Abstract
The genetic component of susceptibility to malaria is complex, both in humans and in the mouse model of infection. Two murine loci on chromosomes 8 (Pchr/Char2) and 9 (Char1) have previously been mapped in F2 crosses, and play an important role in regulating blood parasitemia and survival to infection with Plasmodium chabaudi. These loci explain only part of the interstrain phenotypic variance, and their penetrance and expressivity vary in different inbred strains. Novel loci regulating response to P. chabaudi infection were investigated by using an alternative strategy based on a newly derived set of AcB/BcA recombinant congenic strains bred from malaria-susceptible A/J (A) and resistant C57BL/6J (B6). One of the AcB strains, AcB55, is shown to be highly resistant to infection despite 83% susceptible A genomic composition, including susceptibility alleles at Char1 and Pchr/Char2. Early onset of parasite clearance in AcB55 is associated with lower peak parasitemia and absence of mortality. Linkage analysis in an informative (AcB55 × A)F2 population, using peak parasitemia as a quantitative trait, located a new B6-derived resistance locus on chromosome 3 (lod score = 6.57) that we designate Char4. A second, suggestive linkage on chromosome 10 (lod score = 2.53) shows additive effect with Char4 on peak parasitemia. Char4 maps to a small congenic B6 fragment in AcB55 that should facilitate the search for candidate genes. Our findings provide an entry point for parallel association studies in humans between the syntenic 4q21–4q25 region and susceptibility to disease in endemic areas of malaria.
Malaria caused by Plasmodium parasites is a leading cause of death by infectious diseases worldwide (http://www.who.ch). Despite extraordinary research efforts to better understand the immunobiology of the parasite and to control populations of the mosquito vector in the field, the situation has worsened. Major contributing factors are the widespread emergence of chloroquine/mefloquine resistance in Plasmodium falciparum and resistance to pyrethroid insecticides in the Anopheles vector. In addition, efforts to develop effective antimalarial vaccines have remained unsuccessful (1, 2). A better understanding of the host natural mechanisms of defense against the Plasmodium parasite may provide new targets for therapeutic intervention in this disease. Such mechanisms may manifest themselves as genetic determinants of susceptibility to infection in endemic areas of disease and during epidemics (3). In humans, the influence of genetic factors on onset, progression, pathophysiology, and ultimate outcome of malaria infection has been well documented (reviewed in refs. 4 and 5). In sickle cell anemia, heterozygosity for mutant hemoglobin alleles confers survival advantage over homozygosity for either mutant or wild-type alleles. Likewise, a protective effect against malaria has been associated with α thalassaemia and glucose-6-phosphate dehydrogenase deficiencies (4, 5). Class I and Class II HLA haplotypes as well as levels of the proinflammatory cytokine tumor necrosis factor (TNF) are also associated with differential severity/outcome of disease. In the case of TNF, promoter polymorphisms may influence transcriptional activity of the gene (6) through differential recruitment of specific transcription factors (7). However, the genetic component of malaria susceptibility is acknowledged to be very complex and heterogeneous in humans, and further modified by environmental factors (4).
The mouse is an important complementary model for the genetic analysis of susceptibility to malaria. A murine model allows one not only to control environmental factors such as parasite status, dose, and route of infection, but also enables dissection of complex genetic traits in large crosses generated between resistant and susceptible progenitor strains. Plasmodium chabaudi AS produces an infection in mice that shares many similarities with P. falciparum malaria in humans, including anemia, splenomegaly, hepathomegaly, renal alterations, hypoglycemia, cerebral disease, and ultimately death (reviewed in ref. 8). Inbred mouse strains differ widely in susceptibility to P. chabaudi AS infection, with strains such as A/J (A), C3H, and SJL being susceptible and C57BL/6J (B6) being resistant (9–11). Resistance in B6 mice is associated with robust inflammatory and immune responses, reduced parasitemia at the peak of infection, and increased survival to infection (9). Production of granulocyte–macrophage colony-stimulating factor (12), IFN-γ (13), TNF-α (6), and IL-12 (14) are required for resistance to infection.
Studies in A × B recombinant inbred strains (derived from A and B6) suggested that the genetic basis for interstrain difference in outcome of P. chabaudi AS infection was multigenic, with unlinked loci affecting peak parasitemia and mortality (A.F., M.T., M.M.S., and P.G., unpublished data). Quantitative trait loci (QTL) mapping by whole genome scanning has been used to map loci independently regulating peak parasitemia, parasite clearance, and overall survival to infection. Studies in informative F2 and backcross mice derived from A and B6 parents have mapped a gene (Pchr/Char2) on chromosome 8 [logarithm of odds (lod) score = 3.7] that controls bloodstage replication of the P. chabaudi AS parasite (10). Parallel studies by Foote et al. (1997) in female F2 mice derived from C3H (susceptible), SJL (susceptible), and B6 parents also identified the Chr.8 Char2 locus as regulating peak parasitemia (lod score = 8.8 in C3B6F2 only), but also mapped another locus on distal chromosome 9 (Char1) that determines death or survival to infection with P. chabaudi adami DS (lod scores = 6.6 and 9.1 in SJLB6F2 and C3B6F2, respectively; ref. 11). A third H-2 linked locus (Char3) on chromosome 17 (lod score = 5) was detected during the course of infection and appears to regulate parasite clearance from the blood in C3B6F2 females, immediately following the peak of infection (15). In addition, female mice are found to be more resistant (blood parasitemia and survival) than males, possibly because of hormonal influence (16). Therefore, susceptibility to P. chabaudi is under complex genetic control in the mouse. Because the loci mapped to date explain only a fraction of the phenotypic variance, additional genes are likely to contribute to host response in a strain-specific fashion.
Recently, we reported the construction and characterization of a novel set of AcB/BcA recombinant congenic strains, derived from A and B6 parents by systematic inbreeding of pairs of [(A × B) × A] × A (15 strains) and of [(A × B) × B] × B (22 strains) double backcross (N3) mice (17). Genotyping of these strains with 625 DNA markers [average spacing 2.6 centimorgans (cM)] indicates that in the AcB set (i) individual strains contain on average 13.25% of B6 “resistant” genome, and (ii) that 79% of the total B6 “resistant” genome has been transferred in independent strains of the AcB series. In the present study, AcB strains were used to map novel malaria resistance loci derived from B6 mice, and fixed by inbreeding on the susceptible background of A mice. Our strategy consisted in looking for strains bearing susceptibility alleles at Char1 and Char2, but yet showing a malaria resistance phenotype, possibly caused by the transfer of a small B6 chromosomal segment carrying a novel resistance locus. The AcB55 strain is found to be resistant to P. chabaudi AS infection, despite susceptible alleles at Char1 and Char2. Using 199 animals from an informative (AcB55 × A)F2 cross, we have identified a novel resistance locus on chromosome 3 (Char4).
Materials and Methods
Animals.
C57BL/6J (B6) and A/J (A) inbred strains were purchased from The Jackson Laboratory. The AcB series of recombinant congenic strains of mice (RCS) were generated and maintained at the Montreal General Hospital Research Institute, according to a breeding scheme and a genotyping protocol previously described (17). Strains AcB 51, 52, 53, 54, 55, 56, 57, 58, 60, 61, 62, 64, and 65 were tested for susceptibility to malaria infection (P. chabaudi AS). (AcB55 × A)F1 and F2 mice were bred in the animal facility of the Montreal General Hospital Research Institute. All mice were handled according to guidelines and regulations of the Canadian Council on Animal Care. Male and female mice 8 to 12 weeks of age, were used for infections.
P. chabaudi AS Infection.
An LDH virus-free isolate of P. chabaudi AS, originally obtained from Dr. D. Walliker (University of Edinburgh), was maintained by weekly passage in A mice by i.p. infection with 106 parasitized red blood cells (PRBC). Experimental mice were infected i.p. with 106 PRBC suspended in 1 ml of pyrogen-free saline. The percentage of PRBC was determined daily on duplicate thin blood smears stained with Dif-Quik (American Scientific Products, McGraw Park, IL) on days 4 to 28 postinfection, as described (9). Death was recorded daily or moribund animals were killed.
Genotyping.
Before infection, tail biopsies were obtained from all animals and genomic DNA was isolated by a standard procedure involving proteinase K treatment (17). Primer pairs defining dinucleotide repeats markers informative for the A and B6 mouse strains, and covering the B6-derived congenic segments of AcB55 at a maximum spacing of 10 cM, were purchased from Research Genetics (Huntsville, AL). Sequence length polymorphisms were typed by a standard PCR-based method using [α-32P]dATP labeling, and separation on denaturing 6% polyacrylamide gels. The following markers were used for the genotyping step: D1Mit156, D1Mit282, D1Mit215, D1Mit305, D1Mit136, D1Mit493, D1Mit139, D2Mit117, D2Mit370, D3Mit109, D3Mit14, D3Mit254, D3Mit110, D3Mit349, D3Mit351, D3Mit44, D3Mit128, D4Mit204, D4Mit148, D4Mit344, D6Mit159, D6Mit184, D6Mit284, D6Mit230, D6Mit389, D7Mit130, D8Mit14, D8Mit42, D8Mit156, D10Mit167, D10Mit246, D10Mit189, D10Mit51, D10Mit213, D10Mit106, D10Mit214, D13Mit132, D14Mit98, D14Mit113, D19Mit46, D19Mit66, D19Mit53, D19Mit26, D19Mit137, DXMit74, DXMit16, DXMit172, DXMit37, DXMit197.
Statistical Analysis.
Genetic map construction and genome scan analyses of the (AcB55 × A)F2 mice were performed by using MAPMAKER/EXP Version 3.0 and MAPMAKER/QTL Version 1.1 (18). Peak parasitemia for individual mice was used as a quantitative trait to detect linkage. The distribution of peak parasitemia in the (AcB55 × A)F2 closely followed a normal distribution. The maximum deviation from normality occurred in the female group, where the skewness reached only −0.48 (see Table 1). Overall, the mean differences between males and females were highly significant, with males having higher average parasitemia values (males mean = 50.93, females mean = 44.06, two-tailed t = 5.215, P < 10−5. Individual parasitemia values were not transformed before genetic analysis, and multipoint analysis was performed by using recessive, dominant, additive, and free models of inheritance. Possible linkage to chromosome X was initially detected (lod score = 4.02), suggesting a possible sex effect. Further examination of the chromosome X lod score showed no significant linkage in either males alone (lod score = 0.23) or females alone (lod score = 0.66), suggesting that the original linkage primarily reflects mean differences between genders (Table 1) rather than any chromosome X loci. To compensate for sex effects, the peak parasitemia data were adjusted by subtracting the gender-specific mean from each individual to create a “sex-adjusted parasitemia” value. Linkage models were fitted to raw, females only, males only, and sex-adjusted parasitemia. To assess whether linkage to chromosome 3 was a result of simple mean differences between sexes, a likelihood-ratio heterogeneity test was conducted. The difference between the likelihoods [lod score × 2ln(10)] of the individual male and female data sets (lod score = 2.46 + 4.34 = 6.80) and the combined sample (lod score = 6.57) yields a nested model comparison distributed as χ2 with 4 − 2 = 2 degrees of freedom [i.e., χ2(2) = 2(19.9 + 11.3) − 30.3) = 2.18; P = 0.34]. This test suggests that the chromosome 3 results are robust to other sex-related phenotypic differences in peak parasitemia. A similar test was conducted to distinguish between 2-loci additive effects and epistatic interactions in the chromosome 3 and 10 QTLs. Simultaneous analysis of the Chr.3 and 10 QTLs for the sex-adjusted parasitemia produced a χ2 value of 40.12 (lod score = 8.71), whereas the sum of the χ2 values for the two loci (see Table 2) yielded a χ2 of 41.92. The difference between these values is not significant [χ2(2) = 3.60, P = 0.17)], strongly suggesting additive effects.
Table 1.
Distribution of the phenotype of peak parasitemia in the 199 (AcB55XA)F2 hybrids
| Trait | n | Mean | SD | Skewness | Kurtosis |
|---|---|---|---|---|---|
| Peak parasitemia (pp) | 199 | 47.83 | 11.39 | −0.19 | 0.85 |
| Female pp | 90 | 44.06 | 11.14 | −0.48 | 0.98 |
| Male pp | 109 | 50.93 | 10.64 | −0.13 | 0.28 |
| Sex-adjusted pp | 199 | 0.00 | 10.87 | −0.16 | 0.64 |
Table 2.
Statistical values for linkages obtained in the (AcB55XA)F2 population
| Measure and chromosome | Maximum lod scores under specific model
|
Variance*, % | χ2 (2)* | P | |||
|---|---|---|---|---|---|---|---|
| Free | Dominant | Recessive | Additive | ||||
| Peak parasitemia (pp) | |||||||
| D3Mit109 | 5.10 | 0.76 | 5.08 | 3.80 | 11.1 | 23.49 | 0.000008 |
| D10Mit189 | 3.00 | 2.28 | 1.53 | 2.94 | 6.7 | 13.82 | 0.001 |
| Females pp | |||||||
| D3Mit109 | 2.46 | 0.62 | 2.35 | 2.07 | 11.8 | 11.33 | 0.003 |
| D10Mit189 | 1.54 | 1.52 | 0.08 | 0.80 | 7.6 | 7.09 | 0.03 |
| Males pp | |||||||
| D3Mit109 | 4.34 | 0.38 | 4.33 | 2.88 | 16.8 | 19.99 | 0.00005 |
| D10Mit189 | 1.94 | 0.72 | 1.65 | 1.75 | 8.1 | 8.93 | 0.01 |
| Sex-adjusted pp | |||||||
| D3Mit109 | 6.57 | 0.99 | 6.54 | 4.90 | 14.1 | 30.26 | 0.0000003 |
| D10Mit189 | 2.53 | 2.08 | 1.12 | 2.43 | 5.7 | 11.65 | 0.003 |
Calculated by using free model. The free model has two df; all others have 1 df. The P value is shown for the most conservative free model.
Results
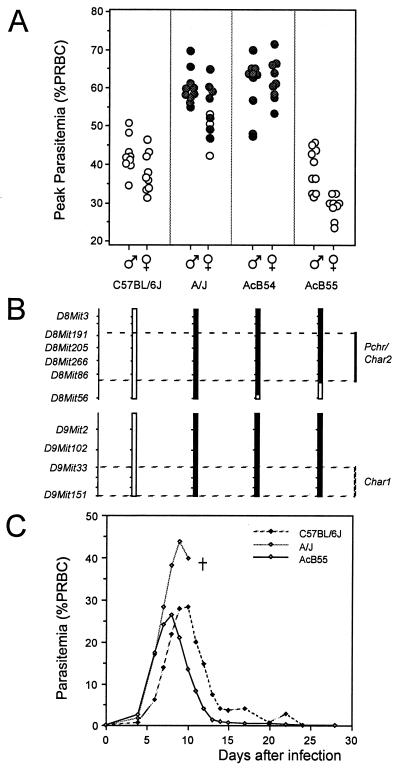
On average, 84.5% of the genome of AcB recombinant congenic strains (RCS) is derived from the malaria-susceptible A strain, whereas 13.25% is derived from the resistant B6 strain in the form of chimeric chromosomes fixed by inbreeding (17). This set of strains offers a unique opportunity to map B6-derived resistance loci that may have segregated in a specific AcB strain. Groups of 8 to 10 male and female mice from 13 AcB strains were infected with P. chabaudi AS, and blood parasitemia were monitored daily in individual mice for a period of 28 days. Blood parasitemia at the peak of infection and mortality were used to phenotype for susceptibility/resistance. As expected from their A background, 8/13 AcB strains were susceptible A-like (for example, AcB54 in Fig. 1A: high parasitemia, 100% mortality), whereas 3/13 showed intermediate phenotypes (peak parasitemia falling between parental susceptible A and resistant B6 controls, along with variable survival rates) and the other two AcB strains were strongly resistant (data not shown). AcB strains not susceptible to infection generally harbored parental B6 haplotypes at the Chr.8 (Pchr/Char2) region (data not shown), with the notable exception of the AcB55 strain. Indeed, AcB55 is completely resistant to infection (low parasitemia, 100% survival) despite the presence of susceptible A haplotypes at the Pchr/Char2 locus as well as at the Chr.9 Char1 locus (Fig. 1 A and B; ref. 17). In addition, AcB55 females were significantly more resistant than males, with respect to peak parasitemia values (Fig. 1A). Further analysis of the course of blood parasitemia during P. chabaudi AS infection (Fig. 1C) showed rapid parasite replication in AcB55 mice during the first 6 days of infection, with initial rates similar to those seen in A and B6 controls. Interestingly, in AcB55 mice, peak parasitemia occurred at day 7–8, 1–2 days earlier than resistant B6 controls (day 9–10), followed by a curative phase of parasite clearance that occurred at a rate similar to that seen in B6 controls (Fig. 1C). These results suggest segregation of a new resistance locus (loci) in AcB55, which is (are) phenotypically expressed as early onset of inhibition of parasite replication and rapid parasite clearance following peak parasitemia.
Figure 1.
Phenotypic response of selected recombinant congenic strains (RCS) to P. chabaudi AS infection. RC strains and parental controls were infected i.p. with 106 RBC infected with P. chabaudi AS, and daily parasitemia (expressed as a percentage of parasitized RBC) as well as death from infection were recorded. (A) Peak parasitemia are shown for strain AcB54 and AcB55 compared with parental strains. Each circle represents one mouse, and filled circles represent mice that succumbed to infection. (B) Schematic representation of mouse Chr.8 and Chr.9 with black and white segments representing haplotypes of A and B6 strains, respectively. Positions of Char1 and Char2/Pchr loci are indicated. (C) Course of infection (blood parasitemia) in AcB55 and parental controls over a 28 day period. Five to ten animals of both sexes were infected per strain, and the results of three separate infections were combined.
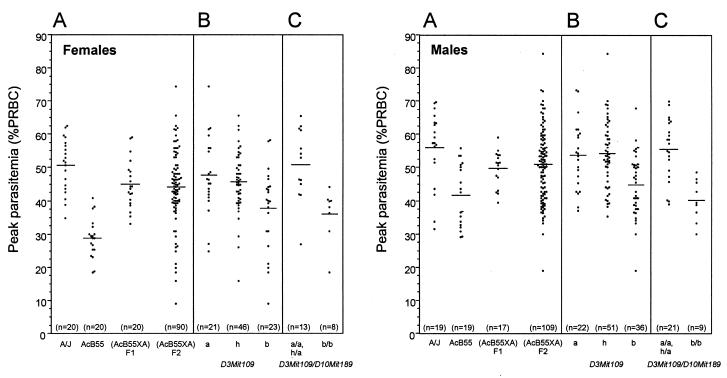
The mode of inheritance of the AcB55 resistance trait was examined in a (AcB55 × A)F1. Following P. chabaudi AS infection, the F1 mice showed an intermediate phenotype with respect to both peak parasitemia (Fig. 2A, shown separately for females and males) and mortality (not shown), consistent with resistance being transmitted in a codominant fashion. To map potential resistance loci expressed in AcB55, 199 animals (90 females, 109 males) of an informative (AcB55 × A)F2 cross were generated and infected with P. chabaudi AS, and parasitemia were followed daily in each mouse. Parasitemia at the height of infection in F1, F2, and parental controls were tabulated and are shown in Fig. 2A, for males and females, and summarized in Table 1. Peak parasitemia values for F2 mice followed a continuous distribution between 9 and 74% for the females and 19 and 84% for the males, and therefore behaved as a quantitative trait amenable to QTL analysis. The distribution of peak parasitemia in the total F2 population showed only minor departures from normality (Table 1), and thus the raw data did not justify modification of the data before genetic analysis.
Figure 2.
Susceptibility of (AcB55 × A)F2 mice to infection with P. chabaudi AS. Individual mice from the F1 and F2 populations as well a parental controls were infected i.p. with 106 parasitized RBC, and daily parasitemia as well as death from infection were recorded. (A) Peaks of parasitemia are shown separately for males and females of each group. To visualize the contribution of the Chr.3 locus on peak parasitemia, and the additive effect of the Chr.10 locus, mice of the F2 were separated according to their genotype at marker D3Mit109 (B) and in combination with marker D10Mit189 (C). Letters a and b represent alleles of A and B6 origins, respectively, and h represents heterozygotes. Each dot represents one mouse; means are indicated by horizontal lines.
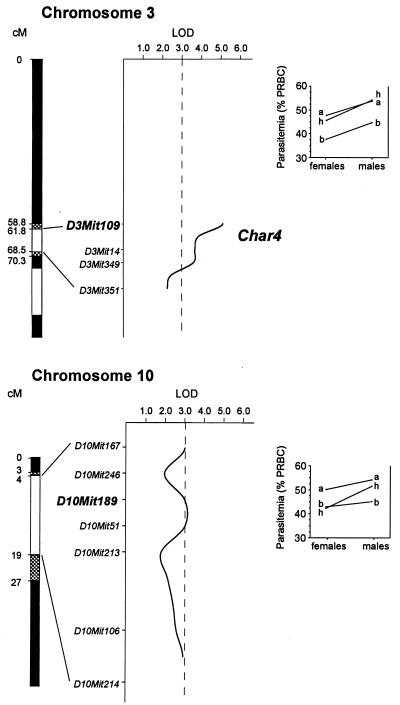
Extensive genotyping of the AcB55 strain shows that this strain contains 17 short chromosomal segments derived from B6 and distributed over 12 chromosomes (1, 2, 3, 4, 6, 7, 8, 10, 13, 14, 19, and X; ref. 17). Our working hypothesis was that P. chabaudi AS resistance in the AcB55 was derived from one or several of these B6 segments, and therefore only those were genotyped for informative microsatellite markers (listed in Materials and Methods). These 49 markers provided a coverage with an average spacing of 5.4 cM, the largest gap being ≈17 cM on chromosomes 19 and X. Multipoint linkage analysis was carried out by using the MAPMAKER/EXP Version 3.0 and MAPMAKER QTL Version 1.1 software packages. Results of this analysis are shown as multipoint lod score traces in Fig. 3, and numerical data for individual intervals are shown in Table 2. No evidence for linkage to mortality rate was obtained in the F2. However, using peak parasitemia as a quantitative phenotypic trait, two linkages were detected. A highly significant locus was identified on a short segment of Chr.3 of ≈10 cM, with maximum linkage to marker D3Mit109 (χ2 = 23.49; lod score = 5.1; P = 0.000008). Evidence for a second weaker linkage was obtained on a portion of Chr.10, with maximal effect at marker D10Mit189 (χ2 = 13.82; lod score = 3.0; P = 0.001).
Figure 3.
Schematic representation of Chr.3 and Chr.10 segments of AcB55. White and black areas depict B6 and A-derived genomic segments, respectively. Shaded areas extend the B6 segments to their maximum length based on the genetic map (17). Markers shown were used to genotype mice of the F2 cross. Intermarker distances are based on the frequency of recombination in the cross, but their absolute positions along the chromosomes are based on the mouse genome database (www.jax.org). The x axes of the graphs represent the lod score (log10 likelihood ratio) values. (Right) The norms of reaction of the genotype at D3Mit109 (Upper) and D10Mit189 (Lower) according to the sex. Letters a and b represent alleles of A and B6 origins, respectively, and h represents heterozygotes.
Because peak parasitemia is influenced by sex (generally higher in males than females; see Figs. 1 and 2), linkage analysis was also performed in a gender-specific fashion (Table 2). In this analysis, a significant lod score for the Chr.3 locus was only seen in males (lod score = 4.34 vs. 2.46 for females). To further explore possible gender specificity in linkage to the Chr.3 QTL, residual peak parasitemia values for F2 mice were examined after controlling for sex effects (Table 1, and see Material and Methods). This sex-adjusted peak parasitemia transformation provides a means to retain all experimental data (and therefore statistical power), while removing the effects of baseline sex differences in parasitemia. Analysis of this sex-adjusted peak parasitemia increased evidence for linkage to Chr.3 (D3Mit109, χ2 = 30.26; lod score = 6.57; P = 0.0000003), whereas it decreased the strength of the Chr.10 QTL (χ2 = 11.65; lod score = 2.53; P = 0.003). A likelihood-ratio χ2 test was constructed by comparing the χ2 values for the sex-adjusted data with the sum of the χ2 for the male and female peak parasitemia values. For the general model (without fixing heterozygotes to a dominant or recessive inheritance pattern), this test showed that males and females were equally linked to the Chr.3 QTL [χ2(2) = 2.18; P = 0.34]. Thus, the observed gender-associated differences in lod score are most likely attributable to unlinked differences in phenotypic means and/or sampling variability.
The Chr.3 locus is estimated to account for 14.1% of the variance in sex-adjusted parasitemia, and the Chr.10 linkage, despite its relatively low lod score, still explains 5.7% of the variance. Simultaneous analysis of the Chr.3 and 10 QTLs yields a lod score of 8.71, explaining 18.3% of the phenotypic variance [χ2(2) = 40.12]. Interaction models showed no significant difference between the joint analysis of Chr.3 and Chr.10 linkages and the sum of the individual analysis, suggesting that the two loci act additively, without epistasis [χ2(2) = 3.60, P = 0.17]. The Chr.3 QTL appears to be inherited in a recessive manner, whereas the weaker linkage to Chr.10 is probably inherited in a codominant or dominant fashion (Table 2, Fig. 3). To further visualize the effect of parental allele combinations in the F2 population, including the additive effect of the Chr.3 and Chr.10 linkages, F2 animals were separated according to their genotypes at markers D3Mit109 alone (Fig. 2B) or in combination with D10Mit189 (Fig. 2C). Results of this analysis showed that animals homozygous for the A allele (Fig. 2B, a) and heterozygous (h) at D3Mit109 present a mean peak parasitemia value of 48% ± 12 (n = 21) and 46% ± 9 (n = 46) for females, and 54% ± 10 (n = 22) and 54% ± 10 (n = 51) for males, respectively. In contrast, mice homozygous for the B6 allele (b) at D3Mit109 present an average peak parasitemia of 38% ± 12 (n = 23) for females and 45% ± 9 (n = 36) for males. These results are in agreement with a recessive mode of inheritance of the Chr.3 QTL, with B6 alleles associated with resistance. Moreover, double homozygosity for B6 alleles at both D3Mit109 and D10Mit189 (Fig. 2C) is associated with further reduction in mean peak parasitemia, with 36% ± 8 (n = 8) for the females group, and 40% ± 6 (n = 9) for males, illustrating the additive effect of B6 alleles at each locus.
Discussion
After diarrheal diseases, malaria is the most important cause of mortality by an infectious disease worldwide, and kills one child every 30 seconds (http://www.who.ch). Chloroquine and mefloquine resistance in Plasmodium parasites and insecticide resistance in the mosquito vector, together with the lack of an efficacious vaccine, have exacerbated the problem (1). Modulation of normal host mechanisms of defense against the parasite may be an alternative method of intervention in this severe disease. Such mechanisms may be identified by standard genetic mapping in segregating populations, followed by positional cloning in the murine model of P. chabaudi infection. This approach has led to the recent identification of two major loci on chromosomes 8 (Pchr/Char2) and 9 (Char1), which control blood stage replication of the parasite at the peak of infection and mortality (10, 11). However, the genetic component of susceptibility to malaria is complex both in man and mouse (4), and alternative mapping tools are required to identify new resistance/susceptibility loci.
RCS are an attractive tool to study complex traits such as differential susceptibility to malaria. By virtue of the breeding scheme used in their derivation, individual RCS contain a small amount (12.5%) of DNA from one parent fixed as a set of discrete congenic segments, on the background (87.5%) of DNA from the other parent (19). Therefore, individual resistance/susceptibility loci may independently segregate in individual RCS. Such strains cannot only be used to map a locus of interest, but also to characterize the specific phenotypic component of disease susceptibility under genetic control. The relatively small size of the congenic segments fixed in individual RCS also facilitates the search and testing of candidate genes. RCS have been used to study many complex traits including metabolic diseases, lung and colon cancer, and infectious diseases caused by Leishmania major (20–24). This research has led to the identification of several “susceptibility” loci, but has also permitted the segregation of some “subphenotypes” of susceptibility previously assumed to be pleiotropic manifestations of the same genetic control (24). However, the usefulness and broad applicability of RCS to the study of complex traits has been limited by the small number of strain pairs available (25). We have recently reported (17) the creation and characterization of a set of 37 AcB/BcA strains derived from malaria-resistant and malaria-susceptible, commonly used strains of laboratory mice, B6 and A, respectively.
To identify novel malaria-resistance loci, our strategy with RCS of the AcB/BcA series was to (i) fix the known susceptibility loci on Chr.8 (Pchr/Char2) and Chr.9 (Char1) in the A susceptible configuration, and (ii) look for individual AcB strains displaying a discordant malaria-resistance phenotype. In this way, we hoped to identify novel, B6-derived resistance loci whose effect may have gone unnoticed in standard QTL mapping experiments involving traditional (A × B6) backcross of F2 progeny (10). The AcB55 strain was identified as resistant, with respect to both peak parasitemia and overall survival to infection, despite A susceptible alleles at Char1 and Pchr/Char2 (Fig. 1 A and B). The phenotypic expression of resistance as evidenced by the kinetics of infection is unique to AcB55 (Fig. 1C). Indeed, early in infection blood parasitemia in AcB55 was similar to that seen in A, with respect to both onset and rate of replication, but clearly distinct from the delayed replication seen in B6 controls. However by day 7, AcB55 have started to control the infection and parasite clearance was initiated between day 8 and 9, a time at which the parasite was still replicating in both A and B6 progenitors. In B6 hosts, parasite clearance started at day 9–10, a time at which AcB55 have already eliminated a good part of their blood parasite load. Although the mechanism by which AcB55 can quickly control parasite replication and initiate clearance is of great interest, it remains unknown. This mechanisms could involve superior inflammatory or early immune responses, and/or increased erythropoietic activity.
Studies in (AcB55 × A) F2 mice identified a novel, B6-derived, significant linkage on Chr.3 (D3Mit109, χ2 = 30.26; lod score = 6.57; P = 0.0000003 for sex-adjusted parasitemia) that we designate Char4 (Chabaudi resistance locus 4). This QTL was detected with equal strength at days 8 and 9 (data not shown), but was not as detectable when parasite burden at other times during infection were used as quantitative phenotypes. This locus explains ≈14% of the variance at peak parasitemia, and B6 alleles at this locus confer resistance to infection that is inherited in a recessive manner in both males and females. This mode of inheritance for Char4 alleles may explain why this locus went undetected in a previous genome scan of an (A × B6) × A informative backcross, using peak parasitemia as a phenotype (10). A second, suggestive linkage was mapped to Chr.10 (D10Mit189, χ2 = 11.65; lod score = 2.53; P = 0.003). Although this QTL is rather weak and the percentage of variance explained by this locus is only ≈6%, the additive effect of this locus on Char4 for controlling peak parasitemia in AcB55 is nevertheless clear (Fig. 2C). The Chr.10 QTL segregated as dominant in females and codominant in males (see norms of reactions in Fig. 3 Lower).
The size of the Chr.3 congenic B6 segment in AcB55 that harbors the Char4 locus is quite small at 6–10 cM (Fig. 3). Transfer of this small segment to create a pure congenic line on A background should allow further dissection of the phenotypic basis of resistance with respect to immune, inflammatory, and erythropoietic responses during infection. This strategy, coupled to tissue-specific, transcriptional profiling for genes in this interval, should shed light on the molecular mechanism underlying malaria resistance of AcB55. The small B6 congenic segment defining Char4 in AcB55 contains several interesting candidates with respect to their known tissue or cell-specific expression or role in immune and/or inflammatory responses. These include (i) Lef1 (lymphoid enhancer binding factor 1; 61.6 cM), a transcription factor part of the Wnt signaling pathway which regulates gene expression in T-lymphocytes and other lineages (26, 27); (ii) Cfi (complement component factor I, 66.6 cM), a regulatory serine protease that activates the cleavage of C3b/C4b, a pathway known to be defective in A mice (28, 29) and responsible for the acute sensitivity of this line to Listeria infection (30); and (iii) the p105 subunit of NfκB1 (68.9 cM), a signal transduction regulator known to be essential for B-cell function, and for nonspecific resistance to infections (31). Other genes regionally mapped to the area, and known to play key roles in macrophage function and lymphocyte migration to tissues include Csf1 (macrophage colony stimulating factor; refs. 32 and 33) and Vcam1 (vascular cell adhesion molecule; refs. 34 and 35), respectively.
The identified Char4 region on mouse Chr.3 can now be used as an entry point to look for possible association of the corresponding human syntenic chromosomal region with susceptibility and/or severity of disease in endemic areas of malaria. The power of this approach is exemplified by the NRAMP1 gene, which was originally isolated by positional cloning of a simple trait regulating resistance to infections in mice (36), and that was subsequently shown to be a determinant of susceptibility to tuberculosis (37) and leprosy (38) in association and linkage studies in humans. Human 4q21-q25 is syntenic with the mouse Chr.3 segment carrying Char4. This region can now be examined as a possible genetic predisposition or outcome predictor for malaria in humans.
Supplementary Material
Acknowledgments
The authors are indebted to Patrick Fortin for skillful technical assistance. This work was supported by a research grant from the Burrough Wellcome Funds (to P.G. and M.M.S.), and by a research grant from the Canadian Genetic Disease Network (to P.G.). P.G. is an International Research Scholar of the Howard Hughes Medical Institute (HHMI) and a senior scientist of the Canadian Institutes of Health Research. L.R.C. is supported by the Wellcome Trust and by NIH Grant EY-12562. A.F. is supported by a studentship from McGill University.
Abbreviations
- QTL
quantitative trait loci
- RCS
recombinant congenic strains of mice
- lod
logarithm of odds
- Chr.n
chromosome n
- cM
centimorgans
- B6
C57BL/6J
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Butler D. Nature (London) 1997;386:535–536. doi: 10.1038/386535a0. [DOI] [PubMed] [Google Scholar]
- 2.Marsh K. Lancet. 1997;349:SIII1–SIII2. [Google Scholar]
- 3.Hill A V. Annu Rev Immunol. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- 4.Kwiatkowski D. Curr Opin Genet Dev. 2000;10:320–324. doi: 10.1016/s0959-437x(00)00087-3. [DOI] [PubMed] [Google Scholar]
- 5.Taylor J G, Ferdig M T, Su X Z, Wellems T E. Curr Opin Genet Dev. 2000;10:314–319. doi: 10.1016/s0959-437x(00)00077-0. [DOI] [PubMed] [Google Scholar]
- 6.Wilson A G, Symons J A, McDowell T L, McDevitt H O, Duff G W. Proc Natl Acad Sci USA. 1997;94:3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knight J C, Udalova I, Hill A V, Greenwood B M, Peshu N, Marsh K, Kwiatkowski D. Nat Genet. 1999;22:145–150. doi: 10.1038/9649. [DOI] [PubMed] [Google Scholar]
- 8.Sherman I W. Malaria. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 401–417. [Google Scholar]
- 9.Stevenson M M, Lyanga J J, Skamene E. Infect Immun. 1982;38:80–88. doi: 10.1128/iai.38.1.80-88.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortin A, Belouchi A, Tam M F, Cardon L, Skamene E, Stevenson M M, Gros P. Nat Genet. 1997;17:382–383. doi: 10.1038/ng1297-382. [DOI] [PubMed] [Google Scholar]
- 11.Foote S J, Burt R A, Baldwin T M, Presente A, Roberts A W, Laural Y L, Lew A M, Marshall V M. Nat Genet. 1997;17:380–381. doi: 10.1038/ng1297-380. [DOI] [PubMed] [Google Scholar]
- 12.Riopel J, Tam M, Mohan K, Marino M W, Stevenson M M. Infect Immun. 2001;69:129–136. doi: 10.1128/IAI.69.1.129-136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su Z, Stevenson M M. Infect Immun. 2000;68:4399–4406. doi: 10.1128/iai.68.8.4399-4406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sam H, Su Z, Stevenson M M. Infect Immun. 1999;67:2660–2664. doi: 10.1128/iai.67.5.2660-2664.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burt R A, Baldwin T M, Marshall V M, Foote S J. Immunogenetics. 1999;50:278–285. doi: 10.1007/s002510050603. [DOI] [PubMed] [Google Scholar]
- 16.Benten W P, Bettenhaeuser U, Wunderlich F, Van Vliet E, Mossmann H. Infect Immun. 1991;59:4486–4490. doi: 10.1128/iai.59.12.4486-4490.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortin A, Diez E, Rochefort D, Laroche L, Malo D, Rouleau G A, Gros P, Skamene E. Genomics. 2001;74:21–35. doi: 10.1006/geno.2001.6528. [DOI] [PubMed] [Google Scholar]
- 18.Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newburg L. Genomics. 1987;1:174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 19.Demant P, Hart A A. Immunogenetics. 1986;24:416–422. doi: 10.1007/BF00377961. [DOI] [PubMed] [Google Scholar]
- 20.Castellani L W, Weinreb A, Bodnar J, Goto A M, Doolittle M, Mehrabian M, Demant P, Lusis A J. Nat Genet. 1998;18:374–377. doi: 10.1038/ng0498-374. [DOI] [PubMed] [Google Scholar]
- 21.Fijneman R J, de Vries S S, Jansen R C, Demant P. Nat Genet. 1996;14:465–467. doi: 10.1038/ng1296-465. [DOI] [PubMed] [Google Scholar]
- 22.Moen C J, Groot P C, Hart A A, Snoek M, Demant P. Proc Natl Acad Sci USA. 1996;93:1082–1086. doi: 10.1073/pnas.93.3.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Wezel T, Stassen A P, Moen C J, Hart A A, van der Valk M A, Demant P. Nat Genet. 1996;14:468–470. doi: 10.1038/ng1296-468. [DOI] [PubMed] [Google Scholar]
- 24.Lipoldova M, Svobodova M, Krulova M, Havelkova H, Badalova J, Nohynkova E, Holan V, Hart A A M, Volf P, Demant P. Genes Immun. 2000;1:170–206. doi: 10.1038/sj.gene.6363660. [DOI] [PubMed] [Google Scholar]
- 25.Stassen A P, Groot P C, Eppig J T, Demant P. Mamm Genome. 1996;7:55–58. doi: 10.1007/s003359900013. [DOI] [PubMed] [Google Scholar]
- 26.Oosterwegel M, van de Wetering M, Timmerman J, Kruisbeek A, Destree O, Meijlink F, Clevers H. Development (Cambridge, UK) 1993;118:439–448. doi: 10.1242/dev.118.2.439. [DOI] [PubMed] [Google Scholar]
- 27.Galceran J, Farinas I, Depew M J, Clevers H, Grosschedl R. Genes Dev. 1999;13:709–717. doi: 10.1101/gad.13.6.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gervais F, Stevenson M, Skamene E. J Immunol. 1984;132:2078–2083. [PubMed] [Google Scholar]
- 29.Gervais F, Desforges C, Skamene E. J Immunol. 1989;142:2057–2060. [PubMed] [Google Scholar]
- 30.Minta J O, Wong M J, Kozak C A, Kunnath-Muglia L M, Goldberger G. Mol Immunol. 1996;33:101–112. doi: 10.1016/0161-5890(95)00116-6. [DOI] [PubMed] [Google Scholar]
- 31.Sha W C, Liou H C, Tuomanen E I, Baltimore D. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 32.Yoshida H, Hayashi S, Kunisada T, Ogawa M, Nishikawa S, Okamura H, Sudo T, Shultz L D. Nature (London) 1990;345:442–444. doi: 10.1038/345442a0. [DOI] [PubMed] [Google Scholar]
- 33.Guleria I, Pollard J W. Infect Immun. 2001;69:1795–1807. doi: 10.1128/IAI.69.3.1795-1807.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurtner G C, Davis V, Li H, McCoy M J, Sharpe A, Cybulsky M I. Genes Dev. 1995;9:1–14. doi: 10.1101/gad.9.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Koni P A, Joshi S K, Temann U A, Olson D, Burkly L, Flavell R A. J Exp Med. 2001;193:741–754. doi: 10.1084/jem.193.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vidal S M, Malo D, Vogan K, Skamene E, Gros P. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 37.Bellamy R, Ruwende C, Corrah T, McAdam K P, Whittle H C, Hill A V. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 38.Abel L, Sanchez F O, Oberti J, Thuc N V, Hoa L V, Lap V D, Skamene E, Lagrange P H, Schurr E. J Infect Dis. 1998;177:133–145. doi: 10.1086/513830. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.