Abstract
Next generation sequencing has accelerated the discovery of a variety of new fusion gene types in clinical breast cancer samples by analyzing cancer genomes and transcriptomes. Although previous studies have focused on a few clinically validated oncogenic fusion genes as diagnostic and therapeutic targets in breast cancer, a perspective consideration has not been given thus far for a plethora of breast cancer fusion genes, which are being newly identified at an overwhelmingly increasing pace. In this perspective review, we discuss diverse fusion gene types recently identified in a variety of breast cancer subtypes, including breast clinical cancer samples in TCGA (The Cancer Genome Atlas) database. This perspective review will confer fresh and promising guidance onto breast cancer surgeons, clinical oncologists, and tumor biologists in determining research directions for seeking and developing novel fusion gene biomarkers for breast cancer diagnostics and therapeutic treatment in upcoming years.
Keywords: fusion gene transcript, breast cancer, biomarker
1. Introduction
Genomic rearrangements are well known to be closely associated with carcinogenesis in diverse types of human tissues [1,2,3]. Genomic rearrangements as diagnostic and therapeutic targets were initially identified in hematological malignancies, such as chronic myelogenous leukemia (CML) [4,5] and Burkitt’s lymphoma [6]. Recurrent translocations were subsequently reported in Ewing’s sarcoma [7,8] and synovial sarcoma [9]. Even though diverse kinds of genomic rearrangements, including conjoined genes (read-through fusion genes) [1], were previously known to be associated with carcinogenesis, a majority of the validated oncogenic fusion genes seem to be generated from the inter-chromosomal and intra-chromosomal rearrangements, thus far. With the advent of high-throughput next generation sequencing technology, substantial discovery of genomic rearrangements is now being accelerated at an unprecedented pace in diverse kinds of tumor types, including breast cancer. For instance, whole genome and whole exome sequencing and RNA-Seq recently enabled discoveries of KIAA1217-RET [2], SEC31A-ALK [3], TPR-ALK [4] and HIP1-ALK [5], all of which are chromosomal translocation-derived novel fusion oncogenes causing lung adenocarcinoma. Such genomic alterations have turned out to be diagnostic and therapeutic targets for treating lung cancer patients.
In contrast to other cancer types, genomic rearrangements leading to the formation of fusion genes in breast cancer are, so far, less well known. In this perspective paper, we discuss fusion genes recently identified in breast cancer patients, including TCGA (The Cancer Genome Atlas) clinical breast cancer samples, addressing what kinds of fusion genes might be potential targets to prioritize in seeking future fusion gene biomarkers in breast cancer.
2. Abundance of Diverse Kinds of Fusion Gene Types in Breast Cancer
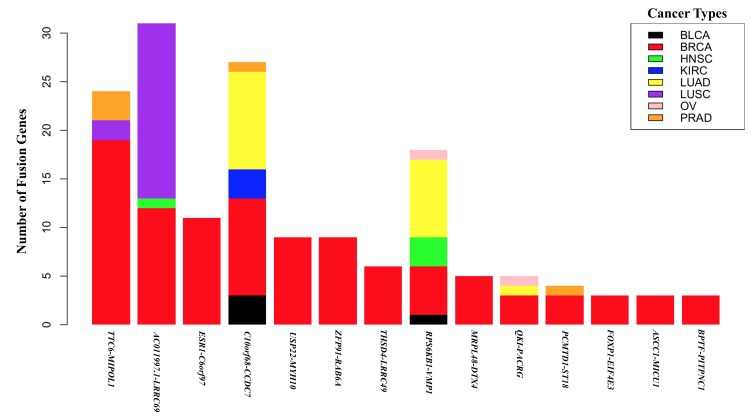
Recent study has shown us an overview of how many fusion genes there are across diverse cancer types in TCGA clinical cancer samples and what kinds of fusion types they are [6]. Fusion gene transcripts showing high recurrence in the breast invasive carcinoma samples are as follows: TTC6-MIPOL1, AC011997.1-LRRC69, ESR1-C6orf97, C10orf68-CCDC7, USP22-MYH10, ZFP91-RAB6A, THSD4-LRRC49, RPS6KB1-VMP1, MRPL48-DTX4, QKI-PACRG, PCMTD1-ST18, FOXP1-EIF4E3, ASCC1-MICU1, and BPTF-PITPNC1. Among the 14 fusion genes recurring in breast invasive carcinoma samples, AC011997.1-LRRC69, C10orf68-CCDC7 and RPS6KB1-VMP1 showed relatively high recurrence not only in breast carcinoma but also in LUSC (Lung squamous cell carcinoma) and in LUAD (lung adenocarcinoma) (Figure 1). The fusion genes in TCGA clinical breast cancer samples diverge greatly from one another in terms of translocation patterns, genomic positions conjoining between upstream and downstream partner genes, and functional classification of those partner genes. Nevertheless, no matter what kinds of genomic translocation might occur in generating such fusion genes, the prerequisite for creating oncogenic translocation and fusion gene transcripts should primarily be in-frame fusion and then the acquisition of optimal oncogenic function in those fusion proteins. Because this perspective review focuses mainly on fusion gene transcripts based on RNA-Seq data analysis, detailed explanations of genomic rearrangements are beyond the scope of our review, and could be found somewhere else.
Figure 1.
Occurrence of recurrent breast cancer transcript fusion genes across diverse cancer types. The stacked bar plot shows occurrence frequencies of the top 14 most recurrent breast cancer transcript fusion genes in 8 cancer types, BLCA (Bladder Urothelial Carcinoma), BRCA (breast Invasive Carcinoma), HNSC (Head and Neck Squamous Cell Carcinoma), KIRC (Kidney Renal Clear Cell Carcinoma), LUAD (Lung Adenocarcinoma), LUSC (Lung Squamous Cell Carcinoma), OV (Ovarian Serous Cystadenocarcinoma), and PRAD (Prostate Adenocarcinoma).
If highly recurrent genomic translocation regions can be associated with frequent fusion events involving oncogenes, we should not neglect the high likelihood of as yet unidentified in-frame fusion events involving the same oncogenes in the genomic regions, and we should consider the genomic regions as target regions for screening oncogenic fusion genes. Recently, several pioneering studies have elucidated the reasonable cause of specific regions in the cancer genomes being more frequently broken and more recurrently involved in translocations than other regions, and how the epigenomic and genomic structural landscapes could affect the genomic structural fragility of these regions [7]. Notably, they found a new explanation in epigenetic aspects regarding such frequent structural fragility at specific genomic regions. The authors demonstrated that histone methylation modification mark H3K4me3 could be closely associated with genomic regions which are very vulnerable to DNA double strand breakage.
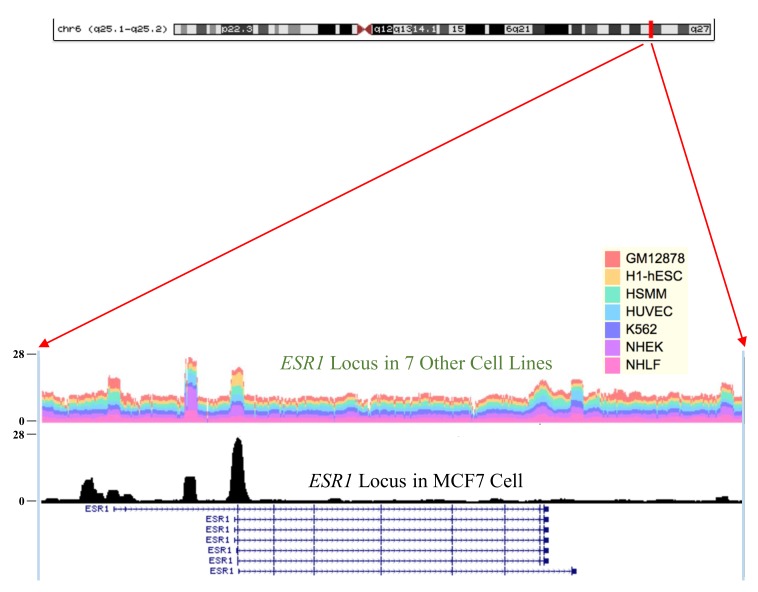
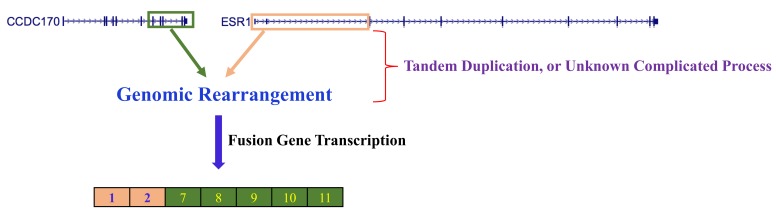
In order to check whether their assertion may also be applicable to the genomes in human breast adenocarcinoma cell line MCF-7, we analyzed ChIP-Seq dataset (GEO accession: GSE35583 [8]) for a genome-wide H3K4me3 landscape in MCF-7 cells. Intriguingly, we found that, in a genomic region harboring the exon 3 and its adjacent intron 2 portion within the ESR1 gene locus, there are the two strongest histone-modified mark peaks for H3K4me3 in MCF-7 cells (Figure 2). More surprisingly, we found that the genomic region with the two strongest H3K4me3 marks is the region most vulnerable to DNA strand breakage; releasing a 5′ portion of ESR1 which has exon 1 and exon 2 intact, which can often conjoin to the intron 6 region of the neighboring gene CCDC170, consequently causes the ESR1-CCDC170 fusion gene (Figure 3) [9]. This finding corroborates their assertion that epigenetic mark H3K4me3 could be associated with genomic DNA breakage and subsequently with genomic rearrangements. In contrast to those epigenetic marks in MCF-7 cell, other human cells (GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK and NHLF) did not exhibit such strong H3K4me3 marks in the genomic regions, explaining why ESR1 could be involved in various fusion gene types only in breast carcinoma, but not in other human cell types (Figure 2).
Figure 2.
Strong H3K4me3 epigenetic modification signals within the ESR1 locus in human breast adenocarcinoma MCF-7 cells. H3K4me3 signals in the MCF-7 ESR1 locus are compared with the stacked spectra of those signals in the seven other cell lines, GM12878 (human lymphoblastoid cell), H1-hESC (H1-human Embryonic Stem Cell), HSMM (Human Skeletal Muscle Myoblast cell), HUVEC (Human Umbilical Vein Endothelial Cell), K562 (human immortalised myelogenous leukemia cell), NHEK (Normal Human Epidermal Keratinocytes), and NHLF (normal human lung fibroblasts). The y-axis indicates the intensity of the epigenetic modification H3K4me3 at each genomic position.
Figure 3.
ESR1-CCDC170 fusion gene transcript formation. The intron 2 region within the ESR1 genomic locus appeared to undergo a DNA breakage, subsequently conjoining with the intron 6 region within the genomic locus of its neighboring gene CCDC170. This genomic rearrangement results in an oncogenic fusion mRNA transcript involving an upstream part (exons 1 and 2) of ESR1 and the C-terminal portion (exons 7–11) of its neighboring gene CCDC170.
Such novel insight based on epigenomics may provide molecular pathologists and clinical tumor biologists with a fascinating and exciting clue for further delving deeper into the molecular mechanism of fusion gene formation in breast cancer and for discovering fusion gene biomarkers in the future.
3. Recurrent in-Frame Fusion Genes across TCGA Clinical Breast Tumor Samples
There were 319 fusion genes in TCGA clinical breast cancer tumors checked in the previous study [6]. Among them, there were 16 recurrent in-frame fusion genes (TTC6-MIPOL1, MRPL48-DTX4, THSD4-LRRC49, ASCC1-MICU1, QKI-PACRG, RPS6KB1-VMP1, RFTN1-DAZL, AFAP1-ABLIM2, SBNO2-C19orf20, MVP-KIF22, GIGYF2-EFHD1, ARFGEF1-CPA6, EFHD1-EIF4E2, SLC39A11-SDK2, BPTF-PITPNC1, and TBC1D4-COMMD6) with frequency ≥2. Interestingly, of the 16 recurrent in-frame fusion genes, 13 (MRPL48-DTX4, THSD4-LRRC49, ASCC1-MICU1, QKI-PACRG, AFAP1-ABLIM2, SBNO2-C19orf20, MVP-KIF22, GIGYF2-EFHD1, ARFGEF1-CPA6, EFHD1-EIF4E2, SLC39A11-SDK2, BPTF-PITPNC1, and TBC1D4-COMMD6) had occurred only in breast cancer samples, but not in other cancer types. These in-frame fusion genes might be suitable candidate targets for screening fusion genes causing breast carcinogenesis in the future. In addition, the in-frame fusion genes generated through a joining between non-coding regions, like the breast cancer-specific oncogenic fusion gene ESR1-CCDC170, should be carefully identified by using whole genome sequencing and RNA-Seq data in the future.
4. Fusion Genes Involving Tumor Suppressor Genes in Breast Cancer
Fusion genes involving tumor suppressor genes as fused partners that have been identified in the breast cancer samples are as follows: TSC2-TMEM204, ARID1A-THBS3, ARID1A-YTHDF2, AXIN1-ITFG3, AXIN1-ITCH, AXIN1-IFT140, CASP8-ALS2CR12, CDH1-CCDC132, CDH1-NPFFR2, CDH1-DEXI, CCDC132-CDH1, ASXL1-TM9SF4, ASXL1-PDRG1 (Tumor suppressor genes are underlined in the names of fusion genes for clarity) (Table 1). Previous investigations had reported that TSC2, ARID1A, AXIN1, CASP8, CDH1, and ASXL1 could play roles in tumor suppression in diverse cancer types, including breast cancer. Intriguingly, we found that the above-mentioned fusion events could break the functional domains of the tumor suppressor genes as the fusion partners, via the involvement of their incomplete functional domain structures in the fusion gene bodies, no matter whether the fusion styles are in-frame or out-of-frame (Table 1). This phenomenon might be another mechanism for causing the inactivity of tumor suppressor proteins, contributing to carcinogenesis of breast cells.
Table 1.
Fusion Gene Transcripts involving Tumor Suppressor Genes in Breast Cancer.
| Gene_A | Gene_B | TSG | A_chr | B_chr | Junction_A | Junction_B | Frame | FusionPair |
|---|---|---|---|---|---|---|---|---|
| TSC2 | TMEM204 | TSC2 | 16 | 16 | 2098754 | 1604783 | Out-of-frame | TSC2__TMEM204 |
| ARID1A | THBS3 | ARID1A | 1 | 1 | 27059283 | 155172928 | In-frame | ARID1A__THBS3 |
| ARID1A | YTHDF2 | ARID1A | 1 | 1 | 27059283 | 29064170 | In-frame | ARID1A__YTHDF2 |
| AXIN1 | ITFG3 | AXIN1 | 16 | 16 | 402369 | 299548 | 5UTR-5UTR | AXIN1__ITFG3 |
| AXIN1 | ITCH | AXIN1 | 16 | 20 | 396148 | 32957200 | CDS-5UTR | AXIN1__ITCH |
| AXIN1 | IFT140 | AXIN1 | 16 | 16 | 354304 | 1576797 | Out-of-frame | AXIN1__IFT140 |
| CASP8 | ALS2CR12 | CASP8 | 2 | 2 | 202131514 | 202195556 | In-frame | CASP8__ALS2CR12 |
| CDH1 | CCDC132 | CDH1 | 16 | 7 | 68857529 | 92952923 | In-frame | CDH1__CCDC132 |
| CDH1 | NPFFR2 | CDH1 | 16 | 4 | 68772314 | 73003757 | In-frame | CDH1__NPFFR2 |
| CDH1 | DEXI | CDH1 | 16 | 16 | 68842751 | 11023416 | CDS-3UTR | CDH1__DEXI |
| CCDC132 | CDH1 | CDH1 | 7 | 16 | 92940584 | 68862077 | In-frame | CCDC132__CDH1 |
| ASXL1 | TM9SF4 | ASXL1 | 20 | 20 | 30956926 | 30720816 | CDS-5UTR | ASXL1__TM9SF4 |
| ASXL1 | PDRG1 | ASXL1 | 20 | 20 | 30959677 | 30536685 | 3UTR-CDS | ASXL1__PDRG1 |
5. Fusion Genes Involving Kinase Partner Genes in Breast Cancer
Among 1019 TCGA breast clinical cancer samples tested in the previous study, 94 (9.2%) harbored in-frame fusion genes conjoined involving protein kinase partner genes. A total of 105 in-frame kinase fusion genes were identified in the breast cancer samples and, of them, the druggable kinase family partner genes involved in those in-frame fusions are RET, NTRK3, FGFR1 and FGFR2. The kinase gene partners involved in those in-frame fusion genes belong to the following kinase gene families: CAMK Ser/Thr protein kinase family (OBSCN-IGSF21, PHF21A-CAMK1D, TRIO-MARCH11, PIM3-CRELD2 and SPG11-CAMK2G), STE Ser/Thr protein kinase family (TAOK1-ACER3 and POLA1-MAP3K15), AGC Ser/Thr protein kinase family (CDC42BPB-KIAA1409, ADRBK2-SF3A1, PRKCA-CRYBA1, MAST4-NLN, MAST2-NASP, ROCK1-LRIG3, RPS6KA6-HDX, PRKCA-CDH8, RPS6KB2-PAAF1, PKN2-COL24A1, ADRBK1-BAIAP2 and ADRBK1-IQGAP1), CMGC Ser/Thr protein kinase family (KHDC1-NLK, AHR-CDK14, PPP2CA-CDKL3, CDK12-FBXL20 and MAPK14-EFHA1), PI3/PI4-kinase family (ATR-DBR1, SMG1-ANO3 and PRKDC-CEP290), Tyr protein kinase family (TMEM87B-MERTK, SOX13-MERTK and ETV6-NTRK3), TKL Ser/Thr protein kinase family (KSR2-P2RX4, LRRK1-ADAMTS17, LIMK2-RUFY1 and TESK2-GPR37L1), ADCK protein kinase family (ADCK2-DPP6), and NEK Ser/Thr protein kinase family (NVL-NEK2). In contrast to most known kinase fusion cases, in which only one partner is a kinase gene, there is another kinase fusion (ERBB2-CDK12), in which both partners are kinase genes. The genomic regions easily vulnerable to the recurrent translocation breaks which generate those kinase fusion genes should be checked with priority given to screening potential candidate kinase oncogenic fusion targets in the coming years.
6. Fusions Involving Chromatin Modifier Genes in Breast Cancer
Among 1019 breast cancer samples tested by Yoshihara et al. in their study [6], 38 (3.7%) harbored in-frame fusion involving chromatin modifier genes. Of those chromatin modifier genes involved in the fusion genes, there are genes encoding histone demethylase (KDM5A-ANO2, USP21-PVRL4, WNK1-KDM5A, KDM5A-NINJ2, KDM4B-UHRF1, MYSM1-APOM, and KDM2A-RHOD), NURD complex components (CHD4-DTX1, XPC-RCOR1, and TRAF3-RCOR1), histone methyltransferases (NSD1-CDH23, CTPS2-RBBP7, CDH23-NSD1, MLL3-CLIP2, B4GALT7-NSD1, NSD1-ZNF346, SETDB1-PKP4, and SCMH1-EIF2B3), SWI/SNF family members (SMARCA4-DNM2, TBC1D9-SMARCA5, ARID1A-THBS3, and ARID1A-YTHDF2), methyl-CpG-binding proteins (MBD2-DOCK2), histone acetyltransferases (CREBBP-NUBP1, ARHGEF17-EP300, and CREBBP-ZNF500), histone deacetylases (HDAC1-KPNA6, and HDAC2-TRDN), methylcytosine hydroxylase (TET3-DGUOK, and TET3-C3ORF78), 5_3_ORF chromatin modifier (MANF-SETD2) and histone demethylase_5_3_ORF chromatin modifier (USP22-MYO19) (Table 2).
Table 2.
Fusion Gene Transcripts involving Chromatin Modifier Genes in Breast Cancer.
| FusionPair | Gene_A | Gene_B | A_chr | B_chr | Junction_A | Junction_B | Frame |
|---|---|---|---|---|---|---|---|
| KDM5A__ANO2 | KDM5A | ANO2 | 12 | 12 | 427272 | 5708795 | In-frame |
| CHD4__DTX1 | CHD4 | DTX1 | 12 | 12 | 6690210 | 113515229 | In-frame |
| CTPS2__RBBP7 | CTPS2 | RBBP7 | X | X | 16716359 | 16871965 | In-frame |
| USP21__PVRL4 | USP21 | PVRL4 | 1 | 1 | 161134040 | 161042675 | In-frame |
| MANF__SETD2 | MANF | SETD2 | 3 | 3 | 51423766 | 47108608 | In-frame |
| XPC__RCOR1 | XPC | RCOR1 | 3 | 14 | 14209757 | 103177272 | In-frame |
| WNK1__KDM5A | WNK1 | KDM5A | 12 | 12 | 939326 | 432947 | In-frame |
| NSD1__CDH23 | NSD1 | CDH23 | 5 | 10 | 176563031 | 73434869 | In-frame |
| CDH23__NSD1 | CDH23 | NSD1 | 10 | 5 | 73501678 | 176618885 | In-frame |
| SMARCA4__DNM2 | SMARCA4 | DNM2 | 19 | 19 | 11136184 | 10930656 | In-frame |
| KDM5A__NINJ2 | KDM5A | NINJ2 | 12 | 12 | 404739 | 675344 | In-frame |
| MLL3__CLIP2 | MLL3 | CLIP2 | 7 | 7 | 151871216 | 73811404 | In-frame |
| B4GALT7__NSD1 | B4GALT7 | NSD1 | 5 | 5 | 177035623 | 176618885 | In-frame |
| MBD2__DOCK2 | MBD2 | DOCK2 | 18 | 5 | 51715244 | 169502951 | In-frame |
| CREBBP__NUBP1 | CREBBP | NUBP1 | 16 | 16 | 3819175 | 10846467 | In-frame |
| ARHGEF17__EP300 | ARHGEF17 | EP300 | 11 | 22 | 73022875 | 41545042 | In-frame |
| HDAC1__KPNA6 | HDAC1 | KPNA6 | 1 | 1 | 32790154 | 32620189 | In-frame |
| TBC1D9__SMARCA5 | TBC1D9 | SMARCA5 | 4 | 4 | 141560240 | 144438508 | In-frame |
| TET3__DGUOK | TET3 | DGUOK | 2 | 2 | 74317174 | 74184252 | In-frame |
| USP22__MYO19 | USP22 | MYO19 | 17 | 17 | 20945978 | 34852250 | In-frame |
| KDM4B__UHRF1 | KDM4B | UHRF1 | 19 | 19 | 5144913 | 4929234 | In-frame |
| MYSM1__APOM | MYSM1 | APOM | 1 | 6 | 59160801 | 31624249 | In-frame |
| HDAC2__TRDN | HDAC2 | TRDN | 6 | 6 | 114292021 | 123545276 | In-frame |
| ARID1A__THBS3 | ARID1A | THBS3 | 1 | 1 | 27059283 | 155172928 | In-frame |
| TET3__C2orf78 | TET3 | C2orf78 | 2 | 2 | 74275538 | 74040604 | In-frame |
| NSD1__ZNF346 | NSD1 | ZNF346 | 5 | 5 | 176619020 | 176489059 | In-frame |
| ARID1A__YTHDF2 | ARID1A | YTHDF2 | 1 | 1 | 27059283 | 29064170 | In-frame |
| SETDB1__PKP4 | SETDB1 | PKP4 | 1 | 2 | 150915164 | 159477502 | In-frame |
| TRAF3__RCOR1 | TRAF3 | RCOR1 | 14 | 14 | 103342862 | 103173688 | In-frame |
| KDM2A__RHOD | KDM2A | RHOD | 11 | 11 | 66888829 | 66833367 | In-frame |
| SCMH1__EIF2B3 | SCMH1 | EIF2B3 | 1 | 1 | 41578955 | 45316675 | In-frame |
| CREBBP__ZNF500 | CREBBP | ZNF500 | 16 | 16 | 3929833 | 4803059 | In-frame |
An interesting thing to be mentioned here is the fusion event between ARHGEF17 and EP300. EP300—a histone acetyltransferase involved in the regulation of transcription via chromatin remodeling—has recently been revealed as a tumor suppressor modulating paclitaxel resistance and stemness against metastatic breast cancer [10]. Intriguingly, by analyzing protein domains of the EP300 region involved in the fusion event, we have found that, by omitting important functional domains (zf-TAZ and KIX) of EP300, the fusion ARHGEF17-EP300 might cause a deficiency of EP300 protein function, likely contributing to breast carcinogenesis and metastasis (Table 2).
Given the fact that epigenomic changes could exert significantly influential roles in malignant transformation, those fusion genes involving chromatin modifier partners should be good candidate targets for screening tumor suppressor or oncogenic fusion genes associated with breast carcinogenesis in the future.
7. Conjoined Genes in Breast Cancer
Conjoined genes, also called read-through fusion genes, have been reported in normal breast and breast cancer tissues by Kim et al. [1]. The authors discovered that those conjoined genes could be formed by a continuous read-through transcription from the upstream partner gene to the downstream partner gene due to a lack of poly-A signal sequence for stopping a transcription in the 3′UTR of the upstream partner gene. They also identified that the conjoined transcripts could exist in a variety of alternative splicing isoforms, among which some could have novel exons that could be formed by new exonization of certain lengths of sequences in the intergenic regions between the upstream and downstream fusion partner genes. Intriguingly, each isoform of the conjoined gene transcripts appeared to show a different tendency in expression level between breast cancer and normal tissues, suggesting that those conjoined gene transcript isoforms might play different roles between breast cancer and normal cells. Moreover, the authors showed that those conjoined genes could have been evolutionarily conserved between human and chimpanzee, suggesting that such conjoined gene formation could occur across a variety of species.
Similarly, Varley et al. found two recurrent read-through fusion transcripts, SCNN1A-TNFRSF1A and CTSD-IFITM10, that were present in breast cancer cell lines and estrogen receptor positive (ER+) breast primary tumor tissues, but not in normal breast tissues [11]. Both read-through fusion transcripts had in-frame open reading frames, whose translated proteins had been confirmed through Western blotting by the authors. They also demonstrated that breast cancer cell proliferation could be reduced by knock-downing CTSD-IFITM10 mRNA expression via targeting its fusion junction region with custom small interfering RNAs, suggesting that the conjoined genes might, in the future, be a diagnostic biomarker and therapeutic target for treating a breast cancer subtype harboring those fusion biomarkers.
An important thing to be addressed here is that not only those conjoined gene transcripts, but also numerous other conjoined gene transcripts could be formed through the read-through transcription between upstream and downstream genes, primarily depending upon the distance between them [12]. There may be numerous ways of forming fusions between neighboring genes, as shown in the fusion between the ESR1 gene and its neighboring gene CCDC170 by a tandem duplication [9]. Based upon this phenomenon, it is estimated that one third of all genes in the human genome might be involved in the formation of such conjoined genes with neighboring partner genes, even though the detection of all such conjoined genes is still difficult due to their extremely low expression level [12]. This fact implies that carcinogenesis might even be a far more complicated phenomenon than we imagined at present if a significant portion of such conjoined genes might be involved in the carcinogenesis. Understanding molecular mechanisms of how such conjoined genes could be implicated in the carcinogenesis may be an interesting research field in future years.
8. Fusion Gene Formation within Recurrent Genomic Copy Number Amplicon Region
Another important thing we should mention here is a very intriguing fact regarding the breast cancer fusion gene, ERBB2-GRB7, which is a fusion between genes located in a close neighborhood in the human genome. Even though the fusion gene had been identified in breast cancer patients, the copy number amplicon (the one harboring the independent ERBB2 and GRB7 genes) had previously been reported in other cancer types, including gastric cancer [13,14]. In addition, the authors demonstrated that the co-amplification of those genes in the amplicons could result in simultaneous up-regulation in their expression and also might contribute to carcinogenesis in an orchestrated, cooperative, and synergistic manner for their oncogenic activities.
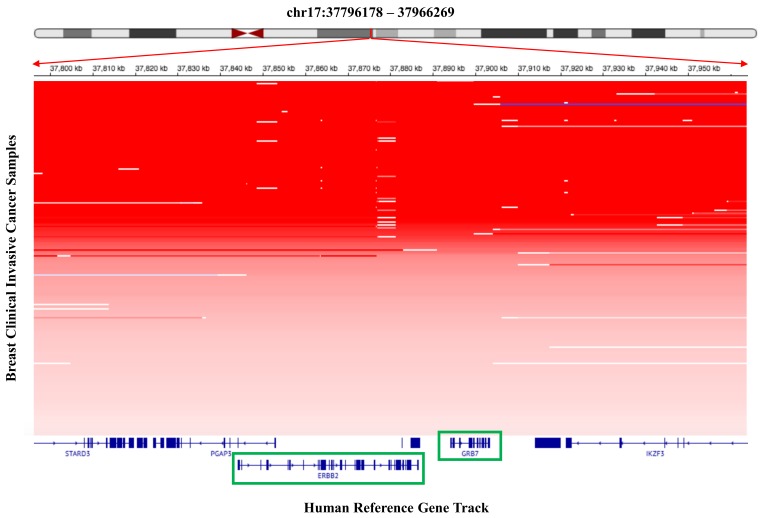
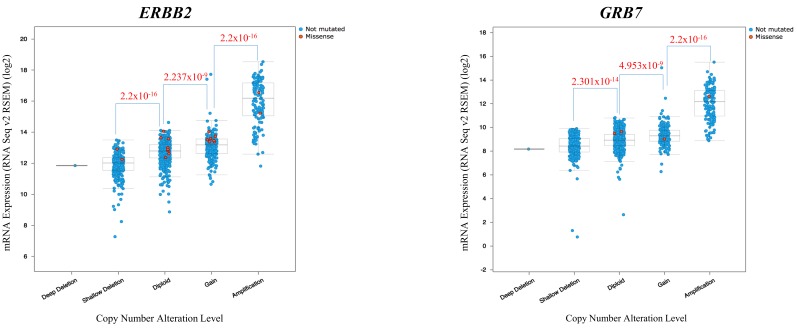
In order to consider whether this might be a relatively generalized phenomenon in other cancer types, we analyzed the copy number amplification status within the close genomic neighborhood for the well-known oncogene, ERBB2, in TCGA clinical breast cancer samples, and also the relationship between copy number alteration and expression for the genes located within the genomic neighborhood. As shown in Figure 4, nearly 18% of clinical breast invasive carcinoma samples in TCGA database had shown high amplification of the genomic region that harbored ERBB2, GRB7 and other neighboring genes. In addition, in those clinical breast invasive carcinoma samples with the amplified genomic region, there are statistically significant differences (p value calculations by Wilcoxon rank sum test) in mRNA expression level among different copy number alteration levels for ERBB2 and GRB7, respectively (Figure 5). The higher their copy numbers are, the higher their mRNA expression levels are, which is statistically significant. This phenomenon implies that either fusion or co-amplification for those genes might be nearly equally suitable structures for their orchestrated and cooperative contribution to carcinogenesis. In the upcoming years, we may see more surprising results in considering how such cooperation between genes located within other recurrently prevalent oncogenic amplicons is and what kinds of structural bridging between them might be suitable for consequently promoting carcinogenesis.
Figure 4.
Co-amplification of GRB7 within ERBB2 genomic amplicon. The top panel shows ERBB2 genomic amplicon region (indicated by a red color line) in a human chromosome ideogram. The two red arrowheads indicate an expanded ERBB2 genomic amplicon region (chr17:37796178-37966269) harboring several co-amplified genes, which is scaled by a nucleotide position interval ruler for the human chromosome 17. The middle panel shows overall copy number amplifications of the ERBB2 genomic amplicon involving several genes within the regions surrounding the ERBB2 locus, including GRB7, in TCGA breast clinical invasive carcinoma samples. The denser the red color is, the higher the degree of the copy number amplification in the genomic region is. The bottom panel shows genomic exon-intron structures of those genes residing within the ERBB2 amplicon.
Figure 5.
Relationships between copy number alteration and mRNA expression for ERBB2 and GRB7 in clinical breast invasive carcinoma samples. p-values were calculated by Wilcoxon rank sum test, except for only one breast carcinoma sample showing deep deletion of both ERBB2 and GRB7.
9. Clinical and Therapeutic Implication of Fusion Genes in Breast Cancers
Even though numerous fusion genes, which are vastly diverse in terms of fusion gene type and fusion structure, are nowadays being rapidly identified in clinical breast cancer samples, validation of their involvement in carcinogenesis and the establishment for their clinical and therapeutic application as useful biomarkers require time-consuming, big money-consuming, painstaking and long-term processes. Nevertheless, in recent years, there has been some convincing progress in this field, especially regarding breast cancer. ETV6-NTRK3, which had been identified as a causative fusion oncogene in over 90% of secretary breast cancer samples, encodes a dimerization domain of the ETV6 transcription factor, conjoined to kinase domain of NTRK3 [15]. Such in-frame fusion enables a constitutive ligand-independent dimerization and auto-phosphorylation, consequently causing an oncogenic transformation of mammary epithelial breast cells [15]. Upon administration, larotrectinib (NTRK1/2/3 inhibitor, LOXO-101) binds to Trk (tropomyosin receptor kinase), causing the prevention of the neurotrophin-Trk interaction and Trk activation, and consequently resulting in both the induction of cellular apoptosis and the inhibition of cell growth in tumors overexpressing Trk [16]. At present, protein kinase inhibitors, such as NTRK1/2/3 inhibitor, LOXO-101, are now being tested for the therapeutic treatment of patients with solid tumors harboring the fusion ETV6-NTRK3 in phase II clinical trials [16].
The recurrent conjoined gene ESR1-CCDC170 occurring in nearly 4% of ER-positive clinical breast cancer patients has been proven to be a causative fusion oncogene underlying breast carcinogenesis [17]. Efforts for identifying inhibitors for therapeutic intervention of this oncogenic fusion are still under investigation. As mentioned in the previous section, it has been substantiated that the proliferation of breast cancer cells harboring the two conjoined genes, SCNN1A-TNFRSF1A and CTSD-IFITM10, could be decreased by targeting them with the corresponding siRNAs. This demonstrates the possibility that siRNA-based therapeutic drugs are suitable to be applied to the clinical treatment of cancers harboring a variety of other oncogenic fusion gene types in the future.
10. Conclusions
All the fusion gene types mentioned above in this paper and all the genomic regions harboring their translocation breakpoints and conjoining points should be chosen as priority targets for screening fusion gene biomarkers representing each of the diverse and extremely heterogeneous breast cancer subtypes in the coming years. For instance, the genomic regions across the translocation breakpoints or conjoining points generating the recurrent fusion genes mentioned in this paper should be chosen for designing forward or reverse primers for RT-PCR (reverse transcriptase-polymerase chain reaction) test and also selected as candidate regions for targeted RNA sequencing. With the advent of a next generation deep sequencing era guaranteed by a decreasing cost, the relationship between conjoined genes and carcinogenesis will become increasingly transparent. Also, the application of the CRISPR-CAS9 system to the high-throughput genome-wide functional validation of those fusion and conjoined genes will make our discovery process of breast cancer fusion gene biomarkers accelerate excitingly far beyond our present imagination.
Acknowledgments
This study has been supported by a research grant (2017R1D1A1B04033856 (Ryong Nam Kim)) funded by the Ministry of Education and the National Research Foundation in Korea.
Author Contributions
Ryong Nam Kim had conceived this research process and analyzed the data, and had written the paper. Hyeong-Gon Moon, Wonshik Han, and Dong-Young Noh had participated in the above-mentioned processes.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Kim R.N., Kim A., Choi S.H., Kim D.S., Nam S.H., Kim D.W., Kim D.W., Kang A., Kim M.Y., Park K.H., et al. Novel mechanism of conjoined gene formation in the human genome. Funct. Integr. Genom. 2012;12:45–61. doi: 10.1007/s10142-011-0260-1. [DOI] [PubMed] [Google Scholar]
- 2.Lee M.S., Kim R.N., I H., Oh D.Y., Song J.Y., Noh K.W., Kim Y.J., Yang J.W., Lira M.E., Lee C.H., et al. Identification of a novel partner gene, KIAA1217, fused to RET: Functional characterization and inhibitor sensitivity of two isoforms in lung adenocarcinoma. Oncotarget. 2016;7:36101–36114. doi: 10.18632/oncotarget.9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim R.N., Choi Y.L., Lee M.S., Lira M.E., Mao M., Mann D., Stahl J., Licon A., Choi S.J., van Vrancken M., et al. SEC31A-ALK Fusion Gene in Lung Adenocarcinoma. Cancer Res. Treat. 2016;48:398–402. doi: 10.4143/crt.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi Y.L., Lira M.E., Hong M., Kim R.N., Choi S.J., Song J.Y., Pandy K., Mann D.L., Stahl J.A., Peckham H.E., et al. A novel fusion of TPR and ALK in lung adenocarcinoma. J. Thorac. Oncol. 2014;9:563–566. doi: 10.1097/JTO.0000000000000093. [DOI] [PubMed] [Google Scholar]
- 5.Hong M., Kim R.N., Song J.Y., Choi S.J., Oh E., Lira M.E., Mao M., Takeuchi K., Han J., Kim J., et al. HIP1-ALK, a novel fusion protein identified in lung adenocarcinoma. J. Thorac. Oncol. 2014;9:419–422. doi: 10.1097/JTO.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 6.Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burman B., Zhang Z.Z., Pegoraro G., Lieb J.D., Misteli T. Histone modifications predispose genome regions to breakage and translocation. Genes Dev. 2015;29:402–1393. doi: 10.1101/gad.262170.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thurman R.E., Rynes E., Humbert R., Vierstra J., Maurano M.T., Haugen E., Sheffield N.C., Stergachis A.B., Wang H., Vernot B., et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veeraraghavan J., Tan Y., Cao X.X., Kim J.A., Wang X., Chamness G.C., Maiti S.N., Cooper L.J., Edwards D.P., Contreras A., et al. Recurrent ESR1-CCDC170 rearrangements in an aggressive subset of oestrogen receptor-positive breast cancers. Nat. Commun. 2014;5:4577. doi: 10.1038/ncomms5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asaduzzaman M., Constantinou S., Min H.X., Gallon J., Lin M.L., Singh P., Raguz S., Ali S., Shousha S., Coombes R.C., et al. Tumour suppressor EP300, a modulator of paclitaxel resistance and stemness, is downregulated in metaplastic breast cancer. Breast Cancer Res. Treat. 2017;163:461–474. doi: 10.1007/s10549-017-4202-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varley K.E., Gertz J., Roberts B.S., Davis N.S., Bowling K.M., Kirby M.K., Nesmith A.S., Oliver P.G., Grizzle W.E., Forero A., et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res. Treat. 2014;146:287–297. doi: 10.1007/s10549-014-3019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prakash T., Sharma V.K., Adati N., Ozawa R., Kumar N., Nishida Y., Fujikake T., Takeda T., Taylor T.D. Expression of Conjoined Genes: Another Mechanism for Gene Regulation in Eukaryotes. PLoS ONE. 2010;5:e13284. doi: 10.1371/journal.pone.0013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross J.S., Wang K., Sheehan C.E., Boguniewicz A.B., Otto G., Downing S.R., Sun J., He J., Curran J.A., Ali S., et al. Relapsed Classic E-Cadherin (CDH1)-Mutated Invasive Lobular Breast Cancer Shows a High Frequency of HER2 (ERBB2) Gene Mutations. Clin. Cancer Res. 2013;19:2668–2676. doi: 10.1158/1078-0432.CCR-13-0295. [DOI] [PubMed] [Google Scholar]
- 14.Kwon M.J., Kim R.N., Song K., Jeon S., Jeong H.M., Kim J.S., Han J., Hong S., Oh E., Choi J.S., et al. Genes co-amplified with ERBB2 or MET as novel potential cancer-promoting genes in gastric cancer. Oncotarget. 2017;8:92209–92226. doi: 10.18632/oncotarget.21150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tognon C., Knezevich S.R., Huntsman D., Roskelley C.D., Melnyk N., Mathers J.A., Becker L., Carneiro F., MacPherson N., Horsman D., et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2:367–376. doi: 10.1016/S1535-6108(02)00180-0. [DOI] [PubMed] [Google Scholar]
- 16.Doebele R.C., Davis L.E., Vaishnavi A., Le A.T., Estrada-Bernal A., Keysar S., Jimeno A., Varella-Garcia M., Aisner D.L., Li Y.L., et al. An Oncogenic NTRK Fusion in a Patient with Soft-Tissue Sarcoma with Response to the Tropomyosin-Related Kinase Inhibitor LOXO-101. Cancer Discov. 2015;5:1049–1057. doi: 10.1158/2159-8290.CD-15-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paratala B.S., Dolfi S.C., Khiabanian H., Rodriguez-Rodriguez L., Ganesan S., Hirshfield K.M. Emerging Role of Genomic Rearrangements in Breast Cancer: Applying Knowledge from Other Cancers. Biomark. Cancer. 2016;8:1–14. doi: 10.4137/BIC.S34417. [DOI] [PMC free article] [PubMed] [Google Scholar]