ABSTRACT
Osteocalcin has recently been shown to regulate energy homeostasis through multiple pathways. Adipose tissue is a main organ of energy metabolism, and administration of recombinant osteocalcin in mice promoted energy consumption, thus counteracting obesity and glucose intolerance. The regulation of osteocalcin in islet β cells has been well documented; however, it is unknown whether osteocalcin can also act on adipocytes and, if it does, how it functions. Here, we provide evidence to demonstrate a specific role for osteocalcin in brown adipocyte thermogenesis. Importantly, expression of the Gprc6a gene encoding a G protein-coupled receptor as an osteocalcin receptor was activated by brown fat-like differentiation. Moreover, Gprc6a expression could be further potentiated by osteocalcin. Meanwhile, overexpression and knockdown experiments validated the crucial role of Gprc6a in osteocalcin-mediated activation of thermogenic genes. For the first time, we identified Tcf7 and Wnt3a as putative targets for osteocalcin signaling. T cell factor 7 (TCF7) belongs to the TCF/LEF1 family of DNA binding factors crucial for the canonical WNT/β-catenin pathway; however, TCF7 modulates Gprc6a and Ucp1 promoter activation independent of β-catenin. Further studies revealed that the thermogenesis coactivator PRDM16 and the histone demethylase LSD1 might be required for TCF7 activity. Hence, our study described a TCF7-dependent feedback control of the osteocalcin-GPRC6A axis in brown adipocyte physiologies.
KEYWORDS: TCF7/TCF1, WNT/β-catenin pathway, osteocalcin, GPRC6A, UCP1, WNT/β-catenin
INTRODUCTION
Osteocalcin encoded by the Bglap (bone gamma-carboxyglumate protein) gene is the most abundant noncollagen protein in bone extracellular matrix; however, its absence in mice causes a series of metabolic disorders, such as obesity, hypoinsulinemia, and glucose intolerance, rather than defects of bone mineral deposition (1–4). Actually, there is evidence pointing toward a crucial role for osteocalcin in energy metabolism, establishing an interesting communication between skeleton and other metabolic organs, including pancreas, liver, and adipose tissue (5–11).
Within osteoblasts, osteocalcin is initially synthesized in the form of precursors sequentially consisting of a signal peptide directing its secretion, a propeptide for recognition by vitamin K-dependent gamma-glutamyl carboxylase, and an osteocalcin chain with 46 to 50 amino acid residues. After multistep posttranslational modification, vitamin K-dependent gamma-carboxylation causes deposition of osteocalcin in bone extracellular matrix, whereas osteocalcin escaping carboxylation is released to the circulation, acting as a functional hormone (3). Although the exact molecular mechanism for the release of osteocalcin from bone extracellular matrix remains unclear, one gene, Ptprv, encoding protein tyrosine phosphatase receptor V, has been suggested to perform a biologically important role in preventing osteocalcin release (5). To initiate its downstream signaling, osteocalcin has been thought to engage G protein-coupled receptor GPRC6A (G protein-coupled receptor family C group 6 member A) to fulfill its signal pathway. The extracellular region of GPRC6A is capable of recognizing a conserved sequence within the C terminus of osteocalcin, and GPRC6A overexpression can restore osteocalcin-mediated signaling transduction in GPRC6A-null HEK293 cells (6, 7). Genetic manipulation of Gprc6a in mice is closely similar to the effect of osteocalcin deficiencies (8, 9). Additionally, CRISPR/Cas9 targeting of GPRC6A in prostate cancer cells attenuates osteocalcin-stimulated cancer cell migration and growth (12). Nevertheless, comprehensive knowledge of the intracellular signaling pathway of osteocalcin has yet to be elucidated.
The canonical WNT/β-catenin pathway is recognized as a key regulator for embryogenesis, tissue specification, and stem cell biology, and dysregulation of this pathway causes numerous diseases, including tumor, type 2 diabetes mellitus, obesity, and Alzheimer's (13–17). Briefly, the canonical WNT/β-catenin signaling pathway is initiated at the cell surface where secreted Wnt glycoproteins bind to the coreceptor complex formed by Frizzled and the low-density lipoprotein receptor-related protein 5/6 (LRP5/6), leading to the stabilization of cytosolic β-catenin. As a consequence, stabilized β-catenin enters the nucleus and thereafter activates downstream target genes. β-Catenin has potent transcription activation domains at the N and C termini, but it has no intrinsic ability to bind to DNA. Thus, β-catenin needs to interact with the T cell factor/lymphoid enhanced binding factor 1 (TCF/LEF1) family of DNA-binding factors to regulate gene transcription. The TCF/LEF1 family in the canonical WNT/β-catenin pathway consists of four members, LEF1, TCF7/TCF1, transcription factor 7-like 1 (TCF7L1/TCF3), and transcription factor 7-like 2 (TCF7L2/TCF4), that bind to the DNA consensus sequence (A/T)(A/T)CAAAG through a high-mobility group (HMG) domain. Although the canonical WNT/β-catenin pathway is valued for its significant role in tumorigenesis, this pathway has recently drawn increasing attention for its positive physiologies in glucose metabolism. Particularly, the single nucleotide polymorphism (SNP) within the TCF7L2 gene, rs7903146, is considered the most significant genetic marker associated with type 2 diabetes risk (18–20). However, the functionality of other components of the canonical WNT/β-catenin pathway in energy metabolism is still not fully understood.
Adipose tissue, as an important energy metabolic organ, is under the control of multiple signaling pathways, including the osteocalcin and canonical WNT/β-catenin signaling pathways (11, 21, 22). There are two categories of adipose tissues identified in rodents and humans: white adipose tissue and brown adipose tissue (WAT and BAT), respectively. WAT deposits excessive energy as triglyceride and resupplies the organism with energy in the form of nonesterified fatty acid during periods of exercise, fasting, or starvation. Chronic and excessive accumulation of WAT causes obesity, associated with increased risks of cardiovascular diseases, diabetes mellitus, and osteoporosis (23–27). In contrast, BAT expends energy as heat through highly expressing uncoupling protein 1 (UCP1), a core molecule for uncoupling respiration from ATP synthesis in the mitochondria (28–30). Since the discovery of metabolically active BAT in adult humans (31, 32), it has been regarded as a highly suitable target for drugs against diabetes and obesity.
In the current study, we uncovered unexpected cross talk between the osteocalcin molecule and the canonical WNT/β-catenin pathway, thus demonstrating a novel intracellular signaling axis accounting for the osteocalcin molecule in brown adipocytes. Using in vitro differentiation models of brown adipocytes, we validated a specific role for osteocalcin in regulating the activation of thermogenic genes and described a hypothetical feedback mechanism by which osteocalcin initiates its downstream signaling by relying on the receptor GPRC6A and further amplifies its signals through promoting GPRC6A expression. For the first time, we identified that the components of the canonical WNT/β-catenin pathway, Wnt3a and TCF7, might be transcriptionally controlled by osteocalcin signaling. Interestingly, TCF7 could directly augment Gprc6a and Ucp1 promoter activation through a β-catenin-independent manner. Furthermore, PRDM16 (PR domain-containing protein 16) and LSD1 (lysine-specific demethylase 1A; KDM1A) were demonstrated to collaborate with TCF7 to mediate Ucp1 and Gprc6a transcription. Taken together, our results suggest that osteocalcin-stimulated Tcf7 expression in brown adipocytes contributes to the feedback control of osteocalcin signaling and also osteocalcin-mediated activation of thermogenic genes.
RESULTS
Osteocalcin stimulates thermogenesis in brown adipocytes.
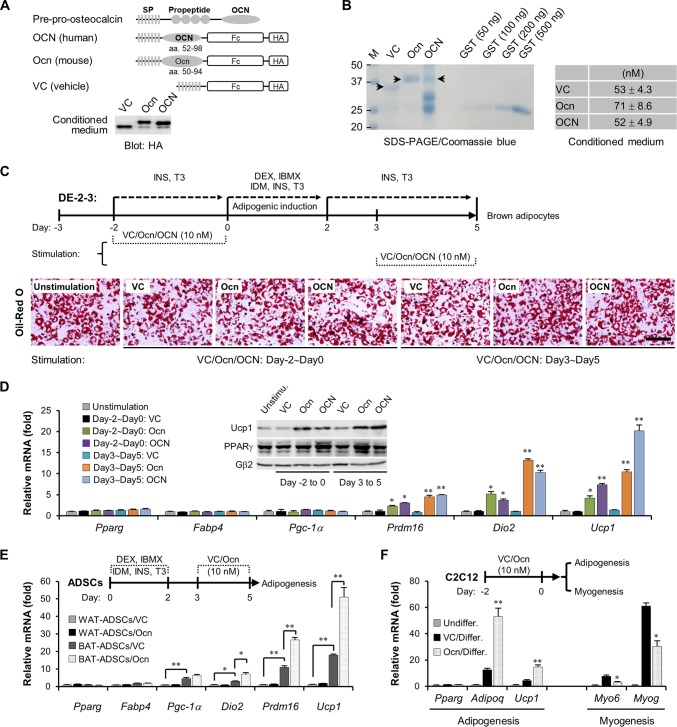
Enhanced thermogenesis indicated by activation of a specific gene program involving Ucp1, Dio2, Pgc-1α, and Prdm16 in brown adipocytes contributes to total energy expenditure (33–37). Several mechanisms, such as prolonged cold exposure, long-term physical exercise, and thiazolidinedione treatment, have testified to this process (38–42). Recently, several studies suggested that circulating osteocalcin diffused from skeleton might be crucial for energy metabolism of adipose tissues (5, 10); however, the signaling mechanism for osteocalcin remains unclear. In the current study, we investigated whether and how osteocalcin can act on brown adipocytes, thus modulating energy consumption. To obtain diffused osteocalcin peptides, osteocalcin chain lacking the propeptide for gamma-glutamyl carboxylase recognition was fused with the immunoglobulin Fc region and a hemagglutinin (HA) tag and then overexpressed in HEK293T cells for 48 h (Fig. 1A). The conditioned medium containing vehicle peptide, human osteocalcin or mouse osteocalcin was harvested and verified by Western blotting (Fig. 1A, OCN and Ocn, respectively), and the concentration of each peptide in conditioned medium was further evaluated through a Coomassie blue-stained SDS-PAGE gel (Fig. 1B).
FIG 1.
Osteocalcin stimulates the thermogenesis in brown adipocytes. (A) The schematic diagram shows the construction maps for human osteocalcin (OCN) and murine osteocalcin (Ocn), with a vehicle peptide as the control (VC, vector control). After being overexpressed in HEK293T cells for 48 h, diffused osteocalcin in conditioned medium was harvested and then verified by Western blotting. Fc, immunoglobulin Fc region; HA, hemagglutinin tag. SP, signal peptide. (B) The measurement of osteocalcin concentrations in conditioned medium. A total of 100 μl of vehicle, Ocn, or OCN conditioned medium was enriched and purified by protein A/G-Plus–agarose, and then the purified peptides were subjected to a Coomassie blue-stained SDS-PAGE gel. Purified glutathione S-transferase (GST) proteins were used as the standard samples. The concentrations of vehicle peptide, Ocn, and OCN are shown in the table at the right. M, size standards. Numbers on the left are in kilodaltons. (C) Schematic diagram showing adipogenesis of DE-2-3 cells and stimulation of osteocalcin peptides. Oil red O staining shows lipid droplet contents in mature brown adipocytes. Scale bar, 100 μm. INS, insulin; T3, l-thyroxine; DEX, dexamethasone; IBMX, isobutyl-1-methylxanthine; IDM, indomethacin. (D) Examination of mRNA levels in DE-2-3-derived brown adipocytes. Pparg and Fabp4 are markers for adipogenesis, and Pgc-1α, Prdm16, Dio2 and Ucp1 are markers for thermogenesis. The inset panel shows UCP1 protein content. (E) Primary adipose-derived stromal cells (ADSCs) were isolated from epididymis white adipose tissues (WAT) or scapular brown adipose tissues (BAT), which then underwent adipogenesis. Osteocalcin stimulation was performed at maturation stage (days 3 to 5). Quantitative PCR analyses were carried out. (F) C2C12 cells were pretreated with osteocalcin for 2 days and then underwent adipogenesis or myogenesis. Adipoq and Ucp1 are markers for brown adipocytes, and Myo6 and Myog are markers for myocytes. Undiffer, undifferentiation; Differ, adipogenic or myogenic differentiation. All data are represented as means ± SD. *, P < 0.05; **, P < 0.01 (Student's t test, n = 3).
Immortalized brown fat preadipocyte from newborn mice DE-2-3 cells (43) that underwent adipogenesis to form mature brown adipocytes in vitro were stimulated with 10 nM osteocalcin at the preadipogenic stage (day −2 to 0) or maturation stage (day 3 to 5); unstimulated cells were used as the control (Fig. 1C). The adipogenic differentiation, indicated by oil red O-stained lipid droplets (Fig. 1C) and expression of Pparg and Fabp4 (Fig. 1D), could not be obviously altered by osteocalcin. However, a typical thermogenic gene program including Ucp1, Prdm16, and Dio2 was significantly activated by the supplement of osteocalcin peptides (Fig. 1D). In particular, increased expression of UCP1, a core molecule in thermogenesis, was validated by both mRNA levels and protein contents (Fig. 1D), suggestive of a latent thermoregulatory role for osteocalcin in brown adipocytes. To further confirm this role for osteocalcin in brown adipocytes, we isolated primary adipose-derived stromal cells (ADSCs) from epididymis white adipose tissues (WAT-ADSCs) or scapular brown adipose tissues (BAT-ADSCs) and then subjected them into adipogenesis (Fig. 1E). At the basal level, BAT-ADSC-derived brown adipocytes exhibited stronger expression levels of thermogenic genes than WAT-ADSC-derived white adipocytes, indicating that in vitro-differentiated brown adipocytes have much stronger thermogenic potential (Fig. 1E). More importantly, we observed a remarkable increase in Ucp1, Prdm16, and Dio2 mRNA levels in BAT-ADSC-derived but not WAT-ADSC-derived adipocytes upon the addition of osteocalcin peptides to the culture medium (Fig. 1E).
In vivo, brown preadipocytes originate from Myf5+ muscle-like precursors. It has been reported that multipotent progenitor C2C12 cells highly express MYF5 proteins and, after undergoing brown fat-like commitment, are able to be differentiated into brown adipocytes, albeit at low efficiency (28). We then verified whether osteocalcin can enhance brown fat-like differentiation of C2C12 cells. C2C12 cells were pretreated with osteocalcin peptides for 48 h and then underwent adipogenesis or myogenesis. The increase in Adipoq and Ucp1 (brown adipocyte markers) mRNA levels and the decrease in Myo6 and MyoG (myocyte markers) mRNA levels clearly demonstrated that osteocalcin could switch C2C12 cells to favor a brown fat-like fate (Fig. 1F). Taking these results together, we conclude that osteocalcin not only modulates the activation of thermogenic genes but may also facilitate brown fat-like differentiation of Myf5+ muscle-like precursors.
Osteocalcin signaling depends on GPRC6A in brown adipocytes.
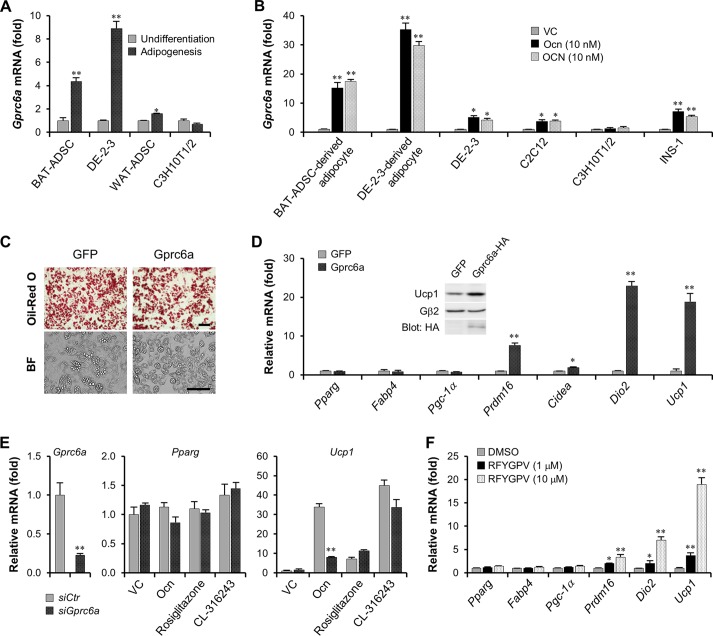
Interestingly, expression of the Gprc6a gene encoding the osteocalcin receptor was strongly enhanced by brown fat-like adipogenesis (DE-2-3 cells and BAT-ADSCs), whereas expression remained constant during white fat-like adipogenesis (WAT-ADSCs and C3H10T1/2 cells) (Fig. 2A). Moreover, expression of the Gprc6a gene could be robustly augmented by osteocalcin in multiple cell types, including brown fat lineages and the rat islet β cell line INS-1 but not in C3H10T1/2 cells (Fig. 2B). Furthermore, these results are also consistent with our findings mentioned above that osteocalcin acts on brown fat-like lineages rather than on white fat lineages. Overexpression of Gprc6a in DE-2-3 cells did not affect their adipogenesis, including the contents of lipid droplets and the expression of Pparg and Fabp4 genes, compared to adipogenesis of the control group expressing green fluorescent protein (GFP) (Fig. 2C and D). However, similar to osteocalcin stimulation, overexpression of the Gprc6a gene remarkably activated key thermogenic genes (Fig. 2D). The abundance of UCP1 proteins in DE-2-3-derived adipocytes was also increased by Gprc6a ectopic expression (Fig. 2D, inset panel). More importantly, knockdown of Gprc6a expression in BAT-ADSC-derived brown adipocytes significantly suppressed osteocalcin-mediated Ucp1 gene activation; Gprc6a knockdown did not affect expression of the osteocalcin-stimulated controls, the peroxisome proliferator-activated receptor gamma (PPARγ) activator rosiglitazone (41, 42) and the selective β3-adrenergic receptor agonist CL316243 (38) (Fig. 2E). The 6-amino-acid peptide RFYGPV derived from the C-terminal sequence of human osteocalcin has been reported to be sufficient for binding to and activating GPRC6A receptor (6). Accordingly, we synthesized this peptide and utilized it to stimulate thermogenic gene activation in brown adipocytes. The analyses of thermogenic gene mRNA levels demonstrated that the RFYGPV peptide could also augment thermogenesis, despite its concentration over 1.0 μM (Fig. 2F). Given these findings, we conclude that osteocalcin signaling in brown adipocytes depends on GPRC6A. Moreover, the finding that osteocalcin stimulates Gprc6a expression also suggests a positive feedback control in the osteocalcin-GPRC6A axis.
FIG 2.
Osteocalcin signaling depends on GPRC6A. (A) The examination of Gprc6a mRNA levels in undifferentiated or adipogenic BAT-ADSCs, DE-2-3 cells, WAT-ADSCs, and C3H10T1/2 cells. (B) Osteocalcin stimulated Gprc6a gene expression for 2 days, and then Gprc6a mRNA levels were examined. (C) Oil red O staining and bright-field (BF) microscopy showed the lipid droplet contents in GFP- or Gprc6a-expressing DE-2-3-derived brown adipocytes. Scale bar, 100 μm. (D) Examination of mRNA levels in GFP- or Gprc6a-expressed DE-2-3-derived brown adipocytes. The inset panel shows the protein contents of UCP1. (E) Knockdown of Gprc6a in BAT-ADSC-derived brown adipocytes inhibits osteocalcin-stimulated Ucp1 activation. Rosiglitazone, 1 μM; CL-316243, 0.1 μM. siCtr, control small interfering RNA; siGprc6a, small interfering RNA targeting Gprc6a. (F) RFYGPV peptide derived from human osteocalcin was synthesized to stimulate thermogenic genes in DE-2-3-derived brown adipocytes. All data are represented as means ± SD. *, P < 0.05; **, P < 0.01 (Student's t test, n = 3). DMSO, dimethyl sulfoxide.
Cross talk of osteocalcin signaling and the canonical WNT/β-catenin pathway.
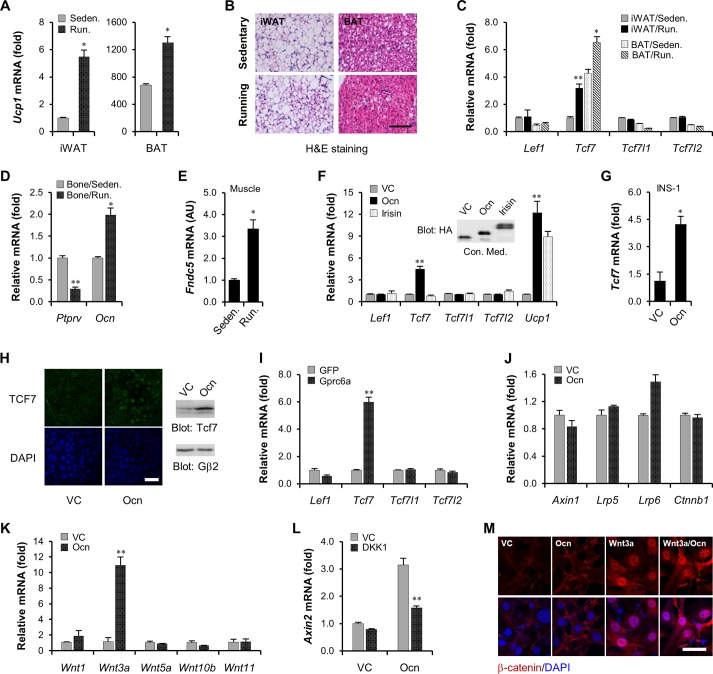
Long-term exercise is beneficial to coordinate metabolic homeostasis of the whole body, for example, by enhancing thermogenesis in adipocytes (40). As shown in Fig. 3A, mice given 4 weeks of free wheel running exhibited a significant elevation of Ucp1 mRNA levels in thermogenic subcutaneous inguinal white adipose tissues (iWAT) and scapular brown adipose tissues (BAT). At the same time, hematoxylin-eosin (H&E) staining also showed smaller lipid droplets and obvious consumption of triglyceride deposition in these adipose depots when mice underwent running exercise (Fig. 3B). Unexpectedly, compared to a sedentary group, running mice displayed a unique activation of the Tcf7 gene in iWAT and BAT (Fig. 3C). TCF7 is a TCF/LEF1 family DNA binding factor crucial for the canonical WNT/β-catenin pathway. In addition to TCF7, there are three other members: LEF1, TCF7L1, and TCF7L2; however, we did not observe any increase in their expression levels in running mice (Fig. 3C), suggesting a specific activation of the Tcf7 gene during long-term exercise. Several studies have demonstrated that long-term exercise is also capable of enhancing production of a variety of metabolic regulation hormones, such as osteocalcin in bone cells (44, 45) and irisin in skeletal muscle (40). Indeed, the group of running mice exhibited upregulated mRNA levels of the osteocalcin gene and downregulated mRNA levels of the Ptprv gene crucial for osteocalcin carboxylation in bone (Fig. 3D), suggesting that circulating osteocalcin might be elevated during long-term exercise. Meanwhile, we also detected activation of the irisin precursor gene Fndc5 in skeletal muscle (Fig. 3E). However, the finding that the Tcf7 gene in brown adipocytes was uniquely induced by osteocalcin rather than by irisin strongly supported a positive role for osteocalcin signaling in modulating Tcf7 gene expression (Fig. 3F). We additionally confirmed this upregulation of the Tcf7 gene by osteocalcin in the islet β cell line INS-1 cells (Fig. 3G). Moreover, the augmented nuclear localization and protein contents of TCF7 in osteocalcin-stimulated brown adipocytes further supported the idea that TCF7 could be transcriptionally controlled by osteocalcin signaling (Fig. 3H). Finally, overexpression of the Gprc6a gene could also uniquely give rise to activation of the Tcf7 gene (Fig. 3I), providing additional evidence for the positive role of osteocalcin signaling in Tcf7 gene transcription.
FIG 3.
Cross talk of osteocalcin signaling and the canonical WNT/β-catenin pathway. (A) The examination of Ucp1 mRNA levels in subcutaneous inguinal white adipose depots (iWAT) and scapular brown adipose depots (BAT) from sedentary (Seden) mice or mice given 4 weeks of free wheel running (Run) (n = 8 from each group). (B) Hematoxylin-eosin (H&E) staining for iWAT and BAT from sedentary or running mice. Scale bar, 100 μm. (C) The examination of Lef1, Tcf7, Tcf7l1, and Tcf7l2 mRNA levels in iWAT and BAT from sedentary or running mice (n = 8 from each group). (D) The examination of Ptprv and osteocalcin mRNA levels in bone from sedentary or running mice (n = 8 from each group). (E) The examination of Fndc5 mRNA levels in skeletal muscle from sedentary or running mice (n = 8 from each group). AU, arbitrary units. (F) Osteocalcin stimulates Tcf7 expression in DE-2-3-derived brown adipocytes. Forty-eight hours of osteocalcin stimulation was carried out. (G) Osteocalcin stimulates Tcf7 expression in rat islet β cell line INS-1. (H) Immunofluorescence for nuclear TCF7 contents (left) and Western blotting for TCF7 protein levels (right) in DE-2-3-derived brown adipocytes. Scale bar, 50 μm. (I) Overexpression of Gprc6a stimulates Tcf7 expression in DE-2-3-derived brown adipocytes. (J) The examination of mRNA levels of canonical WNT/β-catenin signaling components in DE-2-3-derived brown adipocytes. (K) The expression of Wnt genes in DE-2-3-derived brown adipocytes. (L) Osteocalcin stimulates Axin2 expression in the absence or presence of DKK1 (100 ng/ml) in DE-2-3-derived brown adipocytes. (M) Osteocalcin regulates cytosolic β-catenin accumulation in the absence or presence of Wnt3a (100 ng/ml). The stimulation of osteocalcin and Wnt3a was performed for 3 h. Red, β-catenin; blue (4′,6′-diamidino-2-phenylindole [DAPI]), nucleus. All data are represented as means ± SD. *, P < 0.05; **, P < 0.01 (Student's t test, n = 3). Scale bar, 50 μm.
We also determined that the Wnt3a gene, in addition to the Tcf7 gene, was probably controlled by osteocalcin signaling (Fig. 3J and K). Moreover, the transcription of the Axin2 gene, a specific target of the canonical WNT/β-catenin pathway (46, 47), was activated by osteocalcin, and this activation could be inhibited by the Wnt3a antagonist DKK1 (Fig. 3L), thus revealing cross talk between osteocalcin signaling and the canonical WNT/β-catenin pathway. We then investigated whether osteocalcin can modulate canonical WNT/β-catenin signaling transduction directly or only through an indirect manner. As showed in Fig. 3M, transient stimulation with osteocalcin (3 h) could neither induce cytosolic β-catenin accumulation nor enhance Wnt3a-stimulated β-catenin stabilization. Taking these results together, we conclude that osteocalcin takes part in the canonical WNT/β-catenin signaling pathway probably by transcriptionally modulating expression of the Tcf7 and Wnt3a genes.
TCF7 directly modulates Ucp1 and Gprc6a promoter activation.
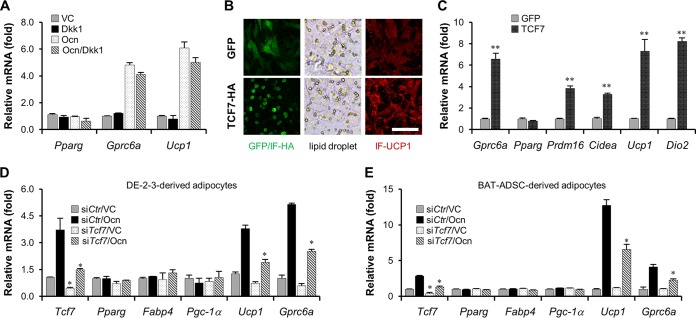
Given the finding that osteocalcin transcriptionally controls the Wnt3a gene, we examined first whether the Wnt3a-initiated canonical WNT/β-catenin pathway is involved in osteocalcin signaling transduction in brown adipocytes. However, the failure of DKK1 to prevent osteocalcin-stimulated activation of Gprc6a and Ucp1 suggested that the Wnt3a-initiated canonical WNT/β-catenin pathway might be dispensable for osteocalcin signaling (Fig. 4A). Although TCF7 is a key transcription factor of the canonical WNT/β-catenin pathway, it can also perform in a β-catenin-independent manner (48). We then verified whether TCF7 regulates adipocyte thermogenesis, thus mediating the intracellular signaling of osteocalcin. The Gfp or Tcf7 gene was delivered into DE-2-3 cells by lentiviruses, and their expression level was examined by immunofluorescence (Fig. 4B). Overexpression of the Tcf7 gene did not alter the formation of lipid droplets or expression of the Pparg gene, but it obviously elevated UCP1 protein content and remarkably stimulated thermogenic gene activation (Fig. 4B and C). More importantly, knockdown of Tcf7 gene expression significantly blocked osteocalcin-induced Ucp1 and Gprc6a transcription in both DE-2-3-derived and BAT-ADSC-derived brown adipocytes (Fig. 4D and E), strongly supporting a downstream regulatory role for TCF7 in osteocalcin signaling. In addition, transcription of the Gprc6a gene was modulated by TCF7 proteins, an observation which provides an unexpected but logical molecular mechanism to elucidate the positive feedback characteristic of the osteocalcin signaling mentioned above.
FIG 4.
TCF7 mediates osteocalcin signaling. (A) DKK1 does not impair osteocalcin-stimulated Gprc6a and Ucp1 activation in DE-2-3-derived brown adipocytes. (B) The contents of lipid droplets (bright field) and UCP1 proteins (red) in GFP- or TCF7-overexpressed DE-2-3-derived brown adipocytes. Scale bar, 100 μm. IF, immunofluorescence. (C) The examination of mRNA levels in GFP- or TCF7-overexpressing DE-2-3-derived brown adipocytes. (D) Knockdown of Tcf7 in DE-2-3-derived brown adipocytes inhibits osteocalcin-stimulated Ucp1 and Gprc6a expression. (E) Knockdown of Tcf7 in BAT-ADSC-derived brown adipocytes inhibits osteocalcin-stimulated Ucp1 and Gprc6a expression. In panels D and E, small interfering RNAs are indicated by “si” prefixes with protein names. All data are represented as means ± SD. *, P < 0.05; **, P < 0.01 (Student's t test, n = 3).
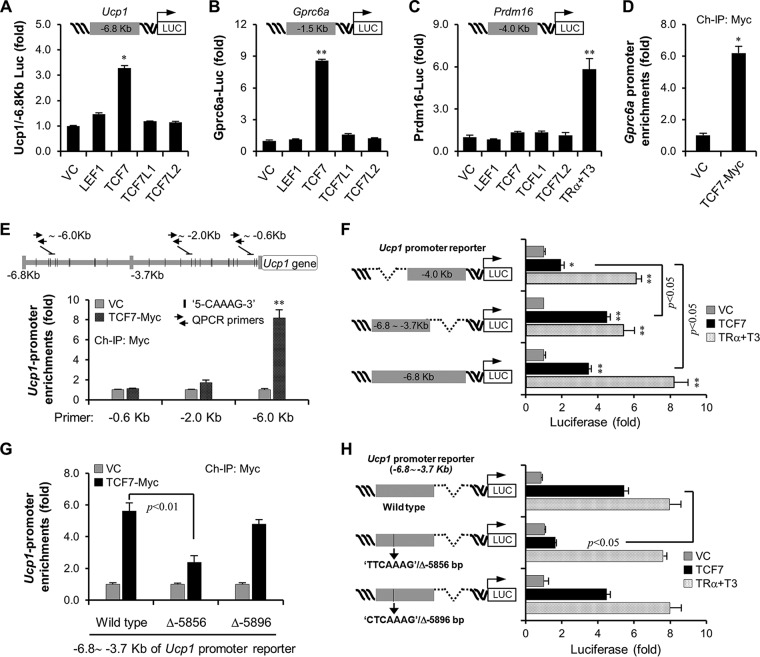
To further access this molecular mechanism, we verified whether TCF7 directly modulates activation of the promoters of the Ucp1 and Gprc6a genes. Although all members of the TCF/LEF1 family share main structural features (49), using in vitro-constructed Ucp1 and Gprc6a promoter-dependent luciferase reporters, we observed that only TCF7 could drive reporter activity of both the Ucp1 and Gprc6a promoter (Fig. 5A and B). In addition, TCF7 appeared to be dispensable for activation of the Prdm16 promoter even though TCF7 can activate the Prdm16 gene in vivo, probably indicating an indirect role of TCF7 in Prdm16 gene transcription or the TCF7 DNA binding site beyond the region of the Prdm16 promoter DNA used in the reporter (Fig. 5C). We then utilized chromatin immunoprecipitation of TCF7 to confirm enrichment of this transcription factor on the Gprc6a and Ucp1 promoters. TCF7 belongs to a high-mobility group (HMG) box-containing family specifically binding to a DNA consensus site with the core sequence CAAAG. It has previously been reported that the optimal binding site for TCF7 is (A/T)(A/T)CAAAG (50). Within the kb −1.5 Gprc6a gene promoter, we found a typical sequence, TTCAAAG, beginning at base −604 on the complementary strand, and then confirmed the enrichment of TCF7 in this region (Fig. 5D). Although the typical sequence TTCAAAG was found at base −649 of the Ucp1 promoter, analyses of chromatin immunoprecipitation showed rare enrichment of TCF7 on this site, which seemed likely to bind to the distal sequence AACAAAG at base −5856 of the Ucp1 promoter (Fig. 5E). In accordance with this finding, TCF7 barely activated the reporter activity driven by the kb −4.0 promoter DNA of Ucp1 but strongly activated a kb −3.7 to −6.8 promoter DNA-dependent reporter (Fig. 5F). As a control, thyroid receptor α could activate both reporters well (Fig. 5F). Deletion of the sequence AACAAAG at base −5856 but not the flanked sequence CTCAAAG at base −5896 could abolish TCF7 occupation on the Ucp1 promoter DNA (Fig. 5G). This deletion also significantly suppressed TCF7 regulation in reporter assays (Fig. 5H). Taken together, these data unequivocally demonstrate that TCF7 proteins can directly modulate Ucp1 and Gprc6a promoter activation.
FIG 5.
TCF7 controls Ucp1 and Gprc6a promoter activation. (A) TCF7 activates a Ucp1 promoter-driven luciferase (Luc) reporter. For reporter assays, Ucp1-luciferase reporter (15 ng/well) and GFP (20 ng/well) were cotransfected into HEK293T cells (48-well) with TCF/LEF1 family transcription factors (50 ng/well). The LacZ plasmid was added to make the total amount of DNA equal (250 ng/well). (B) TCF7 activates a Gprc6a promoter-driven luciferase reporter. (C) TCF7 does not regulate the Prdm16 promoter-driven luciferase reporter. (D and E) Myc-tagged TCF7 was introduced into BAT-ADSCs and then underwent adipogenesis. Adipocytes from two 10-cm dishes were fixed, and then chromatin immunoprecipitation (ChIP) was carried out by a Myc antibody as described in Materials and Methods. The primer pairs flanking kb −0.6 of the Gprc6a promoter DNA (D) and flanking kb −0.6, −2.0, or −6.0 of the Ucp1 promoter DNA (E) were designed for quantitative PCR. (F) The regulation of TCF7 at kb −4.0, −3.7 to −6.8, and −6.8 of the Ucp1 promoter DNA-driven luciferase reporters. Thyroid receptor α (TRα) in the presence of thyroxine (T3, 10 nM) was used as the control. (G) Deletion of the AACAAAG sequence at base −5856 of the kb −3.7 to −6.8 Ucp1 promoter reporter inhibits TCF7 binding to the Ucp1 promoter. (H) Deletion of the AACAAAG sequence at base −5856 of the kb −3.7 to −6.8 Ucp1 promoter reporter inhibits TCF7-mediated reporter activation. All data are represented as means ± SD. *, P < 0.05; **, P < 0.01 (Student's t test, n = 3).
PRDM16 is identified as a coactivator of TCF7.
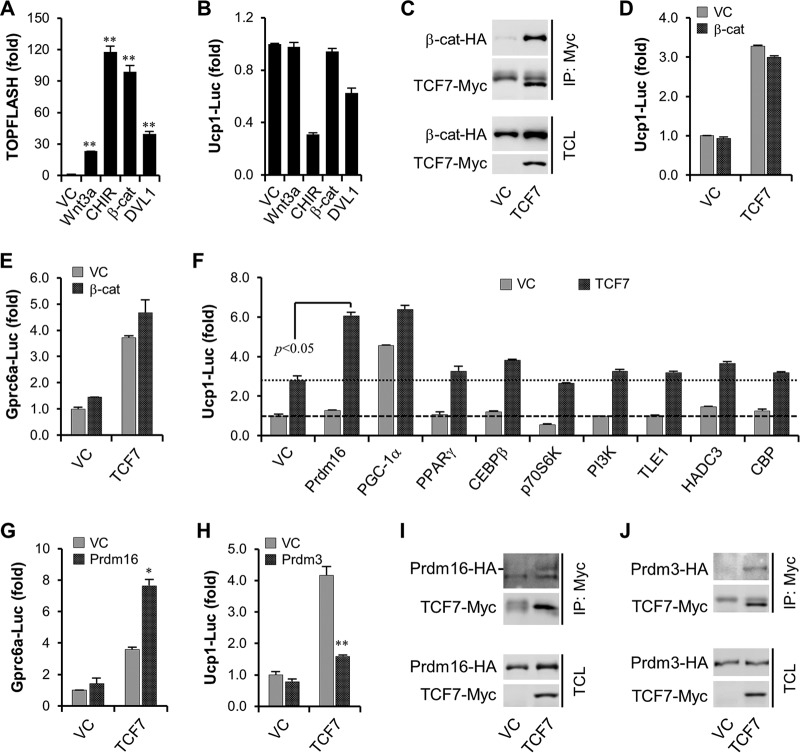
In agreement with the finding that inhibition of Wnt3a activity did not affect osteocalcin signaling in brown adipocytes (Fig. 4A), we found that activation of the canonical WNT/β-catenin pathway by Wnt3a protein, by the glycogen synthase kinase 3β (GSK3β) inhibitor CHIR99021, by overexpression of active β-catenin (Ser-45 deletion), or by DVL1 protein is dispensable for Ucp1 promoter DNA-dependent reporter activation (Fig. 6A and B). Even though β-catenin could efficiently interact with TCF7 (Fig. 6C), it also failed to improve TCF7-mediated activation of both Ucp1 and Gprc6a promoter DNA reporters (Fig. 6D and E). Thus, we hypothesized that TCF7 facilitates activation of Ucp1 and Gprc6a promoters probably independent of the canonical WNT/β-catenin pathway. Since TCF7 itself is not sufficient to activate gene transcription (49), we then tried to identify novel coactivators for TCF7 proteins in brown adipocytes. In this effort, we discovered that PRDM16, crucial for brown adipocyte formation and thermogenesis (28), is a surprising candidate (Fig. 6F). At the same time, PRDM16 also augmented TCF7-activated Gpr6a promoter activation (Fig. 6G). Interestingly, as a homologue of PRDM16 (51), PR domain-containing protein 3 (PRDM3), however, significantly inhibited TCF7 transcriptional activity (Fig. 6H). Coimmunoprecipitation experiments indicated that both proteins could interact with TCF7 (Fig. 6I and J), thus demonstrating a unique role for PRDM16 in TCF7-modulated transcriptional activity.
FIG 6.
PRDM16 is identified as a coactivator of TCF7. (A and B) The effect of activators for the canonical WNT/β-catenin pathway on TOPFlash marking WNT/β-catenin signaling activation (A) and Ucp1 promoter reporter (B). β-Catenin-ΔSer45 was transfected as 25 ng/48-well, and DVL1 was transfected as 50 ng/48 well. Wnt3a, 100 ng/ml; CHIR99021, 5 μM. (C) Coimmunoprecipitation of β-catenin–HA with TCF7-Myc. (D and E) β-Catenin cannot enhance TCF7-mediated activation of Ucp1 (D) and Gprc6a (E) promoter reporters. (F) PRDM16 was identified to regulate TCF7-mediated activation of Ucp1 promoter reporter. PI3K, phosphatidylinositol 3-kinase. (G) PRDM16 augments TCF7-mediated activation of Gprc6a promoter reporter. (H) PRDM3 suppresses TCF7-mediated activation of Ucp1 promoter reporter. (I) Coimmunoprecipitation (IP) of Prdm16-HA with TCF7-Myc. (J) Coimmunoprecipitation of Prdm3-HA with TCF7-Myc. All data are represented as means ± SD. *, P < 0.05; **, P < 0.01 (Student's t test, n = 3). TCL, total cell lysate.
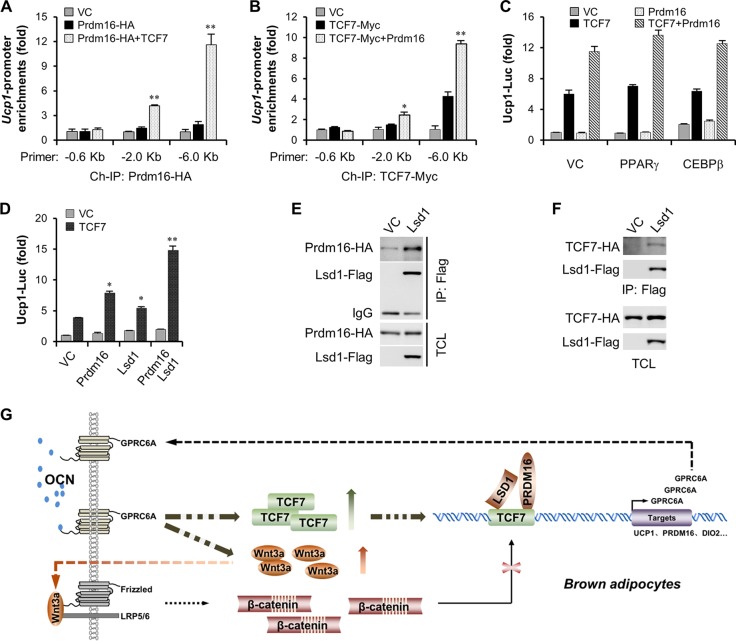
Finally, we investigated how PRDM16 modulates TCF7 transcriptional activity. Chromatin immunoprecipitation experiments revealed that PRDM16 itself had only a slight ability to bind to the Ucp1 promoter, but it could be well recruited by TCF proteins (Fig. 7A). Simultaneously, we found that the presence of PRDM16 could also enhance TCF7 DNA binding activity (Fig. 7B), pointing toward a mutual dependence of two proteins on their DNA binding. Several partners have been suggested to coordinate PRDM16 activation in brown fat formation and thermogenesis, including PPARγ, CEBPβ, and LSD1 (41, 52, 53). We then verified their effects on TCF7-involved transcriptional activity using a Ucp1 promoter reporter. Analyses of reporter activity showed that PPARγ and CEBPβ were dispensable for TCF7 transcriptional activity, but LSD1 could have a prominent enhancement effect on PRDM16 and TCF7-modulated Ucp1 promoter activation. LSD1 is critical for the activation of brown fat-like genes and the suppression of white fat-like genes through interaction with PRDM16 (53). Coimmunoprecipitation experiments validated this interaction of LSD1 with PRDM16 (Fig. 7E) and also revealed a novel interaction between LSD1 and TCF7 (Fig. 7F). Overall, these data suggest a novel nuclear transcription machinery involved in TCF7, PRDM16, and LSD1 in brown adipocyte physiologies.
FIG 7.
LSD1 is involved in TCF7/PRDM16-mediated transcriptional machinery. (A) TCF7 recruits PRDM16 to Ucp1 promoter DNA. Kilobase −6.8 Ucp1 promoter DNA was cotransfected with Prdm16-HA (200 ng/well) in HEK293T (six-well) cells in the absence or presence of TCF7-Myc (200 ng/well) for 24 h, and then chromatin immunoprecipitation was performed. (B) PRDM16 enhances TCF7 DNA binding activity. Kilobase −6.8 Ucp1 promoter DNA was cotransfected with TCF7-Myc (200 ng/well) to HEK293T (six-well) cells in the absence or presence of Prdm16-HA (200 ng/well) for 24 h, and then chromatin immunoprecipitation was performed. (C) PPARγ and CEBPβ do not regulate TCF7 and PRDM16-mediated Ucp1 promoter reporter. (D) LSD1 enhances TCF7 and PRDM16-mediated Ucp1 promoter reporter. (E) Coimmunoprecipitation of Prdm16-HA with Lsd1-Flag. (F) Coimmunoprecipitation of TCF7-HA with Lsd1-Flag. (G) The model of TCF7-mediated feedback control of osteocalcin signaling. All data are represented as means ± SD. *, P < 0.05; **, P < 0.01 (Student's t test, n = 3).
In summary, our study sheds light on a novel molecular mechanism by which osteocalcin regulates metabolic action of brown adipocytes. Taking into account data mentioned above, a hypothetical model is presented (Fig. 7G) according to which osteocalcin signaling is in cross talk with the canonical WNT/β-catenin pathway through transcriptionally controlling Wnt3a and Tcf7, and then TCF7 directly facilitates Gprc6a and Ucp1 promoter activation, thereby describing a positive feedback profile of the osteocalcin signaling pathway. Interestingly, our study demonstrates that TCF7 modulates gene transcription probably by relying on PRDM16 and LSD1 but not β-catenin in brown adipocytes.
DISCUSSION
A growing body of studies has demonstrated that there may be multiple interactions between bone remodeling and adipose metabolic actions. For example, excessive accumulation of white adipose tissues is highly associated with osteoporosis (26, 27), whereas cold-activated brown adipose tissues are conductive to bone mineral deposition (54, 55). Meanwhile, bone can also function as an important endocrine organ acting on various metabolic regulation organs, including adipose tissues (4, 5, 11). Nevertheless, not much is known about the molecular mechanism by which bone and adipose tissue can mutually interact so far. Recent studies had demonstrated that osteocalcin, a noncollagen protein in bone extracellular matrix, could be released into circulation and then modulate the physiologies of a number of tissues such as islet β cells, skeletal muscle, intestine, and adipose tissues (5, 11, 44, 56). In the current study, we set out to explore a novel molecular mechanism for osteocalcin by which healthy bone anabolism can augment energy expenditure of brown adipocytes.
Using multiple in vitro differentiation models of brown adipocytes, for example, lipid droplet formation, and expression of the Pparg and Fabp4 genes, we validated that osteocalcin stimulation could augment thermogenesis of brown adipocytes while failing to facilitate adipogenic differentiation of brown adipocytes. In vivo, brown adipocyte formation requires two differentiation stages sequentially: brown fat-like commitment of Myf5+ muscle-like precursors, giving rise to brown preadipocytes, and adipogenesis of preadipocytes into mature brown adipocytes. Although osteocalcin is dispensable for adipogenesis, its stimulation might tune Myf5+ muscle-like precursors to favor a brown fat-like fate. Interestingly, the gene encoding GPRC6A proteins, previously proved to be osteocalcin receptors in multiple cell types, was remarkably activated by brown fat-like but not white fat-like differentiation. Moreover, the Gprc6a gene in brown fat-like lineages could be stimulated significantly by osteocalcin. Especially, overexpression of the Gprc6a gene, similar to osteocalcin stimulation, could specifically potentiate the thermogenesis of brown adipocytes, whereas knockdown of Gprc6a gene expression inhibited osteocalcin-mediated but not either rosiglitazone- or CL316243-mediated Ucp1 gene activation, strongly suggesting a positive role for an osteocalcin-GPRC6A axis in the thermogenesis of brown adipocytes.
Unexpectedly, we observed a unique activation of the Tcf7 gene encoding a key DNA binding factor crucial for the canonical WNT/β-catenin pathway in osteocalcin-stimulated cells. Moreover, like the Tcf7 gene, the Wnt3a gene encoding a canonical Wnt glycoprotein was also identified as a putative target of osteocalcin signaling. Particularly, the canonical WNT/β-catenin pathway target Axin2 could be activated by osteocalcin, and this activation was obviously impaired by the canonical Wnt glycoprotein antagonist DKK1, pointing toward a previously unknown cross talk between osteocalcin signaling and the canonical WNT/β-catenin pathway. However, we also showed that this cross talk could happen probably indirectly with osteocalcin transcriptionally controlling Wnt3a and Tcf7 gene expression rather than modulating Wnt3a-initiated signaling transduction. Nevertheless, it seems that osteocalcin does not rely on the canonical WNT/β-catenin pathway but, rather, TCF7 proteins to control expression of the receptor Gprc6a and activation of thermogenic genes. Actually, knockdown of Tcf7 expression instead of antagonizing WNT/β-catenin signaling initiation could inhibit osteocalcin-stimulated Gprc6a and Ucp1 expression. Overexpression of the Tcf7 gene contributed to Gprc6a and thermogenic gene activation. More importantly, chromatin immunoprecipitation of TCF7 exhibited the enrichment of this factor within the upstream promoter DNA of the Gprc6a and Ucp1 genes, and TCF7 proteins could activate a Gprc6a and Ucp1 promoter-dependent reporter in vitro, suggesting a key role for TCF7 in osteocalcin signaling.
Except for TCF7, other TCF/LEF1 family members, like Lef1, Tcf7l1, and Tcf7l2, were not regulated by osteocalcin signaling, nor did they control the Ucp1 and Gprc6a promoters. These results might reflect the fact that the TCF/LEF family has only partial redundancy and sometimes even opposite effects, depending on cell or tissue specificity (49). Moreover, although TCF7 proteins are the key DNA binding factors acting in the canonical WNT/β-catenin pathway, TCF7 can still perform in a β-catenin-independent manner (48). Our study clearly showed that the modulation of TCF7 on the Ucp1 and Gprc6a promoters could be independent of the WNT/β-catenin pathway. However, as a DNA binding factor, TCF7 has no intrinsic transcriptional activity. So to access how TCF7 is activated on Ucp1 and Gprc6a promoters, we revealed PRDM16, the key regulator of brown adipocyte commitment and thermogenesis, to be a coactivator of TCF7. Coimmunoprecipitation experiments validated the interaction of PRDM16 and TCF7. Interestingly, PRDM16 may further facilitate the DNA binding activity of TCF7 after its recruitment to the target promoter by TCF7. Further studies revealed that LSD1 rather PPARγ and CEBPβ may be involved in TCF7 transcriptional activity through interaction with PRDM16 and TCF7.
In conclusion, our study reveals a possible signaling axis for osteocalcin in brown adipocytes that seems to be unexpectedly linked with TCF7 protein, a DNA binding factor of the canonical WNT/β-catenin pathway, therefore suggesting a feedback loop formed by TCF7 and the osteocalcin-GPRC6A axis in regulating thermogenesis of brown adipocytes. In addition, the osteocalcin-GPRC6A axis might be implicated in prostate cancer cell progression (12), and abnormal expression of TCF7 is also associated with the tumorigenesis of prostate cancer (57). Therefore, the osteocalcin-GPRC6A-TCF7 feedback loop and also the cross talk of osteocalcin signaling with the canonical WNT/β-catenin pathway, both of which we uncovered in the current study, may provide new perspectives on the pathogenesis of metastatic prostate cancer.
MATERIALS AND METHODS
Materials.
Protein A/G-Plus–agarose beads were purchased from Santa Cruz (Dallas, TX); antibodies to TCF7 (monoclonal antibody [MAb] C63D9, at a dilution of 1:1,000; catalog number 2203) and PPARG (MAb 81B8, 1:1,000; 2443) were from Cell Signaling (Danvers, MA); UCP1 (1:1,000; ab10983) was from Abcam (Cambridge, UK); β-catenin (1:1,000; 610154) was from BD Biosciences (San Jose, CA); Gβ2 (C-16, 1:1,000; sc-380) was from Santa Cruz; HA.11 (16B12, 1:1,000; 901514) and Myc (9E10, 1:1,000; 626802) were from BioLegend (San Diego, CA); Flag M2 (1:1,000, F3165) was from Sigma-Aldrich (St. Louis, MO); recombinant mouse Wnt3a protein (1324-WN) and Dkk1 (5897-DK) were from R&D Systems (Minneapolis, MN); CHIR99021 was from Stemcell Technologies (Canada). Peptide RFYGPV was synthesized by Sangon Biotech (Shanghai, China). The oligonucleotides for gene silence and real-time PCR used in this study are listed in Table S1 in the supplemental material.
Cell culture.
HEK293T, DE-2-3, C3H10T1/2, and C2C12 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) (Life Technologies, Waltham, MA), and 1× GlutaMAX (Life Technologies) at 37°C in 5% CO2. Rat pancreatic islet β cell line INS-1 was maintained in RPMI 1640 medium with 10% FBS, 1 mM sodium pyruvate, 10 mM HEPES, 50 μM β-mercaptoethanol, and 1× GlutaMAX at 37°C in 5% CO2. The adipose-derived stromal cells (ADSCs) of epididymis white adipose and scapular brown adipose were isolated from C57BL/6J mice by collagenase digestion as described previously (58) and cultured in alpha minimal essential medium (α-MEM) with 10% FBS and 1× GlutaMAX at 37°C in 5% CO2.
Animals.
Six-week-old C57BL/6 mice were from Shanghai Laboratory Animal Center of Chinese Academy of Sciences. All experiments were approved by the Animal Care and Use Committee of the Fudan University Shanghai Medical College. For athletic stimulation, mice were housed individually in cages with free access to a running wheel (59). An exercise procedure containing daily 2-h running was applied for 4 weeks.
Viruses.
Lentiviruses were constructed with pLVX-puro carrier vector (Clontech, Mountain View, CA) for gene expression. To prepare viruses, carrier vectors were packaged with helper plasmid dR8.9 and vesicular stomatitis virus G protein (VSV-G), and viruses were purified by ultracentrifugation. Cells were infected with viruses at a multiplicity of infection of 10.
Gene transcription analysis.
Total RNAs were isolated from mouse tissues using a magnetic bead homogenizer in TRIzol reagent or from cultured cells by directly adding TRIzol reagent to the cells. cDNAs were synthesized using a ProtoScript II, First Strand cDNA synthesis kit (BioLabs, Ipswich, MA). Quantitative real-time PCR (qPCR) analysis was performed using Power SYBR green PCR master mix (Life Technologies). 18S RNA levels were used as internal controls in the qPCR analysis.
Reporter assay.
HEK293T cells were transiently transfected with DNA using Lipofectamine LTX from Life Technologies according to the manufacturer's instructions. A LacZ plasmid was added to make the total amount of DNA equal (0.25 μg/well in a 48-well plate). Promoter DNA-driven luciferase reporters were constructed with pGL4.20 vector (Promega, WI). In brief, kb −1.5 murine and rat Gprc6a promoter DNA, kb −6.8 murine Ucp1 promoter DNA, kb −4.0 murine Ucp1 promoter DNA, kb −3.7 to −6.8 murine Ucp1 promoter DNA, and kb −4.0 murine Prdm16 promoter DNA were inserted upstream of the luciferase open reading frame. Luciferase reporter assays were performed, and the luciferase activities presented were normalized against the levels of GFP expression as described previously (60).
Protein interaction and complex assays.
Coimmunoprecipitation experiments were carried out in HEK293T cells. Briefly, cells were transfected with constructs indicated in the figures for 24 h and then lysed in a cell lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.5 mM EDTA, protease inhibitor cocktail [Roche, Indianapolis, IN] and phosphatase inhibitor cocktail [Roche]). After centrifugation, the supernatants were pulled down by the indicated antibodies and then analyzed by immunoblotting.
ChIP.
For chromatin immunoprecipitation (ChIP) assays, cells were fixed with 1% paraformaldehyde (PFA) in culture medium at 37°C for 20 min. After PFA solution was removed, cells were washed with Dulbecco's phosphate-buffered saline (DPBS) and lysed in SDS lysis buffer (50 mM Tris-HCl, pH 8.1, 1% SDS, and 10 mM EDTA); then ChIP was performed using a ChIP assay kit (Millipore) according to the manufacturer's instructions.
Immunofluorescence assay.
Cells on coverslips were washed once with DPBS and then fixed for 20 min in DPBS containing 4% paraformaldehyde at room temperature. Fixed cells were permeabilized by 0.1% Triton X-100 for 5 min and then blocked by 2% bovine serum albumin (BSA) for 30 min. Finally, cells were stained with primary antibodies followed by fluorescein isothiocyanate (FITC)-conjugated secondary antibodies. Immunofluorescence images were captured using a Leica SP5 microscope.
Cell differentiation.
For white adipocyte differentiation, after 2 days of confluence, WAT-ADSC or C3H10T1/2 cells were induced with 1 μM dexamethasone (Sigma, St. Louis, MO), 0.5 mM isobutyl-1-methylxanthine (Sigma), 5 μg/ml insulin (Sigma), and 1 μM rosiglitazone (Sigma). After 2 days, the cells were transferred to maintenance medium containing 5 μg/ml insulin for 3 days (61). For brown adipocyte differentiation, the preadipocyte DE-2-3 or BAT-ADSC cells were cultured with 125 ng/ml insulin and 1 nM 3,3′,5-triido-l-thyronine (Sigma) after they reached 70% confluence. After 2 days, the confluent cells were induced with 1 μM dexamethasone, 0.5 mM isobutyl-1-methylxanthine, 0.125 mM indomethacin, 125 ng/ml insulin, and 1 nM 3,3,5-triido-l-thyronine for 2 days. After induction, cells were transferred to maintenance medium containing 125 ng/ml insulin and 1 nM 3,3′,5-triido-l-thyronine for 3 days (43, 58). For adipogenesis of C2C12 cells, at confluence, cells were stimulated with 1 μM dexamethasone, 0.5 mM isobutyl-1-methylxanthine, 0.125 mM indomethacin, 5 μg/ml insulin, 1 μM rosiglitazone, and 1 nM 3,3′,5-triido-l-thyronine for 2 days, and then cells were cultured in medium containing 1 μg/ml insulin, 1 μM rosiglitazone, and 1 nM 3,3′,5-triido-l-thyronine for a further 4 days. For myogenesis of C2C12 cells, C2C12 cells at 80% of confluence were induced by 2% horse serum (Invitrogen) and 1 mM sodium pyruvate (Invitrogen) for 4 days (28). For oil red O staining, the differentiated cells were fixed with 4% of paraformaldehyde for 30 min and stained with a solution containing 1.8 mg/ml of oil red O (Sigma) and 60% isopropanol for 30 min.
Statistical analysis.
We used two-tailed Student t tests to evaluate statistical significance, and a P value of <0.05 was declared to be statistically significant. We present all values as means ± standard deviations (SD).
Supplementary Material
ACKNOWLEDGMENTS
This work was sponsored by the National Natural Science Foundation of China (grant 31671466).
We are grateful to D. Pan and Q. Tang for providing DE-2-3 and C3H10T1/2 cells. We thank Y. Huang and J. Li for confocal microscopy technical help.
Q.L. did constructs, reporter assays, protein interaction assays, cells differentiation, qPCR assays, and cells staining assays; Y.H. and Y.Y. made mouse tissue samples and performed QPCR assays; X.H. and W.Z. did technical work; J.W. supervised the project and wrote the manuscript; X.G. supervised the project and wrote the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00562-17.
REFERENCES
- 1.Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, Bradley A, Karsenty G. 1996. Increased bone formation in steoclacin-deficient mice. Nature 382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- 2.Wei J, Karsenty G. 2015. An overview of the metabolic functions of osteocalcin. Rev Endocr Metab Disord 16:93–98. doi: 10.1007/s11154-014-9307-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, Zhang H, Yang C, Li Y, Dai Z. 2016. An overview of osteocalcin progress. J Bone Miner Metab 34:367–379. doi: 10.1007/s00774-015-0734-7. [DOI] [PubMed] [Google Scholar]
- 4.Karsenty G, Olson EN. 2016. Bone and muscle endocrine functions: unexpected paradigms of inter-organ communication. Cell 164:1248–1256. doi: 10.1016/j.cell.2016.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee NK, Sowa H, Hinoi E, Ferron M, Ahn JD, Confavreux C, Dacquin R, Mee PJ, McKee MD, Jung DY, Zhang Z, Kim JK, Mauvais-Jarvis F, Ducy P, Karsenty G. 2007. Endocrine regulation of energy metabolism by the skeleton. Cell 130:456–469. doi: 10.1016/j.cell.2007.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pi M, Kapoor K, Ye R, Nishimoto SK, Smith JC, Baudry J, Quarles LD. 2016. Evidence for osteocalcin binding and activation of GPRC6A in β-cells. Endocrinology 157:1866–1880. doi: 10.1210/en.2015-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pi M, Wu Y, Quarles LD. 2011. GPRC6A mediates responses to osteocalcin in β-cells in vitro and pancreas in vivo. J Bone Miner Res 26:1680–1683. doi: 10.1002/jbmr.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pi M, Chen L, Huang MZ, Zhu W, Ringhofer B, Luo J, Christenson L, Li B, Zhang J, Jackson PD, Faber P, Brunden KR, Harrington JJ, Quarles LD. 2008. GPRC6A null mice exhibit osteopenia, feminization and metabolic syndrome. PLoS One 3:e3858. doi: 10.1371/journal.pone.0003858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei J, Hanna T, Suda N, Karsenty G, Ducy P. 2014. Osteocalcin promotes β-cell proliferation during development and adulthood through Gprc6a. Diabetes 63:1021–1031. doi: 10.2337/db13-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du J, Zhang M, Lu J, Zhang X, Xiong Q, Xu Y, Bao Y, Jia W. 2016. Osteocalcin improves nonalcoholic fatty liver disease in mice through activation of Nrf2 and inhibition of JNK. Endocrine 53:701–709. doi: 10.1007/s12020-016-0926-5. [DOI] [PubMed] [Google Scholar]
- 11.Ferron M, Hinoi E, Karsenty G, Ducy P. 2008. Osteocalcin differentially regulates beta cell and adipocyte gene expression and affects the development of metabolic diseases in wild-type mice. Proc Natl Acad Sci U S A 105:5266–5270. doi: 10.1073/pnas.0711119105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye R, Pi M, Cox JV, Nishimoto SK, Quarles LD. 2017. CRISPR/Cas9 targeting of GPRC6A suppresses prostate cancer tumorigenesis in a human xenograft model. J Exp Clin Cancer Res 36:90. doi: 10.1186/s13046-017-0561-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clevers H, Nusse R. 2012. Wnt/β-catenin signaling and disease. Cell 149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Nusse R, Clevers H. 2017. Wnt/β-catenin signaling, disease, and emerging therapeutic modalities. Cell 169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 15.MacDonald BT, Tamai K, He X. 2009. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sherwood V. 2015. WNT signaling: an emerging mediator of cancer cell metabolism? Mol Cell Biol 35:2–10. doi: 10.1128/MCB.00992-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schinner S. 2009. Wnt-signaling and the metabolic syndrome. Horm Metab Res 41:159–163. doi: 10.1055/s-0028-1119408. [DOI] [PubMed] [Google Scholar]
- 18.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A, Styrkarsdottir U, Magnusson KP, Walters GB, Palsdottir E, Jonsdottir T, Gudmundsdottir T, Gylfason A, Saemundsdottir J, Wilensky RL, Reilly MP, Rader DJ, Bagger Y, Christiansen C, Gudnason V, Sigurdsson G, Thorsteinsdottir U, Gulcher JR, Kong A, Stefansson K. 2006. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet 38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 19.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. 2007. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 20.Vaquero AR, Ferreira NE, Omae SV, Rodrigues MV, Teixeira SK, Krieger JE, Pereira AC. 2012. Using gene-network landscape to dissect genotype effects of TCF7L2 genetic variant on diabetes and cardiovascular risk. Physiol Genomics 44:903–914. doi: 10.1152/physiolgenomics.00030.2012. [DOI] [PubMed] [Google Scholar]
- 21.Wright WS, Longo KA, Dolinsky VW, Gerin I, Kang S, Bennett CN, Chiang SH, Prestwich TC, Gress C, Burant CF, Susulic VS, MacDougald OA. 2007. Wnt10b inhibits obesity in ob/ob and agouti mice. Diabetes 56:295–303. doi: 10.2337/db06-1339. [DOI] [PubMed] [Google Scholar]
- 22.Prestwich TC, Macdougald OA. 2007. Wnt/beta-catenin signaling in adipogenesis and metabolism. Curr Opin Cell Biol 19:612–617. doi: 10.1016/j.ceb.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manolopoulos KN, Karpe F, Frayn KN. 2010. Gluteofemoral body fat as a determinant of metabolic health. Int J Obes (Lond) 34:949–959. doi: 10.1038/ijo.2009.286. [DOI] [PubMed] [Google Scholar]
- 24.Mandviwala T, Khalid U, Deswal A. 2016. Obesity and cardiovascular disease: a risk factor or a risk marker? Curr Atheroscler Rep 18:21. doi: 10.1007/s11883-016-0575-4. [DOI] [PubMed] [Google Scholar]
- 25.Abranches MV, Oliveira FC, Conceiçao LL, Peluzio MD. 2015. Obesity and diabetes: the link between adipose tissue dysfunction and glucose homeostasis. Nutr Res Rev 28:121–132. doi: 10.1017/S0954422415000098. [DOI] [PubMed] [Google Scholar]
- 26.Rosen CJ, Bouxsein ML. 2006. Mechanisms of disease: is osteoporosis the obesity of bone? Nat Clin Pract Rheumatol 2:35–43. doi: 10.1038/ncprheum0070. [DOI] [PubMed] [Google Scholar]
- 27.Palermo A, Tuccinardi D, Defeudis G, Watanabe M, D'Onofrio L, Lauria Pantano A, Napoli N, Pozzilli P, Manfrini S. 2016. BMI and BMD: the potential interplay between obesity and bone fragility. Int J Environ Res Public Health 13:E544. doi: 10.3390/ijerph13060544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. 2008. PRDM16 controls a brown fat/skeletal muscle switch. Nature 454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enerback S. 2010. Human brown adipose tissue. Cell Metab 11:248–252. doi: 10.1016/j.cmet.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 30.Cohen P, Spiegelman BM. 2015. Brown and beige fat: molecular parts of a thermogenic machine. Diabetes 64:2346–2351. doi: 10.2337/db15-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cypess A, Sanaz Lehman M, Williams G, Tal I, Rodman D, Goldfine A, Kuo F, Palmer E, Tseng Y, Doria A, Kolodny G, Kahn C. 2009. Identification and importance of brown adipose tissue in adult humans. N Engl J Med 360:1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Virtanen K, Lidell M, Orava J, Heglind M, Westergren R, Niemi T, Taitonen M, Laine J, Savisto N, Enerback S, Nuutila P. 2009. Functional brown adipose tissue in healthy adults. N Engl J Med 360:1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 33.Kontani Y, Wang Y, Kimura K, Inokuma KI, Saito M, Suzuki-Miura T, Wang Z, Sato Y, Mori N, Yamashita H. 2005. UCP1 deficiency increases susceptibility to diet-induced obesity with age. Aging Cell 4:147–155. doi: 10.1111/j.1474-9726.2005.00157.x. [DOI] [PubMed] [Google Scholar]
- 34.Keipert S, Jastroch M. 2014. Brite/beige fat and UCP1—is it thermogenesis? Biochim Biophys Acta 1837:1075–1082. doi: 10.1016/j.bbabio.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Long JZ, Svensson KJ, Tsai L, Zeng X, Roh HC, Kong X, Rao RR, Lou J, Lokurkar I, Baur W, Castellot JJ, Rosen ED, Spiegelman BM. 2014. A smooth muscle-like origin for beige adipocytes. Cell Metab 19:810–820. doi: 10.1016/j.cmet.2014.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, Khandekar M, Virtanen KA, Nuutila P, Schaart G, Huang K, Tu H, van Marken Lichtenbelt WD, Hoeks J, Enerback S, Schrauwen P, Spiegelman BM. 2012. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150:366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen P, Levy JD, Zhang Y, Frontini A, Kolodin DP, Svensson KJ, Lo JC, Zeng X, Ye L, Khandekar MJ, Wu J, Gunawardana SC, Banks AS, Camporez JP, Jurczak MJ, Kajimura S, Piston DW, Mathis D, Cinti S, Shulman GI, Seale P, Spiegelman BM. 2014. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 156:304–316. doi: 10.1016/j.cell.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Penicaud L, Casteilla L. 1992. Occurrence of brown adipocytes in rat white adipose tissue: molecular and morphological characterization. J Cell Sci 103:931–942. [DOI] [PubMed] [Google Scholar]
- 39.Fisher FM, Kleiner S, Douris N, Fox EC, Mepani RJ, Verdeguer F, Wu J, Kharitonenkov A, Flier JS, Maratos-Flier E, Spiegelman BM. 2012. FGF21 regulates PGC-1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev 26:271–281. doi: 10.1101/gad.177857.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bostrom P1, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, Rasbach KA, Bostrom EA, Choi JH, Long JZ, Kajimura S, Zingaretti MC, Vind BF, Tu H, Cinti S, Hojlund K, Gygi SP, Spiegelman BM. 2012. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nature 481:463–468. doi: 10.1038/nature10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohno H, Shinoda K, Spiegelman BM, Kajimura S. 2012. PPARγ agonists induce a white-to-brown fat conversion through stabilization of PRDM16 protein. Cell Metab 15:395–404. doi: 10.1016/j.cmet.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, Accili D. 2012. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of PPARγ. Cell 150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan D, Fujimoto M, Lopes A, Wang YX. 2009. Twist-1 is a PPARδ-inducible, negative-feedback regulator of PGC-1α in brown fat metabolism. Cell 137:73–86. doi: 10.1016/j.cell.2009.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mera P, Laue K, Ferron M, Confavreux C, Wei J, Galan-Diez M, Lacampagne A, Mitchell SJ, Mattison JA, Chen Y, Bacchetta J, Szulc P, Kitsis RN, de Cabo R, Friedman RA, Torsitano C, McGraw TE, Puchowicz M, Kurland I, Karsenty G. 2016. Osteocalcin signaling in myofibers is necessary and sufficient for optimum adaptation to exercise. Cell Metab 23:1078–1092. doi: 10.1016/j.cmet.2016.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee S, Suzuki T, Izawa H, Satoh A. 2016. The influence of the type of continuous exercise stress applied during growth periods on bone metabolism and osteogenesis. J Bone Metab 23:157–164. doi: 10.11005/jbm.2016.23.3.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leung JY, Kolligs FT, Wu R, Zhai Y, Kuick R, Hanash S, Cho KR, Fearon ER. 2002. Activation of AXIN2 expression by beta-catenin-T cell factor. J Biol Chem 277:21657–21665. doi: 10.1074/jbc.M200139200. [DOI] [PubMed] [Google Scholar]
- 47.Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. 2002. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22:1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Campbell JE, Ussher JR, Mulvihill EE, Kolic J, Baggio LL, Cao X, Liu Y, Lamont BJ, Morii T, Streutker CJ, Tamarina N, Philipson LH, Wrana JL, MacDonald PE, Drucker DJ. 2016. TCF1 links GIPR signaling to the control of beta cell function and survival. Nat Med 22:84–90. doi: 10.1038/nm.3997. [DOI] [PubMed] [Google Scholar]
- 49.Arce L, Yokoyama NN, Waterman ML. 2006. Diversity of LEF/TCF action in development and disease. Oncogene 25:7492–7504. doi: 10.1038/sj.onc.1210056. [DOI] [PubMed] [Google Scholar]
- 50.Cadigan K., Waterman M. 2012. TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 4:a007906. doi: 10.1101/cshperspect.a007906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinheiro I, Margueron R, Shukeir N, Eisold M, Fritzsch C, Richter FM, Mittler G, Genoud C, Goyama S, Kurokawa M, Son J, Reinberg D, Lachner M, Jenuwein T. 2012. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 150:948–960. doi: 10.1016/j.cell.2012.06.048. [DOI] [PubMed] [Google Scholar]
- 52.Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. 2009. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeng X, Jedrychowski MP, Chen Y, Serag S, Lavery GG, Gygi SP, Spiegelman BM. 2016. Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation. Genes Dev 30:1822–1836. doi: 10.1101/gad.285312.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee P, Brychta RJ, Collins MT, Linderman J, Smith S, Herscovitch P, Millo C, Chen KY, Celi FS. 2013. Cold-activated brown adipose tissue is an independent predictor of higher bone mineral density in women. Osteoporos Int 24:1513–1518. doi: 10.1007/s00198-012-2110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ponrartana S, Aggabao PC, Hu HH, Aldrovandi GM, Wren TA, Gilsanz V. 2012. Brown adipose tissue and its relationship to bone structure in pediatric patients. J Clin Endocrinol Metab 97:2693–2698. doi: 10.1210/jc.2012-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mizokami A, Yasutake Y, Gao J, Matsuda M, Takahashi I, Takeuchi H, Hirata M. 2013. Osteocalcin induces release of glucagon-like peptide-1 and thereby stimulates insulin secretion in mice. PLoS One 8:e57375. doi: 10.1371/journal.pone.0057375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kypta RM, Waxman J. 2012. Wnt/β-catenin signaling in prostate cancer. Nat Rev Urol 9:418–428. doi: 10.1038/nrurol.2012.116. [DOI] [PubMed] [Google Scholar]
- 58.Tseng Y, Kriauciunas K, Kokkotou E, Kahn C. 2004. Differential roles of insulin receptor substrates in brown adipocyte differentiation. Mol Cell Biol 24:1918–1929. doi: 10.1128/MCB.24.5.1918-1929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koteja P, Garland T Jr, Sax JK, Swallow JG, Carter PA. 1999. Behaviour of house mice artificially selected for high levels of voluntary wheel running. Anim Behav 58:1307–1318. doi: 10.1006/anbe.1999.1270. [DOI] [PubMed] [Google Scholar]
- 60.Gan XQ, Wang JY, Xi Y, Wu ZL, Li YP, Li L. 2008. Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J Cell Biol 180:1087–1100. doi: 10.1083/jcb.200710050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hashimoto Y, Matsuzaki E, Higashi K, Takahashi-Yanaga F, Takano A, Hirata M, Nishimura F. 2015. Sphingosine-1-phosphate inhibits differentiation of C3H10T1/2 cells into adipocyte. Mol Cell Biochem 401:39–47. doi: 10.1007/s11010-014-2290-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.