Abstract
We assessed immunogenicity of a malaria DNA vaccine administered by needle i.m. or needleless jet injection [i.m. or i.m./intradermally (i.d.)] in 14 volunteers. Antigen-specific IFN-γ responses were detected by enzyme-linked immunospot (ELISPOT) assays in all subjects to multiple 9- to 23-aa peptides containing class I and/or class II restricted epitopes, and were dependent on both CD8+ and CD4+ T cells. Overall, frequency of response was significantly greater after i.m. jet injection. CD8+-dependent cytotoxic T lymphocytes (CTL) were detected in 8/14 volunteers. Demonstration in humans of elicitation of the class I restricted IFN-γ responses we believe necessary for protection against the liver stage of malaria parasites brings us closer to an effective malaria vaccine.
No subunit vaccine against an infectious agent as complex as the parasites that cause malaria has been licensed. The extra- and intracellular life cycle in humans, stage-specific expression of proteins, allelic and antigenic variation of the parasites, and variant disease expression based on transmission dynamics and host genetic background led us to speculate that only a similarly complex vaccine will provide sustainable protection against this disease (1, 2). An ideal malaria vaccine would induce antibody and T cell responses against all stages of the parasite life cycle, and protect all recipients against all strains of Plasmodium falciparum (3, 4). Given current vaccine technologies, we concluded that DNA-based vaccines provide the best opportunity for construction of such a vaccine (4), and that the immunogenicity of DNA-based vaccines in humans must be optimized.
Our focus has been induction of CD8+ T cell responses against parasite proteins expressed in infected hepatocytes (1). We reported previously that i.m. needle immunization of volunteers with a single gene P. falciparum circumsporozoite protein (PfCSP) DNA vaccine (VCL-2510) elicited antigen-specific, genetically restricted, CD8+ T cell-dependent cytotoxic T lymphocytes (CTL) (5), but not antibodies (6). We did not report on the IFN-γ responses that we now know are critical to protection in mice immunized with radiation-attenuated sporozoites or DNA vaccines (7–10).
The Biojector (Bioject Inc., Portland, OR) is a needle-free injection device (11, 12) known to enhance antibodies in other systems (12, 13). Studies in rabbits and rhesus monkeys (J. Aguiar, unpublished work; W. O. Rogers, unpublished work) showed that Plasmodium-specific antibodies could be increased by administration of DNA by Biojector as compared with needle. To improve T cell-mediated immunity and elicit antibodies, we assessed the safety and immunogenicity of VCL-2510 administered by Biojector or by needle in healthy human volunteers, focusing on the frequency, magnitude, breadth, and cellular dependence of IFN-γ responses.
Methods
PfCSP DNA Vaccine.
The plasmid VCL-2510 (5, 6, 15, 16) was produced under Good Manufacturing Practice conditions (14). The vaccine was stored as 1.0-ml doses at −20°C and thawed at room temperature for 30 min before injection.
Volunteer Recruitment and Study Design.
Twenty-one 18–30-year-old, healthy, HLA-A*0201-positive and malaria-naive volunteers were recruited. None had antibodies to PfCSP, HIV, HBV core antigen, HCV, vaccinia, or dsDNA. Fifteen volunteers were randomized into three groups of five, and received three injections of 2.5 mg of PfCSP DNA vaccine administered at 0, 4, and 8 weeks by three different methods: Group 1, i.m. needle; group 2, i.m. by needleless Biojector jet; and group 3, i.m./i.d. by Biojector [70% of the total dose (1.75 mg/dose) i.m. by Biojector and 30% (0.75 mg/dose) i.d. by Biojector]. For all groups, the three i.m. doses of DNA were administered in the same deltoid muscle on one side of body, and the three i.d. doses in group 3 were administered in the contralateral posterior upper arm. The six additional HLA-A*0201-positive and three HLA-mismatched volunteers did not receive the PfCSP DNA vaccine and provided specimens used as negative controls in the assays. One volunteer (i.m. needle) withdrew from the study after the first immunization to relocate.
Reagents Used in T Cell Assays.
Recombinant canary-pox (ALVAC) virus expressing the full-length PfCSP (vCP182) used for infection of the CTL stimulators was produced by Virogenetics Inc. (Troy, NY; refs. 5 and 17). Peptides used for CTL and ELISPOT assays were synthesized by Chiron Technologies (Clayton, Victoria, Australia). Nine experimental and two control peptides known to bind to HLA-A or -B alleles (class I) were 8–10 aa in length, and six longer PfCSP-derived and one control peptide known to bind to one or more HLA-DR molecules were 15–25 aa in length (Table 2). Five of the six longer peptides included sequences from the class I binding peptides (Table 2). The positive control was derived from the influenza matrix protein [residues 58–66, GILGFVFTL (ref. 18), HLA-A*0201-restricted], and the negative controls from HIV gag protein [residues 77–85, SLYNTVATL (ref. 19), HLA-A*0201-restricted], or P. falciparum protein, Exp-1 [residues 82–96, sequence AGLLGNVSTVLLGGV (ref. 20), HLA-DR-restricted].
Table 2.
Breadth of DNA-induced IFN-γ responses (fresh and frozen PBMCs)
| Peptide | Residues | HLA restriction | Sequence | Range of net SFCs per 102 PBMCs (geomean) | Number of positive assays/total assays (%) | Number of responders/number tested |
|---|---|---|---|---|---|---|
| Class I | ||||||
| HLA-A2 | ||||||
| A2.386 | 386–394 | A2 supertype | GLIMVLSFL | 11–543 (36.2) | 23/70 (32.9%) | 11/14 |
| A2.7 | 7–16 | A2 supertype | ILSVSSFLFV | 13–163 (39.3) | 12/70 (17.1%) | 10/14 |
| A2.1 | 1–10 | A2.1 | MMRKLAILSV | 12–56 (32.9) | 4/70 (5.7%) | 5/14 |
| A2.319 | 319–327 | A2.1 | YLNKIQNSL | 17.5 | 1/70 (1.4%) | 1/14 |
| Non-HLA-A2 | ||||||
| A1.310 | 310–319 | A1 | EPSDKHIKEY | 19–29.5 (23.7) | 2/14 (14.3%) | 2/7 |
| A3/11.336 | 336–345 | A3/11 supertype | VTCGNGIQVR | 0/12 (0%) | 0/4* | |
| B7.285 | 285–294 | B7 | MPNDPNRNV | 20–23 (21.4) | 2/14 (14.3%) | 1/4 |
| B8.86 | 86–94 | B8 | LRKPKHKKL | 0/12 (0%) | 0/4 | |
| B35.353 | 353–360 | B35 | KPKDELDY | 0/3 (0%) | 0/1 | |
| Class II | ||||||
| DR.1 | 1–20 | DR (A2.1 and A2 supertype) | MMRKLAILSVSSFLFVEALF | 12–177 (30.7) | 24/70 (34.3%) | 11/14 |
| DR.375 | 375–397 | DR (A2 supertype) | SSVFNVVNSSIGLIMVLSFLFLN | 12–519 (40.7) | 31/70 (44.3%) | 11/14 |
| DR.316 | 316–335 | DR (A2.1) | IKEYLNKIQNSLSTEWSPCS | 10–27 (16.2) | 6/48 (12.5%) | 5/14 |
| DR.346 | 346–365 | Th3R (B35) | IKPGSANKPKDELDYANDIE | 10–37.7 (22.2) | 4/50 (8.0%) | 3/14 |
| DR.281 | 281–305 | DR (B7) | QGHNMPNDPNRNVDENANANSAVKN | 22.3 | 1/14 (7.1%) | 1/4 |
| DR.363 | 363–383 | DR | DIEKKICKMEKCSSVFNVVNS | 10–59 (21.0) | 13/58 (22.4%) | 6/14 |
| Positive control | ||||||
| Flu. M A2.1 | 58–66 | A2.1 | GILGFVFTL | 14–837 (88.9) | 118/122 (96.7%) | 20/20 |
PfCSP peptide sequences and residue numbers were based on those of P. falciparum clone 3D7 (GenBank no. X15363). Residues known to vary among isolates are indicated in bold. Nested class I-restricted epitopes within the DR binding peptides are underlined.
None of the four volunteers tested expressed the HLA-A*0301 allele, but all expressed one of the A3-superfamily alleles HLA-A*1101, A*3101, and A*6801.
Cytotoxic T Cell Assays.
Assays were performed as described (5). Percent lysis was defined as (experimental release − medium control release)/(maximum release − medium control release) × 100. Percent specific lysis was determined by subtracting % lysis of targets sensitized with the control HIV gag peptide from % lysis of targets incubated with experimental peptide. CTL responses were considered positive if % specific lysis postimmunization was ≥10% for at least two effector to target ratios in the same assay and if % specific lysis preimmunization was <10%. Spontaneous release values were always <25%.
ELISPOT Assay.
PfCSP-specific IFN-γ- or IL-4-producing cells were determined by ELISPOT. Sterile 96-well polyvinylidene difluoride (PVDF)-backed plates (Millipore) were coated with anti-IFN-γ mAb (1-D1K) or anti-IL-4 mAb (82.4) at 15 μg/ml (MABTECH, Stockholm) overnight at 4°C. Plates were washed six times with RPMI medium 1640 and blocked with medium with 10% human AB serum (ICN) for 2 h at room temperature. Then, 100 μl of experimental or control peptide was added at 10 μg/ml final concentration. Freshly isolated or cryopreserved PBMCs were added in 100 μl at 2.5 and 5.0 × 105 cells per well in quadruplicate. Cultures were incubated for 36 h at 37°C, 5% CO2. Plates were washed six times with PBS/0.05% Tween 20 and incubated with 100 μl of 1 μg/ml biotinylated anti-IFN-γ mAb (7B6–1) or biotinylated anti-IL-4 mAb (12.1) for 3 h. Plates were washed six times and then incubated with 100 μl of 1:1,000 dilution of streptavidin alkaline phosphatase (MABTECH) for 1 h. After washing six times with PBS/0.05% Tween 20, the color was developed by adding 100 μl per well of alkaline phosphatase substrate (Bio-Rad), and terminated after 15 min by washing with tap water, and plates were air-dried. Spot forming cells (SFCs) were enumerated with a spot counting system (Scanalytics, Fairfax, VA). Responses were expressed as mean number of SFCs per 106 PBMCs, and were considered significant if (i) mean number of cells in wells with experimental peptide was significantly greater (P < 0.05, student's t test) than in wells with control peptide, (ii) net SFCs per well (mean SFCs in experimental peptide wells minus mean number of SFCs in control peptide wells) was ≥5 SFCs per well, and (iii) the ratio of mean number of SFCs in experimental peptide wells to mean number of SFCs in control peptide wells was greater than 2.0. Furthermore, in the assays assessing responses in fresh PBMCs, a response to a specific PfCSP peptide was not considered positive if cells obtained before immunization had a positive response as defined above.
Characterization of CTL Effectors and IFN-γ-Producing Cells by Cell Depletion or Enrichment Studies.
In negative depletion experiments, ELISPOT assays were carried out with PBMCs after depletion of T cells using anti-CD4+- or anti-CD8+-coated Dynabeads M-450 (Dynal, Great Neck, NY). In selective enrichment experiments, PBMCs were enriched for CD8+ or CD4+ T cells by using MACS positive selection beads (Miltenyi Biotec, Auburn, CA). Antigen-presenting cells (APC) in these assays were PBMCs after depletion of CD8+, CD4+, and CD56+ cells by passing the PBMCs through all three positive selection columns individually. Flow cytometry confirmed that cell subset depletions or enrichment were >95% in all experiments.
Statistical Analysis.
The prevalence of dichotomous outcome variables (frequency and magnitude of peptide-specific IFN-γ and CTL responses) was assessed by using the Pearson χ2 test (uncorrected) and Fisher's exact test (two-tailed) or Student's t test (two-tailed), respectively. Analysis was conducted by using SPSS Version 8.0 (SPSS, Chicago) or EPI INFO Version 6.04b (Centers for Disease Control, Atlanta). Concordance studies were assessed by using Cohen's Kappa test (SPSS Version 8.0). The level of significance or concordance was P < 0.05.
Results
Frequency and Magnitude of IFN-γ Responses.
IFN-γ responses were assessed using freshly isolated PBMCs incubated with up to 15 PfCSP-specific peptides. To confirm findings with fresh PBMCs (Table 1), frozen specimens obtained pre- and postimmunization were assayed in parallel in a coded and blinded manner using the five peptides against which the best responses were detected with fresh PBMCs. With these five peptides, there were 26.4% (33/125) positive assays with fresh PBMCs versus 32.0% (40/125) positive assays with frozen PBMCs. The concordance between assays conducted with fresh or frozen PBMCs collected at the same time and incubated with the same peptides was significant (P = 0.029, kappa = 0.195; Cohen's Kappa test).
Table 1.
Frequency and magnitude of DNA-induced IFN-γ responses with fresh PBMCs
| Peptide | Number of positive responders | Number of positive peptides/ peptides tested | Number of positive assays/ total assays (% positive assays) | Range of net SFCs per 106 PBMCs (geomean) | P values of frequency compared to control | P values of magnitude compared to control |
|---|---|---|---|---|---|---|
| Needle i.m. | ||||||
| HLA-A2 | 4 /4 | 3/4 | 7/48 (14.6%) | 18–50 (28.5) | 0.043 | 0.01 |
| HLA-DR | 3 /4* | 4/5 | 8/60 (13.3%) | 10.5–69 (22.6) | 0.00037 | 0.00 |
| Total | 4 /4 | 7/9 | 15/108 (13.9%) | 10.5–69 (25.2) | 0.0001 | 0.003 |
| Biojector i.m. | ||||||
| HLA-A2 | 4 /5* | 4/4 | 11/60 (18.3%) | 17.5–156.3 (39.8) | 0.008 | 0.021 |
| HLA-DR | 4 /5* | 5/5 | 24/75 (32.0%) | 12–50 (22.5) | <0.00001 | 0.00 |
| Total | 5 /5 | 9/9 | 35/135 (25.9%) | 12–156.3 (26.9) | <0.00001 | 0.0005 |
| Biojector i.m./i.d. | ||||||
| HLA-A2 | 2 /5 | 3/4 | 3/60 (5.0%) | 10–24 (12.9) | 0.82 | 0.89 |
| HLA-DR | 3 /5 | 3/5 | 10/75 (10.3%) | 16–26.3 (20.8) | 0.00035 | 0.00 |
| Total | 4 /5 | 6/9 | 13/135 (9.6%) | 10–26.3 (18.6) | 0.003 | 0.007 |
| Overall | ||||||
| HLA-A2 | 10 /14 | 4/4 | 21/168 (12.5%) | 10–156.3 (30.3) | 0.048 | 0.005 |
| HLA-DR | 10 /14 | 5/5 | 42/210 (20.0%) | 10.5–69 (22.1) | <0.00001 | 0.00 |
| Total | 13 /14 | 9/9 | 63/378 (16.7%) | 10–156.3 (24.5) | <0.00001 | <0.00001 |
| Controls | ||||||
| HLA-A2 | 3 /6 | 3/4 | 3/72 (4.2%) | 10.6–16.6 (12.6) | ||
| HLA-DR | 0 /6 | 0/5 | 0/90 (0%) | 0 | ||
| Total | 3 /6 | 3/9 | 3/162 (1.9%) | 10.6–16.6 (12.6) |
Two volunteers (BJ13, needle i.m.; BJ3, Biojector i.m./i.d.) were considered nonresponders to one or two DR binding peptides (DR316 and DR.346 or DR.363, respectively), and two volunteers (BJ7, Biojector i.m.; BJ1 Biojector i.m./i.d.) were considered nonresponders to class I binding peptides (peptides A2.386 or A2.7, respectively), because their preimmunization specimens were positive.
Responses of fresh PBMCs to peptides assayed in all immunized and control volunteers (four A2 and five DR peptides) were analyzed (Table 1). Frequency of IFN-γ responses was significantly greater in immunized volunteers as compared with controls, overall (63/378 vs. 3/162; P < 0.00001) and for each route of immunization (P < 0.003–0.0001). Likewise, magnitude of IFN-γ responses was greater overall (P < 0.00001) and for each route (P < 0.007–0.0005; Table 1). Although responses classified as positive were detected in 3/6 controls to 3/9 peptides tested, both the frequency (1.9%) and magnitude (maximum 16.6 SFCs per 106 PBMCs) of responses were low.
Overall, IFN-γ responses were detected by using fresh or frozen PBMCs in 14/14 volunteers to 9/9 peptides in 20.7% [107/518 (number of positive assays per total assays)] of assays (Tables 1 and 2). Simultaneous assessment of coded, frozen PBMCs collected from eight volunteers pre- and postimmunization confirmed that the PfCSP-specific IFN-γ responses were induced by DNA immunization (Fig. 1). In the needle i.m. group, responses were detected in 4/4 volunteers to 7/9 peptides in 17.6% (26/148) of assays; in the Biojector i.m. group, responses were detected in 5/5 volunteers to 9/9 peptides in 26.5% (49/185) of assays; and in the Biojector i.m./i.d. group, responses were detected in 4/5 volunteers to 7/9 peptides in 17.3% (32/185) of assays.
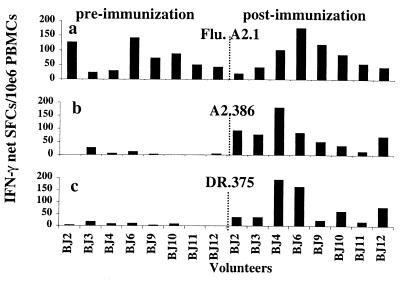
Figure 1.
Antigen-specific IFN-γ responses induced by DNA vaccination. IFN-γ responses against: (a) HLA-A2.1-restricted positive control peptide from influenza matrix protein (Flu. A2.1); (b) HLA-A2.1-restricted peptide from PfCSP (A2.386); or (c) HLA-DR-restricted peptide from PfCSP (DR.375) with coded and double-blinded frozen PBMCs collected from eight volunteers pre- and postimmunization with PfCSP DNA. BJ2, -3, and -4 from Biojector i.m./i.d.; BJ6, -9, and -10 from Biojector i.m.; and BJ11 and -12 from needle i.m.
There was no difference between assays conducted 2 weeks after the second or third immunizations in terms of frequency of positive responders (9/14 vs. 9/14), positive assays per total assays (29/126 vs. 23/126, P = 0.35) or magnitude of the IFN-γ responses [range of SFCs: 10–156 (26.2) vs. 10–86 (24.6), P = 0.29]. There was an apparent decrease in frequency of IFN-γ responses 6 weeks after the third immunization as compared with 2 weeks after the third immunization, but this was not statistically significant (13/126 vs. 23/126, P = 0.072). Twelve to 14 months after the last immunization, PBMCs from 10 of the 14 study subjects were tested in a single assay (in triplicate) against the same panel of class I peptides and DR-binding peptides tested at the earlier time points. IFN-γ responses could not be detected to any class I peptide (total of seven peptides tested), nor to five of the six DR-binding peptides tested. However, IFN-γ responses specific for peptide DR.375 were detected in three of the ten volunteers (range of SFCs per 106 PBMCs: 14.4–20.6); this peptide was one of the two DR-binding peptides to which the best responses were previously detected (Table 2).
Breadth of DNA-Induced IFN-γ Responses.
PfCSP-specific IFN-γ responses to the defined class I peptides were detected in 13/14 volunteers (1/5–6/6 peptides recognized per volunteer), and responses to the DR-binding peptides in 12/14 volunteers (2/5–5/6 peptides per volunteer); 11/14 volunteers produced IFN-γ in response to both class I and DR binding peptides (Table 2). Overall, IFN-γ responses to 4/4 HLA-A2, 2/5 non-A2, and 6/6 DR-binding peptides were detected. Positive responses to the HLA-A2-restricted influenza control peptide were detected in 14/14 volunteers and 6/6 controls (Table 2).
Responses to multiple peptides were detected simultaneously in the same volunteer. One individual recognized 92% (11/12) of peptides studied, five responders recognized 50–60%, six responders recognized 30–40%, and two others recognized 8–18%. Representative data of IFN-γ responses in one volunteer with six class I binding and five DR-binding peptides postimmunization, are presented in Fig. 2.
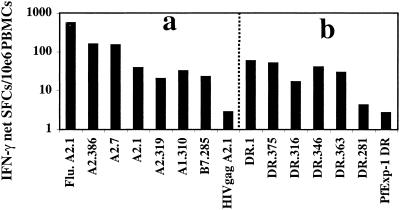
Figure 2.
Breadth of DNA-induced IFN-γ responses. Fresh PBMCs from volunteer BJ10 (Biojector i.m.), who expressed the alleles HLA-A*0101, -A*0201, -B*0701, -DRB1*1501, and -DRB5*0101, were cultured with (a) six class I-restricted PfCSP peptides, an HLA-A2.1-restricted positive control peptide (Flu. A2.1), or an HLA-A2.1-restricted negative control peptide (HIV gag A2.1); or (b) six class II-restricted PfCSP peptides or an HLA-DR restricted negative control peptide (P. falciparum Exp-1 DR).
The best responses were detected with two A2.1-restricted epitopes, A2.386 (11/14 volunteers, 32.9% of assays) and A2.7 (10/14, 17.1%), and two DR-binding peptides, DR.1 (11/14, 34.3%) and DR.375 (11/14, 44.3%; Table 2). The frequency and magnitude of response to these peptides were greater than responses to the other peptides.
The two most antigenic DR-binding peptides overlap the most antigenic class I peptides (Table 2). Specifically, peptide DR.1 (34.3% response) overlaps peptides A2.7 (17.1%) and A2.1 (5.7%), and peptide DR.375 (44.3%) overlaps peptide A2.386 (32.9%). Concordance between assays conducted simultaneously with peptides A2.386 and DR.375 (P = 0.009, kappa = 0.313), or with peptides A2.1/A2.7 and DR.1 (P = 0.02, kappa = 0.265), was significant. Concordance with regard to frequency of responses was paralleled by similarity in magnitude of respective responses (Table 2). Moreover, for the DR.281/B7.285 combination, the only volunteer of the four tested who recognized the DR.281 peptide was also the only volunteer who recognized the B7.285 peptide, and the frequency and magnitude of response were highly comparable between the DR and class I peptide (Table 2). The concordance between the assays conducted with overlapping class I and DR-binding peptides indicates correlation or dependence between the DNA-induced CD8+ and CD4+ T cell responses.
Frequency and Magnitude of CTL Responses.
CTL responses were detected in 8/14 volunteers: in 2/4 volunteers immunized by needle i.m. to 4/7 peptides; 3/5 by Biojector i.m. to 6/8 peptides; and 3/5 by Biojector i.m./i.d. to 6/6 peptides. The frequency of response (fresh and frozen) after immunization (30/458 assays, 6.6%) was significantly greater than before immunization (1/151 assays, 0.7%; P = 0.0052). As compared with controls, the frequency of CTL response was significantly greater, overall (30/458 vs. 2/232; P = 0.00079) and for each route of immunization: needle i.m., P = 0.043; Biojector i.m., P = 0 0.0016; and Biojector i.m./i.d., P = 0.00012. However, the magnitude of CTL response (range of % specific lysis: 12.1–90.6; see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org) was significantly greater for Biojector, but not needle immunization, as compared with controls: needle i.m., P = 0.067; Biojector i.m., P < 0.0001; and Biojector i.m./i.d., P = 0.0002. Simultaneous assessment of coded, frozen PBMC collected pre- and postimmunization confirmed that the CTL were induced by DNA administration (data not shown).
Breadth of DNA-Induced CTL Responses.
CTL responses were detected to 7/9 peptides tested, and were antigen-specific and genetically restricted. CTL of multiple genetic restrictions were induced simultaneously in a given volunteer as reported (ref. 5; data not shown). CTL to at least one HLA-A2-restricted epitope were detected in 7/8 of the CTL responders, to B7.285 in 3/3, to A1.310 in 3/4, and to B8.86 in 2/4 responders. No responses were detected to peptides B35.353 and A3.336, but only one individual expressed HLA-B*3501 and none HLA-A*0301.
Comparison of the Three Different Administration Strategies.
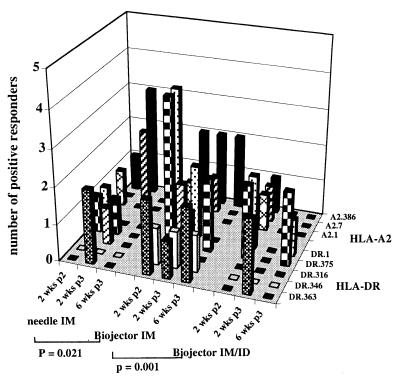
To assess the comparative immunogenicity of the three methods of administration, we compared the frequency and magnitude of IFN-γ responses to the four HLA-A2 and five HLA-DR peptides that were tested in all individuals in all assays using fresh PBMCs. With regard to the frequency of IFN-γ responses, overall, Biojector i.m. was significantly better than either needle i.m. (P = 0.021) or Biojector i.m./i.d. (P = 0.001), and there was no difference between needle i.m. and Biojector i.m./i.d. (Table 1 and Fig. 3). At the epitope level, Biojector i.m. was significantly better than Biojector i.m./i.d. for response to both A2 and DR peptides (P = 0.0023 and P = 0.0063, respectively) and was significantly better than needle i.m. for responses to the DR peptides (P = 0.011), but not the A2 peptides (P = 0.6). With regard to magnitude of IFN-γ responses, overall, Biojector i.m. and needle i.m. were both significantly better than Biojector i.m./i.d. (P = 0.01 and P = 0.043, respectively) and there was no difference between Biojector i.m. and needle i.m. At the epitope level, the responses to A2 peptides, but not DR peptides, was significantly greater with Biojector i.m. as compared with Biojector i.m./i.d. (P = 0.022 and P = 0.12), but not with needle i.m. There was no significant difference among the three groups in regard to frequency or magnitude of CTL responses.
Figure 3.
Comparison of frequency of IFN-γ responses that were induced by the three different routes of immunization: needle i.m., Biojector i.m., and Biojector i.m./i.d. IFN-γ responses were assessed by using fresh PBMCs from all of the volunteers at each of the three time points against the four HLA-A2.1-restricted epitopes and five HLA-DR-restricted epitopes, which were tested in all individuals in all assays. Data are presented as the number of responding volunteers (y axis) at each of the time points (x axis) for each of the peptides (z axis). P values were calculated by using Student's t test (two-tailed), and only the significant differences (P = 0.05) between groups are indicated.
T Cell Subset Dependence of DNA-Induced Immune Responses.
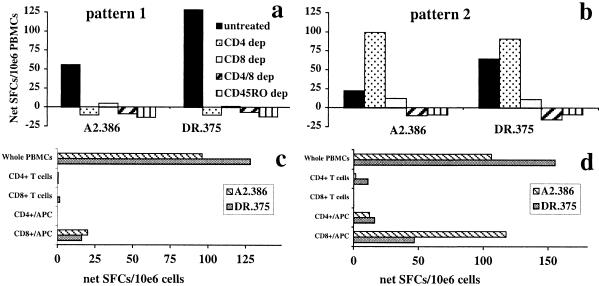
CTL responses were dependent on CD8+ T cells, but not CD4+ T cells, for all five peptides tested in three volunteers (see Fig. 6, which is published as supporting information). Likewise, IFN-γ responses were CD8+ T cell-dependent; depletion of CD8+ T cells immediately before culture of fresh or frozen PBMCs completely abrogated or significantly reduced IFN-γ responses in all cases (Fig. 4). When considering the CD4+ dependence of the IFN-γ response, two profiles were apparent. In eight of the nine volunteers tested (two needle i.m., three Biojector i.m., three Biojector i.m./i.d.), in vitro depletion of CD4+ T cells abrogated or significantly reduced the IFN-γ response to both peptides (Fig. 4a). The IFN-γ responses elicited by both short and long peptides were dependent on both CD8+ and CD4+ T cells (Fig. 4). In one volunteer (BJ7, Biojector i.m./i.d.), depletion of CD4+ T cells did not reduce the IFN-γ response to either peptide (Fig. 4b). The presence of regulatory CD4+ T cells could not be completely excluded, because there was an increase in IFN-γ responses, albeit not statistically significant, to both peptides upon depletion of CD4+ T cells (Fig. 4b).
Figure 4.
CD8+ and CD4+ T cell dependency of IFN-γ responses. Frozen PBMCs from (a) volunteer BJ11 (needle i.m.) or (b) volunteer BJ7 (Biojector i.m.) collected 2 weeks after the second immunization or 6 weeks after the third immunization, respectively, were either untreated or selectively depleted of CD4+, CD8+, both CD4+ and CD8+ T cells, or CD45RO+ cells immediately before culture with peptide A2.386 or peptide DR.375. Alternatively, frozen PBMCs from the same volunteer were either untreated (whole PBMCs), or selectively enriched for CD8+ T cells, CD4+ T cells, or CD8+ plus APCs or CD4+ plus APCs immediately before culture with peptides A2.386 or DR.375. Two distinct patterns are presented. Pattern 1: (a) IFN-γ induced against both A2.386 and DR.375 peptides depended on CD4+, CD8+, and CD45RO+ T cells. (c) Either positively selected CD8+ or CD4+ T cells, with or without APCs, failed to reconstitute the effector response. Pattern 2: (b) IFN-γ induced against peptides A2.386 and DR.375 depended on CD8+ and CD45RO+ T cells, but not CD4+ T cells. (d) Positively selected CD4+ cells with or without APCs, or of CD8+ cells without APCs, failed to reconstitute the effector response. Positively selected CD8+ T cells with APCs completely restored the effector response to the class I peptide A2.386 and partially restored the response to the class II peptide, which contains a nested class I epitope.
These two distinct profiles were identified by in vitro depletion (negative selection) experiments. To further investigate this finding, positively selected enriched populations were tested. Purified populations of CD8+ or CD4+ T cells, combined CD8+ T cells and APC populations, or combined CD4+ T cells and APCs were added to the ELISPOT cultures in numbers reflective of untreated PBMCs. Results of these studies were consistent with the two profiles identified above. Representative examples are presented in Fig. 4 c and d. With pattern 1, studies carried out with three of the eight volunteers showed that the IFN-γ effector response could not be reconstituted by addition of either CD8+ or CD4+ T cells, with or without APCs (Fig. 4c). With pattern two, addition of CD4+ T cells with or without APCs, or of CD8+ cells without APCs, failed to reconstitute the effector response. However, addition of CD8+ T cells with APCs completely restored the effector response to the peptide A2.386 and partially restored the response to the peptide DR.375 (Fig. 4d). If the IFN-γ responses were CD8+ but not CD4+ T cell-dependent, or CD4+ but not CD8+ T cell-dependent, then positively selected experiments with either CD8+/APC or CD4+/APC would be expected to restore the IFN-γ response. In our studies (Fig. 4a, pattern 1), we were not able to restore the IFN-γ response by either coculture of CD4+/APCs or of CD8+/APCs (Fig. 4c), consistent with the CD4+ and CD8+ T cells dependence of the pattern 1 response. However, with one of nine volunteers, the IFN-γ response was eliminated by selective depletion of CD8+ T cells, but not CD4+ T cells (Fig. 4b), and was restored by coculture with CD8+/APCs, but not CD4+/APCs (Fig. 4d), consistent with the CD8+ dependence of the pattern 2 response.
Antigen-Specific IL-4 and Antibody Responses Not Detected Following DNA Vaccination.
Frozen PBMCs from all 14 volunteers, collected preimmunization and 7 weeks after the third immunization, were assessed for production of IL-4 after incubation with DR.375 and DR.363 peptides. We could not detect IL-4 responses in any of 31 assays with the PfCSP peptides (range SFCs: preimmune, 0–20; postimmune, 0–9). Responses to control mitogens PMA and ionomycin were positive (range: 47–351 SFCs per 106 PBMCs). PfCSP-specific antibodies were not detected in any volunteer at any time point.
Discussion
The primary goals of this study were to determine whether DNA immunization of humans would elicit the class I restricted IFN-γ responses considered important for protection against malaria, and whether altering the route or method of administration of DNA would enhance immunogenicity. All 14 volunteers immunized with PfCSP DNA had IFN-γ responses, and 13 of the 14 volunteers had antigen-specific, CD8+ T cell, IFN-γ responses against 20–100% of PfCSP-derived peptides containing defined class I binding epitopes. We have previously reported induction of antigen-specific, CD8+ T cell-dependent, genetically restricted, class I-restricted CTL against the same peptides after DNA immunization (5). However, based on murine studies, we concluded that class I-restricted IFN-γ responses are the primary immune effectors contributing to the long-lasting, preerythrocytic stage-specific protection of mice and humans after immunization with irradiated sporozoites (7, 20). Data presented here establish that these responses can be induced in humans by plasmid DNA expressing the PfCSP. In contrast, immunization of volunteers with a PfCSP recombinant protein (RTS,S) in adjuvant provides short-term protection of naive adults against experimentally induced malaria (21, 22), and semiimmune adults against naturally transmitted malaria (K. Bojang, personal communication), and induces class II-restricted anti-PfCSP IFN-γ responses, but not class I-restricted responses (23).
We also demonstrated induction of antigen-specific IFN-γ responses in 12/14 volunteers against 40–83.3% of HLA-DR-binding peptides tested. Five of the six peptides contained at least one defined class I T cell epitope (Table 2). When tested, concordance between parallel assays with the DR and class I peptides (DR.375/A2.386 and DR.1/A2.1/A2.7) was significant (P < 0.025), and there was similarity in magnitude of the respective responses. Furthermore, where concordance could not be determined, the volunteers who recognized the DR peptide generally recognized the nested class I epitope, and vice versa, and frequency and magnitude of these responses were similar. We speculate that the IFN-γ responses against DR binding peptides may have been directed against the class I epitope nested within, because the IFN-γ responses elicited by the short and long peptides were always dependent on CD8+ T cells (Fig. 4). Taken with the CD4+ T cell dependence of the IFN-γ response seen in most of our assays (Fig. 4), we speculate that CD4+ T cells function in a bystander helper capacity for CD8+ T cell production of IFN-γ following DNA vaccination. Consistent with this interpretation is our inability to detect Th2 (IL-4) responses or antibody responses following PfCSP DNA vaccination, and the failure by others to detect IL-4 responses in normal humans following immunization with an HIV-1 DNA vaccine (24).
Antigen-specific, CD8+ T cell, genetically restricted, class I-restricted CTL were detected in 8/14 volunteers immunized with PfCSP DNA, consistent with our previous study where CTL responses were detected in 11/17 volunteers who expressed HLA alleles for which responses could be assessed (5).
Our data suggested that Biojector i.m. was the most effective route of those assessed for induction of antigen-specific IFN-γ responses, and that Biojector (i.m. and/or i.m./i.d.) was more effective than needle i.m. for induction of CTL. In comparison with i.m. injection by needle, delivery of injectate by needleless jet injection may improve distribution at the site of administration (25, 26). Recently, the induction of antigen-specific lymphoproliferative, supernatant IFN-γ and B-chemokine responses following Biojector i.m. administration of HIV env/rev DNA in normal volunteers was reported (24). In that study, CTL were detected in only one assay and one of five volunteers tested, IFN-γ was not detected by ELISPOT, IL-4 responses were not detected, and antibodies were not assessed. Others have reported induction of CD8+ CTL (27–29) and lymphoproliferative (28–30) responses by needle i.m. (27, 29) or Biojector i.m. (30), and the boosting of antigen-specific antibody responses following needle i.m. (28, 30) administration of HIV-1 DNA in asymptomatic HIV1-infected volunteers. However, these volunteers were HIV infected, complicating interpretation of results.
Despite the induction of IFN-γ responses and CTL by each of three different routes of administration, we believe that the frequency and magnitude of these responses were suboptimal, and can be significantly improved. If we sum the IFN-γ responses by any given volunteer to those individual peptides tested simultaneously (at the same time and with the same PBMC sample) and consider those summed responses reflective of responses induced to a peptide pool containing the same peptides, IFN-γ responses are in the range of 23–572.7 SFCs per 106 cells (geomean 92.7 SFCs per 106 cells) with fresh PBMCs and 30.6–1227.6 SFCs per 106 cells (geomean 144.5 SFCs per 106 cells) with frozen PBMCs.
In this study, criteria used to define positivity in the IFN-γ ELISPOT assay were conservative, and the positive and negative control peptides used have been well characterized. Furthermore, responses to the immunodominant, conserved influenza matrix protein peptide (positive control) and an HIV gag protein or P. falciparum Exp1 peptide (negative controls) were always assessed in parallel, providing internal standardization between volunteers and between assays. We are confident that we are not detecting false positive responses to P. falciparum—derived T cell epitopes elicited by crossreactivity with common bacterial, viral, and nonpathogenic organisms. Thus, we believe that this methodology should be applicable to assessing most vaccine candidates.
As compared with the CTL assay, the ELISPOT assay was more sensitive (14/14 vs. 8/14 responders overall) and reproducible (of 13 IFN-γ responders by ELISPOT using fresh PBMCs, 12 were detected by using frozen PBMCs, whereas of eight CTL responders with fresh cells, only three were reproduced with frozen PBMCs). Furthermore, the ELISPOT assay requires fewer cells than does the CTL assay and is less labor intensive.
The results of this study provide us with an improved method of administration of DNA and a demonstration in humans of elicitation of the class I restricted IFN-γ responses that we believe will be critical for a subunit vaccine to duplicate the excellent protection elicited by the impractical irradiated sporozoite vaccine.
Supplementary Material
Acknowledgments
We thank A. Figer, V. Fallarame, S. Abot, and C. DaCosta for excellent technical assistance; J. Tine (Virogenetics Corporation) for the canary pox; the staff of the Clinical Trial Center at U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) for excellent support; W. O. Rogers for manuscript review; the many individuals at Vical who contributed to the project; and the volunteers, without whom this study would not have been possible. The protocol was approved by the Naval Medical Research Center's Committee for the Protection of Human Subjects, the USAMRIID Human Use Committee, and the Army and Navy Surgeon General's Human Subjects Research Review Board, in accordance with the U.S. Navy regulation (SECNAVINST 3900.39B). This work was supported by the Naval Medical Research and Development Command Work Units 61102A.S13.F.A0009, 62787A.870.F.A0010, 63002A.810.F.A0011, and 603792N.01889.135.A0039 and 60000.000.000.A0062. Funding from the U.S. Agency for International Development and the Office of Naval Research Advanced Technology Demonstration is acknowledged.
Abbreviations
- PfCSP
P. falciparum circumsporozoite protein
- CTL
cytotoxic T lymphocyte
- SFCs
spot forming cells
- APC
antigen-presenting cell
- ELISPOT
enzyme-linked immunospot
- PBMC
peripheral blood mononuclear cell
- i.d.
intradermally
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hoffman S L, Franke E D, Hollingdale M R, Druilhe P. In: Malaria Vaccine Development. Hoffman S L, editor. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 35–75. [Google Scholar]
- 2.Hoffman S L. In: MIM Africa Malaria Conference. Sharp B, Davies K, editors. Multilateral Initiative on Malaria; 1999. pp. 145–152. [Google Scholar]
- 3.Doolan D L, Hoffman S L. Parasitol Today. 1997;13:171–178. doi: 10.1016/s0169-4758(97)01040-5. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman S L, Doolan D L. In: Developments and Clinical Progress of DNA Vaccines. Brown F, Cichutek K, Robertson J, editors. Vol. 104. Basel: Karger; 2000. pp. 121–132. [Google Scholar]
- 5.Wang R, Doolan D L, Le T P, Hedstrom R C, Coonan K M, Charoenvit Y, Jones T R, Hobart P, Margalith M, Ng J, et al. Science. 1998;282:476–480. doi: 10.1126/science.282.5388.476. [DOI] [PubMed] [Google Scholar]
- 6.Le T P, Coonan K M, Hedstrom R C, Charoenvit Y, Sedegah M, Epstein J E, Kumar S, Wang R, Doolan D L, Maguire J D, et al. Vaccine. 2000;18:1893–1901. doi: 10.1016/s0264-410x(99)00407-7. [DOI] [PubMed] [Google Scholar]
- 7.Doolan D L, Hoffman S L. J Immunol. 1999;163:884–892. [PubMed] [Google Scholar]
- 8.Doolan D L, Hoffman S L. J Immunol. 2000;165:1453–1462. doi: 10.4049/jimmunol.165.3.1453. [DOI] [PubMed] [Google Scholar]
- 9.Sedegah M, Weiss W, Sacci J B, Jr, Charoenvit Y, Hedstrom R, Gowda K, Majam V F, Tine J, Kumar S, Hobart P, Hoffman S L. J Immunol. 2000;164:5905–5912. doi: 10.4049/jimmunol.164.11.5905. [DOI] [PubMed] [Google Scholar]
- 10.Schneider J, Gilbert S C, Hannan C M, Degano P, Prieur E, Sheu E G, Plebanski M, Hill A V. Immunol Rev. 1999;170:29–38. doi: 10.1111/j.1600-065x.1999.tb01326.x. [DOI] [PubMed] [Google Scholar]
- 11.Davis H L, Michel M L, Mancini M, Schleef M, Whalen R G. Vaccine. 1994;12:1503–1509. doi: 10.1016/0264-410x(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 12.Williams J, Fox-Leyva L, Christensen C, Fisher D, Schlicting E, Snowball M, Negus S, Mayers J, Koller R, Stout R. Vaccine. 2000;18:1939–1943. doi: 10.1016/s0264-410x(99)00446-6. [DOI] [PubMed] [Google Scholar]
- 13.Hoke C H, Jr, Egan J E, Sjogren M H, Sanchez J, DeFraites R F, MacArthy P O, Binn L N, Rice R, Burke A, Hill J, et al. J Infect Dis. 1995;171, Suppl 1:S53–S60. doi: 10.1093/infdis/171.supplement_1.s53. [DOI] [PubMed] [Google Scholar]
- 14.Parker S E, Borellini F, Wenk M L, Hobart P, Hoffman S L, Hedstrom R, Le T, Norman J A. Hum Gene Ther. 1999;10:741–758. doi: 10.1089/10430349950018508. [DOI] [PubMed] [Google Scholar]
- 15.Hedstrom R C, Doolan D L, Wang R, Kumar A, Sacci J B, Jr, Gardner M J, Aguiar J C, Charoenvit Y, Sedegah M, Tine J A, et al. Int J Mol Med. 1998;2:29–38. doi: 10.3892/ijmm.2.1.29. [DOI] [PubMed] [Google Scholar]
- 16.Wang R, Doolan D L, Charoenvit Y, Hedstrom R C, Gardner M J, Hobart P, Tine J, Sedegah M, Fallarme V, Sacci J B, Jr, et al. Infect Immun. 1998;66:4193–4202. doi: 10.1128/iai.66.9.4193-4202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tine J A, Lanar D E, Smith D M, Wellde B T, Schultheiss P, Ware L A, Kauffman E B, Wirtz R A, Taisne C D, Hui G S N, et al. Infect Immun. 1996;64:3833–3844. doi: 10.1128/iai.64.9.3833-3844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gotch F, Rothbard J, Howland K, Townsend A, McMichael A. Nature (London) 1987;326:881–882. doi: 10.1038/326881a0. [DOI] [PubMed] [Google Scholar]
- 19.Tsomides T J, Aldovini A, Johnson R P, Walker B D, Young R A, Eisen H N. J Exp Med. 1994;180:1283–1293. doi: 10.1084/jem.180.4.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doolan D L, Southwood S, Chesnut R W, Appella E, Gomez E, Richards A, Higashimoto Y, Maewal A, Sidney J, Gramzinski R A, et al. J Immunol. 2000;165:1123–1137. doi: 10.4049/jimmunol.165.2.1123. [DOI] [PubMed] [Google Scholar]
- 21.Stoute J A, Slaoui M, Heppner D G, Momin P, Kester K E, Desmons P, Wellde B T, Garcon N, Krzych U, Marchand M. N Engl J Med. 1997;336:86–91. doi: 10.1056/NEJM199701093360202. [DOI] [PubMed] [Google Scholar]
- 22.Stoute J A, Kester K E, Krzych U, Wellde B T, Hall T, White K, Glenn G, Ockenhouse C F, Garcon N, Schwenk R, et al. J Infect Dis. 1998;178:1139–1144. doi: 10.1086/515657. [DOI] [PubMed] [Google Scholar]
- 23.Lalvani A, Moris P, Voss G, Pathan A A, Kester K E, Brookes R, Lee E, Koutsoukos M, Plebanski M, Delchambre M, et al. J Infect Dis. 1999;180:1656–1664. doi: 10.1086/315074. [DOI] [PubMed] [Google Scholar]
- 24.Boyer J D, Cohen A D, Vogt S, Schumann K, Nath B, Ahn L, Lacy K, Bagarazzi M L, Higgins T J, Baine Y, et al. J Infect Dis. 2000;181:476–483. doi: 10.1086/315229. [DOI] [PubMed] [Google Scholar]
- 25.Figge F H J, Barnett D J. Amer Pract. 1948;III:197–206. [PubMed] [Google Scholar]
- 26.Lemon S M, Scott R M, Bancroft W H. J Med Virol. 1983;12:129–136. doi: 10.1002/jmv.1890120207. [DOI] [PubMed] [Google Scholar]
- 27.Calarota S, Bratt G, Nordlund S, Hinkula J, Leandersson A C, Sandstrom E, Wahren B. Lancet. 1998;351:1320–1325. doi: 10.1016/S0140-6736(97)09440-3. [DOI] [PubMed] [Google Scholar]
- 28.MacGregor R R, Boyer J D, Ugen K E, Lacy K E, Gluckman S J, Bagarazzi M L, Chattergoon M A, Baine Y, Higgins T J, Ciccarelli R B, et al. J Infect Dis. 1998;178:92–100. doi: 10.1086/515613. [DOI] [PubMed] [Google Scholar]
- 29.Calarota S A, Leandersson A C, Bratt G, Hinkula J, Klinman D M, Weinhold K J, Sandstrom E, Wahren B. J Immunol. 1999;163:2330–2338. [PubMed] [Google Scholar]
- 30.MacGregor R R, Boyer J D, Ciccarelli R B, Ginsberg R S, Weiner D B. J Infect Dis. 2000;181:406. doi: 10.1086/315199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.