Abstract
Members of the NF-YB transcription factor gene family play important roles in diverse processes related to plant growth and development, such as seed development, drought tolerance, and flowering time. However, the function of NF-YB genes in cotton remains unclear. A total of 23, 24, and 50 NF-YB genes were identified in Gossypium arboreum (G. arboreum), Gossypium raimondii (G. raimondii), and G. hirsutum, respectively. A systematic phylogenetic analysis was carried out in G. arboretum, G. raimondii, G. hirsutum, Arabidopsis thaliana, cacao, rice and, sorghum, where the 150 NF-YB genes were divided into five groups (α–ε). Of these groups, α is the largest clade, and γ contains the LEC1 type NF-YB proteins. Syntenic analyses revealed that paralogues of NF-YB genes in G. hirsutum exhibited good collinearity. Owing to segmental duplication within the A sub-genome (At) and D sub-genome (Dt), there was an expanded set of NF-YB genes in G. hirsutum. Furthermore, we investigated the structures of exons, introns, and conserved motifs of NF-YB genes in upland cotton. Most of the NF-YB genes had only one exon, and the genes from the same clade exhibited a similar motif pattern. Expression data show that most NF-YB genes were expressed ubiquitously, and only a few genes were highly expressed in specific tissues, as confirmed by quantitative real-time PCR (qRT-PCR) analysis. The overexpression of GhDNF-YB22 gene, predominantly expressed in embryonic tissues, indicates that GhDNF-YB22 may affect embryogenesis in cotton. This study is the first comprehensive characterization of the GhNF-YB gene family in cotton, and showed that NF-YB genes could be divided into five clades. The duplication events that occurred over the course of evolution were the major impetus for NF-YB gene expansion in upland cotton. Collectively, this work provides insight into the evolution of NF-YB in cotton and further our knowledge of this commercially important species.
Keywords: genome-wide analysis, NF-YB transcription factor, Gossypium hirsutum, overexpression, embryogenesis
1. Introduction
Nuclear factor Y (NF-Y), also called heme activator protein (HAP) or CCAAT-binding factor (CBF), can be found in almost all eukaryotes. Genes are normally regulated by transcription factors via the specific interactions between the upstream promoter regions and proteins encoded by transcription factors. The CCAAT-box, a common and conserved eukaryotic promoter element, is associated with large range of trans-acting factors, where only the NF-Y is absolutely required for gene regulation [1]. The NF-Y consists of three different subunits: NF-YA (CBF-B or HAP2), NF-YB (CBF-A or HAP3), and NF-YC (CBF-C or HAP5) [2]. All NF-Y subunits contain a highly conserved core region for subunit interactions, which are vital to the function of the transcription factor [3]. The NF-YB subunit includes an amino-terminal A domain, a B domain, and a carboxyl-terminal C domain [4]. Of these, the B domain is the most essential owing to the presence of amino acid residues necessary for its interaction with NF-YA and NF-YC [5]. Moreover, the NF-YB subunit can be divided into two classes in A. thaliana according to sequence: the LEC1-type and the non-LEC1-type, which differ in the 16 amino acid (aa) residues at equivalent positions in the B domain [6]. The LEC1-type contains LEC1 and LEC1-LIKE (L1L), while the rest belong to the non-LEC1-type [7].
Although, NF-YB is generally encoded by only one gene in animals and yeast, there are multiple genes encoding NF-YB in plants [8]. To date, the NF-YB gene family has been identified and characterized in several plant species. For example, there are 13 annotated NF-YB genes in the model plant A. thaliana [9]. As two representative species of monocotyledons, rice and wheat both have 11 NF-YB genes [10,11]. Moreover, 14, 32, 7, 18, and 29 NF-YB genes have been characterized in canola, soybean, tung tree, grape, and tomato, respectively [12,13,14,15], indicating that the NF-YB gene family has been expanded in plants. This expansion suggests that the function of NF-YB genes are more complex than previously thought owing to the genetic redundancy and functional divergence of the gene family over the course of evolution.
There is a large body of evidence that NF-YB genes have multiple functions. It has been demonstrated that the overexpression of AtLEC1 (AtNF-YB9), a well characterized NF-YB gene in A. thaliana, in lec1 mutant and wild-type A. thaliana can induce embryo-like structures on the leaves [6]. Moreover, AtLEC1 has also been reported to be an essential regulator in zygotic embryogenesis, seed maturation, and fatty acid synthesis [16,17]. In contrast to AtLEC1, other NF-YB genes in A. thaliana have been shown to function in drought tolerance, abscisic acid signalling transduction, flowering, and root elongation [18,19,20,21]. Aside from A. thaliana, the functional characterization of NF-YB genes also have been performed in several other staple crops, and have exhibited varying biological roles. For example, BnLEC1 and ZmLEC1 have been reported to increase oil content in seeds [22,23]. Furthermore, NF-YB genes have been shown to be involved in the process of chloroplast biogenesis in rice, and fruit ripening in the tomato [10,12,24], while the over-expression of a single NF-YB gene in wheat resulted in a 20–30% increase in grain yield [25]. In another study, VfNF-BY genes have been shown to play a vital role in pathogen response in the tung tree [4]. Even though NF-YB genes have been identified and characterized in dozens of plant species, the members and roles of this gene family in cotton, most notably in upland cotton (G. hirsutum), remain unclear. Thanks to the Gossypium sequencing project, many Gossypium species have been sequenced, including upland cotton and its two diploid progenitors (https://www.cottongen.org/). The accessibility of these genome sequences allows us to comprehensively identify and characterize NF-YB genes in cotton [26,27,28,29].
Upland cotton is an economically important crop, which supplies natural and renewable fibre for the textile industry. The aim of the current study was to systematically analyse NF-YB genes in G. hirsutum (GhNF-YBs) using a genome-wide analysis. As a result, 50 members of the NF-YB gene family were identified and further characterized to infer the phylogenetic relationships, chromosome locations, gene structures, and conserved motifs of GhNF-YBs. In addition, we analysed the expression patterns of GhNF-YB genes in different tissues. Lastly, the possible function of GhDNF-YB22 was characterized by overexpression in cotton. Here, our results will provide a foundation for the future study of NF-YB genes in upland cotton and further our understanding of this commercially important species.
2. Results
2.1. Identification of NF-YB Genes in Cotton
The A. thaliana protein sequences of the NF-YB gene family were used as queries to search NF-YB genes in the G. arboretum, G. raimondii, G. hirsutum, rice, sorghum, and cacao genomes. In total, 23, 40, 52, 16, 18, and 21, respectively, putative NF-YB genes were detected. InterProScan 56.0 was used to identify the NF-YB genes, where 23, 24, 50, 12, 15, and 13 NF-YB genes were successfully identified in the G. arboretum, G. raimondii, G. hirsutum, rice, sorghum, and cacao genomes, respectively (Table S1). The cotton NF-YB genes were named based on the distribution locations on the chromosomes (Table S1). We determined that the numbers of gene were very close in the two diploid cotton G. arboreum (AA) and G. raimondii (DD) species, where the total numbers of genes in the two diploid cottons were slightly smaller than that of the allotetraploid cotton G. hirsutum. However, the numbers of NF-YB genes in the two diploid cottons were much greater than in rice, sorghum and cacao, indicating that the NF-YB gene family has expanded during the evolution of Gossypium species. The protein sequence length of GhDNF-YB16 was 746 amino acid (aa), while the length of the orthologue GhANF-YB16 was 173 aa. To further verify the differences in sequences between GhDNF-YB16 and GhANF-YB16, we designed primers (Table S2) for GhANF-YB16 and cloned it from upland cotton. The results showed that the nucleic acid sequence of GhANF-YB16 was shorter than that of GhDNF-YB16 owing to transcription termination. The length of NF-YB protein sequences ranged from 90 to 318 aa in our study.
2.2. Phylogenetic Analysis of the NF-YB Gene Family
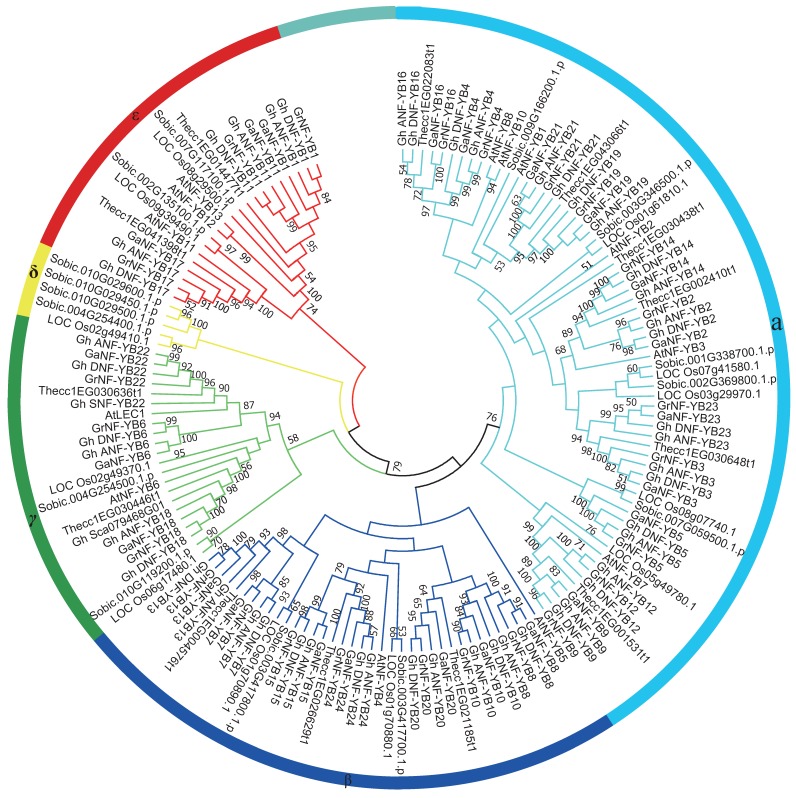
To better understand the evolutionary relationships of NF-YB gene, a neighbour-joining (NJ) phylogenetic tree was constructed using the NF-YB genes from G. hirsutum, G. arboretum, G. raimondii, A. thaliana, rice, sorghum, and cacao. As shown in Figure 1, the NF-YB genes were naturally divided into five clades, designated as α, β, γ, δ, and ε. The α clade was the largest group, containing 65 NF-YB genes, whereas the δ clade was the smallest, consisting of only five members, indicating that NF-YB genes were distributed unevenly in the different clades. The α, β, γ, and ε clades consisted of genes both from dicot and monocot species, while the δ clade only contained genes from monocot species, including four NF-YB genes from sorghum and one from rice. According to the presence of the typical LEC1 motif—consisting of 16 shared residues in the B domain—NF-YB proteins can be classified as either LEC1 type or non-LEC1 proteins. We found that only the members of the γ clade can be classified as LEC1 type proteins. GhA/DNF-YB6, GhA/DNF-YB18, and GhA/DNF-YB22—typical LEC1-type proteins—share a common ancestor with AtLEC1 and AtLEC1-like proteins (Figure S1), and were determined to be important candidate genes for embryogenesis in cotton. Notably, nearly all the orthologous genes from the two monocot species (sorghum and rice) tended to form orthologous gene pairs at the end of branches in the phylogenetic tree, where NF-YB genes from dicots (cotton, cacao, and Arabidopsis) tended to cluster together, indicating that the main function of these members of the gene family diverged prior to the divergence of dicots and monocots. As reported by Wang et al. [26], cotton has been experienced a recent duplication event whereas cacao did not, in agreement with our findings that, in most cases, each cacao gene corresponds to two orthologues in diploid cotton. For example, in the ε clade, cc1EG014477t1 corresponded to two orthologues in both G. arboreum and G. raimondii.
Figure 1.
Phylogenetic relationships of NF-YB gene family. The analysis included full-length protein sequences from Gossypium hirsutum, Gossypium arboretum, Gossypium raimondii, Arabidopsis, Oryza sativa, Sorghum bicolor, and Theobroma cacao. Using MEGA software, the phylogenetic tree was constructed with 1000 bootstrap replicates using the neighbour-joining method, where only bootstrap values >50% are shown. A total of 150 NF-YB proteins were divided into five branches corresponding to subunit type, and are indicated by different colours.
2.3. Chromosomal Distribution and Synteny Analysis of GhNF-YB Genes
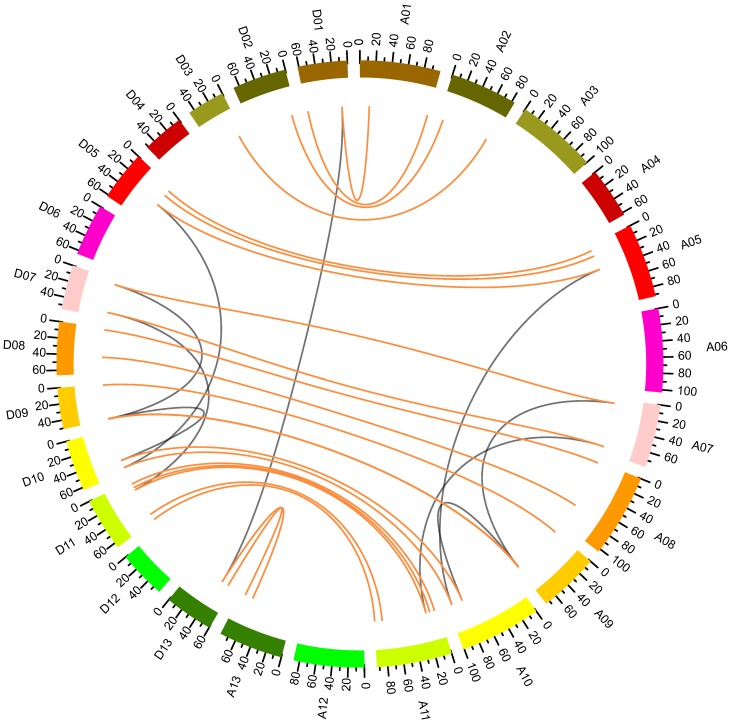
A total of 50 NF-YB genes were detected in G. hirsutum and were unevenly distributed on chromosomes, where 48 of the genes detected were located on nine At chromosomes (A1, A2, A5, A7, A8, A9, A10, A11, and A13) and ten Dt chromosomes (D1, D2, D3, D5, D7, D8, D9, D10, D11 and D13) (Figure 2 and Figure S2). The remaining two genes (GhSNF-YB18, GhSNF-YB22) were distributed on two unoriented scaffolds. The total number of NF-YB genes mapped within At sub-genomes was equal to that of the Dt sub-genomes. We found that the distribution of genes was uneven within each chromosome, and most of the orthologues from the At and Dt sub-genomes were located on homologous chromosomes. Nine chromosomes contained two NF-YB genes, six chromosomes contained three genes, and two chromosomes contained five genes (Figure 2 and Figure S2).
Figure 2.
Collinearity analyses of Gossypium hirsutum NF-YB genes. A01–13 and D01–13 represent chromosomes from the A and D sub-genomes, respectively. The red lines link two genes that were identified to be homologous chromosome pairs from the At and Dt sub-genomes. The grey lines link gene pairs formed by segmental duplication within the At and Dt sub-genomes.
Gossypium hirsutum, as the typical allotetraploid species, was derived from the hybridization of two diploid species resembling the ancestors of G. arboretum and G. raimondii, where the resulting chromosome was doubled [30]. Tandem duplication, segmental duplication, and whole-genome duplication are the main impetus for gene family expansion [31]. As shown in Figure 2, the orthologues maintained good collinearity between the At and Dt sub-genomes. A segmental duplication analysis showed that nine pairs of genes may have been derived from segmental duplication events (Table S3). Eight genes formed four pairs of duplicated genes in the Dt sub-genome, while their orthologues in the At subgenome also formed four pairs of duplicated genes accordingly, indicating that the duplication events happened prior to the doubling of the upland cotton chromosome. The results of our duplication analysis were consistent with those of the phylogenetic analysis, as the duplication pairs clustered closely to each other in the phylogenetic tree (Figure 1 and Figure 2).
Over the course of evolutionary history, duplicated genes have three potential evolutionary fates: non-functionalisation, neo-functionalisation, and sub-functionalisation [32]. In comparing the non-synonymous (Ka) and synonymous substitution (Ks) rates of substitution (Ka/Ks), one could infer the magnitude of selective constraint and positive selection. Generally, Ka/Ks > 1, Ka/Ks = 1, and Ka/Ks < 1 indicate positive selection, neutral evolution, and purifying selection, respectively. In the present study, the Ka, Ks, and Ka/Ks of NF-YB homologous gene pairs were estimated in G. hirsutum (Table 1). We found that the Ka/Ks ratios of NF-YB gene homologous pairs were less than 0.5, and that the ratios of three of these homologous pairs were smaller than 0.1, suggesting that NF-YB genes have undergone purifying selection after segmental and whole genome duplications.
Table 1.
Comparative analysis of Ka, Ks, and Ka/Ks values for homologous pairs in Gossypium hirsutum.
| Homologous Pairs | Ka | Ks | Ka/Ks | |
|---|---|---|---|---|
| Gh_ANF-YB21 | Gh_ANF-YB19 | 0.071 | 0.376 | 0.189 |
| Gh_ANF-YB11 | Gh_ANF-YB1 | 0.025 | 0.563 | 0.044 |
| Gh_ANF-YB14 | Gh_ANF-YB2 | 0.061 | 0.621 | 0.098 |
| Gh_ANF-YB20 | Gh_ANF-YB10 | 0.203 | 1.113 | 0.182 |
| Gh_DNF-YB23 | Gh_DNF-YB3 | 0.067 | 0.486 | 0.139 |
| Gh_DNF-YB21 | Gh_DNF-YB19 | 0.183 | 0.474 | 0.386 |
| Gh_DNF-YB11 | Gh_DNF-YB1 | 0.025 | 0.481 | 0.052 |
| Gh_DNF-YB14 | Gh_DNF-YB2 | 0.083 | 0.500 | 0.165 |
| Gh_DNF-YB20 | Gh_DNF-YB10 | 0.245 | 0.846 | 0.289 |
Transposable elements (TEs) compose a major fraction of eukaryotic genomes, especially in plants, mainly in retrotransposons and DNA transposons, which move around the genome [33]. Transposable elements are expressed and mobilized in order to respond to specific stimuli [34]. To investigate whether TEs played roles in expansion of the NF-YB protein family, TEs close to the NF-YB genes were identified in the present study (Table 2). Only three retroelements—L1 (1) and Copia (2)—were found in the 2000 bp region upstream and downstream of the genes (Table S4). When the scanning region was broadened to 10,000 bp, fifty-four TEs were identified. Of these, only one could be classified as a DNA transposon, while the rest of them were retroelements (i.e., L1 [10], copia [33], and gypsy [10]) (Table S5). Upon further investigation, we found that one L1 was located upstream of GhDNF-YB1, and two Copia were located in the gene region of GhDNF-YB2, within the 2000 bp region. Moreover, within 10,000 bp region, one DNA/hAT-Ac was located downstream of GhDNF-YB6; two L1 elements were located upstream of GhANF-YB6 and downstream of GhDNF-YB3 and GhANF-YB10; one L1 element was located downstream of GhANF-YB3 and upstream of GhANF-YB21, GhDNF-YB21, and GhDNF-YB1; seven Copia were located downstream of GhDNF-YB18; five Copia were located upstream of GhDNF-YB10 and GhDNF-YB14; four Copia elements were located downstream of GhANF-YB19; two Copia elements were located within the gene region of GhANF-YB2 and upstream of GhDNF-YB3, GhANF-YB3, and GhDNF-YB21; one Copia element was located downstream of GhDNF-YB15 and GhDNF-YB20 and upstream of GhANF-YB13 and GhANF-YB1; three gypsy elements were located upstream of GhDNF-YB24 and GhANF-YB23; and one gypsy element was located downstream of GhANF-YB14, GhANF-YB2 and upstream of GhDNF-YB14 and GhDNF-YB1. We noted that most of the TEs were located in the vicinity of duplicated genes, suggesting that TEs contributed to the expansion of the NF-YB gene family. The numbers of simple repeat sequences were more abundant than those of TEs, and their lengths were variable, which could play important roles in functional divergence after duplication.
Table 2.
Transposable elements in the vicinity of the NF-YB gene locus.
| Type | Elements | Number of Elements | Length Occupied (bp) | Percentage of Sequence (%) | Number of Elements | Length Occupied (bp) | Percentage of Sequence (%) |
|---|---|---|---|---|---|---|---|
| 10,000 bp region | 2000 bp region | ||||||
| DNA transposons | 1 | 91 | 0.10 | 0 | 0 | 0 | |
| CMC-EnSpm | 0 | 0 | 0 | 0 | 0 | 0 | |
| MULE-MuDR | 0 | 0 | 0 | 0 | 0 | 0 | |
| PIF-Harbinger | 0 | 0 | 0 | 0 | 0 | 0 | |
| TcMar-Pogo | 0 | 0 | 0 | 0 | 0 | 0 | |
| hAT | 0 | 0 | 0 | 0 | 0 | 0 | |
| hAT-Ac | 1 | 91 | 0.10 | 0 | 0 | 0 | |
| hAT-Charlie | 0 | 0 | 0 | 0 | 0 | 0 | |
| hAT-Tag1 | 0 | 0 | 0 | 0 | 0 | 0 | |
| hAT-Tip100 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Retroelements | 53 | 17,673 | 18.90 | 3 | 1038 | 7.43 | |
| LINE: | 10 | 2923 | 3.13 | 1 | 91 | 0.65 | |
| L1 | 10 | 2923 | 3.13 | 1 | 91 | 0.65 | |
| LTR: | 43 | 14,750 | 15.78 | 2 | 947 | 6.78 | |
| Caulimovirus | 0 | 0 | 0 | 0 | 0 | 0 | |
| Copia | 33 | 12,359 | 13.22 | 2 | 947 | 6.7 | |
| Gypsy | 10 | 2391 | 2.56 | 0 | 0 | 0 | |
| RC: | 0 | 0 | 0 | 0 | 0 | 0 | |
| Helitron | 0 | 0 | 0 | 0 | 0 | 0 | |
| DNA | 1 | 72 | 0.08 | 0 | 0 | 0 | |
| Low_complexity | 166 | 9514 | 10.18 | 47 | 2479 | 17.74 | |
| Simple_repeat | 586 | 25176 | 26.93 | 221 | 8121 | 58.12 | |
| Unspecified | 151 | 49452 | 52.90 | 16 | 3681 | 26.35 | |
| tRNA | 1 | 30 | 0.03 | 0 | 0 | 0 | |
2.4. Gene Structure and Analysis of Conserved Motifs
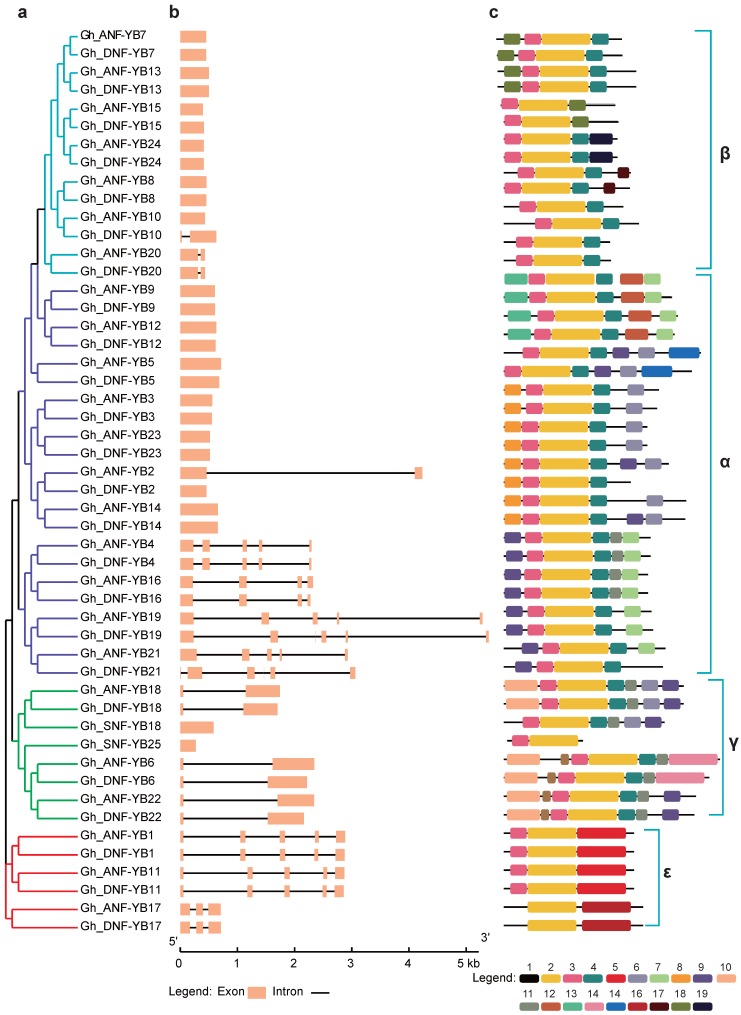
To comprehensively study the phylogenetic relationships between the NF-YB genes, we performed analyses of gene structure and conserved motifs. As shown in Figure 3a, the NF-YB genes were classified into five clades that were consistent with the phylogenetic relationships illustrated in Figure 1. To elucidate the gene structure of the GhNF-YB family, we compared coding sequences to their corresponding genomic sequences to determine positions of the exons and introns position the genomic sequences. As shown in Figure 3b, the numbers of exons ranged from one to six, where genes with one exon accounted for 60% of the total NF-YB genes, most of which were from the α and β clades. In analysing the conserved motifs in the GhNF-Y B genes using MEME, we found that all 50 NF-YB proteins shared motif 2 (yellow box) (Figure 3c), which was contained within the B domain. In addition, most of the NF-YB proteins contained similar motifs. For instance, motifs 3 and 4 were widely distributed. We also found that NF-YB genes with close phylogenetic relationships exhibited similar arrangements of motifs. We also identified the pattern of amino acid residues conservation in the domains of GhNF-YBs (Figure S3).
Figure 3.
Phylogenetic relationships, exon-intron structures, and conserved motifs of NF-YB genes in Gossypium hirsutum. (a) An unrooted tree was constructed in MEGA using the neighbour-joining method, while the four subfamilies are indicated by different colours. (b) The pink boxes and black lines indicate exons and introns, respectively. (c) The distribution of conserved motifs in GhNF-YB family, where motif 2 represents the B domain.
2.5. Analyses of Tissue-Specific Expression Patterns of 50 G. hirsutum NF-YB Genes
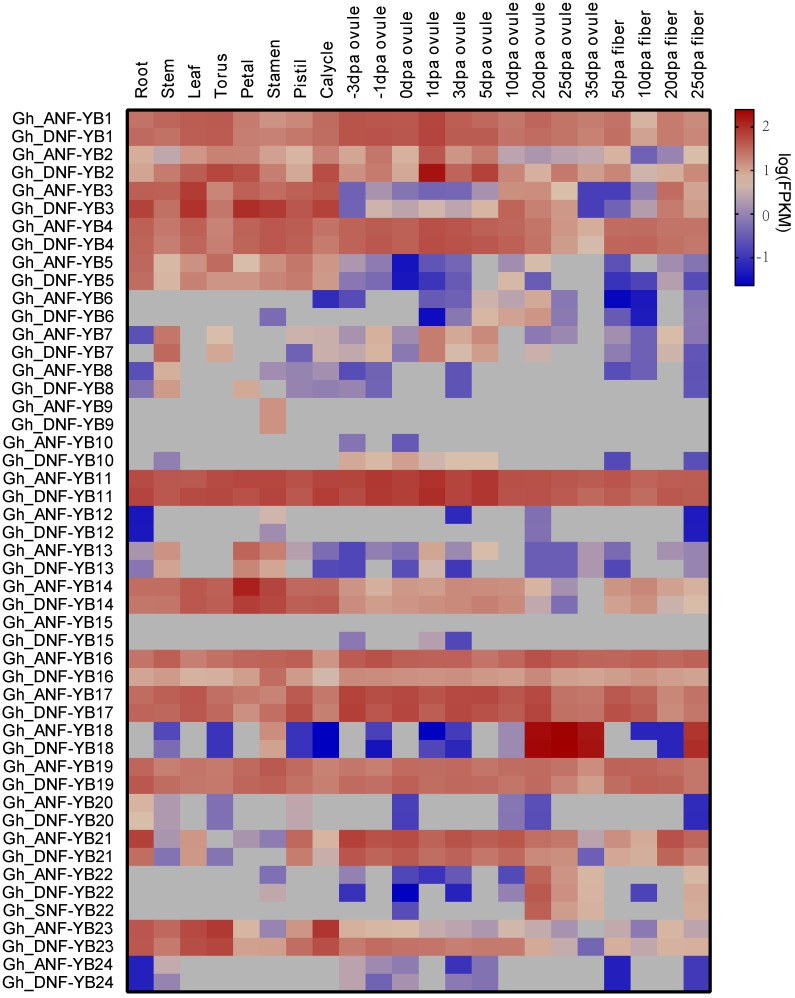
To assess the expression patterns of GhNF-YB genes, RNA-seq data were downloaded from NCBI and analysed. Gene expression patterns of GhNF-YB genes were analysed in a variety of tissues in G. hirsutum, including vegetative tissues (root, stem and leaf), reproductive tissues (some parts of the floral organ), and fibre (5, 10, 20, and 25 d post-anthesis). As shown in Figure 4, we found that some NF-YB genes were widely expressed in all of the aforementioned tissues, indicating that these genes have important biological functions during plant development. For example, GhA/DNF-YB4, GhA/DNF-YB16, and GhA/DNF-YB19 exhibited very high levels of expression in vegetative tissues, reproductive tissues, and fibre. In contrast, other genes exhibited much different expression patterns. Specifically, GhA/DNF-YB9 was expressed in the stamen, while GhA/DNF-YB18 and GhA/DNF-YB22 were preferentially expressed in 20, 25, and 35 days post-anthesis (DPA) ovules and 25 DPA fibres. GhA/DNF-YB1, GhA/DNF-YB11, and GhA/DNF-YB17 not only exhibited phylogenetic relationships (Figure 1 and Figure 3), but also similar expression patterns. An additional investigation revealed that the syntenic duplicates, with the exception of GhA/DNF-YB11/1, were divergent in expression patterns, indicating sub-functionalisation.
Figure 4.
Gene expression patterns of NF-YB genes in a variety of upland cotton tissues. The raw data for RNA-Seq were downloaded from NCBI and analysed using Tophat and Cufflinks [35]. Gene expression levels are depicted with different colour on the scale. Blue and red represent low and high expression, respectively.
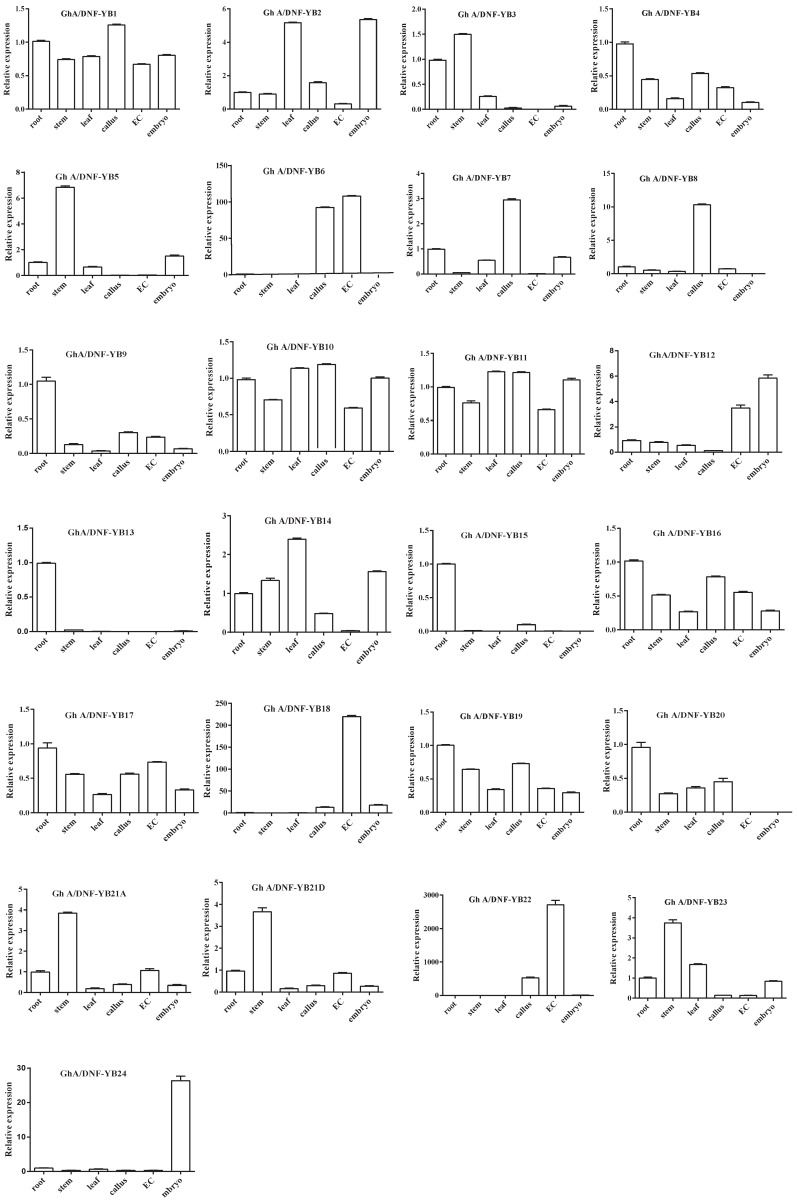
To validate the expression levels of GhNF-YBs, qRT-PCR was used to test gene expression in the root, stem, leaf, callus, embryogenic callus, and embryo. The results of the qRT-PCR were in agreement with expression patterns observed in the analysis of the RNA-seq data (Figure 5). For example, GhA/DNF-YB1, GhA/D NF-YB11, and GhA/DNF-YB17 were expressed in all tissues selected, while GhA/DNF-YB6, GhA/DNF-YB18, and GhA/DNF-YB22 exhibited very high expression levels only in several selected tissues (callus and embryogenic callus). In contrast, GhA/DNF-YB9, GhA/DNF-YB12, GhA/DNF-YB13, and GhA/DNF-YB24 were very lowly expressed in any of the tissues assayed.
Figure 5.
Expression levels of NF-YB genes in different tissues, as determined by qRT-PCR. Error bars represent the standard deviations of three independent experiments.
2.6. Overexpression of GhDNF-YB22 in Cotton Affects Embryogenesis
GhA/DNF-YB6, GhA/DNF-YB18, GhA/DNF-YB22, AtLEC1, and AtNF-YB6 were clustered in the γ clade (Figure 1). In A. thaliana, LEC1 is a main regulator of embryogenesis [36]. To characterize the function of the GhNF-YB gene, GhDNF-YB22, which is highly homologous to AtLEC1, GhDNF-YB22 was transformed into cotton under the control of the CaMV35 promoter. After performing the Agrobacterium-mediated transformation of cotton hypocotyl, hypocotyl somatic cells underwent dedifferentiation and redifferentiation, formed the callus and embryogenic callus, then produced somatic embryo, and lastly developed into new plants. Over the course of these processes, we found that transgenic seedlings exhibited a set of morphological phenotypes. Callus-like structures formed on the leaf-like organ surfaces of seedlings (Figure 6a), while some embryo-like structures developed from the callus-like structures (Figure 6d). Remarkably, some embryo-like structures emerged on the margins of leaf-like organs (Figure 6b), or substituted for growth of leaves (Figure 6c). The transgenic lines of GhDNF-YB22 were determined by kanamycin selection and qRT-PCR test (Figure S4). These resulting morphological phenotypes indicate that GhDNF-YB22 plays an important role in embryogenesis.
Figure 6.
Phenotypes of transgenic cotton seedlings ectopically expressing GhDNF-YB22: (a) seedlings produced a callus-like structure; (b) seedling produced embryo-like organs; (c) embryo-like organs were substituted for leaf growth; and (d) embryo-like structures developed from the callus. Bars: 0.5 mm (a,b); and 0.1 mm (c,d).
3. Discussion
The NF-YB gene family had been previously analysed in several plant species, including A. thaliana, rice, wheat, tung tree, soybean, canola, grape, and tomato. However, a genome-wide identification and characterization of NF-YB genes has not been reported in G. hirsutum, an allotetraploid species. In the present study, we conducted an integrated investigation of the GhNF-YBs, consisting of phylogenetic analyses, an investigation of expression patterns, and transgenic verification.
3.1. Variation in the NF-YB Gene Family in G. hirsutum
In the present study, nearly all of the orthologues from two monocot species (sorghum and rice) and three dicots (cotton, cacao, and Arabidopsis) tended to cluster together, indicating that the main functions of the NF-YB gene family diverged prior to the divergence of dicots and monocots.
The allotetraploid cotton G. hirsutum was derived from the hybridization of an A-genome species resembling G. arboreum and a D-genome species resembling G. raimondii [26], followed by a chromosome doubling event. Because of the whole genome duplication, the upland cotton experienced polyploidisation, which results in an extensive reshuffling of the entire genome [37]. At present, there is much evidence to support the notion that the gain and loss of genes or the expansion or contraction of gene families is common following polyploidisation [38,39]. Thus, the expansion of the GhNF-YB gene family also could be an indication that GhNF-YB genes play roles in additional biological processes or have novel functions, in agreement with the allotetraploid nature of G. hirsutum [40,41,42]. An analysis of collinearity showed that orthologous genes maintained good collinearity between the At and Dt sub-genomes, while segmental duplication analysis showed that nine pairs of genes may be derived from segmental duplication (Figure 2). These results suggest that segmental duplication also played an important role in the expansion of the NF-YB gene family.
In analysing gene structure, we found that many NF-YB genes in G. hirsutum had only one exon with no introns (Figure 3), which is consistent with findings in Arabidopsis and Brassica napus L. [13]. Previous studies have postulated that an intron-rich gene would lose multiple introns simultaneously by retrotransposition, thereby producing intron-less ancestral genes [43]. Thus, some NF-YB genes in G. hirsutum may experience the loss of multiple introns during gene family diversification. Genome-wide analyses have shown that the loss and gain of introns has been extensive during the process of eukaryotic diversification [44,45].
3.2. Expression Patterns of NF-YB Genes in G. hirsutum
Previous studies have reported that NF-YB genes play important roles in plant developmental processes (e.g., in late embryogenesis, flowering time, drought tolerance, etc.) [46]. In the present study, we identified the tissue-specific expression patterns of GhNF-YB genes in a variety of tissues, where the results show that most of the NF-YB genes are expressed ubiquitously, with the exception of a few genes that are expressed in specific tissues (Figure 5). This observation was consistent with previous studies [10], suggesting that NF-YB genes are polyfunctional and are involved in a wide range of biological processes [47].
In phylogenetic analysis, GhNF-YB genes were divided into five clades with several G. hirsutum- and A. thaliana-specific NF-YB genes, with the exception of the δ clade. Of these, NF-YB1, NF-YB2, NF-YB3, NF-YB6, and NF-YB9 have been extensively studied in A. thaliana. Previous studies revealed that NF-YB1 not only regulated drought tolerance [18], but also interacted with CO (CONSTANS) to affect the transcript levels of two key integrators (FT: FLOWERING LOCUS T and SOC1: SUPPRESSOR OF OVEREXPRESSION OF CO1) in the flowering pathway, and therefore adjusted the flowering time [48]. Interestingly, GhA/DNF-YB21 and GhA/DNF-YB19 clustered with AtNF-YB1, where GhA/DNF-YB19 was expressed in all selected tissues, while GhA/DNF-YB21 was mainly expressed in reproductive tissues. These observations indicate that GhA/DNF-YB21 and GhA/DNF-YB19 may have similar functions as AtNF-YB1. Moreover, GhA/DNF-YB2, GhA/DNF-YB3, GhA/DNF-YB14, and GhA/D NF-YB23 were observed to cluster with AtNF-YB2 and AtNF-YB3, which have been reported to regulate the photoperiod-dependent flowering time [20]. In barley, HvNF-YB3 and HvNF-YB1 clustered with AtNF-YB2 and AtNF-YB3, and have been shown to greatly promote early flowering [49]. NF-YB9/LEC1 was the first NF-YB gene identified and studied in A. thaliana, and has been shown to be required for the maintenance embryonic of cell fate, where the ectopic expression of LEC1 can induce somatic embryos from vegetative cells [36]. In addition, LEC1 has also been shown to play an essential role in embryogenesis and seed maturation [6,50]. LEC1 and LEC1-LIKE (NF-YB6) regulated embryo development by activating the expression of genes required for embryogenesis and cellular differentiation [7,36]. In the present study, GhA/DNF-YB6 and GhA/DNF-YB22 were grouped with AtLEC1, while GhA/DNF-YB18 was grouped with AtLEC1-LIKE. Furthermore, GhA/DNF-YB6, GhA/DNF-YB18, and GhA/DNF-YB22 were all highly expressed in the callus and embryogenic callus as evidenced by qRT-PCR. Thus, these three paralogue pairs may be involved in regulating embryonic development.
3.3. Role of GhDNF-YB22 in Embryogenesis
LEC1 has been shown to function in different aspects of embryogenesis, such as embryonic development, the induction of embryogenesis at morphogenesis and maturation phases, the induction of embryonic programs in vegetative cells, and the identification of cotyledons [36,51]. The function of LEC1 is conserved in seed development by regulating distinct genes at different developmental stages in Arabidopsis and soybean [52]. In addition, vegetative or reproductive cells could change their fate and exhibit somatic embryo development via the ectopic expression of LEC [53]. Here, GhDNF-YB22 was ectopically expressed in upland cotton, whereupon callus- and embryo-like structures emerged on the leaf-like organs as a result (Figure 6). This in agreement with 35S/LEC1 seedlings, which produced multiple embryo-like structures on the leaves of Arabidopsis [36]. This indicates that GhDNF-YB22 is functionally similar to LEC1, which promotes the transcription of genes required for embryo morphogenesis. Furthermore, GhA/DNF-YB6, GhA/DNF-YB18 and GhA/DNF-YB22 in γ clade have been revealed conservative exon-intron structures and expression patterns (Figure 3 and Figure 4). These indicate that NF-YB genes in γ clade may have similar biological function in embryogenesis.
4. Materials and Methods
4.1. Identification of the NF-YB Gene Family
The protein sequences of NF-YB in A. thaliana (http://www.arabidopsis.org) were used as queries to search the sequences of G. arboretum, G. raimondii, G. hirsutum, rice, sorghum, and cacao in blastp. Cotton sequences—including G. arboretum, G. raimondii, and G. hirsutum—were downloaded from COTTONGEN (http://www.cottongen.org), while the other aforementioned species here were obtained from phytozome (https://phytozome.jpi.doe.gov/pz/portal.html). In addition, InterProScan 56.0 (http://www.ebi.ac.uk/inerpro/) was used to identify the NF-YB gene family numbers.
4.2. Phylogenetic Analyses
NF-YB proteins from seven plant species (A. thaliana, O. sativa, G. arboreum, G. raimondii, G. hirsutum, T. cacao, and S. bicolor) were used in a multiple alignment in CLUSTAL-X [54]. Subsequently, a phylogenetic tree based on NF-YB protein sequences was constructed via the neighbour-joining method using MEGA 7.0 (http://www.megasoftware.net/) [55]. To establish the reliability of the phylogenetic analysis, the p-distance method with 1000 bootstrap samples was used with pairwise deletion and a Poisson correction.
4.3. Chromosome Locations and Collinearity Analyses
The loci of NF-YB genes were obtained from the genome annotation data. Mapchart was applied to map the chromosome locations [30]. The basic local alignment search tool (BLAST) [56] was used to retrieve the GhNF-YB protein sequences from a local database. Next, these sequences were analysed to identify the collinearity blocks against the whole genome using MCSCAN (http://chibba.agtec.uga.edu/duplication/mcscan/) [30], while CIRCOS software (http://circos.ca/) was used to draw the collinearity map [57].
4.4. Estimating Ka/Ks Rates
Using Clustal X 2.0 (ftp://ftp.ebi.ac.uk/pub/software/clustalw2/) [54], the amino acid sequences from duplicated pairs were aligned and the aligned sequences converted to cDNA using PAL2NAL (http://www.bork.embl.de/pal2nal/). Lastly, the synonymous (Ks) and nonsynonymous (Ka) substitution rates were estimated using the CODEML program of PAML (http://abacus.gene.ucl.ac.uk/software/paml.html) [58].
4.5. Analysis of Transposable Elements
To study the function of transposable elements (TEs) in the NF-YB family, we identified and analysed the different types of TEs in the 2000 and 10,000 bp upstream and downstream regions of the gene. PILER-DF, RepeatModeler, and LTR_FINDER [59,60] were used to predict TEs. Using RepbaseTE (http://www.girinst.org/repbase/), the TEs were identified at the DNA level with RepeatMasker (http://repeatmasker.org/).
4.6. Gene Structure and Conserved Motifs Analysis
The Gene Structure Display Server (GSDS) (http://gsds.cbi.pku.edu.cn/) was employed to analyse the exon-intron structure of GhNF-YB genes using cDNAs and corresponding genomic sequences. The online program Multiple Em for Motif Elicitation (MEME) (http://meme-suite.org/tools/meme) was chosen to identify the conserve motifs in all GhNF-YB proteins according to the following parameters: the optimum width of motifs ranged from 6 to 200 aa, and the maximum number of motifs to find was defined at 20. The annotations of the identified motifs were completed by the program of InterProScan 56.0 (http://www.ebi.ac.uk/interpro/).
4.7. Gene Expression Heat Map
To measure the expression levels of NF-YB family genes, raw data from the RNA-sequencing of various tissues (i.e., root, stem, leaf, torus, petal, stamen, pistil, calycle, ovule and fibre) in G. hirsutum cultivar TM-1 was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA248163/). Then the data were normalized to calculate the expression levels. Subsequently, Genesis software (http://www.gsoft.com.au/) was used to draw the heat map [61].
4.8. RNA Isolation and qRT-PCR Verification
The seeds of G. hirsutum cultivar CCRI24 were grown in a field in Anyang, China. Root, stem, and leaf tissue were sampled and frozen in liquid nitrogen, and subsequently stored at −80 °C. In addition, the seeds of CCRI24 were rinsed with 70% ethanol for 1 min, washed three times with sterile distilled water, and soaked for 24 h in 30% H2O2. The sterilized seeds were germinated on MS medium (PH: 5.8–6.0) for 7 days, and the hypocotyls of aseptic seedlings were cut into approximately 5 mm sections and used as explants. The explants were cultured using different media for the callus, embryogenic callus, and somatic embryos according to previously published methods [62]. The callus, embryogenic callus, and somatic embryos were sampled and frozen at −80% until RNA extraction. Total RNA was extracted from prepared samples using the RNAprep Pure Plant Kit (Tiangen, Beijing, China). The PrimeScript® RT reagent kit (Takara, Dalian, China) was used to synthesize the first strand cDNA using approximately 2 μg of RNA. Gene-specific primers for qRT-PCR were designed using DNAMAN 7.0 (Table S2). The histone 3 gene in G. hirsutum (GenBank accession no.AF024716) was used as an internal control [63,64]. PCR amplifications were performed using SYBR Premix Ex Taq (Takara), according to previously published methods [65]. For each analysis, qRT-PCR assays had three biological replicates, each consisting of three technical replicates. Error bars were standard error of three technical replications. The relative expression levels of GhNF-YB genes were calculated by the 2−ΔΔCt method [66].
4.9. Gene Cloning and Transformation into Cotton
The mixed cDNA of root, stem, leaf, callus, and embryogenic callus tissues from CCRI24 was synthesized as a template to amplify genes based on gene-specific primers. The complete protein-coding region was cloned into the pCAMBIA2301 vector with the cauliflower mosaic virus 35S (CaMV35) promoter, and the constructed vector was transferred into Agrobacterium tumefaciens strain LBA4404 in the subsequent step. Finally, Hypocotyl explants from CCRI24 were transformed using A. tumefaciens-mediated transformation according to previously published methods [67,68].
5. Conclusions
Although the function of some NF-YB genes has been demonstrated clearly in several plant species, especially in Arabidopsis, their roles in G. hirsutum are still elusive. In the current study, we performed a genome-wide analysis of the NF-YB gene family in G. hirsutum, including investigated the evolutionary relationships, gene structure and expression patterns. Fifty NF-YB genes are identified, and whole genome and segmental duplication might be the major ways for the expansion of the NF-YB family in upland cotton. Furthermore, the duplicated genes showed different expression patterns, indicating that the duplicated genes probably have experienced functional divergence. Our results will provide a foundation for further study of NF-YB gene family in upland cotton.
Acknowledgments
We sincerely grateful to Fuguang Li (Cotton Research Institute) for his valuable support, advice and suggestions during the course of this research. To the entire research team, friends and any other person who contributed, we are greatly indebted to you. This research was funded by the National Key R&D Program of China (2016YFD0100505).
Supplementary Materials
Supplementary Materials can be found at http://www.mdpi.com/1422-0067/19/2/483/s1.
Author Contributions
Yanli Chen, Chaojun Zhang, Xianlong Zhang and Fuguang Li conceived and designed the experiments. Yanli Chen, Zhaoen Yang, Yanqing Xiao and Peng Wang performed the experiments and analysed the data. Yanli Chen drafted the manuscript. Yanli Chen, Zhaoen Yang, Ye Wang and Xiaoyang Ge revised the manuscript. All authors have read and approved the final manuscript.
Conflicts of Interest
The authors declare that they have no competing interests.
References
- 1.Dorn A., Bollekens J., Staub A., Benoist C., Mathis D. A multiplicity of CCAAT box-binding proteins. Cell. 1987;50:863–872. doi: 10.1016/0092-8674(87)90513-7. [DOI] [PubMed] [Google Scholar]
- 2.Romier C., Cocchiarella F., Mantovani R., Moras D. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003;278:1336–1345. doi: 10.1074/jbc.M209635200. [DOI] [PubMed] [Google Scholar]
- 3.Gusmaroli G., Tonelli C., Mantovani R. Regulation of the CCAAT-Binding NF-Y subunits in Arabidopsis thaliana. Gene. 2001;264:173–185. doi: 10.1016/S0378-1119(01)00323-7. [DOI] [PubMed] [Google Scholar]
- 4.Yang S., Wang Y., Yin H., Guo H., Gao M., Zhu H., Chen Y. Identification and characterization of NF-YB family genes in tung tree. Mol. Genet. Genom. 2015;290:2187–2198. doi: 10.1007/s00438-015-1073-z. [DOI] [PubMed] [Google Scholar]
- 5.Sinha S., Kim I.S., Sohn K.Y., DeCrombrugghe B., Maity S.N. Three classes of mutations in the A subunit of the CCAAT-Binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol. Cell. Biol. 1996;16:328–337. doi: 10.1128/MCB.16.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee H.S., Fischer R.L., Goldberg R.B., Harada J.J. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. USA. 2003;100:2152–2156. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwong R.W., Bui A.Q., Lee H., Kwong L.W., Fischer R.L., Goldberg R.B., Harada J.J. LEAFY COTYLEDON1-like defines a class of regulators essential for embryo development. Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edwards D., Murray J.A.H., Smith A.G. Multiple genes encoding the conserved CCAAT-box transcription factor complex are expressed in Arabidopsis. Plant Physiol. 1998;117:1015–1022. doi: 10.1104/pp.117.3.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siefers N., Dang K.K., Kumimoto R.W., Bynum W.E., Tayrose G., Holt B.F., III Tissue-specific expression patterns of arabidopsis NF-Y transcription factors suggest potential for extensive combinatorial complexity. Plant Physiol. 2009;149:625–641. doi: 10.1104/pp.108.130591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thirumurugan T., Ito Y., Kubo T., Serizawa A., Kurata N. Identification, characterization and interaction of hap family genes in rice. Mol. Genet. Genom. 2008;279:279–289. doi: 10.1007/s00438-007-0312-3. [DOI] [PubMed] [Google Scholar]
- 11.Stephenson T.J., McIntyre C.L., Collet C., Xue G.-P. Genome-wide identification and expression analysis of the NF-Y family of transcription factors in triticum aestivum. Plant Mol. Biol. 2007;65:77–92. doi: 10.1007/s11103-007-9200-9. [DOI] [PubMed] [Google Scholar]
- 12.Li S., Li K., Ju Z., Cao D.Y., Fu D.Q., Zhu H.L., Zhu B.Z., Luo Y.B. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genom. 2016;17:36. doi: 10.1186/s12864-015-2334-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang M., Yin X., Lin Z., Zheng Q., Liu G., Zhao G. Identification and characterization of NF-Y transcription factor families in canola (Brassica napus L.) Planta. 2014;239:107–126. doi: 10.1007/s00425-013-1964-3. [DOI] [PubMed] [Google Scholar]
- 14.Quach T., Nguyen H., Valliyodan B., Joshi T., Xu D., Nguyen H. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol. Genet. Genom. 2015;290:1095–1115. doi: 10.1007/s00438-014-0978-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren C., Zhang Z., Wang Y., Li S.H., Liang Z.C. Genome-wide identification and characterization of the NF-Y gene family in grape (Vitis vinifera L.) BMC Genom. 2016;17:605. doi: 10.1186/s12864-016-2989-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mu J.Y., Tan H.L., Zheng Q., Fu F.Y., Liang Y., Zhang J.A., Yang X.H., Wang T., Chong K., Wang X.J., et al. LEAFY COTYLEDON1 is a key regulator of fatty acid biosynthesis in arabidopsis. Plant Physiol. 2008;148:1042–1054. doi: 10.1104/pp.108.126342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rider S.D., Henderson J.T., Jerome R.E., Edenberg H.J., Romero-Severson J., Ogas J. Coordinate repression of regulators of embryonic identity by pickle during germination in arabidopsis. Plant J. 2003;35:33–43. doi: 10.1046/j.1365-313X.2003.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson D.E., Repetti P.P., Adams T.R., Creelman R.A., Wu J., Warner D.C., Anstrom D.C., Bensen R.J., Castiglioni P.P., Donnarummo M.G., et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA. 2007;104:16450–16455. doi: 10.1073/pnas.0707193104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warpeha K.M., Upadhyay S., Yeh J., Adamiak J., Hawkins S.I., Lapik Y.R., Anderson M.B., Kaufman L.S. The GCR1, GPA1, PRN1, Nf-Y signal chain mediates both blue light and abscisic acid responses in arabidopsis. Plant Physiol. 2007;143:1590–1600. doi: 10.1104/pp.106.089904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumimoto R.W., Adam L., Hymus G.J., Repetti P.P., Reuber T.L., Marion C.M., Hempel F.D., Ratcliffe O.J. The nuclear factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in arabidopsis. Planta. 2008;228:709–723. doi: 10.1007/s00425-008-0773-6. [DOI] [PubMed] [Google Scholar]
- 21.Ballif J., Endo S., Kotani M., MacAdam J., Wu Y. Over-expression of HAP3b enhances primary root elongation in Arabidopsis. Plant Physiol. Biochem. 2011;49:579–583. doi: 10.1016/j.plaphy.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Shen B., Allen W.B., Zheng P., Li C., Glassman K., Ranch J., Nubel D., Tarczynski M.C. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010;153:980–987. doi: 10.1104/pp.110.157537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan H., Yang X., Zhang F., Zheng X., Qu C., Mu J., Fu F., Li J., Guan R., Zhang H., et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011;156:1577–1588. doi: 10.1104/pp.111.175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miyoshi K., Ito Y., Serizawa A., Kurata N. OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 2003;36:532–540. doi: 10.1046/j.1365-313X.2003.01897.x. [DOI] [PubMed] [Google Scholar]
- 25.Yadav D., Shavrukov Y., Bazanova N., Chirkova L., Borisjuk N., Kovalchuk N., Ismagul A., Parent B., Langridge P., Hrmova M., et al. Constitutive overexpression of the TANF-YB4 gene in transgenic wheat significantly improves grain yield. J. Exp. Bot. 2015;66:6635–6650. doi: 10.1093/jxb/erv370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K., Wang Z., Li F., Ye W., Wang J., Song G., Yue Z., Cong L., Shang H., Zhu S., et al. The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 2012;44:1098. doi: 10.1038/ng.2371. [DOI] [PubMed] [Google Scholar]
- 27.Li F., Fan G., Wang K., Sun F., Yuan Y., Song G., Li Q., Ma Z., Lu C., Zou C., et al. Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 2014;46:567–572. doi: 10.1038/ng.2987. [DOI] [PubMed] [Google Scholar]
- 28.Zhang T., Hu Y., Jiang W., Fang L., Guan X., Chen J., Zhang J., Saski C.A., Scheffler B.E., Stelly D.M., et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015;33:531. doi: 10.1038/nbt.3207. [DOI] [PubMed] [Google Scholar]
- 29.Li F., Fan G., Lu C., Xiao G., Zou C., Kohel R.J., Ma Z., Shang H., Ma X., Wu J., et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015;33:524. doi: 10.1038/nbt.3208. [DOI] [PubMed] [Google Scholar]
- 30.Yang Z., Gong Q., Qin W., Yang Z., Cheng Y., Lu L., Ge X., Zhang C., Wu Z., Li F. Genome-wide analysis of wox genes in upland cotton and their expression pattern under different stresses. BMC Plant Biol. 2017;17:113. doi: 10.1186/s12870-017-1065-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu G., Guo C., Shan H., Kong H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA. 2012;109:1187–1192. doi: 10.1073/pnas.1109047109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynch M., Conery J.S. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 33.Parisod C., Alix K., Just J., Petit M., Sarilar V., Mhiri C., Ainouche M., Chalhoub B., Grandbastien M.-A. Impact of transposable elements on the organization and function of allopolyploid genomes. New Phytol. 2010;186:37–45. doi: 10.1111/j.1469-8137.2009.03096.x. [DOI] [PubMed] [Google Scholar]
- 34.Grandbastien M., Audeon C., Bonnivard E., Casacuberta J.M., Chalhoub B., Costa A.P.P., Le Q.H., Melayah D., Petit M., Poncet C., et al. Stress activation and genomic impact of Tnt1 retrotransposons in solanaceae. Cytogenet. Genome Res. 2005;110:229–241. doi: 10.1159/000084957. [DOI] [PubMed] [Google Scholar]
- 35.Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with tophat and cufflinks. Nat. Protoc. 2012;7:562. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lotan T., Ohto M., Yee K.M., West M.A.L., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in Vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/S0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- 37.Paterson A.H., Bowers J.E., Chapman B.A. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl. Acad. Sci. USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van de Peer Y., Fawcett J.A., Proost S., Sterck L., Vandepoele K. The flowering world: A tale of duplications. Trends Plant Sci. 2009;14:680–688. doi: 10.1016/j.tplants.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 39.Jiao Y., Wickett N.J., Ayyampalayam S., Chanderbali A.S., Landherr L., Ralph P.E., Tomsho L.P., Hu Y., Liang H., Soltis P.S., et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011;473:97–100. doi: 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- 40.Scarpino S.V., Levin D.A., Meyers L.A. Polyploid formation shapes flowering plant diversity. Am. Nat. 2014;184:456–465. doi: 10.1086/677752. [DOI] [PubMed] [Google Scholar]
- 41.Soltis D.E., Burleigh J.G. Surviving the K-T mass extinction: New perspectives of polyploidization in angiosperms. Proc. Natl. Acad. Sci. USA. 2009;106:5455–5456. doi: 10.1073/pnas.0901994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang X., Guo H., Wang J., Lei T., Liu T., Wang Z., Li Y., Lee T.-H., Li J., Tang H., et al. Comparative genomic de-convolution of the cotton genome revealed a decaploid ancestor and widespread chromosomal fractionation. New Phytol. 2016;209:1252–1263. doi: 10.1111/nph.13689. [DOI] [PubMed] [Google Scholar]
- 43.Xie Z.Y., Li X., Glover B.J., Bai S.N., Rao G.Y., Luo J.C., Yang J. Duplication and functional diversification of HAP3 genes leading to the origin of the seed-developmental regulatory gene, LEAFY COTYLEDON1 (LEC1), in nonseed plant genomes. Mol. Biol. Evol. 2008;25:1581–1592. doi: 10.1093/molbev/msn105. [DOI] [PubMed] [Google Scholar]
- 44.Rogozin I.B., Wolf Y.I., Sorokin A.V., Mirkin B.G., Koonin E.V. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 2003;13:1512–1517. doi: 10.1016/S0960-9822(03)00558-X. [DOI] [PubMed] [Google Scholar]
- 45.Roy S.W., Penny D. Patterns of intron loss and gain in plants: Intron loss-dominated evolution and genome-wide comparison of O-sativa and A-thaliana. Mol. Biol. Evol. 2007;24:171–181. doi: 10.1093/molbev/msl159. [DOI] [PubMed] [Google Scholar]
- 46.Zhao H., Wu D., Kong F., Lin K., Zhang H., Li G. The arabidopsis thaliana nuclear factor Y transcription factors. Front. Plant Sci. 2017;8:2045. doi: 10.3389/fpls.2016.02045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmermann P., Hirsch-Hoffmann M., Hennig L., Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cao S., Kumimoto R.W., Gnesutta N., Calogero A.M., Mantovani R., Holt B.F. A distal ccaat nuclear factor Y complex promotes chromatin looping at the flowering locus T promoter and regulates the timing of flowering in arabidopsis. Plant Cell. 2014;26:1009–1017. doi: 10.1105/tpc.113.120352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang M., Hole D., Wu J., Blake T., Wu Y. Expression and functional analysis of nuclear factor-Y, subunit B genes in barley. Planta. 2012;235:779–791. doi: 10.1007/s00425-011-1539-0. [DOI] [PubMed] [Google Scholar]
- 50.Huang M., Hu Y., Liu X., Li Y., Hou X. Arabidopsis LEAFY COTYLEDON1 controls cell fate determination during post-embryonic development. Front. Plant Sci. 2015;6:955. doi: 10.3389/fpls.2015.00955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meinke D.W. A homoeotic mutant of arabidopsis thaliana with LEAFY COTYLEDONS. Science. 1992;258:1647–1650. doi: 10.1126/science.258.5088.1647. [DOI] [PubMed] [Google Scholar]
- 52.Pelletier J.M., Kwong R.W., Park S., Le B.H., Baden R., Cagliari A., Hashimoto M., Munoz M.D., Fischer R.L., Goldberg R.B., et al. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. USA. 2017;114:E6710–E6719. doi: 10.1073/pnas.1707957114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braybrook S.A., Harada J.J. Lecs go crazy in embryo development. Trends Plant Sci. 2008;13:624–630. doi: 10.1016/j.tplants.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 54.Kakar K.U., Nawaz Z., Kakar K., Ali E., Almoneafy A.A., Ullah R., Ren X.-L., Shu Q.-Y. Comprehensive genomic analysis of the CNGC gene family in brassica oleracea: Novel insights into synteny, structures, and transcript profiles. BMC Genom. 2017;18:869. doi: 10.1186/s12864-017-4244-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumar S., Stecher G., Tamura K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGinnis S., Madden T.L. Blast: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32:W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krzywinski M., Schein J., Birol İ., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Z. Paml 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 59.Xu Z., Wang H. LTR_FINDER: An efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:W265–W268. doi: 10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar R.C., Myers E.W. PILER: Identification and classification of genomic repeats. Bioinformatics. 2005;21:i152–i158. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- 61.Sturn A., Quackenbush J., Trajanoski Z. Genesis: Cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 62.Zhang C., Yu S., Fan S., Zhang J., Li F. Inheritance of somatic embryogenesis using leaf petioles as explants in upland cotton. Euphytica. 2011;181:55–63. doi: 10.1007/s10681-011-0380-7. [DOI] [Google Scholar]
- 63.Yang Z., Zhang C., Yang X., Liu K., Wu Z., Zhang X., Zheng W., Xun Q., Liu C., Lu L., et al. PAG1, a cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 2014;203:437–448. doi: 10.1111/nph.12824. [DOI] [PubMed] [Google Scholar]
- 64.Ren Z., Yu D., Yang Z., Li C., Qanmber G., Li Y., Li J., Liu Z., Lu L., Wang L., et al. Genome-wide identification of the MIKC-type MADS-box gene family in Gossypium hirsutum L. Unravels their roles in flowering. Front. Plant Sci. 2017;8:384. doi: 10.3389/fpls.2017.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Z., Ge X., Yang Z., Zhang C., Zhao G., Chen E., Liu J., Zhang X., Li F. Genome-wide identification and characterization of SnRK2 gene family in cotton (Gossypium hirsutum L.) BMC Genet. 2017;18:54. doi: 10.1186/s12863-017-0517-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 67.Yang Z., Li C., Wang Y., Zhang C., Wu Z., Zhang X., Liu C., Li F. Ghagl15s, preferentially expressed during somatic embryogenesis, promote embryogenic callus formation in cotton (Gossypium hirsutum L.) Mol. Genet. Genom. 2014;289:873–883. doi: 10.1007/s00438-014-0856-y. [DOI] [PubMed] [Google Scholar]
- 68.Shang H.-H., Liu C.-L., Zhang C.-J., Li F.-L., Hong W.-D., Li F.-G. Histological and ultrastructural observation reveals significant cellular differences between agrobacterium transformed embryogenic and non-embryogenic calli of cotton. J. Integr. Plant Biol. 2009;51:456–465. doi: 10.1111/j.1744-7909.2009.00824.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.