Abstract
We report here the main characteristics of ‘Collinsella provencensis’ strain Marseille-P3740 (CSUR P3740), ‘Parabacteroides bouchesdurhonensis’ strain Marseille-P3763 (CSUR P3763) and ‘Sutterella seckii’ strain Marseille-P3660 (CSUR P3660), which were isolated using culturomics from the human gut microbiota of healthy individuals living in Marseille.
Keywords: Collinsella provencensis, culturomics, Parabacteroides bouchesdurhonensis, Sutterella seckii
As a part of culturomics study of the human microbiota [1], we isolated in 2016, from stool samples of three different healthy men, three new bacteria that could not be identified by our systematic matrix-assisted laser desorption–ionization time-of-flight mass spectrometry (MALDI-TOF MS) screening on a Microflex spectrometer (Bruker Daltonics, Bremen, Germany) [2]. Each of the three species has been described according to their following characteristics: Gram, oxidase and catalase tests, growth conditions, colony size, phylogeny and MALDI-TOF MS spectra. This study was approved by the ethics committee of the Foundation Méditerranée Infection under number 2016-011.
Characteristics of ‘Collinsella provencensis’ sp. nov.
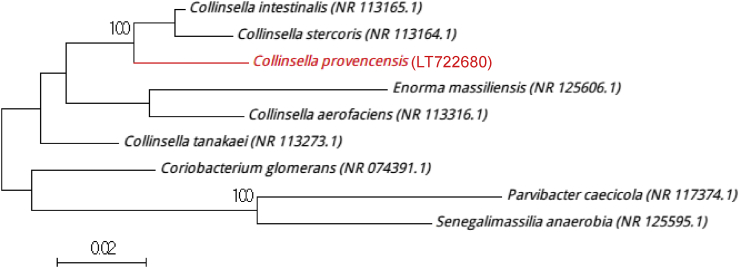
A stool sample from a 32-year-old man living in France was diluted with 1 mL of phosphate-buffered saline and preincubated in a blood culture bottle containing an additional 5 mL of rumen and sheep's blood. The culture bottle was incubated at 37°C for 3 days under anaerobic conditions. On agar plate, colonies were small and translucent, with an average diameter ranging from 0.4 to 0.9 mm. Bacterial cells were Gram positive and ranged in length from 0.5 to 1 mm. Strain Marseille-P3740 was catalase and oxidase negative. The 16S rRNA gene was sequenced using fD1-rP2 primers using a 3130-XL sequencer (Applied Biosciences, Saint Aubin, France) as previously described [3]. Strain Marseille-P3740 exhibited a 96% sequence identity with Collinsella stercoris strain JCM 10641 (GenBank accession no. NR_113164), the phylogenetically closest species with standing in nomenclature (Fig. 1), which putatively classifies it as a member of the genus Collinsella within the family Coriobacteriaceae in the phylum Actinobacteria [4]. This similarity value is below the 16S rRNA gene sequence threshold of 98.65% set by Stackebrandt and Ebers [5] to delineate a new species without carrying out DNA-DNA hybridization. We therefore propose the creation of the new species ‘Collinsella provencensis’ (pro.ven.cen'cis, N.L. fem. adj. provencensis, pertaining to Provence, the region of France where the type strain was isolated). Strain Marseille-P3740T is the type strain of the new species ‘Collinsella provencensis.’
Fig. 1.
Phylogenetic tree showing position of ‘Collinsella provencensis’ strain Marseille-P3740 relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstrap scores of at least 95% were retained. Scale bar indicates 2% nucleotide sequence divergence.
Characteristics of ‘Parabacteroides bouchesdurhonensis’ sp. nov.
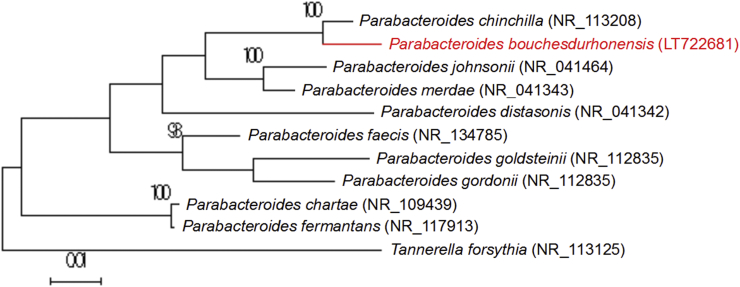
Strain Marseille-P3763 was isolated from a 27-year-old healthy man living in France after 7 days' preincubation in blood culture bottle and then 48 hours' incubation on 5% sheep's blood–enriched Columbia agar (bioMérieux, Marcy l’Etoile, France). Colonies of strain Marseille-P3763 appeared beige, smooth and small, with a mean diameter of 0.4 to 0.8 mm. Bacterial cells were Gram negative and rod shaped, ranging in length from 0.7 to 1.2 mm. Strain Marseille-P3763 was catalase positive and oxydase negative. The 16S rRNA gene was sequenced using fD1-rP2 primers using a 3130-XL sequencer (Applied Biosciences) as previously described [3]. Strain Marseille-P3763 exhibited a 96.7% sequence identity with Parabacteroides chinchillae strain JCM 17104 (GenBank accession no. NR_113208), the phylogenetically closest species with standing in nomenclature (Fig. 2), which putatively classifies it as a member of the genus Parabacteroides within the family Tannerellaceae in the phylum Bacteroidetes. This similarity value is below the 16S rRNA gene sequence threshold of 98.65% set by Stackebrandt and Ebers [5] to delineate a new species without carrying out DNA-DNA hybridization. We thus propose the creation of the new species ‘Parabacteroides bouchesdurhonensis’ (bou.ches.du.rho.nen'sis, N.L. neut. adj. bouchesdurhonensis, pertaining to Bouches du Rhône, the name of the French territory where strain Marseille-P3763 was isolated). Strain Marseille-P3763T is the type strain of the new species ‘Parabacteroides bouchesdurhonensis.’
Fig. 2.
Phylogenetic tree showing position of ‘Parabacteroides bouchesdurhonensis’ strain Marseille-P3763 relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstrap scores of at least 95% were retained. Scale bar indicates 1% nucleotide sequence divergence.
Characteristics of ‘Sutterella seckii’ sp. nov.
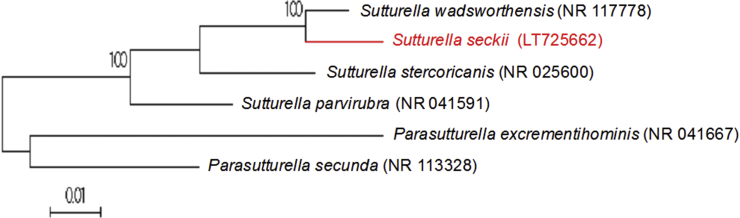
Initial growth was obtained from a stool sample of a 33-year-old healthy man living in France after 3 days' preincubation in blood culture bottle and then 72 hours' incubation on 5% sheep's blood–enriched Columbia agar (bioMérieux) at 37°C in anaerobic conditions. Agar-grown colonies were transparent and flat, with an average diameter between 0.3 and 0.8 mm. Bacterial cells were Gram negative, rod shaped and polymorphic, ranging in length from 0.6 to 0.8 mm. Strain Marseille-P3660 was catalase positive and negative. The 16S rRNA gene was sequenced using fD1-rP2 primers using a 3130-XL sequencer (Applied Biosciences) as previously described [3]. Strain Marseille-P3660 exhibited a 97.5% sequence identity with Sutterella wadsworthensis strain WAL 9799 (GenBank accession no. NR_117778), the phylogenetically closest species with standing in nomenclature (Fig. 3), which putatively classifies it as a member of the genus Sutterella within the family Sutterellaceae in the phylum Firmicutes. This similarity value is below the 16S rRNA gene sequence threshold of 98.65% set by Stackebrandt and Ebers [5] to delineate a new species without carrying out DNA-DNA hybridization. We therefore propose the creation of the new species ‘Sutterella seckii’ (seck.i, N.L. masc. gen. n. seckii, ‘of Seck,’ named after Senegalese PhD student El Hadji Seck). Strain Marseille-P3660T is the type strain of the new species ‘Sutterella seckii.’
Fig. 3.
Phylogenetic tree showing position of ‘Sutterella seckii’ strain Marseille-P3660 relative to other phylogenetically close neighbours. Sequences were aligned using CLUSTALW, and phylogenetic inferences were obtained using maximum-likelihood method within MEGA software. Numbers at nodes are percentages of bootstrap values obtained by repeating analysis 500 times to generate majority consensus tree. Only bootstrap scores of at least 95% were retained. Scale bar indicates 1% nucleotide sequence divergence.
Nucleotide sequence accession number
The 16S rRNA gene sequences of the three species were deposited in GenBank under accession numbers LT722680 (‘Collinsella provencensis’ strain Marseille-P3740), LT722681 (‘Parabacteroides bouchesdurhonensis’ strain Marseille-P3763) and LT725662 (‘Sutterella seckii’ strain Marseille-P3660), respectively.
Deposit in a culture collection
The three bacterial strains described here were deposited in the open Collection de Souches de l’Unité des Rickettsies (CSUR, WDCM 875) under the reference numbers CSUR P3740, P3763 and P3660 respectively for ‘Collinsella provencensis’ strain Marseille-P3740, ‘Parabacteroides bouchesdurhonensis’ strain Marseille-P3763 and ‘Sutterella seckii’ strain Marseille-P3660.
Conflict of interest
None declared.
Acknowledgement
This work has benefited from the French State support, managed by the ‘Agence Nationale pour la Recherche’ including the “Programme d'Investissement d'avenir” under the reference Méditerranée Infection 10-IAHU-03.
References
- 1.Lagier J.C., Khelaifia S., Alou M.T., Ndongo S., Dione N., Hugon P. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol. 2016;1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PubMed] [Google Scholar]
- 2.Seng P., Drancourt M., Gouriet F., La Scola B., Fournier P.E., Rolain J.M. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 3.Drancourt M., Bollet C., Carlioz A., Martelin R., Gayral J.P., Raoult D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. 2000;38:3623–3630. doi: 10.1128/jcm.38.10.3623-3630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kageyama A., Benno Y., Nakase T. Phylogenetic and phenotypic evidence for the transfer of Eubacterium aerofaciens to the genus Collinsella as Collinsella aerofaciens gen. nov., comb. nov. Int J Syst Evol Microbiol. 1999;49:557–565. doi: 10.1099/00207713-49-2-557. [DOI] [PubMed] [Google Scholar]
- 5.Stackebrandt E., Ebers J. Taxonomic parameters revisited: tarnished gold standards. Microbiol Today. 2006;33:152–155. [Google Scholar]