To the Editor:
Mycobacterium tuberculosis (Mtb) is a major global cause of death from infectious disease (1). Rarely contagious, extrapulmonary tuberculosis (EPTB) is neglected in tuberculosis (TB) control strategies. Nonetheless, EPTB contributes significantly to the burden of disease, is frequently difficult to diagnose, and inflicts significant morbidity (2).
After the establishment of infection in the host, it is believed that heterogeneous populations develop, distinguishable by specific growth requirements (3–5), metabolic features (6, 7), and differential susceptibility to antimicrobial agents (3, 8). The application of limiting dilution method to enumerate Mtb in liquid media has previously revealed that human TB sputum was dominated by Mtb populations that could not be detected by standard methods but required addition of culture supernatant (CS) obtained from actively growing Mtb or recombinant Rpf (resuscitation-promoting factor) (3, 4). The importance of Rpf in recovery of these bacteria was further supported by control experiments with Rpf-depleted or Rpf-inactivated culture supernatants (3, 9). However, a recently published study demonstrated the higher complexity of mycobacterial populations in sputum, including bacilli growing only in 7H9 supplemented with CS or Rpf-deficient CS, those growing in 7H9 medium only (nonplateable), and those growing on standard 7H10 agar plates (plateable) Mtb bacilli, collectively designated as differentially culturable tubercule bacteria (DCTB) (4, 5). Furthermore, it was proposed that relative proportions of these populations depended on patient CD4+ T cell counts (4), and it is unknown whether other host factors may hold influence. DCTB can be generated in vitro by application of combined stresses (10) or treatment with antimicrobial agents (11).
There is accumulating experimental evidence to suggest the presence of DCTB has ramifications for clinical and treatment outcomes. CS- and Rpf-dependent Mtb bacilli were apparently enriched in treated patents and showed high tolerance to killing by antimicrobials, including rifampicin (3), isoniazid, and streptomycin (8). Furthermore, the eradication of CS-dependent Mtb prevented relapse in a Cornell mouse model (12). Finally, the application of recombinant Rpf or Rpf-containing CS improved time to positivity in a substantial number of sputum samples and often enhanced bacterial recovery (4, 13). Although three independent studies demonstrated the presence of Rpf- or CS-dependent bacilli in sputum samples (3, 4, 13), it remains unclear whether similar bacilli are commonly generated in EPTB. An understanding of DCTB populations in EPTB has important implications for diagnosis and treatment. Therefore, we examined a broad range of extrapulmonary samples to answer: 1) Are CS-dependent bacilli present in EPTB? 2) Can we identify host factors that may modulate CS-dependent populations?
We used previously developed methodology and assessed the number of Mtb bacilli recoverable on agar plates (colony-forming units [cfu]), in liquid 7H9 medium (most probable number counts on 7H9 medium [MPN_7H9]), or in 7H9 medium supplemented with CS (most probable number counts in culture supernatant [MPN_CS]) (3). The study was approved by the National Research Ethics Service Committee East Midlands Leicester (07/Q2501/58).
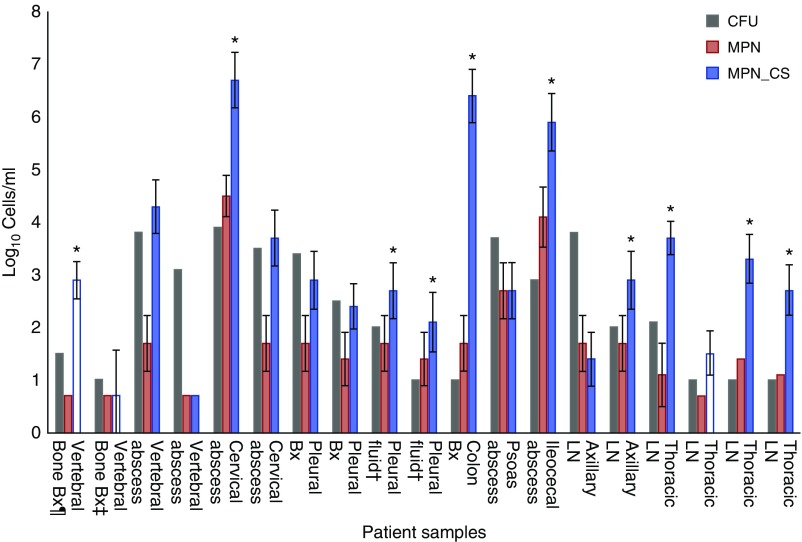
Forty-one patients were recruited before the onset of chemotherapy; 18 were culture positive for Mtb, one patient having two separate positive samples (Figure 1). Most patients were from Indian subcontinent backgrounds (83%), and the remainder were black African (17%); 67% were male, 17% were infected with HIV, and 82% were vitamin D deficient (cutoff, <30 nmol/L). Samples were obtained from varying anatomical locations, including colonic tissue, vertebral bone, abscess pus, and lymph nodes, and Ziehl-Neelsen stained within 24 hours after collection. No bacilli were detected using this protocol in any samples before cultivation, which fits with the typically paucibacillary nature of EPTB disease (14).
Figure 1.
Mycobacterial subpopulations present in extrapulmonary tuberculosis samples. The error bars indicate the 95% confidence intervals for the most probable number (MPN) values. Owing to small sample volumes and the requirement for invasive sampling, only one cfu assay technical replicate per sample was possible. Volumes of liquid samples were in range of 1 to 10 ml. The lymph node (LN) sample sites (from left to right) are left axillary LN; left axillary LN; intrathoracic lymph node (ILN) obtained via mediastinoscopy; ILN station 7; ILN station 4R; ILN station 4R. The MPN assay limit of detection was 5 cells /ml. White bars: samples recovered only with culture supernatant (CS) supplementation. *Samples with dominating CS-dependent populations. ‡Two samples obtained from the same patient separated by 10 days. ¶The cfu assay limit was 10 cfu/ml. Bx = biopsy; cfu = colony-forming units; MPN_CS = MPN with supplementary CS; MPN_7H9 = MPN with standard 7H9 media.
We detected substantial differences in DCTB counts and relative DCTB proportions, with bacillary counts ranging over log10 0.69 to 6.7 for MPN_CS, over log10 0.69 to 4.5 for MPN_7H9, and over log10 1 to 3.9 for cfu counts (Figure 1). In most samples (11 of 19), the MPN_7H9 counts were lower than MPN_CS and cfu counts. In 52% (10 of 19) of samples, CS-dependent Mtb were dominating populations, including three samples that produced Mtb cultures only with CS supplementation. Interestingly, in 6 out of 19 samples (32%), there was no significant difference between cfu and MPN_CS counts (taking into consideration limit of confidence), and, in two samples, plateable Mtb exceeded those grown in liquid media.
The isolated Mtb strains were acid-fast positive and diverse, with eight (42%) of isolates belonging to the Delhi/Central Asian Strain lineage, four (21%) East African-Indian, two (11%) Haarlem, one (5%) Beijing, one Turkish, one S-type, and one H37Rv-like, as determined by mycobacterial interspersed repetitive units–variable number of tandem repeats typing (15). We found no relationship between MPN_CS counts and strain lineage (P = 0.45; Kruskal-Wallis test). Examination of host factors elucidated a significant correlation (P = 0.04) between the host peripheral lymphocyte counts (PLC) and MPN_CS bacillary counts. However, additional experiments are required, because multivariate regression analysis to adjust for confounding was precluded by the sample size. No other host parameters, including HIV, diabetes mellitus, or vitamin D status, were significantly associated with DCTB. Furthermore, there was also no correlation between MPN_CS counts and C-reactive protein levels, neutrophil or monocyte counts, or the monocyte:lymphocyte ratio. All patients responded appropriately to antituberculous therapy, and by the end of June 2017, 83% of patients had completed treatment according to World Health Organization definitions (16); treatment outcome for 17% of patients could not be evaluated because they returned to home countries.
CS-dependent DCTB were isolated from a wide range of anatomically and physiologically distinct sites (Figure 1), indicating that CS-dependent and Rpf-dependent bacilli are not exclusive to sputum. Rather, their formation is an important manifestation of Mtb and host interactions, as previously demonstrated in a mouse model (17). Whether DCTB population distributions do differ between pulmonary and extrapulmonary disease is unknown. We identified CS-dependent DCTB populations in only 52% of EPTB samples, considerably less than detected in sputum studies (3, 4), although the absence of a comparator group prevents clarification of whether a true difference exists.
The significant positive correlation demonstrated between the PLC and MPN_CS bacillary counts supports the contention that the cellular host immune response is a major factor determining population proportions of CS-dependent bacilli (4, 17). Differences in the effectiveness of individual host immune responses and consequently mycobacterial stress may explain the interhost differences in CS-dependent DCTB populations. We examined certain host factors, such as vitamin D deficiency and diabetes mellitus, but they did not influence CS dependency in our study. Although this pilot study was underpowered for multivariate analysis, these factors may impact DCTB populations and should be examined in future studies. At this stage, we also cannot conclude whether the presence of DCTB may influence treatment outcomes; additional studies focused on duration of treatment and its correlation with DCTB should be conducted.
In conclusion, we demonstrated CS-dependent DCTB are common in EPTB, their proportion potentially modulated by the host immune response. The PLC represents a candidate biomarker of host CS-dependent DCTB population burden, potentially replacing highly laborious growth assays and the need for repeat sampling, difficult in EPTB. The relationship between DCTB populations and treatment outcome should be studied. Host biomarkers may represent an easy way to measure the dynamics of populations during treatment. Immunomodulators have been previously proposed as a tool for reduction of persisters (18) and may abrogate DCTB formation. Future studies should examine whether host factor modulation can minimize DCTB generation.
Acknowledgments
Acknowledgment
The authors thank the Centre for Core Biotechnology Services at the University of Leicester for providing access to a level 3 containment laboratory and the assistance of the tuberculosis nursing service with this study.
Footnotes
The PreDiCT-TB project is funded by European Commission the Innovative Medicines Initiative (No. 115337), a joint undertaking of the European Union Seventh Framework Program and The European Federation of Pharmaceutical Industries and Associations (O.T. and G.V.M.); Biotechnology and Biological Sciences Research Council grant BB/K000330/1 (G.V.M.); the Dowager Countess Eleanor Peel Trust (A.R. and G.V.M.); and National Institute for Health Research grant PDF-2015-08-102 (M.P.). The views expressed in this publication are those of the authors and not necessarily those of the National Health Service, the National Institute for Health Research, or the Department of Health.
Author Contributions: Conception and design: A.R., M.P., O.T., M.J.W., and G.V.M.; analysis and interpretation: A.R. and G.V.M.; drafting the manuscript for important intellectual content: A.R., M.P., and G.V.M.
Originally Published in Press as DOI: 10.1164/rccm.201705-1048LE on September 7, 2017
Author disclosures are available with the text of this letter at www.atsjournals.org.
References
- 1.World Health Organisation. Global tuberculosis report 2016. 2016 [accessed 2017 Aug 11]. Available from: http://apps.who.int/iris/bitstream/10665/250441/1/9789241565394-eng.pdf.
- 2.Sandgren A, Hollo V, van der Werf MJ. Extrapulmonary tuberculosis in the European Union and European economic area, 2002 to 2011. Euro Surveill. 2013;18 [PubMed] [Google Scholar]
- 3.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle bacilli in sputum. Am J Respir Crit Care Med. 2010;181:174–180. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chengalroyen MD, Beukes GM, Gordhan BG, Streicher EM, Churchyard G, Hafner R, Warren R, Otwombe K, Martinson N, Kana BD. Detection and quantification of differentially culturable tubercle bacteria in sputum from patients with tuberculosis. Am J Respir Crit Care Med. 2016;194:1532–1540. doi: 10.1164/rccm.201604-0769OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dartois V, Saito K, Warrier T, Nathan C. New evidence for the complexity of the population structure of Mycobacterium tuberculosis increases the diagnostic and biologic challenges. Am J Respir Crit Care Med. 2016;194:1448–1451. doi: 10.1164/rccm.201607-1431ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garton NJ, Waddell SJ, Sherratt AL, Lee SM, Smith RJ, Senner C, Hinds J, Rajakumar K, Adegbola RA, Besra GS, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. Plos Med. 2008;5:e75. doi: 10.1371/journal.pmed.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manina G, Dhar N, McKinney JD. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe. 2015;17:32–46. doi: 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Turapov O, O’Connor BD, Sarybaeva AA, Williams C, Patel H, Kadyrov AS, Sarybaev AS, Woltmann G, Barer MR, Mukamolova GV. Phenotypically adapted Mycobacterium tuberculosis populations from sputum are tolerant to first-line drugs. Antimicrob Agents Chemother. 2016;60:2476–2483. doi: 10.1128/AAC.01380-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaprelyants AS, Mukamolova GV, Ruggiero A, Makarov VA, Demina GR, Shleeva MO, Potapov VD, Shramko PA. Resuscitation-promoting factors (Rpf): in search of inhibitors. Protein Pept Lett. 2012;19:1026–1034. doi: 10.2174/092986612802762723. [DOI] [PubMed] [Google Scholar]
- 10.Saito K, Warrier T, Somersan-Karakaya S, Kaminski L, Mi J, Jiang X, Park S, Shigyo K, Gold B, Roberts J, et al. Rifamycin action on RNA polymerase in antibiotic-tolerant Mycobacterium tuberculosis results in differentially detectable populations. Proc Natl Acad Sci USA. 2017;114:E4832–E4840. doi: 10.1073/pnas.1705385114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loraine J, Pu F, Turapov O, Mukamolova GV. Development of an in vitro assay for detection of drug-induced resuscitation-promoting-factor-dependent mycobacteria. Antimicrob Agents Chemother. 2016;60:6227–6233. doi: 10.1128/AAC.00518-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu Y, Liu A, Ortega-Muro F, Alameda-Martin L, Mitchison D, Coates A. High-dose rifampicin kills persisters, shortens treatment duration, and reduces relapse rate in vitro and in vivo. Front Microbiol. 2015;6:641. doi: 10.3389/fmicb.2015.00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang W, Qi Y, Diao Y, Yang F, Zha X, Ren C, Huang D, Franken KL, Ottenhoff TH, Wu Q, et al. Use of resuscitation-promoting factor proteins improves the sensitivity of culture-based tuberculosis testing in special samples. Am J Respir Crit Care Med. 2014;189:612–614. doi: 10.1164/rccm.201310-1899LE. [DOI] [PubMed] [Google Scholar]
- 14.Chakravorty S, Sen MK, Tyagi JS. Diagnosis of extrapulmonary tuberculosis by smear, culture, and PCR using universal sample processing technology. J Clin Microbiol. 2005;43:4357–4362. doi: 10.1128/JCM.43.9.4357-4362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res. 2010;38:W326–W331. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organisation. Definitions and reporting framework for tuberculosis - 2013 revision (updated December 2014). 2014 [accessed 2017 Aug 11]. Available from: http://apps.who.int/iris/bitstream/10665/79199/1/9789241505345_eng.pdf.
- 17.Turapov O, Glenn S, Kana B, Makarov V, Andrew PW, Mukamolova GV. The in vivo environment accelerates generation of resuscitation-promoting factor-dependent mycobacteria. Am J Respir Crit Care Med. 2014;190:1455–1457. doi: 10.1164/rccm.201407-1289LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nathan C. Fresh approaches to anti-infective therapies. Sci Transl Med. 2012;4:140sr2. doi: 10.1126/scitranslmed.3003081. [DOI] [PMC free article] [PubMed] [Google Scholar]