Abstract
Grass carp is an important aquaculture fish species in China that is affected by severe diseases, especially haemorrhagic disease caused by grass carp reovirus (GCRV). However, the mechanisms of GCRV invasion and infection remain to be elucidated. In the present study, Ctenopharyngodon idellus kidney (CIK) cells were infected with GCRV, harvested at 0, 8, 24, and 72 h post infection, respectively, and then subjected to transcriptomics sequencing. Each sample yielded more than 6 Gb of clean data and 40 million clean reads. To better understand GCRV infection, the process was divided into three phases: the early (0–8 h post infection), middle (8–24 h post infection), and late (24–72 h) stages of infection. A total of 76 (35 up-regulated, 41 down-regulated), 553 (463 up-regulated, 90 down-regulated), and 284 (150 up-regulated, 134 down-regulated) differently expressed genes (DEGs) were identified during the early, middle, and late stages of infection, respectively. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis revealed that DEGs were mainly involved in carbohydrate biosynthesis, transport, and endocytosis in the early stage, phagocytosis and lysosome pathways were mainly enriched in the middle stage, and programmed cell death, apoptosis, and inflammation were largely associated with the late stage. These results suggest GCRV infection is a gradual process involving adsorption on the cell surface, followed by endocytosis into cells, transport by lysosomes, and eventually resulted in cell necrosis and/or apoptosis. Our findings provide insight into the mechanisms of grass carp reovirus infection.
Keywords: grass carp, grass carp reovirus, transcriptomics sequencing, phagosome
1. Introduction
The grass carp (Ctenopharyngodon idellus) has been reared in China for more than 60 years as an important aquaculture species. The production of grass carp reached 5.8 million tons in 2015, accounting for more than 13% of the world’s freshwater aquaculture production [1]. Grass carp haemorrhage disease, caused by grass carp reovirus (GCRV), is one of the most damaging diseases, resulting in huge economic losses to the aquaculture industry of grass carp [2]. GCRV was first isolated in China and belongs to the genus Aquareovirus of the family Reoviridae [3]. GCRV not only infects grass carp but also infects the rare minnow (Gobiocypris rarus), black carp (Mylopharyngodon piceus) and topmouth gudgeon (Pseudorasbora parva), causing haemorrhagic symptoms and death in these species.
Grass carp haemorrhage disease outbreaks are frequent and result in huge economic losses in the aquaculture industry, hence the GCRV is of particular interest to geneticists aiming to develop strategies for breeding disease-resistant fish. Recent studies on GCRV have mainly focused on identification of genes involved in immune responses to GCRV infection [4,5], cloning and characterization of GCRV-encoded proteins [6,7], the development of vaccines against GCRV [8,9], and transcriptomic sequencing of discovered immune-related genes or uncovered the antiviral immunity mechanism of grass carp infected with GCRV [10,11,12,13,14]. However, the mechanisms of grass carp reovirus invasion and infection remain poorly explored.
In the present study, C. idellus kidney (CIK) cells were infected with GCRV, harvested at 0, 8, 24, and 72 h post infection, respectively, and subjected to transcriptomics sequencing. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed on differentially expressed genes (DEGs) identified during different stages of infection. Our findings expand our understanding of the mechanisms underlying grass carp reovirus infection.
2. Results
2.1. GCRV Infection Induces Cytopathic Effects
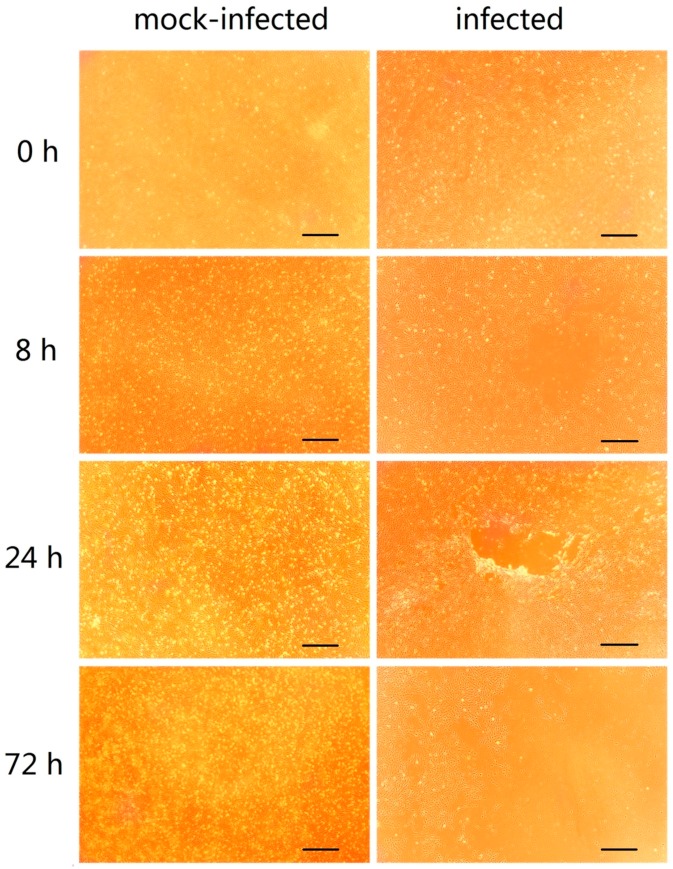
As shown in Figure 1, no cytopathic effect (CPE) was observed in mock-infected cells. However, in cells infected with GCRV, a CPE was observed as early as 8 h post infection, and it became more pronounced with over time. At 72 h post infection, it could be seen that most cells were necrotic.
Figure 1.
(C. idellus kidney) CIK cells mock-infected and infected with grass carp reovirus (GCRV). The images were adjusted for contrast to improve their appearance, scale bar represents 200 μm.
2.2. Cell Counting Kit-8 Assay
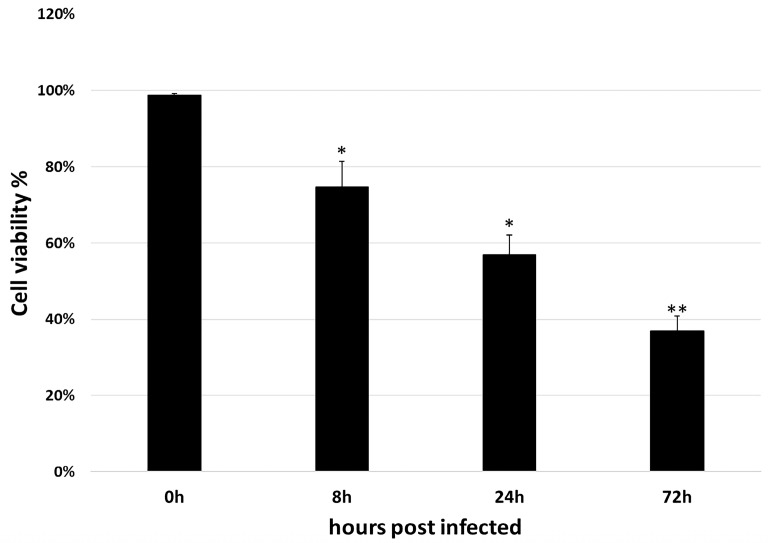
As shown in Figure 2, the mock-infected cells showed the viability near 100%. However, for cells that infected for 8, 24, and 72 h, only 74.66%, 56.88%, and 36.95% of cell viability were observed. Therefore, the result suggested that GCRV infection is efficiency.
Figure 2.
Cell viability assay after infected with GCRV. After 0, 8, 24 and 72 h post infection, cells were evaluated using CCK-8 assay, respectively. The mean of three replicates was shown with the ±standard deviation (S.D.). Significant difference between the control and treated group was indicated with asterisks (*: p < 0.05; **: p < 0.01)
2.3. Preliminary Analysis of Transcriptomics Sequencing Data
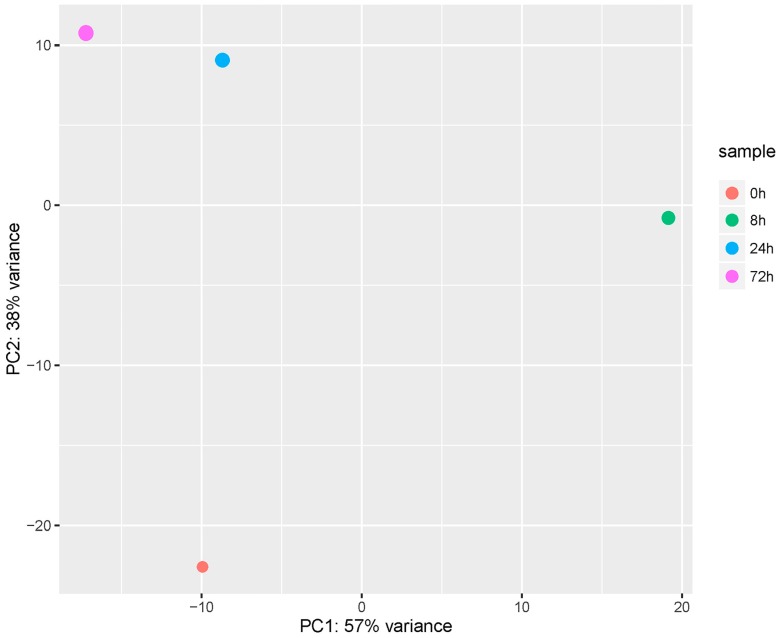
As shown in Table 1, the raw reads, clean reads, clean bases, Q20, Q30, and mapped percentage were recorded for each library. For all libraries, Q20 ≥ 96%, Q30 ≥ 92% and mapped percentage ≥87%. These results showed the high quality of the sequencing data and ensured suitability for further analysis. All sequencing data have been uploaded to the Sequence Read Archive (SRA) of the National Center for Biotechnology Information (NCBI; accession number SRP111072). Moreover, principal component analysis (PCA) was performed on each sample according to the expression level. As shown in Figure 3, mock-infected cells were clearly distinct from cells infected with GCRV. For samples infected with GCRV, correlation values were proportional to the time post-infection; samples from 8 h did not cluster with those from 24 and 72 h. Therefore, the results suggested that infection efficiency and differences existed in each sample post infection.
Table 1.
Summary of sequencing data obtained in this study.
| Sample | Raw Reads | Clean Reads | Clean Data (bp) | Q20 (%) | Q30 (%) | Mapped (%) |
|---|---|---|---|---|---|---|
| C0 | 47,178,538 | 46,732,668 | 6,905,828,036 | 97.24 | 93.27 | 87.96 |
| C8 | 45,666,538 | 45,217,760 | 6,679,552,386 | 96.98 | 92.7 | 87.24 |
| C24 | 41,778,528 | 41,398,026 | 6,111,817,194 | 97.21 | 93.15 | 88.07 |
| C72 | 41,012,162 | 40,628,112 | 6,014,381,958 | 97.15 | 93.07 | 87.40 |
Figure 3.
Principal Component Analysis. The PC1 indicates the differences among infected samples, whereas PC2 shows differences between control and infected samples.
2.4. Identification of Differently Expressed Genes (DEGs)
To better understand GCRV infection, the process was divided into three phases: early (0–8 h post infection), middle (8–24 h post infection), and late (24–72 h) stages. In each phase, data from the latter time point were compared with the former (8 vs. 0, 24 vs. 8, and 72 vs. 24) to identify differentially expressed genes (DEGs). As shown in Table 2, a total of 76 (35 up-regulated, 41 down-regulated), 553 (463 up-regulated, 90 down-regulated), and 284 (150 up-regulated, 134 down-regulated) DEGs were identified during early, middle, and late stages of infection, respectively. Detailed information related to these DEGs is shown in Table S1. DEGs that could not be functionally annotated are listed as ‘unknown’.
Table 2.
Summary of identified DEGs.
| Case | Control | Up-Regulated Genes | Down-Regulated Genes | Total DEGs |
|---|---|---|---|---|
| C8 | C0 | 35 | 41 | 76 |
| C24 | C8 | 463 | 90 | 553 |
| C72 | C24 | 150 | 134 | 284 |
2.5. GO and KEGG Enrichment Analysis
Go enrichment analysis was performed to investigate the putative roles of DEGs. The early stage of infection was mainly related to iron transport, carbohydrate metabolism, and inclusion bodies. For the middle stage of infection, iron ion binding, lysosome categories were significantly enriched. Meanwhile, the late stage was mainly associated with metabolic processes such as lipid biosynthetic, steroid metabolic, and alcohol biosynthetic metabolic. The top five enriched GO terms in each of the three stages are listed in Table 3, and details of the GO terms are included in Table S2.
Table 3.
Results of GO enrichment during the three stages following infection.
| Time | Category | GO_Term | Gene Num. |
|---|---|---|---|
| 0−8 h | Biological_process | Iron ion homeostasis | 3 |
| Cellular_component | Inclusion body | 2 | |
| molecular_function | FAD binding | 2 | |
| biological_process | Transition metal ion homeostasis | 3 | |
| Biological_process | Carbohydrate biosynthetic process | 3 | |
| 8−24 h | Cellular_component | Vacuole | 16 |
| Molecular_function | Iron ion binding | 13 | |
| Biological_process | Organic acid biosynthetic process | 12 | |
| Molecular_function | Oxidoreductase activity | 13 | |
| Cellular_component | Lysosome | 10 | |
| 24−72 h | Biological_process | Lipid biosynthetic process | 19 |
| Biological_process | Steroid metabolic process | 11 | |
| Biological_process | Alcohol biosynthetic metabolic process | 8 | |
| Biological_process | Isoprenoid biosynthetic process | 6 | |
| Biological_process | Steroid metabolic process | 8 |
KEGG enrichment analysis was also performed for DEGs identified in the three stages. During the early stage of infection, DEGs were mainly involved in pathways related to focal adhesion, ECM (extracellular matrix)–receptor interactions, cytokine–cytokine receptor interactions. In the middle stage, pathways related to virus entry into cells and cell recognition were enriched, including the phagosome, lysosome, and hippo signaling pathways. In the late stage of infection, the most enriched pathways were the biosynthesis of steroids and the terpenoid backbone, and degradation of ketone bodies. The top five enriched KEGG terms in each of the three stages are listed in Table 4, and details of the KEGG terms are included in Table S3.
Table 4.
KEGG enrichment results of the three stages following infection.
| Time | Pathway | DEGs | p-Value |
|---|---|---|---|
| 0−8 h | ECM-receptor interaction | 3 | 0.002216 |
| Focal adhesion | 4 | 0.005585 | |
| Mineral absorption | 2 | 0.008391 | |
| Glutathione metabolism | 2 | 0.012446 | |
| Cytokine-cytokine receptor interaction | 3 | 0.01791 | |
| 8−24 h | Collecting duct acid secretion | 4 | 0.0017689 |
| Lysosome | 10 | 0.0019208 | |
| Phagosome | 10 | 0.0023994 | |
| Hippo signaling pathway | 11 | 0.0052756 | |
| Sphingolipid metabolism | 5 | 0.0138641 | |
| 24−72 h | Steroid biosynthesis | 11 | 1.58 × 10−16 |
| Terpenoid backbone biosynthesis | 7 | 7.96 × 10−9 | |
| Metabolism of xenobiotics by cytochrome P450 | 5 | 0.000122 | |
| Sesquiterpenoid and triterpenoid biosynthesis | 2 | 0.000187 | |
| Carbon fixation pathways in prokaryotes | 3 | 0.001499 |
2.6. Expression of DEGs in Key KEGG Pathways
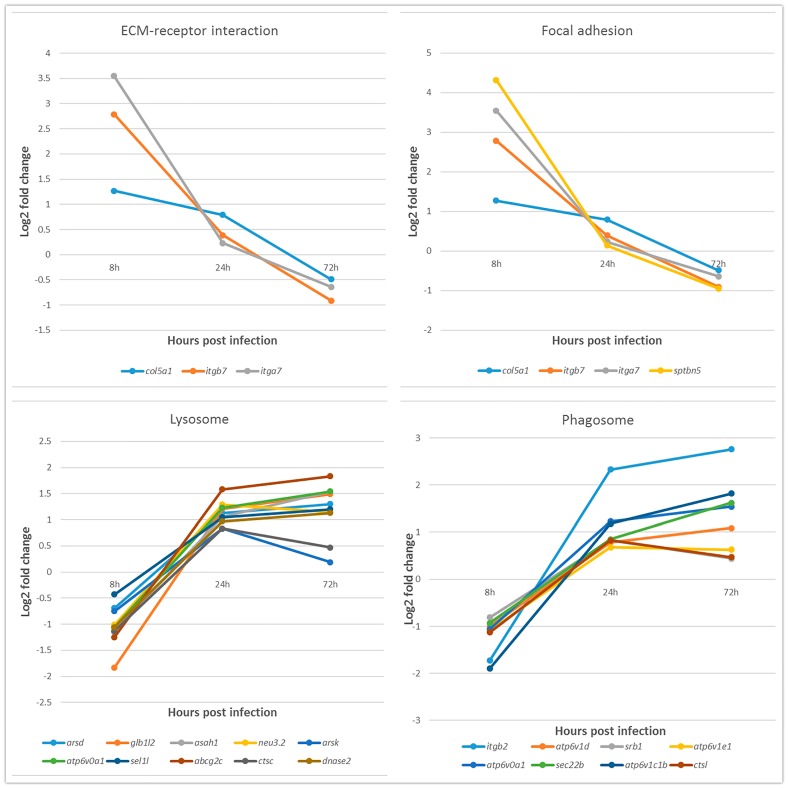
Expression patterns of DEGs in the key KEGG pathways ECM-receptor interactions, focal adhesion, phagosome, and lysosome KEGG pathways were further evaluated. As shown in Table 5, DEGs related to ECM-receptor interactions and focal adhesion, such as col5a1, itgb7, and sptbn5 was up-regulated during the early stage of infection but was down-regulated during the middle and late stages. By contrast, expression of DEGs involved in phagosomes and lysosomes, such as itgb2, srb1, and sel1l, followed the opposite trend, and was down-regulated during the early stage of infection, but up-regulated during the middle and late stages. Comparison of the four KEGG pathways at each time point after infection revealed a significant difference in the expression patterns of these DEGs (Figure 4).
Table 5.
DEGs associated with four major pathways.
| Pathway | Gene Name | Log2 Fold Change | ||
|---|---|---|---|---|
| 8 h | 24 h | 72 h | ||
| ECM-receptor interaction | col5a1 (collagen_alpha-1(V)_chain) | 1.27 | 0.79 | −0.49 |
| itgb7 (integrin_beta-7-like) | 1.52 | −0.40 | −0.42 | |
| itga7 (integrin_alpha-7) | 0.76 | −0.16 | 0.27 | |
| Focal adhesion | col5a1 (collagen_alpha-1(V)_chain) | 1.27 | 0.79 | −0.49 |
| itgb7 (integrin_beta-7-like) | 1.52 | −0.40 | −0.42 | |
| itga7 (integrin_alpha-7) | 0.76 | −0.16 | 0.27 | |
| sptbn5 (spectrin_beta_chain,_non-erythrocytic_5-like) | 0.77 | −0.09 | −0.31 | |
| Lysosome | arsd (arylsulfatase_D_isoform_X2) | −0.69 | 1.13 | 1.30 |
| glb1l2 (beta-galactosidase-1-like_protein_2-like) | −1.83 | 1.21 | 1.49 | |
| Asah1 (N-acylsphingosine_amidohydrolase_1_precursor) | −1.15 | 1.06 | 1.54 | |
| neu3.2 (Sialidase-3-like) | −1.01 | 1.29 | 1.15 | |
| arsk (arylsulfatase_K_isoform_X1) | −0.75 | 0.83 | 0.19 | |
| atp6v0a1 (V-type proton ATPase 116 kDa subunit a-like) | −1.06 | 1.23 | 1.54 | |
| sel1l (Protein sel-1 homolog 1 precursor) | −0.43 | 1.05 | 1.20 | |
| abcg2c (ATP binding cassette subfamily G member 2) | −1.25 | 1.58 | 1.83 | |
| ctsc (dipeptidyl_peptidase_1_precursor) | −1.13 | 0.83 | 0.47 | |
| dnase2 (deoxyribonuclease-2-beta-like) | −1.06 | 0.97 | 1.13 | |
| Phagosome | itgb2 (Integrin beta-2-like) | −1.73 | 2.33 | 2.76 |
| atp6v1d (V-type proton ATPase subunit D) | −1.00 | 0.79 | 1.09 | |
| srb1 (scavenger receptor class B member 1) | −0.81 | 0.83 | 0.44 | |
| atp6v1e1 (V-type proton ATPase subunit E 1-like) | −1.09 | 0.68 | 0.63 | |
| atp6v0a1 (V-type proton ATPase 116 kDa subunit a-like) | −1.06 | 1.23 | 1.54 | |
| sec22b (Vesicle-trafficking protein SEC22b-A-like) | −0.93 | 0.85 | 1.62 | |
| atp6v1c1b (ATPase, H+ transporting, lysosomal, V1 subunit C1b) | −1.90 | 1.18 | 1.82 | |
| ctsl (Cathepsin L1-like) | −1.13 | 0.83 | 0.47 | |
Figure 4.
Line chart of gene expression patterns of differentially expressed genes (DEGs) in selected KEGG pathways. The x-axis is the hours post infection, and the y-axis is the log2 fold change in expression level. Lines of different colors represent different DEGs.
2.7. Key DEGs Identified in the Three Stages of Infection
The more significant DEGs may play important roles in the responses to changes in the environment, hence they were identified and annotated. As shown in Table 6, most of the up-regulated genes between 0 and 8 h post infection such as protein tyrosine phosphatase epsilon (ptpε), integrin alpha 7 (itga7) and periplakin (ppl), are associated with cell signaling, whereas down-regulated genes such as C–X–C motif chemokine 10-like (cxcl10) and interferon alpha-inducible protein 27-like (ifi27l) are associated with immunity. Genes up-regulated between 8 and 24 hpi are primarily immune-related, including viperin, antigen peptide transporter 1 (tap1), toll-like receptor 3 (tlr3), and interferon regulatory factor-1 (irf11). Meanwhile, down-regulated genes such as angiopoietin-related protein 2-like (angptl2), phosphoglycolate phosphatase (pgp), and RAP1 GTPase activating protein 1 (rap1gap1) are mainly related to blood vessels, which may explain why haemorrhage symptoms were observed in grass carp following GCRV infection. Other down-regulated genes include protein damage repair-related genes such as methionine sulfoxide oxidase 3b (mical3b) and heat shock protein beta 11 (hspb11). Finally, between 24 and 72 h, up-regulated genes are mainly related to programmed cell death, apoptosis, and inflammation, consistent with the high level of cell viability reduction observed at 24 and 72 hpi (Figure 2). The main down-regulated genes are involved in steroid synthesis, while others are associated with microtubules and blood vessels. Details of these DEGs in the three stages are included in Table S4.
Table 6.
Top five up-regulated and down-regulated genes of the three stages following infection.
| Time Period | Gene Name | Log2 Fold Change | Description | |
|---|---|---|---|---|
| 0−8 h | Up | ptp ε | 6.01 | Protein tyrosine phosphatase epsilon |
| itga7 | 5.49 | Integrin alpha7 | ||
| hspg2 | 4.64 | Heparan sulfate proteoglycan 2 | ||
| leprel4 | 4.12 | Synaptonemal complex protein SC65 | ||
| ppl | 3.86 | Periplakin | ||
| Down | cxcl10 | −7.11 | C–X–C motif chemokine 10-like | |
| tnfrsf9 | −5.51 | Tumour necrosis factor receptor superfamily member 9-like | ||
| plaur | −5.39 | Urokinase plasminogen activator surface receptor-like | ||
| ubl1 | −4.71 | Ubquitin-like protein 1 | ||
| ifi27 | −4.63 | Interferon alpha-inducible protein 27-like | ||
| 8−24 h | Up | itgb2 | 6.84 | Integrin beta-2 |
| viperin | 4.71 | Virus inhibitory protein | ||
| tap1 | 3.81 | Antigen peptide transporter 1 | ||
| tlr3 | 3.05 | Toll-like receptor 3 | ||
| irf11 | 2.97 | Interferon regulatory factor 11 | ||
| Down | angptl2 | −5.78 | Angiopoietin-related protein 2-like | |
| pgp | −5.22 | Phosphoglycolate phosphatase | ||
| mical3b | −4.9 | Protein-methionine sulfoxide oxidase mical3b | ||
| hspb11 | −4.45 | Heat shock protein beta-11 | ||
| rap1gap1 | −2.73 | rap1 GTPase-activating protein 1 | ||
| 24−72 h | Up | tnfrsf9 | 2.68 | Tumour necrosis factor receptor superfamily member 9 |
| hspa | 2.42 | Heat shock 70 kDa protein | ||
| fbw7 | 2.35 | F-box/WD repeat-containing protein 7 | ||
| nfil3 | 1.87 | Nuclear factor, interleukin 3 regulated | ||
| osbp1 | 1.57 | Oxysterol-binding protein 1 | ||
| Down | lss | −2.95 | Lanosterol synthase | |
| sc5d | −2.19 | Lathosterol oxidase | ||
| cyp51a1 | −1.9 | Lanosterol 14α-demethylase | ||
| ablim1 | −1.11 | Actin-binding LIM protein 1 | ||
| cyr61 | −1.08 | Cyr61 protein | ||
2.8. Expression Patterns of Key Interferon-Related Genes
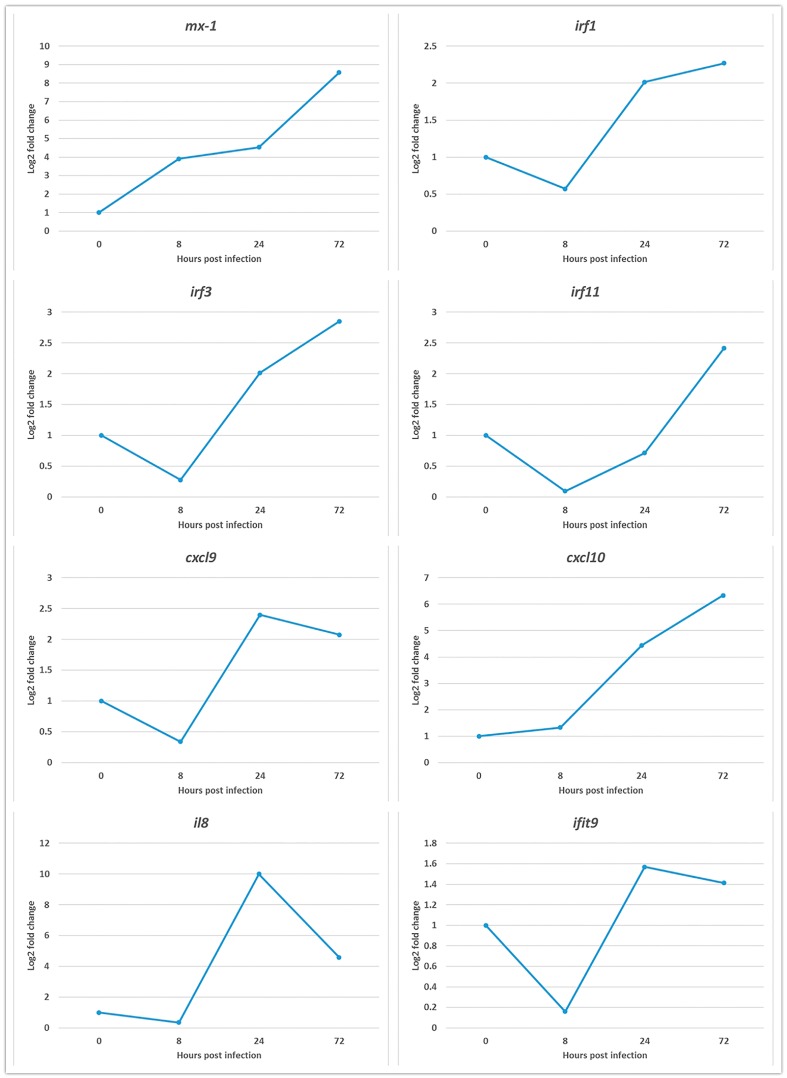
The expression patterns of key interferon-related genes were examined in this study. As shown in Figure 5, except gene mx-1, most of the genes showed log2 fold change ≥2 or even ≥10 at 24 or 72 h post-infection, whereas the expression level decreased at 8 h post-infection. These results suggested that strong immune response induced by GCRV infection.
Figure 5.
Expression patterns of key genes involved in immune responses. Expression pattern of eight interferon-related genes including interferon-inducible Mx protein 1 (mx-1), interferon regulatory factor 1 (irf1), interferon regulatory factor 3 (irf3), interferon regulatory factor 11 (irf11), C–X–C motif chemokine 9 (cxcl9), C–X–C motif chemokine 11 (cxcl10), interleukin 8 (il8), interferon-induced protein with tetratricopeptide repeats 9 (ifit9) were examined.
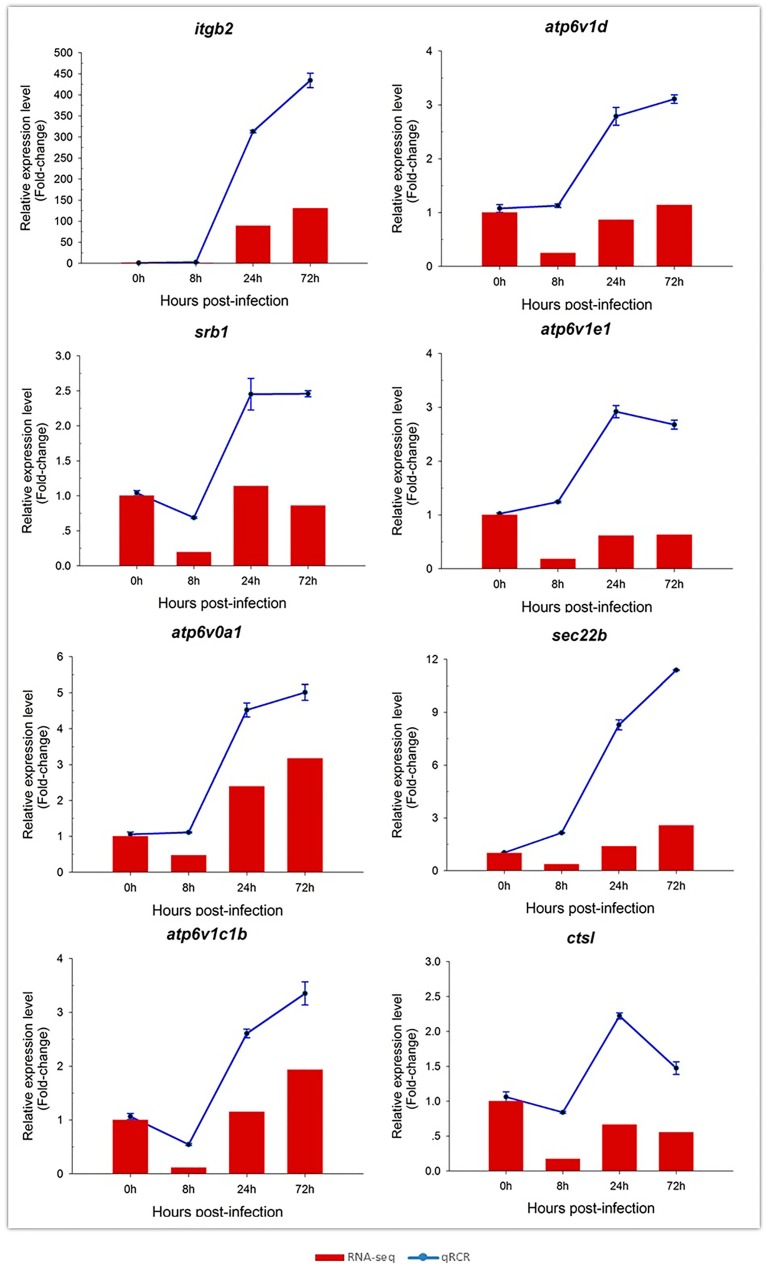
2.9. Validation of Selected DEGs by RT-qPCR
To confirm the reliability of the RNA-Seq data, eight DEGs involved in the phagosome pathway were selected for RT-qPCR analysis. Expression levels of these eight DEGs are shown in Figure 6. Overall, expression patterns of all eight DEGs obtained by qPCR were similar to those from RNA-Seq analysis, although the relative expression levels were not completely consistent. Therefore, the results of the RT-qPCR analysis confirmed the reliability and accuracy of the RNA-Seq data.
Figure 6.
Confirmation of transcriptomics sequencing data by RT-qPCR. Eight DEGs involved in the phagosome pathway were selected for RT-qPCR. These genes included integrin β2 (itgb2), V-type proton ATPase subunit D (atp6v1d), srb1, V-type proton ATPase subunit E 1-like (atp6v1e1), V-type proton ATPase 116 kDa subunit A-like (atp6v0a1), sec22b, ATPase, H+ transporting, lysosomal, V1 subunit C1b (atp6v1c1b), and cathepsin L1-like (ctsl). The relative expression levels of the genes at different time points were calculated as the ratio of gene expression level (qPCR) or normalized RPKM level (RNA-seq) relative to that at 0 days (control) post-infection. All data represent the mean ± S.D. of three replicates.
3. Discussion
At present, previous studies on grass carp transcriptome profiles in response to grass carp reovirus infection have been carried out [10,11,12,13,14]. Most of them involved in the discovery of immune-related genes or uncovered the antiviral immunity mechanism of grass carp. However, the purpose of our study is to understand the whole process of grass carp reovirus infection.
In this study, we used the CIK cells as an in vitro model to explore the molecular changes following reovirus infection. CIK cells were mock-infected or infected with GCRV and harvested at 0 (mock), 8, 24, and 72 hpi. To better understand the process of GCRV entry into cells, it was divided into three stages: early (0–8 h), middle (8–24 h), and late (24–72 h). In each stage, data from the latter time point were compared with that from the former to identify DEGs. The method for DEG identification differed that in the previous study, which used a case-control method [15,16]. DEGs in the 0–8 h were mainly associated with virus adhesion, DEGs in the 8–24 h were largely associated virus endocytosis and transmission, and DEGs in the 24–72 h were involved in cell death and apoptosis. Therefore, the DEGs identified in the present study appear to be representative and accurate.
The CPE appeared as early as 8 h after CIK cells infected with GCRV. DEGs from this stage were mainly enriched with Focal adhesion and ECM-receptor interaction terms. Focal adhesions are sub-cellular structures that mediate the regulatory effects of cells in response to ECM adhesion [17]. In fact, previous studies showed that focal adhesion kinase can interact with rabies virus phosphoprotein P, which is involved in the process of viral infection [18]. During influenza A invasion of cells, inhibition of focal adhesion kinase signaling leads to reduced viral replication [19], suggesting that focal adhesion may be an important pathway during GCRV infection. Moreover, hspg2, ptpε, and ppl were significantly up-regulated during the 0–8 h post infection. HSPG2 plays an important role in mediating virus envelope-target cell interaction during human papillomavirus (HPV) and hepatitis C virus (HCV) infection [20,21]. PTPε plays an important role in the control of macrophage function [22]. And what’s more, tyrosine-protein phosphatase non-receptor type 22 (PTPN22) associates with TNF receptor-associated factor 3 (TRAF3) to augment Toll-like receptor (TLR)-induced type I IFN production [23]. However, PPL acts as a cytolinker between intermediate filament scaffolding and the desmosomal plaque and is involved in epithelial cohesion, intracellular signal transduction and antigen presentation [24]. These results suggest that hspg2 may be involved in cellular binding of GCRV, after which GCRV likely binds to the receptor on the cell membrane, ptpε and ppl maybe as signaling molecules that regulate cellular processes.
During the 8 to 24 hpi period, enriched DEGs were mainly related to phagosome and lysosome pathways. In recent years, more and more studies have shown that the viral infection process is closely related to autophagy of host cells. Autophagy can be used as innate immunity and an adaptive immune response against intracellular pathogens [25]. In the phagosome pathway, many genes are up-regulated, including itgb2, srb1, and sec22b, and it is worth noting that the itgb2 is up-regulated at this period. Cell integrins are commonly used receptors for diverse viral pathogens [26], and reovirus internalization is mediated by integrin β1 proteins, most likely via clathrin-dependent endocytosis [27]. Moreover, integrin β3 proteins serve as receptors of pathogenic hantaviruses [28]. In the phagosome pathway, other receptor genes appear to be up-regulated, such as scavenger receptor srb1, which, along with CD81, is the receptor used for entry of the hepatitis C virus into liver cells [29]. The v-SNARE protein SEC22 is involved in the transport of eukaryotic cell endoplasmic reticulum (ER) and Golgi apparatus [30]. Hepatitis C virus may be assembled in the ER and transported to the Golgi apparatus in COPII vesicles to begin Golgi secretion [31]. However, a recent study shows that SEC22 reduces RNAi efficiency by influencing late endosomal functions, through the promotion of fusion between late endosomes and lysosomes [32]. These results suggest that itgb2 and/or srb1 may be important for cell entry of GCRV. Furthermore, GCRV entry into the endosomal region followed by fusion with lysosomes is likely to be regulated by sec22b.
In the 24–72 h post infection, DEGs were mainly enriched in lipid metabolism, programmed cell death, apoptosis, and inflammation. Genes in the cell death pathway have been studied intensely, and it is interesting to note that genes in the lipid metabolism pathway, such as cyp51a, are enriched. CYP51A1 is a cytochrome P450 enzyme, a conserved group of proteins that serve as key players in the metabolism of organic substances and the biosynthesis of important steroids, lipids, and vitamins in eukaryotes [33], and it plays an essential role in mediating membrane permeability [34]. These sterols, located on the plasma membrane of cells, play an important structural role in regulating the fluidity and permeability of membranes while also affecting the activity of enzymes, ion channels and other cellular components [35]. Therefore, these results suggest that GCRV probably not only causes cell death through cell apoptosis and inflammatory pathways but also changes the permeability of the cell membrane by affecting the structure of steroids in the cell membrane, leading to cell death through this mechanism.
4. Materials and Methods
4.1. Cells, Virus and Virus Infection
C. idellus kidney (CIK) cells purchased from Shenzhen ziker Biological Technology Co., Ltd. (Shenzhen, China) and grown in Medium 199 (Gibco, Gaithersburg, MD, USA) supplemented with 10% fetal bovine serum (FBS), 100 mg/mL penicillin, and 100 mg/mL streptomycin at 28 °C. GCRV was kindly provided by Professor Jianguo Su (Huazhong Agricultural University). CIK cells that grown in 25 cm2 flask were mock-infected or infected with GCRV at a multiplicity of infection (MOI) of 1, then harvested at 0 (mock), 8, 24, and 72 h post infection (pi), respectively. Three flasks of cells were collected for each treatment at each time point.
4.2. CCK-8 Assay
CCK-8 detection kit (Beyotime, Shanghai, China) was used to measure cell viability according to the manufacturer’s instructions. About 4 × 103 CIK cells were seeded in 96 well plates and cultured in M199 supplemented with 10% FBS at 28 °C for 24 h. Wells containing no cells but only culture medium alone were served as blank control. CIK cells were mock-infected or infected with GCRV at an MOI of 1 and maintained for 0, 8, 24, and 72 h at 28 °C, respectively. Then each well was added 10 μL of CCK-8 solution. After 4 h incubation at 28 °C, the plates were measured spectrophotometrically on Microplate Manager 6 (BIO-RAD, Hercules, CA, USA) at wavelength 450 nm. The relative cell viability was calculated as Ainfected-Ablank-i/A mock-Ablank % (Ablank-i represented the wells containing no cells but only medium and GCRV). Data were presented as means of at least three independent experiments ± standard deviation (S.D).
4.3. RNA Isolation, Library Construction, and Sequencing
Cells were collected at 0, 8, 24, and 72 h after infection (C0, C8, C24, and C72 groups, respectively), rapidly frozen in liquid nitrogen, and total RNA was extracted with Trizol reagent (Invitrogen, Waltham, MA, USA). RNase-free DNase treatment was performed in order to remove possible genomic DNA. The DNase was removed by phenol-chloroform re-extraction. mRNA was purified from total RNA using poly-T oligo-attached magnetic beads and segregated into 200–300 bp fragments. The concentration of mRNA used to prepare first strand cDNA is approximate 40 ng/μL and a total of 3 μg mRNA used as input material for first strand DNA synthesis. First strand DNA was synthesized from 6-base random primers and reverse transcriptase using RNA as a template, and second strand cDNA was synthesized using the first strand cDNA as template. In the second strand cDNA synthesis, the base T was replaced by U to generate a chain-specific library. RNA-Seq was performed using chain-specific kits, and chain-specific libraries were used to determine the transcriptional direction of sense and antisense strands to increase the accuracy of subsequent gene function annotation and gene expression analysis.
After library construction, library fragments were enriched by PCR amplification, and selection based on fragment size resulted in a 300–400 bp library. The library was tested using an Agilent 2100 Bioanalyzer (Agilent, Palo Alto, CA, USA), and both the total and effective concentration of the library were tested. Based on the effective concentration and the amount of data required, libraries containing different Index sequences (samples plus different Index sequences, and difference data of samples according to Index) were mixed proportionally. The mixed library was diluted to 2 nM and alkaline degeneration was performed to generate a single-chain library. Next, 150 bp pair-end reads were performed on the library using an Illumina NextSeq500 sequencing platform to carry out Next-Generation Sequencing (NGS) at Personalbio (Personalbio, Shanghai, China). For each treatment in each time point, the sample was sequenced three times as three technical replicates.
4.4. Data Analysis
Clean data were obtained by removing adapters, poly-N sequences and poor-quality data [36]. The Q20, Q30, and GC content of clean data were calculated, and all downstream analysis was performed using high-quality clean data.
Clean data were mapped to the grass carp reference genome [37] using TopHat2 software [38]. The number of reads mapped to each gene was used HTSeq software [39], and the reads per kilobase of the exon model per million mapped reads (RPKM) was calculated as expression values for each gene [40].
4.5. Differential Expression Analysis
Differential expression analysis using data collected at different time points was performed using the DESeq package [41]. The resulting p-values were adjusted using the Benjamini and Hochberg approach to control the false discovery rate. Genes with an adjusted p-value < 0.05 (q-value < 0.05) and fold change > 2 in DESeq analysis were assigned as differentially expressed genes (DEGs).
ClueGO and CluePedia were used to perform Gene Ontology (GO) enrichment analysis [42,43]. In GO enrichment analysis, only categories with a p-value < 0.05 were considered as enriched in the network.
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database was used to indicate the location of the DEGs in the different pathways. In this study, KOBAS software was used to test the statistical enrichment of DEGs in KEGG pathways [44]. Pathways with p-values < 0.05, were considered for as statistical significance.
4.6. Validation of DEGs by RT-qPCR
To confirm the reliability of the data obtained with RNA-Seq, eight DEGs were selected for RT-qPCR using the primers listed in Table S5. Primers were designed using the Primer Premier 5.0 software (PREMIER Biosoft International, Palo Alto, CA, USA) and only with an efficiency of 90–110% were used. First strand cDNAs were obtained from total RNA (>3 μg) using a random hexamer primer and the ReverTra Ace kit (TOYOBO, Osaka, Japan). RT-qPCR was performed using a fluorescence quantitative PCR instrument (BIO-RAD). Each RT-qPCR mixture contained 0.8 μL of each primer, 1 μL template, 10 μL 2× SYBR Green master mix (TOYOBO), and 7.4 μL ddH2O. The program for RT-qPCR was 95 °C for 10 s, followed by 40 cycles at 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 30 s. Three replicates were included for each sample, and the β-actin gene of grass carp was served as an internal control to normalize the expression levels. Relative expression levels of the different genes were calculated using the 2−ΔΔCt method [45]. All data represent the mean ± standard deviation of three replicates.
5. Conclusions
In conclusion, the results of this study provide insight into the mechanisms of grass carp reovirus infection. In the early stage of infection, GCRV interacts with polysaccharides and accumulates on the cell surface, then binds to the GCRV receptor to initiate endocytosis mediated by integrins. In the middle stage of infection, GCRV is likely to be regulated by v-SNARE protein and transported from the endosomal region to lysosomes. In the last stage, GCRV functions through several different pathways, by affecting the structure of steroids in the cell membrane, then leading to cell death in the final stage. These findings broaden our understanding of the invasion of GCRV and may assist the development of strategies to combat diseases caused by GCRV.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (31721005 and 31702332), the Strategic Priority Research Program of Chinese Academy of Sciences (XDA08030203), and the Hubei Provincial Natural Science Foundation of China (2015CFA007). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Supplementary Materials
Supplementary materials can be found at http://www.mdpi.com/1422-0067/19/2/488/s1.
Author Contributions
Yaping Wang, Zuoyan Zhu and Geng Chen conceived and designed the experiments; Geng Chen, Lifei Luo, and Rong Huang performed the experiments; Lanjie Liao and Yongming Li helped to do sample collection; Geng Chen analyzed the data and wrote the paper; Libo He revised the paper. All authors have read and approved the manuscript for publication.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Food and Agriculture Organization of the United Nations . Fishery and Aquaculture Statistics Yearbook. Food and Agriculture Organization of the United Nations; Rome, Italy: 2017. [Google Scholar]
- 2.Rao Y., Su J. Insights into the antiviral immunity against grass carp (Ctenopharyngodon idella) reovirus (GCRV) in grass carp. J. Immunol. Res. 2015;2015:670437. doi: 10.1155/2015/670437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Attoui H., Mertens P.P., Becnel J., Belaganahalli S., Bergoin M., Brussaard C.P., Chappell J.D., Ciarlet M., del Vas M., Dermody T.S., et al. Family Reoviridae. In: King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J., editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; San Diego, CA, USA: 2012. pp. 538–637. [Google Scholar]
- 4.Rao Y., Su J., Yang C., Peng L., Feng X., Li Q. Characterizations of two grass carp Ctenopharyngodon idella hmgb2 genes and potential roles in innate immunity. Dev. Comp. Immunol. 2013;41:164–177. doi: 10.1016/j.dci.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Pei Y.Y., Huang R., Li Y.M., Liao L.J., Zhu Z.Y., Wang Y.P. Characterizations of four toll-like receptor 4s in grass carp Ctenopharyngodon idellus and their response to grass carp reovirus infection and lipopolysaccharide stimulation. J. Fish Biol. 2015;86:1098–1108. doi: 10.1111/jfb.12617. [DOI] [PubMed] [Google Scholar]
- 6.Lu J., Wang H., Zhang Y., Li Y., Lu L. Grass carp reovirus ns26 interacts with cellular lipopolysaccharide-induced tumor necrosis factor-alpha factor, LITAF. Virus Genes. 2016;52:789–796. doi: 10.1007/s11262-016-1370-6. [DOI] [PubMed] [Google Scholar]
- 7.Liu W., Wang H., Yu F., Lu L. Grass carp reovirus outer capsid proteins vp5 and vp7 interact in vitro. Arch. Virol. 2017;162:2375–2380. doi: 10.1007/s00705-017-3354-9. [DOI] [PubMed] [Google Scholar]
- 8.Zhu B., Liu G.L., Gong Y.X., Ling F., Song L.S., Wang G.X. Single-walled carbon nanotubes as candidate recombinant subunit vaccine carrier for immunization of grass carp against grass carp reovirus. Fish Shellfish Immunol. 2014;41:279–293. doi: 10.1016/j.fsi.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 9.Tian Y., Ye X., Zhang L., Deng G., Bai Y. Development of a novel candidate subunit vaccine against grass carp reovirus Guangdong strain (GCRV-GD108) Fish Shellfish Immunol. 2013;35:351–356. doi: 10.1016/j.fsi.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 10.Jin C., Cai L., Rong H., Du F., Liao L., Zhu Z. Transcriptomics analysis of head kidney in grass carp and discovery of immune-related genes. BMC Vet. Res. 2012;8:108. doi: 10.1186/1746-6148-8-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dang Y., Xu X., Shen Y., Hu M., Zhang M., Li L. Transcriptomics analysis of the innate immunity-related complement system in spleen tissue of Ctenopharyngodon idella infected with Aeromonas hydrophila. PLoS ONE. 2016;11:e0157413. doi: 10.1371/journal.pone.0157413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu B.H., Zhong L., Liu Q.L., Xiao T.Y., Su J.M., Chen K.J. Characterization of grass carp spleen transcriptome during GCRV infection. Genet. Mol. Res. GMR. 2016;15 doi: 10.4238/gmr.15026650. [DOI] [PubMed] [Google Scholar]
- 13.Wan Q., Su J. Transcriptome analysis provides insights into the regulatory function of alternative splicing in antiviral immunity in grass carp (Ctenopharyngodon idella) Sci. Rep. 2015;5:12946. doi: 10.1038/srep12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi M., Huang R., Du F., Pei Y., Liao L., Zhu Z. RNA-seq profiles from grass carp tissues after reovirus (GCRV) infection based on singular and modular enrichment analyses. Mol. Immunol. 2014;61:44–53. doi: 10.1016/j.molimm.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Hu X., Fu X., Li N., Dong X., Zhao L., Lan J. Transcriptomic analysis of mandarin fish brain cells infected with infectious spleen and kidney necrosis virus with an emphasis on retinoic acid-inducible gene 1-like receptors and apoptosis pathways. Fish Shellfish Immunol. 2015;45:619–629. doi: 10.1016/j.fsi.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 16.Flori L., Rogelgaillard C., Cochet M., Lemonnier G., Hugot K., Chardon P. Transcriptomic analysis of the dialogue between Pseudorabies virus and porcine epithelial cells during infection. BMC Genom. 2008;9:123. doi: 10.1186/1471-2164-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C.S., Alonso J.L., Ostuni E., Whitesides G.M., Ingber D.E. Cell shape provides global control of focal adhesion assembly. Biochem. Biophys. Res. Commun. 2003;307:355–361. doi: 10.1016/S0006-291X(03)01165-3. [DOI] [PubMed] [Google Scholar]
- 18.Fouquet B., Nikolic J., Larrous F., Bourhy H., Wirblich C., Lagaudrière-Gesbert C., Blondel D. Focal Adhesion Kinase Is Involved in Rabies Virus Infection through Its Interaction with Viral Phosphoprotein P. J. Virol. 2015;89:1640–1651. doi: 10.1128/JVI.02602-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elbahesh H., Cline T., Baranovich T., Govorkova E.A., Schultzcherry S., Russell C.J. Novel roles of focal adhesion kinase in cytoplasmic entry and replication of influenza A viruses. J. Virol. 2014;88:6714–6728. doi: 10.1128/JVI.00530-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barth H., Schafer C., Adah M.I., Zhang F., Linhardt R.J., Toyoda H. Cellular binding of hepatitis c virus envelope glycoprotein e2 requires cell surface heparan sulfate. J. Biol. Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 21.Giroglou T., Florin L., Schäfer F., Streeck R.E., Sapp M. Human papillomavirus infection requires cell surface heparan sulfate. J. Virol. 2001;75:1565–1570. doi: 10.1128/JVI.75.3.1565-1570.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sully V., Pownall S., Vincan E. Functional abnormalities in protein tyrosine phosphatase epsilon-deficient macrophages. Biochem. Biophys. Res. Commun. 2001;286:184–188. doi: 10.1006/bbrc.2001.5360. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y. The autoimmunity-associated gene PTPN22 potentiates toll-like receptor-driven, type 1 interferon-dependent immunity. Immunity. 2013;39:111–122. doi: 10.1016/j.immuni.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taillé C., Grootenboer-Mignot S., Estellat C. Periplakin is a target for autoimmunity in asthma. Respir. Res. 2016;17:126. doi: 10.1186/s12931-016-0441-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deretic V., Levine B. Autophagy, Immunity, and Microbial Adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart P.L., Nemerow G.R. Cell integrins: Commonly used receptors for diverse viral pathogens. Trends Microbiol. 2007;15:500–507. doi: 10.1016/j.tim.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Maginnis M.S., Forrest J.C., Kopecky-Bromberg S.A., Dickeson S.K., Santoro S.A., Zutter M.M., Nemerow G.R., Bergelson J.M., Dermody T.S. β1 Integrin Mediates Internalization of Mammalian Reovirus. J. Virol. 2006;80:2760–2770. doi: 10.1128/JVI.80.6.2760-2770.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raymond T., Gorbunova E., Gavrilovskaya I.N., Mackow E.R. Pathogenic hantaviruses bind plexin-semaphorin-integrin domains present at the apex of inactive, bent alphavbeta3 integrin conformers. Proc. Natl. Acad. Sci. USA. 2005;102:1163–1168. doi: 10.1073/pnas.0406743102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapadia S.B., Barth H., Baumert T., Mckeating J.A., Chisari F.V. Initiation of hepatitis c virus infection is dependent on cholesterol and cooperativity between cd81 and scavenger receptor B type I. J. Virol. 2007;81:374–383. doi: 10.1128/JVI.01134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jahn R., Scheller R.H. SNAREs—Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 31.Syed G.H., Khan M., Yang S., Siddiqui A. Hepatitis C virus lipoviroparticles (HCV-LVP) assemble in the endoplasmic reticulum (ER) and bud off from the ER to Golgi in COPII vesicles. J. Virol. 2017;91:e00499-17. doi: 10.1128/JVI.00499-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Y., Holmgren B.T., Hinas A. The conserved snare sec-22 localizes to late endosomes and negatively regulates RNA interference in Caenorhabditis elegans. RNA. 2017;23:297–307. doi: 10.1261/rna.058438.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lepesheva G.I., Waterman M.R. Sterol 14α-Demethylase Cytochrome P450 (CYP51), a P450 in All Biological Kingdoms. Biochim. Biophys. Acta. 2008;1770:467–477. doi: 10.1016/j.bbagen.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daum G., Lees N.D., Bard M., Dickson R. Biochemistry, Cell Biology and Molecular Biology of Lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 35.Abe F., Usui K., Hiraki T. Fluconazole modulates membrane rigidity, heterogeneity, and water penetration into the plasma membrane in Saccharomyces cerevisiae. Biochemistry. 2009;48:8494–8504. doi: 10.1021/bi900578y. [DOI] [PubMed] [Google Scholar]
- 36.Martin M. Cut adapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 2011;17 doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 37.Wang Y., Lu Y., Zhang Y., Ning Z., Li Y., Zhao Q. The draft genome of the grass carp (Ctenopharyngodon idellus) provides insights into its evolution and vegetarian adaptation. Nat. Genet. 2015;47:625–631. doi: 10.1038/ng.3280. [DOI] [PubMed] [Google Scholar]
- 38.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: Accurate alignment of the transcriptome in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S., Pyl P.T., Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptome by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 41.Anders S., Huber W. Differential expression analysis for sequence count data. Genome Boil. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A. Clue GO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bindea G., Galon J., Mlecnik B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–663. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.