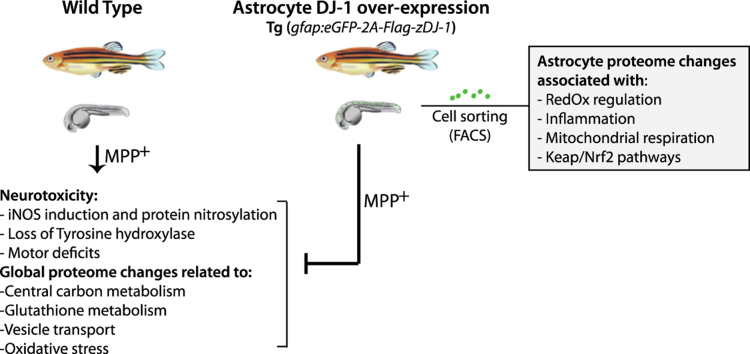
Abstract
DJ-1, a Parkinson's disease-associated protein, is strongly up-regulated in reactive astrocytes in Parkinson's disease. This is proposed to represent a neuronal protective response, although the mechanism has not yet been identified. We have generated a transgenic zebrafish line with increased astroglial DJ-1 expression driven by regulatory elements from the zebrafish GFAP gene. Larvae from this transgenic line are protected from oxidative stress-induced injuries as caused by MPP+, a mitochondrial complex I inhibitor shown to induce dopaminergic cells death. In a global label-free proteomics analysis of wild type and transgenic larvae exposed to MPP+, 3418 proteins were identified, in which 366 proteins were differentially regulated. In particular, we identified enzymes belonging to primary metabolism to be among proteins affected by MPP+ in wild type animals, but not affected in the transgenic line. Moreover, by performing protein profiling on isolated astrocytes we showed that an increase in astrocytic DJ-1 expression up-regulated a large group of proteins associated with redox regulation, inflammation and mitochondrial respiration. The majority of these proteins have also been shown to be regulated by Nrf2. These findings provide a mechanistic insight into the protective role of astroglial up-regulation of DJ-1 and show that our transgenic zebrafish line with astrocytic DJ-1 over-expression can serve as a useful animal model to understand astrocyte-regulated neuroprotection associated with oxidative stress-related neurodegenerative disease.
Keywords: DJ-1, Astrocyte, Parkinson's disease, Oxidative stress, Cell survival
Graphical abstract
Highlights
-
•
Increases astrocytic proteins linked to oxidative stress regulation & inflammation.
-
•
Protects from MPP+-induced changes in central metabolism and protein nitrosylation.
-
•
Protects from MPP+-induced tyrosine hydroxylase loss and motor deficits.
1. Introduction
DJ-1 is a multifunctional redox-sensitive protein that mediates neuroprotection and cell stress protection in general, possibly by regulating anti-oxidant and anti-apoptotic gene expression [1], [2], in addition to pro-survival signalling pathways [3]. Loss of function mutations in the PARK7 gene that encodes DJ-1 cause an early onset familial form of Parkinsonism. DJ-1 is found to be oxidatively modified in both neurons and astrocytes from post-mortem analysis of tissue from Parkinson's disease (PD) patients [4], [5]. DJ-1-deficient mice, on the other hand, do not show neural degeneration [6], but have enhanced sensitivity of dopaminergic neurons to oxidative stress [7]. Both fibroblasts differentiated into dopaminergic neurons and lymphocytes obtained from DJ-1 deficient PD patients show altered mitochondrial morphology and function in addition to increased mitochondrial oxidative stress, which could contribute to the increased sensitivity to oxidative stress-induced cell death [8], [9].
Astroglial function was long considered to be limited to metabolic and structural support to neurons, but astrocytes, which outnumber neurons in the brain, are now recognized to have a key role in neuroprotection including controlling redox homeostasis [10]. Despite neurons being highly dependent on oxidative metabolism they display limited defense mechanisms against oxidative stress compared to astrocytes. The adaptive response of astrocytes to oxidative stress therefore seems indispensable in order to maintain redox homeostasis in the brain [11]. In brain tissue obtained from sporadic PD patients, DJ-1 is strongly up-regulated in reactive astrocytes, but not in neurons [12]. Increased astrocytic DJ-1 is also found adjacent to infarcted brain regions after stroke [13]. Moreover, in astrocyte primary cultures DJ-1 regulates inflammatory responses [14], and in neuron-astrocyte co-cultures astrocytic DJ-1 expression protects neurons from mitochondrial complex I-induced oxidative stress [15]. Astrocytic DJ-1 expression therefore seems to have a major role in protecting neurons from oxidative damage and may produce neuron-protective factors, although these agents have not yet been identified [16].
Zebrafish is an increasingly useful model of human neurodegenerative diseases including PD [17]. Loss of dopaminergic neurons and motor deficits, as observed in PD, can be replicated at the larvae stage of zebrafish exposed to oxidative stressors [18], [19], [20], [21]. We have used zebrafish to generate an in vivo model with increased astroglial DJ-1 expression. Regulatory elements from the zebrafish glial fibrillary acidic protein (GFAP) gene [22] were used to drive the astrocyte specific over-expression of DJ-1 in the transgenic line. In this study we aimed to 1) investigate whether astrocytic over-expression of DJ-1 would protect against MPP+, a neurotoxin causing PD-related symptoms and, if so, 2) identify overall proteomic changes induced by MPP+ and protected by increased astrocytic DJ-1 expression and 3) characterize the astrocytic protein profile underlying the neuroprotective action.
2. Material and methods
2.1. Animal maintenance and exposure to oxidative stressors
Animals were generated, housed and experiments carried out at the zebrafish facility at the Department of Molecular Biology, University of Bergen. The facility is run in agreement with European Convention for Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes. Adult zebrafish were maintained at 26–28 °C on a 14/10 light/dark cycle and fed twice daily. Embryos were obtained by natural mating and raised in E3 buffer (5 mM NaCl, 0.17 mM KCl, 0.33 mM MgSO4) at 28 °C.
Hatched or dechorinated embryos were exposed to 500–600 µM 1-methyl-4-phenylpyridinium iodide (MPP+ iodide) (Sigma-Aldrich: D048) or vehicle from 24 or 48 h post fertilization (hpf).
2.2. DNA constructs and transgenesis
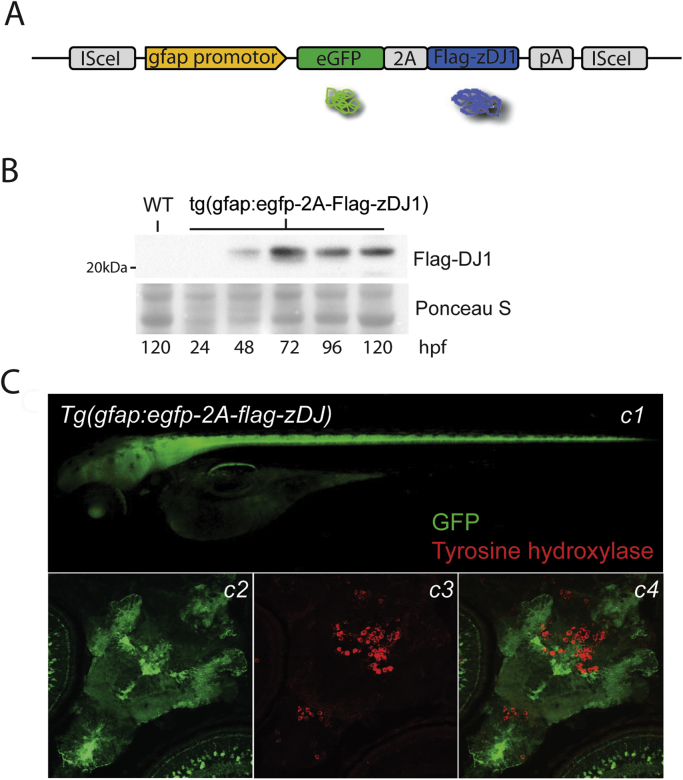
Complimentary oligonucleotides with sequences for viral 2A [23] and Flag-tag with BsrGI/MfeI restriction sites were heated to 95 °C and left to hybridize overnight. Zebrafish DJ-1 [24] was amplified by PCR using primers which included MfeI/KpnI sites. Sequences for viral 2A, Flag-tag, and zebrafish DJ-1 were then ligated into the BsrGI/KpnI sites of pBS-I-Sce1-GFP [25]. The GFAP regulatory elements (intron 1/exon1) were kindly provided by Pamela A. Raymond [22] and inserted upstream from GFP, giving the final construct pBS-I-Sce1-gfap:GFP-2A-Flag-zDJ-1 as shown in Fig. 1A. Restriction digest was prepared on ice: plasmid (0.6 µg), injection dye (0.5% phenol red, 240 mM KCl, 40 mM HEPES pH 7.4) 1 µl, 10× I-Sce1 buffer 0.5 µl, I-SceI (New England Biolabs: R0694S) 1 µl, ddH2O to total 10 µl. Single cell embryos were microinjected with 0.5 nl of digestion mix. Embryos expressing GFP were selected at 48 hpf, raised to adulthood and bred to identify Tg(gfap:egfp-2A-flag-zDJ-1) (F0 chimeras). Founder fish were outcrossed with wild type and progeny embryos (F1) collected. Stable Tg(gfap:egfp-2A-flag-zDJ-1) lines were expanded from single F1 founders. Transgenic lines established from three different F1 founder fish were used in this study. GFP expression in transgenic animals was examined using a fluorescent Zeiss SteREO Lumar microscope.
Fig. 1.
Generation of zebrafish with astroglial over-expression of DJ-1. A, Transgenic zebrafish expressing Flag-tagged DJ-1, Tg(gfap:egfp-2A-flag-zDJ-1), were generated using ISce1-transgenesis. Flag-tagged DJ-1 expression in astrocytes was controlled by Glial Fibrillary Acidic Protein (GFAP) promotor elements. A viral 2A peptide allows expression of GFP and Flag-DJ1 as uncoupled proteins from the same transgene. B, Western blot shows expression of Flag-DJ1 from total protein extracts of larvae 24–120 hpf. Ponceau S staining is used as loading control. C, GFP expression localized astroglia, neural retina and spinal cord was observed from 48 hpf. c1–3, Immunoblots of brain sections (4 dpf) showing astroglial GFP and neuronal tyrosine hydroxylase expression. Tyrosine hydroxylase was used as a marker for dopaminergic cells.
For FACS sorting Tg(gfap:Gal4FF/UAS-mCherry) was used as a control. The line was generated by crossing Tg(gfap:Gal4FF) with Tg(UAS:mCherry). Tg(gfap:Gal4FF) was establish by inserting the GFAP regulatory elements and Gal4FFpoly into pBS-I-SceI followed by transgenesis as explained above. To establish Tg(UAS:mCherry) mCherry was inserted into pT2–5UASMCS (a gift from Dr. Koichi Kawakami, NIG, Japan) and the Tol2 method for transgenesis was used as described in [26].
2.3. Larval motility assay
Spontaneous movement of 5 dpf larvae was recorded for 1 h in 48-well dishes in E3 medium after removal of exposure media. Five dpf larvae were used due to optimal recording, as pigmentation of larvae was required to obtain contrast. Recording was done at 10 fps with a USB 2.0 uEye LE camera (UI-1240LE, IDS Imaging Development Systems GmbH, Germany) fitted with a 50 mm Nikon lens using the IDS software suite (http://en.ids-imaging.com/ids-software-suite.html). Each movie (1024 × 1280 pixel frames) was processed post-acquisition with a custom-written MATLAB tracking software modified from A. Pérez-Escudero (http://www.multiwelltracker.es/). The software identifies the centroid of the larva in each frame and computes its position in the entire movie. Distance travelled by the larval centroid was used as an index of motility.
2.4. Protein extraction
100–200 deyolked embryos (24–48 hpf) or larvae (72–120 hpf) were washed twice in PBS and disrupted by sonication (4 × 5 s) in homogenization buffer (10 mM K2HPO4, 10 mM KH2PO4, 1 mM EDTA, 0.6% CHAPS, 0.2 mM Na3VO4, 50 mM NaF added protease cocktail (Roche Diagnostics GmbH: 11836153001)). Five µl homogenization buffer was added per embryo. Samples were pelleted at 16,000 g for 15 min and supernatant was either used directly or stored at −80 °C.
2.5. Detection of protein nitrosylation and Western blotting
Protein nitrosylation induced by MPP+ was measured using Pierce S-Nitrosylation Western blot kit (Thermo Scientific: 90105) using the supplied protocol. 30–40 larvae were homogenized in HENS buffer (100 mM HEPES, pH 7.8; 1 mM EDTA; 0.1 mM Neocuproine; 1% SDS) and 100 µg protein was used for each condition. Free sulfhydryls were first blocked with methyl methanethiosulfonate followed by selective reduction of S-nitrosocysteines with ascorbate and labeling with iodoTMTzero reagent. Derivatized nitrosylated proteins were detected using Western blotting and anti-TMT.
Samples for S-nitrosylation detection and Western blotting were separated by SDS-PAGE and transferred to PVDF membranes using 14 V over night at 4 °C. The membranes were blocked in 1% BSA for 1 h at room temperature. Membranes were incubated with primary antibodies; anti-Flag (Sigma-Alderich: F3165), anti-TMT (Thermo Scientific: 90105), anti-iNOS (BD Transduction Laboratories: 610431), anti-DJ-1(Novous Biologicals: NB 300-270), anti-Tyrosine hydroxylase (Millipore: MAB318) anti-Glutathione S-transferase m (Developmental Studies Hybridoma Bank (DSHB):CPTC-GSTMu1-1), or anti-Malate dehydrogenase (Sigma-Aldrich: AV48286) followed by the appropriate secondary antibody, both for 1 h at room temperature. Washed membranes were developed using Super Signal West Pico Plus chemiluminescent substrate (Thermo scientific: 34577) and BioRad GelDoc XRS+. Blots were quantitated using Image Lab version 5.1 (BioRad).
2.6. Protein profiling of zebrafish larvae
Larvae from 10 individual pairs of wild type or Tg(gfap:egfp-2A-flag-zDJ-1) were combined and further processed for proteome profiling. Proteins from larvae, 4 days post fertilization (dpf), were extracted as explained previously (see protein extraction). An aliquot was taken for Western blotting and the remaining lysate were snap frozen for later LC-MS/MS analysis.
Sample reduction, alkylation and tryptic digestion for mass spectrometry analysis were performed according to Martens et al. [27]. 50 µl of each sample, corresponding to 50 µg of protein, was washed with in RapiGest at 0.05% (w/v) (Waters Corporation, USA) in 50 mM ammonium bicarbonate in order to hydrolyze the proteins present. The samples were reduced using 100 mM dithitreitol at 60 °C for 15 min and alkylated for 45 min at 25 °C in the presence of 200 mM iodoacetamide. Proteolytic digestion proceeded at a 1:50 (w/w) ratio with an over night incubation at 37 °C by the addition of sequencing grade trypsin (Promega, USA). Following this TFA was added to the samples to hydrolyze the RapiGest and the solutions incubated at 37 °C for 20 min before being vortexed and centrifuged. To be able to estimate the protein amount during analysis the samples were diluted 1:1 with 10 fmol/µl of a tryptic digest standard of yeast alcohol dehydrogenase (Waters Corporation, USA) before analysis.
Peptides were separated by a nanoACQUITY UPLC system (Waters Corporation, USA) equipped with a 25 cm × 75 µm analytical RP column (Waters Corporation, USA) and a trap column (Symmetry C18 5 µm, 2 cm × 180, Waters Corporation, USA). A reversed phase gradient was employed to separate peptides using 5–40% acetonitrile over 90 min, at a flow rate of 300 nL/min and a constant temperature at 35 °C. Mobile phases A and B were water containing 0.1% (v/v) formic acid and acetonitrile containing 0.1% (v/v) formic acid respectively.
The peptides were eluted directly into a SYNAPT G2-S HDMS mass spectrometer (Waters, Manchester, UK), operated in data independent manner coupled with ion mobility (HDMSE) [28]. The mass spectrometer was operated in positive ESI resolution mode with resolution of > 25,000 FWHM. The mass spectrometer was programmed to step between low energy (4 eV) and elevated (14–40 eV) collision energies on the Triwave collision cell, using a scan time of 0.9 s per function over 50–2000 m/z. All samples were analyzed in triplicate.
Protein identification and quantification information were obtained using ProteinLynxGlobal Server v2.5.3 (PLGS) and Progenesis QI by searching the combined UniProKB Zebrafish database (downloaded June 9th, 2017). The matching process required more than three fragment ions per peptide, three fragments per protein, and more than one peptide per protein. Peptides with a mass error greater than 10 ppm were disguarded. One missed trypsin cleavages was accepted, and carbamidomethylation of cysteins and oxidation of methionine were used as fixed and variable modifications respectively. The maximum protein false discovery rate was set to 4% (corresponding to a peptide FDR less than 1%). Only proteins with two or more unique peptides were used for quantitation. The significant threshold was p ≤ 0.05, and only proteins with a max fold value ≥ 2 were regarded as regulated.
2.7. Dissociation of larvae and sorting of astrocytes
Larvae were put on ice and washed twice in Ca2+-free Ringer buffer before incubated in 10 ml 0.25% Trypsin-EDTA (Gibco 15400-054) in PBS for 30 min at 30 °C. Samples were triturated by pipetting until cells were dissociated before added 2 ml fetal calf serum and 2 ml 20 mM CaCl2. Samples were then filtered (40 µm VWR, cat. No 734-0002) before pelleting the cells at 300 g for 30 min at 4 °C. Pellet was washed in PBS and resuspended in 1 ml PBS. Cell sorting was performed using a BD FACS Aria SORP paired with PC-based FACSDiVa analysis software. Both loaded and collected samples were kept at 4̊C during the sorting procedure. Viable cells were gated using Forward Scattered (FSC) and Side Scattered (SSC) light. Further gating for singlets was achieved by correlating FSC signal width and area. Fluorescent cell populations were gated by using either a sample culture from WT larvae or a culture from a line not expressing the sorting-based fluorophore.
2.8. Sample preparation and mass spectrometry of isolated astrocytes
FACS sorted cells (approx. 600,000 sampled from three independent experiments per condition) were disrupted in homogenization buffer and proteins were precipitated in acetone and resuspended in 20 µl homogenization buffer. SDS was added to a final concentration of 4% and heated at 95 °C for 7 min. The SDS was further diluted to 0.5% with the Filter Aided sample preparation (FASP)-urea buffer (8 M urea in 0.1 M Tris-HCl pH8.5). The protein extract were digested using the FASP method [29] followed by reduction, alkylation and peptide up-concentration as described in Frøyset et al. [30].
2.9. Label-free mass spectrometry analysis of isolated astrocytes
Tryptic peptides were dissolved in 2% ACN/ 0.1% TFA to a final concentration of 0.8 µg/µl. In total 3 µg as tryptic peptides were analyzed on a Q-Exactive HF (Thermo Scientific) connected to a Dionex Ultimate NCR-3500RS LC system. The sample was trapped on the pre-column (Dionex, Acclaim PepMap 100, 2 cm × 75 µm i.d, 3 µm C18 beads) in loading buffer (0.1% TFA) at a flowrate of 5 µl/min for 5 min before separation by revers phase chromatography (PepMap RSLC, 50 cm × 75 µm i.d. EASY-spray column, packed with 2 µm C18 beads) at a flow of 200 nL/min. Solvent A and B were 0.1% FA (vol/vol) in water and 100% ACN respectively. The gradient composition was 5% B during trapping (5 min) followed by 5–8% B over 0.5 min, 8–24% B for the next 109.5 min, 24–35% B over 25 min, and 35–90% B over 15 min. Elution of very hydrophobic peptides and conditioning of the column were performed during 15 min isocratic elution with 90% B and 20 min isocratic conditioning with 5% B. The total length of the LC run was 190 min.
MS spectra were acquired as described in Frøyset et al. [30] except that threshold intensity for eluting peptides was increased to 5e4. Three technical repeats were performed.
The raw files were searched in MaxQuant (version 1.5.3.28) against Danio rerio, a combined UniProtKB database (downloaded May 13th, 2016, 58,693 entries). The search parameters used for MaxQuant and criteria used for further analyzing the data in Perseus (1.5.2.6) are described in Frøyset et al. [30]. Three or more valid values and two or more unique peptides were required for further analysis.
3. Results
3.1. Generation of zebrafish with astroglial over-expression of DJ-1
A glial fibrillary acidic protein (GFAP) regulatory element was used to generate a transgenic line with astrocyte-specific expression of Flag-tagged zebrafish DJ-1 (Fig. 1A) [22]. Flag-DJ-1 was separated from GFP by viral 2A peptide sequence [23], which allows stoichiometric translation of unfused protein products (Fig. 1A and B).
Flag-tagged DJ-1 expression was visualized by Western blotting (Fig. 1B) and GFP expression by fluorescent microscopy from 48 hpf (Fig. 1C). GFAP was primarily expressed in astroglia of the central nervous system (Fig. 1Cc1) and distinct from dopaminergic neuronal cells (Fig. 1Cc2–4).
3.2. Astroglial over-expression of DJ-1 protects from PD-related features as induced by MPP+
The mitochondrial complex I inhibitor MPP+ is highly toxic to dopaminergic cells and frequently used as a model to replicate features of Parkinson's disease (PD) including: mitochondria dysfunction, oxidative damage, inflammatory response, degeneration of dopaminergic neurons and motor abnormalities [31], [32]. It should be noted that MPP+ also enters astrocytes even though they seem more resistant to the toxicant [32], [33]. Moreover, the pro-inflammatory neurotoxic mediator inducible nitric oxide (iNOS) has been shown to increase in the substantia nigra of PD patients and in MPP+-exposed mouse and zebrafish models [34], [35], [36].
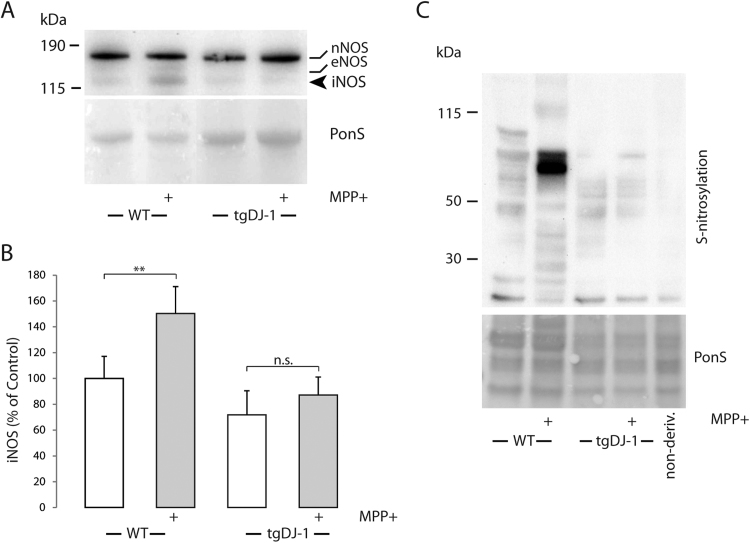
To evaluate the effect of astroglial DJ-1 over-expression in an MPP+ model, we exposed wild type (WT) and Tg(gfap:egfp-2A-flag-zDJ-1)(TgDJ-1) larvae to 600 μM MPP+ over 1–4 dpf and analyzed iNOS expression (Fig. 2A and B) and protein nitrosylation (Fig. 2C) in total extracts from the larvae. Expression of iNOS increased significantly in WT larvae exposed to MPP+, whilst increased astroglial DJ-1 prevented iNOS increase in the transgenic larvae. Increased iNOS activity would lead to increased levels of reactive nitric oxide that can modify cysteine residues by S-nitrosylation. As can be seen in Fig. 2C astroglial DJ-1 over-expression also reflected in the prevention of protein S-nitrosylation in total protein extracts from the larvae (Fig. 2C).
Fig. 2.
Astroglial DJ-1 over-expression inhibits MPP+-induced iNOS up-regulation and protein S-nitrosylation Larvae from wild type or Tg(gfap:egfp-2A-flag-zDJ-1) were exposed to 600 µM MPP (1–4 dpf) or left untreated. At 4 dpf total protein extracts from the larvae were subjected to Western blotting and nitric oxide synthase detection or derivatization and S-nitrosylation determination using anti-TMT. A, Expression of nitric oxide synthase isozymes (nNOS, eNOS, and iNOS). Arrow points at iNOS. B, iNOS abundance. Data are expressed as the means of five-six individual experiments +/-SEM, **p < 0.01. Data were analyzed using Student t-test. C, Representative Western blot showing S-nitrosylation in total extracts. Ponceau-S was used as a loading control.
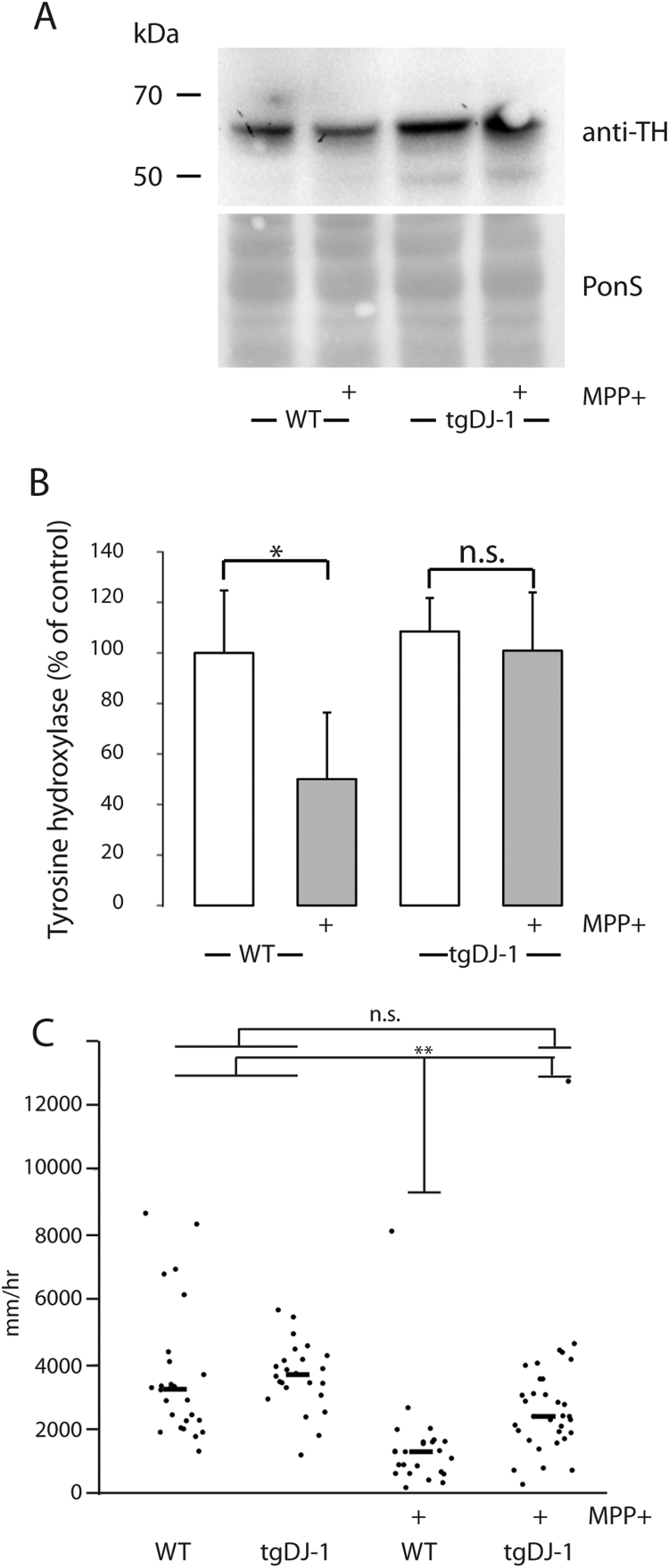
Several studies have shown that MPP+ induces dopaminergic cell loss leading to motility disorder in zebrafish [18], [20], [21], [37]. We analyzed the expression of Tyrosine hydroxylase using Western blotting in order to determine the effect of MPP+ exposure on dopaminergic cells. MPP+ caused a significant reduction in Tyrosine hydroxylase expression (Fig. 3A and B). In MPP+-treated TgDJ-1 on the other hand, no significant effect of MPP+ on Tyrosine hydroxylase expression could be observed (Fig. 3A and B).
Fig. 3.
Astroglial over-expression of DJ-1 protects larvae from MPP+-induced down-regulation of Tyrosine hydroxylase expression and motility dysfunction. A-B, Tyrosine hydroxylase (TH) expression in wild type and Tg(gfap:egfp-2A-flag-zDJ-1) larvae exposed to 600 µM MPP (1–4 dpf) or vehicle. At 4 dpf total protein extracts from the larvae were subjected to Western blotting. Ponceau-S staining was used as loading control. A, A representative Western blot of Tyrosine hydroxylase expression. B, Tyrosine hydroxylase expression values as mean +/-SEM from five individual experiments. C, Scatter plot of larval motility over one hour recording. Tg(gfap:egfp-2A-flag-zDJ-1) and wild type larvae were treated with 500 µM MPP+ or vehicle for 48 h starting at 3 dpf. Graph shows swimming distance (mm/h) at 5 dpf. Plot shows the values and median from three separate experiments. Data were analyzed using Student t-test * p < 0.05, **p < 0.01.
We next evaluated if motor dysfunction caused by MPP+ was prevented in TgDJ-1 (Fig. 3C). By 5 dpf larval size and pigmentation increase the reliability of automated motion tracking, and larval motility is significantly increased and less variable compared with earlier time points [21] so that the signal:noise ratio for spontaneous movement is higher. Consequently, in order to optimize the assay for detection of MPP+-induced phenotypes, we exposed larvae to 500 μM MPP+ from 48 hpf and carried out motor assays at 5 dpf. In the absence of MPP+, the spontaneous motility of TgDJ-1 larvae at 5 dpf was indistinguishable from controls (Fig. 3C). WT larvae exposed to MPP+ showed a significantly reduction in spontaneous motility so their swimming distance was reduced by approximately 50%. In contrast, the motor deficit caused by MPP+ exposure was partially rescued in TgDJ-1 larvae, so that swimming distance was reduced by only 15% following MPP+ exposure.
Together, these data indicate that increased astroglial DJ-1 expression prevented iNOS induction, protein nitrosylation, loss of Tyrosine hydroxylase expression and motor deficits in zebrafish exposed to the PD-relevant toxicant MPP+.
3.3. Global proteome analysis of larvae exposed to MPP+ and the effect of increased astroglial DJ-1 expression
We next asked whether the protective effect of increased astrocytic DJ-1 expression was reflected in the regulation of other proteins or cellular pathways. In particular, to determine changes that might precede PD-related features. We therefore reduced the MPP+ exposed of larvae to 500 μM over 2–4 dpf before performing a label-free quantitative global proteomics analysis of wild type and TgDJ-1 larvae exposed to MPP+ or vehicle. From triplicate nanoLC-HDMSE analyses of transgenic and WT larvae a total of 3418 proteins were identified on the basis of two or more peptides with a mass accuracy ≤ 10 ppm and peptide score ≥ 4. For quantitative analysis a minimum of two unique peptides were required for further statistical analysis. Thereafter we restricted for proteins which showed a 2-fold or above regulation when comparing untreated and treated wild type and transgenic larvae leaving 366 proteins (Supplementary material Table 1). To identify proteins affected by the DJ-1 astrocytic protective response we search for proteins either showing a significantly U-shaped (WT > WT MPP+ < TgDJ-1 MPP+) (Table 1) or N-shaped (WT < WT MPP+ > TgDJ-1 MPP+) profile (Table 2). Table 1 thus shows proteins which were down-regulated in WT larvae exposed to MPP+ exposure, but not in TgDJ-1 exposed larvae. Table 2 shows proteins up-regulated in MPP+ exposed WT, but not in TgDJ-1 exposed animals.
Table 1.
MPP+-induced down-regulation of proteins in wild type larvae counteracted by glial DJ-1 over-expression.
| Normalized abundance | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Accession | Description | Total peptides | Unique peptides | Confidence score | Anova (p) | Max fold change | WT | WT MPP+ | TgDJ-1 | TgDJ-1 MPP+ | Protein characteristics | |
| Metabolic process | ||||||||||||
| F1Q4Q8 | Sarcosine dehydrogenase | 3 | 2 | 17 | 0,004950 | 30,8 | 277 | 134 | 14 | 430 | glycin serine metabolism | |
| Q6PFJ6 | Glutathione S-transferase M | 10 | 6 | 62 | 2,17E−09 | 3,0 | 12466 | 9603 | 28661 | 13751 | oxidative stress response | |
| Q6DH98 | CNDP dipeptidase 2 | 12 | 4 | 75 | 1,55E−08 | 3,4 | 9746 | 8359 | 28567 | 10638 | over-expressed in PD | |
| Q6AZB2 | Succinyl-CoA:3-ketoacid-coenzyme A transferase | 9 | 3 | 46 | 5,61E−05 | 4,0 | 780 | 608 | 197 | 788 | pre-TCA cycle | |
| Q6PBI0 | Succinate-CoA ligase, ADP-forming, beta subunit | 11 | 4 | 67 | 3,46E−06 | 2,8 | 4845 | 1822 | 1724 | 2583 | TCA cycle | |
| Q7T334 | Malate dehydrogenase | 23 | 16 | 208 | 9,94E−09 | 2,1 | 79363 | 56824 | 37543 | 79709 | TCA cycle | |
| Small GTPase mediated signal transduction | ||||||||||||
| A9ULS4 | Rab6a protein | 7 | 2 | 40 | 7,89E−10 | 4,6 | 1497 | 1231 | 5669 | 1821 | ||
| B8JLC8 | RAB1A, member RAS oncogene family a | 15 | 2 | 119 | 1,25E−08 | 2,9 | 3157 | 2594 | 7507 | 3903 | ||
| Nucleic acid binding | ||||||||||||
| B3DIX5 | Lemd3 protein | 2 | 2 | 9 | 1,14E−07 | 8,3 | 3786 | 455 | 1394 | 760 | ||
| A0A0B4J1A6 | Insulin-like growth factor 2 mRNA-binding protein 1 | 9 | 4 | 65 | 9,33E−06 | 2,5 | 4595 | 2981 | 1809 | 4155 | ||
| Q6P271 | ATPase, Na+/K+ transporting, alpha 3a polypeptide | 52 | 2 | 393 | 5,01E−08 | 2,7 | 6859 | 5840 | 15605 | 7124 | locomotor behavior | |
| Q9PW80 | Insulin-like growth factor 2 mRNA-binding protein 3 | 10 | 3 | 54 | 2,44E−07 | 2,4 | 2546 | 2091 | 5064 | 2798 | ||
| Cytoskeletal organization and transport | ||||||||||||
| Q6DG66 | Kertain 95 | 9 | 2 | 82 | 1,31E−06 | 2,4 | 3205 | 2672 | 6338 | 3891 | ||
| E7F0A1 | Profilin | 11 | 7 | 105 | 1,22E−09 | 2,0 | 31848 | 23622 | 15731 | 31795 | ||
| C6K2H9 | Cellular trafficking protein | 10 | 3 | 60 | 8,55E−12 | 3,1 | 5880 | 4242 | 13109 | 5092 | ||
| Q2TTK0 | Cation-independent mannose 6-phosphate receptor | 12 | 2 | 78 | 1,03E−10 | 3,2 | 125632 | 95188 | 308630 | 110380 | ||
| Other | ||||||||||||
| Q804G4 | Annexin | 11 | 4 | 58 | 6,97E−07 | 2,1 | 2701 | 2037 | 1308 | 2364 | Ca2+dependent phospholipid binding | |
| A4QN79 | Si: dkey-251i10.1 protein | 7 | 2 | 65 | 7,18E−06 | 9,7 | 1504 | 156 | 679 | 401 | ||
| Q6P025 | Gnb3 protein | 2 | 2 | 11 | 1,21E−06 | 2,0 | 1760 | 870 | 915 | 1010 | glucagon-mediated signalling | |
| F2Z4S3 | Transcobalamin II | 12 | 5 | 65 | 9,11E−10 | 3,1 | 7415 | 6563 | 20501 | 7603 | vitB12 binding, impaired vitB12 related to PD features | |
| F1R8B4 | GCN1 eIF2 alpha kinase activator homolog | 14 | 6 | 84 | 1,12E−07 | 2,1 | 21900 | 10,372 | 22118 | 13284 | stress response; Nrf2 binding | |
| Q7T358 | G1 to S phase transition 1 | 11 | 7 | 70 | 1,28E−07 | 3,1 | 20620 | 17311 | 53654 | 22955 | G1 to S phase transition 1; astrogliosis | |
| Q1LVD7 | EF-hand domain family, member D2 | 8 | 5 | 40 | 1,37E−06 | 2,2 | 2710 | 2248 | 5012 | 3065 | altered regulation in neurdegeneration | |
Table 2.
MPP+-induced protein up-regulation counteracted by increased glial DJ-1 expression.
| Accession | Description | Peptide count | Unique peptides | Confidence score | Anova (p) | Max fold change | WT | WT MPP+ | TgDJ-1 | TgDJ-1MPP+ | Protein characteristics |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolic process | |||||||||||
| Q503G5 | GMP reductase | 4 | 3 | 19 | 4,66E−05 | 2,3 | 2750 | 3420 | 2777 | 1490 | purine metabolism; redox defense |
| Cytoskeletal organization and transport | |||||||||||
| I3ISP1 | Pericentrin | 43 | 12 | 260 | 1,07E−06 | 2,9 | 28216 | 36753 | 63941 | 34510 | mitotic assembly |
| Q802D5 | Profilin 2 | 16 | 4 | 123 | 1,40E−05 | 2,8 | 3465 | 4539 | 7139 | 3247 | actin-binding, iron homeostasis |
| F1Q5R6 | GN = si: dkey-27m7.4 | 6 | 2 | 44 | 2,87E−05 | 2,7 | 3262 | 4279 | 6539 | 3033 | axon development |
| A2BG19 | Novel protein similar to vertebrate skeletal alpha-actin 1 | 43 | 2 | 518 | 8,43E−08 | 2,2 | 18899 | 23815 | 30240 | 15530 | |
| E7F9V7 | Pleckstrin homology domain-containing, family A member 6 | 11 | 5 | 60 | 6,58E−05 | 2,1 | 2181 | 2492 | 3240 | 1780 | Phosphatidyl inositol metabolism |
| Other | |||||||||||
| B0UYL3 | Poly [ADP-ribose] polymerase (PARP) | 18 | 3 | 106 | 3,15E−06 | 2,6 | 1263 | 2027 | 2352 | 991 | oxidative stress, cell death, nucleic acid binding, inflammation, MPP+ induced |
| B7ZVT7 | F5 protein | 6 | 3 | 31 | 1,03E−07 | 2,6 | 11689 | 17739 | 25189 | 16367 | metal ion binding |
| F1R598 | Histone-lysine N-methyltransferase | 9 | 2 | 50 | 2,07E−08 | 2,3 | 20120 | 32826 | 21587 | 26639 | histone modifying |
| E7F9Z1 | Von Willebrand factor | 6 | 3 | 29 | 1,92E−07 | 2,0 | 8992 | 10442 | 12699 | 7097 | koagulerings faktor, eNOS |
| Q08CD9 | Augmin-like complex subunit 3-like protein alpha | 3 | 3 | 14 | 2,91E−05 | 2,0 | 4013 | 6642 | 7311 | 3962 | mitotic assembly |
It should be noted that as the zebrafish proteome has not yet been comprehensively annotated with gene ontology (GO) terms our data interpretation mostly relies on the knowledge of their mammalian orthologs.
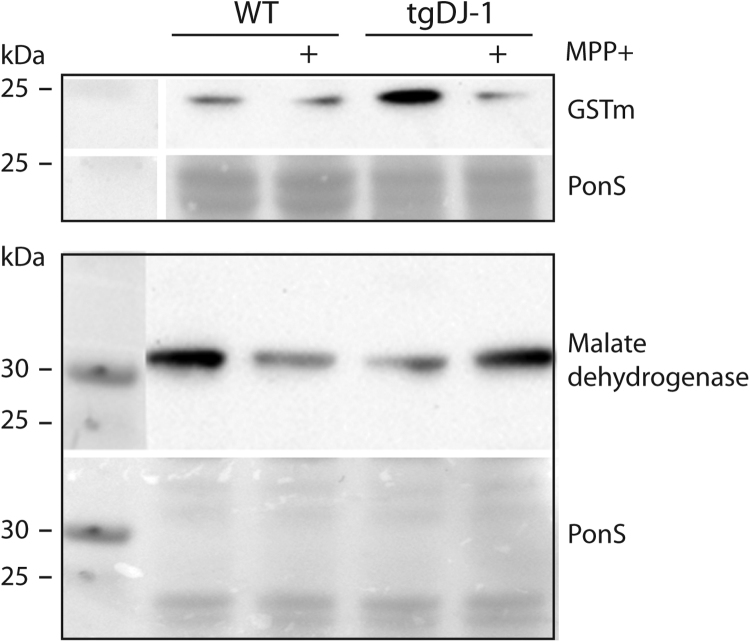
The most pronounced effect of increased astrocytic DJ-1 expression was its inhibitory effect on metabolic stress induced by MPP+ (Table 1). Mitochondrial proteins with rate limiting roles in the tricarboxylic acid (TCA) cycle (Succinate-CoA, Malate dehydrogenase, Succinyl-CoA) affected by MPP+ in WT were shown to be normalized in MPP+ exposed transgenic larvae. The most prominent regulation was observed for Sarcosine dehydrogenase, a mitochondrial enzyme converting sarcosine to glycine, which is linked to both the respiratory chain and the TCA cycle. Glutathione S-transferase M and CNDP dipetidase 2, both known to be up-regulated by cellular stress [38], [39], were up-regulated in the TgDJ-1 larvae, but in MPP+-exposed TgDJ-1 the expression level of these proteins were close to WT controls. The mass spectrometry based observed regulation of Malate dehydrogenase and Glutathione S-transferase M were verified by Western blotting (Fig. 4). Proteins with a U-shaped profile also included proteins with function in small GTPase mediated signal transduction, nucleic acid binding and regulation, cytoskeleton organization and transport (Table 1).
Fig. 4.
Verification of mass spectrometry data. Glutathione S-transferase M (GSTm) and Malate dehydroxylase were selected to verify mass spectrometry observed protein changes. Protein extracts from 4 dpf larvae exposed to 500 mM MPP+ or vehicle for 48 h starting at 3 dpf were subjected to Western blotting. Figure shows representative blots of GSTm and Malate dehydroxylase and the respective Ponceau-S staining used as loading control.
Additionally, the B12 vitamin carrier transcobalamin II showed MPP+-regulated change, which was reduced by astrocytic DJ-1 over-expression, likewise for the stress response associated proteins GCN1 eIF2 alpha kinase activator homologe and G1 to S phase transition 1. The latter, is a recently proposed regulator of astrogliosis [40].
Among the proteins that increased in WT during oxidative stress, but did not show similar increase in MPP+-exposed TgDJ-1 larvae (N-shaped regulation, Table 2), the largest group of proteins belonged to the cytoskeleton and cytoskeleton organizing proteins. Proteins involved in hemostasis (F5 protein and Von Willebrand factor) regulation were also regulated similarly. Additionally, the cell death and oxidative stress related Poly(ADP-ribose) polymerase (PARP) and the GMP reductase were also found to be N-shaped regulated. The latter may link purine metabolism and mitochondrial redox defense [41].
3.4. Protein profiling of astrocytes
To explore the direct cell specific effect of increased astroglial DJ-1 expression we isolated astrocytes from TgDJ-1 and control larvae using FACS-based sorting of 4 dpf larvae. Cells were selected based on the fluorescent tags driven by the inserted gfap promotor elements in TgDJ-1 and Tg(gfap:mCherry). From triplicate technical runs 835 proteins were identified and 407 of these met the criteria for further analysis (See Materials and Methods and Supplementary material Table 2). Of these 315 proteins were only found in astrocytes from TgDJ-1. Presumably,these proteins were below detectable levels in control larvae. Table 3 shows the LFQ intensity (log2 values) of GFP and mCherry detected in TgDJ-1 and Tg(gfap:mCherry), respectively. Astroglial DJ-1 expression was found to be more than 10-fold increased in TgDJ-1 compared to control (Table 3). Increased astrocytic DJ-1 expression induced up-regulation of a number of proteins, of which ten proteins were more than 50-fold up-regulated compared to control (Table 4).
Table 3.
DJ-1 and selectable marker protein abundance in FACS-sorted cells.
Table 4.
Highly up-regulated proteins in glia cells of Tg(gfap:egfp-2A-flag-zDJ-1).
| LFQ intensity (log2 values) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acc.no | Description | Peptides | Unique peptides | Score | Fold change | p-value | Control | TgDJ-1 | Protein characteristics | |||
| RedOx regulation | ||||||||||||
| Q6DGJ6 | Peroxiredoxin 2 | 10 | 6 | 88 | 135 | 7,92E−07 | 20,31 ± 0,23 | 27,38 ± 0,02 | Antioxidant, Nrf2 regulated | |||
| A0A0R4IVY7 | Prothymosin alpha-B | 2 | 2 | 80 | n.d. | 27,37 ± 0,05 | Keap1 partner, pro-regulation of Nrf2 | |||||
| Q7ZUI4 | Thioredoxin | 6 | 6 | 158 | 293 | 1,67E−06 | 18,74 ± 0,32 | 26,94 ± 0,08 | Antioxidant, Nrf2 regulated | |||
| B2GRH9 | Superoxide dismutase [Cu-Zn] | 6 | 6 | 57 | n.d. | 25,34 ± 0,11 | Antioxidant, Nrf2 regulated | |||||
| Q08CQ9 | Nucleolin | 14 | 14 | 146 | 189 | 4,90E−07 | 18,98 ± 0,20 | 26,54 ± 0,10 | ||||
| Q6PFN7 | Protein arginine methyltransferase 1 | 8 | 8 | 68 | n.d. | 24,67 ± 0,03 | Regulator of Nrf2, Upregulated in oxidative stress | |||||
| Q9I8N9 | Brain-type fatty acid binding protein 7 | 7 | 6 | 98 | 51 | 5,26E−08 | 21,85 ± 0,08 | 27,53 ± 0,06 | Up-regulated in PD | |||
| Q7ZVC6 | High-mobility group box 1 | 10 | 6 | 144 | 54 | 7,20E−07 | 20,60 ± 0,17 | 26,36 ± 0,08 | Inflammation, Nrf2 regulated release | |||
| F1QGP8 | High mobility group box 3a | 4 | 4 | 32 | 77 | 2,14E−07 | 18,65 ± 0,11 | 24,93 ± 0,10 | ||||
| Q6PBJ8 | Peptidyl-prolyl cis-trans isomerase | 3 | 3 | 38 | 97 | 7,08E−06 | 19,53a | 26,13 ± 0,02 | ||||
| Q6PC53 | Peptidyl-prolyl cis-trans isomerase/CyclophilinA | 5 | 5 | 75 | 61 | 1,57E−06 | 20,24 ± 0,08 | 26,16 ± 0,22 | Inflammation, redox regulated release, p38/MAPK inhibitor | |||
| Mitochondrial respiration | ||||||||||||
| B8JM34 | ATP synthase-coupling factor 6, mitochondrial | 5 | 5 | 31 | n.d. | 24,72 ± 0,21 | ||||||
| Q7SXM1 | Cytochrome c oxidase subunit | 7 | 7 | 48 | n.d. | 24,61 ± 0,03 | ||||||
| Ubiqitin-Autophagy | ||||||||||||
| F1Q8D2 | NSFL1 (p97) cofactor (p47) | 12 | 12 | 102 | n.d. | 24,56 ± 0,01 | ||||||
| RNA binding, translation | ||||||||||||
| E9QB42 | Anp32a | 10 | 10 | 117 | 153 | 1,89E−05 | 18,64 ± 0,10 | 25,89 ± 0,52 | Up-regulated in Alzheimers disease | |||
| Q6PHJ4 | Hnrpa0 protein | 5 | 5 | 95 | n.d. | 25,53 ± 0,15 | ||||||
| Q6PBK3 | 40 S ribosomal protein S28 | 3 | 3 | 102 | n.d. | 24,83 ± 0,05 | ||||||
| Q6P2A9 | Far upstream element (FUSE) binding protein 1 | 9 | 9 | 102 | n.d. | 24,62 ± 0,03 | Up-regulated in PD and MPTP models | |||||
| Q504C3 | SAP domain containing ribonucleoprotein | 7 | 7 | 47 | 51 | 5,86E−07 | 18,94a | 24,60 ± 0,02 | ||||
n < 3.
The majority of proteins up-regulated or detected only in TgDJ-1 astrocytes were antioxidants (Peroxiredoxin 2, Thioredoxin, and Superoxide dismutase 1) and proteins proposed to be involved in redox regulation (Prothymosin α, Nucleolin, Protein arginine methyltransferase 1, Brain-type fatty acid binding protein 7, High mobility groups box 1/3a, and Peptidyl-propyl cis-trans isomerase) [42], [43], [44], [45], [46].
Proteins involved mitochondrial respiration, and autophagy were identified in the astrocytes from TgDJ-1, but not in astrocytic control cells. The phosphatase inhibitor and transcription regulator Anp32a was found to be highly up-regulated in TgDJ-1 astrocytes compared to control cells. Interestingly, up-regulation of Anp32a is also found in Alzheimer disease [47].
Only three proteins were shown to be down-regulated TgDJ-1 astrocytes (Supplementary material Table 2). One of these was Transitional endoplasmic reticulum ATPase, a protein proposed to regulate cross-talk between autophagy and apoptotic cell death in neuronal cells [48].
4. Discussion
Reactive astroglia show an increased expression level of DJ-1 in Parkinson's disease (PD) and other neurodegenerative diseases [12], [49]. This may be a way not only to protect themselves, but also their neighboring neurons. Thus, studies in primary co-cultures of astrocytes-neurons show that knock-down of astrocytic DJ-1 renders the neurons more susceptible to neurotoxin-induced oxidative stress [15], [50]. Additionally, it has been shown that conditioned media from astrocytic cultures of wild type mice can protect neuronal cells from oxidative insult, whereas media derived from DJ-1 knock-out mice does not [51].
Here, we describe a new transgenic zebrafish model, Tg(gfap:egfp‐2A‐flag‐zDJ‐1 (TgDJ-1)(Fig. 1), which mirrors the increased DJ-1 expression levels found both in PD and its animal and cell culture based models. The transgenic line, TgDJ-1 has an approximately 10-fold increase in astrocyte DJ-1 expression at 4 dpf compared to control (Table 3). To evaluate the neuroprotective effect of increased astrocyte DJ-1 expression we exposed zebrafish embryos and larvae to MPP+. MPP+ and its precursor MPTP are used to induce PD pathology in cell culture and animal models, including zebrafish [18], [31], [34]. These toxicants induce neuroinflammation and dopaminergic cell loss mediated by iNOS [35], an inducible form of nitric oxide synthase shown to be regulated by DJ-1 [52], [53]. We observed an increased in iNOS in MPP+-exposed wild type (WT) larvae (Fig. 2A and B) as also observed previously by others [34]. We also demonstrated an MPP+-induced general S-nitrosylation of proteins (Fig. 2C). In the presence of increased astrocytic DJ-1 expression, both iNOS induction and protein S-nitrosylation were inhibited (Fig. 2). To explore the protective effect of astrocytic DJ-1 up-regulation on dopaminergic cells, we examined the level of Tyrosine hydroxylase in MPP+-exposed larvae. TgDJ-1 larvae showed a small increase in Tyrosine hydroxylase expression compared to WT, but exposure to MPP+ did not have any significant effect on the Tyrosine hydroxylase levels in these transgenes (Fig. 3A and B). On the contrary, MPP+-exposed WTs had a significant decrease in Tyrosine hydroxylase compared to untreated WT. In line with this, MPP+ had a significantly less reductive effect on larvae motility, as monitored by swim distance, in TgDJ-1 compared to WT (Fig. 3C). Our results demonstrate that protection from known PD-related features as induced by MPP+ can be obtained by solely increasing astrocyte DJ-1 expression.
The neuroprotective trait observed by increasing astroglial DJ-1 expression (Fig. 2, Fig. 3) seemed to be reflected in a DJ-1-induced altered astrocytic protein profile (Table 4 and Supplementary material Table 2). From mass spectrometry analysis of isolated astrocytes we could observe a prominent increased expression of proteins associated with the Kelch-like ECH-associating protein 1 (Keap1)-nuclear factor-E2-related factor2 (Nrf2) pathways in TgDJ-1. This included both proteins regulated by Nrf2 (Periredoxin 2, Thioredoxin, Superoxide dismutase [Cu-Zn], High-mobility group box 1), but interestingly, also proteins proposed to regulate Nrf2 activity (Prothymosin α, and Protein arginine methyltransferase 1) [42], [54].
The transcription factor Nrf2 induces up-regulation of a large number of antioxidant proteins in response to oxidative stress [55]. Nrf2 activity is regulated by Keap1, which under normal conditions degrades Nrf2. DJ-1 stabilizes Nrf2 by inhibiting its association to Keap1 [56] and over-expression of DJ-1 increases Nrf2 levels in neuroblastoma cells [57]. Additionally, it has been shown that astrocytic Nrf2 over-expression introduced into Nrf2 deficient mice protects against MPTP-induced Tyrosine hydroxylase-loss and gliosis [58]. This is in line with our results and suggests that astrocytic DJ-1 in TgDJ-1 may act up-stream of Nrf2 in order to protect the surrounding neurons. It would also explain why neuroprotective effects of Nrf2 activators are independent of DJ-1 [59].
Astrocytic neuroprotection by Keap1/Nrf2 pathways is proposed to be mediated by increased astrocytic glutathione release to be available for uptake by neurons [60]. The endogenous antioxidant glutathione is important in limiting damaging nitric oxide action, as S-nitrosylation of proteins, in the brain [61]. Astrocyte DJ-1 mediated neuroprotection through the Keap1/Nrf2 pathways (Table 4) may therefore inhibit MPP+-induced iNOS expression and protein nitrosylation (Fig. 2, Fig. 3) by strengthening the glutathione buffer capacity. Increased astrocyte DJ-1 expression also regulated the global expression of Glutathione S-transferase M (GSTm) and CNDP2 dipeptidase 2 (Table 1) (Fig. 4), both are involved in glutathione metabolism [61], [62] and associated with oxidative stress response [38], [63]. The untreated TgDJ-1 showed a three-fold increase in both GSTm and CNDP2 compared to untreated WT. This possibly reflects an astrocytic DJ-1 regulated preparation for oxidative stress. Thus, both GSTm and CNDP2 were down-regulated by MPP+, but in exposed TgDJ-1 the expression remained above WT levels (Table 1).
MPP+ accumulates in the dopaminergic neurons via uptake by the dopamine transporter [64] rendering the nigral dopaminergic neurons, which depend on oxidative phosphorylation, highly sensitive to this mitochondrial complex I inhibitor. Even though MPP+ also enters astrocytes and induces oxidative stress, these cells seem less vulnerable to MPP+ compared to neurons, presumable due to a faster antioxidant response [32]. Astrocytes from TgDJ-1 seem to have already obtained an antioxidative and inflammatory regulating trait before any oxidative stress had been applied (Table 4). TgDJ-1 astrocytes from untreated larvae also showed up-regulation of proteins involved in mitochondrial respiration possibly rendering these cells even more resistant to MPP+. This effect on mitochondrial respiration may be associated to Nrf2-related signalling since Nrf2 also has a role in controlling mitochondrial bioenergetics [65]. By adding MPP+ this pre-established protective condition may also be further regulated by oxidative modification of the astrocyte DJ-1 [66]. Thus, DJ-1 acts as sensor for oxidative stress involving oxidation of its reactive cysteine residues, in particular C106. From being predominately cytosolic a small fraction oxidatively modified DJ-1 may translocate to the mitochondrial matrix and regulate energy metabolism under stress conditions [67]. On the other hand, DJ-1 may also elicit its function through Nrf2 dependent signalling independently of C106 oxidation [68]. The possibility of astrocytic DJ-1 involvement in both cysteine oxidation dependent and independent MPP+ protective pathways may be reflected in its effect on metabolic enzymes of the TCA cycle in zebrafish larvae (Table 1) (Fig. 4). MPP+-induced a down-regulation of metabolic enzymes of the TCA cycle in WT, presumably as a consequence of complex I inhibition and shift to glycolysis [69]. A similar metabolic shift in central metabolism was also observed in unexposed TgDJ-1, however, when exposed to MPP+ the TCA cycle key enzymes seemed unaffected. Possibly, this astrocytic DJ-1 compensatory action requires an oxidative stress-induced change of DJ-1 function through oxidation of its oxidative sensitive cysteines [66], [70].
In addition to proteins associated to redox regulation the TgDJ-1 astrocyte proteome showed an up-regulation of proteins involved in inflammatory response (Table 4). PD and MPP+ models show an up-regulation of reactive astrocytes [35], [49]. Reactive astrocytes respond to stress and injury by releasing inflammatory factors. These factors can be pro-inflammatory and neurotoxic, but more recently their anti-inflammatory and pro-survival function have become more evident [71]. Interestingly, both High-mobility group box 1 (HMGB1) and Peptidyl-prolyl cis-trans isomerase (Cyclophilin A), which were up-regulated in TgDJ-1 astrocytes have both pro- and anti-inflammatory roles. HMGB1 is a pro-inflammatory protein and high serum levels are associated with PD [45]. The release of HMGB1 is proposed to be regulated both by p38 MAPK and Nrf2 signalling pathways [72]. Even though it is stated as a pro-inflammatory mediator, oxidation of HMGB1 attenuates its pro-inflammatory action [73]. Peptidyl-prolyl cis-trans isomerase (Cyclophilin A) was another DJ-1 regulated protein with a double-edged function being an oxidative stress protector intracellularly and pro-inflammatory extracellularly [74], [75].
Our global and astrocyte specific proteomics analysis of our zebrafish model also provided further insight into the relationship between astrocytic DJ-1 function and previously observed changes PD and its MPP+ disease model. Firstly, we found that MPP+ down-regulated both Rab1a and 6a in WT, but not in TgDJ-1 (Table 1). An underlying pathogenesis of PD is proposed to be defects in vesicle trafficking, with Rab GTPases as the core regulators [76]. Secondly, astrocytic DJ-1 over-expression inhibited PARP increase (Table 2) which has been shown to occur in MPP+-induced Parkinsonism [77]. Thirdly, amongst proteins highly up-regulated in TgDJ-1 astrocytes we found Far upstream element (FUSE) binding protein 1 and Brain-type fatty acid binding protein 7 (Table 4), both of which have been observed to be increased in PD brains and serum, respectively [78], [79].
In conclusion, our results show that selective over-expression of astrocyte DJ-1 in zebrafish larvae protects them from insults induced by the PD-relevant neurotoxin MPP+. The acquired protective trait is reflected in altered global expression of proteins, in particular proteins associated with the central metabolism and glutathione metabolism. An astrocyte specific proteome analysis suggests that the astroglial protective effect is accomplished by an up-regulation of redox and inflammatory regulating proteins, possibly involving Nrf2 activation.
Acknowledgement
This work was supported by the Fulbright foundation (KEF).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.02.010.
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Zhou W., Freed C.R. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J. Biol. Chem. 2005;280:43150–43158. doi: 10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- 2.Fan J., Ren H., Jia N., Fei E., Zhou T., Jiang P., Wu M., Wang G. DJ-1 decreases Bax expression through repressing p53 transcriptional activity. J. Biol. Chem. 2008;283:4022–4030. doi: 10.1074/jbc.M707176200. [DOI] [PubMed] [Google Scholar]
- 3.Yang Y., Gehrke S., Haque M.E., Imai Y., Kosek J., Yang L., Beal M.F., Nishimura I., Wakamatsu K., Ito S., Takahashi R., Lu B. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc. Natl. Acad. Sci. USA. 2005;102:13670–13675. doi: 10.1073/pnas.0504610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonifati V., Oostra B.A., Heutink P. Linking DJ-1 to neurodegeneration offers novel insights for understanding the pathogenesis of Parkinson's disease. J. Mol. Med. 2004;82:163–174. doi: 10.1007/s00109-003-0512-1. [DOI] [PubMed] [Google Scholar]
- 5.Saito Y., Miyasaka T., Hatsuta H., Takahashi-Niki K., Hayashi K., Mita Y., Kusano-Arai O., Iwanari H., Ariga H., Hamakubo T., Yoshida Y., Niki E., Murayama S., Ihara Y., Noguchi N. Immunostaining of oxidized DJ-1 in human and mouse brains. J. Neuropathol. Exp. Neurol. 2014;73:714–728. doi: 10.1097/NEN.0000000000000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen L., Cagniard B., Mathews T., Jones S., Koh H.C., Ding Y., Carvey P.M., Ling Z., Kang U.J., Zhuang X. Age-dependent motor deficits and dopaminergic dysfunction in DJ-1 null mice. J. Biol. Chem. 2005;280:21418–21426. doi: 10.1074/jbc.M413955200. [DOI] [PubMed] [Google Scholar]
- 7.Kim R.H., Smith P.D., Aleyasin H., Hayley S., Mount M.P., Pownall S., Wakeham A., You-Ten A.J., Kalia S.K., Horne P., Westaway D., Lozano A.M., Anisman H., Park D.S., Mak T.W. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc. Natl. Acad. Sci. USA. 2005;102:5215–5220. doi: 10.1073/pnas.0501282102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irrcher I., Aleyasin H., Seifert E.L., Hewitt S.J., Chhabra S., Phillips M., Lutz A.K., Rousseaux M.W., Bevilacqua L., Jahani-Asl A., Callaghan S., MacLaurin J.G., Winklhofer K.F., Rizzu P., Rippstein P., Kim R.H., Chen C.X., Fon E.A., Slack R.S., Harper M.E., McBride H.M., Mak T.W., Park D.S. Loss of the Parkinson's disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 2010;19:3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- 9.Burbulla L.F., Song P., Mazzulli J.R., Zampese E., Wong Y.C., Jeon S., Santos D.P., Blanz J., Obermaier C.D., Strojny C., Savas J.N., Kiskinis E., Zhuang X., Kruger R., Surmeier D.J., Krainc D. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson's disease. Science. 2017;357:1255–1261. doi: 10.1126/science.aam9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez-Fernandez S., Almeida A., Bolanos J.P. Antioxidant and bioenergetic coupling between neurons and astrocytes. Biochem. J. 2012;443:3–11. doi: 10.1042/BJ20111943. [DOI] [PubMed] [Google Scholar]
- 11.Liddell J.R. Are astrocytes the predominant cell type for activation of Nrf2 in aging and neurodegeneration? Antioxidants (Basel) 2017;6 doi: 10.3390/antiox6030065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bandopadhyay R., Kingsbury A.E., Cookson M.R., Reid A.R., Evans I.M., Hope A.D., Pittman A.M., Lashley T., Canet-Aviles R., Miller D.W., McLendon C., Strand C., Leonard A.J., Abou-Sleiman P.M., Healy D.G., Ariga H., Wood N.W., de Silva R., Revesz T., Hardy J.A., Lees A.J. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson's disease. Brain. 2004;127:420–430. doi: 10.1093/brain/awh054. [DOI] [PubMed] [Google Scholar]
- 13.Mullett S.J., Hamilton R.L., Hinkle D.A. DJ-1 immunoreactivity in human brain astrocytes is dependent on infarct presence and infarct age. Neuropathol.: Off. J. Jpn. Soc. Neuropathol. 2009;29:125–131. doi: 10.1111/j.1440-1789.2008.00955.x. [DOI] [PubMed] [Google Scholar]
- 14.Waak J., Weber S.S., Waldenmaier A., Gorner K., Alunni-Fabbroni M., Schell H., Vogt-Weisenhorn D., Pham T.T., Reumers V., Baekelandt V., Wurst W., Kahle P.J. Regulation of astrocyte inflammatory responses by the Parkinson's disease-associated gene DJ-1. FASEB J. 2009;23:2478–2489. doi: 10.1096/fj.08-125153. [DOI] [PubMed] [Google Scholar]
- 15.Mullett S.J., Hinkle D.A. DJ-1 deficiency in astrocytes selectively enhances mitochondrial Complex I inhibitor-induced neurotoxicity. J. Neurochem. 2011;117:375–387. doi: 10.1111/j.1471-4159.2011.07175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Booth H.D.E., Hirst W.D., Wade-Martins R. The role of astrocyte dysfunction in Parkinson's disease pathogenesis. Trends Neurosci. 2017;40:358–370. doi: 10.1016/j.tins.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin-Jimenez R., Campanella M., Russell C. New zebrafish models of neurodegeneration. Curr. Neurol. Neurosci. Rep. 2015;15:33. doi: 10.1007/s11910-015-0555-z. [DOI] [PubMed] [Google Scholar]
- 18.Sallinen V., Torkko V., Sundvik M., Reenila I., Khrustalyov D., Kaslin J., Panula P. MPTP and MPP+ target specific aminergic cell populations in larval zebrafish. J. Neurochem. 2009;108:719–731. doi: 10.1111/j.1471-4159.2008.05793.x. [DOI] [PubMed] [Google Scholar]
- 19.Bretaud S., Lee S., Guo S. Sensitivity of zebrafish to environmental toxins implicated in Parkinson's disease. Neurotoxicol. Teratol. 2004;26:857–864. doi: 10.1016/j.ntt.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Lam C.S., Korzh V., Strahle U. Zebrafish embryos are susceptible to the dopaminergic neurotoxin MPTP. Eur. J. Neurosci. 2005;21:1758–1762. doi: 10.1111/j.1460-9568.2005.03988.x. [DOI] [PubMed] [Google Scholar]
- 21.Farrell T.C., Cario C.L., Milanese C., Vogt A., Jeong J.H., Burton E.A. Evaluation of spontaneous propulsive movement as a screening tool to detect rescue of Parkinsonism phenotypes in zebrafish models. Neurobiol. Dis. 2011;44:9–18. doi: 10.1016/j.nbd.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bernardos R.L., Raymond P.A. GFAP transgenic zebrafish. Gene Expr. Patterns. 2006;6:1007–1013. doi: 10.1016/j.modgep.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Provost E., Rhee J., Leach S.D. Viral 2A peptides allow expression of multiple proteins from a single ORF in transgenic zebrafish embryos. Genesis. 2007;45:625–629. doi: 10.1002/dvg.20338. [DOI] [PubMed] [Google Scholar]
- 24.Bai Q., Mullett S.J., Garver J.A., Hinkle D.A., Burton E.A. Zebrafish DJ-1 is evolutionarily conserved and expressed in dopaminergic neurons. Brain Res. 2006;1113:33–44. doi: 10.1016/j.brainres.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 25.Bai Q., Garver J.A., Hukriede N.A., Burton E.A. Generation of a transgenic zebrafish model of tauopathy using a novel promoter element derived from the zebrafish eno2 gene. Nucleic Acids Res. 2007;35:6501–6516. doi: 10.1093/nar/gkm608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suster M.L., Kikuta H., Urasaki A., Asakawa K., Kawakami K. Transgenesis in zebrafish with the tol2 transposon system. Methods Mol. Biol. 2009;561:41–63. doi: 10.1007/978-1-60327-019-9_3. [DOI] [PubMed] [Google Scholar]
- 27.Martens G.A., Jiang L., Verhaeghen K., Connolly J.B., Geromanos S.G., Stange G., Van Oudenhove L., Devreese B., Hellemans K.H., Ling Z., Van Schravendijk C., Pipeleers D.G., Vissers J.P., Gorus F.K. Protein markers for insulin-producing beta cells with higher glucose sensitivity. PLoS One. 2010;5:e14214. doi: 10.1371/journal.pone.0014214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez-Suarez E., Hughes C., Gethings L., Giles K., Wildgoose J., Stapels M., Fadgen K.E., Geromanos S.J., Vissers J.P.C., Elortza F., Langridge J.I. An ion mobility assisted data independent LC-MS strategy for the analysis of complex biological samples. Curr. Anal. Chem. 2013;9:199–211. [Google Scholar]
- 29.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 30.Frøyset A.K., Khan E.A., Fladmark K.E. Quantitative proteomics analysis of zebrafish exposed to sub-lethal dosages of beta-methyl-amino-L-alanine (BMAA) Sci. Rep. 2016;6:29631. doi: 10.1038/srep29631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schildknecht S., Di Monte D.A., Pape R., Tieu K., Leist M. Tipping points and endogenous determinants of nigrostriatal degeneration by MPTP. Trends Pharmacol. Sci. 2017;38:541–555. doi: 10.1016/j.tips.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 32.Yu X., Song N., Guo X., Jiang H., Zhang H., Xie J. Differences in vulnerability of neurons and astrocytes to heme oxygenase-1 modulation: implications for mitochondrial ferritin. Sci. Rep. 2016;6:24200. doi: 10.1038/srep24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alarcon-Aguilar A., Luna-Lopez A., Ventura-Gallegos J.L., Lazzarini R., Galvan-Arzate S., Gonzalez-Puertos V.Y., Moran J., Santamaria A., Konigsberg M. Primary cultured astrocytes from old rats are capable to activate the Nrf2 response against MPP+ toxicity after tBHQ pretreatment. Neurobiol. Aging. 2014;35:1901–1912. doi: 10.1016/j.neurobiolaging.2014.01.143. [DOI] [PubMed] [Google Scholar]
- 34.Diaz-Casado M.E., Lima E., Garcia J.A., Doerrier C., Aranda P., Sayed R.K., Guerra-Librero A., Escames G., Lopez L.C., Acuna-Castroviejo D. Melatonin rescues zebrafish embryos from the parkinsonian phenotype restoring the parkin/PINK1/DJ-1/MUL1 network. J. Pineal Res. 2016;61:96–107. doi: 10.1111/jpi.12332. [DOI] [PubMed] [Google Scholar]
- 35.Liberatore G.T., Jackson-Lewis V., Vukosavic S., Mandir A.S., Vila M., McAuliffe W.G., Dawson V.L., Dawson T.M., Przedborski S. Inducible nitric oxide synthase stimulates dopaminergic neurodegeneration in the MPTP model of Parkinson disease. Nat. Med. 1999;5:1403–1409. doi: 10.1038/70978. [DOI] [PubMed] [Google Scholar]
- 36.Hunot S., Boissiere F., Faucheux B., Brugg B., Mouatt-Prigent A., Agid Y., Hirsch E.C. Nitric oxide synthase and neuronal vulnerability in Parkinson's disease. Neuroscience. 1996;72:355–363. doi: 10.1016/0306-4522(95)00578-1. [DOI] [PubMed] [Google Scholar]
- 37.McKinley E.T., Baranowski T.C., Blavo D.O., Cato C., Doan T.N., Rubinstein A.L. Neuroprotection of MPTP-induced toxicity in zebrafish dopaminergic neurons. Brain Res. Mol. Brain Res. 2005;141:128–137. doi: 10.1016/j.molbrainres.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 38.Licker V., Cote M., Lobrinus J.A., Rodrigo N., Kovari E., Hochstrasser D.F., Turck N., Sanchez J.C., Burkhard P.R. Proteomic profiling of the substantia nigra demonstrates CNDP2 overexpression in Parkinson's disease. J. Proteom. 2012;75:4656–4667. doi: 10.1016/j.jprot.2012.02.032. [DOI] [PubMed] [Google Scholar]
- 39.Kumar A., Dhull D.K., Gupta V., Channana P., Singh A., Bhardwaj M., Ruhal P., Mittal R. Role of Glutathione‐S‐transferases in neurological problems. Expert Opin. Ther. Pat. 2017;27:299–309. doi: 10.1080/13543776.2017.1254192. [DOI] [PubMed] [Google Scholar]
- 40.Ishii T., Ueyama T., Shigyo M., Kohta M., Kondoh T., Kuboyama T., Uebi T., Hamada T., Gutmann D.H., Aiba A., Kohmura E., Tohda C., Saito N. A novel Rac1-GSPT1 signaling pathway controls astrogliosis following central nervous system injury. J. Biol. Chem. 2017;292:1240–1250. doi: 10.1074/jbc.M116.748871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kristal B.S., Vigneau-Callahan K.E., Moskowitz A.J., Matson W.R. Purine catabolism: links to mitochondrial respiration and antioxidant defenses? Arch. Biochem. Biophys. 1999;370:22–33. doi: 10.1006/abbi.1999.1387. [DOI] [PubMed] [Google Scholar]
- 42.Karapetian R.N., Evstafieva A.G., Abaeva I.S., Chichkova N.V., Filonov G.S., Rubtsov Y.P., Sukhacheva E.A., Melnikov S.V., Schneider U., Wanker E.E., Vartapetian A.B. Nuclear oncoprotein prothymosin alpha is a partner of Keap1: implications for expression of oxidative stress-protecting genes. Mol. Cell. Biol. 2005;25:1089–1099. doi: 10.1128/MCB.25.3.1089-1099.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jun M.H., Ryu H.H., Jun Y.W., Liu T., Li Y., Lim C.S., Lee Y.S., Kaang B.K., Jang D.J., Lee J.A. Sequestration of PRMT1 and Nd1-L mRNA into ALS-linked FUS mutant R521C-positive aggregates contributes to neurite degeneration upon oxidative stress. Sci. Rep. 2017;7:40474. doi: 10.1038/srep40474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Rosa A., Pellegatta S., Rossi M., Tunici P., Magnoni L., Speranza M.C., Malusa F., Miragliotta V., Mori E., Finocchiaro G., Bakker A. A radial glia gene marker, fatty acid binding protein 7 (FABP7), is involved in proliferation and invasion of glioblastoma cells. PLoS One. 2012;7:e52113. doi: 10.1371/journal.pone.0052113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santoro M., Maetzler W., Stathakos P., Martin H.L., Hobert M.A., Rattay T.W., Gasser T., Forrester J.V., Berg D., Tracey K.J., Riedel G., Teismann P. In-vivo evidence that high mobility group box 1 exerts deleterious effects in the 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine model and Parkinson's disease which can be attenuated by glycyrrhizin. Neurobiol. Dis. 2016;91:59–68. doi: 10.1016/j.nbd.2016.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Y., Jinchuan Y., Yi L., Jun W., Zhongqun W., Cuiping W. Antiapoptotic and proapoptotic signaling of cyclophilin A in endothelial cells. Inflammation. 2013;36:567–572. doi: 10.1007/s10753-012-9578-7. [DOI] [PubMed] [Google Scholar]
- 47.Tanimukai H., Grundke-Iqbal I., Iqbal K. Up-regulation of inhibitors of protein phosphatase-2A in Alzheimer's disease. Am. J. Pathol. 2005;166:1761–1771. doi: 10.1016/S0002-9440(10)62486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeo B.K., Hong C.J., Chung K.M., Woo H., Kim K., Jung S., Kim E.K., Yu S.W. Valosin-containing protein is a key mediator between autophagic cell death and apoptosis in adult hippocampal neural stem cells following insulin withdrawal. Mol. Brain. 2016;9:31. doi: 10.1186/s13041-016-0212-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rizzu P., Hinkle D.A., Zhukareva V., Bonifati V., Severijnen L.A., Martinez D., Ravid R., Kamphorst W., Eberwine J.H., Lee V.M., Trojanowski J.Q., Heutink P. DJ-1 colocalizes with tau inclusions: a link between parkinsonism and dementia. Ann. Neurol. 2004;55:113–118. doi: 10.1002/ana.10782. [DOI] [PubMed] [Google Scholar]
- 50.Mullett S.J., Hinkle D.A. DJ-1 knock-down in astrocytes impairs astrocyte-mediated neuroprotection against rotenone. Neurobiol. Dis. 2009;33:28–36. doi: 10.1016/j.nbd.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lev N., Ickowicz D., Melamed E., Offen D. Oxidative insults induce DJ-1 upregulation and redistribution: implications for neuroprotection. Neurotoxicology. 2008;29:397–405. doi: 10.1016/j.neuro.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 52.Waak J., Weber S.S., Gorner K., Schall C., Ichijo H., Stehle T., Kahle P.J. Oxidizable residues mediating protein stability and cytoprotective interaction of DJ-1 with apoptosis signal-regulating kinase 1. J. Biol. Chem. 2009;284:14245–14257. doi: 10.1074/jbc.M806902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meiser J., Delcambre S., Wegner A., Jager C., Ghelfi J., d'Herouel A.F., Dong X., Weindl D., Stautner C., Nonnenmacher Y., Michelucci A., Popp O., Giesert F., Schildknecht S., Kramer L., Schneider J.G., Woitalla D., Wurst W., Skupin A., Weisenhorn D.M., Kruger R., Leist M., Hiller K. Loss of DJ-1 impairs antioxidant response by altered glutamine and serine metabolism. Neurobiol. Dis. 2016;89:112–125. doi: 10.1016/j.nbd.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 54.Morales Y., Nitzel D.V., Price O.M., Gui S., Li J., Qu J., Hevel J.M. Redox control of protein arginine methyltransferase 1 (PRMT1) activity. J. Biol. Chem. 2015;290:14915–14926. doi: 10.1074/jbc.M115.651380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. doi: 10.1016/j.tibs.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 56.Clements C.M., McNally R.S., Conti B.J., Mak T.W., Ting J.P.-Y. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. USA. 2006;103:15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Im J.-Y., Lee K.-W., Woo J.-M., Junn E., Mouradian M.M. DJ-1 induces thioredoxin 1 expression through the Nrf2 pathway. Hum. Mol. Genet. 2012;21:3013–3024. doi: 10.1093/hmg/dds131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen P.C., Vargas M.R., Pani A.K., Smeyne R.J., Johnson D.A., Kan Y.W., Johnson J.A. Nrf2-mediated neuroprotection in the MPTP mouse model of Parkinson's disease: critical role for the astrocyte. Proc. Natl. Acad. Sci. USA. 2009;106:2933–2938. doi: 10.1073/pnas.0813361106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gan L., Johnson D.A., Johnson J.A. Keap1-Nrf2 activation in the presence and absence of DJ-1. Eur. J. Neurosci. 2010;31:967–977. doi: 10.1111/j.1460-9568.2010.07138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vargas M.R., Johnson D.A., Sirkis D.W., Messing A., Johnson J.A. Nrf2 activation in astrocytes protects against neurodegeneration in mouse models of familial amyotrophic lateral sclerosis. J. Neurosci. 2008;28:13574–13581. doi: 10.1523/JNEUROSCI.4099-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aquilano K., Baldelli S., Ciriolo M.R. Glutathione: new roles in redox signaling for an old antioxidant. Front. Pharmacol. 2014;5:196. doi: 10.3389/fphar.2014.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaur H., Kumar C., Junot C., Toledano M.B., Bachhawat A.K. Dug1p Is a Cys-Gly peptidase of the gamma-glutamyl cycle of Saccharomyces cerevisiae and represents a novel family of Cys-Gly peptidases. J. Biol. Chem. 2009;284:14493–14502. doi: 10.1074/jbc.M808952200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cho S.G., Lee Y.H., Park H.S., Ryoo K., Kang K.W., Park J., Eom S.J., Kim M.J., Chang T.S., Choi S.Y., Shim J., Kim Y., Dong M.S., Lee M.J., Kim S.G., Ichijo H., Choi E.J. Glutathione S-transferase mu modulates the stress-activated signals by suppressing apoptosis signal-regulating kinase 1. J. Biol. Chem. 2001;276:12749–12755. doi: 10.1074/jbc.M005561200. [DOI] [PubMed] [Google Scholar]
- 64.Javitch J.A., D'Amato R.J., Strittmatter S.M., Snyder S.H. Parkinsonism-inducing neurotoxin, N-methyl-4-phenyl-1,2,3,6 -tetrahydropyridine: uptake of the metabolite N-methyl-4-phenylpyridine by dopamine neurons explains selective toxicity. Proc. Natl. Acad. Sci. USA. 1985;82:2173–2177. doi: 10.1073/pnas.82.7.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holmstrom K.M., Baird L., Zhang Y., Hargreaves I., Chalasani A., Land J.M., Stanyer L., Yamamoto M., Dinkova-Kostova A.T., Abramov A.Y. Nrf2 impacts cellular bioenergetics by controlling substrate availability for mitochondrial respiration. Biol. Open. 2013;2:761–770. doi: 10.1242/bio.20134853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Canet-Aviles R.M., Wilson M.A., Miller D.W., Ahmad R., McLendon C., Bandyopadhyay S., Baptista M.J., Ringe D., Petsko G.A., Cookson M.R. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cali T., Ottolini D., Soriano M.E., Brini M. A new split-GFP-based probe reveals DJ-1 translocation into the mitochondrial matrix to sustain ATP synthesis upon nutrient deprivation. Hum. Mol. Genet. 2015;24:1045–1060. doi: 10.1093/hmg/ddu519. [DOI] [PubMed] [Google Scholar]
- 68.Moscovitz O., Ben-Nissan G., Fainer I., Pollack D., Mizrachi L., Sharon M. The Parkinson's-associated protein DJ-1 regulates the 20S proteasome. Nat. Commun. 2015;6:6609. doi: 10.1038/ncomms7609. [DOI] [PubMed] [Google Scholar]
- 69.Mazzio E., Soliman K.F. The role of glycolysis and gluconeogenesis in the cytoprotection of neuroblastoma cells against 1-methyl 4-phenylpyridinium ion toxicity. Neurotoxicology. 2003;24:137–147. doi: 10.1016/s0161-813x(02)00110-9. [DOI] [PubMed] [Google Scholar]
- 70.Requejo-Aguilar R., Lopez-Fabuel I., Jimenez-Blasco D., Fernandez E., Almeida A., Bolanos J.P. DJ1 represses glycolysis and cell proliferation by transcriptionally up-regulating Pink1. Biochem. J. 2015;467:303–310. doi: 10.1042/BJ20141025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liddelow S.A., Barres B.A. Reactive astrocytes: production, function, and therapeutic potential. Immunity. 2017;46:957–967. doi: 10.1016/j.immuni.2017.06.006. [DOI] [PubMed] [Google Scholar]
- 72.Yu Y., Tang D., Kang R. Oxidative stress-mediated HMGB1 biology. Front. Physiol. 2015;6:93. doi: 10.3389/fphys.2015.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu A., Fang H., Dirsch O., Jin H., Dahmen U. Oxidation of HMGB1 causes attenuation of its pro-inflammatory activity and occurs during liver ischemia and reperfusion. PLoS One. 2012;7:e35379. doi: 10.1371/journal.pone.0035379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kim H., Oh Y., Kim K., Jeong S., Chon S., Kim D., Jung M.H., Pak Y.K., Ha J., Kang I., Choe W. Cyclophilin A regulates JNK/p38-MAPK signaling through its physical interaction with ASK1. Biochem. Biophys. Res. Commun. 2015;464:112–117. doi: 10.1016/j.bbrc.2015.06.078. [DOI] [PubMed] [Google Scholar]
- 75.Xue C., Sowden M., Berk B.C. Extracellular cyclophilin A, especially acetylated, causes pulmonary hypertension by stimulating endothelial apoptosis, redox stress, and inflammation. Arterioscler. Thromb. Vasc. Biol. 2017;37:1138–1146. doi: 10.1161/ATVBAHA.117.309212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi M.M., Shi C.H., Xu Y.M. Rab GTPases: the key players in the molecular pathway of Parkinson's disease. Front. Cell. Neurosci. 2017;11:81. doi: 10.3389/fncel.2017.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mandir A.S., Przedborski S., Jackson-Lewis V., Wang Z.Q., Simbulan-Rosenthal C.M., Smulson M.E., Hoffman B.E., Guastella D.B., Dawson V.L., Dawson T.M. Poly(ADP-ribose) polymerase activation mediates 1-methyl-4-phenyl-1, 2,3,6-tetrahydropyridine (MPTP)-induced parkinsonism. Proc. Natl. Acad. Sci. USA. 1999;96:5774–5779. doi: 10.1073/pnas.96.10.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ko H.S., Kim S.W., Sriram S.R., Dawson V.L., Dawson T.M. Identification of far upstream element-binding protein-1 as an authentic Parkin substrate. J. Biol. Chem. 2006;281:16193–16196. doi: 10.1074/jbc.C600041200. [DOI] [PubMed] [Google Scholar]
- 79.Teunissen C.E., Veerhuis R., De Vente J., Verhey F.R., Vreeling F., van Boxtel M.P., Glatz J.F., Pelsers M.A. Brain-specific fatty acid-binding protein is elevated in serum of patients with dementia-related diseases. Eur. J. Neurol. 2011;18:865–871. doi: 10.1111/j.1468-1331.2010.03273.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material