Abstract
Acute myeloid leukemias (AML) are clonal disorders of hematopoietic progenitor cells which are characterized by relevant heterogeneity in terms of phenotypic, genotypic, and clinical features. Among the genetic aberrations that control disease development there are microRNAs (miRNAs). miRNAs are small non-coding RNAs that regulate, at post-transcriptional level, translation and stability of mRNAs. It is now established that deregulated miRNA expression is a prominent feature in AML. Functional studies have shown that miRNAs play an important role in AML pathogenesis and miRNA expression signatures are associated with chemotherapy response and clinical outcome. In this review we summarized miRNA signature in AML with different cytogenetic, molecular and clinical characteristics. Moreover, we reviewed the miRNA regulatory network in AML pathogenesis and we discussed the potential use of cellular and circulating miRNAs as biomarkers for diagnosis and prognosis and as therapeutic targets.
Keywords: microRNAs, acute myeloid leukemia, biomarkers, therapeutic targets
1. Introduction
Acute myeloid leukemia (AML) is an aggressive hematological malignancy characterized by the abnormal proliferation and differentiation of myeloid progenitor cells [1,2]. This neoplasm is the most common acute leukemia in adult patients and can arise “de novo” or as a secondary event [3]. In particular, secondary AML can derive from a previous clonal disorder of hematopoiesis, such as myelodysplastic syndrome (MDS) or chronic myeloproliferative neoplasm (MPN), or following a previous therapy (chemotherapy or radiotherapy, immunosuppressive drugs or environmental leukemogenic agents) [4,5]. These myeloid malignancies are characterized by acquired common recurrent mutations in hematopoietic progenitors. Gene alterations can contribute to hematopoietic transformation conferring a growth advantage or modifying the expression of key transcriptional targets in myelopoiesis. These mutations occur in genes with known roles in regulating chromatin and/or methylation states in hematopoietic progenitors (e.g., mixed-lineage leukemia (MLL)), or exert a role in altering epigenetic pattern in myeloid malignancies (e.g., TET methylcytosine dioxygenase 2 (TET2), DNA methyltransferase 3A (DNMT3A), sex combs-like 1 (ASXL1), isocitrate dehydrogenase (IDH) 1 and IDH2). They contribute to the pathogenesis, and could represent attractive targets for directed therapy in myeloid malignancies [6].
AML is a highly heterogeneous disease at molecular and clinical levels; well-identified genetic and cytogenetic aberrations hold a pathogenetic and prognostic relevance in this neoplasm [3]. The new 2016 WHO classification defined six major AML entities incorporating genetic alterations with morphology, immunophenotype, and clinical presentation: (i) AML with recurrent genetic abnormalities; (ii) AML with myelodysplasia-related changes; (iii) therapy-related AML; (iv) AML not otherwise specified; (v) myeloid sarcoma; (vi) myeloid proliferation related to Down syndrome [7]. On the basis of cytogenetic and molecular profile, AML patients can be divided into favorable, intermediate, and adverse prognostic risk groups [3]. AML treatment is currently based on high-intensity induction chemotherapy in eligible patients to achieve complete remission (CR), followed by consolidation regimen, including conventional chemotherapy, as well as hematopoietic cell transplantation [1]. However, patients achieving CR can relapse due to the persistence of minimal residual disease (MRD) [1,8].
It is important to underline, beyond, that AML is perpetuated and maintained by a small population of leukemia initiating cells or leukemia stem cells (LSCs) [9,10]. These cells exhibit similar characteristics of hematopoietic stem cells (HSCs), including the ability to generate identical daughter cells as well as differentiated cells. On the contrary, LSCs have different properties than the AML bulk population [11,12]. These features make LSCs difficult to eliminate with standard chemotherapy, and therefore, result in a source of malignancy resistance and relapse.
MicroRNAs (miRNAs) are evolutionary conserved short non-coding single-stranded RNAs (19–22 nucleotides) [13,14] that negatively regulate mRNA stability [14,15,16]. They play an essential role in many biological functions, such as cell growth, proliferation, differentiation, and apoptosis [13,17]. Moreover, miRNAs can act as oncogenes or tumor suppressors, contributing to malignant transformation in solid and hematological tumors, including AML [16,18,19]. In addition, miRNAs regulate different mRNA targets, and their modulation can represent a potential therapeutic target in leukemic progenitors, as well as in LSCs. In this context, the identification of miRNA expression patterns in HSCs, LSCs, and leukemic progenitors is required.
miRNA expression is frequently deregulated in AML by different mechanisms, like: (i) copy number alterations; (ii) epigenetic changes; (iii) miRNA location in proximity of oncogenic genomic region due to chromosomal translocation or overexpression of protein-coding gene; (iv) aberrant targeting of miRNA promoter regions by altered transcription factors or oncoproteins; and finally (v) deregulated miRNA processing [14].
During the last 15 years, microarray was a widely used platform to investigate miRNA expression in AML. In particular, many studies identified the expression signatures associated with different AML subtypes, especially with specific molecular and cytogenetic alterations [20]. Recently, the emerging RNA-Sequencing technologies were used to evaluate miRNA abundance and to discover new miRNAs in AML [21].
In this review, we summarized miRNAs differentially expressed in AML subtypes, LSCs, and HSCs, their role in AML pathogenesis, their potential use as disease biomarkers for diagnosis/prognosis and as therapeutic targets.
2. Distinctive Pattern of miRNAs in AML
Molecular and cytogenetic features are currently defined as entities of clinical significance, and are included in diagnostic criteria of AML, according to 2016 WHO classification [7].
Numerous studies demonstrated the existence of distinct miRNA profiles in different AML subtypes, indicating that miRNA signature contributed to AML heterogeneity, and suggesting its potential inclusion in clinical setting [22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43]. In Table 1 were reported all major studies comparing miRNA profile between AML blasts and normal cells, and among AML with recurrent genetic abnormalities [22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43]. Most of the reported studies exhibited high concordance in AML miRNA profile. For example, in patients with NPM1 mutation, different authors found the upregulation of miR-10a, miR-10b, and miR-196b [26,37,38], and the downregulation of miR-192 [36,38]. A common signature was also reported in patients with t(8;21) showing the upregulation of miR-126 [22,23] and miR-146a [24,25], while overexpression of miR-155 was strongly associated with FLT3-internal tandem duplication (ITD) alteration [26,34,36,39].
Table 1.
MicroRNA (miRNA) expression in acute myeloid leukemias (AML) with recurrent genetic abnormalities.
| Genetic Abnormalities | miRNAs | |||
|---|---|---|---|---|
| Upregulated | Downregulated | |||
| t(8;21)(q22;q22.1) RUNX1-RUNX1T1 | miR-126, miR-320, miR-328, miR-449b; | [22] | miR-9, miR-10a, miR-125b, miR-133a, miR-135a, miR-148a, miR-196a, miR-221; | [22] |
| miR-126/126*; | [23] | miR-133a; | [24] | |
| miR-146a; | [24] | let-7b, let-7c; | [26] | |
| miR-146a, miR-181a, miR-181b | [25] | miR-221, miR-222, miR-223; | [27] | |
| miR-223; | [28] | |||
| inv(16)(p13.1q22) or t(16;16)(p13.1;q22) CBFB-MYH11 | miR-126/126*; | [23] | miR-29c, miR-149; | [22] |
| miR-99a, miR-100, miR-224; | [24] | miR-92; | [23] | |
| miR-212 | [29] | miR-127, let-7b, let-7c; | [26] | |
| miR-221, miR-222, miR-223 | [27] | |||
| PML-RARA | miR-100, miR-181a, miR-181b, miR-181c, miR-181d, miR-224, miR-368, miR-382, miR-424; | [23] | miR-17-5p, miR-20a, miR-126/126*, miR-150; | [23] |
| miR-127, miR-154, miR-154*, miR-299, miR-323, miR-368, miR-370; | [24] | miR-17-3p, miR-185, miR-187, miR-194, miR-200a, miR-200b, miR-200c, miR-330, miR-339; | [24] | |
| miR-100, miR-125b, miR-146-5p, miR-181a, miR-181b; | [25] | miR-18b, miR-22, miR-24, miR-27a, miR-126-3p, miR-150, miR-223, miR-342-3p, miR-378 let-7b, let-7c; | [25] | |
| miR-127, miR-134, miR-299-5p, miR-323, miR-376a, miR-382; | [26] | miR-107, miR-342, let-7c | [30] | |
| miR-15a, miR-15b, miR-16, miR-142-3p, miR-142-5p, miR-223, let-7a, let-7d; | [30] | |||
| miR-128a | [31] | |||
| t(9;11)(p21.3;q23.3) MLLT3-KMT2A | miR-196b; | [32] | miR-29a, miR-29b, miR-29c; | [34] |
| miR-196b; | [33] | miR-495 | [35] | |
| miR-326 | [34] | |||
| t(6;9)(p23;q34.1);DEK-NUP214 | - | - | ||
| inv(3)(q21.3q26.2) or t(3;3)(q21.3;q26.2); GATA2, MECOM | - | miR-30a-3p, miR-95, miR-101, miR-145, miR-199b, miR-422a miR-618 | [22] | |
| t(1;22)(p13.3;q13.3) RBM15-MKL1 | - | - | ||
| BCR-ABL1 | - | - | ||
| mutated NPM1 | miR-10a, miR-10b, miR-196a, miR-196b; | [26] | miR-22, miR-127, miR-128a, miR-135a, miR-139, miR-145, miR-192, miR-193b, miR-198, miR-204, miR-299, miR-324, miR-326, miR-373, miR-373*, miR-383, miR-429, miR-486, miR-493, miR-498; | [36] |
| miR-9, miR-10a, miR-10b, miR-15a, miR-16a, miR-16b, miR-16-1, miR-16-2, miR-17, miR-18a, miR-19a, miR-19b, miR-20, miR-21, miR-24, miR-29a, miR-29b, miR-29c, miR-100, miR-98, miR-102, miR-106, miR-142, miR-152, miR-155, miR-195, miR-369, miR-378, miR-374, let-7a-1, let-7a-2, let-7a-3, let-7c, let-7d, let-7f, let-7g; | [36] | miR-126, miR-130a, miR-424, miR-450a, miR-451, miR-486-5p; | [37] | |
| miR-9, miR-10a, miR-10b, miR-100, miR-196a, miR-196b, let-7a, let7c; | [37] | miR-192, miR-495; | [38] | |
| miR-7, miR-10a, miR-10b, miR-15a, miR-15b, miR-16, miR-17, miR-18a, miR-28-5p, miR-19a, miR-19b, miR-20a, miR-21, miR-23a, miR-23b, miR-27a, miR-27b, miR-29a, miR-29b, miR-29c, miR-30a, miR-30b, miR-30c, miR-30e, miR-92a, miR-98, miR-105, miR-106a, miR-124, miR-125a-5p, miR-125b, miR-136, miR-142-5p, miR-146a, miR-151-5p, miR-155, miR-195, miR-196b, miR-199a-3p, miR-200b, miR-221, miR-223, miR-224, miR-320, miR-339-5p, miR-340*, miR-361-5p, miR-369-3p, miR-373, miR-379, miR-380, miR-381, miR-382, miR-491-5p, miR-493*, miR-494, miR-518c*, miR-526b, let-7a, let-7b, let-7c, let-7d, let-7e, let-7f | [38] | miR-424 | [39] | |
| biallelic mutations of CEBPA | miR-128, miR-181a, miR-181a*, miR-181b, miR-181c, miR-181d, miR-192, miR-219-1-3p, miR-224, miR-335, miR-340 | [40] | miR-34a, miR-194; | [40] |
| miR-34a | [41] | |||
| mutated RUNX1 | miR-21, miR-220, miR-595 | [42] | miR-99a, miR-100, miR-223, let-7a, let-7f | [42] |
| FLT3-ITD | miR-10b, miR-155; | [26] | miR-144, miR-451, miR-486-5p, miR-488 | [43] |
| miR-128a; | [31] | |||
| miR-10a, miR-10b, miR-155; | [34] | |||
| miR-155; | [36] | |||
| miR-155; | [39] | |||
| miR-125b2*; | [43] | |||
*: complementary strand to microRNA.
Therefore, some studies showed partial overlap in miRNA profile, and this is probably due to different microarray platforms, sample type, or comparison analysis among AML subtypes.
According to cytogenetic and molecular alterations, European LeukemiaNet divided AML patients into three risk groups: favorable, intermediate, or adverse [44]. Each subtype seems to exhibit a unique miRNA signature that distinguishes it from other AML subtypes. For example, Marcucci G et al. reported the increased expression of all members of miR-181 family in patients with favorable risk (cytogenetically normal AML (CN-AML) with CEBPA mutations) [40] and its decreased expression in a high risk subgroup (CN-AML with FLT3-ITD and/or wild-type NPM1) [45]. Similarly, in other studies, miR-181 was also observed increased in cytogenetically abnormal AML (CA-AML) with favorable cytogenetic abnormalities, such as t(15;17) (favorable risk), and decreased in CA-AML with unfavorable cytogenetic alterations, such as MLL-rearrangements (adverse risk) [23,46]. Garzon et al. compared CN-AML with CA-AML patients, identifying a signature in CN-AML of 10 upregulated (miR-10a, miR-10b, miR-16-2, miR-21, miR-26a, miR-30c, miR-181b, miR-192, miR-368, and let-7a-2) and 13 downregulated miRNAs (miR-126, miR-145, miR-182, miR-183, miR-191, miR-193, miR-194, miR-196b, miR-199a, miR-200c, miR-203, miR-204, and miR-299) [34]. miR-139-5p was found downregulated in CN-AML with mutated FLT3, and acted as tumor suppressor in a primary AML transplant model [47]. Dixon-McIver et al. reported that the expression of miR-9 and let7b was low in the favorable groups, and high in intermediate and adverse AML groups [24]. They also found miR-125b mainly expressed in AML with a normal karyotype [24].
miRNA expression was also correlated with bone marrow (BM) morphology. Chen et al. demonstrated that mature miR-181 expression was detectable in BM undifferentiated progenitor cells [48]. Moreover, its expression was strongly correlated with the AML morphological subtype, resulting in elevated levels in samples with M1 or M2 French–American–British (FAB) classification, compared with the samples with M4 or M5 morphology [49]. By contrast, in normal BM, miR-181a has been reported to be preferentially expressed in B cells, T cells, monocytes, and granulocytes, which are more closely allied to M4 and M5 subtypes [50]. Another study, instead, compared M1 with M5 miRNA profile, indicating the higher expression of not only miR-181a and miR-181b, but also miR-181a*, miR-181d, miR130a, miR-135b, miR-146a, miR-146b, and miR-663 in FAB M1. Instead, miR-21, miR-193a, and miR-370 were overexpressed in FAB M5 samples [51]. Recently, de Leeuw et al. identified the highest expression of miR-551b in the most primitive cell populations, the HSC and multipotent progenitors; instead, differentiated progenitors, monocytes and lymphocytes, showed decreased and absent miR-551b expression, respectively. This expression pattern might indicate a miR-551b role in early hematopoiesis and stem cells. Therefore, authors hypothesized that its expression in AML might be indicative of an immature leukemia with stem cell features. In fact, they found a high miR-551b expression in undifferentiated AML (FAB M0); on the contrary, miR-551b was not expressed in patients with favorable cytogenetics [52].
Yang et al. demonstrated that miR-122 was downregulated in AML with respect to normal controls, and its decreased expression was more frequently observed in FAB M7 with respect to M1–M6 subtypes, and in unfavorable cytogenetic abnormalities, acting as tumor suppressor [53].
In a recent paper, we evaluated miR-128a expression in AML patients stratified for morphologic features, observing high expression levels of this miRNA in AML with maturation and in acute promyelocytic leukemia cases compared to healthy subject CD34+ [31].
In any case, the majority of reported studies, based on the FAB classification system, needs to be revised according to the 2016 WHO classification [7].
Thus, the all cited literature data highlighted that the aberrant expression of miRNAs contributes to AML heterogeneity.
3. miRNAs Involved in AML Pathogenesis
MicroRNAs alterations are recognized, through various mechanisms, to be involved in AML pathogenesis [54]. In particular, literature data indicate that miRNAs can induce leukemogenesis, altering numerous biological processes, including self-renewal, survival, proliferation, differentiation, and epigenetic regulation. Their involvement in leukemic development and progression is due to their collaboration with deregulated protein (oncogene or tumor suppressor), either by direct control of protein translation, or by working jointly with them to promote malignancy [14,54].
To summarized these concepts, we have illustrated in Table 2 the most important miRNAs found to play a role in AML, specifying for each miRNA the altered expression (up or downregulation), the mechanisms of dysregulation, their targets, and their functional effects in AML pathogenesis [23,28,31,32,33,41,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102,103,104,105]. However, literature data revealed that miRNA dysregulation in AML can have different effects on the basis of their expression levels. miR-155, for example, is upregulated respect to healthy subjects having an oncogenic function in AML FLT3-ITD subgroup [91,95]; by contrast, in different AML subsets, it was demonstrated to have a tumor suppress function [106,107].
Table 2.
miRNAs involved in AML pathogenesis.
| miRNA | Altered Expression | Mechanism of Dysregulation | Targets | Functional Effect of Mirna Altered Expression | Refs |
|---|---|---|---|---|---|
| miR-9 | Up: in MLL-AML | Promoter targeted by MLL-fusion proteins | RHOH, RYBP | Increased proliferation, survival and leukemogenesis in mice | [55] |
| Down: in t(8;21) AML | HMGA2, LIN28B | Increased proliferation and decreased monocytic differentiation | [56] | ||
| Down: in EVI1-induced AML | EVI1 hypermethylates promoter | FOXO1, FOXO3 | Increased proliferation and decreased monocytic differentiation | [57] | |
| miR-17-92 cluster | Up: in LSCs in MLL-AML | Activated by MYC | P21 | Increased proliferation, survival, differentiation, self-renewal, colony forming capacity and leukemogenesis in mice | [58] |
| Genomic amplification and upregulation by MLL-fusion proteins | Inhibited differentiation and apoptosis, promoted cell proliferation | [59] | |||
| miR-22 | Up: in MDS/MDS-derived AML | TET2 | Increased proliferation, survival, self-renewal and decreased differentiation. miR-22 overexpression led to myeloid malignancy in mice | [60] | |
| Down: in de novo AML | Downregulated via TET1/GFI1/EZH2/SIN3A-mediated epigenetic repression and DNA copy-number loss | CRTC1, FLT3, MYCBP | Increased AML blast cell growth. Decreased differentiation and increased leukemic progression in mice | [61] | |
| Down: in de novo AML | Increased with loss of PU.1 | MECOM | Increased AML blast cell growth. Decreased differentiation and increased leukemic progression | [62] | |
| miR-29b | Down: in various subtypes of AML | Repressed by c-MYC | AKT2, CCND2 | Increased cell growth, leukemic progression in vivo | [63] |
| Down: in various subtypes of AML | MCL-1, CXXC6, CDK6 | Increased cell growth, decreased apoptosis, leukemic progression in vivo | [64] | ||
| Down: in t(8;21) AML | Repressed by MYC and NF-κB | SP1 | KIT upregulation contributing to malignant proliferation | [65] | |
| Down: in CEBPA mutated AML | Downregulated via loss of CEBPA; chromosome 7q deletions | [66] | |||
| Down: in various subtypes of AML | SP1, DNMT3A, DNMT3B | miR-29b downregulation led global DNA hypermethylation | [75] | ||
| miR-34a | Down: in CEBPA mutated AML | Downregulated via loss of CEBPA | E2F3 | Increased proliferation and decreased differentiation | [41] |
| Down: in de novo AML | Downregulated via loss of MUC1 | PDL1 | Immune dysregulation | [67] | |
| Down: in de novo AML | PDL1 | Immune dysregulation | [68] | ||
| Down: in CEBPA mutated AML cell lines | HMGB1 | Inhibited cell apoptosis and increased autophagy | [69] | ||
| miR-99 | Up: in initial diagnosis and relapse | Regulated self-renewal, inhibiting differentiation and cell cycle entry | [70] | ||
| Up: in AML-AF9 | SMARCA5, HS2ST3, HOXA1 | Increased proliferation, colony formation, cell survival, inhibited differentiation | [71] | ||
| Up: in pediatric-onset AML (M1–M5) | CTDSPL TRIB2 | Increased proliferation, colony formation, cell survival | [72] | ||
| miR-125b | Up: in MDS and AML with t(2;11)(p21;q23) | Increased by t(2;11)(p21;q23) | Inhibited differentiation | [73] | |
| Up: in AML | LIN28A | Uncontrolled generation of myeloid cells | [74] | ||
| miR-125b overexpression led to AML in mice | [76] | ||||
| IRF4 | miR-125b overexpression induced myeloid leukemia in mice by inducing immortality, self-renewal and tumorigenesis in myeloid progenitors | [77] | |||
| Up: in pediatric AML | miR-125b was associated with PML/RARA status | [78] | |||
| FES, PU.1 | Blocked monocytic differentiation of AML in vitro | [105] | |||
| miR-126 | Up: in t(8;21) and inv(16) AML | Epigenetic regulation | PLK2 | Inhibited cell apoptosis and increased cell viability | [23] |
| Up: in LSC of CN-AML | Epigenetic regulation | Increased LSC maintenance and self-renewal | [79] | ||
| Up: in t(8;21) AML | ERRFI1, SPRED1, FZD7 | Both gain and loss of function of miR-126 promoted leukemogenesis in vivo through targeting distinct gene signaling | [80] | ||
| Up: in LSCs of AML | Epigenetic regulation | ADAM9, ILK, GOLPH3, CDK3, TOM1 | Increased LSC maintenance and self-renewal, quiescence, chemotherapy resistance in vivo | [81] | |
| Up: in LSCs of AML | Increased leukemic growth, and survival of leukemic stem and progenitor cells in vivo | [82] | |||
| miR-128 | Up: in AML LSC, highest in FLT3-ITD and PML-RARalfa | LIN28A | Increased proliferation and decreased differentiation | [31] | |
| miR-145 miR-146a | Down: in AML with normal karyotype | Resulted in AML in vivo | [83] | ||
| miR-146a | Down: in del(5q) MDS | TIRAP, TRAF6 | Inappropriate activation of innate immune signaling in HSPCs and megakaryocytic abnormalities | [84] | |
| Deleted: in del(5q) MDS/AML | Deletion in del(5q) MDS/AML | Co-deletion of TIFAB and miR-146a may cooperate to induce TRAF6 signaling contributing to ineffective hematopoiesis | [85] | ||
| IRAK1 | miR-146a knockout mice developed myeloid and lymphoid malignancies | [86] | |||
| Deleted: in del(5q) MDS/AML | Deletion in del(5q) MDS/AML | Increased cell survival and proliferation of propagating cells through the TRAF6/p62/NF-κB complex | [87] | ||
| miR-146a deletion led to myeloproliferation in mice | [88] | ||||
| miR-150 | Down: in various subtypes of AML | NANOG | Increased proliferation, colony and sphere formation, increased tumor growth in vivo | [89] | |
| Down: in various subtypes of AML | EIF4B, FOXO4, PRKCA, TET3 | Increased cell growth and inhibited apoptosis in vitro and in vivo | [90] | ||
| miR-155 | Up: in FLT3-ITD+ AML | Targeted by STAT5 and NF-κB | PU.1 | Inhibited myeloid differentiation. Proliferation and survival of FLT3-ITD leukemic cells | [91] |
| Up: in CN-AML | Negative impact on outcome | [92] | |||
| Up: in MLL AML | Upregulated by MLL fusion genes via MEIS1 | No effect in MLL-AF9 mouse model | [93] | ||
| Up: in FLT3-ITD+ AML | CEBPB, SHIP1 | miR-155 overexpression led to myeloproliferative neoplasm in mice | [94] | ||
| Up: in AML (FAB M4-M5) | Enforced expression of miR-155 in HSCs caused a myeloproliferative disorder in mice BM | [95] | |||
| Up: in FLT3-ITD+ AML | Upregulated by IL3 | [96] | |||
| Up: in various subtypes of AML | Epigenetically regulated | PU.1 | Leukemic cell growth of blast cells | ||
| miR-182 | Down: in AML with CEBPA mutation, t(8;21) and t(15;17) | Upregulated via loss of CEBPA | CEBPA | Decreased myeloid differentiation | [97] |
| miR-192 | Down: in various subtype of AML | CCNT2 | Increased proliferation and cell cycling, decreased differentiation | [98] | |
| miR-193a | Down: in t(8;21) AML | Epigenetically silenced by AML1/ETO | AML1/ETO, DNMT3A, HDAC3, KIT, CCND1, MDM2 | Decreased apoptosis and differentiation | [99] |
| Down: in various subtypes of AML | Epigenetically repressed by promoter hypermethylation | KIT | Increased cell growth. decreased apoptosis and differentiation, and increased KIT expression | [100] | |
| miR-194-5p | Down: in AML cell line | Increased by HDACi SAHA treatment in AML cells | BCLAF1 | Decreased apoptosis, differentiation | [101] |
| miR-196b | Up: in MLL-associated AML | Activated by MLL-fusion proteins | Increased proliferation and survival. Decreased differentiation and replating potential. | [32] | |
| Up: in MLL-associated AML | Activated by MLL-fusion proteins; Co-expressed with HOXA9 in MLL rearranged leukemia | HOXA9, MEIS1, FAS | Inhibited differentiation, promoted cell proliferation via inhibiting apoptosis. Induced leukemic progression in mice | [33] | |
| miR-223 | Down: in t(8;21) AML | Epigenetically silenced by AML1/ETO | Myeloid differentiation block | [28] | |
| Down: in various subtypes of AML | Activated by CEBPA and repressed by E2F1 transcription factors | E2F1 | Increased proliferation. Decreased differentiation/granulopoiesis | [102] | |
| Down: in various subtypes of AML | FBXW7 | Increased cell proliferation and enhanced apoptosis | [103] | ||
| Down: in AML with adverse prognosis | Impaired differentiation | [104] |
Abbreviations: AML, acute myeloid leukemia; LSCs, leukemia stem cells; MDS, myelodysplastic syndrome; CN-AML, cytogenetically normal AML.
Narayan et al. demonstrated in three different murine models of AML (HoxA9/Meis1, MLL-ENL, MLL-AF9) that this is due to different expression levels of miR-155. High doses of this miRNA repressed clonal proliferation, instead, intermediate levels of miR-155 showed an oncogenic function, leading to an increased proliferation and enhancing colony formation [108]. Moreover, this group confirmed that the intermediate miR-155 expression level was associated with poor prognosis in pediatric AML patients, suggesting a prevalent oncogenic function in this hematological neoplasm [34,95,108].
miRNAs exert a functional role in leukemogenesis both in vivo and in vitro controlling their mRNA targets. Generally, miRNA–mRNA relationships are related to AML subtypes, and some targets play an important role than others, depending on the AML cytogenetic and molecular signatures [109], as reported in Table 2.
One of the most studied miRNAs in AML, miR-155, for example, negatively regulated the transcription factor PU.1, involved in hematopoietic cell determination [110]. Basova et al. confirmed this data in AML pathogenesis, and reported miR-155 cooperation with Myb. This last acted as an activator of cell proliferation and inhibitor of myeloid maturation, with a hierarchical order of Myb/miR-155/PU.1. Moreover, p53 regulated this axis. Its loss caused the upregulation of the two oncogenes Myb and miR-155, concomitantly with progressive PU.1 downregulation and characterized aggressive AML. The inhibition of the oncogenes Myb and miR-155, or by rescue of PU.1, led to leukemia blast differentiation [111]. miR-155 was able to induce proliferation of myeloid cells, causing a myeloproliferative syndrome after ectopic expression in myeloid progenitors [95]. Ghani et al. suggested that PU.1 activated miR-155 expression inducing quiescent HSC to entry into cell cycle, and thus activated their initial differentiation into a transient myeloid progenitor stage. When PU.1 was removed from its binding site at pre–miR-155, the lack of expression of miR-155 blocked proliferation, thus preventing leukemia, and initiated terminal myeloid maturation. Thus, PU.1 was needed to initiate the expression of this miRNA, but not for its maintenance [112].
miR-17 polycistronic miRNA (miR-17-92 cluster) has been shown to be highly expressed in human MLL-rearranged leukemia [58,59]. Recently, Wong et al. demonstrated that the cyclin-dependent kinase inhibitor p21 was a critical downstream target of this microRNA. Leukemic cells expressing increased levels of miR-17 polycistron displayed a higher frequency of LSCs, a block of differentiation, and increased proliferation associated with reduced expression of p21. Moreover, c-myc was shown to be a critical upstream regulator of miR-17 expression in MLL transformation [58]. Notably, downregulation of the miR-17p-92 cluster can promote myeloid lineage fate in normal cord blood CD34+ HPCs [113].
MiR-125b was found upregulated in AML. Chaudhuri et al. demonstrated that miR-125b overexpression led to uncontrolled generation of myeloid progenitors and mature myeloid cells that subsequently caused a highly invasive myeloid leukemia. The authors also showed that Lin28A, an induced pluripotent stem cell gene, was a bona fide primary target of miR-125b in hematopoietic cells. Lin28A downregulation mimicked the preleukemic state induced by miR-125b. [74]. MiR-125b physiologically regulated hematopoiesis [114]. Studies of miRNA expression profiles in multiple hematopoietic subpopulations demonstrated that miR-125b was one of the most expressed miRNAs in HSCs, compared with all other progenitor populations. Moreover, miR-125b was specifically enriched in the long-term HSC population [76]. While its level significantly decreased in committed progenitors, miR-125b was more highly expressed in common lymphoid progenitors than in common myeloid progenitors [115].
It is known that leukemic cells release extracellular vesicles (EVs) that modify target cells in BM microenvironments by transferring their metabolic, protein, and genetic content, such as mRNA, DNA, and miRNA [116,117]. EV miRNAs act to create a favorable BM microenvironment that sustains and support leukemia [117,118,119]. In this context, Hornick et al. showed a direct effect of AML exosome (Exo) miRNAs compromising hematopoiesis of healthy hematopoietic stem and progenitor cells (HSPCs) [120]. This study suggested that leukemia-derived Exo were sufficient to induce systemic impairment of hematopoiesis, demonstrating that miR-150 and miR-155 and their regulatory network were sufficient to suppress HSPC clonogenicity. [120]. Similarly, Razmkhah et al. demonstrated that AML BM microvesicles (MVs) induced colony formation and increased expression of two leukemic oncomiRs, miR-21 and miR-29a, in healthy HSPCs [121]. In addition, Horiguchi et al. revealed that miR-7977 in EVs from AML/myelodysplastic syndrome induced a reduction of hematopoietic-supportive capacity of normal BM mesenchymal stem cells [122].
4. Differential Expression of MiRNAs in HSCs and LSCs
It is well established that miRNAs play a role in the regulation of normal hematopoiesis and in HSC maintenance. Similarly, miRNAs regulated the behavior of LSCs, their self-renewal and development [11,54]. In fact, LSCs showed a differential expression of miRNAs with respect to HSC. Therefore, modulation of miRNA aberrantly expressed in LSC and, consequently, the regulation of their targets could determine the elimination of resistant LSCs, by inducing apoptosis or by sensitizing them to chemotherapy [123].
Although many studies reported miRNA profiles of primary AML cell bulk, as described above, little is known about miRNA expression in LSCs.
Recently, de Leeuw et al. compared LSCs (CD34+ CD38−) versus HSCs, and LSCs versus leukemic progenitors (CD34+ CD38+), all from the same AML BM, identifying multiple differentially expressed miRNAs. In particular, they found miR-551b, miR-10a, miR-151-5p, miR-29b, and miR-125b were higher expressed in HSCs than in LSCs, while miR-181b, miR-221, miR-21, and miR-22 were expressed at lower levels in HSCs than in LSCs [82]. Information about their role in leukemia pathogenesis are reported in Table 2.
It was also reported miR-126 increases in LSCs and HSC compared to leukemic progenitors, suggesting miR-126 as a stem cell-associated miRNA [82]. It was found to restrain cell cycle progression, prevent differentiation, and increase self-renewal of LSCs, in vivo, by regulating the phosphatidylinositol-3-kinases/Protein Kinase B/mechanistic target of rapamycin (PI3K/AKT/MTOR) signaling pathway that drives LSC self-renewal and chemotherapy resistance. Interestingly, it also exerted an opposite functional effect in LSCs with respect to HSC; in fact, reduced miR-126 levels induced HSC expansion in vivo, while impaired LSC maintenance [81]. Finally, miR-126 inhibition caused apoptosis and decreased clonogenic capacity of LSCs [79,82].
Other altered miRNAs in LSCs were involved in development of leukemia. Wong et al. showed that miR-17 polycistron maintained LSC potential in a mouse model of MLL-AML by modulating the expression of the cyclin-dependent kinase inhibitor p21 [58]. Moreover, in MLL-AML, the inhibition of miR-196 and miR-21 reduced LSCs in vivo [124].
Han et al. reported that miR-29a was highly expressed in HSC and primary human LSC, but downregulated in more committed progenitors; this work suggested miR-29a role in the regulation of early hematopoiesis and in the initiation of AML by converting myeloid progenitors into self-renewing LSC [125].
Comparing LSCs versus leukemic progenitors, it was found miR-1274a, miR-886, and miR-1305 were more lowly expressed in LSCs with respect to leukemic progenitors, while miR-126-5p, miR-126-3p, miR-22, miR-335, and mir-150 were more highly expressed in LSCs than in leukemic progenitors [82].
Interestingly, MVs released by LSC were able to promote proliferation and migration, and to inhibit apoptosis of AML cells. Restoration of miR-34a in LSC not only inhibited LSC proliferation, but also generated MVs containing a high level of miR-34a, which could revert the effects of LSC-MVs on AML cells [126].
Collectively, the reported studies suggested that miRNA targeting in LSC could be a potential strategy to eradicate leukemic cells. Further studies are necessary to define this aspect.
5. miRNAs as Biomarkers in AML
miRNAs have several properties of good biomarkers, such as their presence in various biological fluids (e.g., serum, plasma, urine, saliva, etc.), sequence conservation between human and animal models, and available sensitive measurement methodologies [127].
Moreover, they show high stability and survive in unfavorable conditions, such as extreme variations in pH, boiling, multiple freeze–thaw cycles, and extended storage [128].
The majority of miRNAs are intracellularly localized; however, they are also cell-free: (i) protected into apoptotic bodies and EVs; (ii) associated with proteins of the Argonaute (AGO) family; (iii) joined with high-density lipoprotein [128]. These miRNAs are generally defined as circulating miRNAs. Emerging evidence shows that circulating miRNAs have been gaining importance for their ability to provide early, sensitive, and non-invasive biomarkers in solid and hematologic cancers [129,130,131]. In particular, different studies have identified as candidates specific circulating miRNAs as new biomarkers in AML disease [132,133].
As mentioned above, miRNA expression profile is aberrant in AML and, besides being associated with genetic alterations, hold a diagnostic and prognostic relevance. The evaluation of a single or panel of miRNAs with potential diagnostic/prognostic impact could implement data from cytogenetic, gene mutations, and gene expression profiles.
In Table 3 it’s reported cellular and circulating miRNAs with diagnostic power, selected from the most significant diagnostic studies based on receiver operator characteristic (ROC) curve analysis [134,135,136,137,138,139,140].
Table 3.
miRNAs as diagnostic biomarkers in AML.
| miRNA | Expression | Specimen | Refs |
|---|---|---|---|
| miR-29a, miR-142-3p | under-expression | PBMCs | [134] |
| circulating miR-10a-5p | over-expression | serum | [135] |
| circulating miR-10a-5p, miR-93-5p, miR-129-5p, miR-155-5p, miR-181b-5p, miR-320d | over-expression | serum | [136] |
| circulating miR-92a, miR-143, miR-342 | under-expression | plasma | [137] |
| circulating miR-150, miR-342 | under-expression | plasma | [138] |
| circulating miR-155 | over-expression | serum EVs | [139] |
| circulating miR-370 | under-expression | serum | [140] |
Abbreviations: PBMCs, peripheral blood mononuclear cells; EVs, extracellular vesicles.
In our recent work, we demonstrated that EV miR-155 was highly expressed in AML with respect to healthy subjects; notably, ROC curve analysis revealed that this miRNA could be a potential new biomarker in AML [139].
We also summarized, in Table 4, miRNAs involved in AML prognosis, reporting their expression level, origin, and their impact in predicting relapse or progression, as well as their relationship with overall survival (OS) and relapse free survival (RFS) [29,33,34,45,46,70,135,136,140,141,142,143,144,145,146,147,148,149,150,151,152,153,154,155,156,157,158,159,160,161,162]
Table 4.
miRNAs as prognostic biomarkers in AML.
| miRNA | Expression | Prognostic Impact | Specimen | Refs |
|---|---|---|---|---|
| miR-9 | overexpression | unfavorable OS and RFS | BMMCs | [141] |
| miR-24 | overexpression | shorter OS | BMMCs/PBMCs | [142] |
| miR-26a, miR-29b, miR-146a | overexpression | shorter OS | BMMCs | [143] |
| miR-29a | underexpression | shorter OS and RFS | BMMCs | [144] |
| miR-29b | underexpression | poorer OS | BMMCs/PBMCs | [145] |
| miR-96 | underexpression | lower OS and RFS | BMMCs/PBMCs | [146] |
| miR-99a | overexpression | worse OS and EFS | BMMCs | [70] |
| miR-124-1 | underexpression | longer OS RFS | BMMCs | [147] |
| miR-135a, miR-409-3p | underexpression | higher cumulative incidence of relapse | BMMCs/PBMCs | [148] |
| miR-181a, miR-181b | overexpression | decreased risk of an event (i.e., lack of complete remission, relapse, or death) | BMMCs | [45] |
| miR-181a | overexpression | favorable prognosis | BMMCs | [149] |
| miR-181a, miR-181b, miR-181d | overexpression | longer OS | not specified | [46] |
| miR-181a | overexpression | higher CR rate, longer OS | BMMCs | [150] |
| miR-181b | overexpression | lower CR rates, shorter RFS and OS | BMMCs | [151] |
| miR-181b | overexpression | better prognosis and lower probability of relapse | BMMCs | [152] |
| miR-188-5p | underexpression | longer OS and EFS | not specified | [153] |
| miR-191, miR-199a | overexpression | worse prognosis (worse overall and event free survival) | BMMCs | [34] |
| miR-196b | overexpression | shorter OS | leukemic blasts | [33] |
| miR-196b, miR-644 | overexpression | shorter OS | BMMCs/PBMCs | [148] |
| miR-212 | overexpression | better OS, higher CR rate, better EFS and RFS | BMMCs/PBMCs | [29] |
| miR-328 | underexpression | poor OS and shorter RFS | plasma | [154] |
| miR-331 | overexpression | worse response to therapy and shorter OS | BMMCs | [155] |
| miR-375 | overexpression | shorter RFS and OS | BMMCs | [156] |
| miR-378 | overexpression | shorter RFS | BMMCs | [157] |
| miR-3151 | overexpression | shorter DFS and OS | PBMCs | [158] |
| let-7a-3 | overexpression | shorter OS and RFS | BMMCs | [159] |
| let-7a-2-3p | overexpression | longer OS and EFS | not specified | [153] |
| circulating miR-10a-5p | overexpression | shorter OS | serum | [135] |
| circulating miR-181b-5p | over-expression | worse OS | serum | [136] |
| circulating miR-183 | overexpression | shorter OS and RFS | serum | [160] |
| circulating miR-210 | overexpression | shorter OS and RFS | serum | [161] |
| circulating miR-335 | overexpression | shorter OS and RFS | serum | [162] |
| circulating miR-370 | underexpression | shorter RFS and OS | serum | [140] |
Abbreviations: BMMCs, bone marrow mononuclear cells; PBMCs, peripheral blood mononuclear cells; OS, overall survival; RFS: relapse free survival; EFS: event free survival; DFS, disease free survival; CR: complete remission.
Of note, discovery of new sensitive and specific biomarkers may be clinically useful in MRD monitoring. Notably, circulating miRNAs could be exerting an important relevance in this context, and their quantification could be more accurate over conventional biomarkers. In this setting, Hornick et al. investigated Exo miRNAs as minimally invasive biomarkers in AML. They developed a xenograft model for high-risk AML. In mouse serum Exo, they detected a panel of biomarkers, miR-150, miR-155, and miR-1246, that resulted in equal sensitivity for detection of MRD, when compared to conventional immuno-phenotypic cell evaluation [163].
Despite the appealing use of circulating miRNAs as novel biomarkers, optimization and standardization of purification and quantification are needed to translate basic research into clinical practice. Further studies are required to promote the future use of EV analysis in clinical applications. In particular, accurate procedures concerning sample collection, EV isolation and storage, as well as EV-miRNA quantification, are necessary to improve the use of EVs as biomarkers [127,164].
6. miRNAs as Therapeutic Targets
Drug resistance against conventional chemotherapy in AML is one of the major reasons of treatment failure [165]. Current therapies in AML are able to rapidly target dividing blast populations, but have limited capacity to eradicate the functionally distinct LSCs, responsible for disease resistance and relapse [79]. Hence, the identification of new therapeutic strategies in AML is of paramount importance. In this context, miRNA-based therapeutic approaches may provide a novel promising strategy.
The principal approach to target miRNAs was the manipulation of their expression, by the replacing of cancer suppressive miRNAs or by the inhibition of overexpressed oncogenic ones [166]. The replacement therapy employs miRNA mimics (double stranded oligonucleotides) delivered by viral [167] or synthetic vectors, such as lipid-based nanocarriers, amphiphilic star copolymer, inorganic nanoparticles polymeric and dendrimer-based vectors [166,168,169]. These vectors are essential to avoid degradation and to increase cellular uptake of synthetic miRNAs [169].
A consistent number of reports have shown encouraging results of miRNA-based therapeutic approaches, in both in vitro and in vivo studies. For example, miR-29b, an AML tumor suppressor [64], was overexpressed in AML blasts, by nanoparticle-based delivery system, decreasing leukemic cell growth and improving the survival of AML-xenograft mouse model [169]. These data were confirmed in another in vivo study in which miR-29 family member restoration caused a dramatic reduction of AML engraftment in mouse BM, associated with relieved splenomegaly, reduced neoplastic infiltration, decreased cell proliferation, and increased apoptosis activities in spleens [63]. Xu et al., instead, demonstrated that the overexpression of miR-150 in vitro inhibited proliferation and clonogenic growth, and attenuated tumorigenic activity of LSCs, downregulating the expression of cancer stem cell factors like Nanog, Notch2, and CTNNB1. Moreover, an in vivo study confirmed that miR-150 overexpression progressively abrogated tumor growth [89]. The inhibition of leukemia progression was also demonstrated by forced expression of miR-22 and miR-193a. In particular, Jiang et al. demonstrated that the overexpression of miR-22, by nanoparticles, significantly suppressed leukemic cell viability and growth in vitro, and repressed leukemia development and maintenance in vivo [61]. Instead, Li et al. showed that miR-193a restoration by synthetic miRNA contributed to the blockage of malignant-cell proliferation in t(8;21) AML xenograft model [99]. Moreover, Yang et al. described miR-122 as tumor suppressor miRNA in childhood AML, and showed that a miR-122 mimic efficiently inhibited cell proliferation and cell cycle of HL-60 AML cell line [53]. Another potential candidate for miRNA therapy in AML is miR-34. Wang et al. showed that overexpression of this miRNA in AML cell lines reduced both mRNA and cell surface protein expression of PD-L1, an inhibitory immune checkpoint molecule, suggesting the potential role of miR-34a mimic also in AML immunotherapy [68].
The second miRNA-based therapeutic approach is the silencing of aberrantly expressed miRNAs using anti-miRNA oligonucleotides (AMOs), miRNA sponges, or miRNA masking [170]. These synthetic oligonucleotides have sequences complementary to endogenous mature miRNA, and competitively prevent the interaction between miRNAs and their targets [166]. MiR-126, for example, high expressed in CN-AML, was silenced in in vivo models, causing a reduction of LSCs and showing no toxic effect on normal hematopoietic functions. In this study, Dorrance et al. identified a potential novel therapeutic approach to treat AML patients and prevent relapse [79]. Literature data also indicated that miR-181 inhibition could provide a new strategy for AML therapy [171], increasing the differentiation of myeloid progenitors, reducing the engraftment and the infiltration of leukemic HSPCs into BM and spleen, and ameliorating symptoms of leukemia in AML CD34+ HSPC xenograft mice [171]. Another miRNA involved in leukemogenesis seemed to be miR-9; Tian et al. demonstrated that its knockdown in vivo suppressed malignant cell proliferation, decreased leukemic cell counts both in blood and BM, and reduced splenomegaly [172]. Velu et al., instead, studied the combination of miR-21 and miR-196b silencing in murine models of MLL-AF9 leukemia [124]. They demonstrated that co-treatment with both antagomirs improved the outcome of MLL-AF9 leukemia, and did not show toxicity in vivo. In this study, they tested both antagomir with or without a cholesterol modification, demonstrating highest efficacy of anti-miRNAs with lipoprotein particles (the commercial ones) in vivo [124].
Emerging evidence revealed that modification of miRNA expression levels can increase the sensitivity to chemotherapy or other drugs. In this context, Chen et al. demonstrated that overexpression of let-7a, in xenograft mouse of primary human AML cells, increased the cytarabine (Ara-C) efficacy respect to single chemotherapeutic treatment and extended survival of mice [173]. Also, the overexpression of miR-150 in vitro reduced cell viability of two AML cell lines treated with Ara-C [89]. Moreover, Lu et al. reported that, also, forced miR-181b expression rendered leukemic cells more sensitive to doxorubicin and Ara-C in vitro [174]. Gao et al., instead, demonstrated a synergistic apoptotic effect of 5-azacytidine and miR-193a mimic combination in AML cell line, Kasumi-1, suggesting miR-193a restoration as a potential adjuvant therapy in c-KIT-positive AML [100]. In a recent study, it was also demonstrated that the upregulation of miR-217, a tumor suppressor in AML, induced in vitro chemosensitivity to doxorubicin [165]. Additionally, it was reported that ectopic expression of miR-133, a tumor suppressor for EVI1-overexpressing AML cells, increased drug sensitivity specifically in this subset of AML [175].
Instead, Lechman et al. suggested miR-126 inhibition as a strategy to overcome LSC chemoresistance, and to increase the sensitivity of anti-proliferative drugs [81]. Furthermore, targeting of miR-21 and miR-196b increased the effect of induction chemotherapy in xenograft model of MLL leukemia [124].
Another interesting concept concerns the effects of existing drugs on miRNA expression. For example, lenalidomide, a drug approved for the treatment of multiple myeloma and MDS, induced endogenous expression of miR-181 with significant tumor growth inhibition in AML xenograft mouse models [176]. Additionally, Khalife et al. reported that MLN4924 drug (Pevonedistat), decreasing NF-κB activation, downregulated oncogenic miR-155 levels in FLT3-ITD+ AML cell lines and reduced leukemic phenotypes, in both in vitro and in vivo models [177].
The identification of miRNA target genes can help to understand the multiple mechanisms involved in AML [165], and can represent another important tool to provide targeted AML treatment strategies [166]. For example, miRNA activity can be regulated by blocking its access to the target mRNA. A recent study showed that the activity of two oncomiRs, miR-9 and miR-17, could be antagonized by the induction of two RNA binding proteins, RBM38 and DND1, whose function is to protect mRNAs from miRNA-mediated cleavage [178].
MVs represent another important tumor suppressive miRNA vehicle. In a recent work, it was hypothesized that miRNA contents of LSC-derived MVs (LMVs) can be reprogrammed, altering the effects of LMVs on cancer cells [126]. In particular, MVs isolated from human AML cell line KG-la, previously transfected with miR-34a mimic, were used to inhibit proliferation and migration of AML cell by the modulation of caspase-3 and Tim-3 levels [126].
Despite the above described miRNA-therapeutic approach advantages, there are still many challenges to be solved before introducing it into the clinic. Furthermore, although the literature suggests limited toxicity of some of these miRNAs in various model systems, clinical trials are necessary to evaluate whether this preclinical promise will be recapitulated in human patients. Likewise, concentration and dose plans need to be evaluated for efficacy and toxicity, and the long-term effects following miRNA therapy must be assessed. Additionally, as the therapeutic potential of different miRNAs has been investigated, the combination of multiple miRNAs could be evaluated as a possible miRNA therapy in AML.
Nevertheless, a major limitation preventing the development of miRNA-based therapies is the difficulty to deliver drugs to BM, and the necessity of using higher doses to elicit a therapeutic effect [14].
7. Long Non-Coding RNAs Can Interfere with miRNA Function in AML
Although a broad range of non-coding RNAs has been discovered, miRNAs and long non-coding RNAs (lncRNAs) are the best studied. LncRNAs are RNAs longer than 200 nucleotides. Based on the genomic locations where lncRNAs are transcribed, they can be classified into sense, overlapping with at least part of another gene in the same strand; antisense, overlapping with at least part of another gene on the opposite strand; intronic, originating from the intron of another gene; intergenic, not overlapping with any gene; and chimeric, resulting from fusion products due to chromosomal rearrangements [179].
LncRNAs could promote the strength of specific enhancer–promoter looping, contributing to gene activation [179].
Their expression is tightly controlled, and exhibits higher cell specificity than proteins, including lineage-determining transcription factors [180]. LncRNAs play an important role in several biological processes, including cell development, differentiation, proliferation, invasion, and migration [181]. LncRNAs regulate gene expression at the epigenetic, transcriptional, and post-transcriptional level [182].
Aberrant expression of miRNAs and lncRNAs has been described in all types of cancers [183,184].
In AML, aberrant expression of lncRNAs contributes to initiation, maintenance, and development of leukemogenesis [179]. They are involved in chromatin remodeling, as well as transcriptional and post-transcriptional regulation, through a variety of chromatin-based mechanisms and via cross-talk with other RNA species. LncRNAs can function as decoys, scaffolds, and enhancer RNAs [185]. Moreover, lncRNAs competed with endogenous miRNAs in AML.
A lncRNA, CCAT1, for example, was found overexpressed in AML; it repressed monocytic differentiation, and promoted AML cell growth by sequestering miR-155 [186]. In accord with Palma’s study [107], Chen’s work showed that miR-155 has a pro-differentiation and anti-proliferation role in FLT3 wild-type AML cells [186], in contrast to its oncogenic function reported in FLT3-ITD mutated AML [91,187]. These data suggested that the gene regulation by miR-155 is complex, as it targets both tumor suppressor genes or oncogenes, and its function in a specific subtype of AML is likely subject to disease context and cell type [107,186].
An oncogenic activity of lncRNA was also shown by HOTAIR that competitively binds miR-193a, an important tumor-suppressor miRNA, thus modulating the expression of c-Kit in AML cells [188]. The lncRNA HOTAIRM1, instead, was associated with myeloid differentiation; it also had pivotal roles in the degradation of oncoprotein PML-RARA, by acting as a microRNA sponge, sequestering miR-20a, miR-106b, and miR-125b, whose targets are autophagy-associated genes [189].
Moreover, Mangiavacchi et al. showed that the host non-coding transcript of miR-223, linc-223, is a functional lncRNA that controls proliferation and differentiation of AML cells by binding miR-125-5p. This lncRNA is an oncogenic miRNA upregulated in primary AML where linc-223 was instead downregulated [190].
In conclusion, lncRNA functions introduce an extra layer of complexity in the miRNA-target network, making it necessary to study the interactions between miRNAs and lncRNAs, both to shed light on AML pathogenesis and on therapy.
8. Conclusions
miRNAs have emerged as a class of gene expression pivotal regulators contributing to AML pathogenesis and as potential biomarkers. Actually, miRNA analysis has not yet been included in AML clinical practice. Specific miRNA expression could help clinicians to classify subtypes, to determine prognosis, and to predict the response of treatment in AML.
Particular attention should be given to circulating miRNA, both free or contained in EVs, which are easy to collect with a non-invasive peripheral blood sample. In addition, while malignant cells are usually reduced after treatment, circulating RNAs can still be detected. Finally, miRNA analysis, through advanced next generation sequencing, will provide more details on miRNAs, and also lncRNAs involved in leukemia onset and progression. This information supports the concept that circulating miRNAs, in addition to current clinical AML parameters, could have significant value as new AML biomarkers for detection of disease progression. They can also serve as indicators of therapeutic response, and in MRD detection in a future clinical practice.
Another approach to improving AML outcomes could be the potential use of miRNAs as therapeutic targets. miRNA modulation could be obtained through new strategies by using miRNA inhibitors or mimics.
Moreover, miRNA therapy in combination with current chemotherapy could potentially eradicate LSCs.
Actually, the main problems in developing miRNA therapy are the identification of the best miRNA candidates and the design of miRNA delivery vehicles that confer higher stability to the therapeutic candidate, avoiding potential toxicities and off target effects [191]. Many complexities and mysteries remain in miRNA biology; miRNA studies will be key to our understanding of how to improve their use in an AML clinical setting.
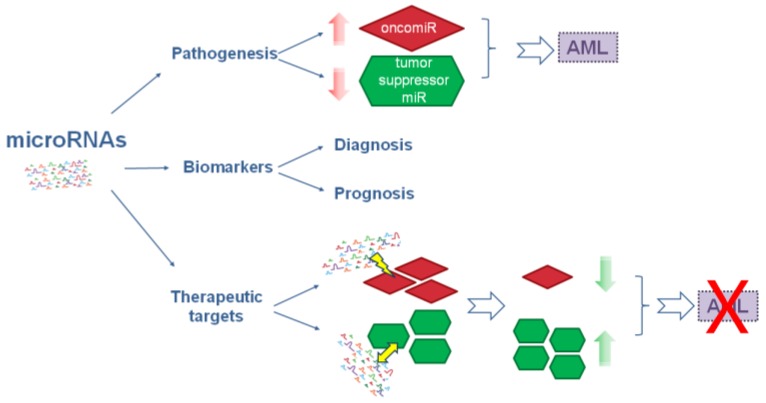
The role of miRNAs in AML is reported in a schematic diagram in Figure 1.
Figure 1.
Schematic diagram of microRNAs role in AML. MicroRNAs are involved in pathogenesis and are considered as biomarkers and therapeutic targets in AML. Red arrows indicate overexpression of oncomiR and downexpression of tumor suppressor miR which cause AML; lightning and two-headed arrow indicate targeting and restore, respectively; green arrows indicate decreased oncomiR or increased tumor suppressor miR levels that block AML; red cross indicates AML block.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Dohner H., Weisdorf D.J., Bloomfield C.D. Acute myeloid leukemia. N. Engl. J. Med. 2015;373:1136–1152. doi: 10.1056/NEJMra1406184. [DOI] [PubMed] [Google Scholar]
- 2.Papaemmanuil E., Dohner H., Campbell P.J. Genomic classification in acute myeloid leukemia. N. Engl. J. Med. 2016;375:900–901. doi: 10.1056/NEJMoa1516192. [DOI] [PubMed] [Google Scholar]
- 3.De Kouchkovsky I., Abdul-Hay M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J. 2016;6:e441. doi: 10.1038/bcj.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sperling A.S., Gibson C.J., Ebert B.L. The genetics of myelodysplastic syndrome: From clonal haematopoiesis to secondary leukaemia. Nat. Rev. Cancer. 2017;17:5–19. doi: 10.1038/nrc.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sill H., Olipitz W., Zebisch A., Schulz E., Wolfler A. Therapy-related myeloid neoplasms: Pathobiology and clinical characteristics. Br. J. Pharmacol. 2011;162:792–805. doi: 10.1111/j.1476-5381.2010.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shih A.H., Abdel-Wahab O., Patel J.P., Levine R.L. The role of mutations in epigenetic regulators in myeloid malignancies. Nat. Rev. Cancer. 2012;12:599–612. doi: 10.1038/nrc3343. [DOI] [PubMed] [Google Scholar]
- 7.Arber D.A., Orazi A., Hasserjian R., Thiele J., Borowitz M.J., Le Beau M.M., Bloomfield C.D., Cazzola M., Vardiman J.W. The 2016 revision to the world health organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–2405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 8.Zebisch A., Hatzl S., Pichler M., Wolfler A., Sill H. Therapeutic resistance in acute myeloid leukemia: The role of non-coding RNAs. Int. J. Mol. Sci. 2016;17:2080. doi: 10.3390/ijms17122080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venton G., Perez-Alea M., Baier C., Fournet G., Quash G., Labiad Y., Martin G., Sanderson F., Poullin P., Suchon P., et al. Aldehyde dehydrogenases inhibition eradicates leukemia stem cells while sparing normal progenitors. Blood Cancer J. 2016;6:e469. doi: 10.1038/bcj.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crews L.A., Balaian L., Delos Santos N.P., Leu H.S., Court A.C., Lazzari E., Sadarangani A., Zipeto M.A., La Clair J.J., Villa R., et al. RNA splicing modulation selectively impairs leukemia stem cell maintenance in secondary human AML. Cell Stem Cell. 2016;19:599–612. doi: 10.1016/j.stem.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung S.S., Hu W., Park C.Y. The role of microRNAs in hematopoietic stem cell and leukemic stem cell function. Ther. Adv. Hematol. 2011;2:317–334. doi: 10.1177/2040620711410772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roden C., Lu J. MicroRNAs in control of stem cells in normal and malignant hematopoiesis. Curr. Stem Cell Rep. 2016;2:183–196. doi: 10.1007/s40778-016-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitsios D.M., Davis M.P., van Dongen S., Enright A.J. Large-scale analysis of microRNA expression, epi-transcriptomic features and biogenesis. Nucleic Acids Res. 2017;45:1079–1090. doi: 10.1093/nar/gkw1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallace J.A., O’Connell R.M. MicroRNAs and acute myeloid leukemia: Therapeutic implications and emerging concepts. Blood. 2017;130:1290–1301. doi: 10.1182/blood-2016-10-697698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez N., Cordiner R.A., Young R.S., Hug N., Macias S., Caceres J.F. Genetic variation and RNA structure regulate microRNA biogenesis. Nat. Commun. 2017;8:15114. doi: 10.1038/ncomms15114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Svoronos A.A., Engelman D.M., Slack F.J. Oncomir or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pichiorri F., De Luca L., Aqeilan R.I. MicroRNAs: New players in multiple myeloma. Front. Genet. 2011;2:22. doi: 10.3389/fgene.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simeon V., Todoerti K., La Rocca F., Caivano A., Trino S., Lionetti M., Agnelli L., De Luca L., Laurenzana I., Neri A., et al. Molecular classification and pharmacogenetics of primary plasma cell leukemia: An initial approach toward precision medicine. Int. J. Mol. Sci. 2015;16:17514–17534. doi: 10.3390/ijms160817514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X., Chen H., Bai J., He A. MicroRNA: An important regulator in acute myeloid leukemia. Cell Biol. Int. 2017;41:936–945. doi: 10.1002/cbin.10770. [DOI] [PubMed] [Google Scholar]
- 20.O’Connell R.M., Zhao J.L., Rao D.S. MicroRNA function in myeloid biology. Blood. 2011;118:2960–2969. doi: 10.1182/blood-2011-03-291971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shivarov V., Bullinger L. Expression profiling of leukemia patients: Key lessons and future directions. Exp. Hematol. 2014;42:651–660. doi: 10.1016/j.exphem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Cammarata G., Augugliaro L., Salemi D., Agueli C., La Rosa M., Dagnino L., Civiletto G., Messana F., Marfia A., Bica M.G., et al. Differential expression of specific microRNA and their targets in acute myeloid leukemia. Am. J. Hematol. 2010;85:331–339. doi: 10.1002/ajh.21667. [DOI] [PubMed] [Google Scholar]
- 23.Li Z., Lu J., Sun M., Mi S., Zhang H., Luo R.T., Chen P., Wang Y., Yan M., Qian Z., et al. Distinct microRNA expression profiles in acute myeloid leukemia with common translocations. Proc. Natl. Acad. Sci. USA. 2008;105:15535–15540. doi: 10.1073/pnas.0808266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dixon-McIver A., East P., Mein C.A., Cazier J.B., Molloy G., Chaplin T., Andrew Lister T., Young B.D., Debernardi S. Distinctive patterns of microRNA expression associated with karyotype in acute myeloid leukaemia. PLoS ONE. 2008;3:e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daschkey S., Rottgers S., Giri A., Bradtke J., Teigler-Schlegel A., Meister G., Borkhardt A., Landgraf P. MicroRNAs distinguish cytogenetic subgroups in pediatric AML and contribute to complex regulatory networks in AML-relevant pathways. PLoS ONE. 2013;8:e56334. doi: 10.1371/journal.pone.0056334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jongen-Lavrencic M., Sun S.M., Dijkstra M.K., Valk P.J., Lowenberg B. MicroRNA expression profiling in relation to the genetic heterogeneity of acute myeloid leukemia. Blood. 2008;111:5078–5085. doi: 10.1182/blood-2008-01-133355. [DOI] [PubMed] [Google Scholar]
- 27.Brioschi M., Fischer J., Cairoli R., Rossetti S., Pezzetti L., Nichelatti M., Turrini M., Corlazzoli F., Scarpati B., Morra E., et al. Down-regulation of microRNAs 222/221 in acute myelogenous leukemia with deranged core-binding factor subunits. Neoplasia. 2010;12:866–876. doi: 10.1593/neo.10482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fazi F., Racanicchi S., Zardo G., Starnes L.M., Mancini M., Travaglini L., Diverio D., Ammatuna E., Cimino G., Lo-Coco F., et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell. 2007;12:457–466. doi: 10.1016/j.ccr.2007.09.020. [DOI] [PubMed] [Google Scholar]
- 29.Sun S.M., Rockova V., Bullinger L., Dijkstra M.K., Dohner H., Lowenberg B., Jongen-Lavrencic M. The prognostic relevance of miR-212 expression with survival in cytogenetically and molecularly heterogeneous AML. Leukemia. 2013;27:100–106. doi: 10.1038/leu.2012.158. [DOI] [PubMed] [Google Scholar]
- 30.Careccia S., Mainardi S., Pelosi A., Gurtner A., Diverio D., Riccioni R., Testa U., Pelosi E., Piaggio G., Sacchi A., et al. A restricted signature of miRNAs distinguishes APL blasts from normal promyelocytes. Oncogene. 2009;28:4034–4040. doi: 10.1038/onc.2009.255. [DOI] [PubMed] [Google Scholar]
- 31.De Luca L., Trino S., Laurenzana I., Tagliaferri D., Falco G., Grieco V., Bianchino G., Nozza F., Campia V., D’Alessio F., et al. Knockdown of miR-128a induces Lin28a expression and reverts myeloid differentiation blockage in acute myeloid leukemia. Cell Death Dis. 2017;8:e2849. doi: 10.1038/cddis.2017.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Popovic R., Riesbeck L.E., Velu C.S., Chaubey A., Zhang J., Achille N.J., Erfurth F.E., Eaton K., Lu J., Grimes H.L., et al. Regulation of miR-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113:3314–3322. doi: 10.1182/blood-2008-04-154310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z., Huang H., Chen P., He M., Li Y., Arnovitz S., Jiang X., He C., Hyjek E., Zhang J., et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and fas tumour suppressor in MLL-rearranged leukaemia. Nat. Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garzon R., Volinia S., Liu C.G., Fernandez-Cymering C., Palumbo T., Pichiorri F., Fabbri M., Coombes K., Alder H., Nakamura T., et al. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang X., Huang H., Li Z., He C., Li Y., Chen P., Gurbuxani S., Arnovitz S., Hong G.M., Price C., et al. miR-495 is a tumor-suppressor microRNA down-regulated in MLL-rearranged leukemia. Proc. Natl. Acad. Sci. USA. 2012;109:19397–19402. doi: 10.1073/pnas.1217519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garzon R., Garofalo M., Martelli M.P., Briesewitz R., Wang L., Fernandez-Cymering C., Volinia S., Liu C.G., Schnittger S., Haferlach T., et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc. Natl. Acad. Sci. USA. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker H., Marcucci G., Maharry K., Radmacher M.D., Mrozek K., Margeson D., Whitman S.P., Wu Y.Z., Schwind S., Paschka P., et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: A cancer and leukemia group B study. J. Clin. Oncol. 2010;28:596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Russ A.C., Sander S., Luck S.C., Lang K.M., Bauer M., Rucker F.G., Kestler H.A., Schlenk R.F., Dohner H., Holzmann K., et al. Integrative nucleophosmin mutation-associated microrna and gene expression pattern analysis identifies novel microRNA-target gene interactions in acute myeloid leukemia. Haematologica. 2011;96:1783–1791. doi: 10.3324/haematol.2011.046888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faraoni I., Laterza S., Ardiri D., Ciardi C., Fazi F., Lo-Coco F. miR-424 and miR-155 deregulated expression in cytogenetically normal acute myeloid leukaemia: Correlation with NPM1 and FLT3 mutation status. J. Hematol. Oncol. 2012;5:26. doi: 10.1186/1756-8722-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marcucci G., Maharry K., Radmacher M.D., Mrozek K., Vukosavljevic T., Paschka P., Whitman S.P., Langer C., Baldus C.D., Liu C.G., et al. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: A cancer and leukemia group B study. J. Clin. Oncol. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pulikkan J.A., Peramangalam P.S., Dengler V., Ho P.A., Preudhomme C., Meshinchi S., Christopeit M., Nibourel O., Muller-Tidow C., Bohlander S.K., et al. C/ebpalpha regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010;116:5638–5649. doi: 10.1182/blood-2010-04-281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mendler J.H., Maharry K., Radmacher M.D., Mrozek K., Becker H., Metzeler K.H., Schwind S., Whitman S.P., Khalife J., Kohlschmidt J., et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and microRNA expression signatures. J. Clin. Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitman S.P., Maharry K., Radmacher M.D., Becker H., Mrozek K., Margeson D., Holland K.B., Wu Y.Z., Schwind S., Metzeler K.H., et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: A cancer and leukemia group B study. Blood. 2010;116:3622–3626. doi: 10.1182/blood-2010-05-283648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dohner H., Estey E., Grimwade D., Amadori S., Appelbaum F.R., Buchner T., Dombret H., Ebert B.L., Fenaux P., Larson R.A., et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129:424–447. doi: 10.1182/blood-2016-08-733196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marcucci G., Radmacher M.D., Maharry K., Mrozek K., Ruppert A.S., Paschka P., Vukosavljevic T., Whitman S.P., Baldus C.D., Langer C., et al. MicroRNA expression in cytogenetically normal acute myeloid leukemia. N. Engl. J. Med. 2008;358:1919–1928. doi: 10.1056/NEJMoa074256. [DOI] [PubMed] [Google Scholar]
- 46.Li Z., Huang H., Li Y., Jiang X., Chen P., Arnovitz S., Radmacher M.D., Maharry K., Elkahloun A., Yang X., et al. Up-regulation of a HOXA-PBX3 homeobox-gene signature following down-regulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood. 2012;119:2314–2324. doi: 10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krowiorz K., Ruschmann J., Lai C., Ngom M., Maetzig T., Martins V., Scheffold A., Schneider E., Pochert N., Miller C., et al. MiR-139-5p is a potent tumor suppressor in adult acute myeloid leukemia. Blood Cancer J. 2016;6:e508. doi: 10.1038/bcj.2016.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 49.Debernardi S., Skoulakis S., Molloy G., Chaplin T., Dixon-McIver A., Young B.D. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia. 2007;21:912–916. doi: 10.1038/sj.leu.2404605. [DOI] [PubMed] [Google Scholar]
- 50.Ramkissoon S.H., Mainwaring L.A., Ogasawara Y., Keyvanfar K., McCoy J.P., Jr., Sloand E.M., Kajigaya S., Young N.S. Hematopoietic-specific microRNA expression in human cells. Leukemia Res. 2006;30:643–647. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 51.Lutherborrow M., Bryant A., Jayaswal V., Agapiou D., Palma C., Yang Y.H., Ma D.D. Expression profiling of cytogenetically normal acute myeloid leukemia identifies microRNAs that target genes involved in monocytic differentiation. Am. J. Hematol. 2011;86:2–11. doi: 10.1002/ajh.21864. [DOI] [PubMed] [Google Scholar]
- 52.De Leeuw D.C., Verhagen H.J., Denkers F., Kavelaars F.G., Valk P.J., Schuurhuis G.J., Ossenkoppele G.J., Smit L. MicroRNA-551b is highly expressed in hematopoietic stem cells and a biomarker for relapse and poor prognosis in acute myeloid leukemia. Leukemia. 2016;30:742–746. doi: 10.1038/leu.2015.160. [DOI] [PubMed] [Google Scholar]
- 53.Yang J., Yuan Y., Yang X., Hong Z., Yang L. Decreased expression of microRNA-122 is associated with an unfavorable prognosis in childhood acute myeloid leukemia and function analysis indicates a therapeutic potential. Pathol. Res. Pract. 2017;213:1166–1172. doi: 10.1016/j.prp.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 54.Liao Q., Wang B., Li X., Jiang G. miRNAs in acute myeloid leukemia. Oncotarget. 2017;8:3666–3682. doi: 10.18632/oncotarget.12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen P., Price C., Li Z., Li Y., Cao D., Wiley A., He C., Gurbuxani S., Kunjamma R.B., Huang H., et al. miR-9 is an essential oncogenic microRNA specifically overexpressed in mixed lineage leukemia-rearranged leukemia. Proc. Natl. Acad. Sci. USA. 2013;110:11511–11516. doi: 10.1073/pnas.1310144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emmrich S., Katsman-Kuipers J.E., Henke K., Khatib M.E., Jammal R., Engeland F., Dasci F., Zwaan C.M., den Boer M.L., Verboon L., et al. miR-9 is a tumor suppressor in pediatric AML with t(8;21) Leukemia. 2014;28:1022–1032. doi: 10.1038/leu.2013.357. [DOI] [PubMed] [Google Scholar]
- 57.Senyuk V., Zhang Y., Liu Y., Ming M., Premanand K., Zhou L., Chen P., Chen J., Rowley J.D., Nucifora G., et al. Critical role of miR-9 in myelopoiesis and EVI1-induced leukemogenesis. Proc. Natl. Acad. Sci. USA. 2013;110:5594–5599. doi: 10.1073/pnas.1302645110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wong P., Iwasaki M., Somervaille T.C., Ficara F., Carico C., Arnold C., Chen C.Z., Cleary M.L. The miR-17-92 microrna polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70:3833–3842. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mi S., Li Z., Chen P., He C., Cao D., Elkahloun A., Lu J., Pelloso L.A., Wunderlich M., Huang H., et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc. Natl. Acad. Sci. USA. 2010;107:3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song S.J., Ito K., Ala U., Kats L., Webster K., Sun S.M., Jongen-Lavrencic M., Manova-Todorova K., Teruya-Feldstein J., Avigan D.E., et al. The oncogenic microRNA miR-22 targets the TET2 tumor suppressor to promote hematopoietic stem cell self-renewal and transformation. Cell Stem Cell. 2013;13:87–101. doi: 10.1016/j.stem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang X., Hu C., Arnovitz S., Bugno J., Yu M., Zuo Z., Chen P., Huang H., Ulrich B., Gurbuxani S., et al. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat. Commun. 2016;7:11452. doi: 10.1038/ncomms11452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen C., Chen M.T., Zhang X.H., Yin X.L., Ning H.M., Su R., Lin H.S., Song L., Wang F., Ma Y.N., et al. The PU.1-modulated microRNA-22 is a regulator of monocyte/macrophage differentiation and acute myeloid leukemia. PLoS Genet. 2016;12:e1006259. doi: 10.1371/journal.pgen.1006259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gong J.N., Yu J., Lin H.S., Zhang X.H., Yin X.L., Xiao Z., Wang F., Wang X.S., Su R., Shen C., et al. The role, mechanism and potentially therapeutic application of microRNA-29 family in acute myeloid leukemia. Cell Death Differ. 2014;21:100–112. doi: 10.1038/cdd.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Garzon R., Heaphy C.E., Havelange V., Fabbri M., Volinia S., Tsao T., Zanesi N., Kornblau S.M., Marcucci G., Calin G.A., et al. MicroRNA 29b functions in acute myeloid leukemia. Blood. 2009;114:5331–5341. doi: 10.1182/blood-2009-03-211938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu S., Wu L.C., Pang J., Santhanam R., Schwind S., Wu Y.Z., Hickey C.J., Yu J., Becker H., Maharry K., et al. Sp1/NFκB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17:333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Eyholzer M., Schmid S., Wilkens L., Mueller B.U., Pabst T. The tumour-suppressive miR-29a/b1 cluster is regulated by CEBPA and blocked in human AML. Br. J. Cancer. 2010;103:275–284. doi: 10.1038/sj.bjc.6605751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pyzer A.R., Stroopinsky D., Rosenblatt J., Anastasiadou E., Rajabi H., Washington A., Tagde A., Chu J.H., Coll M., Jiao A.L., et al. MUC1 inhibition leads to decrease in PD-L1 levels via upregulation of miRNAs. Leukemia. 2017;31:2780–2790. doi: 10.1038/leu.2017.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang X., Li J., Dong K., Lin F., Long M., Ouyang Y., Wei J., Chen X., Weng Y., He T., et al. Tumor suppressor miR-34a targets PD-L1 and functions as a potential immunotherapeutic target in acute myeloid leukemia. Cell. Signal. 2015;27:443–452. doi: 10.1016/j.cellsig.2014.12.003. [DOI] [PubMed] [Google Scholar]
- 69.Liu L., Ren W., Chen K. MiR-34a promotes apoptosis and inhibits autophagy by targeting HMGB1 in acute myeloid leukemia cells. Cell. Physiol. Biochem. 2017;41:1981–1992. doi: 10.1159/000475277. [DOI] [PubMed] [Google Scholar]
- 70.Si X., Zhang X., Hao X., Li Y., Chen Z., Ding Y., Shi H., Bai J., Gao Y., Cheng T., et al. Upregulation of miR-99a is associated with poor prognosis of acute myeloid leukemia and promotes myeloid leukemia cell expansion. Oncotarget. 2016;7:78095–78109. doi: 10.18632/oncotarget.12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khalaj M., Woolthuis C.M., Hu W., Durham B.H., Chu S.H., Qamar S., Armstrong S.A., Park C.Y. miR-99 regulates normal and malignant hematopoietic stem cell self-renewal. J. Exp. Med. 2017;214:2453–2470. doi: 10.1084/jem.20161595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L., Li X., Ke Z., Huang L., Liang Y., Wu J., Zhang X., Chen Y., Zhang H., Luo X. MiR-99a may serve as a potential oncogene in pediatric myeloid leukemia. Cancer Cell Int. 2013;13:110. doi: 10.1186/1475-2867-13-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bousquet M., Quelen C., Rosati R., Mansat-De Mas V., La Starza R., Bastard C., Lippert E., Talmant P., Lafage-Pochitaloff M., Leroux D., et al. Myeloid cell differentiation arrest by miR-125b-1 in myelodysplastic syndrome and acute myeloid leukemia with the t(2;11)(p21;q23) translocation. J. Exp. Med. 2008;205:2499–2506. doi: 10.1084/jem.20080285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaudhuri A.A., So A.Y., Mehta A., Minisandram A., Sinha N., Jonsson V.D., Rao D.S., O’Connell R.M., Baltimore D. Oncomir miR-125b regulates hematopoiesis by targeting the gene Lin28a. Proc. Natl. Acad. Sci. USA. 2012;109:4233–4238. doi: 10.1073/pnas.1200677109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Garzon R., Liu S., Fabbri M., Liu Z., Heaphy C.E., Callegari E., Schwind S., Pang J., Yu J., Muthusamy N., et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.O’Connell R.M., Chaudhuri A.A., Rao D.S., Gibson W.S., Balazs A.B., Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. USA. 2010;107:14235–14240. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.So A.Y., Sookram R., Chaudhuri A.A., Minisandram A., Cheng D., Xie C., Lim E.L., Flores Y.G., Jiang S., Kim J.T., et al. Dual mechanisms by which miR-125b represses IRF4 to induce myeloid and B-cell leukemias. Blood. 2014;124:1502–1512. doi: 10.1182/blood-2014-02-553842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang H., Luo X.Q., Zhang P., Huang L.B., Zheng Y.S., Wu J., Zhou H., Qu L.H., Xu L., Chen Y.Q. MicroRNA patterns associated with clinical prognostic parameters and CNS relapse prediction in pediatric acute leukemia. PLoS ONE. 2009;4:e7826. doi: 10.1371/journal.pone.0007826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dorrance A.M., Neviani P., Ferenchak G.J., Huang X., Nicolet D., Maharry K.S., Ozer H.G., Hoellarbauer P., Khalife J., Hill E.B., et al. Targeting leukemia stem cells in vivo with antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia. 2015;29:2143–2153. doi: 10.1038/leu.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Z., Chen P., Su R., Li Y., Hu C., Wang Y., Arnovitz S., He M., Gurbuxani S., Zuo Z., et al. Overexpression and knockout of miR-126 both promote leukemogenesis. Blood. 2015;126:2005–2015. doi: 10.1182/blood-2015-04-639062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lechman E.R., Gentner B., Ng S.W.K., Schoof E.M., van Galen P., Kennedy J.A., Nucera S., Ciceri F., Kaufmann K.B., Takayama N., et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29:214–228. doi: 10.1016/j.ccell.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.De Leeuw D.C., Denkers F., Olthof M.C., Rutten A.P., Pouwels W., Schuurhuis G.J., Ossenkoppele G.J., Smit L. Attenuation of microRNA-126 expression that drives CD34+38− stem/progenitor cells in acute myeloid leukemia leads to tumor eradication. Cancer Res. 2014;74:2094–2105. doi: 10.1158/0008-5472.CAN-13-1733. [DOI] [PubMed] [Google Scholar]
- 83.Starczynowski D.T., Morin R., McPherson A., Lam J., Chari R., Wegrzyn J., Kuchenbauer F., Hirst M., Tohyama K., Humphries R.K., et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood. 2011;117:595–607. doi: 10.1182/blood-2010-03-277012. [DOI] [PubMed] [Google Scholar]
- 84.Starczynowski D.T., Kuchenbauer F., Argiropoulos B., Sung S., Morin R., Muranyi A., Hirst M., Hogge D., Marra M., Wells R.A., et al. Identification of miR-145 and miR-146a as mediators of the 5q- syndrome phenotype. Nat. Med. 2010;16:49–58. doi: 10.1038/nm.2054. [DOI] [PubMed] [Google Scholar]
- 85.Varney M.E., Niederkorn M., Konno H., Matsumura T., Gohda J., Yoshida N., Akiyama T., Christie S., Fang J., Miller D., et al. Loss of Tifab, a del(5q) MDS gene, alters hematopoiesis through derepression of Toll-like receptor-TRAF6 signaling. J. Exp. Med. 2015;212:1967–1985. doi: 10.1084/jem.20141898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhao J.L., Rao D.S., Boldin M.P., Taganov K.D., O’Connell R.M., Baltimore D. NF-κB dysregulation in microRNA-146a-deficient mice drives the development of myeloid malignancies. Proc. Natl. Acad. Sci. USA. 2011;108:9184–9189. doi: 10.1073/pnas.1105398108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fang J., Barker B., Bolanos L., Liu X., Jerez A., Makishima H., Christie S., Chen X., Rao D.S., Grimes H.L., et al. Myeloid malignancies with chromosome 5q deletions acquire a dependency on an intrachromosomal NF-κB gene network. Cell Rep. 2014;8:1328–1338. doi: 10.1016/j.celrep.2014.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Boldin M.P., Taganov K.D., Rao D.S., Yang L., Zhao J.L., Kalwani M., Garcia-Flores Y., Luong M., Devrekanli A., Xu J., et al. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J. Exp. Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu D.D., Zhou P.J., Wang Y., Zhang Y., Zhang R., Zhang L., Chen S.H., Fu W.Y., Ruan B.B., Xu H.P., et al. miR-150 suppresses the proliferation and tumorigenicity of leukemia stem cells by targeting the nanog signaling pathway. Front. Pharmacol. 2016;7:439. doi: 10.3389/fphar.2016.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fang Z.H., Wang S.L., Zhao J.T., Lin Z.J., Chen L.Y., Su R., Xie S.T., Carter B.Z., Xu B. miR-150 exerts antileukemia activity in vitro and in vivo through regulating genes in multiple pathways. Cell Death Dis. 2016;7:e2371. doi: 10.1038/cddis.2016.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gerloff D., Grundler R., Wurm A.A., Brauer-Hartmann D., Katzerke C., Hartmann J.U., Madan V., Muller-Tidow C., Duyster J., Tenen D.G., et al. NF-κB/STAT5/miR-155 network targets PU.1 in FLT3-ITD-driven acute myeloid leukemia. Leukemia. 2015;29:535–547. doi: 10.1038/leu.2014.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marcucci G., Maharry K.S., Metzeler K.H., Volinia S., Wu Y.Z., Mrozek K., Nicolet D., Kohlschmidt J., Whitman S.P., Mendler J.H., et al. Clinical role of microRNAs in cytogenetically normal acute myeloid leukemia: miR-155 upregulation independently identifies high-risk patients. J. Clin. Oncol. 2013;31:2086–2093. doi: 10.1200/JCO.2012.45.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schneider E., Staffas A., Rohner L., Krowiorz K., Heuser M., Dohner K., Bullinger L., Dohner H., Fogelstrand L., Rouhi A., et al. MicroRNA-155 is upregulated in MLL-rearranged AML but its absence does not affect leukemia development. Exp. Hematol. 2016;44:1166–1171. doi: 10.1016/j.exphem.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 94.Wallace J.A., Kagele D.A., Eiring A.M., Kim C.N., Hu R., Runtsch M.C., Alexander M., Huffaker T.B., Lee S.H., Patel A.B., et al. miR-155 promotes FLT3-ITD-induced myeloproliferative disease through inhibition of the interferon response. Blood. 2017;129:3074–3086. doi: 10.1182/blood-2016-09-740209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O’Connell R.M., Rao D.S., Chaudhuri A.A., Boldin M.P., Taganov K.D., Nicoll J., Paquette R.L., Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J. Exp. Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sadras T., Kok C.H., Perugini M., Ramshaw H.S., D’Andrea R.J. miR-155 as a potential target of IL-3 signaling in primary AML cells. Leukemia Res. 2017;57:57–59. doi: 10.1016/j.leukres.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 97.Wurm A.A., Zjablovskaja P., Kardosova M., Gerloff D., Brauer-Hartmann D., Katzerke C., Hartmann J.U., Benoukraf T., Fricke S., Hilger N., et al. Disruption of the C/EBPα-miR-182 balance impairs granulocytic differentiation. Nat. Commun. 2017;8:46. doi: 10.1038/s41467-017-00032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ke S., Li R.C., Lu J., Meng F.K., Feng Y.K., Fang M.H. MicroRNA-192 regulates cell proliferation and cell cycle transition in acute myeloid leukemia via interaction with CCNT2. Int.J. Hematol. 2017;106:258–265. doi: 10.1007/s12185-017-2232-2. [DOI] [PubMed] [Google Scholar]
- 99.Li Y., Gao L., Luo X., Wang L., Gao X., Wang W., Sun J., Dou L., Li J., Xu C., et al. Epigenetic silencing of microRNA-193a contributes to leukemogenesis in t(8;21) acute myeloid leukemia by activating the PTEN/PI3K signal pathway. Blood. 2013;121:499–509. doi: 10.1182/blood-2012-07-444729. [DOI] [PubMed] [Google Scholar]
- 100.Gao X.N., Lin J., Li Y.H., Gao L., Wang X.R., Wang W., Kang H.Y., Yan G.T., Wang L.L., Yu L. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene. 2011;30:3416–3428. doi: 10.1038/onc.2011.62. [DOI] [PubMed] [Google Scholar]
- 101.Dell’Aversana C., Giorgio C., D’Amato L., Lania G., Matarese F., Saeed S., Di Costanzo A., Belsito Petrizzi V., Ingenito C., Martens J.H.A., et al. miR-194-5p/BCLAF1 deregulation in AML tumorigenesis. Leukemia. 2017;31:2315–2325. doi: 10.1038/leu.2017.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pulikkan J.A., Dengler V., Peramangalam P.S., Peer Zada A.A., Muller-Tidow C., Bohlander S.K., Tenen D.G., Behre G. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115:1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xiao Y., Su C., Deng T. miR-223 decreases cell proliferation and enhances cell apoptosis in acute myeloid leukemia via targeting FBXW7. Oncol. Lett. 2016;12:3531–3536. doi: 10.3892/ol.2016.5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gentner B., Pochert N., Rouhi A., Boccalatte F., Plati T., Berg T., Sun S.M., Mah S.M., Mirkovic-Hosle M., Ruschmann J., et al. MicroRNA-223 dose levels fine tune proliferation and differentiation in human cord blood progenitors and acute myeloid leukemia. Exp. Hematol. 2015;43 doi: 10.1016/j.exphem.2015.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hu J., Zheng L., Shen X., Zhang Y., Li C., Xi T. MicroRNA-125b inhibits AML cells differentiation by directly targeting Fes. Gene. 2017;620:1–9. doi: 10.1016/j.gene.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 106.Forrest A.R., Kanamori-Katayama M., Tomaru Y., Lassmann T., Ninomiya N., Takahashi Y., de Hoon M.J., Kubosaki A., Kaiho A., Suzuki M., et al. Induction of microRNAs, miR-155, miR-222, miR-424 and miR-503, promotes monocytic differentiation through combinatorial regulation. Leukemia. 2010;24:460–466. doi: 10.1038/leu.2009.246. [DOI] [PubMed] [Google Scholar]
- 107.Palma C.A., Al Sheikha D., Lim T.K., Bryant A., Vu T.T., Jayaswal V., Ma D.D. MicroRNA-155 as an inducer of apoptosis and cell differentiation in acute myeloid leukaemia. Mol. Cancer. 2014;13:79. doi: 10.1186/1476-4598-13-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Narayan N., Morenos L., Phipson B., Willis S.N., Brumatti G., Eggers S., Lalaoui N., Brown L.M., Kosasih H.J., Bartolo R.C., et al. Functionally distinct roles for different miR-155 expression levels through contrasting effects on gene expression, in acute myeloid leukaemia. Leukemia. 2017;31:808–820. doi: 10.1038/leu.2016.279. [DOI] [PubMed] [Google Scholar]
- 109.Havelange V., Stauffer N., Heaphy C.C., Volinia S., Andreeff M., Marcucci G., Croce C.M., Garzon R. Functional implications of microRNAs in acute myeloid leukemia by integrating microRNA and messenger rna expression profiling. Cancer. 2011;117:4696–4706. doi: 10.1002/cncr.26096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vigorito E., Perks K.L., Abreu-Goodger C., Bunting S., Xiang Z., Kohlhaas S., Das P.P., Miska E.A., Rodriguez A., Bradley A., et al. MicroRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Basova P., Pospisil V., Savvulidi F., Burda P., Vargova K., Stanek L., Dluhosova M., Kuzmova E., Jonasova A., Steidl U., et al. Aggressive acute myeloid leukemia in PU.1/p53 double-mutant mice. Oncogene. 2014;33:4735–4745. doi: 10.1038/onc.2013.414. [DOI] [PubMed] [Google Scholar]
- 112.Ghani S., Riemke P., Schonheit J., Lenze D., Stumm J., Hoogenkamp M., Lagendijk A., Heinz S., Bonifer C., Bakkers J., et al. Macrophage development from hscs requires PU.1-coordinated microRNA expression. Blood. 2011;118:2275–2284. doi: 10.1182/blood-2011-02-335141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fontana L., Pelosi E., Greco P., Racanicchi S., Testa U., Liuzzi F., Croce C.M., Brunetti E., Grignani F., Peschle C. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 2007;9:775–787. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 114.Shaham L., Binder V., Gefen N., Borkhardt A., Izraeli S. miR-125 in normal and malignant hematopoiesis. Leukemia. 2012;26:2011–2018. doi: 10.1038/leu.2012.90. [DOI] [PubMed] [Google Scholar]
- 115.Ooi A.G., Sahoo D., Adorno M., Wang Y., Weissman I.L., Park C.Y. MicroRNA-125b expands hematopoietic stem cells and enriches for the lymphoid-balanced and lymphoid-biased subsets. Proc. Natl. Acad. Sci. USA. 2010;107:21505–21510. doi: 10.1073/pnas.1016218107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.De Luca L., Trino S., Laurenzana I., Simeon V., Calice G., Raimondo S., Podesta M., Santodirocco M., Di Mauro L., La Rocca F., et al. miRNAs and piRNAs from bone marrow mesenchymal stem cell extracellular vesicles induce cell survival and inhibit cell differentiation of cord blood hematopoietic stem cells: A new insight in transplantation. Oncotarget. 2016;7:6676–6692. doi: 10.18632/oncotarget.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pando A., Reagan J.L., Quesenberry P., Fast L.D. Extracellular vesicles in leukemia. Leukemia Res. 2017;64:52–60. doi: 10.1016/j.leukres.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 118.Caivano A., Laurenzana I., De Luca L., La Rocca F., Simeon V., Trino S., D’Auria F., Traficante A., Maietti M., Izzo T., et al. High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumour Biol. 2015;36:9739–9752. doi: 10.1007/s13277-015-3741-3. [DOI] [PubMed] [Google Scholar]
- 119.Hong C.S., Sharma P., Yerneni S.S., Simms P., Jackson E.K., Whiteside T.L., Boyiadzis M. Circulating exosomes carrying an immunosuppressive cargo interfere with cellular immunotherapy in acute myeloid leukemia. Sci. Rep. 2017;7:14684. doi: 10.1038/s41598-017-14661-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hornick N.I., Doron B., Abdelhamed S., Huan J., Harrington C.A., Shen R., Cambronne X.A., Chakkaramakkil Verghese S., Kurre P. AML suppresses hematopoiesis by releasing exosomes that contain microRNAs targeting c-MYB. Sci. Signal. 2016;9:ra88. doi: 10.1126/scisignal.aaf2797. [DOI] [PubMed] [Google Scholar]
- 121.Razmkhah F., Soleimani M., Mehrabani D., Karimi M.H., Amini Kafi-Abad S., Ramzi M., Iravani Saadi M., Kakoui J. Leukemia microvesicles affect healthy hematopoietic stem cells. Tumour Biol. 2017;39 doi: 10.1177/1010428317692234. [DOI] [PubMed] [Google Scholar]
- 122.Horiguchi H., Kobune M., Kikuchi S., Yoshida M., Murata M., Murase K., Iyama S., Takada K., Sato T., Ono K., et al. Extracellular vesicle miR-7977 is involved in hematopoietic dysfunction of mesenchymal stromal cells via poly(rC) binding protein 1 reduction in myeloid neoplasms. Haematologica. 2016;101:437–447. doi: 10.3324/haematol.2015.134932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Martianez Canales T., de Leeuw D.C., Vermue E., Ossenkoppele G.J., Smit L. Specific depletion of leukemic stem cells: Can microRNAs make the difference? Cancers. 2017;9:74. doi: 10.3390/cancers9070074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Velu C.S., Chaubey A., Phelan J.D., Horman S.R., Wunderlich M., Guzman M.L., Jegga A.G., Zeleznik-Le N.J., Chen J., Mulloy J.C., et al. Therapeutic antagonists of microRNAs deplete leukemia-initiating cell activity. J. Clin. Investig. 2014;124:222–236. doi: 10.1172/JCI66005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Han Y.C., Park C.Y., Bhagat G., Zhang J., Wang Y., Fan J.B., Liu M., Zou Y., Weissman I.L., Gu H. MicroRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J. Exp. Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang Y., Cheng Q., Liu J., Dong M. Leukemia stem cell-released microvesicles promote the survival and migration of myeloid leukemia cells and these effects can be inhibited by microRNA34a overexpression. Stem Cells Int. 2016;2016:9313425. doi: 10.1155/2016/9313425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.He Y., Lin J., Kong D., Huang M., Xu C., Kim T.K., Etheridge A., Luo Y., Ding Y., Wang K. Current state of circulating microRNAs as cancer biomarkers. Clin. Chem. 2015;61:1138–1155. doi: 10.1373/clinchem.2015.241190. [DOI] [PubMed] [Google Scholar]
- 128.Turchinovich A., Samatov T.R., Tonevitsky A.G., Burwinkel B. Circulating miRNAs: Cell-cell communication function? Front. Genet. 2013;4:119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Caivano A., La Rocca F., Laurenzana I., Trino S., De Luca L., Lamorte D., Del Vecchio L., Musto P. Extracellular vesicles in hematological malignancies: From biology to therapy. Int. J. Mol. Sci. 2017;18:1183. doi: 10.3390/ijms18061183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.De Luca L., Trino S., Laurenzana I., Lamorte D., Caivano A., Del Vecchio L., Musto P. Mesenchymal stem cell derived extracellular vesicles: A role in hematopoietic transplantation? Int. J. Mol. Sci. 2017;18:1022. doi: 10.3390/ijms18051022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.De Luca L., D’Arena G., Simeon V., Trino S., Laurenzana I., Caivano A., La Rocca F., Villani O., Mansueto G., Deaglio S., et al. Characterization and prognostic relevance of circulating microvesicles in chronic lymphocytic leukemia. Leuk. Lymphoma. 2017;58:1424–1432. doi: 10.1080/10428194.2016.1243790. [DOI] [PubMed] [Google Scholar]
- 132.Yeh C.H., Moles R., Nicot C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol. Cancer. 2016;15:37. doi: 10.1186/s12943-016-0518-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Grasedieck S., Sorrentino A., Langer C., Buske C., Dohner H., Mertens D., Kuchenbauer F. Circulating microRNAs in hematological diseases: Principles, challenges, and perspectives. Blood. 2013;121:4977–4984. doi: 10.1182/blood-2013-01-480079. [DOI] [PubMed] [Google Scholar]
- 134.Wang F., Wang X.S., Yang G.H., Zhai P.F., Xiao Z., Xia L.Y., Chen L.R., Wang Y., Wang X.Z., Bi L.X., et al. miR-29a and miR-142-3p downregulation and diagnostic implication in human acute myeloid leukemia. Mol. Biol. Rep. 2012;39:2713–2722. doi: 10.1007/s11033-011-1026-5. [DOI] [PubMed] [Google Scholar]
- 135.Zhi Y., Xie X., Wang R., Wang B., Gu W., Ling Y., Dong W., Zhi F., Liu Y. Serum level of miR-10-5p as a prognostic biomarker for acute myeloid leukemia. Int. J. Hematol. 2015;102:296–303. doi: 10.1007/s12185-015-1829-6. [DOI] [PubMed] [Google Scholar]
- 136.Zhi F., Cao X., Xie X., Wang B., Dong W., Gu W., Ling Y., Wang R., Yang Y., Liu Y. Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia. PLoS ONE. 2013;8:e56718. doi: 10.1371/journal.pone.0056718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Elhamamsy A.R., El Sharkawy M.S., Zanaty A.F., Mahrous M.A., Mohamed A.E., Abushaaban E.A. Circulating miR-92a, miR-143 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. Int. J. Mol. Cell. Med. 2017;6:77–86. doi: 10.22088/acadpub.BUMS.6.2.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fayyad-Kazan H., Bitar N., Najar M., Lewalle P., Fayyad-Kazan M., Badran R., Hamade E., Daher A., Hussein N., ElDirani R., et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J. Transl. Med. 2013;11:31. doi: 10.1186/1479-5876-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Caivano A., La Rocca F., Simeon V., Girasole M., Dinarelli S., Laurenzana I., De Stradis A., De Luca L., Trino S., Traficante A., et al. MicroRNA-155 in serum-derived extracellular vesicles as a potential biomarker for hematologic malignancies—A short report. Cell. Oncol. 2017;40:97–103. doi: 10.1007/s13402-016-0300-x. [DOI] [PubMed] [Google Scholar]
- 140.Lin X., Wang Z., Wang Y., Feng W. Serum microRNA-370 as a potential diagnostic and prognostic biomarker for pediatric acute myeloid leukemia. Int. J. Clin. Exp. Pathol. 2015;8:14658–14666. [PMC free article] [PubMed] [Google Scholar]
- 141.Maki K., Yamagata T., Sugita F., Nakamura Y., Sasaki K., Mitani K. Aberrant expression of miR9 indicates poor prognosis in acute myeloid leukaemia. Br. J. Haematol. 2012;158:283–285. doi: 10.1111/j.1365-2141.2012.09118.x. [DOI] [PubMed] [Google Scholar]
- 142.Organista-Nava J., Gomez-Gomez Y., Illades-Aguiar B., Del Carmen Alarcon-Romero L., Saavedra-Herrera M.V., Rivera-Ramirez A.B., Garzon-Barrientos V.H., Leyva-Vazquez M.A. High miR-24 expression is associated with risk of relapse and poor survival in acute leukemia. Oncol. Rep. 2015;33:1639–1649. doi: 10.3892/or.2015.3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wang Y., Li Z., He C., Wang D., Yuan X., Chen J., Jin J. MicroRNAs expression signatures are associated with lineage and survival in acute leukemias. Blood Cells Mol. Dis. 2010;44:191–197. doi: 10.1016/j.bcmd.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhu C., Wang Y., Kuai W., Sun X., Chen H., Hong Z. Prognostic value of miR-29a expression in pediatric acute myeloid leukemia. Clin. Biochem. 2013;46:49–53. doi: 10.1016/j.clinbiochem.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 145.Xiong Y., Li Z., Ji M., Tan A.C., Bemis J., Tse J.V., Huang G., Park J., Ji C., Chen J., et al. MIR29B regulates expression of MLLT11 (AF1Q), an mll fusion partner, and low MIR29B expression associates with adverse cytogenetics and poor overall survival in AML. Br. J. Haematol. 2011;153:753–757. doi: 10.1111/j.1365-2141.2011.08662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao J., Lu Q., Zhu J., Fu J., Chen Y.X. Prognostic value of miR-96 in patients with acute myeloid leukemia. Diagn. Pathol. 2014;9:76. doi: 10.1186/1746-1596-9-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chen X.X., Lin J., Qian J., Qian W., Yang J., Ma J.C., Deng Z.Q., Xie D., An C., Tang C.Y., et al. Dysregulation of miR-124-1 predicts favorable prognosis in acute myeloid leukemia. Clin. Biochem. 2014;47:63–66. doi: 10.1016/j.clinbiochem.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 148.Diaz-Beya M., Brunet S., Nomdedeu J., Tejero R., Diaz T., Pratcorona M., Tormo M., Ribera J.M., Escoda L., Duarte R., et al. MicroRNA expression at diagnosis adds relevant prognostic information to molecular categorization in patients with intermediate-risk cytogenetic acute myeloid leukemia. Leukemia. 2014;28:804–812. doi: 10.1038/leu.2013.281. [DOI] [PubMed] [Google Scholar]
- 149.Zhu Y.D., Wang L., Sun C., Fan L., Zhu D.X., Fang C., Wang Y.H., Zou Z.J., Zhang S.J., Li J.Y., et al. Distinctive microRNA signature is associated with the diagnosis and prognosis of acute leukemia. Med. Oncol. 2012;29:2323–2331. doi: 10.1007/s12032-011-0140-5. [DOI] [PubMed] [Google Scholar]
- 150.Schwind S., Maharry K., Radmacher M.D., Mrozek K., Holland K.B., Margeson D., Whitman S.P., Hickey C., Becker H., Metzeler K.H., et al. Prognostic significance of expression of a single microRNA, miR-181a, in cytogenetically normal acute myeloid leukemia: A cancer and leukemia group B study. J. Clin. Oncol. 2010;28:5257–5264. doi: 10.1200/JCO.2010.29.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Xiang L., Li M., Liu Y., Cen J., Chen Z., Zhen X., Xie X., Cao X., Gu W. The clinical characteristics and prognostic significance of MN1 gene and MN1-associated microRNA expression in adult patients with de novo acute myeloid leukemia. Ann. Hematol. 2013;92:1063–1069. doi: 10.1007/s00277-013-1729-x. [DOI] [PubMed] [Google Scholar]
- 152.Butrym A., Rybka J., Baczynska D., Poreba R., Mazur G., Kuliczkowski K. Expression of microRNA-181 determines response to treatment with azacitidine and predicts survival in elderly patients with acute myeloid leukaemia. Oncol. Lett. 2016;12:2296–2300. doi: 10.3892/ol.2016.4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jinlong S., Lin F., Yonghui L., Li Y., Weidong W. Identification of let-7a-2-3p or/and miR-188-5p as prognostic biomarkers in cytogenetically normal acute myeloid leukemia. PLoS ONE. 2015;10:e0118099. doi: 10.1371/journal.pone.0118099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Liu L., Chen R., Zhang Y., Fan W., Xiao F., Yan X. Low expression of circulating microRNA-328 is associated with poor prognosis in patients with acute myeloid leukemia. Diagn. Pathol. 2015;10:109. doi: 10.1186/s13000-015-0345-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Butrym A., Rybka J., Baczynska D., Tukiendorf A., Kuliczkowski K., Mazur G. Expression of microRNA-331 can be used as a predictor for response to therapy and survival in acute myeloid leukemia patients. Biomark. Med. 2015;9:453–460. doi: 10.2217/bmm.14.112. [DOI] [PubMed] [Google Scholar]
- 156.Wang Z., Hong Z., Gao F., Feng W. Upregulation of microRNA-375 is associated with poor prognosis in pediatric acute myeloid leukemia. Mol. Cell. Biochem. 2013;383:59–65. doi: 10.1007/s11010-013-1754-z. [DOI] [PubMed] [Google Scholar]
- 157.Qian J., Lin J., Qian W., Ma J.C., Qian S.X., Li Y., Yang J., Li J.Y., Wang C.Z., Chai H.Y., et al. Overexpression of miR-378 is frequent and may affect treatment outcomes in patients with acute myeloid leukemia. Leukemia Res. 2013;37:765–768. doi: 10.1016/j.leukres.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 158.Diaz-Beya M., Brunet S., Nomdedeu J., Cordeiro A., Tormo M., Escoda L., Ribera J.M., Arnan M., Heras I., Gallardo D., et al. The expression level of baalc-associated microRNA miR-3151 is an independent prognostic factor in younger patients with cytogenetic intermediate-risk acute myeloid leukemia. Blood Cancer J. 2015;5:e352. doi: 10.1038/bcj.2015.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Li Y., Lin J., Yang J., Qian J., Qian W., Yao D.M., Deng Z.Q., Liu Q., Chen X.X., Xie D., et al. Overexpressed let-7a-3 is associated with poor outcome in acute myeloid leukemia. Leukemia Res. 2013;37:1642–1647. doi: 10.1016/j.leukres.2013.09.022. [DOI] [PubMed] [Google Scholar]
- 160.Wang X., Zuo D., Yuan Y., Yang X., Hong Z., Zhang R. MicroRNA-183 promotes cell proliferation via regulating programmed cell death 6 in pediatric acute myeloid leukemia. J. Cancer Res. Clin. Oncol. 2017;143:169–180. doi: 10.1007/s00432-016-2277-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tang X., Chen L., Yan X., Li Y., Xiong Y., Zhou X. Overexpression of miR-210 is associated with poor prognosis of acute myeloid leukemia. Med. Sci. Monitor. 2015;21:3427–3433. doi: 10.12659/MSM.894812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lin X., Wang Z., Zhang R., Feng W. High serum microRNA-335 level predicts aggressive tumor progression and unfavorable prognosis in pediatric acute myeloid leukemia. Clin. Transl. Oncol. 2015;17:358–364. doi: 10.1007/s12094-014-1237-z. [DOI] [PubMed] [Google Scholar]
- 163.Hornick N.I., Huan J., Doron B., Goloviznina N.A., Lapidus J., Chang B.H., Kurre P. Serum exosome microRNA as a minimally-invasive early biomarker of AML. Sci. Rep. 2015;5:11295. doi: 10.1038/srep11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Boyiadzis M., Whiteside T.L. Plasma-derived exosomes in acute myeloid leukemia for detection of minimal residual disease: Are we ready? Expert Rev. Mol. Diagn. 2016;16:623–629. doi: 10.1080/14737159.2016.1174578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xiao Y., Deng T., Su C., Shang Z. MicroRNA 217 inhibits cell proliferation and enhances chemosensitivity to doxorubicin in acute myeloid leukemia by targeting KRAS. Oncol. Lett. 2017;13:4986–4994. doi: 10.3892/ol.2017.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McDermott A.M., Heneghan H.M., Miller N., Kerin M.J. The therapeutic potential of microRNAs: Disease modulators and drug targets. Pharm. Res. 2011;28:3016–3029. doi: 10.1007/s11095-011-0550-2. [DOI] [PubMed] [Google Scholar]
- 167.Brown B.D., Venneri M.A., Zingale A., Sergi Sergi L., Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat. Med. 2006;12:585–591. doi: 10.1038/nm1398. [DOI] [PubMed] [Google Scholar]
- 168.Wang H., Jiang Y., Peng H., Chen Y., Zhu P., Huang Y. Recent progress in microRNA delivery for cancer therapy by non-viral synthetic vectors. Adv. Drug Deliv. Rev. 2015;81:142–160. doi: 10.1016/j.addr.2014.10.031. [DOI] [PubMed] [Google Scholar]
- 169.Huang X., Schwind S., Yu B., Santhanam R., Wang H., Hoellerbauer P., Mims A., Klisovic R., Walker A.R., Chan K.K., et al. Targeted delivery of microRNA-29b by transferrin-conjugated anionic lipopolyplex nanoparticles: A novel therapeutic strategy in acute myeloid leukemia. Clin. Cancer Res. 2013;19:2355–2367. doi: 10.1158/1078-0432.CCR-12-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Weiler J., Hunziker J., Hall J. Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs implicated in human disease? Gene Ther. 2006;13:496–502. doi: 10.1038/sj.gt.3302654. [DOI] [PubMed] [Google Scholar]
- 171.Su R., Lin H.S., Zhang X.H., Yin X.L., Ning H.M., Liu B., Zhai P.F., Gong J.N., Shen C., Song L., et al. miR-181 family: Regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. 2015;34:3226–3239. doi: 10.1038/onc.2014.274. [DOI] [PubMed] [Google Scholar]
- 172.Tian C., You M.J., Yu Y., Zhu L., Zheng G., Zhang Y. MicroRNA-9 promotes proliferation of leukemia cells in adult CD34-positive acute myeloid leukemia with normal karyotype by downregulation of Hes1. Tumour Biol. 2016;37:7461–7471. doi: 10.1007/s13277-015-4581-x. [DOI] [PubMed] [Google Scholar]
- 173.Chen Y., Jacamo R., Konopleva M., Garzon R., Croce C., Andreeff M. Cxcr4 downregulation of let-7a drives chemoresistance in acute myeloid leukemia. J. Clin. Investig. 2013;123:2395–2407. doi: 10.1172/JCI66553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lu F., Zhang J., Ji M., Li P., Du Y., Wang H., Zang S., Ma D., Sun X., Ji C. miR-181b increases drug sensitivity in acute myeloid leukemia via targeting HMGB1 and Mcl-1. Int. J. Oncol. 2014;45:383–392. doi: 10.3892/ijo.2014.2390. [DOI] [PubMed] [Google Scholar]
- 175.Yamamoto H., Lu J., Oba S., Kawamata T., Yoshimi A., Kurosaki N., Yokoyama K., Matsushita H., Kurokawa M., Tojo A., et al. miR-133 regulates evi1 expression in AML cells as a potential therapeutic target. Sci. Rep. 2016;6:19204. doi: 10.1038/srep19204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hickey C.J., Schwind S., Radomska H.S., Dorrance A.M., Santhanam R., Mishra A., Wu Y.Z., Alachkar H., Maharry K., Nicolet D., et al. Lenalidomide-mediated enhanced translation of C/EBPα-p30 protein up-regulates expression of the antileukemic microRNA-181a in acute myeloid leukemia. Blood. 2013;121:159–169. doi: 10.1182/blood-2012-05-428573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Khalife J., Radomska H.S., Santhanam R., Huang X., Neviani P., Saultz J., Wang H., Wu Y.Z., Alachkar H., Anghelina M., et al. Pharmacological targeting of miR-155 via the NEDD8-activating enzyme inhibitor MLN4924 (pevonedistat) in FLT3-ITD acute myeloid leukemia. Leukemia. 2015;29:1981–1992. doi: 10.1038/leu.2015.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wampfler J., Federzoni E.A., Torbett B.E., Fey M.F., Tschan M.P. The RNA binding proteins RBM38 and DND1 are repressed in AML and have a novel function in APL differentiation. Leukemia Res. 2016;41:96–102. doi: 10.1016/j.leukres.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 179.Wei S., Wang K. Long noncoding RNAs: Pivotal regulators in acute myeloid leukemia. Exp. Hematol. Oncol. 2015;5:30. doi: 10.1186/s40164-016-0059-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Schwarzer A., Emmrich S., Schmidt F., Beck D., Ng M., Reimer C., Adams F.F., Grasedieck S., Witte D., Kabler S., et al. The non-coding RNA landscape of human hematopoiesis and leukemia. Nat. Commun. 2017;8:218. doi: 10.1038/s41467-017-00212-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Li Q., Pan X., Wang X., Jiao X., Zheng J., Li Z., Huo Y. Long noncoding RNA MALAT1 promotes cell proliferation through suppressing miR-205 and promoting SMAD4 expression in osteosarcoma. Oncotarget. 2017;8:106648–106660. doi: 10.18632/oncotarget.20678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol. Proced. Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ling H., Vincent K., Pichler M., Fodde R., Berindan-Neagoe I., Slack F.J., Calin G.A. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34:5003–5011. doi: 10.1038/onc.2014.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pichler M., Calin G.A. MicroRNAs in cancer: From developmental genes in worms to their clinical application in patients. Br. J. Cancer. 2015;113:569–573. doi: 10.1038/bjc.2015.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Fang Y., Fullwood M.J. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genom. Proteom. Bioinform. 2016;14:42–54. doi: 10.1016/j.gpb.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen L., Wang W., Cao L., Li Z., Wang X. Long non-coding RNA CCAT1 acts as a competing endogenous RNA to regulate cell growth and differentiation in acute myeloid leukemia. Mol. Cells. 2016;39:330–336. doi: 10.14348/molcells.2016.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Xue H., Hua L.M., Guo M., Luo J.M. SHIP1 is targeted by miR-155 in acute myeloid leukemia. Oncol. Rep. 2014;32:2253–2259. doi: 10.3892/or.2014.3435. [DOI] [PubMed] [Google Scholar]
- 188.Xing C.Y., Hu X.Q., Xie F.Y., Yu Z.J., Li H.Y., Bin Z., Wu J.B., Tang L.Y., Gao S.M. Long non-coding RNA hotair modulates c-KIT expression through sponging miR-193a in acute myeloid leukemia. FEBS Lett. 2015;589:1981–1987. doi: 10.1016/j.febslet.2015.04.061. [DOI] [PubMed] [Google Scholar]
- 189.Chen Z.H., Wang W.T., Huang W., Fang K., Sun Y.M., Liu S.R., Luo X.Q., Chen Y.Q. The lncRNA HOTAIRM1 regulates the degradation of PML-RARA oncoprotein and myeloid cell differentiation by enhancing the autophagy pathway. Cell Death Differ. 2017;24:212–224. doi: 10.1038/cdd.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Mangiavacchi A., Sorci M., Masciarelli S., Larivera S., Legnini I., Iosue I., Bozzoni I., Fazi F., Fatica A. The miR-223 host non-coding transcript linc-223 induces IRF4 expression in acute myeloid leukemia by acting as a competing endogenous RNA. Oncotarget. 2016;7:60155–60168. doi: 10.18632/oncotarget.11165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Rupaimoole R., Slack F.J. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]