Abstract
Lipoteichoic acid (LTA) is an anionic surface pol-ymer that is essential for normal growth of Staphylococcus aureus, making the LTA polymerase, LTA synthase (LtaS), a proposed drug target for combating Staphylococcal infections. LtaS is a polytopic membrane protein with five membrane-spanning helices and an extracellular domain, and it uses phosphatidyl glycerol to assemble a glycerol phosphate chain on a glycosylated diacylglycerol membrane anchor. We report here the first reconstitution of LtaS polymerization activity and show that the azo dye Congo red inhibits this enzyme both in vitro and in cells. Related azo dyes and the previously reported LtaS inhibitor 1771 have weak or no in vitro inhibitory activity. Synthetic lethality with mutant strains known to be non-viable in the absence of LTA confirms selective inhibition by Congo red. As the only validated LtaS inhibitor, Congo red can serve as a probe to understand how inhibiting lipoteichoic acid biosynthesis affects cell physiology and may also guide the discovery of more potent inhibitors for use in treating S. aureus infections.
TOC graphic
The use of dyes in biomedical research and drug discovery has a long history. Indeed, the origins of the pharmaceutical industry can be traced to the repurposing of dyes and dye precursors for therapeutic use.1 Examples include the forerunner of phenothiazine antipsychotics, methylene blue, which was once used to treat malaria;2 prontosil, an azo dye that led to the discovery of the “sulfa drug” family of antibiotics;3 suramin, a 100-year old compound derived from a family of dyes found to have anti-trypanosomal activity, which is still used to treat African sleeping sickness;4 and the acridine-based anticancer drugs.5 Here we report the discovery that Congo red (1, Figure 1A), a dye used to stain amyloid fibrils6 and detect biofilm-forming bacterial strains,7 inhibits a Staphylococcus aureus enzyme involved in assembly of the cell envelope. This enzyme, lipoteichoic acid synthase (LtaS), forms the glycerol phosphate polymer of lipoteichoic acid, an important cell surface polymer. LtaS is a proposed target for new antibiotics,8 and Congo red, as the first compound shown to directly block its activity, may serve as a useful probe to guide the discovery of therapeutically useful inhibitors.9
Figure 1.
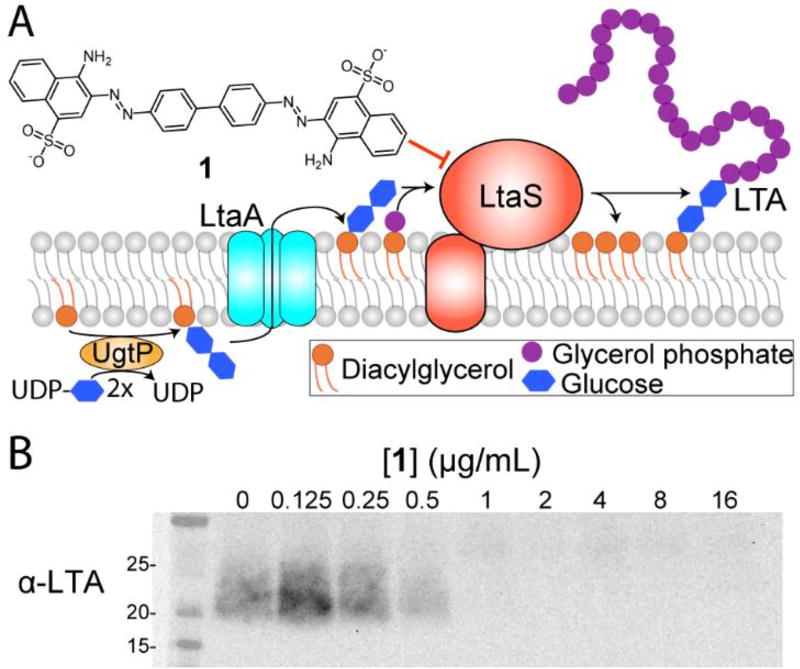
LTA is biosynthesized at the cell membrane and Congo red (1) abolishes its production in cells. (A) Diacylglycerol (DAG) is converted to diglucosyl DAG (Glc2-DAG) by UgtP, then flipped to the outer membrane by LtaA. Using phosphatidyl glycerol (PG) as a substrate, LtaS transfers glycerol phosphate repeats to the Glc2-DAG anchor to produce LTA and DAG. (B) 1 inhibits LTA polymerization in S. aureus RN4220 in a dose-dependent manner.
The S. aureus cell envelope is a protective barrier comprising the cell membrane, the surrounding cell wall, and myriad proteins and anionic polymers anchored to these structures.10 After cell wall peptidoglycan, teichoic acids are the most important polymers for cell envelope integrity.11 S. aureus makes two distinct, highly abundant classes of teichoic acids produced by separate biosynthetic pathways: lipoteichoic acids, anchored to the cell membrane and-consisting of glycerol phosphate repeats,12 and wall teichoic acids, covalently attached to peptidoglycan and composed mainly of ribitol phosphate repeats.13 These cell envelope polymers have been implicated in resistance to cationic antibiotics,14 autolysin regulation,15 and cell division,8 among other processes.16
The glycerol phosphate polymer of lipoteichoic acid is made by lipoteichoic acid synthase (LtaS), a polytopic membrane protein with five N-terminal membrane spanning helices and a C-terminal extracellular domain,8 which transfers glycerol phosphate units from phosphatidyl glycerol (PG) to a diglucosyl diacylglycerol (Glc2-DAG) membrane anchor (Figure 1A).17 The C-terminal extracellular domain uses manganese to facilitate the transfer of glycerol phosphate from PG onto a threonine residue (T300 in S. aureus LtaS), which is then transferred to the growing LTA polymer.9a, 11, 18 The LTA polymer is further modified by covalent attachment of D-alanine residues, which play a role in resistance to cationic antimicrobial peptides and other toxins.19 The Glc2-DAG anchor of LTA is synthesized on the inner leaflet of the membrane by the glucosyltransferase UgtP20 and is then flipped to the outer leaflet by the flippase LtaA.17a Mutants lacking either ugtP or ltaA can still make lipoteichoic acid polymers, but LtaS then uses PG instead of Glc2-DAG as the membrane anchor and the polymers are longer than normal.16a, 17a
LtaS in an important enzyme for cell viability. Mutants lacking LtaS are susceptible to osmotic lysis and can only grow in osmotically stabilizing conditions, at low temperatures, or after acquisition of compensatory mutations that suppress lysis.17b, 21 The most common suppressor mutations prevent expression of an intracellular phosphodiesterase, GdpP, resulting in high concentrations of cyclic-di-AMP, a bacterial second messenger implicated in transport of potassium and other ions.22 These suppressor mutations or osmotically stabilizing conditions are not sufficient to maintain viability if a mutant lacking lipoteichoic acid synthase is also impaired in wall teichoic acid biosynthesis.23
We speculated that Congo red might have a specific target in S. aureus after discovering that it had potent activity against strains lacking wall teichoic acids. At 30°C, the MIC of Congo red decreased from over 1000 µg/mL against wild-type S. aureus to ~2 µg/mL against a strain lacking the earliest acting gene in the wall teichoic acid pathway, tarO.24 This finding suggested a functional connection between the unknown target of Congo red and the wall teichoic acid pathway. Including LtaS, only about a dozen genes become essential when wall teichoic acids are depleted in S. aureus (Table S1).17b, 19c, 23 We subsequently found that gdpP transposon mutants are selected on Congo red agar plates incubated at 37°C,25 leading us to hypothesize that lipoteichoic acid synthase was the target of this azo dye.
To test the hypothesis, we employed a cell-based assay that uses an α-LTA antibody to detect lipoteichoic acid polymers via immunoblotting.17a Treatment of wild-type S. aureus with increasing concentrations of Congo red (1) resulted in a dose-dependent decrease in LTA, consistent with the hypothesis that 1 inhibits LtaS (Figure 1B).
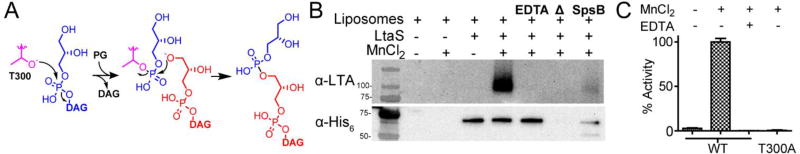
While cell-based assays are powerful ways to evaluate inhibition of a pathway, inhibition may be indirect. Although the purified C-terminal extracellular domain of LtaS, eLtaS,26 was shown to hydrolyze a fluorescent phosphatidyl glycerol analog,27 lipoteichoic acid synthesis has not been reconstituted to enable inhibitor testing. Therefore, we purified S. aureus LtaS bearing a C-terminal His6-tag from E. coli and then exploited the ability of LtaS to use phosphatidyl glycerol as an alternative membrane anchor to reconstitute the enzyme into liposomes containing 23% phosphatidyl glycerol (Figure 2A, supplementary methods, Figures S1 and S2). We detected polymer using the α-LTA antibody only in reactions with proteoliposomes containing LtaS and Mn2+ (Figure 2B). Excess EDTA, boiling the proteoliposomes prior to initiation with manganese, or incubating them with the S. aureus SpsB signal peptidase, which cleaves eLtaS from the membrane domain,28 abolished polymer synthesis (Figure 2B). The metal ion dependence of polymer formation is consistent with previous findings that eLtaS requires a divalent cation to promote hydrolysis of fluorescent phosphatidyl glycerol.27
Figure 2.
LtaS activity was reconstituted in proteoliposomes. (A) Scheme for LtaS-mediated transfer of glycerol phosphate onto phosphatidyl glycerol. Glycerol phosphate from PG (blue) is loaded onto T300 (purple) of LtaS, which is then attacked by the glycerol end of another molecule of PG (red). This process repeats in a cycle in which PG is loaded onto T300 and is then offloaded onto the glycerol unit at the tip of the growing LTA polymer. (B) In vitro reconstitution of LTA biosynthesis in proteoliposomes shows that the LTA polymer is produced only when manganese is present. SpsB-catalyzed cleavage of the extracellular domain from the membrane domain abolishes LTA synthesis. (C) DAG production for WT LtaS and a mutant, T300A, containing an active site mutation. Activity is normalized to DAG produced by WT LtaS in the presence of manganese. LtaS T300A did not produce DAG or LTA polymer (Figure S10).
We also established an assay to detect the other product of the reaction, diacylglycerol (DAG) (Figure 1A). The previously described assay that detects hydrolysis of a fluorescent DAG analogue was not suitable for use in proteoliposomes containing large amounts of native phosphatidyl glycerol.27 We therefore measured diacylglycerol (DAG) produced in the proteoliposome reactions using a commercially available enzyme-coupled assay that converts DAG to a detectable fluorescent signal (Figure S3). As with polymer formation, we found that DAG production was dependent on the presence of manganese and required an active catalytic domain (Figure 2C).9a Our studies showing LTA polymer formation with concomitant release of DAG confirm the first successful reconstitution of LtaS polymerase activity. These new biochemical assays for LtaS activity allow us to test possible inhibitors. We therefore added increasing concentrations of 1 to the proteoliposome assay and blotted for polymer formation.
1 inhibited LTA polymerization with an IC50 of ~2 µM (Figure 3A, Figure S4), which is similar to its MIC against WTA-deficient S. aureus.24 1 also decreased DAG production by over 90% (Figure S5). Adding empty liposomes to the reaction did not significantly affect the IC50 (Figure S6), showing that the decrease in LTA formation is due to LtaS inhibition and not to general disruption of the proteoliposomes. Because 1 has previously been identified as a promiscuous inhibitor and is known to aggregate, we also tested a range of conditions that affect its promiscuous inhibitory activity, resulting in substantial increases in IC50 values.29 Adding detergent or increasing ionic strength had no effect on the IC50; adding micromolar concentrations of BSA (a 10-fold excess relative to LtaS) resulted in only a two-fold shift in the IC50 (Figure S7).29a, 30 These negligible changes in inhibitory potency are not consistent with promiscuous inhibition. Finally, we also measured LtaS activity in the presence of three other azo dyes, Direct Red 7 (2), Acid Red 88 (3) and Sudan Red 7B (4) (Figure 3B).24 Although 2 partially inhibited DAG production at the highest concentration tested, the other dyes had no effect (Figure 3C, Figures S5 and S8). While we cannot exclude the possibility that these dyes affect LtaS activity at higher concentrations,31 the in vitro results show that 1 selectively inhibits the enzyme. These findings are also consistent with in cellulo data showing that only 1 is lethal to the ΔtarO strain (Figure 3D, Figure S11).
Figure 3.
Congo red, but not related azo dyes, inhibits LtaS in vitro and against WTA-deficient S. aureus strains. (A) 1 inhibits in vitro LTA polymerization in proteoliposomes in a dose-dependent manner. (B) Structures of azo dyes tested against S. aureus strains and proteoliposomes. (C) Replicate dot blots of assays containing the indicated concentrations of 1 and 2 (for 3, 4, and 1771, see Figure S8). (D) 1 selectively inhibited the growth of a Newman ΔtarO strain, but not Newman (Figure S9).
We also used our assay system to test the only previously proposed LtaS inhibitor, a compound known as 1771 (Figures S12 and S13). This compound was reported to inhibit lipoteichoic acid polymer synthesis in cells, but was not fully validated because LtaS polymerization activity had not been reconstituted.9c We found that 1771 did not inhibit either LTA polymer formation or the produc-tion of DAG in vitro (Figures S5 and S8). Because an LtaS inhibitor should be synthetically lethal with WTA-deficient strains, we also compared the activity of 1771 against S. aureus wildtype and ΔtarO strains.24 Unlike 1, which was lethal to ΔtarO but not wildtype strains at 30°C, compound 1771 was lethal to both strains (Figure S14). 1771 also inhibited growth of a gdpP mutant, a known suppressor of LtaS loss. In contrast, deletion of gdpP suppresses temperature-dependent lethality of Congo red.21, 25, 32 We have concluded that compound 1771 does not inhibit LtaS directly, and suggest that its effects on lipoteichoic acid abundance in cells reflect inhibition of another enzyme required for polymer production (e.g., in the phosphatidylglycerol biosynthesis pathway) or a more general toxicity.
In summary, we have reconstituted activity of lipoteichoic acid synthase in proteoliposomes for the first time, paving the way to characterize its full structure and its polymerization mechanism. We have also shown that Congo red (1) inhibits LtaS both in cellulo and in vitro.33 Although Congo red aggregates have been shown to inhibit several soluble enzymes in vitro, the in cellulo data for this compound – including lethality at 30°C against WTA-deficient strains and temperature-dependent lethality that is suppressed by gdpP deletion – support the conclusion that it will be a useful chemical probe for understanding the effects of perturbing LtaS on cell physiology. We expect 1 will be useful for genome-wide mapping of synthetic interactions between lipoteichoic acids and other cellular components, and the results of these experiments may facilitate the discovery of more potent LtaS inhibitors to treat multidrug resistant bacterial infections.19c, 23
Supplementary Material
Acknowledgments
We thank Fredrick Rubino and Anthony Hesser for help making proteoliposomes, Sunia Trauger and Jennifer Wang at the Harvard Small Molecule Mass Spectrometry core for MS work, and Charles Sheahan and Anne Rachupka at the HMS East Quad NMR Facility for help with NMR. This work was supported by the NIH (P01 AI083214, U19 AI109764, and R01 AI099144).
Footnotes
ASSOCIATED CONTENT
The Supporting Information is available free of charge on the ACS Publications website.
Experimental methods; supplemental figures and tables (PDF).
The authors declare no competing financial interests.
References
- 1.Volwiler EH. Ind. Eng. Chem. 1926;18(12):1336–1337. [Google Scholar]
- 2.(a) Guttmann P, Ehrlich P. Berlin. Klin. Woch. 1891;28:953–956. [Google Scholar]; (b) Howland RH. J Psychosoc Nurs Ment Health Serv. 2016;54(9):21–24. doi: 10.3928/02793695-20160818-01. [DOI] [PubMed] [Google Scholar]
- 3.Domagk G. Dtsch med Wochenschr. 1935;61(07):250–253. [Google Scholar]
- 4.(a) Fourneau E, Tréfouel J, Vallée J. Ann Inst Pasteur. 1924;38:81–114. [Google Scholar]; (b) Morgan HP, McNae IW, Nowicki MW, Zhong W, Michels PAM, Auld DS, Fothergill-Gilmore LA, Walkinshaw MD. Journal of Biological Chemistry. 2011;286(36):31232–31240. doi: 10.1074/jbc.M110.212613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassileth PA, Gale RP. Leukemia Research. 1986;10(11):1257–1265. doi: 10.1016/0145-2126(86)90331-0. [DOI] [PubMed] [Google Scholar]
- 6.Puchtler H, Sweat F. J Histochem Cytochem. 1965;13(8):693–694. doi: 10.1177/13.8.693. [DOI] [PubMed] [Google Scholar]
- 7.(a) Teather RM, Wood PJ. Appl. Environ. Microbiol. 1982;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Freeman DJ, Falkiner FR, Keane CT. J Clin Pathol. 1989;42(8):872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gründling A, Schneewind O. PNAS. 2007;104(20):8478–8483. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Lu D, Wörmann ME, Zhang X, Schneewind O, Gründling A, Freemont PS. PNAS. 2009;106(5):1584–1589. doi: 10.1073/pnas.0809020106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Pasquina LW, Santa Maria JP, Walker S. Curr Opin Microbiol. 2013;16(5):531–7. doi: 10.1016/j.mib.2013.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Richter SG, Elli D, Kim HK, Hendrickx AP, Sorg JA, Schneewind O, Missiakas D. Proc Natl Acad Sci U S A. 2013;110(9):3531–6. doi: 10.1073/pnas.1217337110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rajagopal M, Walker S. Envelope Structures of Gram-Positive Bacteria. In: Bagnoli F, Rappuoli R, editors. Protein and Sugar Export and Assembly in Gram-positive Bacteria. Springer International Publishing; Cham: 2017. pp. 1–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Percy MG, Grundling A. Annu Rev Microbiol. 2014;68:81–100. doi: 10.1146/annurev-micro-091213-112949. [DOI] [PubMed] [Google Scholar]
- 12.Fischer W, Mannsfeld T, Hagen G. Biochem. Cell Biol. 1990;68(1):33–43. doi: 10.1139/o90-005. [DOI] [PubMed] [Google Scholar]
- 13.Brown S, Santa Maria JP, Jr, Walker S. Annu Rev Microbiol. 2013;67:313–36. doi: 10.1146/annurev-micro-092412-155620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. Journal of Biological Chemistry. 1999;274(13):8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 15.Bierbaum G, Sahl H-G. Arch. Microbiol. 1985;141(3):249–254. doi: 10.1007/BF00408067. [DOI] [PubMed] [Google Scholar]
- 16.(a) Xia G, Kohler T, Peschel A. International Journal of Medical Microbiology. 2010;300(2):148–154. doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]; (b) Percy MG, Karinou E, Webb AJ, Gründling A. J. Bacteriol. 2016;198(15):2029–2042. doi: 10.1128/JB.00116-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Schneewind O, Missiakas D. Lipoteichoic Acid Synthesis and Function in Gram-Positive Bacteria. In: Geiger O, editor. Biogenesis of Fatty Acids Lipids and Membranes. Springer International Publishing; Cham: 2017. pp. 1–18. [Google Scholar]
- 17.(a) Grundling A, Schneewind O. J Bacteriol. 2007;189(6):2521–30. doi: 10.1128/JB.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Oku Y, Kurokawa K, Matsuo M, Yamada S, Lee B-L, Sekimizu K. J. Bacteriol. 2009;191(1):141–151. doi: 10.1128/JB.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.(a) Schirner K, Marles-Wright J, Lewis RJ, Errington J. The EMBO Journal. 2009;28(7):830–842. doi: 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Campeotto I, Percy MG, MacDonald JT, Förster A, Freemont PS, Gründling A. Journal of Biological Chemistry. 2014;289(41):28054–28069. doi: 10.1074/jbc.M114.590570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.(a) Perego M, Glaser P, Minutello A, Strauch MA, Leopold K, Fischer W. Journal of Biological Chemistry. 1995;270(26):15598–15606. doi: 10.1074/jbc.270.26.15598. [DOI] [PubMed] [Google Scholar]; (b) Reichmann NT, Cassona CP, Grundling A. Microbiology. 2013;159(Pt 9):1868–77. doi: 10.1099/mic.0.069898-0. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Pasquina L, Maria JPS, Wood BM, Moussa SH, Matano L, Santiago M, Martin SES, Lee W, Meredith TC, Walker S. Nature chemical biology. 2016;12(1):40–45. doi: 10.1038/nchembio.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiriukhin MY, Debabov DV, Shinabarger DL, Neuhaus FC. J. Bacteriol. 2001;183(11):3506–3514. doi: 10.1128/JB.183.11.3506-3514.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corrigan RM, Abbott JC, Burhenne H, Kaever V, Grundling A. PLoS Pathog. 2011;7(9):e1002217. doi: 10.1371/journal.ppat.1002217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corrigan RM, Gründling A. Nat Rev Micro. 2013;11:513. doi: 10.1038/nrmicro3069. [DOI] [PubMed] [Google Scholar]
- 23.Maria JPS, Sadaka A, Moussa SH, Brown S, Zhang YJ, Rubin EJ, Gilmore MS, Walker S. PNAS. 2014;111(34):12510–12515. doi: 10.1073/pnas.1404099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki T, Campbell J, Kim Y, Swoboda JG, Mylonakis E, Walker S, Gilmore MS. J. Antimicrob. Chemother. 2012;67(9):2143–2151. doi: 10.1093/jac/dks184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeFrancesco AS, Masloboeva N, Syed AK, DeLoughery A, Bradshaw N, Li G-W, Gilmore MS, Walker S, Losick R. PNAS. 2017;114(29):E5969–E5978. doi: 10.1073/pnas.1704544114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wörmann ME, Reichmann NT, Malone CL, Horswill AR, Gründling A. J. Bacteriol. 2011;193(19):5279–5291. doi: 10.1128/JB.00369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karatsa-Dodgson M, Wörmann ME, Gründling A. J. Bacteriol. 2010;192(20):5341–5349. doi: 10.1128/JB.00453-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao S, Bockstael K, Nath S, Engelborghs Y, Anné J, Geukens N. FEBS Journal. 2009;276(12):3222–3234. doi: 10.1111/j.1742-4658.2009.07037.x. [DOI] [PubMed] [Google Scholar]
- 29.(a) McGovern SL, Caselli E, Grigorieff N, Shoichet BK. J. Med. Chem. 2002;45(8):1712–1722. doi: 10.1021/jm010533y. [DOI] [PubMed] [Google Scholar]; (b) Shoichet BK. J. Med. Chem. 2006;49(25):7274–7277. doi: 10.1021/jm061103g. [DOI] [PubMed] [Google Scholar]
- 30.Giannetti AM, Koch BD, Browner MF. J. Med. Chem. 2008;51(3):574–580. doi: 10.1021/jm700952v. [DOI] [PubMed] [Google Scholar]
- 31.We did not test higher dye concentrations in vitro due to solubility concerns.
- 32.Congo Red is commonly used in agar plates to detect S. aureus strains capable of forming biofilms, but these results suggest it may select for genetic suppressors of LtaS inhibition, with consequences on biofilm forming potential at long time points.
- 33.It is worth nothing that we did not find LtaS inhibition with Congo red to be toxic to wildtype strains at 37°C except at high concentrations. Low levels of LTA biosynthesis may be sufficient to support growth under the conditions used. Work to assess these possibilities is ongoing.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.