Abstract
Human papillomavirus (HPV) is a common viral infection worldwide associated with a variety of cancers. The integration of the HPV genome in these patients causes chromosomal instability and triggers carcinogenesis. The aim of this study was to investigate the HPV-16 genome physical status in four major cancers related to HPV infection. Formalin-fixed paraffin-embedded blocks from our previous projects on head and neck, colorectal, penile, and cervical cancers were collected, and HPV-16–positive specimens were used for further analysis. The DNA extraction copy number of E2 and E7 genes was calculated by qualitative real-time PCR method. Serially diluted standards that were cloned in PUC57 plasmid were used. Standard curve and melting curve analysis was used for quantification. Of the 672 specimens studied, 76 (11.3%) were HPV-16 positive. We found that 35.6% (16/45) were integrated. Statistical analysis showed that there were significant correlations between integration of HPV-16 and cervical cancer end-stage carcinogenesis (P < .0001), episomal form, and ASCUS lesions (P = .045). Significant correlation in penile cancer patients was seen between the episomal form and high-grade cancer stage (P = .037). Integration is a major factor in the carcinogenesis mechanism of HPV and has different prevalence in various cancers with a higher rate in progression except in penile cancer.
Introduction
Cancer is one of the major public health challenges with a high mortality worldwide. Human papillomavirus (HPV) is a major cause of some human cancers such as cervical, penile, colorectal, esophagus, oropharyngeal, and head and neck cancers [1], [2], [3], [4]. The risk of malignancy in HPV-positive cases is related to infection with specific types of HPVs [1], [5]. HPVs are of numerous types (≥100), and they are divided into two major groups: high risk (HR) and low risk (LR) [6], [7]. The HR types are more often detected in association with cancers such as cervical cancer. The major HR types include HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82, and LR types are 6, 11, 40, 43, 44, and 54. Some other types such as 26, 53, and 66 are defined as probable high-risk types, and 69, 71, and 74 are defined as unknown-risk types [8], [9]. Over 40 HPV types have been detected in genital tract infections, and 12 of them are related to carcinogenesis. HPV-16 is responsible for about 60% of cervical cancers [10].
It seems that HPV DNA of some HR types such as 16, 18, and 31 in cancerous tissue has been integrated to host genome and causes higher production of cancerous cells [11], [12]. In HPV oncogenesis, it is highly accepted that, in the neoplastic lesions, HPV genome is in the integrated form, and in the vegetative cycle inside differentiated epithelial cells of benign lesions, it is in an episomal form. In the integrated form, the viral genome self-replication has been interrupted [13], [14], [15]. In the cancerous cells, HPV genes with E6, E7, and LCR (long control region) have been reported to integrate with the host genome, and the other viral genes are removed. One of the roles of these viral regions is the disruption in the functioning of the major cellular tumor suppressor genes such as Rb and p53; consequently, uncontrolled cell growth occurs [13], [14].
The frequency of HPV genome integration varies based on several risk factors; the HPV types, specific cancers, and other personal or geographical factors could influence the genome integration [16], [17], [18], [19]. For instance, HPV-16 integration has been reported to vary from 21% to 43% in oropharyngeal cancers, with the occurrence in 50%, 56%, and 100% of the Japanese, Pakistani, and Colombian esophageal cancer populations, respectively [17], [20].
The determination of HPV genome status in cancerous and precancerous lesions is very important in tumor progress prognosis and estimation of survival or response to therapy in these patients [15], [17], [21], [22]. Furthermore, in the HPV mix infection, there are no adequate data about the HPV genome status.
In this regard, we aimed to design a preliminary study of HPV-16 genome status investigation in Iranian patients suffering from head and neck squamous cell carcinoma (HNSCC) and colorectal, cervical, and penile cancers.
Materials and Methods
Patients
Patients were selected from our previous projects on HPV survey in different cancers including cervical cancer [23], HNSCC [24], and colorectal and penile cancer (not published) (n=672). The patients were referred to the referral hospitals affiliated to Iran University of Medical Sciences, Tehran, Iran. For the HPV detection, INNOLiPA HPV Genotyping assay and PCR-sequencing methods were used. The included patients had a positive result for HPV-16, and some had a mixed infection by other types of HPVs.
Ethics Statement
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki as reflected in the prior approval by the institution's human research committee. A written, informed consent was obtained from each patient included in the study. All human subjects were adult, and Institutional Review Board approval code was IR.IUMS.FMD.REC 1396.92215402.
Sampling
In this study with a retrospective design, the archived formalin-fixed paraffin-embedded (FFPE) blocks were collected from HNSCC and cervical, colorectal, and penile cancer populations [23], [24]. Clinical databases and patient health records were used for retrieving pathological and clinical data from March 2011 to January 2017. The FFPE blocks were fixed in 10% buffered formalin before paraffin embedment, according to the documentation of the tissue repository.
DNA Extraction
A thin 20-μm tissue section was obtained from the blocks, and deparaffinization was performed with xylene. Then, series of distilled water and graded ethanol solutions were used for rehydration, according to previous works [2], [24], [25]. The DNA purification was performed by using QIAamp DNA FFPE Tissue Kit (QIAGEN, Hilden, Germany), according to the manufacturer's protocol. NanoDrop ND-1000 (Thermo Fisher Scientific Inc., Waltham, MA) spectrophotometry was used for the evaluation of the isolated DNA. The purified DNA was kept at −20°C.
Physical Status of HPV DNA
In the integrated form of HPV DNA, the E6, E7, and LCR regions were found, and the other HPV genes were removed. E2 and E6 or E7 genes quantification is the way to evaluate the HPV integration [13], [14], [26]. In this regard, by the quantitative real-time PCR (qRT-PCR) method, the viral loads of these genes (E2 and E6/E7) can be counted separately to distinguish the episomal, integrated, or a mixture of the two forms. In this study, we used the E2:E7 ratio to define the HPV-16 genome status. If the E2:E7 ratio was 0, the genome was in an integrated form; if >1, it was episomal; and if >0 and <1, it was a mix of the two forms [9], [13], [27].
Real-Time PCR Assay
The Rotor-Gene-Q 6000 thermocycler (Corbett, Australia) was used for HPV-16 E2 and E7 quantification. The total volume of the reaction mix was 15 μl that was composed of 7.5 μl of 2× Amplicon III mix (Odense M, Denmark), 0.5 μl of each forward and reverse primers corresponding to 0.5-μM concentration, and 3 μl each sample or control corresponding to 0.2 to 0.5 μM concentration, and rest of the total volume was obtained by adding distilled water. HPV16-E2 gene forward primer was 5′-TGC CAA CGT TTA AAT GTG TG-3′ (2767-2786), reverse primer was 5′-CGC ATG AAC TTC CCA TAC TT-3′ (3298-3318), and the amplicon length was 551 base pairs; HPV16-E7 gene forward primer was 5′-TGC AAC CAG AGA CAA CTG AT-3′ (605-624), reverse primer was 5′-TGT CTA CGT GTG TGC TTT GT-3′ (768-787), and the amplicon length was 183 base pairs [13]. The E7 gene was heated at 95°C for 5 minutes followed by 40 cycles at 95°C for 30 seconds and 60°C for 30 seconds, and acquiring was in the annealing/extension step. The E2 gene was heated at 95°C for 5 minutes followed by 45 cycles at 95°C for 30 seconds, 58°C for 30 seconds, and 72°C for 30 seconds, and acquiring was in the extension step. Melting curve program (50°C-95°C with a heating rate of 2°C per second and a continuous fluorescence measurement) was used.
RT-PCR standardization and Interpretation
Full-length HPV-16 E2 and E7 genes were cloned with PUC57 plasmid as a standard control for quantification. Cloning was done using a commercially available cloning system (TA Cloning kit of Invitrogen, San Diego, CA) according to manufacturer’s instructions. The serial dilution manner was used for each gene, and DNase RNase-free water was used as the solvent. Efficient standard curve (E = 0.95-1.05) of standards was used for further quantification.
Statistical Analysis
SPSS software version 22 was used for statistical analyses of anatomic and demographic variables. The P values more than .05 were not regarded as statistically significant.
Results
Participants
Based on the previous HPV surveys (n = 672) on patients with colorectal cancer (n = 66), HNSCC (n = 156), cervical cancer (n = 436), and penile cancer (n = 14), we detected 76 (11.3%) HPV-16 positive specimens by INNOLiPA HPV genotyping assay and PCR-sequencing–based methods. Brief details of the HPV investigations are shown in Table 1.
Table 1.
Descriptive Summary of HPV Survey Studies (n=672)
| HNSCC |
CrCa |
PeCa |
CxCa |
|||||
|---|---|---|---|---|---|---|---|---|
| HPV16+ | Total | HPV16+ | Total | HPV16+ | Total | HPV16+ | Total | |
| Total (%) | 2 (1.3) | 156 (23.7) | 3 (4.5) | 66 (10) | 6 (54) | 11 | 65 (14.9) | 436 (66.2) |
| Men (%) | 2 (1.6) | 121 (77.6) | 2 (66.7) | 38 (57.6) | 6 () | 11 | 0 | 0 |
| Women (%) | 0 | 35 (22.4) | 1 (33.3) | 28 (42.4) | 0 | 0 | 65 (14.9) | 436 (100) |
| Mean age±SD | 65.5±12.6 | 60.5±12.6 | 64.3±13.2 | 59.3±14.4 | 35.0±5.5 | 35.8±5.7 | 38.4±13.6 | 35.3±10.1 |
| Range | 54-77 | 28-86 | 50-76 | 27-85 | 28-40 | 26-44 | 23-62 | 18-73 |
| Other HPVsa | - | - | - | - | - | - | 25 (38.5) | 198 (45.4) |
Co-infected with other HPVs. HNSCC, head and neck squamous cell carcinoma; CrCa, colorectal cancer; PeCa, penile cancer; CxCa, cervical cancer; SD, standard deviation.
Totally, 45 HPV-16 positive specimens, based on the clinical status and availability of the samples, were randomly selected for genome status analysis. Brief results of the cytological and histological analysis of the 45 HPV-16 positive cases are shown in Table 2.
Table 2.
Pathologic Characteristics of our HPV-16–Positive Cases (n=45)
| HNSCC (%) | CrCa (%) | PeCa (%) | CxCa (%) | |
|---|---|---|---|---|
| High grade | 1 (50) | 1 (33.3) | 2 (33.3) | - |
| Low grade | 1 (50) | 2 (66.7) | 4 (66.7) | - |
| ASCUS | - | - | - | 8 (23.5) |
| LSIL | - | - | - | 6 (17.6) |
| HSIL | - | - | - | 11 (32.3) |
| Cancerous | - | - | - | 9 (26.5) |
| Total | 2 (4.4) | 3 (6.7) | 6 (13.3) | 34 (75.6) |
ASCUS, atypical squamous cells of undetermined significance; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion.
qRT-PCR
Standard curves were drawn for HPV E7 and E2 genes by using standards diluted five to seven times. The efficiency of each E7 and E2 standard curve was 97% and 117%, respectively. The melting curve analysis confirms our standard interpretations (Supplementary Figure 1). During 40 real-time PCR amplification cycles, there were no primer-dimer generation.
Then, a total of 45 HPV-16 specimens were subjected to qRT-PCR, and the viral load of each of the E2 and E7 genes was reported based on the melting analysis and appropriate positive (standard) and negative controls (Supplementary Figure 2). A negative control (water or DNA extracted from previous works that was negative for HPV) was used in each run, and it did not have nonspecific yielded fluorescence signals above the background.
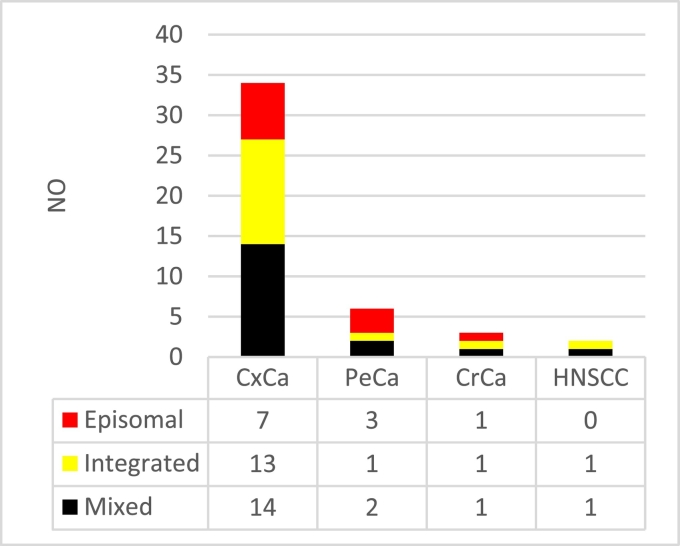
In general, 16 (35.6%) of the 45 specimens had undetectable levels for one or more of the E2 gene, and all of them had E7 viral load ranging from 1,270,709,529 to 1098 copies/μL. For the E2 gene, it was 13,193,565 copies/μL to undetectable levels. The E2:E7 ratio was calculated for the interpretation of the viral genome status (Figure 1). The HPV-16 integrated form was detected in 35.6% (16/45), episomal form in 24.4% (11/45), and mixed form in 40% (18/45) of the specimens. HPV-16 physical genome status investigation details by pathologic status of specimens are shown in Table 3.
Figure 1.
Frequency of HPV-16 genome status in our 45 cancerous patients. HNSCC, head and neck squamous cell carcinoma; CrCa, colorectal cancer; PeCa, penile cancer; CxCa, cervical cancer.
Table 3.
Demographic and Pathologic Genome Status of Our Included Patients (n=45)
| Genome Status | No (%) | Sex | Age Mean ± SD | Grade |
Pathologic Feature |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | ASCUS | LSIL | HSIL | Cancerous | |||||
| HNSCC | Integrated | 1 (50) | Man | 77 | 0 | 1 | - | - | - | - |
| Mixed | 1 (50) | Man | 54 | 1 | 0 | - | - | - | - | |
| CrCa | Episomal | 1 (33) | Man | 76 | 1 | 0 | - | - | - | - |
| Integrated | 1 (33) | Woman | 67 | 0 | 1 | - | - | - | - | |
| Mixed | 1 (33) | Man | 50 | 1 | 0 | - | - | - | - | |
| PeCa | Episomal | 3 (50) | Men | 32.0±6.9 | 3 | 0 | - | - | - | - |
| Integrated | 1 (17) | Man | 40 | 1 | 0 | - | - | - | - | |
| Mixed | 2 (33) | Men | 35.5±2.1 | 1 | 1 | - | - | - | - | |
| CxCa | Episomal | 7 (21) | Women | 30.7±3.4 | - | - | 4 | 2 | 0 | 1 |
| Integrated | 13 (38) | Women | 29.6±3.9 | - | - | 1 | 3 | 5 | 4 | |
| Mixed | 14 (41) | Women | 31.1±10.5 | - | - | 3 | 1 | 6 | 4 | |
Statistical Analysis
The analysis of the different variables showed that there was a significant association between the integration of HPV-16 and end stages of carcinogenesis (higher-stage lesions (HSIL) and cancerous lesions [n = 9]) (P < .0001). This finding suggests that the integration of HPV-16 in high-grade lesions in cervical cancer is more common than the other types of lesions, about 69.2% (9/13) in our study. There was a significant association between HPV-16 episomal form and ASCUS lesions (P = .045). This highlights the primary stage of HPV infection in these patients in which the HPV genome is episomal in the vegetative stage. The analysis of variables from penile cancer patients showed a significant result only in the correlation of the episomal form with high-grade cancer stage (P = .037). The analysis of age with the genome status showed that there was no significant relationship between age and integration of HPV-16 genome. The results for the other variables were not significant (P > .05).
Discussion
HPV is one of the common viral infections, affecting nearly 80 million people currently in the United States (about 25% of the population) [28], [29]. There are several HPV-related cancers including cancer of the cervix, vulva, vagina, penis, anus, rectum, tongue, and tonsils (oropharynx) as well as colorectal cancer [4], [23], [24], [30], [31], [32], [33], [34]. The best described prevention method for HPV infection is vaccination [35], [36], [37]. Oropharyngeal and penile cancers are HPV-related cancers that are more prevalent in men and cervical cancer in women [18], [28], [29], [38]. HPV genome integration plays a crucial role in HPV-related carcinogenesis and can trigger instability in the chromosomes [19], [29]. The severity of the disease after integration of the HPV genome increases and may cause a reduction in the viral load. The E6, E7, and LCR regions of the HPV genome have been reported to be integrated into the cancerous cells, and other genes such as E2 and E1 are eliminated [9], [13], [27]. The viral DNA of HR-HPVs such as HPV-16 and HPV-18 often integrates into the host cell genome and causes overexpression of E6 and E7 regions that disrupt cellular proto-oncogenes such as pRb and p53; consequently, it causes malignancy [21], [39]. The HPV genome status can be used as a prognostic marker in HPV-associated cancers [15], [17], [21], [22]. In this regard, we aimed to investigate the HPV-16 genome status in different HPV-related cancers including HNSCC, colorectal, penile, and cervical cancer specimens which were positive for HPV-16 genome detected by using INNO-LiPA Genotyping and PCR-sequencing methods from the previous studies [23], [24]. Our study was designed to count the HPV-16 E2 and E7 gene viral load using qRT-PCR [13]. Among the 45 HPV-16 infected patient samples analyzed, we found that 11 (24.4%) were episomal, 16 (35.6%) were integrated, and 18 (40%) were mixed form.
A study by Cheung et al. [13] showed that there were integrated, episomal, and mixed forms in 32.4% (11/34), 32.4% (11/34), and 35.3% (12/34) in European and 47% (31/66), 28.8% (19/66), and 24.2% (16/66) in Asian HPV-16–positive cervical specimens, respectively. They found the episomal form in 33.3% of CIN3 and 28% of SCC lesions. They emphasized the challenges in the use of the integration status as an adjunctive marker in the management of HPV-positive patients. In the current study, qRT-PCR was designed for viral load evaluation, and we found that 35.6% of our total specimens and 38% of our cervical specimens had integrated form of HPV-16 genome; furthermore, integration in HSIL lesions was 45.4% (5/11), and in cancerous mass, it was 44.4% (4/9). The statistics showed that there was a significant relationship between the integration of HPV-16 DNA in HSIL and cancerous vs. ASCUS and LSIL) that indicates a higher risk for these patients for cancer progression and chromosomal instability. Our study showed differences between the frequencies of HPV episomal form in cervical specimens similar to that described by Cheung et al. [13] (33% and 28% vs. 5%). HPV episomal forms were seen in only 5% (1/20) of the high-grade cervical cancer specimens (HSIL and cancerous) in our study, which is different from that reported in other studies. This could be due to the impact of other risk factors on the integration status of HPV, such as alcohol consumption or smoking. Meanwhile, there is evidence that indicates that the pure episomal form of HPV associates with some invasive cervical cancers [13], [40], [41]. This association has not been proven by our study data. It may be due to the cancer therapies that were used to suppress the tumor progression. Studies have also shown a substantial proportion of HPV genome integration in low grade and primary stage of HPV infection [13], [42]. We detected 28.6% (4/14) integrated form in ASCUS/LSIL lesions that were in the primary stages. The integration in early stages shows a probable role of other risk factors such as smoking, alcohol consumption, or UV radiation.
A study by Liu et al. [43] of HPV-16–positive cervical carcinoma specimens showed integration in 63% (19/30). We found that the overall integration frequency was 35.6% (16/45), and in cervical specimens, it was 38%. In patients with only cervical cancer, the frequency was 44.4% (4/9), which is different to that mentioned in the report by Liu et al. [43]; this could be due to the environmental, viral, or host factor effects. A study by Bodelon et al. [10] of 38 HPV-16–positive CIN3 (17/38) and cervical cancer (21/38) specimens showed that there were 60.5% (23/38) integrated and 39.5% (15/38) episomal form, respectively. The episomal form was detected in 53.9% (9/17) and 28.6% (6/21) and the integrated form in 47.1% (8/17) and 71.4% (15/21) of CIN3 and cancer patients, respectively. The authors concluded that the instability of chromosomes in the absence of integration might play a crucial role in facilitating integration. Our study showed that 24.4% (11/34) were episomal, 35.6% (16/34) were integrated, and the rest of them were mixed form. These differences may be due to our pooled population of four cancers and the presence of mixed form including episomal and integrated forms. In the pure cervical cancer specimens, the episomal and integrated form frequency was 20.6% (7/34) and 38.2% (13/34), which by using pure HSIL and cancer patients became 5% (1/20) and 45% (9/20), respectively. Our current study showed that the integration occurred more in high-grade and cancerous cervical lesions, 45% (9/20), and it was significant in higher-grade malignant lesions (HSIL and cancerous vs. ASCUS and LSIL). These findings could be related to the different populations or detection methods. It is simple to understand the higher prevalence of integration in the higher stages of cancer due to the higher chromosomal instability in these lesions.
The study by Castillo et al. [20] in Japan, Pakistan, and Colombia of the HPV genome status in patients with tonsil, tongue, other oral cancers, and esophageal carcinoma reported HPV-16 genome integration in 60% (6/10), 80% (16/20), 95% (18/19), and 71% (17/24) and mixed form in 40% (4/10), 10% (2/20), 5% (1/19), and 21% (5/24), respectively; episomal form in tongue mass was 10% (2/20) and in esophagus was 8% (2/24). This emphasizes the frequent integration of HPV-16 genome in SCCs of the oral cavity, oropharynx, and esophagus. Our study showed that the frequency of integration of HPV-16 genome in HNSCC patients is 50% (1/2), this being seen in a patient with tonsil cancer. It confirms the findings of the study by Castillo et al. in three countries. Further study with a larger sample size is needed to obtain comprehensive results. A study by Olthof et al. [44] in 75 patients with HPV-16–positive OSCC analyzed for the physical status of the viral genome showed integration in 39% (29/75) and episomal form in 61% (46/75). Our total integration frequency in four surveys is similar to this study, but in HNSCC patients, it was 50%, and its differences could be due to the limited population in the present study.
The association of HPV with different cancers has been reported frequently. Colorectal cancer is one of the challenging issues, and its association with HPV infection is under investigation [45], [46]. In the study by Bernabe-Dones et al. [45] of 12 colorectal cancer patients infected with HPV-16, the E2 gene evaluation for genome status by nested PCR method found that the genome in all the specimens was in an integrated form. They conclude that the integration of HPV-16 genome into the host genome could be a proof of the probable role of HPV in colorectal carcinogenesis. In the present study, we used the previous study (data not published) specimens that were HPV-16 positive. In a preliminary study, we analyzed the three HPV-16 genomes in colorectal cancer for the physical status and found that 33% (1/3) of them were in an integrated form, 33% (1/3) were episomal, and 33% (1/3) were mixed. Based on the histological and pathological data, 67% (2/3) of the specimens were of high-grade cancerous stage with episomal and mixed forms of HPV-16. Interestingly, integration was seen in low-grade cancerous tissue. This indicates the probable role of HPV in the initial stages of chromosomal instability and its potential to trigger the neoplasia in colorectal cancer patients, although there was no association between the pathologic stage of cancer and the physical status of the HPV genome. Further studies are required to clarify the subject.
Penile cancer is one of the major genital cancers in men that could be multifactorial, with smoking, phimosis, and poor hygiene being some of them. Its association with HPV infection has been investigated, and its frequency has been reported to be 20% to 50% [33], [47], [48]. The study by Djajadiningrat et al. [47] in 212 patients with penile cancer showed that they were infected with the HR HPV types with a frequency of 25%, and of them, 79% were HPV-16 types. Our previous study (data not published) showed the presence of HPV-16 genome in 54% (6/11) of penile cancer patients (6/11). Few studies have reported the HPV genome status in these patients. In the present study, we found the prevalence of the integrated form of HPV-16 in these patients to be 17% (1/6), episomal form 50% (3/6), and mixed form 33% (2/6). It seems that the episomal form of HPV is common in penile cancer patients, and high grade of penile cancer lesions has lower chromosomal instability or insertion of a foreign gene (HPV) compared with that in cervical cancer patients. This finding is different from our understanding about carcinogenesis of HPV infection and needs further analysis for better comprehension.
Our study limitations were the lack of another confirmatory or more accurate method such as TaqMan qRT-PCR to calculate the copy numbers, although relative calculation facilitates our study. For further studies, we recommended using a larger number of specimens and studying the other major HR HPV types’ behavior in the host genome using more demographic data from the history and risk factors involved in the patients.
In conclusion, there is a significant risk of HPV integration in a higher level of carcinogenesis compared with lower stages, although in a group of penile cancer specimens in this study, we found the episomal form to be more common in higher grade of cancer. Higher replication of cancerous and precancerous cells causes chromosomal instability and could facilitate the deletion/insertion of foreign genes such as HPV genome. As we observed in a majority of higher grade of cancerous lesions, the risk of HPV genome integration is inevitable, although other environmental and personal risk factors should not be neglected. The mixed form of HPV genome at cancerous and precancerous levels declares the primary phase of pure integration in the subsequent progressive stages.
The following are the supplementary data related to this article.
Standard curve and melting curve analysis of HPV-16 E7 (part a) and E2 (part b) genes.
qRT-PCR result for some included specimens. (a) Analyses of E7 gene; (b) analyses of E2 gene. Some dimer peaks were for negative results and removed.
Acknowledgement
We are all thankful for the kind assistance of Keyvan Laboratory personnel, especially Mrs. Zohrebandian, and the kind cooperation of Dr. Mahshid Panahi and Dr. Seyed Dawood Mousavi Nasab. This study was a part of the Ph.D. dissertation by the grant number 922154020 in Iran University of Medical Sciences, Tehran, Iran.
References
- 1.de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int J Cancer. 2017;141:664–670. doi: 10.1002/ijc.30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karbalaie Niya MH, Basi A, Koochak A, Safarnezhad Tameshkel F, Rakhshani N. Sensitive high-resolution melting analysis for screening of KRAS and BRAF mutations in Iranian human metastatic colorectal cancers. Asian Pac J Cancer Prev. 2016;17(12):5147. doi: 10.22034/APJCP.2016.17.12.5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lajer CB, Buchwald CV. The role of human papillomavirus in head and neck cancer. APMIS. 2010;118(6-7):510–519. doi: 10.1111/j.1600-0463.2010.02624.x. [DOI] [PubMed] [Google Scholar]
- 4.Jalilvand S, Shoja Z, Hamkar R. Human papillomavirus burden in different cancers in Iran: a systematic assessment. Asian Pac J Cancer Prev. 2014;15(17):7029–7035. doi: 10.7314/apjcp.2014.15.17.7029. [DOI] [PubMed] [Google Scholar]
- 5.Gondim DD, Haynes W, Wang X, Chernock RD, El-Mofty SK, Lewis JS., Jr. Histologic typing in oropharyngeal squamous cell carcinoma: a 4-year prospective practice study with p16 and high-risk HPV mRNA testing correlation. Am J Surg Pathol. 2016;40(8):1117–1124. doi: 10.1097/PAS.0000000000000650. [DOI] [PubMed] [Google Scholar]
- 6.Gelwan E, Malm I-J, Khararjian A, Fakhry C, Bishop JA, Westra WH. Nonuniform distribution of high-risk human papillomavirus in squamous cell carcinomas of the oropharynx: rethinking the anatomic boundaries of oral and oropharyngeal carcinoma from an oncologic HPV perspective. Am J Surg Pathol. 2017;41(12):1722–1728. doi: 10.1097/PAS.0000000000000929. [DOI] [PubMed] [Google Scholar]
- 7.Yahyapour Y, Shamsi-Shahrabadi M, Mahmoudi M, Motevallian A, Siadati S, Shefaii S, Shirvani JS, Mollaie H, Monavari SHR, Keyvani H. High-risk and low-risk human papillomavirus in esophageal squamous cell carcinoma at Mazandaran, Northern Iran. Pathol Oncol Res. 2013;19(3):385–391. doi: 10.1007/s12253-012-9590-0. [DOI] [PubMed] [Google Scholar]
- 8.Dareng E, Ma B, Famooto A, Akarolo-Anthony S, Offiong R, Olaniyan O, Dakum P, Wheeler C, Fadrosh D, Yang H. Prevalent high-risk HPV infection and vaginal microbiota in Nigerian women. Epidemiol Infect. 2016;144(1):123–137. doi: 10.1017/S0950268815000965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abreu AL, Souza RP, Gimenes F, Consolaro ME. A review of methods for detect human Papillomavirus infection. Virol J. 2012;9(1):262. doi: 10.1186/1743-422X-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bodelon C, Vinokurova S, Sampson JN, den Boon JA, Walker JL, Horswill MA, Korthauer K, Schiffman M, Sherman ME, Zuna RE. Chromosomal copy number alterations and HPV integration in cervical precancer and invasive cancer. Carcinogenesis. 2015;37(2):188–196. doi: 10.1093/carcin/bgv171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodelon C, Untereiner ME, Machiela MJ, Vinokurova S, Wentzensen N. Genomic characterization of viral integration sites in HPV-related cancers. Int J Cancer. 2016;139(9):2001–2011. doi: 10.1002/ijc.30243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speel EJM. HPV Infection in Head and Neck Cancer. Springer; 2017. HPV Integration in Head and Neck Squamous Cell Carcinomas: Cause and Consequence; pp. 57–72. [DOI] [PubMed] [Google Scholar]
- 13.Cheung JL, Lo KW, Cheung TH, Tang JW, Chan PK. Viral load, E2 gene disruption status, and lineage of human papillomavirus type 16 infection in cervical neoplasia. J Infect Dis. 2006;194(12):1706–1712. doi: 10.1086/509622. [DOI] [PubMed] [Google Scholar]
- 14.Cheung JL, Cheung T-H, Ng CW, Mei YY, Wong MC, Siu S-SN, Yim S-F, Chan PK. Analysis of human papillomavirus type 18 load and integration status from low-grade cervical lesion to invasive cervical cancer. J Clin Microbiol. 2009;47(2):287–293. doi: 10.1128/JCM.01531-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McBride AA, Warburton A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017;13(4):e1006211. doi: 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Network CGAR Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543(7645):378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ragin C, Modugno F, Gollin S. The epidemiology and risk factors of head and neck cancer: a focus on human papillomavirus. J Dent Res. 2007;86(2):104–114. doi: 10.1177/154405910708600202. [DOI] [PubMed] [Google Scholar]
- 18.Dunne EF, Markowitz LE, Saraiya M, Stokley S, Middleman A, Unger ER, Williams A, Iskander J. CDC grand rounds: reducing the burden of HPV-associated cancer and disease. Morb Mortal Wkly Rep. 2014;63(4):69–72. [PMC free article] [PubMed] [Google Scholar]
- 19.Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P, Kisseljov F, Dürst M, Schneider A, von Knebel Doeberitz M. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68(1):307–313. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
- 20.Castillo A, Koriyama C, Higashi M, Anwar M, Bukhari MH, Carrascal E, Mancilla L, Okumura H, Matsumoto M, Sugihara K. Human papillomavirus in upper digestive tract tumors from three countries. World J Gastroenterol. 2011;17(48):5295. doi: 10.3748/wjg.v17.i48.5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernádi Z, Szarka K, Sápy T, Krasznai Z, Veress G, Póka R. The prognostic significance of HPV-16 genome status of the lymph nodes, the integration status and p53 genotype in HPV-16 positive cervical cancer: a long term follow up. Int J Obstet Gynaecol. 2003;110(2):205–209. [PubMed] [Google Scholar]
- 22.Das P, Thomas A, Kannan S, Deodhar K, Shrivastava SK, Mahantshetty U, Mulherkar R. Human papillomavirus (HPV) genome status & cervical cancer outcome—a retrospective study. Indian J Med Res. 2015;142(5):525. doi: 10.4103/0971-5916.171276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salehi-Vaziri M, Sadeghi F, Hashemi FS, Haeri H, Bokharaei-Salim F, Monavari SH, Keyvani H. Distribution of human papillomavirus genotypes in Iranian women according to the severity of the cervical lesion. Iran Red Crescent Med J. 2016;18(4) doi: 10.5812/ircmj.24458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karbalaie Niya HM, Safarnezhad Tameshkel F, Panahi M, Bokharaei Salim F, Monavari SHR, Keyvani H. Human papillomavirus investigation in head and neck squamous cell carcinoma: initial report from the low risk HPV types associations. Asian Pac J Cancer Prev. 2017;18(9):2573–2579. doi: 10.22034/APJCP.2017.18.9.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koochak A, Rakhshani N, Karbalaie Niya MH, Safarnezhad Tameshkel F, Sohrabi MR, Babaee MR, Bahar B, Imanzade F, Zamani F, Khonsari MR. Mutation analysis of KRAS and BRAF genes in metastatic colorectal cancer: a first large scale study from Iran. Asian Pac J Cancer Prev. 2016;17(2):603–608. doi: 10.7314/apjcp.2016.17.2.603. [DOI] [PubMed] [Google Scholar]
- 26.Olthof NC, Huebbers CU, Kolligs J, Henfling M, Ramaekers F, Cornet I, Lent‐Albrechts JA, Stegmann A, Silling S, Wieland U. Viral load, gene expression and mapping of viral integration sites in HPV16-associated HNSCC cell lines. Int J Cancer. 2015;136(5) doi: 10.1002/ijc.29112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peitsaro P, Johansson B, Syrjänen S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol. 2002;40(3):886–891. doi: 10.1128/JCM.40.3.886-891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 29.Viens LJ. Human papillomavirus–associated cancers—United States, 2008-2012. MMWR Morb Mortal Wkly Rep. 2016;65 doi: 10.15585/mmwr.mm6526a1. [DOI] [PubMed] [Google Scholar]
- 30.Eftekhaar NS, Niya MHK, Izadi F, Teaghinezhad-S S, Keyvani H. Human papillomavirus (HPV) genotype distribution in patients with recurrent respiratory papillomatosis (RRP) in Iran. Asian Pac J Cancer Prev. 2017;18(7):1973. doi: 10.22034/APJCP.2017.18.7.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghasemian E, Monavari S, Irajian GR, Jalali Nodoshan M, Roudsari RV, Yahyapour Y. Evaluation of human papillomavirus infections in prostatic disease: a cross-sectional study in Iran. Asian Pac J Cancer Prev. 2013;14(5):3305–3308. doi: 10.7314/apjcp.2013.14.5.3305. [DOI] [PubMed] [Google Scholar]
- 32.Salehi-Vaziri M, Sadeghi F, Alamsi-Hashiani A, Haeri H, Monavari SH, Keyvani H. Merkel cell polyomavirus and human papillomavirus infections in cervical disease in Iranian women. Arch Virol. 2015;160(5):1181–1187. doi: 10.1007/s00705-015-2368-4. [DOI] [PubMed] [Google Scholar]
- 33.Salehi-Vaziri M, Sadeghi F, Bokharaei-Salim F, Younesi S, Alinaghi S, Monavari SH, Keyvani H. The prevalence and genotype distribution of human papillomavirus in the genital tract of males in Iran. Jundishapur J Microbiol. 2015;8(12) doi: 10.5812/jjm.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yahyapour Y, Shamsi-Shahrabadi M, Mahmoudi M, Siadati S, Shahryar SS, Shokri-Shirvani J, Mollaei H, Monavari SHR. Evaluation of human papilloma virus infection in patients with esophageal squamous cell carcinoma from the Caspian Sea area, north of Iran. Asian Pac J Cancer Prev. 2012;13(4):1261–1266. doi: 10.7314/apjcp.2012.13.4.1261. [DOI] [PubMed] [Google Scholar]
- 35.Hildesheim A, Gonzalez P, Kreimer AR, Wacholder S, Schussler J, Rodriguez AC, Porras C, Schiffman M, Sidawy M, Schiller JT. Impact of human papillomavirus (HPV) 16 and 18 vaccination on prevalent infections and rates of cervical lesions after excisional treatment. Am J Obstet Gynecol. 2016;215(2):212.e211–212.e215. doi: 10.1016/j.ajog.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stokley S, Jeyarajah J, Yankey D, Cano M, Gee J, Roark J, Curtis R, Markowitz L. Human papillomavirus vaccination coverage among adolescents, 2007-2013, and postlicensure vaccine safety monitoring, 2006-2014—United States. Morb Mortal Wkly Rep. 2014;63(29):620–624. [PMC free article] [PubMed] [Google Scholar]
- 37.Beachler DC, Kreimer AR, Schiffman M, Herrero R, Wacholder S, Rodriguez AC, Lowy DR, Porras C, Schiller JT, Quint W. Multisite HPV16/18 vaccine efficacy against cervical, anal, and oral HPV infection. J Natl Cancer Inst. 2015;108(1):djv302. doi: 10.1093/jnci/djv302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dahlstrom KR, Sturgis EM. Cancers in People with HIV and AIDS. Springer; 2014. Other HPV-Associated Cancers (Oropharyngeal and Penile) pp. 289–297. [Google Scholar]
- 39.Kim SH, Koo BS, Kang S, Park K, Kim H, Lee KR, Lee MJ, Kim JM, Choi EC, Cho NH. HPV integration begins in the tonsillar crypt and leads to the alteration of p16, EGFR and c-myc during tumor formation. Int J Cancer. 2007;120(7):1418–1425. doi: 10.1002/ijc.22464. [DOI] [PubMed] [Google Scholar]
- 40.Ho C-M, Chien T-Y, Huang S-H, Lee B-H, Chang S-F. Integrated human papillomavirus types 52 and 58 are infrequently found in cervical cancer, and high viral loads predict risk of cervical cancer. Gynecol Oncol. 2006;102(1):54–60. doi: 10.1016/j.ygyno.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 41.Arias-Pulido H, Peyton CL, Joste NE, Vargas H, Wheeler CM. Human papillomavirus type 16 integration in cervical carcinoma in situ and in invasive cervical cancer. J Clin Microbiol. 2006;44(5):1755–1762. doi: 10.1128/JCM.44.5.1755-1762.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagao S, Yoshinouchi M, Miyagi Y, Hongo A, Kodama J, Itoh S, Kudo T. Rapid and sensitive detection of physical status of human papillomavirus type 16 DNA by quantitative real-time PCR. J Clin Microbiol. 2002;40(3):863–867. doi: 10.1128/JCM.40.3.863-867.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Lu Z, Xu R, Ke Y. Comprehensive mapping of the human papillomavirus (HPV) DNA integration sites in cervical carcinomas by HPV capture technology. Oncotarget. 2016;7(5):5852. doi: 10.18632/oncotarget.6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olthof NC, Speel E-JM, Kolligs J, Haesevoets A, Henfling M, Ramaekers FC, Preuss SF, Drebber U, Wieland U, Silling S. Comprehensive analysis of HPV16 integration in OSCC reveals no significant impact of physical status on viral oncogene and virally disrupted human gene expression. PLoS One. 2014;9(2):e88718. doi: 10.1371/journal.pone.0088718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bernabe-Dones RD, Gonzalez-Pons M, Villar-Prados A, Lacourt-Ventura M, Rodríguez-Arroyo H, Fonseca-Williams S, Velazquez FE, Diaz-Algorri Y, Lopez-Diaz SM, Rodríguez N. High prevalence of human papillomavirus in colorectal cancer in Hispanics: a case-control study. Gastroenterol Res Pract. 2016;2016 doi: 10.1155/2016/7896716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen H, Chen XZ, Waterboer T, Castro FA, Brenner H. Viral infections and colorectal cancer: a systematic review of epidemiological studies. Int J Cancer. 2015;137(1):12–24. doi: 10.1002/ijc.29180. [DOI] [PubMed] [Google Scholar]
- 47.Djajadiningrat RS, Jordanova ES, Kroon BK, van Werkhoven E, de Jong J, Pronk DT, Snijders PJ, Horenblas S, Heideman DA. Human papillomavirus prevalence in invasive penile cancer and association with clinical outcome. J Urol. 2015;193(2):526–531. doi: 10.1016/j.juro.2014.08.087. [DOI] [PubMed] [Google Scholar]
- 48.Stratton KL, Culkin DJ. A contemporary review of HPV and penile cancer. Oncology. 2016;30(3):245–249. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Standard curve and melting curve analysis of HPV-16 E7 (part a) and E2 (part b) genes.
qRT-PCR result for some included specimens. (a) Analyses of E7 gene; (b) analyses of E2 gene. Some dimer peaks were for negative results and removed.