Abstract
Removal of introns from precursor messenger RNA (pre-mRNA) and some noncoding transcripts is an essential step in eukaryotic gene expression. In the nucleus, this process of RNA splicing is carried out by the spliceosome, a multi-megaDalton macromolecular machine whose core components are conserved from yeast to humans. In addition to many proteins, the spliceosome contains five uridine-rich small nuclear RNAs (snRNAs) that undergo an elaborate series of conformational changes to correctly recognize the splice sites and catalyze intron removal. Decades of biochemical and genetic data, along with recent cryo-EM structures, unequivocally demonstrate that U6 snRNA forms much of the catalytic core of the spliceosome and is highly dynamic, interacting with three snRNAs, the pre-mRNA substrate, and >25 protein partners throughout the splicing cycle. This review summarizes the current state of knowledge on how U6 snRNA is synthesized, modified, incorporated into snRNPs and spliceosomes, recycled, and degraded.
Keywords: U6 snRNA, U6 gene transcription, spliceosome, U6 snRNP
U6 snRNA IS A KEY COMPONENT OF THE SPLICEOSOME
U6 snRNA was discovered by virtue of its immunoprecipitation from mouse cell nuclear extract, along with U1, U2, U4, and U5 snRNAs, by anti-Sm patient serum (Lerner and Steitz 1979), and U6 from rat cells was subsequently sequenced (Epstein et al. 1980). It was later found that human U6 snRNA is stably base-paired with U4 snRNA (Bringmann et al. 1984; Hashimoto and Steitz 1984). Using human cell nuclear extract competent for pre-mRNA splicing, U4 and U6 snRNAs were found to be associated with affinity-purified spliceosomes (Grabowski and Sharp 1986), and oligo ablation studies revealed that both snRNAs are required for splicing (Black and Steitz 1986). Soon thereafter, U4 and U6 were found to be highly conserved and essential in yeast (Siliciano et al. 1987; Brow and Guthrie 1988), and both snRNAs were identified in yeast spliceosomes (Cheng and Abelson 1987), implying a central function in splicing.
The spliceosome is an intricate macromolecular machine that consists of five snRNAs, U1, U2, U4, U5, and U6, and their associated proteins (small nuclear ribonucleoprotein complexes or snRNPs), as well as a protein-only NineTeen Complex (NTC) and a number of accessory proteins (Wahl et al. 2009; Will and Lührmann 2011). A small percentage of human introns, as well as those of certain other metazoan and protozoan taxa, have noncanonical splice sites and are recognized and removed by the minor “U12-type” spliceosome. The minor spliceosome shares U5 snRNA with the major spliceosome, but contains the U11, U12, U4atac, and U6atac snRNAs, which replace U1, U2, U4, and U6 snRNAs, respectively (Tarn and Steitz 1996a,b). Both the major and minor spliceosomes assemble de novo on each intron and, after splicing, dissociate into their component snRNPs and accessory proteins. The dynamic nature of the spliceosome makes its study challenging.
U6 snRNA sits at the heart of the spliceosome, where it coordinates the magnesium ions required for splicing chemistry and, with help from U2 and U5 snRNAs, positions the substrate for the splicing reaction. The pre-mRNA splicing reaction consists of two transesterification steps and closely resembles the reaction carried out by group II self-splicing introns. First, the 2′-oxygen of a nucleotide near the 3′ end of the intron (the branch site, most often an adenosine) is activated for nucleophilic attack on the 5′ exon–intron junction (the 5′ splice site) to form a branched “lariat” intron–3′ exon intermediate. Subsequently, nucleophilic attack of the released 5′ exon on the intron–3′ exon junction (the 3′ splice site) produces a ligated 5′ exon–3′ exon product and free lariat intron (Wahl et al. 2009; Hoskins and Moore 2012; Shi 2017).
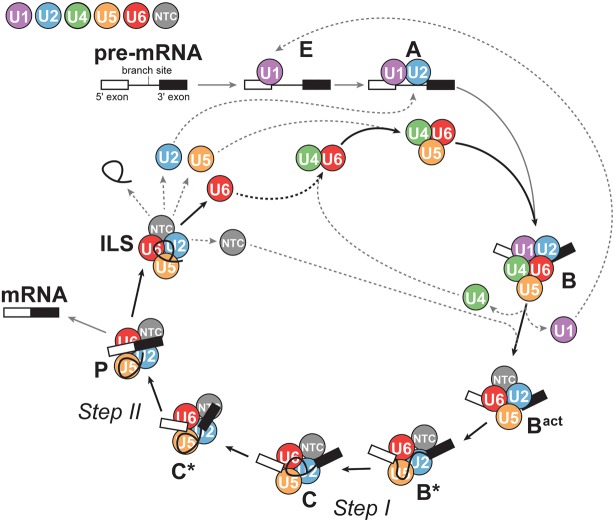
Early steps in recognition of the intron include base-pairing of U1 snRNA in the U1 snRNP to the 5′ splice site and U2 snRNA in the U2 snRNP to the branch site, forming the A complex (also known as the prespliceosome). The A complex is joined by a preassembled U4/U6.U5 triple (tri)-snRNP to form the B complex (Fig. 1). In tri-snRNP nomenclature, the slash indicates base-pairing between U4 and U6 snRNAs and the period conveys that U5 snRNP associates with the U4/U6 di-snRNP through protein–protein and RNA–protein interactions. During activation of the assembled spliceosome for the first catalytic step, large-scale conformational rearrangements cause U1 and U4 snRNPs to be ejected, leaving the U2/U6.U5 complex together with the NTC (Bact) and accessory factors to catalyze the chemical steps (Will and Lührmann 2011). U6 must undergo several RNA base-pairing exchanges in this transition: disruption of U4/U6 in favor of a mutually exclusive interaction with U2, and base-pairing to the 5′SS. The timing of these events and the interactions controlling activation are still being elucidated (Shcherbakova et al. 2013; Hoskins et al. 2016).
FIGURE 1.
U6 snRNA is a core component of the spliceosome. The spliceosome is composed of small nuclear ribonucleoprotein particles (snRNPs) and a protein-only complex called the NTC, which are represented as colored circles. Additional transiently bound proteins (not shown) are also necessary for progression through the splicing cycle. The U1, U2, U4, U5, and U6 snRNPs consist of the small nuclear RNA (snRNA) for which they are named and associated proteins. The snRNPs and NTC undergo ordered assembly on the pre-mRNA and experience both conformational and compositional changes throughout the cycle. After splicing is complete, the snRNPs and NTC are released and reused (dotted lines).
Recent cryo-EM structures of spliceosomal complexes, including the U4/U6.U5 tri-snRNP (Agafonov et al. 2016; Nguyen et al. 2016; Wan et al. 2016b), the precatalytic B complex (Bertram et al. 2017a; Plaschka et al. 2017), and the catalytic U2/U6.U5 spliceosome at different stages of splicing (Yan et al. 2015, 2016, 2017; Galej et al. 2016; Rauhut et al. 2016; Wan et al. 2016a, 2017; Bertram et al. 2017b; Fica et al. 2017; Zhang et al. 2017), have provided near-atomic level views of the extensive structural rearrangements that must take place during splicing. After each round of splicing, the snRNPs and NTC dissociate from the intron and are used again. U6, however, is released as a free RNA and so must bind its own snRNP proteins again before assembling with the U4 and U5 snRNPs to reconstitute the tri-snRNP (Fourmann et al. 2013). The splicing cycle is described in greater detail in recent reviews (Will and Lührmann 2011; Hoskins and Moore 2012).
A role for U6 in the catalytic center of the spliceosome had long been proposed based on a number of lines of evidence. The discovery that group II self-splicing introns use the same two-step chemical mechanism as the spliceosome (Peebles et al. 1986; van der Veen et al. 1986) suggested that the two enzymes share catalytic RNA elements (Sharp 1985; Cech 1986; Madhani and Guthrie 1992). The high degree of conservation in the U6 snRNA sequence (Brow and Guthrie 1988) and the presence of introns in the most conserved region of U6 in some organisms (Tani and Ohshima 1989, 1991), potentially due to reverse splicing, hinted at the proximity of U6 to the active site of the spliceosome (Brow and Guthrie 1989). Subsequent crosslinking and genetic studies indicated that the strictly conserved “ACAGA-box” sequence of U6 base pairs to the intron 5′ splice site in the catalytically active spliceosome (Sawa and Abelson 1992; Sawa and Shimura 1992; Wassarman and Steitz 1992; Kandels-Lewis and Séraphin 1993; Lesser and Guthrie 1993; Sontheimer and Steitz 1993), and mutational studies identified functional similarities between Domain V of group II introns and the U2/U6 base-paired region (Madhani and Guthrie 1992; Chanfreau and Jacquier 1994; Peebles et al. 1995; Shukla and Padgett 2002; Mefford and Staley 2009). Elegant biochemical experiments confirmed that U6 snRNA is responsible for coordinating catalytic metal ions essential for both transesterification steps (Yean et al. 2000; Fica et al. 2013).
At just over 100 nt in length, U6 snRNA is highly economical, as nearly every part of the RNA possesses one or more essential functions. In addition to its catalytic role at the heart of the spliceosome, U6 snRNA is notable for undergoing extensive structural rearrangements, including unwinding and reformation of stable internal secondary structure, and for directly interacting with >25 proteins during a single round of splicing. Herein, we summarize the entire life cycle of the U6 snRNA (Fig. 2), and highlight differences found among the eukaryotes. Commonalities in the U6 snRNA life cycle illustrate conservation across more than a billion years of eukaryotic evolution. We focus primarily on U6 snRNA from the best-studied organisms: yeast (S. cerevisiae, and to a lesser extent, S. pombe) and human.
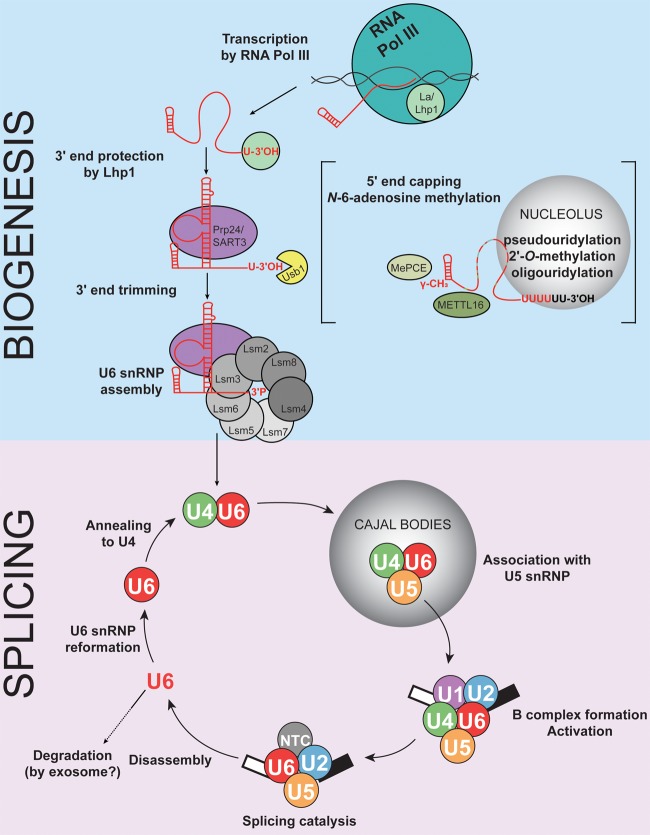
FIGURE 2.
Summary of the U6 lifecycle. Key steps in U6 snRNA biogenesis and assembly (top) and incorporation into the splicing cycle (bottom) are conserved in eukaryotes. Several additional modification steps, represented in brackets, occur in S. pombe and humans.
U6 snRNA GENE TRANSCRIPTION
Number of U6 genes
S. cerevisiae has a single genomic locus for U6 snRNA, the SNR6 gene on chromosome XII (Brow and Guthrie 1988). However, a survey of 145 fungal genomes identified species with up to 20 U6 gene copies and an average of 2.3 per genome (Canzler et al. 2016). In contrast, there are >900 copies of U6 distributed throughout the human genome, although the majority of these are likely pseudogenes and not transcriptionally active (Doucet et al. 2015). At least four human U6 genes encoding identical RNAs are transcriptionally active to various degrees (Domitrovich and Kunkel 2003). Additionally, a variant of human U6 snRNA with nine substitutions and one nucleotide deletion is expressed under the control of an internal promoter, unlike other transcriptionally active human U6 genes (Tichelaar et al. 1994, 1998). The presence of multiple U6 genes of varying transcriptional activity has complicated their individual study, and whether paralogous but divergent U6 snRNAs exhibit differences in modification, localization or function is poorly understood. The U6atac RNA is a paralog of U6 that functions in the minor spliceosome and is even further diverged in sequence from the other transcribed U6 snRNAs (Tarn and Steitz 1996a).
Transcription of U6 genes by RNA polymerase III
Unlike the other spliceosomal snRNAs, which are synthesized by RNA polymerase II (Pol II), U6 is synthesized by RNA polymerase III (Pol III) (Reddy et al. 1987; Moenne et al. 1990). While the sequence of U6 snRNA is highly conserved between yeast and humans, its Pol III promoter structure is divergent. In yeast, the U6 promoter region is similar to tRNA gene promoters (Eschenlauer et al. 1993) in that it contains A and B block elements (Brow and Guthrie 1990), as well as a TATA box that is bound by TATA-binding protein (TBP) (Fig. 3A; Margottin et al. 1991). In S. cerevisiae, the B block is located downstream from the U6 coding region. Consequently, the distance between the A and B blocks is atypical (∼200 bp vs. the normal 30–90 bp in tRNA genes), and this extended spacing is important for transcription in vivo (Kaiser et al. 2004).
FIGURE 3.
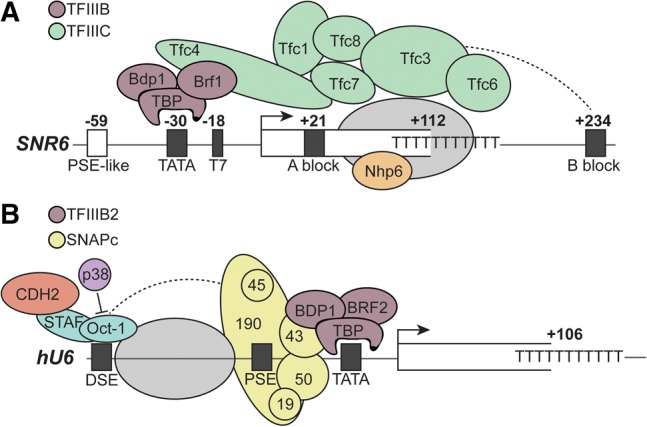
U6 snRNA gene promoter structure is divergent in eukaryotes. (A) U6 gene promoter structure in S. cerevisiae. The U6 gene is under the control of a Pol III Type II promoter, with an upstream TATA box, an internal A block, and downstream B block. The TFIIIC complex recognizes the A and B blocks and directs binding of the TFIIIB complex to the TATA box. Nhp6 also promotes transcription, but its binding site is uncertain. A possible nucleosome is indicated by a gray oval. (B) U6 promoter structure in humans. U6 synthesis is under the control of a Pol III Type III promoter, where promoter elements (in black) are exclusively upstream of the transcription start site. The TATA box is recognized by the TFIIIB2 complex, while the PSE is recognized by the SNAPc complex. Factors including OCT1, STAF, and CDH2 interact with the DSE. p38 inhibits Oct-1 binding to the DSE. A nucleosome between the DSE and PSE enhances Oct-1 and SNAPc binding.
The B block is essential for transcription of the yeast U6 gene in vivo and is the binding site for transcription initiation factor TFIIIC, which also contacts the A block (Burnol et al. 1993; Eschenlauer et al. 1993; Kaiser and Brow 1995). TFIIIC recruits TFIIIB (composed of TBP, Bdp1, and Brf1), which binds over the TATA box and in turn recruits Pol III (Gerlach et al. 1995). Transcription is promoted by but not dependent on the presence of the TATA box and a stretch of thymidines just downstream from it (Burnol et al. 1993; Eschenlauer et al. 1993; Martin et al. 2001).
The nonhistone chromatin protein Nhp6 is also important for transcription of SNR6 (Kruppa et al. 2001; Martin et al. 2001). In a heterologous, in vitro chromatin assembly system, a nucleosome positioned between the A and B blocks brings the regions close together for optimal binding by TFIIIC (Shivaswamy et al. 2004), but the micrococcal nuclease footprint of native chromatin assembled between the A and B blocks in vivo is shorter than expected for an intact nucleosome (Gerlach et al. 1995). It is possible that Nhp6 modifies the structure of a nucleosome bound to SNR6 (Fig. 3A; Stillman 2010). Alternatively, Nhp6 may favor a bent conformation of DNA that promotes TFIIIB binding (Braglia et al. 2007).
U6 gene promoter structure in fungi is flexible and can include or exclude identifiable TATA boxes, intragenic A blocks, and downstream B blocks (Canzler et al. 2016). S. pombe has a similar promoter structure to S. cerevisiae, with the exception that the B block element is located intragenically in an intron (Frendewey et al. 1990).
In humans, U6 RNA is transcribed from a Pol III Type III promoter, a promoter architecture that also drives 7SK RNA and RNase P RNA transcription. Most characterized mammalian U6 genes do not contain intragenic promoter elements, but rather exclusively contain upstream promoter elements (Das et al. 1988). These U6 genes have a TATA box 30 base pairs upstream of the transcription start site, a proximal sequence element (PSE) 50 base pairs upstream, and a distal sequence element (DSE; also called an OCT site) 250 base pairs upstream, which closely resembles the promoter structure for Pol II-transcribed genes (Fig. 3B). Interestingly, the PSE and DSE are similar to, and can be switched with, corresponding elements from the Pol II-synthesized snRNAs (Kunkel and Pederson 1988), and it is inclusion of the TATA box that specifies Pol III rather than Pol II transcription for snRNAs (Lobo and Hernandez 1989). Other organisms have divergent methods of specifying Pol II vs III transcription, such as the sequence of the PSE or the distance between the PSE and the TATA box (for review, see Hernandez 2001).
The PSE is required for basal transcription while the DSE is required for efficient transcription. The PSE is bound by the small nuclear RNA activating protein complex (SNAPc), which also binds the promoters of Pol II-transcribed snRNAs. The DSE is bound by the Oct-1 and Staf proteins (Fig. 3B). Cooperative binding of Oct-1 and SNAPc is enhanced by a nucleosome positioned between the DSE and PSE (Zhao et al. 2001). Oct-1 binding is negatively regulated by p38 kinase (Lin and Natarajan 2012), while Staf binding is influenced by chromatin-modifying enzymes (Yuan et al. 2007). The TATA box is bound by TFIIIB2, composed of TBP, Bdp1, and Brf2. TFIIIC is not required as there is no A or B block. Interestingly, yeast also possess an upstream PSE-like sequence that is not required for transcription (Eschenlauer et al. 1993) and an upstream positioned nucleosome (Arimbasseri and Bhargava 2008), suggesting that while some similarities remain, the promoter structure in yeast and humans has diverged significantly.
Chromatin immunoprecipitation detects very low levels of Pol II over the S. cerevisiae U6 gene, similar to that detected over the Pol II-silenced rDNA and telomeres (Steinmetz et al. 2006). Furthermore, a hypomorphic mutation in the Sen1 helicase increases Pol II levels at all these loci (Steinmetz et al. 2006), and an anti-sense transcript of the U6 gene contains a high-affinity Nrd1 binding site that promotes Sen1-dependent Pol II termination (Steinmetz and Brow 1998). Thus, like the rDNA and telomeres (Vasiljeva et al. 2008), the S. cerevisiae U6 gene may be silenced for Pol II by an unknown mechanism coupled to Sen1-dependent termination of an anti-sense transcript.
Interestingly, in humans, transcription of U6 is dependent upon the interaction of Pol II at a site ∼300 bp upstream of the gene, a phenomenon shown to be generally true for Pol III-transcribed genes (Listerman et al. 2007; Barski et al. 2010; Oler et al. 2010). This may result from the influence of chromatin remodeling through recruitment of Pol II transcription factors that are also used in Pol III transcription (Raha et al. 2010). Transcription of U6atac is also dependent on both Pol II and Pol III (Younis et al. 2013). Thus, there is a complex interplay of Pol II and Pol III at U6 genes in a variety of organisms.
Transcription termination of U6 is caused by a stretch of dA's in the template strand at the end of the gene, although the number of dA's for efficient termination vary in eukaryotes (Arimbasseri et al. 2013). The La protein (Lhp1 in yeast) binds the 3′ end of newly transcribed U6 RNAs (Rinke and Steitz 1985; Pannone et al. 1998) and has been implicated in transcription termination, RNA polymerase recycling, and transcription reinitiation (Gottlieb and Steitz 1989; Maraia et al. 1994; Maraia 1996; French et al. 2008). However, Pol III transcription in vitro is terminated efficiently without the help of additional termination factors. Weak base-pairing between the nascent oligo(rU) and the template strand oligo(dA) stretch, as well as interactions between the nontemplate strand oligo(dT) stretch and a subunit of Pol III, terminate transcription (Bogenhagen and Brown 1981; Hamada et al. 2000; Arimbasseri and Maraia 2015). Yeast U6 is terminated in a stretch of ten dAs, leaving an RNA product with a heterogeneous U-tail length consisting of 4–7 uridines (Brow and Guthrie 1990).
How U6 is transcriptionally regulated during the cell cycle, development, in different tissues, and in disease states has been poorly studied. It has been noted that tissue-specific differences of U6 expression levels exist (Spaniel et al. 2013). Whether this difference is due to tissue-specific stability or transcriptional activity is not clear (Spaniel et al. 2013). Tissue-specific differences in snRNP distribution have also been reported (Hamm and Mattaj 1989). Transcriptional activity represents the first step of regulation in the lifecycle of U6, yet little is known about the cellular regulation of U6 snRNA level and its effect on splicing efficiency or fidelity.
LOCALIZATION DURING BIOGENESIS
It is generally thought that U6 remains in the nucleus after transcription and for all biogenesis steps (Vankan et al. 1990; Pessa et al. 2008), while the Pol II-synthesized snRNAs have a cytoplasmic snRNP assembly step (for review, see Matera and Wang 2014). At some point after transcription termination, the La/Lhp1 protein is displaced from the 3′ end of U6 snRNA. Binding by La/Lhp1 is not sufficient for complete nuclear retention in Xenopus oocytes (Boelens et al. 1995). Instead, binding of the Lsm2-8 heteroheptameric protein ring acts as the primary nuclear retention signal (Spiller et al. 2007a,b).
U6 is modified (pseudouridylated and 2′-O-methylated) in the nucleolus by snoRNPs (Tycowski et al. 1998; Ganot et al. 1999; Lange and Gerbi 2000), while the Poll II-synthesized snRNAs are modified in Cajal bodies by scaRNPs (Darzacq et al. 2002). U6 is not retained in the nucleolus, but rather transiently passes through to obtain modifications (Lange and Gerbi 2000). Nucleolar localization of Xenopus U6 does not depend on base-pairing interactions with U4 or U2, or formation of the U6 5′SL, but both nucleolar localization and Cajal body localization do depend on the 3′ oligo(U) tail (Gerbi and Lange 2002). Furthermore, localization does not depend on post-transcriptional modifications such as pseudouridylation or 2′-O-methylation. In S. cerevisiae, U6 is not constitutively pseudouridylated and is not 2′-O-methylated, and therefore is thought to be entirely nucleoplasmic (Bertrand et al. 1998).
The presence of snRNPs in subnuclear organelles was first identified in the early 1990s (Carmo-Fonseca et al. 1991a,b; Matera and Ward 1993). Work over the past decade has revealed that U6 can be assembled into the U4/U6 di-snRNP and U4/U6.U5 tri-snRNP in subnuclear compartments called Cajal bodies (CBs) (Staněk et al. 2003; Staněk and Neugebauer 2004; Klingauf et al. 2006; Novotńy et al. 2011, 2015). CBs are also the compartment in which the Sm class snRNAs are post-transcriptionally modified by scaRNPs (Jády et al. 2003). In humans, U6 snRNA is localized to CBs through its interaction with the U6 snRNP protein SART3 (aka hPrp24, p110, or Tip110) (Staněk et al. 2003). The N-terminal half-a-tetratricopeptide (HAT) domains of SART3 are necessary for U6 localization to CBs (Staněk et al. 2003). Localization of SART3 depends upon its interaction with coilin, the major structural CB protein (Xu et al. 2005). Interestingly, U4/U6•SART3, rather than U6•SART3, accumulates in CBs, suggesting that CBs may be a site for snRNP assembly (Staněk and Neugebauer 2004). While formation of U4/U6 is enhanced ∼10-fold in CBs, in part due to the increased local concentration of snRNAs (Klingauf et al. 2006; Novotńy et al. 2011), snRNP formation can also occur elsewhere in the nucleus as CBs are nonessential (Lemm et al. 2006). SART3 localizes within CBs transiently (on the order of a few seconds) (Dundr et al. 2004) and re-enters new CBs after a period in the nucleoplasm (Staněk et al. 2008), suggesting that SART3 (and U6 along with it) localizes to CBs during both initial biogenesis and recycling after a round of splicing. Depletion of factors needed for tri-snRNP formation (hPrp6 and hPrp8) induces formation of CBs and results in accumulation of U4/U6 in CBs (Novotńy et al. 2011, 2015). Because SART3 interacts with U6 and U4/U6 but is displaced upon formation of U4/U6.U5 (Bell et al. 2002), it can anchor immature U4/U6 di-snRNPs to CBs through its interaction with coilin (Novotńy et al. 2015). Localization of free U6 snRNP in subnuclear CBs may prevent incorporation of immature splicing components into active spliceosomes. Retention of U6 snRNP in CBs is a convenient checkpoint for the cell to ensure that mature U6 (in the tri-snRNP) is used in splicing.
In yeast, the U6 gene is localized near the nucleolar periphery (Belagal et al. 2016). Although yeast also have nucleolar bodies akin to CBs (Verheggen et al. 2002), little work has been done to determine if and how subnuclear localization plays a role in U6 snRNP biogenesis in yeast. By analogy, work on the biogenesis of the U3 snoRNP has demonstrated that there are differences in localization during biogenesis in yeast and humans, but that overall the processes are similar (Verheggen et al. 2002). It remains to be determined if and why yeast U6 localizes to any subnuclear compartments, and if the U4/U6.U5 tri-snRNP is assembled in a specific region in yeast nuclei.
POST-TRANSCRIPTIONAL MODIFICATION
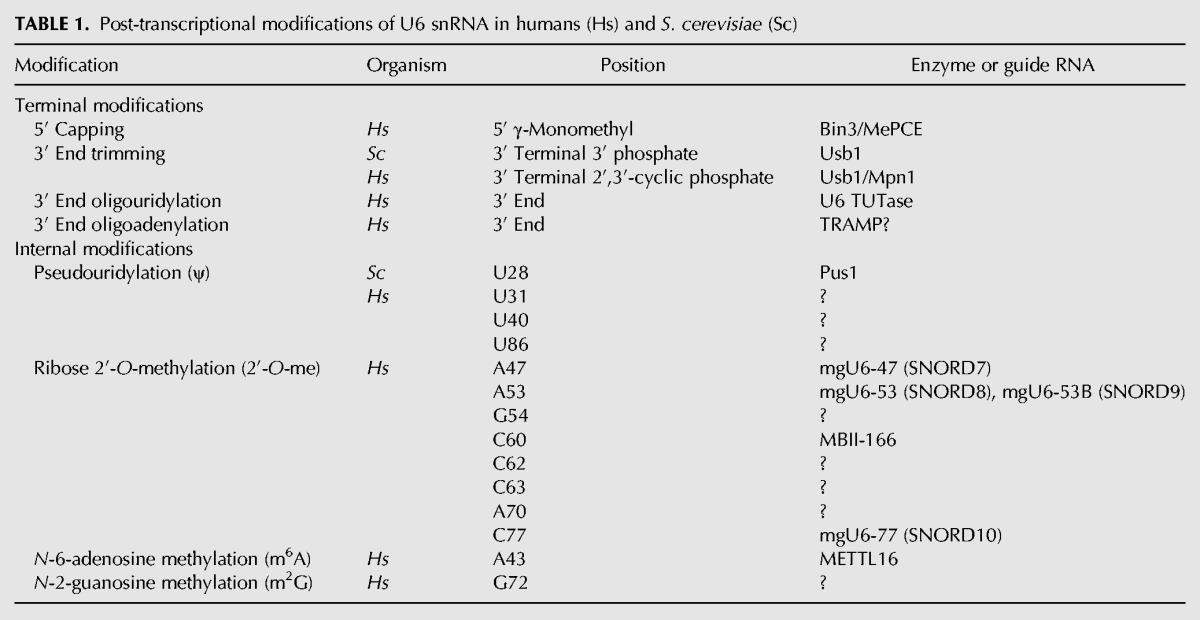
U6 snRNA is extensively modified in metazoans, and many of these post-transcriptional modifications are evolutionarily conserved even in S. pombe. In contrast, S. cerevisiae U6 snRNA contains very few modifications, and only one known obligate modification (3′ end processing). The known post-transcriptional modifications of U6 in S. cerevisiae and humans are summarized in Table 1 and discussed below. Post-transcriptional modifications of U6 are likely to be involved in modulation of U6 interactions with other RNAs and proteins throughout the splicing cycle. However, the precise effect that modifications have on U6 RNA structure or interactions with protein and other snRNAs is poorly understood, as is the timing and cellular location of where these modifications occur.
TABLE 1.
Post-transcriptional modifications of U6 snRNA in humans (Hs) and S. cerevisiae (Sc)
5′ Capping
Instead of the 2,2,7-trimethylguanosine (TMG) cap that the other snRNAs possess or the 7-methylguanosine (m7G/cap-0) cap of mRNAs, human U6 possesses a γ-monomethyl phosphate 5′ modification (Singh and Reddy 1989). The γ-monomethyl cap is shared by several other Pol III transcripts, including the noncoding 7SK RNA (Gupta et al. 1990a). However, capping of U6 small nuclear RNA in vitro is not dependent upon transcription, suggesting that capping of U6 is not obligatorily cotranscriptional in vivo (Gupta et al. 1990b). Capping is dependent upon the 5′ stem loop and is also sequence-dependent (Singh et al. 1990), unlike cotranscriptional capping of Pol II transcripts. It is not known if S. cerevisiae U6 has a γ-monomethyl cap, although deletion of its first 11 nt results in installation of a TMG cap despite still being transcribed by RNA Pol III (Kwan et al. 2000). This finding implies that yeast U6 normally receives a 5′ modification that prevents TMG capping, but that disruption of conserved sequences at the base of the 5′-stem, which are the determinant for γ-monomethyl capping in humans (Singh et al. 1990), blocks this modification.
The function of the U6 cap is not known, although it may be important for stability (Shumyatsky et al. 1993). TMG capping of other snRNAs is important for transport into the nucleus, but the γ-monomethyl cap of U6 likely does not impact transport (Shumyatsky et al. 1993) or retention in the nucleus (Spiller et al. 2007a). The γ-monomethyl cap could also play a role in displacing La/Lhp1 from U6 RNA, as La interacts with the 5′ triphosphate prior to capping (Bhattacharya et al. 2002).
A 130 kDa protein responsible for cap formation in U6 and 7SK was isolated in 1994 (Shimba and Reddy 1994), but the gene was not identified until 2007 (Jeronimo et al. 2007). Bin3 (renamed methyl phosphate capping enzyme, MePCE) is responsible for installation of the γ-monomethyl cap on 7SK and U6 RNA. While U6 RNA copurified with MePCE, knockdown of MePCE reduced levels of 7SK RNA but not U6 RNA (Jeronimo et al. 2007). It is currently not clear how or to what degree the 5′ cap of U6 contributes to stability or function. Interestingly, S. cerevisiae does not have a clear homolog of MePCE, and the methylation status of the 5′ triphosphate is unknown. For a comprehensive review on the enzyme Bin3/MePCE, see Cosgrove et al. (2012).
3′ End modifications
After synthesis by Pol III, U6 is left with a oligo(U) tail of variable length with a terminal 2′,3′ cis diol. The oligo(U) tail and terminal 2′ and 3′ hydroxyl groups are bound by the La protein (Lhp1 in yeast) (Stefano 1984; Rinke and Steitz 1985; Terns et al. 1992). While the length of the oligo(U) tail is initially heterogeneous in part due to Pol III termination, U6 is both post-transcriptionally shortened and extended (oligouridylated) (Reddy et al. 1987).
U6 snRNA is 3′ oligouridylated in many eukaryotes, including S. pombe and humans. A U6 terminal uridylyl transferase (TUTase) activity was identified in 1998 (Trippe et al. 1998) and the gene, TUT1, was identified in 2006 (Trippe et al. 2006). Characterization of this enzyme revealed that it requires the presence of uridine at the 3′ end of U6 and that it can add three additional uridines in vitro (Trippe et al. 2003). Interestingly, TUT1 may recognize additional secondary structure in U6 and therefore may preferentially oligouridylate U6 while the RNA is in a certain conformation (Yamashita et al. 2017). TUT1 localizes to the nucleolus, suggesting that oligouridylation may occur alongside other modifications such as pseudouridylation and 2′-O-methylation (discussed in the “Pseudouridylation” and “Ribose 2′-O-methylation” sections) (Trippe et al. 2006).
The enzyme responsible for 3′ tail shortening was characterized in the late 1990's (Booth and Pugh 1997), but the gene was not identified for an additional 15 years (Mroczek et al. 2012; Shchepachev et al. 2012; Hilcenko et al. 2013). The enzyme was named Usix biogenesis protein 1 (Usb1), and is a 3′–5′ exonuclease (Mroczek et al. 2012; Shchepachev et al. 2012; Hilcenko et al. 2013). Mutations in Usb1 (also called Mpn1) are associated with the disease poikiloderma with neutropenia in humans (for review, see Mroczek and Dziembowski 2013). Usb1 trims back the oligo(U) tail and leaves a phosphate group on the terminal nucleotide. In humans, the terminal nucleotide has a 2′,3′-cyclic phosphate, while in yeast it has a 3′ noncyclic phosphate (Lund and Dahlberg 1992; Didychuk et al. 2017). The majority of human U6 snRNAs end with five terminal uridines and a 2′,3′-cyclic phosphate (Shchepachev et al. 2015).
Some metazoans do not have a homolog of Usb1. These organisms, including C. elegans, do not possess the phosphoryl or cyclic phosphate modifications, but instead have either a 2′,3′-cis diol in their mature form or an intriguing “blocked” modification (Lund and Dahlberg 1992). Interestingly, C. elegans does not have a Usb1 homolog, but rather has a TUTase called USIP-1 (Rüegger et al. 2015). The USIP-1 enzyme interacts with the U6 snRNP protein SART3 (discussed in the “Prp24/SART3” section). The chemical identity of the 3′ end produced by USIP-1 is not known.
The 3′ tail of U6 can also be adenylated (Chen et al. 2000). Adenylation inhibits uridylation and may target U6 for degradation. In cells deficient for Usb1 activity, U6 with adenylated tails accumulate (Hilcenko et al. 2013; Shchepachev et al. 2015). Polyadenylation may result in targeting U6 to the nuclear exosome. Human Usb1 is active on polyadenylate tails, and therefore can counteract polyadenylation (Hilcenko et al. 2013). It is unclear what the polyadenylating enzyme is, or if it can function on the product of human Usb1, a 2′,3′-cyclic phosphate. An additional phosphatase enzyme may be necessary to remove the cyclic phosphate and allow the polyadenylating enzyme to function.
The timing and location of 3′ end modification by Usb1 is not known. An early study suggested that the length and modification of U6 changed during the splicing cycle (Tazi et al. 1993). Tazi et al. made the intriguing observation that 3′ end modification of U6 required pre-mRNA, raising the possibility that U6 may not be processed until after a round of splicing is complete. In this way, after a successful first round of splicing, the cell could mark functional U6 to be protected and kept for additional rounds of splicing. However, a later study showed that depletion of U1 and U2 snRNAs had no effect on U6 processing (Gu et al. 1997). It is possible that the 3′ end of U6 is altered throughout the splicing cycle, as the 3′ end binding Lsm2-8 complex (discussed in the “Lsm2-8” section) dissociates during spliceosome activation, which may render the U6 3′ end accessible for modification. Interestingly, Usb1 localizes to the nucleus but not the nucleolus (Mroczek et al. 2012). This finding implies that in humans, Usb1 recognizes a TUT1-extended U6 in the nucleoplasm after modification in the nucleolus but before it has traveled to Cajal bodies for di- and tri-snRNP assembly. It is unclear if TUT1 can function on the terminal 2′,3′-cyclic phosphate of mature, Usb1-processed U6, and it is also unclear if uridylation occurs only once after transcription termination, or if U6 is trimmed and extended repeatedly throughout its life cycle.
Modification of the U6 3′ end is important for recognition by U6 3′ end binding proteins. The La protein binds U6 immediately after transcription termination and specifically recognizes the 2′,3′-cis diol. Indeed, while a subset of U6 molecules found in the cell have cis diols and are bound by La (Rinke and Steitz 1985), maturation of U6 and incorporation into the U6 snRNP requires the 3′ modification by Usb1. Modification of the 3′ end reduces the ability of La to bind U6 (Terns et al. 1992) and is important for binding of the Lsm2-8 ring (Licht et al. 2008). Interestingly, human Lsm2-8 preferentially binds U6 containing a 2′,3′-cyclic phosphate modification (Licht et al. 2008), while S. cerevisiae Lsm2-8 prefers to bind a noncyclic phosphate, suggesting that Usb1 and Lsm2-8 coevolved to allow for tight binding by Lsm2-8 on the product of Usb1 activity (Didychuk et al. 2017).
Pseudouridylation
Pseudouridylation of RNA is thought to be structurally stabilizing because of increased base stacking (Davis 1995) as well as the added hydrogen bonding potential and conformational flexibility due to the C–C glycosidic bond (Charette and Gray 2000). The effects of pseudouridylation on structure and RNA–protein interactions in U2 snRNA have been well-studied (Yu et al. 1998; Newby and Greenbaum 2001; Wu et al. 2016; van der Feltz et al. 2017), but the effects of pseudouridylation on U6 RNA have not. In S. cerevisiae, U6 is not constitutively pseudouridylated (Massenet et al. 1999). Recently, position U28 was found to be inducibly pseudouridylated during filamentous growth by the pseudouridine synthase Pus1 (Basak and Query 2014). Interestingly, mutations within U6 far from U28 (U36C and G50U) increased pseudouridylation of U28. These mutations likely disrupt interactions of U6 with U6 snRNP protein Prp24 (Montemayor et al. 2014) and would also likely alter interactions within the activated spliceosome. It is not clear at which step in the U6 lifecycle U6–U28 is pseudouridylated, or how changing RNA–protein interactions elsewhere in the RNA increases pseudouridylation of U6–U28. Furthermore, it is not clear how pseudouridylation of U6–U28 would affect RNA–RNA or RNA–protein interactions in different splicing complexes. In the U4/U6.U5 tri-snRNP, U6–U28 interacts with U5–U99 (Nguyen et al. 2016), whereas in the C complex, U6–U28 is within 5 Å of Bud31 and Ecm2 (Galej et al. 2016). Understanding how pseudouridylation of U6–U28 affects RNP structure, and understanding how pseudouridylation of U6 induces filamentous growth in yeast, could reveal important new insights into spliceosome biology.
In contrast, human U6 and U6atac are constitutively pseudouridylated at several positions (Massenet and Branlant 1999; Massenet et al. 1999). These positions (U31, U40, and U86 in U6; U83 in U6atac) (Fig. 4; Massenet and Branlant 1999) are in functionally important parts of the RNA, although it is unclear if or how modifications in U6 can stabilize its structure. Recently solved cryo-EM structures have allowed for structural analysis of splicing complexes; however, they lack the resolution to unambiguously identify post-transcriptional modifications. Improved resolution may lead to a better understanding of how modifications in U6 function.
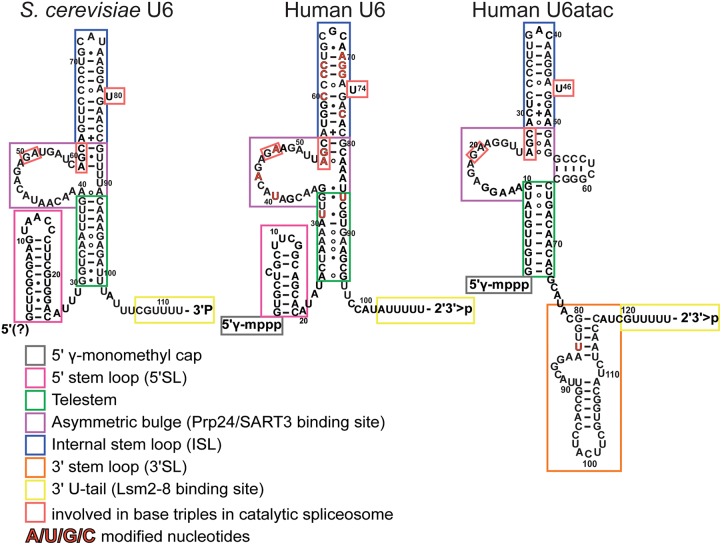
FIGURE 4.
Sequence and putative secondary structure of S. cerevisiae U6, human U6, and U6atac. The secondary structure of U6 from S. cerevisiae (left) is based on the structure of the U6 snRNP core (Montemayor et al. 2014), and includes the 5′SL, telestem, asymmetric bulge, ISL, and 3′ tail. The secondary structure of human U6 and U6atac within the U6 snRNP has not been experimentally determined, and is shown with secondary structure to mimic that of yeast U6. Human U6 contains a 5′SL, the ISL, and 3′ tail, and may contain an asymmetric bulge and telestem region. U6atac lacks a 5′SL but contains an additional 3′SL. Constitutively modified nucleotides are highlighted in red. Residues involved in base triples in the catalytic spliceosome (U2/U6.U5) are boxed in red.
Localization in the nucleolus is sufficient for formation of both the three pseudouridines in human U6 and the eight 2′-O-methyl groups (discussed in the “Ribose 2′-O-methylation” section), suggesting that all of the machinery required for these modifications is present in the nucleolus (Ganot et al. 1999). Localization of U6 during its biogenesis is discussed in the “Localization During Biogenesis” section.
Ribose 2′-O-methylation
Ribose 2′-O-methylation is a ubiquitous modification that can significantly stabilize RNA, either structurally (Sashital et al. 2007) or metabolically (by reducing hydrolytic or nucleolytic cleavage). Like pseudouridylation, ribose methylation occurs in the nucleolus (Ganot et al. 1999). Human U6 contains eight ribose methylations, at positions A47, A53, G54, C60, C62, C63, A70, and C77, which could stabilize secondary structure either in the U6 snRNP (the internal stem loop; ISL) or in U4/U6 (Stem I and Stem II) (Fig. 5). Ribose methylation requires a methyltransferase enzyme and “methylation guide” (mg) snoRNAs, including mgU6-47 and mgU6-77 (Tycowski et al. 1998), mgU6-53 (Ganot et al. 1999), and MBII-166 (which modifies U6–C60) (Hüttenhofer et al. 2001). Modification of many of the U6 ribose groups are conserved in plants (Kiss et al. 1987) and in S. pombe (Gu et al. 1996). The mgU6-47 snoRNA is conserved in S. pombe and is required for methylation. However, while disruption of the mgU6-47 gene resulted in complete loss of methylation, only a small cold-sensitive splicing defect was observed (Zhou et al. 2002). Interestingly, human U6atac has no ribose 2′-O-methyl groups, despite forming a homologous ISL in which ribose methylations in U6 are concentrated (Massenet and Branlant 1999).
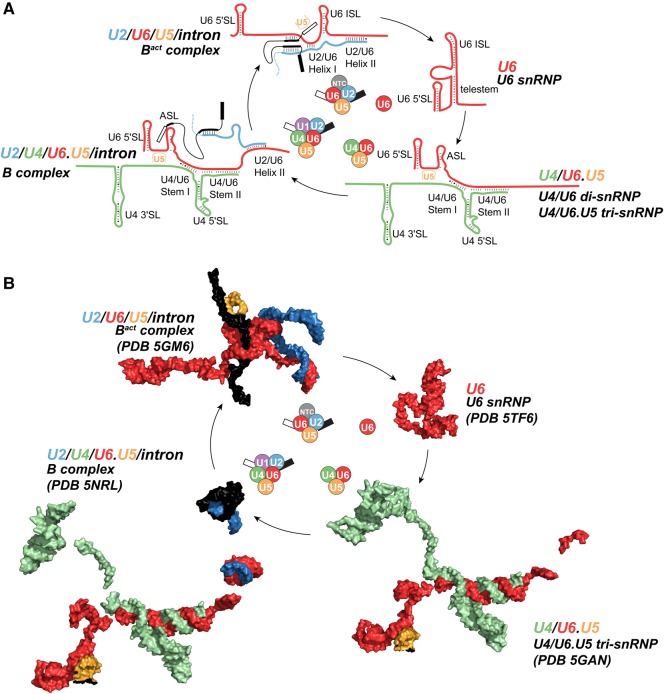
FIGURE 5.
U6 undergoes large conformational changes during the splicing cycle. (A) Cartoon of base-pairing throughout the splicing cycle. For simplicity, minimal interacting sequences of U2 and U5 snRNAs are shown. (B) Structure of U6 and its partners in different splicing complexes.
Recent RiboMeth-seq results show that the level of ribose modification varies in different cell types (Krogh et al. 2017). While ribose methylation may not be essential, its conservation across species suggests it (along with pseudouridylation and other modifications) may play a role in fine-tuning the stability of U6 and its complexes to modulate splicing, perhaps in response to variation in cellular temperatures that occur even in homeotherms.
N-6-adenosine and N-2-guanosine base methylation
Human U6 contains a single N-6-methyladenosine modification (m6A) at position A43 (Shimba et al. 1995). Study of m6A modification has recently undergone a renaissance due to transcriptome-wide mapping studies that show it is a ubiquitous and regulated modification in mRNA (for review, see Meyer and Jaffrey 2014). Most mRNA m6A residues are found within a consensus sequence (GAC or AAC) and are modified by the METTL3 complex. U6–A43 does not lie within a consensus sequence and is not modified by METTL3, but rather by a recently identified methyltransferase, METTL16 (Pendleton et al. 2017; Warda et al. 2017). Interestingly, METTL16 activity is dependent on both sequence and secondary structure. This is consistent with earlier reports that modification of U6–A43 depends on formation of the adjacent ISL secondary structure within U6 (Shimba et al. 1995). It is unclear when A43 is modified to m6A, but in vitro studies suggest that the conversion is rapid (Shimba et al. 1995). A recent report showed that METTL16 associates on U6 alongside the capping enzyme MePCE (see the “5′ Capping” section) and the 3′ end binding protein La (Warda et al. 2017). Determining the order of U6 modifications and the effect of multiple modifying enzymes competing for the same substrate may yield interesting insights into U6 biogenesis.
m6A-modified U6 is incorporated into U4/U6 snRNPs, suggesting it is functional (Bringmann and Lührmann 1987). The modification at position A43 lies in the center of the ACAGAGA motif, a perfectly conserved and essential motif that base pairs to the intron adjacent to the 5′ splice site and helps organize the spliceosome active site through formation of base triple interactions (Fica et al. 2014). Biophysical studies of m6A modification within double-stranded RNA indicate that it destabilizes base-pairing, but at the ends of helices is stabilizing when stacked and unpaired (Roost et al. 2015). In a recent structure of a human C* complex, U6–A43 is unpaired across from an adenosine in the intron (Bertram et al. 2017b). The methylation state of the nucleotide could not be determined in the structure.
Human U6 also contains a single N-2-guanosine methylation (m2G) at position G72 (Epstein et al. 1980). The role of this modification is unknown, as G72 is expected to fall within a G–C base pair in both the U6 ISL and within U4/U6 Stem II (Figs. 4, 5) and because m2G is expected to be isoenergetic with guanosine in G–C base pairs (Rife et al. 1998). Like base methylation of A43, methylation of G72 may also depend on secondary structure (Ganot et al. 1999).
Splicing of U6 snRNA
The sole copy of the U6 gene in S. pombe has an intron that is removed by the spliceosome (Tani and Ohshima 1989). The intron is located immediately adjacent to the highly conserved catalytic AGC triad that is essential for splicing catalysis, leading to the hypothesis that the intron in S. pombe U6 RNA arose from reverse-splicing of an excised intron into a U6 molecule in the active spliceosome, followed by reverse transcription and incorporation into the genome (Brow and Guthrie 1989; Tani and Ohshima 1991). This observation suggested that U6 (and specifically this region of U6) is near the active site of the spliceosome and that splicing is reversible. Indeed, both of these hypotheses have been tested using biochemistry and structural biology. Splicing is reversible under the appropriate conditions (Tseng and Cheng 2008), and it is now well-established both biochemically and in near-atomic structural detail that this region of U6 forms the catalytic core of the spliceosome. Recent bioinformatics analysis shows that many yeast species have evolutionarily unrelated introns and that these introns are distributed throughout the U6 sequence (Canzler et al. 2016), although intron insertion sites are more concentrated near the catalytic core (U6 ISL and ACAGA box) and in a region upstream of the ACAGA box known to function as a branch acceptor for in vitro trans-splicing in nematodes (Yu et al. 1993). These introns (up to four introns in a single fungal U6 gene) possess canonical 5′ splice sites, branch sites, and 3′ splice sites, but are otherwise not conserved in sequence. Phylogenetic analysis suggests that while the presence of some introns in related species likely arose from a common ancestor, many of the U6 introns may be species-specific insertions (Canzler et al. 2016). Experimental validation that U6 genes containing introns are expressed and spliced is necessary. It is possible that species with multiple U6 paralogs of which some contain introns may use splicing as a method to regulate U6 levels.
Spliceosomal introns can also be found in U2 (Takahashi et al. 1993) and U5 snRNAs (Takahashi et al. 1996), supporting the hypothesis that RNAs close to the active site of the spliceosome can be targets of reverse splicing. However, spliceosomal introns have also been identified in U1 snRNA (Takahashi et al. 1996), which departs before the active site of the spliceosome is formed, and in rRNA genes (Bhattacharya et al. 2000). It is not clear what the determinants of novel spliceosomal intron insertion in ncRNAs are, but possibilities include RNA abundance, structure, or proximity to the spliceosomal active site.
THE U6 SNRNP
U6 snRNA secondary structure
U6 is the most conserved of the snRNAs in sequence and length (Brow and Guthrie 1988). The least conserved region is the 5′ stem–loop (5′SL), which varies in both sequence and length across eukarya despite maintaining a stem–loop structure (Roiha et al. 1989). C. merolae, a moderate thermophile, contains a minimal spliceosome that lacks U1 snRNA, yet has an unusually stable 5′SL with 22 base pairs (versus 10 base pairs in S. cerevisiae) (Stark et al. 2015). The 5′SL persists throughout the various U6-containing splicing complexes. It does not have any known catalytic function, but does correspond to part of the A-block promoter element in S. cerevisiae and is necessary for efficient 5′ capping in humans (discussed in the “5′ Capping” section). The 5′ capping determinants are conserved in yeast phyla (Roiha et al. 1989), further evidence that 5′ capping is also conserved.
The remaining portion of U6 forms several mutually exclusive structures. While in the U6 snRNP, U6 consists of the 5′SL, the telestem, and the internal stem–loop (ISL, previously called the 3′ stem–loop or intramolecular stem–loop) (Figs. 4, 5). While the 5′SL likely remains stably base-paired throughout the splicing cycle, the less stable telestem and ISL helices make dynamic transitions into other secondary structures during spliceosome assembly and activation. Incorporation of U6 into the spliceosome requires unwinding of the ISL and subsequent base-pairing to U4, a mutually exclusive interaction. As observed in the U4/U6.U5 tri-snRNP structure (Agafonov et al. 2016; Nguyen et al. 2016; Wan et al. 2016b), the telestem is unwound when U6 is paired to U4 snRNA. However, in vitro data suggest the telestem can form transiently in free U4/U6 RNA (Brow and Vidaver 1995; Rodgers et al. 2016). During spliceosome activation, U4 is unwound from U4/U6 by the helicase Brr2 (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998a), U6 base pairs with U2 (Madhani and Guthrie 1992), and the U6 ISL reforms (Fig. 5). The telestem is mutually exclusive with formation of U2/U6 helix II (Fig. 5). The telestem enhances Prp24 affinity and Prp24-mediated annealing (Didychuk et al. 2016) and also destabilizes U4/U6 (Brow and Vidaver 1995; Rodgers et al. 2016). The dynamic telestem and U6 ISL structures may help U6 molecules avoid kinetic or thermodynamic folding traps during spliceosome assembly and activation.
Essential nucleotides in the U6 ACAGAGA box and the AGC triad were identified early on (Fabrizio and Abelson 1990; Madhani et al. 1990). During activation, the ACAGAGA box interacts with the 5′ splice site (Kandels-Lewis and Séraphin 1993; Lesser and Guthrie 1993; Johnson and Abelson 2001; Rauhut et al. 2016; Yan et al. 2016). The AGC triad is important for intramolecular base-pairing within U6 as well as pairing to U2 (in U2/U6 helix I) and U4 (in U4/U6 Stem I) (Fig. 5). The AGC triad makes intramolecular base-triple tertiary contacts with the last 2 nucleotides of the ACAGAGA sequence (Hilliker and Staley 2004; Fica et al. 2014, 2017; Galej et al. 2016; Rauhut et al. 2016; Wan et al. 2016a; Yan et al. 2016, 2017; Bertram et al. 2017b; Zhang et al. 2017). The AGC triad and base triple interactions are conserved in the self-splicing Group II introns, which share a common evolutionary ancestor with the spliceosome (Pyle 2016). A conserved bulged residue in the ISL (U80 in S. cerevisiae) is involved in catalysis by coordinating a catalytic metal ion and by forming a base triple with the C of the AGC triad (Fica et al. 2013, 2014). Mutation of U80 to C or A has no obvious effect on yeast growth, while mutation to a G is lethal unless U6–C67 is mutated to prevent pairing across the ISL and U4–G14 is mutated to maintain pairing with U6 residue 67 (McManus et al. 2007). Thus, the tertiary structure of this region of the ISL is more important than the primary structure.
U6atac lacks a 5′SL, but contains a similar central secondary structure (Fig. 4). Indeed, the yeast or human U6 ISL can functionally replace the U6atac ISL (Shukla and Padgett 2001). U6atac contains an additional stem loop 3′ of the ISL, which is important for targeting to the minor spliceosome (Dietrich et al. 2009).
Prp24/SART3
The yeast Prp24 protein was discovered through a genetic selection for suppressors of mutations in U4 RNA that disrupt U4/U6 pairing (Shannon and Guthrie 1991) as well as a screen for heat-sensitive mutations in splicing (Vijayraghavan et al. 1989). Mutations in Prp24 were also isolated in a selection for suppressors of a mutation that stabilizes the U6 ISL (Vidaver et al. 1999; Montemayor et al. 2014), underscoring its importance for the structural transitions of U6 snRNA. In S. cerevisiae, Prp24 contains three canonical RNA recognition motifs (RRMs) and a fourth “occluded” RRM in which the typical RNA binding surface, a four-stranded β-sheet, is masked by additional helices (Rader and Guthrie 2002; Kwan and Brow 2005; Bae et al. 2007; Martin-Tumasz et al. 2011; Montemayor et al. 2014). The human homolog of Prp24, SART3, has only two RRMs (which may correspond to yeast RRMs 1 and 2 or 2 and 3), and instead has multiple N-terminal half-a-tetratricopeptide (HAT) repeats (Bell et al. 2002; Rader and Guthrie 2002; Kwan and Brow 2005). SART3 has recently been shown to dimerize through these HAT repeats (Park et al. 2016; Zhang et al. 2016), suggesting that Prp24 and SART3 may both require four RRMs to function efficiently. Prp24 from S. pombe may be an evolutionary intermediate in that it has four RRMs plus the HAT domains. All Prp24 homologs contain a highly conserved short peptide at the C terminus called the SNFFL box, which is important for interactions between Prp24 and Lsm2-8 (Fromont-Racine et al. 2000; Rader and Guthrie 2002). Prp24 is essential for unwinding the U6 ISL and annealing U4 to U6 (discussed in the “U4/U6 Annealing” section) (Ghetti et al. 1995; Rader and Guthrie 2002; Didychuk et al. 2016).
Lsm2-8
The Lsm proteins are paralogs of the Sm proteins that bind to U1, U2, U4, and U5 snRNAs, and are related to bacterial Hfq and Sm-like archaeal proteins (Mura et al. 2013). Lsm proteins are small (9–21 kDa) and assemble into at least 2 different heteroheptameric, ring-shaped complexes, Lsm1-7 and Lsm2-8. The Lsm1-7 ring localizes to the cytosol and is involved in the 5′–3′ mRNA decay pathway (Tharun et al. 2000). The Lsm2-8 ring localizes to the nucleus where it binds the 3′ U-tail of U6 (Achsel et al. 1999; Mayes et al. 1999). The S. cerevisiae Lsm2-8 complex preferentially binds U6 3′ ends modified with a terminal phosphate (Didychuk et al. 2017), while the metazoan Lsm2-8 complex preferentially binds a terminal 2′,3′-cyclic phosphate (Licht et al. 2008), as discussed above in the “3′ End Modifications” section. Structures of the Lsm1-7 and Lsm2-8 complexes revealed that the two rings are exceedingly similar, with <1 Å rmsd between subunits Lsm2-7 in each structure and between the core Sm folds of Lsm1 and Lsm8 (Sharif and Conti 2013; Zhou et al. 2014a,b). The crystal structure of the S. cerevisiae Lsm2-8 bound to a short RNA oligo was determined and revealed that the 3′ end of U6 is recognized in the center of the ring in a manner distinct from Sm-RNA binding (Zhou et al. 2014a). The Lsm2-8 complex can also be observed in cryo-EM structures of the U4/U6.U5 tri-snRNP, albeit at lower resolutions (Agafonov et al. 2016; Nguyen et al. 2016; Wan et al. 2016b).
Unlike the Sm ring, the Lsm2-8 ring is stable in the absence of RNA (Achsel et al. 1999) and assembles without assistance from other proteins. The Sm ring, in contrast, requires the SMN complex and Gemin proteins to assemble three Sm subcomplexes into a ring that encircles RNA in vivo (for reviews, see Battle et al. 2006; Matera and Wang 2014). The Lsm2-8 ring is likely imported into the nucleus via importin β/KAP95 (Spiller et al. 2007a), where the N- and C-terminal domains of Lsm8 act as nuclear retention signals for U6 (Spiller et al. 2007a,b; Reijns et al. 2009).
Structure of the U6 snRNP
The structure of the core of the U6 snRNP, with the majority of both U6 snRNA (nucleotides 30–101) and Prp24 (all four RRMs), has been determined (Montemayor et al. 2014). To date, this is the only structure of a protein containing four RRMs bound to RNA. It reveals how three of the RRMs cooperate to specifically and tightly bind U6 RNA. RRMs typically bind 3–4 single-stranded nucleotides across a β sheet surface using consensus RNP1 and RNP2 motifs (Afroz et al. 2015). In contrast, the Prp24 protein directly contacts 20 nt of U6 RNA via interactions with RRM domains 2–4. The first RRM does not contact U6 snRNA, but forms a large protein–protein interface with RRM2. RRMs 2 and 3 bind the asymmetric bulge in a relatively canonical RRM fashion, targeting the ACAGA box and sequences immediately upstream, respectively. The fourth RRM binds primarily to the ISL and telestem via the two amphipathic helices that occlude the β-sheet face of the RRM (Martin-Tumasz et al. 2011). Surprisingly, the RRM3/RRM4 linker, a single aspartate residue, passes through the asymmetric bulge and RRM4 makes tertiary interactions with RRM2, forming an entwined topology consisting of interlocked rings of protein and RNA (Montemayor et al. 2014). The entwined topology is further closed off by the ISL and telestem and is exceptionally stable, as the complex is resistant to 2 M monovalent salt (Shannon and Guthrie 1991). RRMs 1, 2, and 4 together form a large (∼20 Å wide), positively charged groove. In the crystal lattice, the U6 ISL of a neighboring complex is packed into the groove, leading to the hypothesis that this groove might stabilize U4/U6 pairing during Prp24-mediated stimulation of U4/U6 di-snRNP formation (Montemayor et al. 2014).
The U6•Prp24 structure contained three substitutions to stabilize the RNA (Montemayor et al. 2014) including A62G, which stabilizes the base of the ISL and causes a cold-sensitive phenotype in vivo (Fortner et al. 1994). Interestingly, suppressors of U6–A62G cold sensitivity localize to the RNA–protein interface, suggesting that detrimental overstabilization of the U6 ISL structure can be neutralized by destabilization of the RNA–protein interface in vivo (Montemayor et al. 2014). A second structure with the wild-type ISL crystallized in a different space group, yet exhibits the same interlocked topology (Montemayor et al. 2017). Stabilization of the telestem with U100C/U101C mutations, which increase both Prp24 affinity for U6 as well as Prp24-mediated U4/U6 annealing activity (discussed in the “U4/U6 Annealing” section), likely also stabilizes the interlocked topology (Didychuk et al. 2016). Together, these data strongly suggest that the interlocked topology is a biologically functional structure.
The structure of the entire snRNP, with full-length RNA and including both Prp24 and Lsm2-8, remains unsolved. It is known that Prp24 and Lsm2-8 have at least one protein–protein interaction (via the Prp24 SNFFL box), but where this interaction occurs on the Lsm2-8 ring and how the individual components are oriented with respect to one another has not been determined. Uncovering how Prp24 and Lsm2-8 interact will undoubtedly be vital to understanding the mechanism of Prp24-mediated U4/U6 annealing.
U4/U6 DI-SNRNP AND U4/U6.U5 TRI-SNRNP
U4/U6 annealing
In order to be incorporated into the assembling spliceosome, the ISL of U6 must be completely unwound and base-paired to U4 snRNA via the RNA chaperone activity of Prp24. This assembly step may proofread U6 structure by ensuring that only U6 that can bind Prp24 and base pair to U4 is incorporated into the spliceosome. In addition, U4 may act as an “antisense negative regulator” of the catalytic residues of U6 by sequestering them in the double-stranded helixes of U4/U6 prior to spliceosome activation (Guthrie 1991).
It has long been known that the U6 snRNP protein Prp24 can anneal U4 and U6 RNAs in an ATP-independent manner (Ghetti et al. 1995; Raghunathan and Guthrie 1998b). The 20 Å wide electropositive groove of Prp24 is critical for U4/U6 annealing but not binding of U6, suggesting that it is the “active site” for U4/U6-annealing (Didychuk et al. 2016). Stabilization of the telestem in U6 also contributes to efficient annealing, possibly by promoting formation of the electropositive groove (Didychuk et al. 2016). The Lsm2-8 ring enhances Prp24-mediated annealing in vitro, but does not facilitate U4/U6 annealing by itself (Didychuk et al. 2016). Interestingly, Prp24 binds the U4/U6 product complex in vitro (Ghetti et al. 1995; Didychuk et al. 2016) yet does not stay associated with the U4/U6 di-snRNP in yeast in vivo (Shannon and Guthrie 1991). How the interlocked topology of U6•Prp24 is resolved during annealing, and how Prp24 is displaced from the U4/U6 di-snRNP, is unclear.
The upper ISL is not contacted by protein in either of the determined U6 snRNP core structures. In the ISL-stabilized (A62G) structure, the top of the ISL is disordered and not visible (Montemayor et al. 2014). In the wild-type U6 snRNP core structure, there are two complexes in the asymmetric unit (Montemayor et al. 2017). In one, the ISL is largely disordered and in the other, the ISL displays a 20° bend relative to the A62G ISL (Montemayor et al. 2017). These observations, along with NMR measurements and MD simulations (Blad et al. 2005; Venditti et al. 2009), suggest that the U6 ISL is inherently dynamic. Since the ISL must be completely unwound during U4/U6 annealing, dynamics likely play an important role in the annealing pathway. The stabilized A62G ISL in isolation has a melting temperature of ∼63°C in 200 mM KCl (Sashital et al. 2003) while the wild-type ISL is less stable. Binding of Prp24 may facilitate U4/U6 formation by allowing the ISL to remain in a dynamic, meta-stable conformation while adjacent to the electropositive groove in Prp24. It is unlikely that the ISL needs to fully melt in order for annealing to be initiated, as local unwinding of a few base pairs could allow formation of a nascent U4/U6 duplex to invade the ISL and facilitate the subsequent exchange of intra- for intermolecular base pairs via branch migration (Meselson 1972; Sigal and Alberts 1972).
In humans, SART3 (hPrp24) functions in a similar role in both U4/U6 annealing (Bell et al. 2002; Medenbach et al. 2004) and U4atac/U6atac annealing (Damianov et al. 2004). Although the HAT domains are dispensable for binding U6, both the HAT domains and the two RRMs are required for efficient U4/U6 recycling (Bell et al. 2002; Medenbach et al. 2004). Surprisingly, the C-terminal tail (the SNFFL box), which is known to interact with the Lsm2-8 ring (Rader and Guthrie 2002), is not necessary for efficient recycling in vitro, but an interaction with the U4 snRNP protein 90K (hPrp3) mediated by the HAT domains of SART3 is necessary (Medenbach et al. 2004). It has also been reported that human Lsm2-8 can recycle U4/U6 in the absence of other factors (Achsel et al. 1999). As monomeric SART3 has only two RRMs, it is unlikely to form an electropositive groove as observed in the yeast U6•Prp24 structure, although it may potentially do so via the aforementioned dimerization of the HAT domains (Park et al. 2016). Alternatively, the role of the electropositive groove of Prp24 may have been functionally replaced by protein–protein interactions between SART3 and hPrp3 (Medenbach et al. 2004).
The U4 snRNP proteins Snu13 and Prp31 and U4/U6 di-snRNP proteins Prp3/Prp4 (plus CypH in humans) may also contribute to the annealing process. Snu13 and Prp31 influence the conformation of the U4 5′SL, which may hold U4 snRNA in a conformation amenable to annealing with U6 (Hardin et al. 2015). Indeed, the structure of protein-free U4/U6 (Cornilescu et al. 2016) is significantly different from its structure while in U4/U6.U5 tri-snRNP (Nguyen et al. 2016). Prp3 binds U4/U6 Stem II and a region on U6 3′ of the stem, suggesting that its binding site does not exist until U4/U6 is formed (Liu et al. 2015; Nguyen et al. 2016). However, it may transiently capture this single-stranded region of U6 from U6•Prp24 to accelerate U4/U6 annealing (Liu et al. 2015). The contribution of additional proteins to Prp24-mediated annealing has not been measured, but may reveal additional RNA–protein or protein–protein interactions that accelerate the assembly process. Understanding how the mechanism of Prp24-mediated annealing in both yeast and humans could give new insight into conserved versus divergent mechanisms of RNA chaperones.
Formation and structure of the U4/U6.U5 tri-snRNP
The tri-snRNP is formed through protein–protein and protein–RNA interactions between the U4/U6 di-snRNP and the U5 snRNP. Prp31 is necessary for assembly of both major and minor U4/U6.U5 tri-snRNPs in humans (Schneider et al. 2002). Knockdown of both hPrp31 and the tri-snRNP-specific protein hPrp6 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies (Novotńy et al. 2015) (discussed in the “Localization During Biogenesis” section). The structure of the tri-snRNP from yeast and humans has revealed myriad protein–protein contacts that hold the U4/U6 di-snRNP and U5 snRNP together (Agafonov et al. 2016; Nguyen et al. 2016; Wan et al. 2016b). Prp6 is the “glue” holding the tri-snRNP together, as its N-terminal region contacts U5 snRNP component Prp8 (and Brr2 in the human tri-snRNP) while its C-terminal region contacts U4 snRNP components Snu13, Prp31, Prp3, and Prp4 (Galisson and Legrain 1993; Agafonov et al. 2016; Nguyen et al. 2016; Wan et al. 2016b). In the presence of mutations that destabilize U4/U6 pairing and U6-Prp24 binding, tri-snRNP still assembles in the apparent absence of U4/U6 di-snRNP, suggesting there may be an alternate pathway for incorporating U4 and U6 into the tri-snRNP (Burke et al. 2015).
The structure of the U4/U6.U5 tri-snRNP revealed an unexpected short internal stem loop in U6 5′ of U4/U6 Stem I consisting of nucleotides 35–46 (Nguyen et al. 2016). Interestingly, this stem loop is immediately upstream of the ACAGAGA sequence, which must pair to the 5′ SS. This ACAGAGA stem loop (ASL) is stabilized by Dib1 and Prp8 and is also present in the B complex structure (Plaschka et al. 2017). Consistent with prior genetic and biochemical data indicating that the intron 5′ SS binds first to an upstream ACA sequence present in the loop of the ASL prior to binding the ACAGA box (Li and Brow 1996; Johnson and Abelson 2001), the 5′ exon–intron junction interacts with the ASL loop in the B complex (Plaschka et al. 2017). As Dib1 is not observed in Bact or subsequent catalytic structures, it likely departs during activation and the ASL unwinds, promoting recognition of the intron by the ACAGA box and allowing catalysis to occur. The ASL was not resolved in the structure of the human tri-snRNP, despite Dim1 (the homolog of Dib1) binding near this region (Agafonov et al. 2016). The RNA–RNA and protein–RNA contacts in the tri-snRNP effectively sequester all of the catalytic elements of U6: The ACAGAGA sequence is partially occluded by the ASL and its associated proteins, the AGC triad is base-paired in U4/U6 Stem I, and U80 is base-paired in U4/U6 Stem II.
The most significant difference in the structures of U4/U6.U5 tri-snRNP from human and yeast is in the placement of the helicase Brr2. In the yeast tri-snRNP, Brr2 binds U4 in a single stranded region between the U4 3′SL and U4/U6 Stem I, suggesting that it is poised for unwinding U4/U6 (Nguyen et al. 2016). Indeed, tri-snRNP isolated from yeast cell extract is disassembled in the presence of ATP (Raghunathan and Guthrie 1998a; Nguyen et al. 2015). In contrast, Brr2 in the human tri-snRNP is on the opposite end of the complex ∼20 nm away, far from its U4/U6 Stem I substrate, and the tri-snRNP is insensitive to ATP-dependent disassembly (Agafonov et al. 2016). This difference is likely due to the presence of Sad1 in the human tri-snRNP, where Sad1 both stabilizes the interaction between U4/U6 and U5 snRNPs and tethers Brr2 in a “preactivation” position away from its substrate (Agafonov et al. 2016). Yeast Sad1, in contrast, does not copurify with the tri-snRNP (Stevens et al. 2001; Nguyen et al. 2016). Despite interacting weakly with the tri-snRNP in yeast, Sad1 still prevents Brr2-mediated unwinding in whole cell extract (Huang et al. 2014).
The minor spliceosome tri-snRNP (U4atac/U6atac.U5) contains many of the same components as the major spliceosome U4/U6.U5 (Schneider et al. 2002). The additional 3′ stem loop in U6atac (Fig. 4) may differentiate the major and minor spliceosomes, as this stem loop is both important for targeting U6atac to the minor spliceosome (Dietrich et al. 2009) and for interacting with protein 65K, a U11/U12-specific protein (Singh et al. 2016). It remains to be seen if the major and minor spliceosomes have additional conformational or compositional differences, and how the structures of the major and minor tri-snRNP differ.
U6 IN THE CATALYTIC SPLICEOSOME
Activation
U6 base pairs with the 5′SS after remodeling of the U1-5′SS interaction (Sawa and Abelson 1992) by the DEAD-box helicase Prp28 (Staley and Guthrie 1999). This is an important proof-reading step to ensure that stable and correct U6-5′SS duplexes are formed (Yang et al. 2013). U2 base pairs to U6 via formation of U2/U6 helix II (Fig. 5; Wassarman and Steitz 1992; Schneider et al. 2010). Then, U4/U6 Stems I and II are completely unwound in order for the U6 ISL to reform and for U6 to base pair with U2 (Fig. 5). Ejection of U4 depends on the U5-snRNP component helicase Brr2, which unwinds U4/U6 (Raghunathan and Guthrie 1998a; for review, see Absmeier et al. 2016). Brr2 activity is regulated by protein–protein contacts with itself and Prp8, which can both up- and down-regulate its unwinding activity (Maeder et al. 2009; Mozaffari-Jovin et al. 2012, 2013; Absmeier et al. 2015). The spliceosomal GTPase, Snu114, influences regulation of Brr2 activity by Prp8 (Small et al. 2006). Intramolecular folding of U6 contributes to efficient unwinding (Rodgers et al. 2016; Theuser et al. 2016). Once unwound, U4 snRNA departs with Snu13 and Prp31 (and likely the U4 Sm ring), while Prp3/Prp4 are released separately (Theuser et al. 2016).
Brr2-mediated unwinding and U4 snRNP departure typically occur before arrival of the NTC (Hoskins et al. 2016). Recent data on human spliceosomes chemically stalled between the transition from B to Bact reveal that the Lsm2-8 ring is present on U6 after U4 departure (Sidarovich et al. 2017). The NTC is required for dissociation of the Lsm ring in yeast (Chan et al. 2003) and promotes interactions between U5, U6, and the 5′SS prior to first step chemistry (Chan and Cheng 2005). Recent structures of the spliceosome after activation show that the 3′ end of U6 (the binding site of the Lsm ring) is partially occluded by Syf1 of the NTC, suggesting that the NTC may directly displace the Lsm ring. It is not clear if 3′ end modifying enzymes such as Usb1 and U6-TUTase can access the 3′ end of U6 in these complexes, but high-throughput studies suggest that Usb1 may interact with components of the NTC (Hazbun et al. 2003; Krogan et al. 2006; Yu et al. 2008).
Conformational changes during catalysis
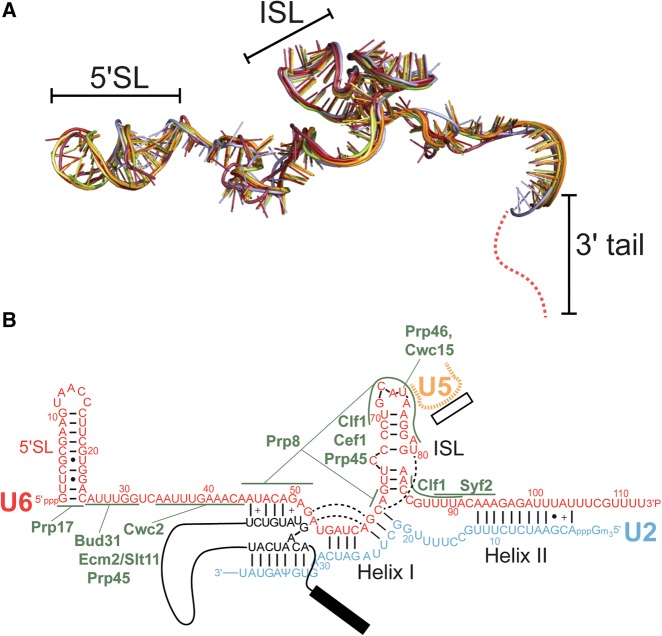
In the past two years, a multitude of spliceosomal structures have been determined at different catalytic steps. Structures of spliceosomes paused before activation (B complex) (Bertram et al. 2017a; Plaschka et al. 2017) the first catalytic step (Bact complex) (Rauhut et al. 2016; Yan et al. 2016), after first step chemistry (C complex) (Galej et al. 2016; Wan et al. 2016a), remodeled before the second catalytic step (C* complex) (Bertram et al. 2017b; Fica et al. 2017; Yan et al. 2017; Zhang et al. 2017), and after ligated exon departure (ILS) (Yan et al. 2015; Wan et al. 2017) reveal that, despite large-scale conformational changes elsewhere in the spliceosome (Table 2), the structure of U6 snRNA changes very little during these transitions. Indeed, the structure of U6 snRNA at each of the steps after activation of B complex is nearly indistinguishable (Fig. 6A), even when comparing structures isolated from S. cerevisiae and S. pombe. U2/U6 Helix Ia and Ib (composed of nucleotides 55–61 in U6) and Helix II (nucleotides 93–102 in U6) remain base-paired and unchanged throughout the structures (Fig. 6B). The ISL (nucleotides 63–84) is also structurally identical. Yeast U6 residue C66 is bulged from the ISL and sits in a pocket composed of the NTC and NTC-associated proteins Clf1, Cef1, Prp45, and Ecm2. The structure of the ISL within the spliceosomal complexes is significantly different from the free RNA or when within the U6 snRNP due to the many protein contacts surrounding it. The 3′ tail of U6 (nucleotides 105–112) is unstructured in all available cryo-EM structures and is not modeled in any of the structures. Small local base-pairing changes do occur between the ACAGA box and the intron, especially U6 nucleotides G50 and A51. However, local changes in accessibility, flexibility, or structure may occur in solution, with important implications for activation and catalysis (Bao et al. 2017).
TABLE 2.
RNA–RNA and protein–RNA contacts in U6-containing complexes
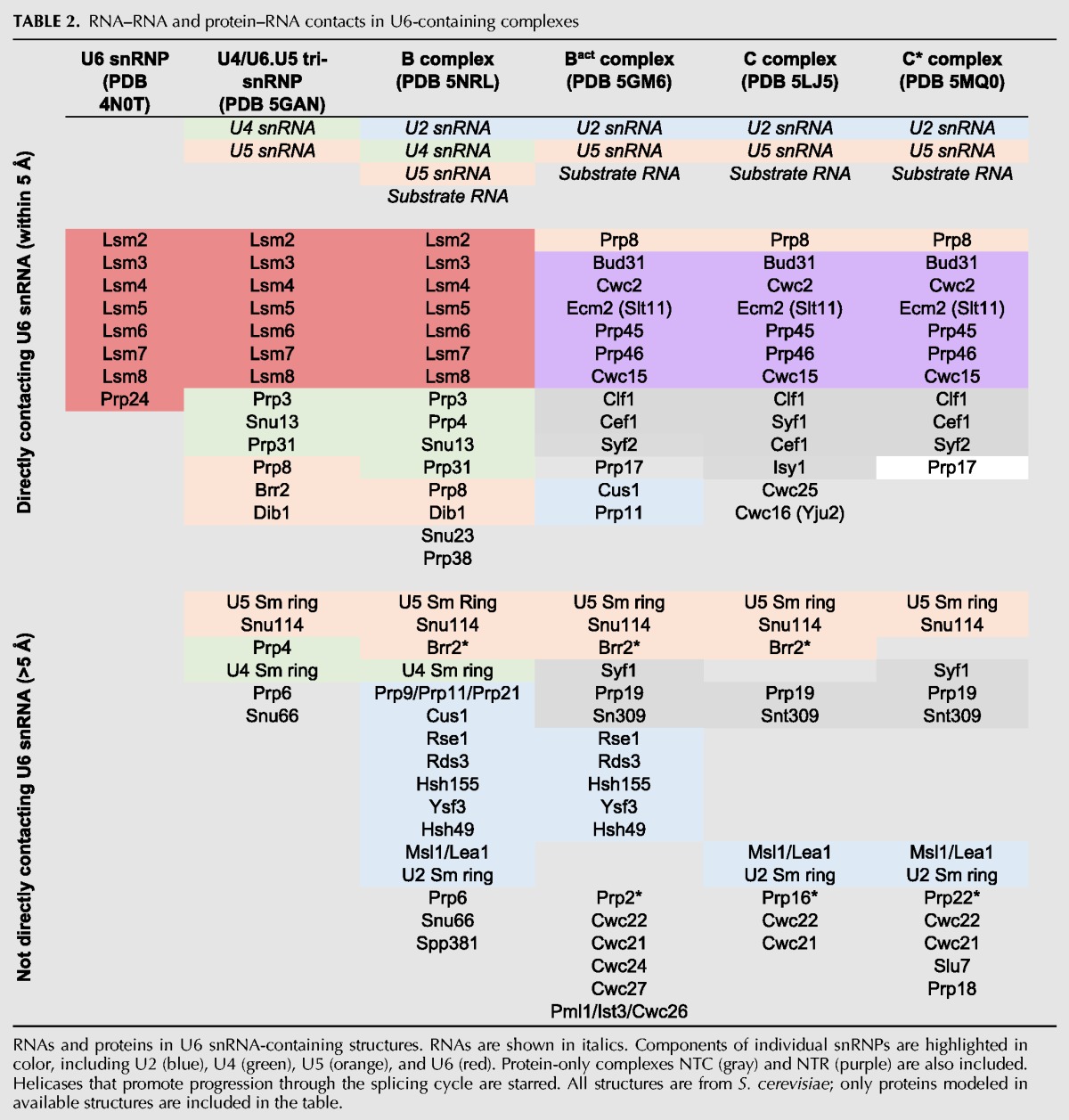
FIGURE 6.
S. cerevisiae U6 does not undergo conformational changes during the transitions from Bact to ILS complexes. (A) Superimposition of U6 snRNA structures from the following S. cerevisiae spliceosomal complexes: Bact (PDB 5GM6, red and PDB 5LQW, salmon), C (PDB 5GMK, orange and PDB 5LJ5, light orange), C* (PDB 5WSG, lemon and PDB 5MQ0, lime), and the S. pombe ILS (PDB 3JB9, blue). (B) Secondary structure of U2/U6 in C* complex (PDB 5MQ0). Protein contacts to U6 are shown in green. Base triple interactions are shown by dashed lines.
In contrast, U2 snRNA and the branch site helix, along with many protein partners, undergo >50 Å motions and large rotations. Analogy to group II introns, along with detailed genetic and biochemical studies, suggested that the active site for first and second step chemistry are the same (Chanfreau and Jacquier 1994; Marcia and Pyle 2012; Fica et al. 2013, 2014), and that large conformational changes are required in order to move the first step product (the 2′–5′ branch site) out of the single active site, followed by positioning of the 3′ exon in the active site for second step chemistry (exon ligation). The first hints of these large-scale motions came from single molecule experiments (Abelson et al. 2010; Crawford et al. 2013). Recent cryo-EM structures of splicing complexes before and after the first and second steps of splicing corroborate the hypothesis that U6 forms a rigid active site in the spliceosome and that its substrates must be positioned within it for catalysis to occur (Yan et al. 2015, 2016, 2017; Galej et al. 2016; Rauhut et al. 2016; Wan et al. 2016a, 2017; Bertram et al. 2017b; Fica et al. 2017; Zhang et al. 2017).
Metal ion binding in the spliceosome active site
A two-metal mechanism for RNA-mediated splicing catalysis was proposed in 1993 (Steitz and Steitz 1993). This model stated, by analogy to protein phosphoryl transfer mechanisms, that two divalent metal ions situated 3.9 Å apart stabilize the nucleophile and the leaving group during transesterification. Two groups used phosphorothioate substitutions to identify functionally important nonbridging oxygen atoms in the U6 phosphoribose backbone (Fabrizio and Abelson 1992; Yu et al. 1995), and subsequent thiophilic metal ion rescue experiments revealed that the U6 ISL coordinates the two Mg2+ ions required for catalysis via five nonbridging phosphate oxygens (Yean et al. 2000; Fica et al. 2013). Electron density in the active site of recent spliceosome cryo-EM structures has been modeled as metal ions (Wan et al. 2016a; Yan et al. 2016, 2017; Fica et al. 2017; Zhang et al. 2017). However, the current resolution of these cryo-EM structures precludes the unambiguous placement of metal ions into the electron density, such that the exact location of magnesium ions in the active site, and positioning and identity of other structural metal ions, remains to be determined.
RECYCLING AND DEGRADATION
Recycling
After a round of splicing, the spliceosome undergoes an active disassembly process in which the snRNPs and NTC are recycled for a new round of splicing. The U2/U6.U5-intron lariat complex is disassembled by the ATP-dependent DEAH-box helicase Prp43 with cofactors Ntr1 and Ntr2 (Martin et al. 2002; Tsai et al. 2005). In in vitro assays, U6 is released as the free snRNA (Tsai et al. 2005; Fourmann et al. 2013, 2016). In the cell, U6 is presumably recognized during or after release from the ILS by Prp24 and/or Lsm2-8. Prp24 is required in yeast even when mutations in U4 and U6 allow for bypass of stable U4/U6 base-pairing, indicating that Prp24 has an essential function other than U4/U6 annealing, perhaps in driving ILS dissociation by capturing U6 (Burke et al. 2015). It is not known if recycling of U6 after a round of splicing follows the same pathway as initial assembly of the U6 snRNP, i.e., U6 is bound by Prp24 and Lsm2-8, then assembled into the U4/U6 di-snRNP.
Degradation
The degradation pathway of U6 is poorly understood. The balance between uridylation, polyadenylation, and trimming by Usb1 (discussed in the “3′ End modification” section) affects the half-life of U6 in metazoans (Shchepachev et al. 2012, 2015; Hilcenko et al. 2013). End processing by Usb1 has a protective effect against degradation in S. cerevisiae, S. pombe, and human cells, suggesting that the modification may protect against other cellular nucleases (Mroczek et al. 2012; Shchepachev et al. 2012, 2015; Hilcenko et al. 2013). However, it is not known which nucleases are responsible for degrading U6 in Usb1-depleted cells or during the normal U6 life cycle. Additionally, other cellular deadenylases can interact with and influence the 3′ sequence of U6 snRNA (Shukla and Parker 2017). How the activities of different 3′ modifying enzymes affect the stability and functionality of U6 is poorly understood.
Recent data suggest that the exosome may play a role in normal turnover of spliceosomal RNAs, as knockdown of the exosome exonuclease Rrp44 resulted in higher levels of U6 (Zhang et al. 2015). The presence of a terminal phosphate (in yeast) or 2′,3′-cyclic phosphate (in humans) on U6 may inhibit degradation by the exosome, as the nuclear exosome-associated exonuclease Rrp6 is inactive on a 3′ phosphate-terminated RNA (Burkard and Butler 2000). Additional enzymes may be necessary to dephosphorylate the 3′ end of U6 prior to adenylation and decay. It has been demonstrated that Mtr4 and Trf4 (components of the TRAMP complex) as well as Rrp6 interact with components of the U4/U6.U5 tri-snRNP (Nag and Steitz 2012). While the primary reason for recruitment of the exosome machinery may be to degrade improperly spliced mRNAs or excised introns, it may also have a role in degrading “expired” snRNAs. The TRAMP complex is recruited to the intron even before splicing is complete and may in fact help stimulate splicing (Kong et al. 2014). Thus, the components of the nuclear decay machinery are temporally and spatially near the spliceosomal RNAs. Supporting this idea, in S. cerevisiae, deletion of the poly(A) polymerase from the TRAMP complex, Trf4, results in a small increase in U6 levels (Copela et al. 2008). Moreover, Rrp44, Rrp6, and Trf4 can be crosslinked to U6 (Schneider et al. 2012). However, deletion of Trf4 in S. pombe does not affect polyadenylation of U6, suggesting that other poly(A) polymerases are involved or can complement loss of Trf4 (Shchepachev et al. 2015). Further study on the determinants of U6 decay, including the adenylation state of the 3′ tail, will help to define how this metabolically stable RNA is monitored, which may provide another mechanism for spliceosome fidelity.
CONCLUSIONS AND PERSPECTIVES
X-ray crystallography and cryo-EM approaches have led to all-atom models of multiple spliceosomal complexes that contain U6 snRNA. These structures allow us to directly visualize the vast conformational changes that U6 must undergo during spliceosome assembly and activation, and highlight the large number of dynamic steps that must occur to form such complex structures. Each assembly and processing step represents an opportunity for quality control, ensuring that functional spliceosomes are prepared to carry out a step of gene expression that is essential in all eukaryotes. U6, U2, and U5 snRNA as well as many proteins must come together to form the catalytic core of the spliceosome, where U6 snRNA coordinates the magnesium ions required for splicing catalysis. Regulation and monitoring of U6 throughout its lifetime is crucial for retaining the spliceosome's catalytic activity and to ensure that only full-length, properly modified U6 molecules are incorporated into the spliceosome. Research in the coming years will further illuminate how U6 is regulated (via transcriptional activity, post-transcriptional modification, RNA–protein interactions, and subnuclear localization) and assembled into the spliceosome.
ACKNOWLEDGMENTS
We thank Aaron Hoskins and Tucker Carrocci for insightful and illuminating discussions and for critical reading of the manuscript. We apologize to our colleagues for work that was not cited due to space limitations. S.E.B. and D.A.B. are supported by grants from the US National Institutes of Health (R35 GM118131 to S.E.B. and R35 GM118075 to D.A.B.).
Author contributions: A.L.D., S.E.B., and D.A.B. wrote the article.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.065136.117.
REFERENCES
- Abelson J, Blanco M, Ditzler MA, Fuller F, Aravamudhan P, Wood M, Villa T, Ryan DE, Pleiss JA, Maeder C, et al. 2010. Conformational dynamics of single pre-mRNA molecules during in vitro splicing. Nat Struct Mol Biol 17: 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absmeier E, Wollenhaupt J, Mozaffari-Jovin S, Becke C, Lee CT, Preussner M, Heyd F, Urlaub H, Lührmann R, Santos KF, et al. 2015. The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev 29: 2576–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absmeier E, Santos KF, Wahl MC. 2016. Functions and regulation of the Brr2 RNA helicase during splicing. Cell Cycle 15: 3362–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J 18: 5789–5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afroz T, Cienikova Z, Cléry A, Allain FH. 2015. One, two, three, four! How multiple RRMs read the genome sequence. Methods Enzymol 558: 235–278. [DOI] [PubMed] [Google Scholar]
- Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Lührmann R, Stark H. 2016. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 351: 1416–1420. [DOI] [PubMed] [Google Scholar]
- Arimbasseri AG, Bhargava P. 2008. Chromatin structure and expression of a gene transcribed by RNA polymerase III are independent of H2A.Z deposition. Mol Cell Biol 28: 2598–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Maraia RJ. 2015. Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol Cell 58: 1124–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Rijal K, Maraia RJ. 2013. Transcription termination by the eukaryotic RNA polymerase III. Biochim Biophys Acta 1829: 318–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae E, Reiter NJ, Bingman CA, Kwan SS, Lee D, Phillips GN Jr, Butcher SE, Brow DA. 2007. Structure and interactions of the first three RNA recognition motifs of splicing factor Prp24. J Mol Biol 367: 1447–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao P, Höbartner C, Hartmuth K, Lührmann R. 2017. Yeast Prp2 liberates the 5′ splice site and the branch site adenosine for catalysis of pre-mRNA splicing. RNA 23: 1770–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. 2010. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol 17: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak A, Query CC. 2014. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep 8: 966–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G. 2006. The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol 71: 313–320. [DOI] [PubMed] [Google Scholar]
- Belagal P, Normand C, Shukla A, Wang R, Léger-Silvestre I, Dez C, Bhargava P, Gadal O. 2016. Decoding the principles underlying the frequency of association with nucleoli for RNA polymerase III-transcribed genes in budding yeast. Mol Biol Cell 27: 3164–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A. 2002. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J 21: 2724–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, Urlaub H, Kastner B, Lührmann R, Stark H. 2017a. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell 170: 701–713.e11. [DOI] [PubMed] [Google Scholar]
- Bertram K, Agafonov DE, Liu WT, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2017b. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542: 318–323. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. 1998. Nucleolar localization of early tRNA processing. Genes Dev 12: 2463–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Lutzoni F, Reeb V, Simon D, Nason J, Fernandez F. 2000. Widespread occurrence of spliceosomal introns in the rDNA genes of ascomycetes. Mol Biol Evol 17: 1971–1984. [DOI] [PubMed] [Google Scholar]
- Bhattacharya R, Perumal K, Sinha K, Maraia R, Reddy R. 2002. Methylphosphate cap structure in small RNAs reduces the affinity of RNAs to La protein. Gene Expr 10: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL, Steitz JA. 1986. Pre-mRNA splicing in vitro requires intact U4/U6 small nuclear ribonucleoprotein. Cell 46: 697–704. [DOI] [PubMed] [Google Scholar]
- Blad H, Reiter NJ, Abildgaard F, Markley JL, Butcher SE. 2005. Dynamics and metal ion binding in the U6 RNA intramolecular stem-loop as analyzed by NMR. J Mol Biol 353: 540–555. [DOI] [PubMed] [Google Scholar]
- Boelens WC, Palacios I, Mattaj IW. 1995. Nuclear retention of RNA as a mechanism for localization. RNA 1: 273–283. [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen DF, Brown DD. 1981. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell 24: 261–270. [DOI] [PubMed] [Google Scholar]
- Booth BL Jr, Pugh BF. 1997. Identification and characterization of a nuclease specific for the 3′ end of the U6 small nuclear RNA. J Biol Chem 272: 984–991. [DOI] [PubMed] [Google Scholar]
- Braglia P, Dugas SL, Donze D, Dieci G. 2007. Requirement of Nhp6 proteins for transcription of a subset of tRNA genes and heterochromatin barrier function in Saccharomyces cerevisiae. Mol Cell Biol 27: 1545–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann P, Lührmann R. 1987. Antibodies specific for N 6-methyladenosine react with intact snRNPs U2 and U4/U6. FEBS Lett 213: 309–315. [DOI] [PubMed] [Google Scholar]
- Bringmann P, Appel B, Rinke J, Reuter R, Theissen H, Lührmann R. 1984. Evidence for the existence of snRNAs U4 and U6 in a single ribonucleoprotein complex and for their association by intermolecular base pairing. EMBO J 3: 1357–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. 1988. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature 334: 213–218. [DOI] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. 1989. Splicing a spliceosomal RNA. Nature 337: 14–15. [DOI] [PubMed] [Google Scholar]
- Brow DA, Guthrie C. 1990. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev 4: 1345–1356. [DOI] [PubMed] [Google Scholar]
- Brow DA, Vidaver RM. 1995. An element in human U6 RNA destabilizes the U4/U6 spliceosomal RNA complex. RNA 1: 122–131. [PMC free article] [PubMed] [Google Scholar]
- Burkard KT, Butler JS. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol Cell Biol 20: 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JE, Butcher SE, Brow DA. 2015. Spliceosome assembly in the absence of stable U4/U6 RNA pairing. RNA 21: 923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnol AF, Margottin F, Schultz P, Marsolier MC, Oudet P, Sentenac A. 1993. Basal promoter and enhancer element of yeast U6 snRNA gene. J Mol Biol 233: 644–658. [DOI] [PubMed] [Google Scholar]
- Canzler S, Stadler PF, Hertel J. 2016. U6 snRNA intron insertion occurred multiple times during fungi evolution. RNA Biol 13: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Pepperkok R, Sproat BS, Ansorge W, Swanson MS, Lamond AI. 1991a. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. EMBO J 10: 1863–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino SM, Merdes A, Brunner C, Zamore PD, Green MR, Hurt E, Lamond AI. 1991b. Mammalian nuclei contain foci which are highly enriched in components of the pre-mRNA splicing machinery. EMBO J 10: 195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR. 1986. The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell 44: 207–210. [DOI] [PubMed] [Google Scholar]
- Chan SP, Cheng SC. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J Biol Chem 280: 31190–31199. [DOI] [PubMed] [Google Scholar]
- Chan SP, Kao DI, Tsai WY, Cheng SC. 2003. The Prp19p-associated complex in spliceosome activation. Science 302: 279–282. [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Jacquier A. 1994. Catalytic site components common to both splicing steps of a group II intron. Science 266: 1383–1387. [DOI] [PubMed] [Google Scholar]
- Charette M, Gray MW. 2000. Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49: 341–351. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sinha K, Perumal K, Reddy R. 2000. Effect of 3′ terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. RNA 6: 1277–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng SC, Abelson J. 1987. Spliceosome assembly in yeast. Genes Dev 1: 1014–1027. [DOI] [PubMed] [Google Scholar]
- Copela LA, Fernandez CF, Sherrer RL, Wolin SL. 2008. Competition between the Rex1 exonuclease and the La protein affects both Trf4p-mediated RNA quality control and pre-tRNA maturation. RNA 14: 1214–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu G, Didychuk AL, Rodgers ML, Michael LA, Burke JE, Montemayor EJ, Hoskins AA, Butcher SE. 2016. Structural analysis of multi-helical RNAs by NMR-SAXS/WAXS: application to the U4/U6 di-snRNA. J Mol Biol 428: 777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove MS, Ding Y, Rennie WA, Lane MJ, Hanes SD. 2012. The Bin3 RNA methyltransferase targets 7SK RNA to control transcription and translation. Wiley Interdiscip Rev RNA 3: 633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford DJ, Hoskins AA, Friedman LJ, Gelles J, Moore MJ. 2013. Single-molecule colocalization FRET evidence that spliceosome activation precedes stable approach of 5′ splice site and branch site. Proc Natl Acad Sci 110: 6783–6788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damianov A, Schreiner S, Bindereif A. 2004. Recycling of the U12-type spliceosome requires p110, a component of the U6atac snRNP. Mol Cell Biol 24: 1700–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Jády BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. 2002. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J 21: 2746–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das G, Henning D, Wright D, Reddy R. 1988. Upstream regulatory elements are necessary and sufficient for transcription of a U6 RNA gene by RNA polymerase III. EMBO J 7: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR. 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didychuk AL, Montemayor EJ, Brow DA, Butcher SE. 2016. Structural requirements for protein-catalyzed annealing of U4 and U6 RNAs during di-snRNP assembly. Nucleic Acids Res 44: 1398–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didychuk AL, Montemayor EJ, Carrocci TJ, DeLaitsch AT, Lucarelli SE, Westler WM, Brow DA, Hoskins AA, Butcher SE. 2017. Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities. Nat Commun 8: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich RC, Padgett RA, Shukla GC. 2009. The conserved 3′ end domain of U6atac snRNA can direct U6 snRNA to the minor spliceosome. RNA 15: 1198–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domitrovich AM, Kunkel GR. 2003. Multiple, dispersed human U6 small nuclear RNA genes with varied transcriptional efficiencies. Nucleic Acids Res 31: 2344–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet AJ, Droc G, Siol O, Audoux J, Gilbert N. 2015. U6 snRNA pseudogenes: markers of retrotransposition dynamics in mammals. Mol Biol Evol 32: 1815–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dundr M, Hebert MD, Karpova TS, Stan D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. 2004. In vivo kinetics of Cajal body components. J Cell Biol 164: 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein P, Reddy R, Henning D, Busch H. 1980. The nucleotide sequence of nuclear U6 (4.7 S) RNA. J Biol Chem 255: 8901–8906. [PubMed] [Google Scholar]
- Eschenlauer JB, Kaiser MW, Gerlach VL, Brow DA. 1993. Architecture of a yeast U6 RNA gene promoter. Mol Cell Biol 13: 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J. 1990. Two domains of yeast U6 small nuclear RNA required for both steps of nuclear precursor messenger RNA splicing. Science 250: 404–409. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J. 1992. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res 20: 3659–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. 2013. RNA catalyses nuclear pre-mRNA splicing. Nature 503: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Mefford MA, Piccirilli JA, Staley JP. 2014. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat Struct Mol Biol 21: 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai XC, Newman AJ, Nagai K. 2017. Structure of a spliceosome remodelled for exon ligation. Nature 542: 377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortner DM, Troy RG, Brow DA. 1994. A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes Dev 8: 221–233. [DOI] [PubMed] [Google Scholar]
- Fourmann JB, Schmitzova J, Christian H, Urlaub H, Ficner R, Boon KL, Fabrizio P, Lührmann R. 2013. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 27: 413–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourmann JB, Dybkov O, Agafonov DE, Tauchert MJ, Urlaub H, Ficner R, Fabrizio P, Lührmann R. 2016. The target of the DEAH-box NTP triphosphatase Prp43 in Saccharomyces cerevisiae spliceosomes is the U2 snRNP-intron interaction. Elife 5: e15564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French SL, Osheim YN, Schneider DA, Sikes ML, Fernandez CF, Copela LA, Misra VA, Nomura M, Wolin SL, Beyer AL. 2008. Visual analysis of the yeast 5S rRNA gene transcriptome: regulation and role of La protein. Mol Cell Biol 28: 4576–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frendewey D, Barta I, Gillespie M, Potashkin J. 1990. Schizosaccharomyces U6 genes have a sequence within their introns that matches the B box consensus of tRNA internal promoters. Nucleic Acids Res 18: 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M, Mayes AE, Brunet-Simon A, Rain JC, Colley A, Dix I, Decourty L, Joly N, Ricard F, Beggs JD, et al. 2000. Genome-wide protein interaction screens reveal functional networks involving Sm-like proteins. Yeast 17: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galisson F, Legrain P. 1993. The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the [U4/U6.U5] tri-snRNP. Nucleic Acids Res 21: 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Jády BE, Bortolin ML, Darzacq X, Kiss T. 1999. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol 19: 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi SA, Lange TS. 2002. All small nuclear RNAs (snRNAs) of the [U4/U6.U5] Tri-snRNP localize to nucleoli; Identification of the nucleolar localization element of U6 snRNA. Mol Biol Cell 13: 3123–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach VL, Whitehall SK, Geiduschek EP, Brow DA. 1995. TFIIIB placement on a yeast U6 RNA gene in vivo is directed primarily by TFIIIC rather than by sequence-specific DNA contacts. Mol Cell Biol 15: 1455–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghetti A, Company M, Abelson J. 1995. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA 1: 132–145. [PMC free article] [PubMed] [Google Scholar]
- Gottlieb E, Steitz JA. 1989. Function of the mammalian La protein: evidence for its action in transcription termination by RNA polymerase III. EMBO J 8: 851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski PJ, Sharp PA. 1986. Affinity chromatography of splicing complexes: U2, U5, and U4 + U6 small nuclear ribonucleoprotein particles in the spliceosome. Science 233: 1294–1299. [DOI] [PubMed] [Google Scholar]
- Gu J, Patton JR, Shimba S, Reddy R. 1996. Localization of modified nucleotides in Schizosaccharomyces pombe spliceosomal small nuclear RNAs: modified nucleotides are clustered in functionally important regions. RNA 2: 909–918. [PMC free article] [PubMed] [Google Scholar]
- Gu J, Shumyatsky G, Makan N, Reddy R. 1997. Formation of 2′,3′-cyclic phosphates at the 3′ end of human U6 small nuclear RNA in vitro. Identification of 2′,3′-cyclic phosphates at the 3′ ends of human signal recognition particle and mitochondrial RNA processing RNAs. J Biol Chem 272: 21989–21993. [DOI] [PubMed] [Google Scholar]
- Gupta S, Busch RK, Singh R, Reddy R. 1990a. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of γ-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J Biol Chem 265: 19137–19142. [PubMed] [Google Scholar]
- Gupta S, Singh R, Reddy R. 1990b. Capping of U6 small nuclear RNA in vitro can be uncoupled from transcription. J Biol Chem 265: 9491–9495. [PubMed] [Google Scholar]
- Guthrie C. 1991. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science 253: 157–163. [DOI] [PubMed] [Google Scholar]
- Hamada M, Sakulich AL, Koduru SB, Maraia RJ. 2000. Transcription termination by RNA polymerase III in fission yeast. A genetic and biochemically tractable model system. J Biol Chem 275: 29076–29081. [DOI] [PubMed] [Google Scholar]
- Hamm J, Mattaj IW. 1989. An abundant U6 snRNP found in germ cells and embryos of Xenopus laevis. EMBO J 8: 4179–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin JW, Warnasooriya C, Kondo Y, Nagai K, Rueda D. 2015. Assembly and dynamics of the U4/U6 di-snRNP by single-molecule FRET. Nucleic Acids Res 43: 10963–10974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto C, Steitz JA. 1984. U4 and U6 RNAs coexist in a single small nuclear ribonucleoprotein particle. Nucleic Acids Res 12: 3283–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun TR, Malmström L, Anderson S, Graczyk BJ, Fox B, Riffle M, Sundin BA, Aranda JD, McDonald WH, Chiu CH, et al. 2003. Assigning function to yeast proteins by integration of technologies. Mol Cell 12: 1353–1365. [DOI] [PubMed] [Google Scholar]
- Hernandez N. 2001. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J Biol Chem 276: 26733–26736. [DOI] [PubMed] [Google Scholar]
- Hilcenko C, Simpson PJ, Finch AJ, Bowler FR, Churcher MJ, Jin L, Packman LC, Shlien A, Campbell P, Kirwan M, et al. 2013. Aberrant 3′ oligoadenylation of spliceosomal U6 small nuclear RNA in poikiloderma with neutropenia. Blood 121: 1028–1038. [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Staley JP. 2004. Multiple functions for the invariant AGC triad of U6 snRNA. RNA 10: 921–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Moore MJ. 2012. The spliceosome: a flexible, reversible macromolecular machine. Trends Biochem Sci 37: 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins AA, Rodgers ML, Friedman LJ, Gelles J, Moore MJ. 2016. Single molecule analysis reveals reversible and irreversible steps during spliceosome activation. Elife 5: e14166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Chung CS, Kao DI, Kao TC, Cheng SC. 2014. Sad1 counteracts Brr2-mediated dissociation of U4/U6.U5 in tri-snRNP homeostasis. Mol Cell Biol 34: 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A, Kiefmann M, Meier-Ewert S, O'Brien J, Lehrach H, Bachellerie JP, Brosius J. 2001. RNomics: an experimental approach that identifies 201 candidates for novel, small, non-messenger RNAs in mouse. EMBO J 20: 2943–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 22: 1878–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Thérien C, Bergeron D, Bourassa S, Greenblatt J, et al. 2007. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 27: 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TL, Abelson J. 2001. Characterization of U4 and U6 interactions with the 5′ splice site using a S. cerevisiae in vitro trans-splicing system. Genes Dev 15: 1957–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser MW, Brow DA. 1995. Lethal mutations in a yeast U6 RNA gene B block promoter element identify essential contacts with transcription factor-IIIC. J Biol Chem 270: 11398–11405. [DOI] [PubMed] [Google Scholar]
- Kaiser MW, Chi J, Brow DA. 2004. Position-dependent function of a B block promoter element implies a specialized chromatin structure on the S. cerevisiae U6 RNA gene, SNR6. Nucleic Acids Res 32: 4297–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandels-Lewis S, Séraphin B. 1993. Involvement of U6 snRNA in 5′ splice site selection. Science 262: 2035–2039. [DOI] [PubMed] [Google Scholar]
- Kiss T, Antal M, Solymosy F. 1987. Plant small nuclear RNAs. II. U6 RNA and a 4.5SI-like RNA are present in plant nuclei. Nucleic Acids Res 15: 543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingauf M, Stan D, Neugebauer KM. 2006. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell 17: 4972–4981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong KY, Tang HM, Pan K, Huang Z, Lee TH, Hinnebusch AG, Jin DY, Wong CM. 2014. Cotranscriptional recruitment of yeast TRAMP complex to intronic sequences promotes optimal pre-mRNA splicing. Nucleic Acids Res 42: 643–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643. [DOI] [PubMed] [Google Scholar]
- Krogh N, Kongsbak-Wismann M, Geisler C, Nielsen H. 2017. Substoichiometric ribose methylations in spliceosomal snRNAs. Org Biomol Chem 15: 8872–8876. [DOI] [PubMed] [Google Scholar]
- Kruppa M, Moir RD, Kolodrubetz D, Willis IM. 2001. Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae. Mol Cell 7: 309–318. [DOI] [PubMed] [Google Scholar]
- Kunkel GR, Pederson T. 1988. Upstream elements required for efficient transcription of a human U6 RNA gene resemble those of U1 and U2 genes even though a different polymerase is used. Genes Dev 2: 196–204. [DOI] [PubMed] [Google Scholar]
- Kwan SS, Brow DA. 2005. The N- and C-terminal RNA recognition motifs of splicing factor Prp24 have distinct functions in U6 RNA binding. RNA 11: 808–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan S, Gerlach VL, Brow DA. 2000. Disruption of the 5′ stem-loop of yeast U6 RNA induces trimethylguanosine capping of this RNA polymerase III transcript in vivo. RNA 6: 1859–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Lührmann R. 1998. The human U5–200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci 95: 4188–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange TS, Gerbi SA. 2000. Transient nucleolar localization Of U6 small nuclear RNA in Xenopus Laevis oocytes. Mol Biol Cell 11: 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonné R, Lührmann R. 2006. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 17: 3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Steitz JA. 1979. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc Natl Acad Sci 76: 5495–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C. 1993. Mutations in U6 snRNA that alter splice site specificity: implications for the active site. Science 262: 1982–1988. [DOI] [PubMed] [Google Scholar]
- Li Z, Brow DA. 1996. A spontaneous duplication in U6 spliceosomal RNA uncouples the early and late functions of the ACAGA element in vivo. RNA 2: 879–894. [PMC free article] [PubMed] [Google Scholar]
- Licht K, Medenbach J, Lührmann R, Kambach C, Bindereif A. 2008. 3′-cyclic phosphorylation of U6 snRNA leads to recruitment of recycling factor p110 through LSm proteins. RNA 14: 1532–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BR, Natarajan V. 2012. Negative regulation of human U6 snRNA promoter by p38 kinase through Oct-1. Gene 497: 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listerman I, Bledau AS, Grishina I, Neugebauer KM. 2007. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet 3: e212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Mozaffari-Jovin S, Wollenhaupt J, Santos KF, Theuser M, Dunin-Horkawicz S, Fabrizio P, Bujnicki JM, Lührmann R, Wahl MC. 2015. A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing. Elife 4: e07320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo SM, Hernandez N. 1989. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell 58: 55–67. [DOI] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. 1992. Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science 255: 327–330. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Bordonné R, Guthrie C. 1990. Multiple roles for U6 snRNA in the splicing pathway. Genes Dev 4: 2264–2277. [DOI] [PubMed] [Google Scholar]
- Maeder C, Kutach AK, Guthrie C. 2009. ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase requires the C terminus of Prp8. Nat Struct Mol Biol 16: 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ. 1996. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc Natl Acad Sci 93: 3383–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ, Kenan DJ, Keene JD. 1994. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol 14: 2147–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcia M, Pyle AM. 2012. Visualizing group II intron catalysis through the stages of splicing. Cell 151: 497–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margottin F, Dujardin G, Gérard M, Egly JM, Huet J, Sentenac A. 1991. Participation of the TATA factor in transcription of the yeast U6 gene by RNA polymerase C. Science 251: 424–426. [DOI] [PubMed] [Google Scholar]
- Martin MP, Gerlach VL, Brow DA. 2001. A novel upstream RNA polymerase III promoter element becomes essential when the chromatin structure of the yeast U6 RNA gene is altered. Mol Cell Biol 21: 6429–6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A, Schneider S, Schwer B. 2002. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem 277: 17743–17750. [DOI] [PubMed] [Google Scholar]
- Martin-Tumasz S, Richie AC, Clos LJ II, Brow DA, Butcher SE. 2011. A novel occluded RNA recognition motif in Prp24 unwinds the U6 RNA internal stem loop. Nucleic Acids Res 39: 7837–7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S, Branlant C. 1999. A limited number of pseudouridine residues in the human atac spliceosomal U snRNAs as compared to human major spliceosomal UsnRNAs. RNA 5: 1495–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. 1999. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol 19: 2142–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Wang Z. 2014. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 15: 108–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Ward DC. 1993. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol 121: 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes AE, Verdone L, Legrain P, Beggs JD. 1999. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J 18: 4321–4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus CJ, Schwartz ML, Butcher SE, Brow DA. 2007. A dynamic bulge in the U6 RNA internal stem-loop functions in spliceosome assembly and activation. RNA 13: 2252–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J, Schreiner S, Liu S, Lührmann R, Bindereif A. 2004. Human U4/U6 snRNP recycling factor p110: mutational analysis reveals the function of the tetratricopeptide repeat domain in recycling. Mol Cell Biol 24: 7392–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford MA, Staley JP. 2009. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA 15: 1386–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. 1972. Formation of hybrid DNA by rotary diffusion during genetic recombination. J Mol Biol 71: 795–798. [DOI] [PubMed] [Google Scholar]
- Meyer KD, Jaffrey SR. 2014. The dynamic epitranscriptome: N 6-methyladenosine and gene expression control. Nat Rev Mol Cell Biol 15: 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moenne A, Camier S, Anderson G, Margottin F, Beggs J, Sentenac A. 1990. The U6 gene of Saccharomyces cerevisiae is transcribed by RNA polymerase C (III) in vivo and in vitro. EMBO J 9: 271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montemayor EJ, Curran EC, Liao HH, Andrews KL, Treba CN, Butcher SE, Brow DA. 2014. Core structure of the U6 small nuclear ribonucleoprotein at 1.7-Å resolution. Nat Struct Mol Biol 21: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montemayor EJ, Didychuk AL, Liao H, Hu P, Brow DA, Butcher SE. 2017. Structure and conformational plasticity of the U6 small nuclear ribonucleoprotein core. Acta Crystallogr D Struct Biol 73: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffari-Jovin S, Santos KF, Hsiao HH, Will CL, Urlaub H, Wahl MC, Lührmann R. 2012. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev 26: 2422–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozaffari-Jovin S, Wandersleben T, Santos KF, Will CL, Lührmann R, Wahl MC. 2013. Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science 341: 80–84. [DOI] [PubMed] [Google Scholar]
- Mroczek S, Dziembowski A. 2013. U6 RNA biogenesis and disease association. Wiley Interdiscip Rev RNA 4: 581–592. [DOI] [PubMed] [Google Scholar]
- Mroczek S, Krwawicz J, Kutner J, Lazniewski M, Kuciński I, Ginalski K, Dziembowski A. 2012. C16orf57, a gene mutated in poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible for the U6 snRNA 3′ end modification. Genes Dev 26: 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mura C, Randolph PS, Patterson J, Cozen AE. 2013. Archaeal and eukaryotic homologs of Hfq: a structural and evolutionary perspective on Sm function. RNA Biol 10: 636–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A, Steitz JA. 2012. Tri-snRNP-associated proteins interact with subunits of the TRAMP and nuclear exosome complexes, linking RNA decay and pre-mRNA splicing. RNA Biol 9: 334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. 2001. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 7: 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K. 2015. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 523: 47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TH, Galej WP, Bai XC, Oubridge C, Newman AJ, Scheres SH, Nagai K. 2016. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature 530: 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotńy I, Blažíková M, Staněk D, Herman P, Malinsky J. 2011. In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol Biol Cell 22: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotńy I, Malinová A, Stejskalová E, Matěju˚ D, Klimešová K, Roithová A, Švéda M, Knejzlík Z, Staněk D. 2015. SART3-dependent accumulation of incomplete spliceosomal snRNPs in Cajal bodies. Cell Rep 10: 429–440. [DOI] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. 2010. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol 17: 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone BK, Xue D, Wolin SL. 1998. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J 17: 7442–7495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Das T, Song EJ, Kim EE. 2016. Structural basis for recruiting and shuttling of the spliceosomal deubiquitinase USP4 by SART3. Nucleic Acids Res 44: 5424–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peebles CL, Perlman PS, Mecklenburg KL, Petrillo ML, Tabor JH, Jarrell KA, Cheng HL. 1986. A self-splicing RNA excises an intron lariat. Cell 44: 213–223. [DOI] [PubMed] [Google Scholar]
- Peebles CL, Zhang M, Perlman PS, Franzen JS. 1995. Catalytically critical nucleotide in domain 5 of a group II intron. Proc Natl Acad Sci 92: 4422–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton KE, Chen B, Liu K, Hunter OV, Xie Y, Tu BP, Conrad NK. 2017. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell 169: 824–835.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessa HK, Will CL, Meng X, Schneider C, Watkins NJ, Perälä N, Nymark M, Turunen JJ, Lührmann R, Frilander MJ. 2008. Minor spliceosome components are predominantly localized in the nucleus. Proc Natl Acad Sci 105: 8655–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaschka C, Lin PC, Nagai K. 2017. Structure of a pre-catalytic spliceosome. Nature 546: 617–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. 2016. Group II intron self-splicing. Annu Rev Biophys 45: 183–205. [DOI] [PubMed] [Google Scholar]
- Rader SD, Guthrie C. 2002. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA 8: 1378–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. 1998a. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 8: 847–855. [DOI] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. 1998b. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science 279: 857–860. [DOI] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. 2010. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci 107: 3639–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, et al. 2016. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353: 1399–1405. [DOI] [PubMed] [Google Scholar]
- Reddy R, Henning D, Das G, Harless M, Wright D. 1987. The capped U6 small nuclear RNA is transcribed by RNA polymerase III. J Biol Chem 262: 75–81. [PubMed] [Google Scholar]
- Reijns MA, Auchynnikava T, Beggs JD. 2009. Analysis of Lsm1p and Lsm8p domains in the cellular localization of Lsm complexes in budding yeast. FEBS J 276: 3602–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rife JP, Cheng CS, Moore PB, Strobel SA. 1998. N 2-methylguanosine is iso-energetic with guanosine in RNA duplexes and GNRA tetraloops. Nucleic Acids Res 26: 3640–3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke J, Steitz JA. 1985. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res 13: 2617–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers ML, Didychuk AL, Butcher SE, Brow DA, Hoskins AA. 2016. A multi-step model for facilitated unwinding of the yeast U4/U6 RNA duplex. Nucleic Acids Res 44: 10912–10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roiha H, Shuster EO, Brow DA, Guthrie C. 1989. Small nuclear RNAs from budding yeasts: phylogenetic comparisons reveal extensive size variation. Gene 82: 137–144. [DOI] [PubMed] [Google Scholar]
- Roost C, Lynch SR, Batista PJ, Qu K, Chang HY, Kool ET. 2015. Structure and thermodynamics of N 6-methyladenosine in RNA: a spring-loaded base modification. J Am Chem Soc 137: 2107–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegger S, Miki TS, Hess D, Großhans H. 2015. The ribonucleotidyl transferase USIP-1 acts with SART3 to promote U6 snRNA recycling. Nucleic Acids Res 43: 3344–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Allmann AM, Van Doren SR, Butcher SE. 2003. Structural basis for a lethal mutation in U6 RNA. Biochemistry 42: 1470–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Venditti V, Angers CG, Cornilescu G, Butcher SE. 2007. Structure and thermodynamics of a conserved U2 snRNA domain from yeast and human. RNA 13: 328–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H, Abelson J. 1992. Evidence for a base-pairing interaction between U6 small nuclear RNA and 5′ splice site during the splicing reaction in yeast. Proc Natl Acad Sci 89: 11269–11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa H, Shimura Y. 1992. Association of U6 snRNA with the 5′-splice site region of pre-mRNA in the spliceosome. Genes Dev 6: 244–254. [DOI] [PubMed] [Google Scholar]
- Schneider C, Will CL, Makarova OV, Makarov EM, Lührmann R. 2002. Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol Cell Biol 22: 3219–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Will CL, Anokhina M, Tazi J, Urlaub H, Lührmann R. 2010. Exon definition complexes contain the tri-snRNP and can be directly converted into B-like precatalytic splicing complexes. Mol Cell 38: 223–235. [DOI] [PubMed] [Google Scholar]
- Schneider C, Kudla G, Wlotzka W, Tuck A, Tollervey D. 2012. Transcriptome-wide analysis of exosome targets. Mol Cell 48: 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon KW, Guthrie C. 1991. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev 5: 773–785. [DOI] [PubMed] [Google Scholar]
- Sharif H, Conti E. 2013. Architecture of the Lsm1-7-Pat1 complex: a conserved assembly in eukaryotic mRNA turnover. Cell Rep 5: 283–291. [DOI] [PubMed] [Google Scholar]
- Sharp PA. 1985. On the origin of RNA splicing and introns. Cell 42: 397–400. [DOI] [PubMed] [Google Scholar]
- Shchepachev V, Wischnewski H, Missiaglia E, Soneson C, Azzalin CM. 2012. Mpn1, mutated in poikiloderma with neutropenia protein 1, is a conserved 3′-to-5′ RNA exonuclease processing U6 small nuclear RNA. Cell Rep 2: 855–865. [DOI] [PubMed] [Google Scholar]
- Shchepachev V, Wischnewski H, Soneson C, Arnold AW, Azzalin CM. 2015. Human Mpn1 promotes post-transcriptional processing and stability of U6atac. FEBS Lett 589: 2417–2423. [DOI] [PubMed] [Google Scholar]
- Shcherbakova I, Hoskins AA, Friedman LJ, Serebrov V, Corrêa IR Jr, Xu MQ, Gelles J, Moore MJ. 2013. Alternative spliceosome assembly pathways revealed by single-molecule fluorescence microscopy. Cell Rep 5: 151–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2017. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol 18: 655–670. [DOI] [PubMed] [Google Scholar]
- Shimba S, Reddy R. 1994. Purification of human U6 small nuclear RNA capping enzyme. Evidence for a common capping enzyme for γ-monomethyl-capped small RNAs. J Biol Chem 269: 12419–12423. [PubMed] [Google Scholar]
- Shimba S, Bokar JA, Rottman F, Reddy R. 1995. Accurate and efficient N 6-adenosine methylation in spliceosomal U6 small nuclear RNA by HeLa cell extract in vitro. Nucleic Acids Res 23: 2421–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaswamy S, Kassavetis GA, Bhargava P. 2004. High-level activation of transcription of the yeast U6 snRNA gene in chromatin by the basal RNA polymerase III transcription factor TFIIIC. Mol Cell Biol 24: 3596–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA. 2001. The intramolecular stem-loop structure of U6 snRNA can functionally replace the U6atac snRNA stem-loop. RNA 7: 94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla GC, Padgett RA. 2002. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol Cell 9: 1145–1150. [DOI] [PubMed] [Google Scholar]
- Shukla S, Parker R. 2017. PARN modulates Y RNA stability and its 3′-end formation. Mol Cell Biol 37: e00264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumyatsky G, Wright D, Reddy R. 1993. Methylphosphate cap structure increases the stability of 7SK, B2 and U6 small RNAs in Xenopus oocytes. Nucleic Acids Res 21: 4756–4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidarovich A, Will CL, Anokhina MM, Ceballos J, Sievers S, Agafonov DE, Samatov T, Bao P, Kastner B, Urlaub H, et al. 2017. Identification of a small molecule inhibitor that stalls splicing at an early step of spliceosome activation. Elife 6: e23533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigal N, Alberts B. 1972. Genetic recombination: the nature of a crossed strand-exchange between two homologous DNA molecules. J Mol Biol 71: 789–793. [DOI] [PubMed] [Google Scholar]
- Siliciano PG, Brow DA, Roiha H, Guthrie C. 1987. An essential snRNA from S. cerevisiae has properties predicted for U4, including interaction with a U6-like snRNA. Cell 50: 585–592. [DOI] [PubMed] [Google Scholar]
- Singh R, Reddy R. 1989. γ-monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci 86: 8280–8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Gupta S, Reddy R. 1990. Capping of mammalian U6 small nuclear RNA in vitro is directed by a conserved stem-loop and AUAUAC sequence: conversion of a noncapped RNA into a capped RNA. Mol Cell Biol 10: 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Sikand K, Conrad H, Will CL, Komar AA, Shukla GC. 2016. U6atac snRNA stem-loop interacts with U12 p65 RNA binding protein and is functionally interchangeable with the U12 apical stem-loop III. Sci Rep 6: 31393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small EC, Leggett SR, Winans AA, Staley JP. 2006. The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell 23: 389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA. 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996. [DOI] [PubMed] [Google Scholar]
- Spaniel C, Honda M, Selitsky SR, Yamane D, Shimakami T, Kaneko S, Lanford RE, Lemon SM. 2013. microRNA-122 abundance in hepatocellular carcinoma and non-tumor liver tissue from Japanese patients with persistent HCV versus HBV infection. PLoS One 8: e76867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller MP, Boon KL, Reijns MA, Beggs JD. 2007a. The Lsm2-8 complex determines nuclear localization of the spliceosomal U6 snRNA. Nucleic Acids Res 35: 923–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiller MP, Reijns MA, Beggs JD. 2007b. Requirements for nuclear localization of the Lsm2-8p complex and competition between nuclear and cytoplasmic Lsm complexes. J Cell Sci 120: 4310–4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64. [DOI] [PubMed] [Google Scholar]
- Staněk D, Neugebauer KM. 2004. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol 166: 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staněk D, Rader SD, Klingauf M, Neugebauer KM. 2003. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol 160: 505–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staněk D, Pridalová-Hnilicová J, Novotńy I, Huranová M, Blazíková M, Wen X, Sapra AK, Neugebauer KM. 2008. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell 19: 2534–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark MR, Dunn EA, Dunn WS, Grisdale CJ, Daniele AR, Halstead MR, Fast NM, Rader SD. 2015. Dramatically reduced spliceosome in Cyanidioschyzon merolae. Proc Natl Acad Sci 112: E1191–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano JE. 1984. Purified lupus antigen La recognizes an oligouridylate stretch common to the 3′ termini of RNA polymerase III transcripts. Cell 36: 145–154. [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Brow DA. 1998. Control of pre-mRNA accumulation by the essential yeast protein Nrd1 requires high-affinity transcript binding and a domain implicated in RNA polymerase II association. Proc Natl Acad Sci 95: 6699–6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell 24: 735–746. [DOI] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA. 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci 90: 6498–6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. 2001. Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA 7: 1543–1553. [PMC free article] [PubMed] [Google Scholar]
- Stillman DJ. 2010. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim Biophys Acta 1799: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Urushiyama S, Tani T, Ohshima Y. 1993. An mRNA-type intron is present in the Rhodotorula hasegawae U2 small nuclear RNA gene. Mol Cell Biol 13: 5613–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Tani T, Ohshima Y. 1996. Spliceosomal introns in conserved sequences of U1 and U5 small nuclear RNA genes in yeast Rhodotorula hasegawae. J Biochem 120: 677–683. [DOI] [PubMed] [Google Scholar]
- Tani T, Ohshima Y. 1989. The gene for the U6 small nuclear RNA in fission yeast has an intron. Nature 337: 87–90. [DOI] [PubMed] [Google Scholar]
- Tani T, Ohshima Y. 1991. mRNA-type introns in U6 small nuclear RNA genes: implications for the catalysis in pre-mRNA splicing. Genes Dev 5: 1022–1031. [DOI] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. 1996a. Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT-AC introns. Science 273: 1824–1832. [DOI] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA. 1996b. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 84: 801–811. [DOI] [PubMed] [Google Scholar]
- Tazi J, Forne T, Jeanteur P, Cathala G, Brunel C. 1993. Mammalian U6 small nuclear RNA undergoes 3′ end modifications within the spliceosome. Mol Cell Biol 13: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns MP, Lund E, Dahlberg JE. 1992. 3′-end-dependent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevis oocyte nuclei. Mol Cell Biol 12: 3032–3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. 2000. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 404: 515–518. [DOI] [PubMed] [Google Scholar]
- Theuser M, Höbartner C, Wahl MC, Santos KF. 2016. Substrate-assisted mechanism of RNP disruption by the spliceosomal Brr2 RNA helicase. Proc Natl Acad Sci 113: 7798–7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichelaar JW, Knerer B, Vrabel A, Wieben ED. 1994. Transcription of a variant human U6 small nuclear RNA gene is controlled by a novel, internal RNA polymerase III promoter. Mol Cell Biol 14: 5450–5457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichelaar JW, Wieben ED, Reddy R, Vrabel A, Camacho P. 1998. In vivo expression of a variant human U6 RNA from a unique, internal promoter. Biochemistry 37: 12943–12951. [DOI] [PubMed] [Google Scholar]
- Trippe R, Sandrock B, Benecke BJ. 1998. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res 26: 3119–3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe R, Richly H, Benecke BJ. 2003. Biochemical characterization of a U6 small nuclear RNA-specific terminal uridylyltransferase. Eur J Biochem 270: 971–980. [DOI] [PubMed] [Google Scholar]
- Trippe R, Guschina E, Hossbach M, Urlaub H, Lührmann R, Benecke BJ. 2006. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 12: 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19: 2991–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CK, Cheng SC. 2008. Both catalytic steps of nuclear pre-mRNA splicing are reversible. Science 320: 1782–1784. [DOI] [PubMed] [Google Scholar]
- Tycowski KT, You ZH, Graham PJ, Steitz JA. 1998. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell 2: 629–638. [DOI] [PubMed] [Google Scholar]
- van der Feltz C, DeHaven AC, Hoskins AA. 2017. Stress-induced pseudouridylation alters the structural equilibrium of yeast U2 snRNA stem II. J Mol Biol 430: 524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen R, Arnberg AC, van der Horst G, Bonen L, Tabak HF, Grivell LA. 1986. Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell 44: 225–234. [DOI] [PubMed] [Google Scholar]
- Vankan P, McGuigan C, Mattaj IW. 1990. Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J 9: 3397–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasiljeva L, Kim M, Terzi N, Soares LM, Buratowski S. 2008. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol Cell 29: 313–323. [DOI] [PubMed] [Google Scholar]
- Venditti V, Clos L II, Niccolai N, Butcher SE. 2009. Minimum-energy path for a U6 RNA conformational change involving protonation, base-pair rearrangement and base flipping. J Mol Biol 391: 894–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonné R, Bertrand E. 2002. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. EMBO J 21: 2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidaver RM, Fortner DM, Loos-Austin LS, Brow DA. 1999. Multiple functions of Saccharomyces cerevisiae splicing protein Prp24 in U6 RNA structural rearrangements. Genetics 153: 1205–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan U, Company M, Abelson J. 1989. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev 3: 1206–1216. [DOI] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Huang G, Shi Y. 2016a. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353: 895–904. [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y. 2016b. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: insights into spliceosome assembly and catalysis. Science 351: 466–475. [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Lei J, Shi Y. 2017. Structure of an intron lariat spliceosome from Saccharomyces cerevisiae. Cell 171: 120–132. [DOI] [PubMed] [Google Scholar]
- Warda AS, Kretschmer J, Hackert P, Lenz C, Urlaub H, Hobartner C, Sloan KE, Bohnsack MT. 2017. Human METTL16 is a N 6-methyladenosine m6A methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep 18: 2004–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA. 1992. Interactions of small nuclear RNA's with precursor messenger RNA during in vitro splicing. Science 257: 1918–1925. [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Adachi H, Ge J, Stephenson D, Query CC, Yu YT. 2016. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J 35: 654–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schümperli D, Matera AG. 2005. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 114: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S, Takagi Y, Nagaike T, Tomita K. 2017. Crystal structures of U6 snRNA-specific terminal uridylyltransferase. Nat Commun 8: 15788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CC, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-Å resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2016. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353: 904–911. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2017. Structure of a yeast step II catalytically activated spliceosome. Science 355: 149–155. [DOI] [PubMed] [Google Scholar]
- Yang F, Wang XY, Zhang ZM, Pu J, Fan YJ, Zhou J, Query CC, Xu YZ. 2013. Splicing proofreading at 5′ splice sites by ATPase Prp28p. Nucleic Acids Res 41: 4660–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yean SL, Wuenschell G, Termini J, Lin RJ. 2000. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 408: 881–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younis I, Dittmar K, Wang W, Foley SW, Berg MG, Hu KY, Wei Z, Wan L, Dreyfuss G. 2013. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. Elife 2: e00780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Maroney PA, Nilsen TW. 1993. Functional reconstitution of U6 snRNA in nematode cis- and trans-splicing: U6 can serve as both a branch acceptor and a 5′ exon. Cell 75: 1049–1059. [DOI] [PubMed] [Google Scholar]
- Yu YT, Maroney PA, Darzynkiwicz E, Nilsen TW. 1995. U6 snRNA function in nuclear pre-mRNA splicing: a phosphorothioate interference analysis of the U6 phosphate backbone. RNA 1: 46–54. [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA. 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J 17: 5783–5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. 2008. High-quality binary protein interaction map of the yeast interactome network. Science 322: 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CC, Zhao X, Florens L, Swanson SK, Washburn MP, Hernandez N. 2007. CHD8 associates with human Staf and contributes to efficient U6 RNA polymerase III transcription. Mol Cell Biol 27: 8729–8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wan Y, Huang G, Wang D, Yu X, Huang G, Guo J. 2015. The exosome controls alternative splicing by mediating the gene expression and assembly of the spliceosome complex. Sci Rep 5: 13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Harding R, Hou F, Dong A, Walker JR, Bteich J, Tong Y. 2016. Structural basis of the recruitment of ubiquitin-specific protease USP15 by spliceosome recycling factor SART3. J Biol Chem 291: 17283–17292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Yan C, Hang J, Finci LI, Lei J, Shi Y. 2017. An atomic structure of the human spliceosome. Cell 169: 918–929. [DOI] [PubMed] [Google Scholar]
- Zhao X, Pendergrast PS, Hernandez N. 2001. A positioned nucleosome on the human U6 promoter allows recruitment of SNAPc by the Oct-1 POU domain. Mol Cell 7: 539–549. [DOI] [PubMed] [Google Scholar]
- Zhou H, Chen YQ, Du YP, Qu LH. 2002. The Schizosaccharomyces pombe mgU6-47 gene is required for 2′-O-methylation of U6 snRNA at A41. Nucleic Acids Res 30: 894–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Hang J, Zhou Y, Wan R, Lu G, Yin P, Yan C, Shi Y. 2014a. Crystal structures of the Lsm complex bound to the 3′ end sequence of U6 small nuclear RNA. Nature 506: 116–120. [DOI] [PubMed] [Google Scholar]
- Zhou L, Zhou Y, Hang J, Wan R, Lu G, Yan C, Shi Y. 2014b. Crystal structure and biochemical analysis of the heptameric Lsm1-7 complex. Cell Res 24: 497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]