteria. Controls were recruited from patients undergoing minor surgical procedures at the general hospitals or from local community centers. Detailed information of the samples, genotyping methods, and statistical analyses can be found in the original report (Converge consortium, 2015).
PGC GWAS sample
The PGC GWAS included 9240 patients and 9519 controls (Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al, 2013). Cases had diagnoses of DSM-IV lifetime MDD using structured diagnostic instruments through direct interviews by trained interviewers or clinician-administered DSM-IV checklists. Most samples ascertained cases from clinical sources, and most controls were randomly selected from the population and screened for their lifetime history of MDD. Detailed descriptions of the samples, data quality, genomic controls, and statistical analyses can be found in the original publication (Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al, 2013).
MDD Replication Data Set
23andMe Replication sample
The 23andMe Replication sample included 45 773 SDD cases and 106 354 controls independent from the discovery cohort (Hyde et al, 2016). The protocols and criteria of participants’ recruitment of the 23andMe Replication sample were the same as 23andMe GWAS sample, although the subjects were totally independent as previously described (Hyde et al, 2016). The 23andMe Replication sample was also used as the validation sample in the original 23andMe MDD GWAS study (Hyde et al, 2016).
Chinese MDD sample
This sample contained 732 MDD cases and 2318 controls as previously described (Zhang et al, 2014, 2016). All patients were diagnosed with MDD strictly according to the DSM-IV criteria. Standard diagnostic assessments were supplemented with clinical information obtained by a review of medical records and interviews with family informants. Patients were excluded either when they had a lifetime diagnosis of bipolar disorder, schizoaffective disorder, schizophrenia, or another psychotic disorder, or when they were female and were pregnant, planning to become pregnant, or breast-feeding during the study period. Control subjects were recruited from local volunteers without any history of mental disorders. All participants provided written informed consents.
SNP Selection and Statistical Analysis
SNPs highlighted in 23andMe GWAS sample were first subject to replication analyses in CONVERGE and PGC GWAS samples, and meta-analytic study was then conducted combining samples from all three data sets. Significant associations identified through the above meta-analysis were then replicated in the 23andMe Replication sample and Chinese MDD sample. In each sample, logistic regression was applied to test the association between phenotypes and SNP dosages under an additive model, and covariates included sample grouping and principal components reflecting ancestry. For the meta-analysis, we used odds ratio (OR) and standard error (SE) to estimate heterogeneity between individual samples and to calculate the pooled OR and 95% confidence interval (CI). The calculation was conducted using the classical inverse variance weighted methods with PLINK v1.07 (Purcell et al, 2007) in consistency with our previous study (Xiao et al, 2017). In the current study, we conducted two-tailed tests for discovery analysis and meta-analysis, and applied one-tailed tests for replication analyses as described before (Psychiatric GWAS Consortium Bipolar Disorder Working Group, 2011). A two-tailed p-value<5.00 × 10−8 was considered genome-wide statistically significant in the combined samples; in the replication sample, a one-tailed p-value<0.05 was considered nominally significant.
Analyses on MDD-Related Phenotypes
Neuroticism
Neuroticism is commonly defined as the proneness to negative emotions (including irritability, anger, sadness, anxiety, worry, hostility, self-consciousness, and vulnerability) usually in response to stress-inducing events (Kotov et al, 2010; Lahey, 2009). Neuroticism is a pervasive risk factor for different psychiatric conditions including MDD and entailed emotional dysregulation (Fanous and Kendler, 2004; Kotov et al, 2010). The neuroticism data (n=170 911) were obtained from a previous GWAS by Okbay et al (2016a). Detailed information of the samples, genotyping methods, and statistical analyses can be found in the original report (Okbay et al, 2016a).
Depressive symptoms (DS)
The DS data (n=180 866) were retrieved from a previous GWAS by Okbay et al (2016a). The authors analyzed the summary statistics from a GWAS of MDD (Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al, 2013) performed by the PGC (9240 cases and 9519 controls) in combination with data from two additional cohorts: the UKB (105 739 subjects) and the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort (7231 cases and 49 137 controls). Details on the samples, genotyping methods, and statistical analyses can be found in the original report (Okbay et al, 2016a).
Preschool internalizing problems (INTs)
INTs are heritable traits with moderate genetic stability from childhood into adulthood, and are found to be highly prevalent in the offspring of MDD patients (Olino et al, 2008). The INT data (n=4596) were taken from a previous GWAS by Benke et al (2014). They investigated the effects of SNPs on INT in a total of 4596 children (3 cohorts) with the Child Behavior Checklist (CBCL). Of the 36 items for INT scale in the most recent version of the CBCL 11/2 – 5, 34 were measured in all 3 cohorts. Example items include ‘Acts too young for age,’ ‘Worries a lot,’ and ‘Clings to adults or too dependent.’ For each item, the rater selected a score of 0 (not true), 1 (somewhat or sometimes true), or 2 (very true or often true), resulting in a potential score range of 0 to 68 for each child. Detailed information of the samples, genotyping methods, and statistical analyses can be found in the original study (Benke et al, 2014).
Anxiety phenotype
Anxiety disorders (ADs), namely generalized AD (GAD), panic disorder (PD), and phobias, are relatively common and often disabling conditions with the lifetime prevalence of over 20% (Kessler et al, 2005). The anxiety data (n=15 299) were from a previous GWAS by Otowa et al (2016). The anxiety phenotype was diagnosed based on DSM with some exceptions, and assessed using quantitative phenotypic factor scores (FSs) derived from a multivariate analysis combining information across the clinical phenotypes. Detailed information of the samples, genotyping methods, and statistical analyses can be found in the original paper (Otowa et al, 2016).
Analyses on Cognitive Function
Educational attainment
We used educational attainment as a ‘proxy phenotype’ for cognitive function. Although educational attainment is not a direct cognitive measure, it is correlated with cognitive ability (r~0.5) as well as personality traits related to persistence and self-discipline (Rietveld et al, 2013). Educational attainment is strongly associated with social outcomes, and there is a well-documented health-education gradient. It was estimated that ~40% of the variance in educational attainment is explained by genetic factors (Rietveld et al, 2013). For analysis of this phenotype, we used the data from a recent GWAS performed in 293 723 European individuals in whom education attainment was quantified with the well-characterized measurement ‘EduYears’ (an individual’s years of schooling) (Okbay et al, 2016b). Briefly, educational attainment was measured at an age at which participants were very likely to have completed their education (>95% of the samples were at least 30 years old). On average, participants had 13.3 years of schooling, and 23.1% had a College degree. Details on the samples, genotyping methods, and statistical analyses can be found in the original report (Okbay et al, 2016b).
Childhood intelligence
Intelligence, a quantifiable index of cognition, has been widely used in relevant genetic analyses given its great heritability and genetic stability, both in an individual’s life course and across generations (Deary et al, 2009). Moreover, childhood intelligence is a strong predictor of many important life outcomes including educational attainment (Deary, 2012), and is also associated with various psychiatric disorders such as schizophrenia and MDD (Batty et al, 2005; Koenen et al, 2009). As a result, we focused on the childhood intelligence phenotype measured with psychometric cognitive tests (Intelligence Quotient (IQ)-type tests). We utilized a recent GWAS of childhood intelligence including 12 441 children of European ancestry (Benyamin et al, 2014). In brief, the age of the participants ranged between 6 and 18 years. The best available measure of general cognitive ability (g) or intelligence quotient (IQ) derived from diverse tests assessing both verbal and nonverbal ability was used. Detailed information of the cohorts, intelligence measurements, genotyping methods, and statistical analyses can be found in the original study (Benyamin et al, 2014).
Healthy Subjects for Expression Quantitative Trait Loci (eQTL) Analysis
To identify the impact of MDD risk SNPs on mRNA expression, we utilized the well-characterized gene expression database BrainCloud (http://braincloud.jhmi.edu/) (Colantuoni et al, 2011). BrainCloud presents data regarding gene expression regulation in the human brain that guides functional studies of disease-associated variants. The BrainCloud sample comprises 261 postmortem dorsolateral prefrontal cortex (DLPFC) tissues of nonpsychiatric individuals, including 113 Caucasian subjects and 148 African-American individuals at various ages across the lifespan. As PCDH9 is differentially expressed across different age groups, we retrieved the genotype and expression data of 152 adult individuals (age >18 years; 64 Caucasians and 88 African Americans) from BrainCloud. The statistical analysis was conducted using linear regression, with RNA integrity number (RIN), sex, race, brain PH, postmortem interval (PMI), and age as covariates.
Diagnostic Analysis of PCDH9 Expression between Patients with MDD and Healthy Controls
We obtained the microarray mRNA expression data of the frontal cortex from 103 adult controls (age >18 years) and 131 adult MDD patients of European ancestry from dbGaP (accession number: phs000979.v1.p1). For the expression data, log2 ratios were normalized across mean log2 florescent intensities using loess correction (Colantuoni et al, 2002). After normalization, surrogate variable analysis (SVA) was conducted on the log2 ratios to optimize the signal/noise ratio and minimize potential impact from known and unknown sources of systematic confounders (Leek and Storey, 2007). The above data analyses were conducted using R with codes and tools retrieved from the Bioconductor project (http://www.bioconductor.org/).
We also collected hippocampal RNA-sequencing (RNA-seq) data of 15 adult MDD cases and 15 adult controls from the Stanley Medical Research Institute (SMRI) data set (http://sncid.stanleyresearch.org/) in the FASTQ file format. The RNA-seq reads underwent adaptation and low-quality filtering using btrim64 (Kong, 2011), and were then aligned to human reference genome (Human GRCh38 (hg38), http://asia.ensembl.org/index.html) using splice-read mapper (Tophat2 v2.0.14) (Kim et al, 2013). The map of known transcripts extracted from Ensembl Build GRCh38. Cufflinks v2.2.1 (Trapnell et al, 2012) was applied to call new transcripts as well as to assemble and quantify both the novel and known transcripts with default parameters. For each subject, accepted hits bam files from Tophat2 alignment were merged by Samtools v0.1.18 (Li et al, 2009) for the following Cufflinks quantification: (1) reads that were uniquely mapped to genes were used to calculate the gene expression level; (2) to quantify mRNA expression, FPKM (Fragments per Kilobase per Million mapped reads) was calculated to measure the gene-level expression according to the formula FPKM=F × 103/L × 106/N (F is the number of fragments mapping to the gene annotation, L is the length of the gene structure in nucleotides, and N is the total number of sequence reads mapped to the genome).
Statistical analyses of mRNA expression associated with diagnosis were conducted in R 3.0.1 using an analyses of covariance (ANCOVA) model. The diagnostic status was applied as independent variables, whereas age, sex, RIN, brain pH, and PMI were set as covariates.
Results
Rs9540720 in PCDH9 Shows Genome-Wide Significant Association with MDD
In this study, the top 10 000 most significant SNPs (two-tailed p⩽5.31 × 10–5) in the 23andMe GWAS (Hyde et al, 2016), which contained the largest sample size, were first subject to replication analyses in the CONVERGE and PGC GWAS data sets (Converge consortium, 2015; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al, 2013). Briefly, SNPs meeting the following criteria were collected for further analyses: (1) the SNPs showed a one-tailed p-value <0.05 in both CONVERGE and PGC GWAS data sets (Converge consortium, 2015; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al, 2013), and (2) the SNPs showed the same direction of allelic effects across all three GWAS data sets (Converge consortium, 2015; Hyde et al, 2016; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al, 2013). This replication investigation returned a total of 33 SNPs (Table 1).
Table 1. Association of 33 SNPs with Major Depressive Disorder.
| SNP | CHR | POS | Location | A1 | A2 | Frequency_A1 |
23andMe |
CONVERGE |
PGC |
Meta-analysis |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EUR | CHI | OR | P-value | OR | P-value | OR | P-value | OR | P-value | Q-value | I2 | ||||||
| rs12127723 | 1 | 73141767 | Intergenic | A | G | 0.539 | 0.163 | 1.029 | 4.04E–06 | 1.091 | 4.68E–03 | 1.051 | 1.25E–02 | 1.032 | 9.63E–08 | 0.207 | 36.6 |
| rs12145482 | 1 | 73227348 | Intergenic | G | A | 0.540 | 0.157 | 1.028 | 6.35E–06 | 1.093 | 3.80E–03 | 1.056 | 7.10E–03 | 1.031 | 1.29E–07 | 0.141 | 49.0 |
| rs1525986 | 1 | 73237902 | Intergenic | T | C | 0.539 | 0.156 | 1.027 | 1.02E–05 | 1.087 | 5.73E–03 | 1.056 | 6.68E–03 | 1.030 | 1.70E–07 | 0.164 | 44.7 |
| rs1358041 | 1 | 73241582 | Intergenic | C | G | 0.539 | 0.156 | 1.027 | 1.06E–05 | 1.087 | 5.74E–03 | 1.056 | 6.69E–03 | 1.030 | 1.85E–07 | 0.163 | 45.0 |
| rs1358042 | 1 | 73241894 | Intergenic | C | G | 0.539 | 0.156 | 1.027 | 1.06E–05 | 1.087 | 5.74E–03 | 1.056 | 6.60E–03 | 1.030 | 1.85E–07 | 0.163 | 45.0 |
| rs11210056 | 1 | 73242477 | Intergenic | G | T | 0.539 | 0.156 | 1.027 | 1.06E–05 | 1.088 | 5.31E–03 | 1.056 | 6.39E–03 | 1.030 | 1.77E–07 | 0.154 | 47.0 |
| rs7549440 | 1 | 73244475 | Intergenic | C | T | 0.539 | 0.156 | 1.027 | 1.07E–05 | 1.088 | 5.31E–03 | 1.056 | 6.48E–03 | 1.030 | 1.78E–07 | 0.155 | 46.4 |
| rs12117265 | 1 | 73251154 | Intergenic | A | G | 0.539 | 0.156 | 1.027 | 1.10E–05 | 1.089 | 4.78E–03 | 1.057 | 6.35E–03 | 1.030 | 1.67E–07 | 0.144 | 48.4 |
| rs4116074 | 1 | 73256388 | Intergenic | G | A | 0.539 | 0.154 | 1.027 | 1.07E–05 | 1.088 | 5.35E–03 | 1.057 | 6.44E–03 | 1.031 | 2.24E–07 | 0.155 | 46.4 |
| rs10889992 | 1 | 73264835 | Intergenic | C | T | 0.539 | 0.157 | 1.027 | 1.07E–05 | 1.093 | 3.52E–03 | 1.061 | 3.99E–03 | 1.031 | 1.38E–07 | 0.105 | 55.7 |
| rs12119161 | 1 | 73489496 | Intergenic | G | A | 0.522 | 0.150 | 1.026 | 2.86E–05 | 1.079 | 2.60E–02 | 1.038 | 4.56E–02 | 1.028 | 2.43E–06 | 0.357 | 2.9 |
| rs12138334 | 1 | 73551507 | Intergenic | A | G | 0.522 | 0.150 | 1.025 | 3.80E–05 | 1.071 | 4.21E–02 | 1.038 | 4.66E–02 | 1.027 | 2.86E–06 | 0.449 | 0 |
| rs4233103 | 1 | 73579799 | Intergenic | G | T | 0.521 | 0.150 | 1.025 | 3.45E–05 | 1.075 | 3.56E–02 | 1.039 | 4.18E–02 | 1.027 | 2.33E–06 | 0.393 | 0 |
| rs4622015 | 1 | 73590062 | Intergenic | A | G | 0.521 | 0.150 | 1.026 | 3.14E–05 | 1.067 | 4.83E–02 | 1.039 | 3.99E–02 | 1.027 | 2.32E–06 | 0.498 | 0 |
| rs4582739 | 1 | 73602381 | Intergenic | T | C | 0.520 | 0.150 | 1.026 | 2.28E–05 | 1.067 | 4.81E–02 | 1.040 | 3.81E–02 | 1.028 | 1.58E–06 | 0.496 | 0 |
| rs6674404 | 1 | 73631632 | Intergenic | T | C | 0.510 | 0.150 | 1.025 | 5.23E–05 | 1.067 | 4.62E–02 | 1.040 | 3.62E–02 | 1.027 | 4.02E–06 | 0.465 | 0 |
| rs12128855 | 1 | 73634013 | Intergenic | G | A | 0.521 | 0.154 | 1.026 | 2.08E–05 | 1.067 | 4.63E–02 | 1.040 | 3.72E–02 | 1.028 | 1.45E–06 | 0.498 | 0 |
| rs7996220 | 13 | 66915901 | PCDH9 | T | A | 0.479 | 0.418 | 1.027 | 1.40E–05 | 1.058 | 2.24E–02 | 1.036 | 4.98E–02 | 1.029 | 8.31E–07 | 0.556 | 0 |
| rs2875472 | 13 | 66921954 | PCDH9 | T | A | 0.500 | 0.601 | 1.030 | 1.07E–06 | 1.077 | 2.96E–03 | 1.039 | 3.90E–02 | 1.033 | 2.17E–08 | 0.288 | 19.8 |
| rs9540720 | 13 | 66922705 | PCDH9 | G | A | 0.500 | 0.600 | 1.030 | 1.07E–06 | 1.077 | 3.00E–03 | 1.041 | 2.75E–02 | 1.033 | 1.69E–08 | 0.276 | 22.3 |
| rs9571576 | 13 | 66936133 | PCDH9 | C | T | 0.510 | 0.607 | 1.026 | 1.56E–05 | 1.077 | 3.14E–03 | 1.035 | 4.95E–02 | 1.029 | 3.88E–07 | 0.234 | 31.3 |
| rs7319883 | 13 | 66939588 | PCDH9 | C | T | 0.496 | 0.601 | 1.029 | 3.12E–06 | 1.076 | 3.54E–03 | 1.039 | 3.36E–02 | 1.031 | 5.14E–08 | 0.269 | 23.8 |
| rs1831972 | 13 | 66939722 | PCDH9 | A | C | 0.496 | 0.603 | 1.029 | 3.21E–06 | 1.076 | 3.55E–03 | 1.040 | 3.08E–02 | 1.031 | 4.93E–08 | 0.266 | 24.4 |
| rs9592461 | 13 | 66941792 | PCDH9 | A | G | 0.498 | 0.696 | 1.029 | 1.78E–06 | 1.081 | 4.84E–03 | 1.042 | 2.50E–02 | 1.032 | 2.63E–08 | 0.247 | 28.5 |
| rs1444387 | 13 | 66944131 | PCDH9 | C | T | 0.500 | 0.696 | 1.030 | 1.28E–06 | 1.080 | 4.92E–03 | 1.040 | 2.96E–02 | 1.032 | 3.13E–08 | 0.277 | 22.2 |
| rs9540728 | 13 | 66944321 | PCDH9 | C | A | 0.496 | 0.603 | 1.029 | 2.96E–06 | 1.075 | 3.82E–03 | 1.041 | 2.96E–02 | 1.031 | 4.50E–08 | 0.279 | 21.7 |
| rs1158646 | 13 | 66946304 | PCDH9 | C | T | 0.496 | 0.603 | 1.029 | 3.01E–06 | 1.075 | 3.82E–03 | 1.040 | 3.32E–02 | 1.031 | 5.26E–08 | 0.282 | 21.0 |
| rs9540731 | 13 | 66949370 | PCDH9 | C | T | 0.499 | 0.696 | 1.029 | 1.96E–06 | 1.080 | 4.93E–03 | 1.040 | 3.35E–02 | 1.032 | 5.23E–08 | 0.272 | 23.1 |
| rs1413573 | 13 | 66951829 | PCDH9 | A | G | 0.496 | 0.603 | 1.029 | 3.23E–06 | 1.079 | 2.71E–03 | 1.041 | 2.89E–02 | 1.032 | 5.93E–08 | 0.225 | 33.0 |
| rs9540734 | 13 | 66965411 | PCDH9 | A | G | 0.496 | 0.608 | 1.029 | 2.48E–06 | 1.058 | 2.20E–02 | 1.044 | 2.03E–02 | 1.031 | 5.59E–08 | 0.516 | 0 |
| rs166500 | 13 | 66965826 | PCDH9 | G | A | 0.671 | 0.850 | 1.027 | 4.90E–05 | 1.101 | 1.78E–02 | 1.045 | 2.41E–02 | 1.029 | 2.82E–06 | 0.226 | 32.8 |
| rs4883777 | 13 | 66974706 | PCDH9 | T | C | 0.606 | 0.652 | 1.028 | 1.13E–05 | 1.054 | 3.66E–02 | 1.043 | 2.45E–02 | 1.030 | 5.20E–07 | 0.593 | 0 |
| rs8066520 | 17 | 27376591 | PIPOX | A | G | 0.150 | 0.255 | 1.037 | 1.18E–05 | 1.050 | 4.23E–02 | 1.055 | 3.55E–02 | 1.039 | 8.13E–07 | 0.820 | 0 |
Abbreviations: EUR, European; CHI, Chinese; CHR, chromosome; OR, odds ratio; POS, position; SNP, single-nucleotide polymorphism. Genome-wide significant p-values (p < 5.00E-08) were marked in bold.
These 33 SNPs then underwent the meta-analysis using the three GWAS data sets. An intronic SNP rs9540720 in PCDH9 at 13q21.32, which showed nominal associations with MDD in 23andMe GWAS (two-tailed p=1.07 × 10–6, OR=1.030), CONVERGE GWAS (one-tailed p=3.00 × 10–3, OR=1.077), and PGC GWAS (one-tailed p=2.75 × 10–2, OR=1.041), was genome-wide significantly associated with the illness in the meta-analysis of all three data sets (a total of 89 610 cases and 246 603 controls, two-tailed p=1.69 × 10–8, OR=1.033, Table 1). Five SNPs (rs2875472, rs1831972, rs9592461, rs1444387, and rs9540728) in high linkage disequilibrium (LD) with rs9540720 in both European and Chinese populations also showed genome-wide significance in the meta-analysis (Table 1). A schematic presentation of the PCDH9 gene and the locations of rs9540720 and other tested SNPs in this gene are shown in Supplementary Figure S1; the LD relationship and haplotype structure of the PCDH9 risk SNPs in European and Chinese populations are also displayed in Supplementary Figure S1. Intriguingly, rs9540720 was not related to any of the 15 genome-wide significant SNPs previously reported in the 23andMe GWAS study (Hyde et al, 2016).
We further investigated the associations of rs9540720 with MDD in the 23andMe Replication sample and the Chinese MDD sample. Intriguingly, rs9540720 was also nominally associated with MDD in both the 23andMe Replication sample (one-tailed p=4.85 × 10–2, OR=1.014) (Hyde et al, 2016) and the Han Chinese MDD sample (one-tailed p=2.74 × 10–2, OR=1.125). In addition, meta-analysis combining these two replication samples consistently yielded a nominal association between rs9540720 and MDD (one-tailed p=2.88 × 10–2, OR=1.016). When all the discovery and replication samples were merged (a total of 136 115 cases and 355 275 controls), the association was further strengthened (two-tailed p=1.20 × 10–8, OR=1.027). Although the effect size for rs9540720 (ie, OR=1.027) was relatively small, it was still similar to the other previously reported GWAS significant SNPs in 23andMe GWAS (OR ranges from 1.028 to 1.051, Supplementary Table S1) (Hyde et al, 2016). This observation is likely attributed to the fact that MDD is polygenic with numerous alleles each accounting for a limited share of genetic risk for the illness (Peterson et al, 2017).
Taken together, rs9540720 shows significant associations with MDD in both Europeans and Han Chinese, in line with its stable allelic frequency across those two populations (Supplementary Figure S2a), suggesting that it is a possible common risk SNP for MDD among various ethnic groups.
Rs9540720 Is Associated with MDD-Related Phenotypes and Cognitive Functions
Given the strong implication of phenotypes such as neuroticism and depressive symptoms (Okbay et al, 2016a), preschool internalizing problems (Benke et al, 2014), as well as anxiety (Otowa et al, 2016) in MDD patients, we hypothesized that these phenotypes were also associated with rs9540720. We utilized the public GWAS resources on these MDD-related phenotypes, and found that this SNP was nominally associated with neuroticism (n=170 911, two-tailed p=7.84 × 10–3), preschool internalizing problems (n=4596, two-tailed p=3.94 × 10–2), and anxiety scores (n=15 299, two-tailed p=2.18 × 10–2), and was also marginally associated with depressive symptoms (n=180 866, two-tailed p=6.12 × 10–2). Intriguingly, the rs9540720 risk [G] allele carriers tended to show more vulnerable personality traits (or worse symptoms) compared with protective allele [A] carries in these series of analyses. It is hypothesized that MDD shares substantial genetic risk components with schizophrenia and bipolar disorder (Ding et al, 2015), and patients with these psychiatric disorders sometimes exhibit overlapping symptoms. We therefore assessed the associations of rs9540720 with schizophrenia and bipolar disorder in the large-scale GWAS samples to understand its role in these two illnesses. However, this SNP was not associated with schizophrenia (34 241 cases and 45 604 controls, two-tailed p=0.459) (Schizophrenia Working Group of the Psychiatric Genomics Consortium, 2014) or bipolar disorder (10 410 cases and 10 700 controls, two-tailed p=0.983) (Ruderfer et al, 2014), suggesting that it is likely a MDD-specific risk variant.
Besides MDD-related phenotypes, it is hypothesized that MDD risk-associated SNP may also affect cognitive abilities. To test the influence of rs9540720 on cognitive functions, we utilized public GWAS data sets of educational attainment (Okbay et al, 2016b) and childhood intelligence (Benyamin et al, 2014), respectively. In this explorative analysis, rs9540720 was significantly associated with educational attainment (two-tailed p=1.09 × 10–2) and childhood intelligence (two-tailed p=1.20 × 10–4). Notably, the MDD risk allele indicated lower education levels and lower scores in standard IQ tests.
Collectively, these data consistently demonstrated negative impact of rs9540720 risk allele on a variety of MDD-related phenotypes and cognitive abilities, further supporting our hypothesis that rs9540720 was a susceptibility SNP for MDD.
Risk Genotypes in rs9540720 and MDD Diagnosis Predict PCDH9 mRNA Expression
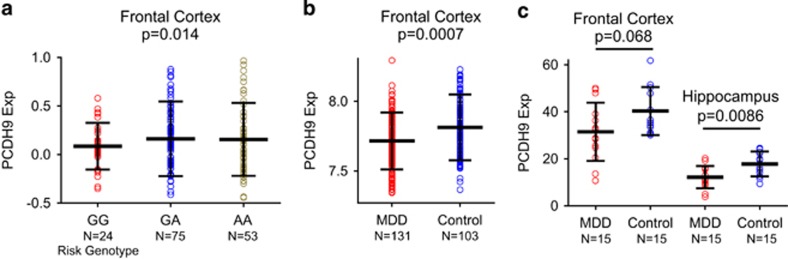
The associations of rs9540720 with MDD and related phenotypes in multiple independent samples lend statistical and biological support for the involvement of this genomic locus in the risk of the illness. However, the exact causal variant and underlying molecular mechanisms remained to be determined. This mission is often difficult in genetic association studies because an associated SNP most likely points to a larger region containing numerous correlated variants with a high degree of LD (Li et al, 2016). For this reason, we explored the LD between rs9540720 and surrounding SNPs to investigate whether there were SNPs linked with rs9540720. A proxy search for SNPs of LD with rs9540720 was performed on the SNAP website with the European panel from the 1000-Human-Genomes (pilot 1) data set (http://archive.broadinstitute.org/mpg/snap/ldplot.php), and returned quite a few SNPs in relatively high LD (r2>0.80) with rs9540720 (Supplementary Figure S2b and Supplementary Table S2). Notably, all the rs9540720-linked SNPs were located in the PCDH9 intron region, and we therefore searched for potential functional SNPs through bioinformatics predictive analyses via synthesizing annotation information of noncoding elements and genomic properties regarding GC content, evolutionary conservation, and so on using the GWAVA data set (http://www.sanger.ac.uk/sanger/StatGen_Gwava) (Ritchie et al, 2014). This functional prediction showed that those SNPs in high LD with rs9540720 were unlikely located in the DNA segments binding to transcription factors or histone markers (eg, H3K4me1, H3K4me3, H3K9ac, and H3K27ac) (Supplementary Table S2) (Ritchie et al, 2014), and thus we were unable to identify the causative variant without performing further functional assays. However, there was also the possibility that rs9540720 was an eQTL of a specific gene. According to the data presented in Supplementary Figure S2b, there was only PCDH9 gene within 400 kb around rs9540720 and its LD SNPs, and hence we examined the association between rs9540720 and PCDH9 expression using data from BrainCloud (Colantuoni et al, 2011). Interestingly, the risk SNP rs9540720 was associated with PCDH9 mRNA expression in human frontal cortex in 152 healthy adult individuals (two-tailed p=0.014, Figure 1a), with the risk allele predicting lower PCDH9 mRNA levels. In addition, most other MDD risk SNPs in this genomic region (with genotypes available in BrainCloud) were also associated with lower PCDH9 mRNA expression in the frontal cortex of these subjects (Supplementary Table S3).
Figure 1.
Risk genotype and diagnosis predict PCDH9 expression. (a) Association of rs9540720 with PCDH9 mRNA expression in the 152 adult subjects from frontal cortex in the BrainCloud data set (Colantuoni et al, 2011). Number of individuals: GG, N=24; GA, N=75; AA, N=53. (b) Diagnostic analysis of PCDH9 expression in adult samples from frontal cortex in dbGaP using microarray data. MDD, major depressive disorder. Number of individuals: MDD, N=131; Control, N=103. (c) Diagnostic analysis of PCDH9 expression in the frontal cortex and hippocampus of adult samples from Stanley center using RNA-seq data. N=15 in each group.
To gain further insights into the potential pathophysiological roles of PCDH9, we assessed the effects of diagnostic status on PCDH9 mRNA expression. According to the frontal cortex microarray mRNA expression data from dbGaP (103 adult controls and 131 adult MDD patients of European ancestry), PCDH9 expression was significantly decreased in MDD patients than in control subjects (two-tailed p=0.0007, Figure 1b). By analyzing the RNA-seq data of frontal cortex and hippocampus tissues from 15 MDD cases and 15 healthy controls from SMRI, we observed a consistent reduction of PCDH9 expression in MDD patients compared with healthy controls in both the hippocampus (two-tailed p=0.0086, Figure 1c) and the frontal cortex (two-tailed p=0.068, Figure 1c). In addition, in a previous transcriptome study using peripheral blood from The Netherlands Study of Depression and Anxiety cohort (Jansen et al, 2016), PCDH9 mRNA expression was also significantly decreased in 882 patients with current MDD vs 331 healthy controls (two-tailed p=0.00402, Supplementary Table S2 in the original study (Jansen et al, 2016)). Moreover, our observations were also partly in concordance with another earlier study (Klempan et al, 2009) showing reduced PCDH9 expression in the DLPFC and inferior frontal gyrus of suicides with MDD compared with healthy controls (Table 2 in their original study). Interestingly, no differences of PCDH9 expression between nondepressed suicides and healthy controls were reported (Table 2 in their original study) (Klempan et al, 2009). These diagnostic results were consistent with our eQTL analysis results that individuals carrying the MDD risk allele at rs9540720 (and other risk LD SNPs) had lower mRNA levels. Notably, PCDH9 expression was enriched in brain tissues compared with other organs (Supplementary Figure S3, PCDH9 expression in brain tissues is marked in yellow box), highlighting its potential role in brain disorders (eg, MDD).
In summary, we have presented convergent and consistent evidence suggesting that PCDH9 is a MDD susceptibility gene, and the reduced expression of this gene might contribute to the pathogenesis of the illness.
Discussion
MDD is a major complex mental disorder affecting millions of people worldwide. The advent of genetic studies, especially the GWASs, has greatly promoted our understanding of this illness (Ding et al, 2015). Although several promising candidate genes associated with the risk of MDD have been identified, studies clarifying more genetic risk factors and the underlying mechanisms for this illness are urgently needed. We believe that if a SNP exhibits (at least) nominal associations with a particular illness among multiple individual samples, and presents genome-wide significance in the comprehensive meta-analysis combining all the above samples, it is then very likely to be an authentic disease relevant locus. To this end, we explored several independent GWAS data sets and report that a locus rs9540720 predicting the mRNA expression of PCDH9 possessed genome-wide significant association with MDD. In addition to showing that rs9540720 was associated with MDD in 23andMe, CONVERGE, and PGC as well as in the combined meta-analysis, we confirmed that the risk allele of this locus was associated with multiple phenotypes related to MDD pathogenesis and cognitive abilities using a series of convergent analyses. Taken one step further, we examined the roles of rs9540720 and PCDH9 in clinical diagnosis of MDD, and observed a consistent pattern of allelic associations that risk genotype at this SNP was linked to MDD diagnosis and lower PCDH9 mRNA expression. Although the analyses on MDD-related phenotypes and cognition failed to achieve genome-wide significance, it was unlikely that the risk allele predicted abnormality associated with the illness in each of those independent phenotypes across diverse samples purely by chance. Moreover, expression of the PCDH9 gene in brain tissues was also in line with the direction of allelic association, indicating that aberrant regulation of PCDH9 transcription was the likely molecular mechanism underlying the genetic risk conferred by rs9540720 or its LD SNPs. Taken together, PCDH9 is likely a bona fide MDD susceptibility locus.
In addition, one feature of the current study is the utilization of transethnic data that are believed to leverage LD structural differences across various ethnic groups and thus promote the resolution of fine-mapping causal variants (Li et al, 2017; Morris, 2011). In fact, this idea has also been applied in a recent MDD study (using CONVERGE and PGC data sets) (Bigdeli et al, 2017), in which the authors successfully demonstrated a partially shared polygenic basis of the illness between Han Chinese and European populations (the trans-ancestry genetic correlation of lifetime MDD was 0.33, whereas female-only and recurrent MDD yielded estimates of 0.40 and 0.41, respectively); however, the PCDH9 region was not highlighted in their study, probably because of the limited sample size (Bigdeli et al, 2017). Although some population-specific risk loci may fail to be identified, this strategy warrants higher detection power for complex trait loci. In our study, rs9540720 showed nominal associations with MDD in both Europeans and Asians, in line with its stable allelic frequency across those two populations (Supplementary Figure S2a), suggesting that it is a possible common risk SNP for MDD among various ethnic groups.
The SNP rs9540720 locates on the chromosome 13q21.32, a chromosomal region that was not recognized as a major locus in the few GWASs of MDD (Lewis et al, 2010; Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium et al, 2013; Muglia et al, 2010; Rietschel et al, 2010; Shi et al, 2011; Wray et al, 2012), probably because of the limited sample sizes. Our meta-analysis combining multiple GWASs significantly boosted the sample size for the genetic analyses and successfully detected associations of PCDH9 SNPs. The PCDH9 gene encodes protocadherin 9, a protein of the protocadherin family and cadherin superfamily (transmembrane proteins containing cadherin domains). Interestingly, we have recently identified another gene encoding a protein of the same family, PCDH17 (locates on 13q21.1), to confer risk of MDD and bipolar disorder (Chang et al, 2017). Both the short distance between these two genes on the chromosome (∼8.57 Mb, but there is no LD between risk SNPs at PCDH9 and PCDH17) and structural similarities shared by their protocadherin protein products suggest potential correlated biological roles. Although the mechanisms by which PCDH9 contributes to MDD pathogenesis are unclear, PCDH17 has been demonstrated to significantly affect synaptic development (Chang et al, 2017; Hoshina et al, 2013). Intriguingly, a recent quantitative analysis of basic mouse behavior uncovered the role of Pcdh9 in specific cognitive functions required for long-term recognition (Bruining et al, 2015). Therefore, the protein encoded by PCDH9 may also affect signaling at neuronal synaptic junctions (Asahina et al, 2012), and further studies investigating this hypothesis are necessary.
In addition to PCDH9, we have also observed marginal genome-wide significant associations between MDD and the SNPs in chromosome 1 region in this meta-analysis (lowest SNP rs12127723, p=9.63 × 10–8, Table 1). Further validation of the risk associations in additional samples is therefore necessary. It should be noted that this chromosome 1 region contains large-scale number of genomic variants that are in high LD with the risk SNPs (eg, rs12127723 and rs12128855) observed here (r2>0.8, Supplementary Figure S4), and these risk SNPs may tag one or more potential functional loci that needs to be identified. However, there are no protein-coding genes within this genomic region (Supplementary Figure S4), adding difficulties to further investigations of the molecular mechanisms underlying this potential genetic risk locus.
Although we herein present a comprehensive report of rs9540720 in the risk of MDD, there are certain limitations to be acknowledged. For example, the present analyses are primarily based on publicly available databases, and the full-scale analyses thus depend on the accessibility of the complete data set. In this case, only a selection of 10 000 SNPs are available from the 23andMe data set (Hyde et al, 2016), and the current analysis therefore falls short of the data from 23andMe (indicated as well by 1 vs 15 GWAS hits). Although the purpose of a meta-analysis is normally to maximize the power to detect true effects, the maximal power is not available in our study because of this limitation. Further genome-wide meta-analyses are thus necessary to reflect additional risk loci. Another caveat lies in the eQTL analysis utilizing both Europeans and African-American subjects to maximize the statistical power, even though the allelic frequencies of rs9540720 did not differ significantly between these two ethnic groups (A allele, 0.506 in Europeans and 0.602 in African Americans), and the eQTL effect sizes were also similar among different populations. Future eQTL analyses utilizing larger samples of Europeans and Han Chinese are needed to confirm the current observations.
In sum, using the existing GWAS data sets on MDD, MDD-related phenotypes plus cognitive function, as well as databases of brain tissue gene expression, we have identified a novel genome-wide significant MDD risk gene PCDH9 through a convergent meta-analysis. Although potential concerns remain to be addressed, our study highlights the necessity and importance of excavating the public data sets to explore undiscovered risk genes for MDD and other complex diseases.
Funding and Disclosure
The authors declare no conflict of interest.
Acknowledgments
This work was supported by grants from the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant No., XDB13000000), CAS Pioneer Hundred Talents Program (to ML), the National Natural Science Foundation of China (81601176 to FZ), the Strategic Priority Research Program (B) of the Chinese Academy of Sciences (XDB02020003 to Y-GY), and the Bureau of Frontier Sciences and Education, Chinese Academy of Sciences (QYZDJ-SSW-SMC005 to Y-GY). The microarray data in MDD sample used for the expression analyses described in this manuscript were obtained from dbGaP accession number phs000979.v1.p1. This research was supported by the Intramural Research Program of the NIMH(NCT00001260, 900142).
Footnotes
Supplementary Information accompanies the paper on the Neuropsychopharmacology website (http://www.nature.com/npp)
Supplementary Material
References
- Asahina H, Masuba A, Hirano S, Yuri K (2012). Distribution of protocadherin 9 protein in the developing mouse nervous system. Neuroscience 225: 88–104. [DOI] [PubMed] [Google Scholar]
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD et al (2012). Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry 17: 887–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batty GD, Mortensen EL, Osler M (2005). Childhood IQ in relation to later psychiatric disorder: evidence from a Danish birth cohort study. Br J Psychiatry 187: 180–181. [DOI] [PubMed] [Google Scholar]
- Benke KS, Nivard MG, Velders FP, Walters RK, Pappa I, Scheet PA et al (2014). A genome-wide association meta-analysis of preschool internalizing problems. J Am Acad Child Adolesc Psychiatry 53: 667–676 e667. [DOI] [PubMed] [Google Scholar]
- Benyamin B, Pourcain B, Davis OS, Davies G, Hansell NK, Brion MJ et al (2014). Childhood intelligence is heritable, highly polygenic and associated with FNBP1L. Mol Psychiatry 19: 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdeli TB, Ripke S, Peterson RE, Trzaskowski M, Bacanu SA, Abdellaoui A et al (2017). Genetic effects influencing risk for major depressive disorder in China and Europe. Transl Psychiatry 7: e1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D et al (2011). Poor replication of candidate genes for major depressive disorder using genome-wide association data. Mol Psychiatry 16: 516–532. [DOI] [PubMed] [Google Scholar]
- Bruining H, Matsui A, Oguro-Ando A, Kahn RS, Van't Spijker HM, Akkermans G et al (2015). Genetic mapping in mice reveals the involvement of Pcdh9 in long-term social and object recognition and sensorimotor development. Biol Psychiatry 78: 485–495. [DOI] [PubMed] [Google Scholar]
- Chang H, Hoshina N, Zhang C, Ma Y, Cao H, Wang Y et al (2017). The protocadherin 17 gene affects cognition, personality, amygdala structure and function, synapse development and risk of major mood disorders. Mol Psychiatry doi:10.1038/mp.2016.231. [DOI] [PMC free article] [PubMed]
- Colantuoni C, Henry G, Zeger S, Pevsner J (2002). SNOMAD (Standardization and NOrmalization of MicroArray Data): web-accessible gene expression data analysis. Bioinformatics 18: 1540–1541. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT et al (2011). Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Converge consortium (2015). Sparse whole-genome sequencing identifies two loci for major depressive disorder. Nature 523: 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ (2012). Intelligence. Annu Rev Psychol 63: 453–482. [DOI] [PubMed] [Google Scholar]
- Deary IJ, Johnson W, Houlihan LM (2009). Genetic foundations of human intelligence. Hum Genet 126: 215–232. [DOI] [PubMed] [Google Scholar]
- Demirkan A, Penninx BW, Hek K, Wray NR, Amin N, Aulchenko YS et al (2011). Genetic risk profiles for depression and anxiety in adult and elderly cohorts. Mol Psychiatry 16: 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Chang LC, Wang X, Guilloux JP, Parrish J, Oh H et al (2015). Molecular and genetic characterization of depression: overlap with other psychiatric disorders and aging. Mol Neuropsychiatry 1: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson N, Macpherson JM, Tung JY, Hon LS, Naughton B, Saxonov S et al (2010). Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet 6: e1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanous AH, Kendler KS (2004). The genetic relationship of personality to major depression and schizophrenia. Neurotox Res 6: 43–50. [DOI] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T et al (2010). Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet 42: 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium (2013). The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin DS, Goodwin RD, Stinson FS, Grant BF (2005). Epidemiology of major depressive disorder: results from the National Epidemiologic Survey on Alcoholism and Related Conditions. Arch Gen Psychiatry 62: 1097–1106. [DOI] [PubMed] [Google Scholar]
- Hoshina N, Tanimura A, Yamasaki M, Inoue T, Fukabori R, Kuroda T et al (2013). Protocadherin 17 regulates presynaptic assembly in topographic corticobasal Ganglia circuits. Neuron 78: 839–854. [DOI] [PubMed] [Google Scholar]
- Hyde CL, Nagle MW, Tian C, Chen X, Paciga SA, Wendland JR et al (2016). Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 48: 1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Penninx BW, Madar V, Xia K, Milaneschi Y, Hottenga JJ et al (2016). Gene expression in major depressive disorder. Mol Psychiatry 21: 339–347. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE (2005). Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry 62: 593–602. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Bromet EJ (2013). The epidemiology of depression across cultures. Annu Rev Public Health 34: 119–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Webster MJ (2009). Postmortem brain tissue for drug discovery in psychiatric research. Schizophr Bull 35: 1031–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Webster MJ (2010). The Stanley neuropathology consortium integrative database: a novel, web-based tool for exploring neuropathological markers in psychiatric disorders and the biological processes associated with abnormalities of those markers. Neuropsychopharmacology 35: 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klempan TA, Sequeira A, Canetti L, Lalovic A, Ernst C, ffrench-Mullen J et al (2009). Altered expression of genes involved in ATP biosynthesis and GABAergic neurotransmission in the ventral prefrontal cortex of suicides with and without major depression. Mol Psychiatry 14: 175–189. [DOI] [PubMed] [Google Scholar]
- Koenen KC, Moffitt TE, Roberts AL, Martin LT, Kubzansky L, Harrington H et al (2009). Childhood IQ and adult mental disorders: a test of the cognitive reserve hypothesis. Am J Psychiatry 166: 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli MA, Lucae S, Saemann PG, Schmidt MV, Demirkan A, Hek K et al (2011). The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron 70: 252–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y (2011). Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98: 152–153. [DOI] [PubMed] [Google Scholar]
- Kotov R, Gamez W, Schmidt F, Watson D (2010). Linking "big" personality traits to anxiety, depressive, and substance use disorders: a meta-analysis. Psychol Bull 136: 768–821. [DOI] [PubMed] [Google Scholar]
- Lahey BB (2009). Public health significance of neuroticism. Am Psychol 64: 241–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, McFarland MJ, Mamidipalli S, Ogden CA, Kuczenski R, Kurian SM et al (2007). Convergent Functional Genomics of bipolar disorder: from animal model pharmacogenomics to human genetics and biomarkers. Neurosci Biobehav Rev 31: 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le-Niculescu H, Patel SD, Niculescu AB (2010). Convergent integration of animal model and human studies of bipolar disorder (manic-depressive illness). Curr Opin Pharmacol 10: 594–600. [DOI] [PubMed] [Google Scholar]
- Leek JT, Storey JD (2007). Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3: 1724–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF (2006). The genetics of depression: a review. Biol Psychiatry 60: 84–92. [DOI] [PubMed] [Google Scholar]
- Levinson DF, Mostafavi S, Milaneschi Y, Rivera M, Ripke S, Wray NR et al (2014). Genetic studies of major depressive disorder: why are there no genome-wide association study findings and what can we do about it? Biol Psychiatry 76: 510–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K et al (2010). Genome-wide association study of major recurrent depression in the U.K. population. Am J Psychiatry 167: 949–957. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N et al (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Chang H, Peng T, Li M, Xiao X (2017). Evidence of AS3MTd2d3-Associated Variants within 10q24.32-33 in the Genetic Risk of Major Affective Disorders. Mol Neuropsychiatry 2: 213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jaffe AE, Straub RE, Tao R, Shin JH, Wang Y et al (2016). A human-specific AS3MT isoform and BORCS7 are molecular risk factors in the 10q24.32 schizophrenia-associated locus. Nat Med 22: 649–656. [DOI] [PubMed] [Google Scholar]
- Luo XJ, Li M, Huang L, Steinberg S, Mattheisen M, Liang G et al (2014). Convergent lines of evidence support CAMKK2 as a schizophrenia susceptibility gene. Mol Psychiatry 19: 774–783. [DOI] [PubMed] [Google Scholar]
- Major Depressive Disorder Working Group of the Psychiatric GWAS Consortium, Ripke S, Wray NR, Lewis CM, Hamilton SP, Weissman MM et al (2013). A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 18: 497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AP (2011). Transethnic meta-analysis of genomewide association studies. Genet Epidemiol 35: 809–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ et al (2010). Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Mol Psychiatry 15: 589–601. [DOI] [PubMed] [Google Scholar]
- Niculescu AB (2005). Genomic studies of mood disorders — the brain as a muscle? Genome Biol 6: 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB et al (2004). Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry 9: 1007–1029. [DOI] [PubMed] [Google Scholar]
- Okbay A, Baselmans BM, De Neve JE, Turley P, Nivard MG, Fontana MA et al (2016. a). Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat Genet 48: 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA et al (2016. b). Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olino TM, Pettit JW, Klein DN, Allen NB, Seeley JR, Lewinsohn PM (2008). Influence of parental and grandparental major depressive disorder on behavior problems in early childhood: a three-generation study. J Am Acad Child Adolesc Psychiatry 47: 53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otowa T, Hek K, Lee M, Byrne EM, Mirza SS, Nivard MG et al (2016). Meta-analysis of genome-wide association studies of anxiety disorders. Mol Psychiatry 21: 1391–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson RE, Cai N, Bigdeli TB, Li Y, Reimers M, Nikulova A et al (2017). The genetic architecture of major depressive disorder in Han Chinese women. JAMA Psychiatry 74: 162–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psychiatric GWAS Consortium Bipolar Disorder Working Group (2011). Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R et al (2014). Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci 17: 1418–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietschel M, Mattheisen M, Frank J, Treutlein J, Degenhardt F, Breuer R et al (2010). Genome-wide association-, replication-, and neuroimaging study implicates HOMER1 in the etiology of major depression. Biol Psychiatry 68: 578–585. [DOI] [PubMed] [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW et al (2013). GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340: 1467–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie GR, Dunham I, Zeggini E, Flicek P (2014). Functional annotation of noncoding sequence variants. Nat Methods 11: 294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderfer DM, Fanous AH, Ripke S, McQuillin A, Amdur RL, Schizophrenia Working Group of Psychiatric Genomics Consortium et al (2014). Polygenic dissection of diagnosis and clinical dimensions of bipolar disorder and schizophrenia. Mol Psychiatry 19: 1017–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511: 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher A, Lennertz L, Wagner M, Hofels S, Pfeiffer U, Guttenthaler V et al (2013). A variant of the neuronal amino acid transporter SLC6A15 is associated with ACTH and cortisol responses and cognitive performance in unipolar depression. Int J Neuropsychopharmacol 16: 83–90. [DOI] [PubMed] [Google Scholar]
- Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA et al (2011). Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry 16: 193–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souery D, Oswald P, Massat I, Bailer U, Bollen J, Demyttenaere K et al (2007). Clinical factors associated with treatment resistance in major depressive disorder: results from a European multicenter study. J Clin Psychiatry 68: 1062–1070. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS (2000). Genetic epidemiology of major depression: review and meta-analysis. Am J Psychiatry 157: 1552–1562. [DOI] [PubMed] [Google Scholar]
- Taylor Tavares JV, Clark L, Cannon DM, Erickson K, Drevets WC, Sahakian BJ (2007). Distinct profiles of neurocognitive function in unmedicated unipolar depression and bipolar II depression. Biol Psychiatry 62: 917–924. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR et al (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung JY, Do CB, Hinds DA, Kiefer AK, Macpherson JM, Chowdry AB et al (2011). Efficient replication of over 180 genetic associations with self-reported medical data. PLoS ONE 6: e23473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP et al (2010). Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijsen JN, Tendolkar I, Arias-Vasquez A, Franke B, Schene AH, Fernandez G et al (2015). Interaction of the 5-HTTLPR and childhood trauma influences memory bias in healthy individuals. J Affect Disord 186: 83–89. [DOI] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR et al (2012). Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Mol Psychiatry 17: 36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Yu H, Li J, Wang L, Li L, Chang H et al (2017). Further evidence for the association between LRP8 and schizophrenia. Schizophr Res doi:10.1016/j.schres.2017.05.002. [DOI] [PubMed]
- Zeng Y, Navarro P, Xia C, Amador C, Fernandez-Pujals AM, Thomson PA et al (2016). Shared genetics and couple-associated environment are major contributors to the risk of both clinical and self-declared depression. EBioMedicine 14: 161–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Wu Z, Hong W, Wang Z, Peng D, Chen J et al (2014). Influence of BCL2 gene in major depression susceptibility and antidepressant treatment outcome. J Affect Disord 155: 288–294. [DOI] [PubMed] [Google Scholar]
- Zhang C, Wu Z, Zhao G, Wang F, Fang Y (2016). Identification of IL6 as a susceptibility gene for major depressive disorder. Sci Rep 6: 31264. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.