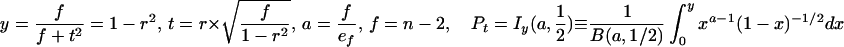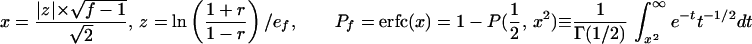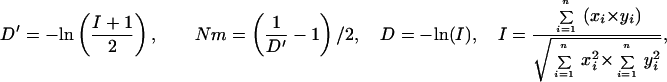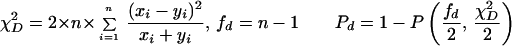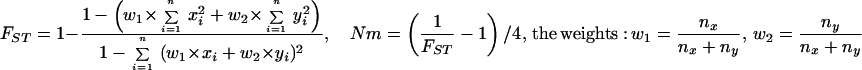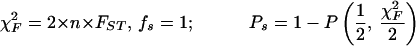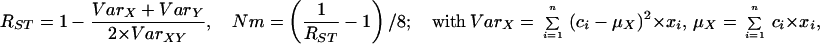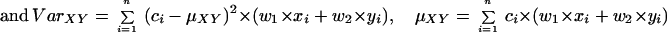Abstract
Genetic distance measurements are an important tool to differentiate field populations of disease vectors such as the mosquito vectors of malaria. Here, we have measured the genetic differentiation between Anopheles arabiensis and Anopheles gambiae, as well as between proposed emerging species of the latter taxon, in whole genome scans by using 23–25 microsatellite loci. In doing so, we have reviewed and evaluated the advantages and disadvantages of standard parameters of genetic distance, FST, RST, (δμ)2, and D. Further, we have introduced new parameters, D′ and DK, which have well defined statistical significance tests and complement the standard parameters to advantage. D′ is a modification of D, whereas DK is a measure of covariance based on Pearson's correlation coefficient. We find that A. gambiae and A. arabiensis are closely related at most autosomal loci but appear to be distantly related on the basis of X-linked chromosomal loci within the chromosomal Xag inversion. The M and S molecular forms of A. gambiae are practically indistinguishable but differ significantly at two microsatellite loci from the proximal region of the X, outside the Xag inversion. At one of these loci, both M and S molecular forms differ significantly from A. arabiensis, but remarkably, at the other locus, A. arabiensis is indistinguishable from the M molecular form of A. gambiae. These data support the recent proposal of genetically differentiated M and S molecular forms of A. gambiae.
Many major infectious diseases, such as malaria, leishmaniasis, and sleeping sickness, are transmitted by insect vectors. Molecular genetic markers have become powerful tools for elucidating the population biology and evolution of such vectors, topics that are highly relevant to disease transmission in the field (1–4). Genetic variation in vector populations contributes to their susceptibility to infection by the pathogen, their degree of anthropophily, their daily survival and reproductive rates, and the epidemiology of the disease in the human host (5). A case in point is the African mosquito of the Anopheles gambiae (sensu latu) complex (5). These include the most important vector of human malaria, A. gambiae (sensu strictu), as well as closely related species that are significant vectors in specific areas (e.g., Anopheles arabiensis) or are altogether unable to serve as vectors (Anopheles quadriannulatus). Furthermore, even within A. gambiae s.s., cytologically defined chromosomal forms (e.g., Mopti, Savanna, and Bamako) are reproductively isolated in the northern dry areas of West Africa, including Mali and Burkina Faso, and may represent emerging species with different disease transmission characteristics (5, 6). Although many DNA regions have been recently analyzed to examine genetic differentiation within A. gambiae s.s, the only fixed molecular differences found so far that consistently discriminate chromosomal forms are in the X-linked ribosomal (r)DNA region (1–4, 7). In Mali and Burkina Faso, these markers distinguish Mopti from Savanna and Bamako chromosomal forms; however, when the analysis is extended to additional populations in West Africa, two nonpanmictic units are identified even in the absence of chromosomal differentiation. This observation recently led to the definition of “molecular forms M and S” (1) or “molecular types I and II” (2), on the basis of fixed differences in the intergenic spacer or internal transcribed spacer rDNA regions, respectively. Because the repetitive nature of rDNA raises doubt as to its reliability as a marker of incipient speciation processes, much interest is now focused on possible new evidence of genetic distinctness between the forms/types.
Among molecular genetic markers, highly polymorphic microsatellites have been used extensively for population studies in humans (8), mammals (9), fruit flies (10), and anopheline mosquitoes (11–13). Various statistical models have been proposed for evaluating genetic differentiation (14–17), but additional theoretical and empirical comparisons regarding their efficacy would be helpful. For microsatellites, FST and D (14) are closely tied to the infinite allele model of mutation (IAM), where each mutation can produce an allele of any size (18). RST (16) and (δμ)2 (15) are related to the stepwise-mutation model (SMM), which assumes that each allele mutates to either one of the immediately neighboring alleles with equal probability (19).
The standard genetic distance D (14) is an often used and popular parameter for classification and evolutionary studies. It was originally defined as an average value over all loci examined, but it can also be defined at each locus separately. Several variations of D have been used, for example, DC (20), DA, Dm (14), DSW (17), and DLR (9). In a bear study (9), D and DLR were comparably satisfactory but failed to resolve the most distantly related pairs of species: when loci have no alleles shared between two populations, D and DLR are not defined or, as has been proposed by Nei (14), take an infinite value that is problematical for any quantitative comparison. As part of our ongoing studies of A. gambiae taxa and populations, here we compare the performance of presently used parameters of genetic distance [e.g., D, FST, RST, and (δμ)2], and we introduce and compare new parameters, D′ and DK. By using a battery of four parameters (FST, RST, D′, and DK), we identify intriguing differences in genetic distance between A. arabiensis and the M and S molecular forms of A. gambiae, at loci representing different chromosomal regions.
Materials and Methods
Origin of Mosquitoes.
Field-collected female mosquitoes were species-identified with molecular markers (21). A total of 268 A. gambiae were collected in July 1996 in Mali, West Africa: 95 from Selenkenyi (Sel) and 92 and 81 from Soulouba (Soul) and Kokouna (Kn). Twenty of the 81 A. arabiensis were collected from the same villages in Mali at the same time as A. gambiae (1, 4, and 15 from Sel, Soul, and Kn, respectively). The remaining 61 A. arabiensis mosquitoes were collected from Kilifi, Kenya, in June 1998. A. gambiae mosquitoes from the villages Sel and Soul were also subjected to karyotyping on the basis of polytene chromosome inversions, but because of technical limitations, only 28, 24, and 11 mosquitoes were identified definitively as Mopti, Savanna, and Bamako (6). Use of a PCR restriction fragment length polymorphism marker (7) unambiguously classified the A. gambiae specimens as M or S molecular forms, with an efficiency of 91%. All mosquitoes were genotyped at microsatellite loci by previously described high-throughput methods (22). All 81 available A. arabiensis were used for Figs. 1–3. Because some parameters are sensitive to differences in sample size, we introduced sample weights for FST and partly for RST (Table 1) and also used a number of A. gambiae comparable to that of A. arabiensis. The percentages of M and S molecular form A. gambiae were 73/27 in Sel, 7/93 in Soul, and 17/83 in Kn, respectively. Figs. 1A and 2A are based on all A. gambiae from Sel; Figs. 1B, 2B, and 3 are based on all M- and S-form mosquitoes from Sel and Soul and an additional individuals 36 from Kn to make the sample sizes comparable.
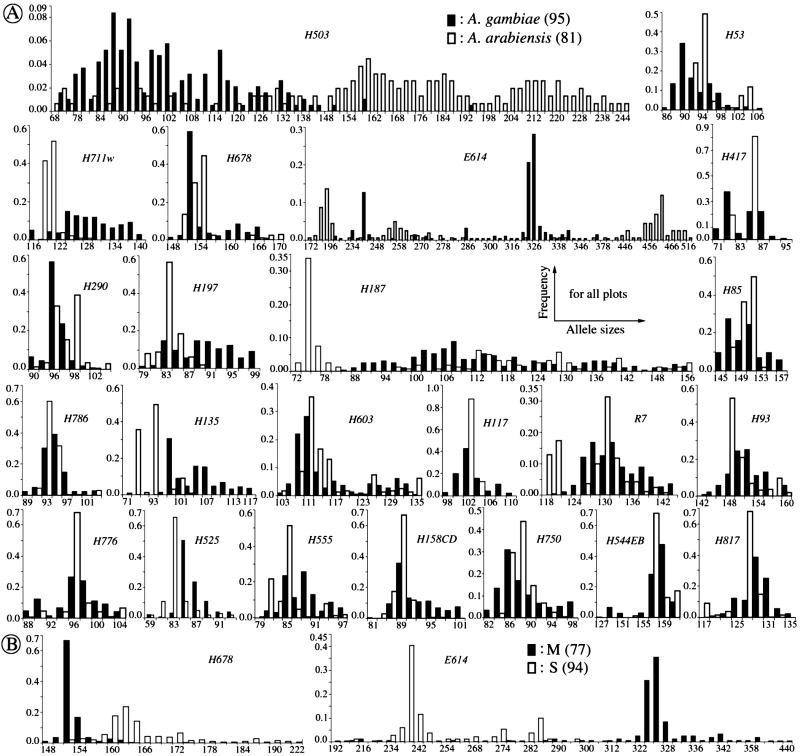
Figure 1.
Comparison of frequencies of allele sizes at 23 microsatellite loci, in 95 A. gambiae and 81 A. arabiensis mosquitoes (A), as well as at two loci in 77 M- and 94 S-form A. gambiae (B). Because of space limitations, allele spacing has been shortened, and alleles at tails have been combined. The data are presented in full with helpful color views on our web site (http://www.embl-heidelberg.de/ExternalInfo/kafatos/publications/PROG/).
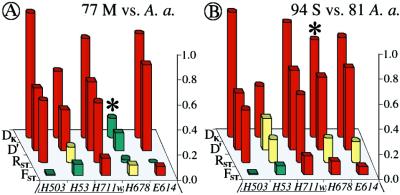
Figure 3.
Comparison of 77 M and 94 S molecular form A. gambiae with 81 A. arabiensis (A and B, respectively) at five X-linked loci, both within and proximal to the Xag inversion. Compare to Fig. 2B.
Table 1.
Measures of genetic differentiation
| DK: Covariance of the deviation of allele frequency (IAM): direct statistical significance tests (Pt, Pf) | ||
| ||
| ||
| ||
| D′: Covariance of allele frequency (IAM): indirect statistical significance tests (Pd) | ||
| ||
| ||
| FST: Variance of allele frequency (IAM): direct statistical significance tests (Ps) | ||
| ||
| ||
| RST: Variance of allele size (SMM): indirect statistical significance test (Nm) | ||
|
Statistical concepts, emphasis, and significance tests for four parameters of genetic differentiation. Consider two populations X and Y with nX and nY individuals, and let xi and yi denote the frequencies of the ith (i = 1, … , n) allele in populations X and Y, respectively, μ1 and μ2 are the mean allele frequencies, μX, μY are the mean allele sizes, and VarX and VarY are the variance of allele sizes in populations X and Y, respectively. The total number of alleles existing at a locus in both populations conbined is n, and alleles are numbered consistently in both populations. DK:r is Pearson's correlation coefficient that varies from −1 to 1. Fisher's z-transformation value is z, and erfc(x) is the complementary error function. The degree of freedom is f, and ef is the integer rounded upwards when f is divided by 10. The probabilities Ptand Pf are based on IY(a, 1/2) and P(1/2, x2), the specific incomplete β and Γ functions, respectively, whereas B(a,1/2) and Γ(1/2) are the actual β and Γ functions, respectively: D′. The same linear transformation as for (r + 1)/2 connects the genetic identity I to (I + 1)/2); correspondingly, Nei's D (14) is transformed to D′. pdis the probability of a χ2 test with the degree of freedom fd. FST, RST: Parameters are as previously defined (see Material and Methods). With both populations combined, μXY and VarXY are the mean and variance of allele sizes, respectively, and w1 and w2 represent the fraction of individuals in the two populations. For FST, PS is the probability of a χ2 test with the degree of freedom fS. Nm is the effective migration rate estimated from the values of DK, D′, FST, and RST. N is the effective population size and m the migration rate. Note that the statistical tests are direct for DK and FST(they include r and FST values) but only indirect for D′ and RST. The potential range of values for DK is 0 to +∞, for D′, 0 to 0.693, for FST, 0 to 1, and for RST, −∞ to +∞.
Figure 2.
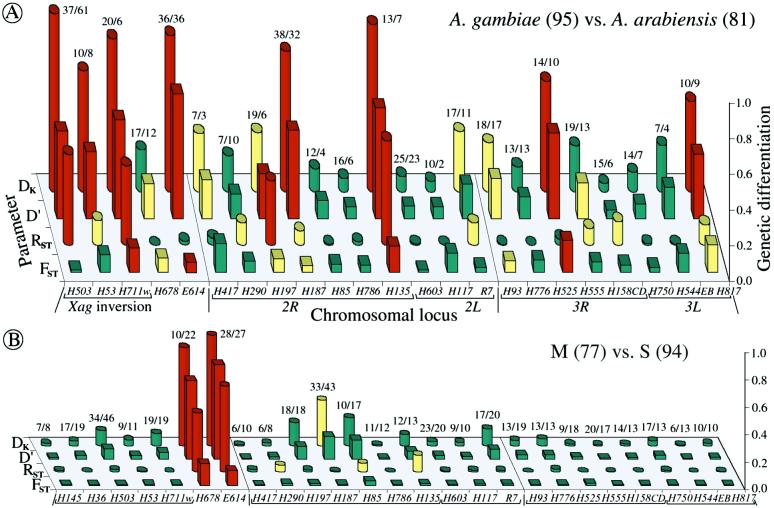
Genetic differentiation at 23 microsatellite loci across the genome on the basis of FST, RST, D′, and DK. A compares A. gambiae and A. arabiensis, whereas B compares the M and S molecular forms of A. gambiae at 25 loci, the first two of which cannot be amplified for A. arabiensis. Note that the two A. gambiae forms are practically indistinguishable except at loci H678 and E614, where they are clearly distinct (see Fig. 1B). The numbers of sampled alleles are shown at each locus. Bars represent genetic distance values in red (“clearly different”), yellow (“marginal”), or green (“indistinguishable”), according to the following criteria. For DK, Pt and Pf are at >10%, between 2.5 and 10% or at <2.5% probability, respectively; for D′ and FST, Pd and Ps are at <2.5%, between 2.5 and 10% or at >10% probability, respectively; for RST, the value of Nm is <0.5, between 0.5 and 3 or >3, respectively.
Statistical Parameters and Significance Tests.
We have introduced DK as a normalized measure of differentiation on the basis of Pearson's correlation coefficient, r, which considers the distribution of alleles in two populations around their respective mean allele frequency (Table 1). Depending on the degree of freedom f, two direct statistical significance tests, Pt and Pf, can be applied. Pt is a modified version of Student's t test, which was originally introduced by Gosset in 1908 (23) to evaluate the difference between two means. However, it can also be used to evaluate the covariance of allele frequencies in two populations around their mean frequencies, which are assumed to be identical. The null hypothesis r = 0 supposes, with regard to population comparisons, that two analyzed populations are independent (23–25). In fact, Student's t test is related to the β function, and t serves only as an intermediate parameter; the parameter actually tested is y = 1 − r2 in the specific incomplete β function Iy (a, 1/2). A condition imposed originally on the t test is that the degree of freedom f is not large, ≈30–60 (23). However, the polymorphism of microsatellites is large and variable between loci; the degree of freedom f varied from 5 to 79 when comparing A. gambiae and A. arabiensis (see below). We have introduced a necessary modification, defining a not as f/2 but as f/ef, where ef is the integer corresponding to f/10 rounded upwards. For example, ef is 2 for 10 < f ≤ 20. Pt is the probability that the null hypothesis holds: two compared populations are certainly independent if Pt = 1 and indistinguishable if Pt < 0.05.
A different approach and significance estimate of r was proposed by Fisher, in particular to analyze statistical correlation in data with small degrees of freedom (23). The two populations are treated as measures of the same entity, and a complementary error function erfc(x) is used to quantify the deviation (or error) of the two data sets. erfc(x) is based on Fisher's z-transformation, which associates each measured r with a corresponding z. Similar to the t test, we have introduced a modified coefficient ef to extend the range of f even below 10. The significance level Pf, at which the null hypothesis (r = 0) holds, is given by erfc(x) (23), which is related to the specific incomplete Γ function P(1/2, x2). It should be noted that the significance tests address the null hypothesis of complete independence in the case of DK (r = 0) and the null hypothesis of identity in the case of D′, FST, and RST.
The standard genetic distance D was defined by Nei (14) as the negative logarithm of the genetic identity I, which also reflects allele frequencies; I ranges from 1 when the two populations have identical allelic frequencies to zero when they share no alleles. In this paper, we introduce a modified D′ based on the same linear transformation we have used for DK, (I + 1)/2 (Table 1). Several indirect statistical significance tests have been proposed for D, and we adopt the χ2 test for allele frequency differences at each locus (14, 26). Pd is the probability that the null hypothesis (D′ = 0) holds: if Pd = 1, the observed and expected (e.g., the two compared) populations are certainly the same.
On the basis of IAM and the statistical significance tests, the effective migration rate Nm can be estimated from the values of D′ and DK (Table 1). When these values are high, Nm becomes much smaller than 1, indicating that no gene flow is occurring between the populations.
The well-known parameter FST defined by Wright (14) and elaborated by Nei (14) measures the degree of genetic differentiation between two populations by using allele frequencies; Goldstein's (δμ)2 (15) is the square of the difference between mean allele sizes, and Slatkin's RST (16) focuses on the variance of allele sizes rather than frequencies (Table 1). A direct statistical significance test for FST is the contingency χ2 test (27, 28), which includes the value of FST and n (which for microsatellites is the number of total alleles in both populations). Ps is the probability that the null hypothesis (FST = 0) holds: if Ps = 1, the two populations are certainly the same. A statistical significance test of (δμ)2 is not available. For RST, the estimated value of Nm is used as an indirect test (16); in this study, Nm ≤ 0.5 is taken to indicate that no statistically significant gene flow occurs between the two populations, whereas Nm ≥ 3 indicates that the two compared populations are indistinguishable.
Results and Discussions
Statistical Parameters.
Genetic differentiation of populations on the basis of microsatellites is often measured by using one of four standard parameters, D, FST, RST, and (δμ)2. It is difficult to select a single adequate measure of differentiation (8, 9) because of uncertainly concerning the underlying mutation processes (IAM and SMM). Furthermore, it can be argued a priori as well as empirically from the literature that different parameters have different drawbacks. In a human evolution study, two parameters based on SMM, RST, and (δμ)2, gave results very different from those recognized from other genetic evidence (8). Although the SMM is often considered more appropriate for microsatellite loci, it appears that their mutational patterns can be often irregular (29); in a honeybee study, IAM produced a better overall fit than SMM (30). As recommended (11, 16), it is prudent to measure differentiation with parameters based on both models. A priori, the least satisfactory parameter is (δμ)2, because it is based on the differences between means, ignoring the allele distribution in the data sets, and has no defined statistical significance test. RST focuses on the variance of allele sizes and, if the distribution is not normal, RST can minimize inappropriately the differences between quite disparate populations that happen to approach the same mean size; the value of RST will then approach zero.
FST is based on the analysis of variance of allele frequencies. An advantage of FST is that it can be weighed to take sample size differences into account. We have introduced a similar partial weighing for RST to accommodate data from samples of different size (Table 1). A human evolution study (8) concluded that FST is the best parameter when compared with RST, (δμ)2, and DSW. A disadvantage of FST might be uncertainty concerning the statistical significance tests, of which four have been used over several decades (27, 28, 31–33). In mosquito studies, the contingency χ2 test is commonly used with the degree of freedom fixed to 1 when comparing two populations.
The standard genetic distance D is based on the analysis of covariance of allele frequencies. It and several proposed variants can fail to resolve distant relationships if loci have no shared alleles. To address this problem and further limitations of these measures (see Materials and Methods), we have introduced a linear transformation of D, D′ (Table 1), which has a defined value (−ln 0.5 = 0.693) when no alleles are shared. A χ2 test of allele frequencies can evaluate the similarity of two populations and serve as an indirect test for D′. It uses the actual degree of freedom to define the statistical significance levels (Table 1) and, in this respect, represents an improvement over the contingency χ2 test used for FST.
We have introduced a new parameter DK (Table 1 and Materials and Methods) that uses Pearson's correlation coefficient r, a well-established measure of correlation in statistics. DK is based on the analysis of covariance of the deviations of allele frequencies around the mean frequency. Importantly, its statistical significance can be tested directly in a robust manner by two mathematically distinct tests of significance. As is true for FST and RST, D′ and DK can also be used to determine the effective migration rate Nm between populations, permitting the detection of gene flow.
Analysis of Mosquito Microsatellite Data with Four Parameters.
We have studied genetic differentiation between A. gambiae and A. arabiensis field populations on the basis of a systematic whole-genome scan. Microsatellite data were collected from 23 different chromosomal loci (25 for A. gambiae alone) across the genome (Figs. 1 and 2). This and a larger analysis, to be reported elsewhere, extending to the more distantly related species Anopheles merus and Anopheles melas (34), showed that the two most commonly used parameters for mosquito studies (11–13), FST and RST, can lead to significantly different results at several loci. After extensive trials of multiple parameters, we came to recommend the use of a panel of four parameters, also including D′ and DK, for the analysis of population biology and evolution by using microsatellites. Additional parameters gave no significant advantage. For example, (δμ)2 failed in our study by showing an unreasonably wide range of values (across 8 orders of magnitude). Software was developed to calculate all of the parameters mentioned in this paper, as well as to support additional useful calculations, for example, observed and expected heterozygosity, Wright's FIS and FIT (14), etc. This software is available on our web site (http://www.embl-heidelberg.de/ExternalInfo/kafatos/publications/PROG/).
The allele distributions in these collections of A. gambiae and A. arabiensis are plotted in Fig. 1A, and the genetic differentiation values at each locus are shown in the bar graph of Fig. 2A. For a visual display of statistical significance, the bars are colored: red, yellow and green indicate loci where the two compared populations are significantly different, marginal in terms of similarity or clearly similar (indistinguishable), respectively.
It is worth noting from Fig. 1 that, at many loci, the allele frequencies follow decidedly not normal distributions, which in some cases are bimodal; this is especially true for A. arabiensis, even for mosquito collections from the same region (data not shown). In many cases, visual comparison of the allele distributions can serve as a common-sense test for the efficacy of the four parameters in detecting obvious differences in allele distribution in the two species. Thus, four of the five sex-linked loci, H503, H53, H711w, and E614, have clearly disparate allele distributions (Fig. 1A), and all are scored as statistically different in the two species by both DK and D′ (Fig. 2A). In contrast, only one of these loci, H711w, is scored as significantly different by both RST and FST. At two other loci with very high polymorphism, H503 and E614, RST and FST give exactly opposite results. Evidently, at these four loci of the X-chromosome, the use of multiple parameters, and DK and D′ in particular, is highly advantageous for detecting clear differences.
Interspecies differences are less prominent among the 18 autosomal loci (Fig. 2A). Only five of these show differences that are validated as statistically significant by two or more parameters. In one of these loci (H135), all four parameters indicate a statistically significant difference; in three loci (H197, H187, and H817), two parameters indicate a significant and two a marginal difference, and in the fifth locus (H525), three parameters detect a clear difference, but RST indicates identity. It may be relevant that in H525, 29 of 81 A. arabiensis gave null alleles; these alleles were evidently mutated in a primer sequence and suggest that this locus may indeed be differentiated in the two species.
It is interesting to see how concordant are the three parameters that are based on the same mutation model, IAM (Fig. 2A). DK and D′ are nonconcordant at only four loci (three marginal/indistinguishable, and one marginal/different). In contrast, D′ and DK are each nonconcordant with FST at seven loci, at two of which FST gives opposite results (significantly different/indistinguishable). Failure of FST to detect clear differences often occurs when allele numbers are either very large (H503, H187) or quite small (H53). However, at E614, despite the large number of alleles, FST is able to detect a clear disparity between the species. The availability of two independent statistical tests for DK proved valuable: both Pt and Pf show the same results for 10 < f < 40. Fisher's Pf should be used for f ≤ 10 and also appears more suitable for f ≥ 40.
Two biologically important conclusions emerge from this analysis: that the X chromosome shows substantially greater disparities between A. gambiae and A. arabiensis than do the autosomes and, in particular, that all three microsatellite loci that map to the Xag inversion of the X chromosome show large differences in allele frequency distribution. In fact, two additional A. gambiae microsatellite loci within this inversion, H145 and H36, could not be amplified in any of the 81 A. arabiensis (data not shown), reinforcing the conclusion of substantial molecular differences between the two species in this larger inversion. The inversion is present in A. gambiae but absent in A. arabiensis. These data are consistent with the observation that the effective migration rate (and estimated gene flow) Nm between A. gambiae and A. arabiensis is lower on the X as compared with the autosomes (12); they lend support to the notion of Coluzzi and coworkers that fixed inversion polymorphisms that discriminate between species of the A. gambiae complex are ancient and associated with local genetic divergence (5, 35).
It is thought that A. gambiae s.s. actually encompasses two or more emerging species, and we examined whether these taxa show different microsatellite profiles. The Mopti, Savanna, and Bamako chromosomal forms can be distinguished by their patterns of chromosomal inversions in the northern drier areas of West Africa (5, 6), but in the more humid southern coastal areas, the Forest chromosomal form is prevalent, and fixed differentiation at the rDNA locus, outside the Xag inversion, is a more robust indicator of two nonpanmictic molecular forms, M and S (1). Molecular typing of our samples yielded 77 M and 94 S individuals of A. gambiae, which were compared directly (Figs. 1B and 2B).
Interestingly, the M and S molecular forms were largely indistinguishable by microsatellites across the genome, except at the base of the X, outside the Xag inversion, where the two forms were unambiguously different according to all four parameters, at both H678 and E614 (cytogenetic divisions 5D and 6, respectively). At H678, most M mosquitoes have short alleles, and S have long alleles, whereas the opposite is true at E614 (Fig. 1B). The rDNA molecular marker distinguishing the M and S forms lies in the same region around cytogenetic division 6 (F. H. Collins, personal communication). Differentiation of the M and S forms on the basis of the tandemly repetitive rDNA locus alone could be ascribed to concerted evolution (1, 2), but the additional observation of clear differences at two nearby microsatellite loci provides strong evidence that the M and S forms are indeed genetically differentiated. Thus, our results to date lend strong support to the concept of emergent M and S taxa of A. gambiae s.s., which are of major taxonomic significance for studying the hypothesized incipient speciation process for which A. gambiae is a uniquely favorable model. Our results provide microsatellite tools to distinguish these forms, at least in Mali. In a preliminary analysis, we have obtained and genotyped 28 and 24 mosquitoes that were karyotyped as Mopti and Savanna, respectively. The results revealed that Mopti differs from Savanna at these two loci in the same way that M differs from S (data not shown); this is not surprising, as all Mopti are M and all Savanna are S in Mali (1).
A remarkable observation came from separate comparisons of M and S forms of A. gambiae with A. arabiensis (Fig. 3). Like the original pooled sample of A. gambiae (Fig. 2A), M-form mosquitoes are clearly different from A. arabiensis within the Xag inversion and at locus E614 but resemble A. arabiensis at locus H678. In sharp contrast, S-form mosquitoes are very clearly different from A. arabiensis in locus H678 as well. This observation raises the interesting possibility of introgression between A. gambiae (M) and A. arabiensis in cytogenetic division 5, where H678 maps. More extensive studies will be necessary to follow up this possibility, as well as to explore further the apparent mosaicism of the autosomes with respect to localized A. gambiae/A. arabiensis differences (36).
Field studies of genetic differentiation within vector populations can yield important information relating to evolution and population biology. Such studies are fundamentally important for understanding the epidemiology of malaria in Africa, where A. gambiae is, overall, the most important vector of the disease. Our work points out the advantages of a systematic whole-genome scan with a larger number of microsatellite loci for detecting chromosomally localized genetic differentiation in field populations. It is notable that this systematic study has detected two genetic differences at microsatellite loci, despite the failure of several previous attempts to find molecular markers specific for the M and S molecular forms in regions different from the rDNA locus (1–4, 7). Systematic genotyping is greatly facilitated by high-throughput methods (22). We have found it is important to subject the data to analysis with multiple parameters of genetic differentiation, including those that correspond to different mutational models. We have offered the modified D′ parameter and the new normalized parameter DK to complement the parameters FST and RST, which are most commonly used in this field. The diversity of allele profiles at different loci, including nonnormal allele distributions with very high and low levels of polymorphism, have highlighted some problems encountered with individual parameters. We strongly suggest that all four parameters be used, together with appropriate statistical tests, at least until an extensive body of studies further clarifies the relative merits and limitations of the different parameters.
Acknowledgments
R.W. is grateful to S. Sherwood for her help in writing the early versions and to Y. Yuan, Y. Li, and R. Saffrich for helpful suggestions on statistics and C programming. We are grateful to C. Mbogo for kindly providing mosquitoes, G. Lanzaro for an earlier collaboration that supported the collection of mosquitoes, A. della Torre for a very helpful review, and M. Coluzzi, J. Powell, C. Taylor, and members of the Kafatos laboratory for comments. In situ hybridizations of the two microsatellites H678 and E614 were performed by C. Blass and E. Kokoza, respectively. This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB 544/B2/C1) (D.T. and F.C.K.), by National Institutes of Health Grant R01A43053 (L.Z.), and by a grant from the John D. and Catherine T. MacArthur Foundation (F.C.K.).
Abbreviations
- IAM
infinite allele model of mutation
- SMM
stepwise mutation model
- rDNA
ribosomal DNA
References
- 1.della Torre A, Fanello C, Akogbeto M, Dossou-yovo J, Favia G, Petrarca V, Coluzzi M. Insect Mol Biol. 2001;10:9–18. doi: 10.1046/j.1365-2583.2001.00235.x. [DOI] [PubMed] [Google Scholar]
- 2.Gentile G, Slotman M, Ketmaier V, Powell J R, Caccone A. Insect Mol Biol. 2001;10:25–32. doi: 10.1046/j.1365-2583.2001.00237.x. [DOI] [PubMed] [Google Scholar]
- 3.Favia G, Lanfrancotti A, Spanos L, Siden-Kiamos I, Louis C. Insect Mol Biol. 2001;10:19–23. doi: 10.1046/j.1365-2583.2001.00236.x. [DOI] [PubMed] [Google Scholar]
- 4.Mukabayire O, Caridi J, Wang X, Touré Y T, Coluzzi M, Besansky N J. Insect Mol Biol. 2001;10:33–46. doi: 10.1046/j.1365-2583.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- 5.Coluzzi M, Sabatini A, Petrarca V, Di Deco M A. Trans R Soc Trop Med Hyg. 1979;73:483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- 6.Touré Y T, Petrarca V, Traore S F, Coulibaly A, Maiga H M, Sankare O, Sow M, Di Deco M A, Coluzzi M. Parassitologia. 1998;40:477–511. [PubMed] [Google Scholar]
- 7.Favia G, della Torre A, Bagayoko M, Lanfrancotti A, Sagnon N, Touré Y T, Coluzzi M. Insect Mol Biol. 1997;6:377–383. doi: 10.1046/j.1365-2583.1997.00189.x. [DOI] [PubMed] [Google Scholar]
- 8.Pérez-Lezaun A, Calafell F, Mateu E, Comas D, Ruiz-Pacheco R, Bertranpetit J. Hum Genet. 1997;99:1–7. doi: 10.1007/s004390050299. [DOI] [PubMed] [Google Scholar]
- 9.Paetkau D, Waits L P, Clarkson P L, Craighead L, Strobeck C. Genetics. 1997;147:1943–1957. doi: 10.1093/genetics/147.4.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harr B, Weiss S, David J R, Brem G, Schlotterer C. Curr Biol. 1998;8:1183–1186. doi: 10.1016/s0960-9822(07)00490-3. [DOI] [PubMed] [Google Scholar]
- 11.Lehmann T, Hawley W A, Grebert H, Collins F H. Mol Biol Evol. 1998;15:264–276. doi: 10.1093/oxfordjournals.molbev.a025923. [DOI] [PubMed] [Google Scholar]
- 12.Lanzaro G C, Touré Y T, Carnahan J, Zheng L, Dolo G, Traore S, Petrarca V, Vernick K D, Taylor C E. Proc Natl Acad Sci USA. 1998;95:14260–14265. doi: 10.1073/pnas.95.24.14260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamau L, Mukabana W R, Hawley W A, Lehmann T, Irungu L W, Orago A A, Collins F H. Insect Mol Biol. 1999;8:287–297. doi: 10.1046/j.1365-2583.1999.820287.x. [DOI] [PubMed] [Google Scholar]
- 14.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ. Press; 1987. [Google Scholar]
- 15.Goldstein D B, Ruiz Linares A, Cavalli-Sforza L L, Feldman M W. Genetics. 1995;139:463–471. doi: 10.1093/genetics/139.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slatkin M. Genetics. 1995;139:457–462. doi: 10.1093/genetics/139.1.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shriver M D, Jin L, Boerwinkle E, Deka R, Ferrell R E, Chakraborty R. Mol Biol Evol. 1995;12:914–920. doi: 10.1093/oxfordjournals.molbev.a040268. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M, Ohta T. Genetics. 1964;49:725–738. doi: 10.1093/genetics/49.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimura M, Ohta T. Proc Natl Acad Sci USA. 1978;75:2868–2872. doi: 10.1073/pnas.75.6.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavalli-Sforza L L, Edwards A W. Am J Hum Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- 21.Scott J A, Brogdon W G, Collins F H. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- 22.Wang R, Kafatos F C, Zheng L. Parasitol Today. 1999;15:33–37. doi: 10.1016/s0169-4758(98)01360-x. [DOI] [PubMed] [Google Scholar]
- 23.Press W H, Teukolsky S A, Vetterling W T, Flannery B P. Numerical Recipes in C. New York: Cambridge Univ. Press; 1992. [Google Scholar]
- 24.Bronstein I N, Semendjajew K A, Musiol G, Muehlig H. Taschenbuch der Mathematik. Frankfurt am Main: Deutsch; 1993. [Google Scholar]
- 25.Sokal R R, Rohlf F J. Biometry. New York: Freeman; 1969. [Google Scholar]
- 26.Sanghvi L D. Am J Phys Anthropol. 1953;11:385–404. doi: 10.1002/ajpa.1330110313. [DOI] [PubMed] [Google Scholar]
- 27.Workman P L, Niswander J D. Am J Hum Genet. 1970;22:24–49. [PMC free article] [PubMed] [Google Scholar]
- 28.Black IV W C, Krafsur E S. Theor Appl Genet. 1985;70:484–490. doi: 10.1007/BF00305980. [DOI] [PubMed] [Google Scholar]
- 29.Takezaki N, Nei M. Genetics. 1996;144:389–399. doi: 10.1093/genetics/144.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Estoup A, Garnery L, Solignac M, Cornuet J M. Genetics. 1995;140:679–695. doi: 10.1093/genetics/140.2.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hudson R R, Boos D D, Kaplan N L. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 32.Weir B S. Genetic Data Analysis II. Sunderland, MA: Sinauer; 1996. [Google Scholar]
- 33.Raymond M, Rousset F. Evolution (Lawrence, KS) 1995;49:1280–1283. doi: 10.1111/j.1558-5646.1995.tb04456.x. [DOI] [PubMed] [Google Scholar]
- 34.Wang R. Ph.D. thesis. Heidelberg: Heidelberg Univ.; 2000. p. 131. [Google Scholar]
- 35.della Torre A, Merzagora L, Powell J R, Coluzzi M. Genetics. 1997;146:239–244. doi: 10.1093/genetics/146.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caccone A, Min G S, Powell J R. Genetics. 1998;150:807–814. doi: 10.1093/genetics/150.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]