Abstract
The aim of this dataset is to identify and collect compounds that are known for being detectable by a living cell, through the action of a genetically encoded biosensor and is centred on bacterial transcription factors. Such a dataset should open the possibility to consider a wide range of applications in synthetic biology. The reader will find in this dataset the name of the compounds, their InChI (molecular structure), the publication where the detection was reported, the organism in which this was detected or engineered, the type of detection and experiment that was performed as well as the name of the biosensor. A comment field is also provided that explains why the compound was included in the dataset, based on quotes from the reference publication or the database it was extracted from. Manual curation of ACS Synthetic Biology abstracts (Volumes 1 to 6 and Volume 7 issue 1) was performed as well as extraction from the following databases: Bionemo v6.0 (Carbajosa et al., 2009) [1], RegTransbase r20120406 (Cipriano et al., 2013) [2], RegulonDB v9.0 (Gama-Castro et al., 2016) [3], RegPrecise v4.0 (Novichkov et al., 2013) [4] and Sigmol v20180122 (Rajput et al., 2016) [5].
Specifications Table
| Subject area | Biology |
| More specific subject area | Synthetic biology |
| Type of data | Table |
| How data was acquired | Database extraction from Bionemo v6.0, RegTransbase r20120406, RegulonDB v9.0, RegPrecise v4.0 and Sigmol v20180122 as well as manual curation ACS Synthetic Biology abstracts (Volumes 1 to 6 and Volume 7 issue 1) |
| Data format | Analysed |
| Experimental factors | Not applicable |
| Experimental features | Not applicable |
| Data source location | https://github.com/brsynth/detectable_metabolites |
| Data accessibility | Data is with this article and on GitHub athttps://github.com/brsynth/detectable_metabolites |
Value of the data
-
•
This dataset provides a basis for the development of new biosensing circuits for synthetic biology and metabolic engineering applications, e.g. the design of whole-cell biosensor, high-throughput screening experiments, dynamic regulation of metabolic pathways, transcription factor engineering or creation of sensing-enabling pathways.
-
•
This dataset provides a unique source of a broad number of compounds that can be detected and acted upon by a cell, increasing the possibility of orthogonal circuit design from the few usual compounds used in those applications.
-
•
The manually curated section provides information on where the biosensor has been first reported and successfully used, enabling the reader to select trustworthy information for his application of choice.
-
•
Detectable compounds can be searched by both by name and chemical similarity.
-
•
This dataset is an update of [10.6084/m9.figshare.3144715.v1].
1. Data
The aim of this dataset is to identify and collect compounds that are known for being detectable by a living cell, through the action of a genetically encoded biosensor and is centred on bacterial transcription factors. The dataset should allow the synthetic biology community to consider a wide range of applications. The reader will find in this dataset the name of the compounds, their InChI (molecular structure), the publication where the detection was reported, the organism in which this was detected or engineered, the type of detection and experiment that was performed as well as the name of the biosensor. A comment field is also provided that explains why the compound was included in the dataset, based on quotes from the reference publication or the database it was extracted from. Manual curation of ACS Synthetic Biology abstracts (Volumes 1 to 6 and Volume 7 issue 1) was performed as well as extraction from the following databases: Bionemo v6.0 [1], RegTransbase r20120406 [2], RegulonDB v9.0 [3], RegPrecise v4.0 [4] and Sigmol v20180122 [5].
This dataset is available online on GitHub to allow for further updates as well as community contributions.
2. Experimental design, materials and methods
-
•
Manual curation of ACS Synthetic Biology (Volume 1–6 and Volume 7 issue 1):
All abstracts of ACS Synthetic Biology (Volume 1–6 and Volume 7 issue 1) were read and information relevant to this dataset was extracted from those abstracts. The aim of this manual curation was to establish a list of detectable compounds whose detection method was already successfully implemented in a synthetic circuit, providing a good basis for further implementation for synthetic biologists.
-
•
Bionemo v6.0 [1]:
The SQL request used to create this dataset is:
SELECT DISTINCT substrate.id_substrate, minesota_code, name FROM substrate
INNER JOIN complex_substrate ON complex_substrate.id_substrate=substrate.id_substrate
INNER JOIN complex ON complex.id_complex=complex_substrate.id_complex
WHERE activity='REG';
-
•
RegTransbase r20120406 [2]:
The SQL request used to create this dataset is:
SELECT DISTINCT a.pmid, e.name, r.name
FROM regulator2effectors AS re
INNER JOIN exp2effectors AS ee ON ee.effector_guid=re.effector_guid
INNER JOIN dict_effectors AS e ON e.effector_guid=ee.effector_guid
INNER JOIN regulators AS r ON r.regulator_guid=re.regulator_guid
INNER JOIN articles AS a ON a.art_guid=ee.art_guid
ORDER BY e.name;
RegTransbase was not maintained anymore at the time of writing of this manuscript.
-
•
RegulonDB v9.0 [3]:
The SQL request used to create this dataset is:
SELECT c.conformation_id, c.final_state, e.effector_id, e.effector_name, tf.transcription_factor_id, tf.transcription_factor_name, p.reference_id, xdb.external_db_name
FROM effector AS e
INNER JOIN conformation_effector_link AS mm_ce ON mm_ce.effector_id=e.effector_id
LEFT JOIN conformation AS c ON c.conformation_id=mm_ce.conformation_id
LEFT JOIN transcription_factor AS tf ON tf.transcription_factor_id=c.transcription_factor_id
LEFT JOIN object_ev_method_pub_link AS x ON x.object_id=c.conformation_id OR x.object_id=tf.transcription_factor_id OR x.object_id=e.effector_id
LEFT JOIN publication AS p ON p.publication_id=x.publication_id
LEFT JOIN external_db AS xdb ON xdb.external_db_id=p.external_db_id
WHERE c.interaction_type IS Null OR c.interaction_type!='Covalent';
-
•
RegPrecise v4.0 [4]:
The RegPrecise website was accessed (version v4.0) and all relevant data was extracted from the effector pages of the website.
-
•
Sigmol v20170216 [5]:
Sigmol was accessed on 16/02/2017 and all effector data was retrieved from the unique Quorum Sensing Signaling Molecule page. In the “detected by” column, we provide the class of signaling compounds the compound belongs to. The comment field reads ‘Extracted from Sigmol v20170216 – Uniq_QSSM_“number”’.
2.1. Data overview
In Table 1 are presented some characteristics of each data source: number of compounds without a structure from this source, total number of compounds with a structure from this source and number of compounds with a structure found only in this source. The last column in particular shows that around half the compounds are found in more than one data source.
Table 1.
Contribution of each data source.
| Source | Compounds without structure | Compounds with structure | Unique compounds with structure |
|---|---|---|---|
| RegPrecise | 136 | 418 | 73 |
| BioNemo | 5 | 499 | 8 |
| RegTransBase | 683 | 2057 | 63 |
| RegulonDB | 12 | 245 | 23 |
| Sigmol | 2 | 175 | 135 |
| ACS Synthetic Biology | 44 | 287 | 73 |
| All sources | 882 | 3681 | 729 |
The first column contains the data source, the second column the number of compounds found without a structure in that source, the third column the number of compounds with a structure (InChI) and the last column the number of compounds with a structure found only in that source.
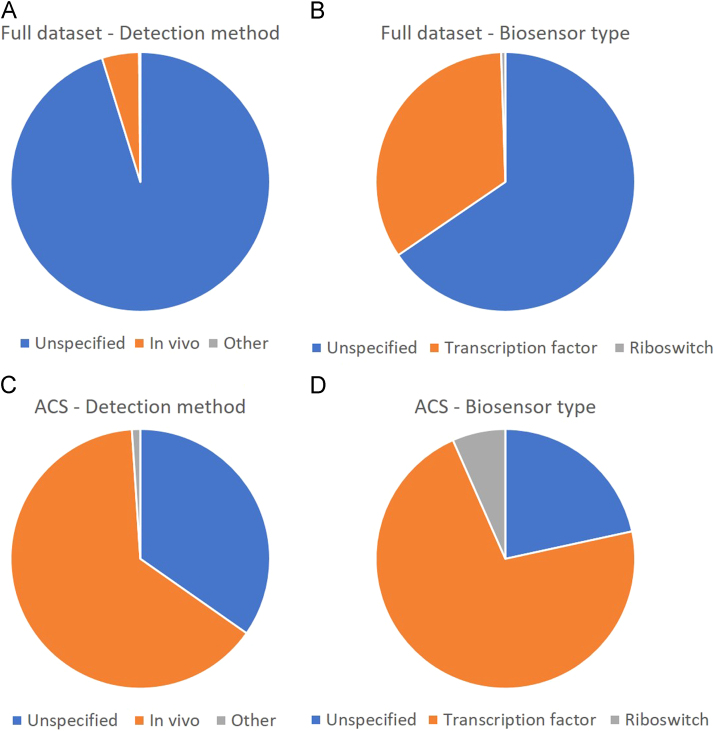
Fig. 1 shows the repartition of the type of experiment (in vivo, unspecified or other), as well as the repartition of Biosensor type (Transcription factor, riboswitch or unspecified) in the full dataset and the manually curated dataset from ACS Synthetic Biology.
Fig. 1.
Type of experiment and biosensor type in the full dataset and the manually curated dataset. A: Full dataset – detection method. B: Full dataset – biosensor type. C: ACS dataset – detection method. D: ACS dataset – biosensor type. A and C: other in detection method corresponds to in silico, in vivo and cell-free detections. C and D: ACS dataset is the dataset obtained from manual curation of ACS Synthetic Biology with compounds that have available structures.
Acknowledgements
This work was supported by the French National Research Agency (ANR-15-CE21-0008), the Biotechnology and Biological Sciences Research Council, Centre for Synthetic Biology of Fine and Speciality Chemicals (BB/M017702/1); Synthetic Biology Applications for Protective Materials (EP/N025504/1. M.K is supported by DGA (French Ministry of Defense) and Ecole Polytechnique. BD was supported by Structure et Dynamique des Systemes Vivants Doctoral School, Universite Paris Saclay.
Acknowledgments
Competing financial interests
None declared.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2018.02.061.
Transparency data associated with this article can be found in the online version at 10.1016/j.dib.2018.02.061.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Carbajosa G., Trigo A., Valencia A., Cases I. Bionemo: molecular information on biodegradation metabolism. Nucleic Acids Res. 2009;37:D598–D602. doi: 10.1093/nar/gkn864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cipriano M.J., Novichkov P.N., Kazakov A.E., Rodionov D.A., Arkin A.P., Gelfand M.S., Dubchak I. RegTransBase – a database of regulatory sequences and interactions based on literature: a resource for investigating transcriptional regulation in prokaryotes. BMC Genom. 2013;14:213. doi: 10.1186/1471-2164-14-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muñiz-Rascado L., García-Sotelo J.S., Alquicira-Hernández K., Martínez-Flores I., Pannier L., Castro-Mondragón J.A., Medina-Rivera A., Solano-Lira H., Bonavides-Martínez C., Pérez-Rueda E., Alquicira-Hernández S., Porrón-Sotelo L., López-Fuentes A., Hernández-Koutoucheva A., Del Moral-Chávez V., Rinaldi F., Collado-Vides J. RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2016;44:D133–D143. doi: 10.1093/nar/gkv1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novichkov P.S., Kazakov A.E., Ravcheev D.A., Leyn S.A., Kovaleva G.Y., Sutormin R.A., Kazanov M.D., Riehl W., Arkin A.P., Dubchak I., Rodionov D.A. RegPrecise 3.0 – a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genom. 2013;14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rajput A., Kaur K., Kumar M. SigMol: repertoire of quorum sensing signaling molecules in prokaryotes. Nucleic Acids Res. 2016;44:D634–D639. doi: 10.1093/nar/gkv1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material