Abstract
Chromosomal microarray analysis (CMA) is performed either by array comparative genomic hybridization (aCGH) or by using a SNP array. In the prenatal setting, CMA is on par with traditional karyotyping for detection of major chromosomal imbalances such as aneuploidy and unbalanced rearrangements. CMA offers additional diagnostic benefits by revealing sub-microscopic imbalances or copy number changes (CNVs) that are too small to be seen on a standard G-banded chromosome preparation. These submicroscopic imbalances are also referred to as microdeletions and microduplications, particularly when they include specific genomic regions that are associated with clinical sequelae. Not all microdeletions/duplications are associated with adverse clinical phenotypes and in many cases, their presence is benign. In other cases, they are associated with a spectrum of clinical phenotypes that may range from benign to severe, while in some situations, the clinical significance may simply be unknown. These scenarios present a challenge for prenatal diagnosis and genetic counseling prior to prenatal CMA greatly facilitates delivery of complex results. In prenatal diagnostic samples with a normal karyotype, chromosomal microarray will diagnose a clinically significant subchromosomal deletion or duplication in approximately 1% of structurally normal pregnancies and 6% with a structural anomaly. Pre-test counseling is also necessary to distinguish the primary differences between the benefits, limitations and diagnostic scope of CMA versus the powerful but limited screening nature of non-invasive prenatal diagnosis using cell-free fetal DNA.
Keywords: chromosomal microarray, prenatal diagnosis, amniocentesis, chorionic villus sampling, microdeletion, microduplication, copy number variation, CNV, VOUS
INTRODUCTION
Prenatal diagnosis of chromosome abnormalities has been offered since the mid 1960’s (1). For the bulk of the past 50 years, cytogenetic testing of the fetus has been accomplished by standard G-banded karyotyping. The diagnostic yield using conventional cytogenetic analysis by karyotype is dependent on the indication. For the most common indications such as advanced maternal age and positive biochemical screening, the diagnostic yield at the time of CVS and amniocentesis is approximately 6% and 3% respectively [Data from additional analysis of the NICHD microarray data set (2)]. For fetuses with structural anomalies, the diagnostic yield is approximately 49% in the first trimester and 17% in the second [Data from additional analysis of the NICHD microarray data set (2)].
The advent of newer molecular cytogenomic technologies such as chromosomal microarray analysis (CMA) brought about the prospect of greater diagnostic resolution. CMA, which detects imbalances in the kilobase range, readily demonstrates its superiority over standard karyotyping which is limited to imbalances greater than 7–10 million bases. In postnatal studies of children with congenital abnormalities, developmental delay or intellectual disability, CMA will have an additional diagnostic yield of clinically relevant sub-chromosomal abnormalities of about 12 to 15% (3, 4). In 2013, Wapner and colleagues published a large multicenter NICHD sponsored study that demonstrated the clinical utility of CMA in prenatal diagnosis (2). The prospective cohort study demonstrated that in pregnancies with fetal structural anomalies and a normal karyotype there was an incremental diagnostic yield of about 6% above what a karyotype would detect. For all other indications this was about 1.7% (2).
CMA works by detecting imbalances in DNA copy number. These imbalances are referred to as copy number variants (CNVs), which in and of itself, does not imply an abnormal or pathogenic phenotype. In fact, a significant number of CNVs are clinically insignificant and are found in apparently normal individuals (5–9). The majority of these “benign” CNVs are very small in size (<50 Kb) and do not have clinically significant coding regions (5–9). CNVs are often referred to as microdeletions (sub-microscopic losses) or microduplications (sub-microscopic gains) and are undetectable by conventional karyotype. The medical relevance of CNVs relates to the functional impact of the micro-deletion/duplication which is more likely to have a phenotypic effect when the region of imbalance occurs in critical gene/s or an important regulatory region.
CMA TECHNIQUES
There are two CMA techniques used in identifying submicroscopic imbalances: comparative genomic hybridization (CGH) and single nucleotide polymorphisms (SNP).
CGH based arrays (aCGH) compare a patient’s DNA to a normal control DNA sample to identify areas that are either over- or under-represented in the patient sample (10). In the aCGH approach, the patient and control DNA samples are cut into fragments then labeled with different fluorescent colors (usually green and red). They are mixed together in equal proportions and placed onto an array (glass slide) containing multiple probes from representative sequences from across the human genome. The DNA mixture binds (hybridizes) in a competitive manner to complimentary sequences located within the probe DNA on the array. The fluorescence intensity of every probe is measured using digital imaging software. After a normalization process, a ratio of the fluorescence intensities between the patient and the control sample is calculated. A ratio of 1 indicates equal contributions from the patient and control sample which in turn represents a normal copy number at that locus. A ratio that is significantly greater than 1 indicates that more of the patient’s DNA hybridized at a particular location compared to the control DNA. This represents a gain of patient chromosomal material (a duplication or trisomy). Conversely, a loss of genetic material (a deletion or monosomy) in the patient would yield a ratio that is significantly less than 1 due to more hybridization of the control DNA sequences compared to patient’s DNA. The location and size of the gain/loss can be determined by the number of consecutive probes that show a ratio above or below 1. A typical clinical CGH array contains a few hundred thousand probes while the number of probes on research CGH arrays may reach into the millions. The resolution and diagnostic capability of aCGH depends on the number and types of probes used and their distribution across the entire genome (11). Most clinical laboratories performing aCGH will report clinically significant imbalances in the range of 50–100 Kb in postnatal studies. The reporting size range is usually larger in prenatal studies and may vary according to the indication for testing.
Single Nucleotide Polymorphisms (SNP) Microarray Analysis (SOMA) uses high-density oligonucleotide-based arrays in which target probes are chosen from DNA locations known to vary between individuals by a single base pair (i.e. SNPs)(12). In the SOMA approach, only the patient’s DNA (fetal) is labelled and hybridized to the SNP array. Copy number changes are determined by measuring the absolute fluorescence probe intensities of the patient sample compared with the intensities of multiple normal controls that were independently hybridized (in silico comparison) (Figure 1). Most SNP arrays used in a clinical setting are in fact hybrid arrays that contain both SNP probes and copy number probes. The density of probes on some of these hybrid arrays may be as high as 2.7 million probes. Clinical laboratories performing SOMA usually report CNVs of known clinical significance in the range of 50–100 Kb and higher. In addition to detecting CNVs, other clinically useful information may be extracted from the genotype plots generated from the SNPs. This includes uniparental disomy (UPD), mosaicism, zygosity, maternal cell contamination, parent of origin and consanguinity (Figure 2). Lastly, triploidy which cannot be detected by aCGH, can easily be identified by SOMA by assessing the SNP allele patterns on the array (Figure 4) (13, 14).
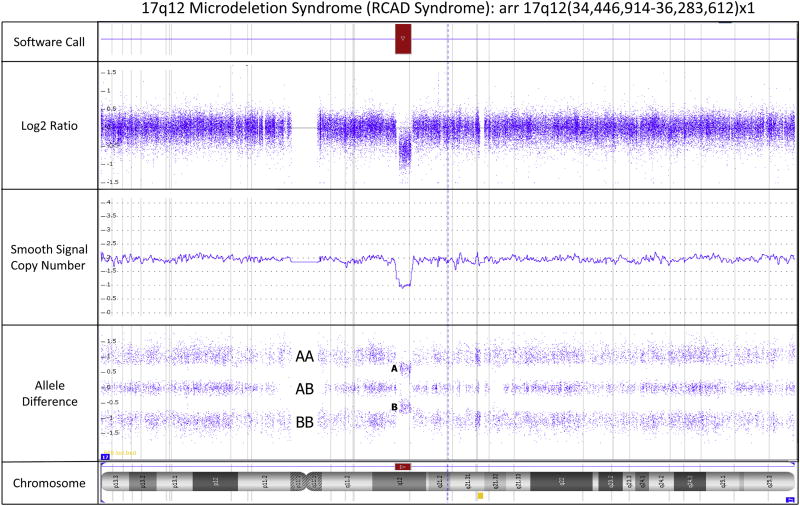
Figure 1.
Screenshot from the Affymetrix Chromosome Analysis Suite Software (Version 3.1) showing an 1.837 Mb interstitial deletion of the proximal long arm region (17q12) of chromosome 17 which is associated with a clinical diagnosis of Renal Cysts and Diabetes Syndrome (OMIM#137920) and is caused by a loss of the HNF1B gene. The precise coordinates of the deletion correspond to chr17: 34,446,914–36,283,612 using Human Genome Build Hg19. The gene content within the deleted region can be ascertained using the genome coordinates. A deletion is indicated in the software call panel by the presence of a red bar. The deletion is identified by a decrease in the Log2 ratio from zero as seen in the Log2 Ratio panel. The smooth signal copy number panel indicates the exact copy number of each probe. This panel is helpful in identifying mosaicism which is evident when the smooth signal for multiple consecutive probes lies between an integer, e.g. between 2 and 3 indicates trisomy mosaicism. The allele difference panel indicates the genotype for each SNP probe. For normal copy number of 2, there are only 3 possible SNP combinations, AA, AB and BB which are plotted on the allele difference graph. When there is a deletion (copy number of 1), the genotype options are either A or B and thus only two distinct tracks are visible on the allele difference graph. The chromosome ideogram in the chromosome panel highlights the position (breakpoints) on the chromosome where copy number imbalances are present. The red bar in the chromosome panel represents a deletion. Gains would typically be shown as blue bars.
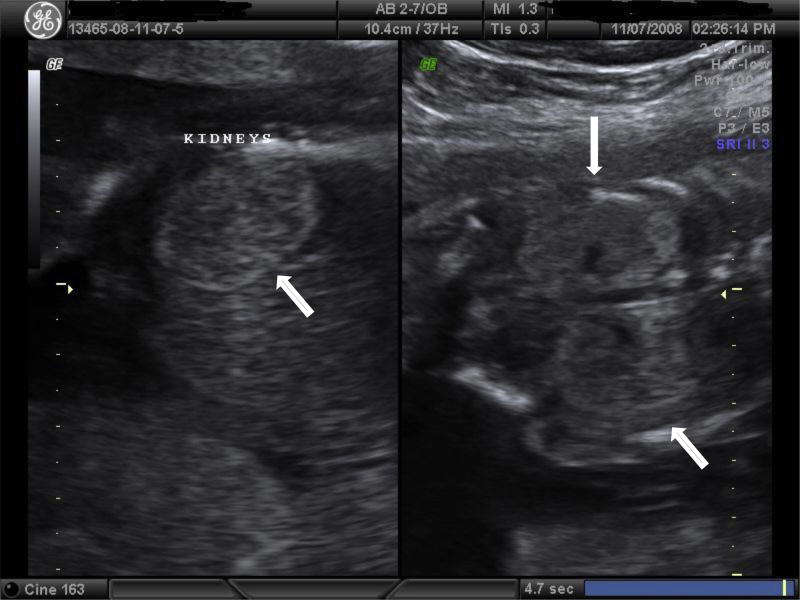
Figure 2.
16 week ultrasound image of fetus diagnosed by CVS with deletion of 17q12. Noted in the scan is bilateral enlargement and hyperechogenicity of kidneys. Kidneys appeared normal at the time of CVS at 12 weeks gestation
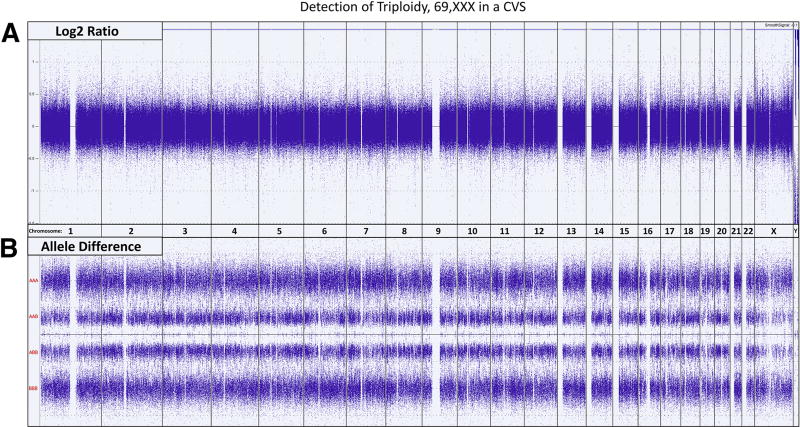
Figure 4.
Whole Genome view showing the Log2 Ratio and Allele Difference for every chromosome. [A] Since the intensities of the probes are normalized, the Log2 ratios for a normal diploid sample and a triploid sample are indistinguishable, with both indicating a DNA copy number of 2. Genotypes, extracted from the SNP data must be utilized to identify triploidy. [B] The allele difference plots show the various SNP genotypes for each SNP locus. In the presence of 2 chromosomes, there are only 3 possible SNP combinations: AA, AB and BB, see Figures 1 and 2. In the presence of 3 chromosomes, there are 4 possible SNP combinations: AAA, AAB, ABB and BBB which results in four distinct tracks on the allele difference graph. A normal diploid female would show the characteristic three tracks for all the chromosomes while the triploid, 69,XXX fetus in this example displays the four characteristic tracks for every autosome as well as the X chromosome. Triploid fetuses with a 69,XXY constitution would show the four characteristic tracks for every autosome, three tracks representing the two X chromosomes and two tracks representing the single Y chromosome.
THE DIAGNOSTIC YIELD OF CMA AND RATIONAL FOR ITS UTILIZATION OVER STANDARD KARYOTYPE
CMA in Fetuses without Ultrasound Anomalies
The biggest advantage to using CMA over classic cytogenetic and FISH techniques for prenatal genetic diagnosis of chromosomal abnormalities lies with CMA’s ability to detect much smaller imbalances. Typical karyotype analysis by G-banding may be able to delineate deletions and duplications that are 5–10 Mb in size (15). However, given the variation in banding resolution from one prenatal preparation to the next, 10–20 Mb and greater is a more realistic threshold of detection for conventional karyotype analysis. Standard FISH for microdeletion/duplication syndromes usually targets imbalances in the 100–200 Kb range but requires clinical features to guide probe selection, a challenging task for prenatal samples. It is possible to multiplex FISH probes but the limited number of spectrally unique commercial fluorophores that can simultaneously be used to interrogate multiple diseases is limited to a handful. To increase the diagnostic yield, one could perform sequential testing of many FISH probes but this is inefficient, time-consuming and very expensive. CMA offers the benefit of detecting submicroscopic imbalances (<< 5 Mb) anywhere in the genome in a single test and its resolution is only limited by the probes present on the chip. CMA is 100% accurate in identifying the common aneuploidies in prenatal specimens compared to karyotype (2, 16, 17) and in the NICHD study, it demonstrated an increased diagnostic yield over standard karyotyping of 1.7% in patients referred for advanced maternal, parental anxiety and positive serum screening (2). A recent meta-analysis assessing CMA on 10,614 fetuses from 10 large studies found a pathogenic, clinically significant CNV in 0.84% (1:119) of cases referred for AMA and parental anxiety (18).
A recent meta-analysis evaluated the onset/penetrance of genomic disorders diagnosed by CMA. 10,314 fetuses from 8 large studies showed that CNVs associated with early onset syndromic disorders occurred in 1:270 (0.37%) pregnancies (18). Approximately 1:909 (0.11%) cases involved late onset diseases and a susceptibility CNV was observed in 1:333 (0.3%) cases (18). By adding the individual risk for pathogenic CNVs to the individual risk for cytogenetically visible chromosome aberrations, Srebniak and colleagues showed that overall a pregnant women has a risk higher than 1:180 for a clinically significant cytogenetic aberration (18). Furthermore, pregnant women younger than 36 years of age have a higher risk for pathogenic CNVs than for Down syndrome (18).
The incremental yield of CMA over karyotype in fetuses without ultrasound identifiable anomalies shows considerable variability in the literature. An earlier large systematic review showed a clinically significant finding on CMA in 1% of case while single reports have been as low as 0.4% in one study (19) and as high as 2.0% in another (20). This variation is likely due to variances in the array platforms utilized, the resolution of the arrays used as well as the differences in reporting practices at each individual clinical laboratory. Certainly, the local definitions of the pathogenicity of specific results varies from laboratory to laboratory and these in turn have changed over time. With new knowledge and greater sharing of results in public databases, the number of genomic regions definitively associated with disease has increased and the incidence of variants of uncertain significance (VUS) has decreased over time. For example, the initial NICHD study reported a pathogenic CNV and VUS incidence of 0.9% and 2.5% respectively. Review of the same dataset on an annual basis facilitated an increase in pathogenic cases to 1.8% and a reduction in the VUS cases to 0.9% based on new literature and public data sharing (21).
CMA in Fetuses with Ultrasound Anomalies
The association between fetal anomalies and genomic imbalance has been recognized for decades, particularly with the classic structural anomalies associated with Down syndrome (trisomy 21), Edward syndrome (trisomy 18) and Patau syndrome (trisomy 13). Microdeletions and microduplications that involve clinically significant genomic regions are also associated with specific genetic syndromes; many of which have congenital abnormalities as part of the phenotype. For example, cardiac defects are observed in about 77% of fetuses with DiGeorge syndrome which is caused by a submicroscopic deletion of the proximal long arm region of chromosome 22 (aka 22q11.2 deletion syndrome) (22). Microdeletions of the terminal region of the short arm of chromosome 17 leads to a congenital lack or underdevelopment of the gyri of the cerebral cortex (lissencephaly) and is associated with the Miller-Dieker Syndrome (17p13.3 deletion). Imprinting disorders such Beckwith-Wideman syndrome and Russell-Silver syndrome can present with specific ultrasound anomalies and when caused by UPD, may only be identified by SOMA (23, 24).
Several large-scale studies evaluating the incremental yield of CMA in fetuses with ultrasound anomalies have been published. The 2012 NICHD study reported clinically significant CNVs in 6% of fetuses with a normal karyotypes and ultrasound anomalies (2). In a SOMA study of 1,033 fetuses with ultrasound anomalies, Srebniak and colleagues reported pathogenic CNVs in 5.5% of cases (25). A larger study of 5,000 fetuses revealed an incidence of 6.6% in 2,462 cases with ultrasound anomalies (26). Two meta-analyses published in 2013 identified an increased diagnostic yield of 7–10% over karyotype in pregnancies with structural fetal abnormalities (17, 27). Overall, the literature clearly demonstrates that CMA will provide additional information over karyotype in about 6–7% of pregnancies when the fetus has an anomaly identified on ultrasound. Based on this, The American Congress of Obstetricians and Gynecologists (ACOG) now recommends CMA as the first tier test in the diagnostic evaluation of fetal structural anomalies (28).
The incidence of pathogenic CNVs in fetuses with ultrasound anomalies can be further refined by the organ system involved and the number of anomalies observed. Approximately 5.6% of fetuses with a normal karyotype and an ultrasound-detected abnormality in a single organ system will have a potentially significant CNV (29). When cases with abnormalities of the nuchal area are excluded, this number increases to 6.7% (29). This indicates that the primary genomic abnormality associated with increased nuchal translucency remains aneuploidy (trisomies 21, 18, 13 and monosomy X) and that the incremental yield from CMA in such cases is not expected to be greater than patients referred for AMA (29).
The organ systems most commonly associated with abnormal CMA results are cardiac, renal, skeletal, urogenital, and central nervous system (26, 29–31). Isolated renal and cardiac anomalies show a 15.0% and 10.6% incremental yield of CMA over karyotyping and represent the greatest diagnostic yield in single organ system fetal anomalies (29). Even more striking is the finding that isolated cardiac outflow tract abnormalities have an incremental benefit of 30.0% (P=.005) (29). The incidence of pathogenic CNVs for the most common congenital heart defect at birth, ventricular septal defect (VSD), is around 7.3% (32). An important observation from the Donnelly et al study is that 66.7% of patients with cardiac defects had CNVs other than a 22q11.2 deletion (29). Although It has previously been common practice to order FISH for the 22q11.2 deletion (DiGeorge syndrome) when a cardiac abnormality is seen on ultrasound, limiting genetic studies to FISH for DiGeorge syndrome in such cases would result in more than 2/3 of genomic abnormalities being missed. For specific cardiac anomalies, the diagnostic rate using FISH for DiGeorge may be significantly lower as shown by a recent study on fetal right aortic arch where a 22q11.2 deletion was only observed in about 5% of cases (33). Prenatal identification of a 22q11.2 deletion has a significant impact on parental counseling since children with this disorder can have multiple other structural anomalies, immune deficiency, and neurocognitive disorders. Young adults with this deletion have almost a 25% occurrence of schizophrenia or other severe psychiatric disorders (22).
When ultrasound anomalies are observed in multiple organ systems and nuchal abnormalities are excluded, the frequency of non-benign CNVs increases to 13.6% (29). Tables 1 and 2 list the most frequent copy number changes observed in association with/without fetal anomalies detected by ultrasound. The 22q11.2 imbalance appears to be the most common submicroscopic imbalance observed in cases with and without fetal anomalies emphasizing the phenotypic heterogeneity that may be associated with CNVs. Such clinical heterogeneity also emphasizes the inability to screen for clinically significant CNVs by ultrasound alone.
Table 1.
Most frequent copy number changes observed in association with fetal anomalies detected by ultrasound.
| Fetal Anomalies | |
|---|---|
| CNV Region | Frequencya (Deletion + Duplication) |
| 10q21.1 | 3.3% |
| 15q13.3 | 3.3% |
| 1q21.1 | 4.9% |
| 16p13.11 | 9.8% |
| 17q12 | 9.8% |
| 22q11.21 | 18.0% |
| Single Occurrence | 50.8% |
Data from Donnelly et al (29)
Table 2.
Most frequent copy number changes observed in fetuses without structural anomalies.
| Without Fetal Anomalies | |
|---|---|
| CNV Region | Frequencyb (Deletion + Duplication) |
| 15q11.2 | 5.8% |
| Xp22.3 | 7.7% |
| Xp21.1 | 7.7% |
| 16p11.2 | 7.7% |
| 1q21.1 | 9.6% |
| 17p12 | 9.6% |
| 16p13.11 | 13.5% |
| 22q11.21 | 15.4% |
| Single Occurrence | 23.1% |
Additional benefits of CMA versus Karyotyping
One of the primary benefits of CMA is its ability to precisely define a region of imbalance. This means that the boundaries (breakpoints) of the region involved can accurately be delineated and the genetic sequence of that region can be further assessed. This is important not only for sub-microscopic imbalances but also for visible cytogenetic abnormalities that are of unknown origin. Examples of the latter include marker chromosomes and unbalanced rearrangements. For chromosomal abnormalities of unknown origin, the resulting clinical phenotype and prognosis depend on the chromosomal origin of the extra or missing material as well as the amount of clinically significant coding and regulatory sequences contained within the region in question. CMA allows for rapid assessment of the clinical relevance of such regions by integrating specific cytogenetic data with phenotypic information available in genome browsers and public databases.
Size alone does not determine pathogenicity of a CNV. For example, a deletion that is 1.6 Mb in size but located in a gene desert may only result in the removal of a few genes and may be less clinically relevant than a smaller deletion (600 Kb in size) that is located within a gene-rich region. CMA may also be helpful in cases with apparently balanced de-novo rearrangements which may have an empirical risk of 6.7% of having an abnormal phenotype (34). As opposed to karyotype analysis, CMA has the potential to reveal small gains or losses around the breakpoints of the structural rearrangements better defining the clinical consequences (35–39). Performing CMA without simultaneous karyotyping removes the possibility of identifying the balanced rearrangement. This in turn would deny the patient the opportunity of pursuing additional molecular investigations such as next generation sequencing to rule out further genomic complexity that may be associated with the apparently balanced rearrangement (40, 41). Finally, since CMA requires DNA and is not reliant on cell culturing, it offers the opportunity for a faster turnaround time as it can be performed on DNA extracted directly from uncultured villi, amniotic fluid, and fetal blood/tissue.
Disadvantages of CMA
CMA works by comparison of patient DNA to normal controls. As such, only differences reflected by imbalances in patients compared to controls will be identified. Balanced rearrangements will escape detection. In addition, while the presence of a balanced rearrangement carries no clinical significance for the majority of pregnancies, there are still reproductive ramifications for future pregnancies if one of the parent is a carrier.
PRE-TEST COUNSELING
The complexity of genomic testing precipitates the requirement for counseling patients prior to prenatal testing using microarrays. Pre-test counseling provides patients an opportunity to understand the benefits and limitations of CMA technology and thus make informed decisions regarding the appropriateness of testing based on their personal beliefs and attitudes (42). When CMA Is performed for prenatal diagnosis after CVS or amniocentesis counseling by a genetic counselor, geneticist, or other health care provider with appropriate expertise in genetic counseling is recommended. (43) When CMA is used to screen embryos to determine suitability for transfer credentials of those providing counseling are not specified by professional organizations.
There are multiple areas that should be covered in the pre-test counseling session. With the explosion of non-invasive prenatal testing (NIPT) using cell-free fetal DNA, it is likely that a portion of the counseling session will be devoted to explaining the differences between the limited but powerful screening potential of NIPT versus the vast scope of genomic abnormalities that CMA can diagnose. Instead of reviewing an extensive list of conditions detectable by CMA, the focus should lie on the types of conditions with specific examples. While the majority of CMA results will be straightforward, a small proportion of results will not be clear-cut and pre-test knowledge of these issues greatly facilitates post-test delivery (44). The concept of phenotypic heterogeneity (due to variable penetrance and variable expressivity), where the severity of a condition can range from apparently normal to severe, should be explained. When such a condition is encountered, patients should be aware that the clinical spectrum of the disorder may not be predictable in their fetus and that they themselves could possibly carry the same CNV, despite showing no obvious clinical phenotype. Additional discussions should also mention that certain CNVs are associated with susceptibility to a variety of neurocognitive conditions such as autism and schizophrenia as well as late onset conditions which in some cases may be unrelated to the indication for testing. Most importantly, the patient should understand the likelihood of finding a clinically significant CNV based on their reason for indication. The likelihood of a CNV finding of uncertain clinical significance (~1.0%) should also be discussed as well as the potential for identification of consanguinity and non-paternity (2, 27).
Sensitivity of Prenatal CMA
As with any clinical test, CMA’s sensitivity relies on the quality of the sample and the extracted DNA. Each CMA platform is limited by the resolution of probe coverage (the number of probes, their location as well as their spacing) such that genomic imbalances that fall within areas with low or no probe coverage may be missed. In reality, most clinical laboratories use arrays that have optimal probe coverage for clinically significant regions associated with known genetic disorders or syndromes. From a counseling perspective, patients should be informed that microarray analysis cannot detect all genetic alterations and that a normal CMA result does not diminish the risk for other genetic conditions or birth defects (2, 27). The types of genetic alterations that escape detection by routine prenatal CMA primarily include balanced rearrangements (translocations/inversions/insertions) and single gene mutations. In addition, triploidy cannot be detected by aCGH because the additional copy of all chromosomes is masked by the normalization procedure run by the software analysis. This limitation can be overcome by specifically using SNP arrays (SOMA) to analyze additional genotypes from the extra chromosomes (Figure 4). Even when CMA is positive, it will not always provide the chromosomal mechanism causing the imbalance. For example, CMA will detect the presence of trisomy 13 in a prenatal sample but cannot discern whether it resulted from a non-disjunction event or whether it was due to a translocation. In such cases, karyotype analysis of the fetus and the parents is essential for determining reproductive risk for future offspring. Lastly, mosaicism, below a certain level (~10–20%) may also be difficult to detect. This limitation is not unique to CMA and is in fact, a universal challenge for all genetic testing. For example, standard karyotyping of 20 cells is only sensitive enough to detect mosaicism of 14% with 95% confidence (45). When CMA is performed on DNA extracted directly from uncultured cell, it has the potential to reveal mosaicism that escapes detection in cultured cells due to preferential growth of a normal cell line (See Figure 3) (46).
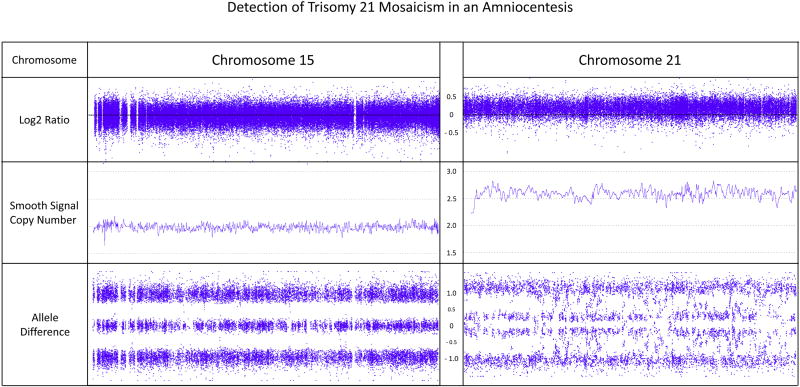
Figure 3.
Screenshot from the Affymetrix Chromosome Analysis Suite Software (Version 3.1) showing mosaic trisomy 21. The Log2 Ratio of chromosome 21 is clearly shifted upwards compared to chromosome 15 (shown for comparison). The smooth signal copy number panel indicates the exact copy number of each probe. This panel is helpful in identifying mosaicism which is evident when the smooth signal for multiple consecutive probes lies between an integer, e.g. An integer between 2 and 3 indicates trisomy mosaicism. In this example, the median copy number state across chromosome 21 is 2.62, indicating the level of trisomy 21 mosaicism to be around 62%. The allele difference panel indicates the genotype for each SNP probe. For normal copy number of 2, there are only 3 possible SNP combinations, AA, AB and BB (see Figure1) which are plotted on the allele difference graph. When there is mosaicism, additional genotypes will be visible representing those present in both normal and abnormal cell lines.
CMA Versus NIPT
With much of the current focus in the prenatal world centering around NIPT, it is important to emphasize that NIPT, in its current form, is a screening test while CMA is a diagnostic test. As a screening test, NIPT offers superior detection of Down syndrome and the other common aneuploidies compared to traditional biochemical markers and nuchal translucency (NT) measurements (47, 48). The attractiveness of a non-invasive blood test coupled with sensitivities and specificities for the common aneuploidies that approach 100% (49) have made NIPT one of the fastest-adopted genetic tests. However, as a screening test, it does not cover the vast scope of genomic abnormalities that are detectable by CMA. A recent study of 2,779 fetuses discerned that approximately half of the abnormal aCGH results reported would not have been detectable by standard NIPT assays (i.e. did not involve trisomies 21, 18, 13, monosomy X, or a sex chromosome trisomy) (50). This is an important consideration when a pregnant woman is deciding between screening and diagnostic testing. In addition, women over 35 and those with a positive biochemical/NT test still retain a risk of CNVs or rare trisomy results despite a negative cell-free DNA screen.
It is also important to understand that despite high detection rates, the positive predictive value (PPV) of NIPT tests will vary depending on the patient’s a priori risk for a chromosomal abnormality. A priori risk is primarily determined by the prevalence of the disorder. For chromosomal aneuploidy, prevalence is dictated by maternal age while for microdeletions/duplications it is unaffected by parental age and is governed by the population incidence in a given ethnic group. The higher the a priori risk, the greater the PPV. PPVs for trisomy 21 have been reported to be as high as 98% in studies of high risk patients (51, 52) and as low as 45.5% in a general low risk population (47). This indicates the potential for a false positive result in about half of the patients who fall into the low category group. Given the screening nature of NIPT, all positive NIPT results should be followed up with an invasive test to confirm the diagnosis (53). Indeed, confined placental mosaicism, maternal copy number variation, vanished twin, maternal cancer and true fetal mosaicism have all been reported as reasons for discordant NIPT results (54).
In response to the growing appreciation of the incidence and a better understanding of the importance of submicroscopic CNVs and cytogenetic abnormalities other than the common aneuploidies, laboratories have begun developing the ability to identify smaller cytogenetic changes using cell free DNA. There are two current approaches; one which targets a handful of clinically significant microdeletions (55) and the other which sets a size-cutoff threshold of >7 Mb for genome-wide imbalances and <7 Mb for select microdeletions (56). NIPT using cell-free fetal DNA is currently the only reliable noninvasive method for screening for microdeletions/duplications. As such, NIPT for these submicroscopic CNVs greatly expands the prenatal diagnosis screening menu. However, given the very low a priori risk for each one of these disorders, there are a considerable number of screen positive cases that are found to be negative upon follow-up with CVS or amniocentesis (57). A recent NIPT study by a large referral genetic diagnostic laboratory using massively parallel sequencing reported PPVs as low as 0% for the Cri-du-Chat syndrome and Prader-Willi/Angelman syndrome; 14% for the 1p36 deletion syndrome and 21% for the 22q11.2 deletion syndrome (57). In comparison, using a SNP-based NIPT assay, Martin et.al projected PPVs with their revised screening protocol to be 9.1% for the Prader-Willi/Angelman syndrome; 66.7% for the Cri-du-Chat syndrome; 50.0% for the 1p36 deletion syndrome and 44.2% for the 22q11.2 deletion syndrome (58). It is therefore apparent that the detection rates of cell-free DNA screening for CNVs vary dramatically depending on the platform, laboratory, region, and size of the deletion or duplication. It is also important to remember that all current approaches to NIPT microdeletion screening target only a small percentage of the microdeletions/duplications commonly observed in routine CMA screening (2, 26, 29, 59–61). As seen in Tables 1 and 2, of the common microdeletions included on commercial NIPT panels (1p36, 4p16, 5p15, 8q24, 11q23, 15q11.2, 22q11.2) only 22q11.2 is found in the top 5 and 10 most common CNVs observed in fetuses with and without structural anomalies respectively (2, 26, 29, 59–61). This is significant information that should be conveyed to patients who are considering NIPT over CVS/amniocentesis with CMA.
There is currently a lack of adequate information regarding the performance of cell-free DNA screening for microdeletions when used for population screening. Consequently, the American Congress of Obstetricians and Gynecologists (ACOG), the Society for Maternal Fetal Medicine recommend (SMFM), the American Society for Human Genetics (ASHG), and the European Society for Human Genetics (ESHG) do not recommend using NIPT for detection of microdeletions (53, 62). Alternatively, the American College of Medical Genetics and Genomics (ACMGG) recommends informing women about the availability of NIPT screening for select microdeletions (but not whole genome) when specific conditions are met by both the healthcare provider and the performing laboratory (63).
An added complexity from screening all 24 chromosomes by whole genome NIPT has been the finding of rare autosomal trisomies. A recent study of 89,817 pregnancies screened by whole genome NIPT found rare autosomal trisomies in approximately 0.44% of cases (64). Adverse pregnancy outcomes were observed in approximately 75% of single rare autosomal trisomy cases for which follow up was available (64, 65). Poor outcomes were associated primarily with an increased risk for feto-placental disease, including intrauterine growth restriction, intrauterine fetal death, miscarriage, true fetal mosaicism and UPD (64). The frequency of these single rare trisomies appears similar to those reported after short-term culture karyotype analysis of cytotrophoblast cells from CVS (64, 65). Furthermore, the finding of multiple rare aneuploidies has, in some rare instances, been associated with maternal cancer (66–69). The suspicion of maternal cancer based on NIPT screening has been confirmed in some patients who were already aware of their neoplasia (66–69). However in some cases, patients were either asymptomatic or presymptomatic, prompting further investigations such as maternal whole-body imaging and assessment of tumor markers. While a proportion of presymptomatic women were subsequently identified with maternal neoplasms, the presence of maternal cancer could not be confirmed in other asymptomatic women (64, 66–69). While it may appear to be a lifesaving byproduct of NIPT, the finding of multiple rare aneuploidies indicative of a maternal cancer may generate unwarranted anxiety in an asymptomatic woman who remains negative following appropriate follow-up investigations. Additional research into the appropriate management and the clinical significance of rare autosomal trisomies and its association with maternal cancer is clearly needed. The unanticipated potential for whole genome NIPT to detect maternal cancer highlights the importance of appropriate genetic counseling when consenting women for NIPT using cell-free fetal DNA.
Despite its obvious superiority over current NIPT, the number of patients taking advantage of prenatal CMA following a CVS/amniocentesis has dramatically dropped in the NIPT/CMA era (70–72). Many women who previously might have considered an invasive procedure with CMA analysis are now opting for non-invasive screening by nuchal translucency measurement/biochemistry or increasingly by cell-free DNA screening, even though they are, in most cases, limited to the common aneuploidies. For some women, the noninvasive option is readily preferred as their inclination is to avoid any risks associated with an invasive diagnostic procedure. Such a choice should only be made after being appropriately informed of the risks, benefits and limitations of all prenatal diagnostic options. Contemporary literature now shows a remarkably small risk of pregnancy loss from either CVS or amniocentesis which in experienced centers is between 1 in 700 and 1 in 1000 (73). The subsequent choice to sacrifice the additional information that CMA offers, which may be as high as 1% or more, must be made in the context of their desire to minimize all risks. This is especially true for couples with infertility that have struggled to achieve an ongoing pregnancy. However, many women are unaware of the incremental information that CMA might provide. This may be in part due to the belief that screening for Down syndrome is synonymous with screening for all causes of intellectual disability. The primary focus of genetic screening for the past 50 years or so has been Down syndrome which could potentially explain the general assumptions made by many pregnant women. In reality, the lack of awareness of the differences between what NIPT offers and what CMA offers stems more from inadequate or nonexistent pretest counseling by health care providers who often have a limited knowledge of genetics and lack effective educational resources pertaining to NIPT (74, 75). In a 2016 survey of OB/GYNs and MFMs, only about one-half of respondents (n=136) indicated that they provided in-depth pretest NIPT counseling while only one-third of the remaining physicians referred their patients to a genetic counselor (75).
PROFESSIONAL SOCIETY CMA GUIDELINES AND REPORTING CRITERIA
The presence of CNVs is not correlated with maternal age (76) which effectively means that younger women are more likely to have a fetus with a microdeletion or microduplication than with Down syndrome. Consequently, ACOG and SMFM recommend that all pregnant women should be offered the option of diagnostic testing regardless of maternal age (77, 78). The finding by Wapner and colleagues (2) that clinically significant CNVs are seen in approximately 6% of pregnancies with ultrasound anomalies, led to the 2013 ACOG/SMFM recommendation that CMA replace or supplement karyotype for prenatal evaluation of fetuses with major structural anomalies (76).
The American College of Medical Genetics (ACMG) in collaboration with the Association for Molecular Pathology (AMP) published guidelines for the interpretation of variants and suggested a five-tier system of reporting CNVs (79). Under this system, CNVs are interpreted as “pathogenic”, “likely pathogenic”, “uncertain significance”, “likely benign”, or “benign”. A CMA report is expected to guide the referring clinician by providing the following specific information relating to the CNV being reported (80): [1] The cytogenetic location (chromosome and bands); [2] the CNV category, i.e., gain or loss as well as a mechanism if known; [3] CNV size and coordinates with genome build (i.e. hg19); [4] statement of significance as defined by the five-tier system (including evidence and references); [5] genes involved (specify genes related to condition if the CNV is associated with a known syndrome; list all RefSeq genes for all other scenarios); [6] recommendations for appropriate clinical follow-up.
While aCGH and SOMA have the potential to detect CNVs that are in the kilobase range, in prenatal cases, most clinical laboratories will only report out kilobase-sized CNVs (< 1Mb) in regions that are well characterized and known to be pathogenic. CNVs that fall outside these clinically significant regions (backbone region) are usually reported only if they meet certain criteria. Such criteria vary from laboratory to laboratory but usually include a size threshold (> 1–3 Mb) as well as the requirement that the CNV contains OMIM genes.
Variants of Uncertain Significance (VOUS)
When chromosomal abnormalities are observed on a karyotype they are frequently associated with clinical sequelae, primarily since visible imbalances involve hundreds to thousands of genes. With CMAs ability to detect submicroscopic imbalances, it soon became evident from population-based studies of normal individuals, that not all CNVs are associated with an adverse outcome (5, 7). The challenge for a clinical laboratory offering diagnostic CMA is to discern what is pathogenic and what is simply a benign polymorphism. Variants of unknown significance (VOUS) are genetic changes that are not commonly seen in the population and thus have little or no clinical evidence available to assess their pathogenicity. VOUS may represent benign familial variants that produce no clinical features, or may be rare deleterious changes resulting in a clinical phenotype. The size, gene content, and inheritance pattern can help to discern whether VOUS are more likely to be benign or pathogenic.
Chromosomal imbalances seen on karyotype are usually fully penetrant due to changes in a large number of genes. Therefore, if the same finding is seen in a normal parent the prognosis is usually good for the fetus. However, since CMA can detect smaller changes in the genotype, that is not always the case. Interpretation of smaller copy number changes identified by CMA can be more challenging. Public databases are available to help identify common, benign CNVs in the population, and every year new reports identify recurrent pathogenic CNVs to help with prenatal diagnosis. In general, a CNV that is inherited from a normal parent is less likely to be clinically relevant; however, emerging data shows that a growing number of CNVs have incomplete penetrance and/or variable expressivity. While studies support the pathogenicity of these CNVs, the patients fall along a spectrum of clinical outcomes, some severely affected while others are mildly affected or normal. Many of the CNVs that fall into this category affect neurocognitive development, and some evidence suggests that a second hit or increased mutational burden plays a role in the observed phenotype. Interpretation and counseling in these situations is particularly challenging because it is not always possible to predict the clinical outcome for the fetus.
Compared to the postnatal setting when candidates for testing are ascertained because of specific clinical features, in the prenatal setting, many phenotypic features such as neurocognitive ability are not readily observed, or only become apparent at a later gestational age. As utilization of CMA increases for prenatal diagnosis, better classification of the pathogenicity of rare copy number variants and their clinical outcomes will be possible and provide better guidance for clinical care. Parameters during CMA analysis can be set to reduce the chance of finding copy number variants of uncertain clinical significance. However, VUS will on occasion be identified and this will require followup parental testing to aide in assessing if the CNV is likely benign, likely pathogenic or simply unknown. Therefore, counseling patients before testing is important so they understand the possibility of receiving results that provide unexpected information about themselves, or that do not have clearly established or specific clinical outcomes.
Follow-up for positives and VOUS
For pregnancies in which abnormal CMA results are found, including those of uncertain significance, post-test counseling should be in-depth and conducted by someone with expertise in the field such as a genetic counselor or clinical geneticist. The session should review the finding, associated abnormalities and disease course, available published information and case reports, additional testing in the parents or other family members if appropriate, additional testing in the fetus such as fetal echocardiogram and ultrasound, as indicated, and a discussion of the pregnancy options available. Referrals to pediatric genetics or other specialists with specific knowledge about the condition and introduction to support groups as well as other families with a child that has the same condition can be useful in helping the patient comprehend the diagnosis and make a decision about the pregnancy. Follow-up with the patient either over the phone or in person is important. Many patients feel abandoned when this doesn’t occur (81).
In the United States, VOUS results are generally reported to patients but in other countries they may be withheld at the discretion of the geneticists. The likelihood of such a result varies depending on the coverage of the platform, especially across the backbone. Hillman et al. report a VOUS rate of 1.4% when all testing indications are considered (27). This is consistent with the initial NIH experience in which the incidence of variants of uncertain significance was initially 2.5% but over the next 7 years with time and accumulated data the implications of many of these have now been better defined. Rereview of the interpretation of the CNVs in this study revealed only a 0.9% VOUS rate; similar to that seen with standard karyotype (21, 82). Counseling a patient about a VOUS result requires research in advance of the appointment and allotment of additional counseling time. Recent SMFM guidelines suggest that patients with results of uncertain significance should receive counseling from an expert that can review and discuss all potential genotype-phenotype correlations as found on available databases (43).
Initial studies at the time of introduction of CMA showed that many women chose microarray because the increased information available was an offer “too good to pass up”. However, in cases in which uncertain or abnormal results were discovered, some felt “blindsided: and “unprepared” and one felt the knowledge was “toxic” (81). Most informative was their comment that they needed support beyond the time of the initial diagnosis. These comments support the importance of the post-test care.
ETHICAL CONSIDERATIONS
Like any genomic test, CMA comes with its own ethical considerations. Many of these relate to some of the less severe conditions CMA can identify. For example, some microdeletions are associated with a strong predisposition for the child to develop autism. However, these same findings can also lead to more benign learning disorders or ADHD (83). Likewise, some microdeletions such as the deletion of 22q11.2 associated with DiGeorge syndrome can be associated with a fairly high risk of later onset schizophrenia or other forms of mental illness (22).
Conclusion
CMA is one of many examples of our increasing ability to interrogate the human genome. In most cases this information will improve health but in others it can lead to clinical and ethical dilemmas. This is especially the case with prenatal testing and the associated reproductive options. It is important to point out however, that as opposed to a fetal diagnosis of a severe aneuploidy, many CMA findings are amenable to postnatal modification such as early intervention programs in which a presymtomatic diagnosis may further improve the outcome. We are now transitioning to prenatal precision medicine and are learning how to use these new tools for the benefit of our patients and their families.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steele MW, Breg WR., Jr Chromosome analysis of human amniotic-fluid cells. Lancet. 1966;1:383–5. doi: 10.1016/s0140-6736(66)91387-0. [DOI] [PubMed] [Google Scholar]
- 2.Wapner RJ, Martin CL, Levy B, Ballif BC, Eng CM, Zachary JM, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. The New England journal of medicine. 2012;367:2175–84. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manning M, Hudgins L, Professional P, Guidelines C. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genetics in medicine : official journal of the American College of Medical Genetics. 2010;12:742–5. doi: 10.1097/GIM.0b013e3181f8baad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. American journal of human genetics. 2010;86:749–64. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–51. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 6.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 8.Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, et al. Segmental duplications and copy-number variation in the human genome. American journal of human genetics. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 10.Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–4. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 11.Shearer BM, Thorland EC, Gonzales PR, Ketterling RP. Evaluation of a commercially available focused aCGH platform for the detection of constitutional chromosome anomalies. American journal of medical genetics Part A. 2007;143A:2357–70. doi: 10.1002/ajmg.a.31954. [DOI] [PubMed] [Google Scholar]
- 12.Beaudet AL, Belmont JW. Array-based DNA diagnostics: let the revolution begin. Annu Rev Med. 2008;59:113–29. doi: 10.1146/annurev.med.59.012907.101800. [DOI] [PubMed] [Google Scholar]
- 13.Bignell GR, Huang J, Greshock J, Watt S, Butler A, West S, et al. High-resolution analysis of DNA copy number using oligonucleotide microarrays. Genome research. 2004;14:287–95. doi: 10.1101/gr.2012304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, et al. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–71. doi: 10.1158/0008-5472.can-03-3308. [DOI] [PubMed] [Google Scholar]
- 15.Shaffer LG, Bejjani BA. A cytogeneticist's perspective on genomic microarrays. Human reproduction update. 2004;10:221–6. doi: 10.1093/humupd/dmh022. [DOI] [PubMed] [Google Scholar]
- 16.Breman A, Pursley AN, Hixson P, Bi W, Ward P, Bacino CA, et al. Prenatal chromosomal microarray analysis in a diagnostic laboratory; experience with >1000 cases and review of the literature. Prenatal diagnosis. 2012;32:351–61. doi: 10.1002/pd.3861. [DOI] [PubMed] [Google Scholar]
- 17.Callaway JL, Shaffer LG, Chitty LS, Rosenfeld JA, Crolla JA. The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: a review of the literature. Prenatal diagnosis. 2013;33:1119–23. doi: 10.1002/pd.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srebniak MI, Joosten M, Knapen M, Arends LR, Polak M, van Veen S, et al. Frequency of submicroscopic chromosome aberrations in pregnancies without increased risk for structural chromosome aberrations: a systematic review of literature and meta-analysis. Ultrasound in obstetrics & gynecology : the official journal of the International Society of Ultrasound in Obstetrics and Gynecology. 2017 doi: 10.1002/uog.17533. [DOI] [PubMed] [Google Scholar]
- 19.Bornstein E, Berger S, Cheung SW, Maliszewski KT, Patel A, Pursley AN, et al. Universal Prenatal Chromosomal Microarray Analysis: Additive Value and Clinical Dilemmas in Fetuses with a Normal Karyotype. Am J Perinatol. 2017;34:340–8. doi: 10.1055/s-0036-1586501. [DOI] [PubMed] [Google Scholar]
- 20.Van Opstal D, de Vries F, Govaerts L, Boter M, Lont D, van Veen S, et al. Benefits and burdens of using a SNP array in pregnancies at increased risk for the common aneuploidies. Human mutation. 2015;36:319–26. doi: 10.1002/humu.22742. [DOI] [PubMed] [Google Scholar]
- 21.Wapner RJ, Zachary J, Clifton R. Change in classification of prenatal microarray anaylsis copy number variants over time. Prenatal diagnosis. 2015;35(Supplement S1) [Google Scholar]
- 22.McDonald-McGinn DM, Sullivan KE. Chromosome 22q11.2 deletion syndrome (DiGeorge syndrome/velocardiofacial syndrome) Medicine (Baltimore) 2011;90:1–18. doi: 10.1097/MD.0b013e3182060469. [DOI] [PubMed] [Google Scholar]
- 23.Le Vaillant C, Beneteau C, Chan-Leconte N, David A, Riteau AS. Beckwith-Wiedemann syndrome: What do you search in prenatal diagnosis? About 14 cases. Gynecol Obstet Fertil. 2015;43:705–11. doi: 10.1016/j.gyobfe.2015.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Wax JR, Burroughs R, Wright MS. Prenatal sonographic features of Russell-Silver syndrome. J Ultrasound Med. 1996;15:253–5. doi: 10.7863/jum.1996.15.3.253. [DOI] [PubMed] [Google Scholar]
- 25.Srebniak MI, Diderich KE, Joosten M, Govaerts LC, Knijnenburg J, de Vries FA, et al. Prenatal SNP array testing in 1000 fetuses with ultrasound anomalies: causative, unexpected and susceptibility CNVs. Eur J Hum Genet. 2016;24:645–51. doi: 10.1038/ejhg.2015.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shaffer LG, Dabell MP, Fisher AJ, Coppinger J, Bandholz AM, Ellison JW, et al. Experience with microarray-based comparative genomic hybridization for prenatal diagnosis in over 5000 pregnancies. Prenatal diagnosis. 2012;32:976–85. doi: 10.1002/pd.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hillman SC, McMullan DJ, Hall G, Togneri FS, James N, Maher EJ, et al. Use of prenatal chromosomal microarray: prospective cohort study and systematic review and meta-analysis. Ultrasound in obstetrics & gynecology : the official journal of the International Society of Ultrasound in Obstetrics and Gynecology. 2013;41:610–20. doi: 10.1002/uog.12464. [DOI] [PubMed] [Google Scholar]
- 28.Committee Opinion No. 682: Microarrays and Next-Generation Sequencing Technology: The Use of Advanced Genetic Diagnostic Tools in Obstetrics and Gynecology. Obstetrics and gynecology. 2016;128:e262–e8. doi: 10.1097/AOG.0000000000001817. [DOI] [PubMed] [Google Scholar]
- 29.Donnelly JC, Platt LD, Rebarber A, Zachary J, Grobman WA, Wapner RJ. Association of copy number variants with specific ultrasonographically detected fetal anomalies. Obstetrics and gynecology. 2014;124:83–90. doi: 10.1097/AOG.0000000000000336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Faas BH, van der Burgt I, Kooper AJ, Pfundt R, Hehir-Kwa JY, Smits AP, et al. Identification of clinically significant, submicroscopic chromosome alterations and UPD in fetuses with ultrasound anomalies using genome-wide 250k SNP array analysis. Journal of medical genetics. 2010;47:586–94. doi: 10.1136/jmg.2009.075853. [DOI] [PubMed] [Google Scholar]
- 31.Kleeman L, Bianchi DW, Shaffer LG, Rorem E, Cowan J, Craigo SD, et al. Use of array comparative genomic hybridization for prenatal diagnosis of fetuses with sonographic anomalies and normal metaphase karyotype. Prenatal diagnosis. 2009;29:1213–7. doi: 10.1002/pd.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sukenik-Halevy R, Sukenik S, Koifman A, Alpert Y, Hershkovitz R, Levi A, et al. Clinical aspects of prenatally detected congenital heart malformations and the yield of chromosomal microarray analysis. Prenatal diagnosis. 2016;36:1185–91. doi: 10.1002/pd.4954. [DOI] [PubMed] [Google Scholar]
- 33.Peng R, Xie HN, Zheng J, Zhou Y, Lin MF. Fetal right aortic arch: associated anomalies, genetic anomalies with chromosomal microarray analysis, and postnatal outcome. Prenatal diagnosis. 2017;37:329–35. doi: 10.1002/pd.5015. [DOI] [PubMed] [Google Scholar]
- 34.Warburton D. De novo balanced chromosome rearrangements and extra marker chromosomes identified at prenatal diagnosis: clinical significance and distribution of breakpoints. American journal of human genetics. 1991;49:995–1013. [PMC free article] [PubMed] [Google Scholar]
- 35.Astbury C, Christ LA, Aughton DJ, Cassidy SB, Kumar A, Eichler EE, et al. Detection of deletions in de novo "balanced" chromosome rearrangements: further evidence for their role in phenotypic abnormalities. Genetics in medicine : official journal of the American College of Medical Genetics. 2004;6:81–9. doi: 10.1097/01.gim.0000117850.04443.c9. [DOI] [PubMed] [Google Scholar]
- 36.Shanske AL, Edelmann L, Kardon NB, Gosset P, Levy B. Detection of an interstitial deletion of 2q21–22 by high resolution comparative genomic hybridization in a child with multiple congenital anomalies and an apparent balanced translocation. American journal of medical genetics Part A. 2004;131:29–35. doi: 10.1002/ajmg.a.30311. [DOI] [PubMed] [Google Scholar]
- 37.Simovich MJ, Yatsenko SA, Kang SH, Cheung SW, Dudek ME, Pursley A, et al. Prenatal diagnosis of a 9q34.3 microdeletion by array-CGH in a fetus with an apparently balanced translocation. Prenatal diagnosis. 2007;27:1112–7. doi: 10.1002/pd.1841. [DOI] [PubMed] [Google Scholar]
- 38.Tabet AC, Verloes A, Pilorge M, Delaby E, Delorme R, Nygren G, et al. Complex nature of apparently balanced chromosomal rearrangements in patients with autism spectrum disorder. Mol Autism. 2015;6:19. doi: 10.1186/s13229-015-0015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higgins AW, Alkuraya FS, Bosco AF, Brown KK, Bruns GA, Donovan DJ, et al. Characterization of apparently balanced chromosomal rearrangements from the developmental genome anatomy project. American journal of human genetics. 2008;82:712–22. doi: 10.1016/j.ajhg.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ordulu Z, Kammin T, Brand H, Pillalamarri V, Redin CE, Collins RL, et al. Structural Chromosomal Rearrangements Require Nucleotide-Level Resolution: Lessons from Next-Generation Sequencing in Prenatal Diagnosis. American journal of human genetics. 2016;99:1015–33. doi: 10.1016/j.ajhg.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Redin C, Brand H, Collins RL, Kammin T, Mitchell E, Hodge JC, et al. The genomic landscape of balanced cytogenetic abnormalities associated with human congenital anomalies. Nat Genet. 2017;49:36–45. doi: 10.1038/ng.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fonda Allen J, Stoll K, Bernhardt BA. Pre- and post-test genetic counseling for chromosomal and Mendelian disorders. Seminars in perinatology. 2016;40:44–55. doi: 10.1053/j.semperi.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Society for Maternal-Fetal Medicine. Electronic address pso. Dugoff L, Norton ME, Kuller JA. The use of chromosomal microarray for prenatal diagnosis. American journal of obstetrics and gynecology. 2016;215:B2–9. doi: 10.1016/j.ajog.2016.07.016. [DOI] [PubMed] [Google Scholar]
- 44.Riedijk S, Diderich KE, van der Steen SL, Govaerts LC, Joosten M, Knapen MF, et al. The Psychological Challenges of Replacing Conventional Karyotyping with Genomic SNP Array Analysis in Prenatal Testing. J Clin Med. 2014;3:713–23. doi: 10.3390/jcm3030713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hook EB. Exclusion of chromosomal mosaicism: tables of 90%, 95% and 99% confidence limits and comments on use. American journal of human genetics. 1977;29:94–7. [PMC free article] [PubMed] [Google Scholar]
- 46.Hall GK, Mackie FL, Hamilton S, Evans A, McMullan DJ, Williams D, et al. Chromosomal microarray analysis allows prenatal detection of low level mosaic autosomal aneuploidy. Prenatal diagnosis. 2014;34:505–7. doi: 10.1002/pd.4333. [DOI] [PubMed] [Google Scholar]
- 47.Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, Das AF, et al. DNA sequencing versus standard prenatal aneuploidy screening. The New England journal of medicine. 2014;370:799–808. doi: 10.1056/NEJMoa1311037. [DOI] [PubMed] [Google Scholar]
- 48.Norton ME, Jacobsson B, Swamy GK, Laurent LC, Ranzini AC, Brar H, et al. Cell-free DNA analysis for noninvasive examination of trisomy. The New England journal of medicine. 2015;372:1589–97. doi: 10.1056/NEJMoa1407349. [DOI] [PubMed] [Google Scholar]
- 49.Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound in obstetrics & gynecology : the official journal of the International Society of Ultrasound in Obstetrics and Gynecology. 2017;50:302–14. doi: 10.1002/uog.17484. [DOI] [PubMed] [Google Scholar]
- 50.Sotiriadis A, Papoulidis I, Siomou E, Papageorgiou E, Eleftheriades M, Papadopoulos V, et al. Non-invasive prenatal screening versus prenatal diagnosis by array comparative genomic hybridization: a comparative retrospective study. Prenatal diagnosis. 2017;37:583–92. doi: 10.1002/pd.5051. [DOI] [PubMed] [Google Scholar]
- 51.McLennan A, Palma-Dias R, da Silva Costa F, Meagher S, Nisbet DL, Scott F. Noninvasive prenatal testing in routine clinical practice--an audit of NIPT and combined first-trimester screening in an unselected Australian population. Aust N Z J Obstet Gynaecol. 2016;56:22–8. doi: 10.1111/ajo.12432. [DOI] [PubMed] [Google Scholar]
- 52.Porreco RP, Garite TJ, Maurel K, Marusiak B, Obstetrix Collaborative Research N. Ehrich M, et al. Noninvasive prenatal screening for fetal trisomies 21, 18, 13 and the common sex chromosome aneuploidies from maternal blood using massively parallel genomic sequencing of DNA. American journal of obstetrics and gynecology. 2014;211:365, e1–12. doi: 10.1016/j.ajog.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 53.Committee Opinion Summary No. 640: Cell-Free DNA Screening For Fetal Aneuploidy. Obstetrics and gynecology. 2015;126:691–2. doi: 10.1097/01.AOG.0000471171.86798.ac. [DOI] [PubMed] [Google Scholar]
- 54.Hartwig TS, Ambye L, Sorensen S, Jorgensen FS. Discordant non-invasive prenatal testing (NIPT) - a systematic review. Prenatal diagnosis. 2017;37:527–39. doi: 10.1002/pd.5049. [DOI] [PubMed] [Google Scholar]
- 55.Wapner RJ, Babiarz JE, Levy B, Stosic M, Zimmermann B, Sigurjonsson S, et al. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. American journal of obstetrics and gynecology. 2015;212:332, e1–9. doi: 10.1016/j.ajog.2014.11.041. [DOI] [PubMed] [Google Scholar]
- 56.Lefkowitz RB, Tynan JA, Liu T, Wu Y, Mazloom AR, Almasri E, et al. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. American journal of obstetrics and gynecology. 2016;215:227, e1–e16. doi: 10.1016/j.ajog.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 57.Petersen AK, Cheung SW, Smith JL, Bi W, Ward PA, Peacock S, et al. Positive predictive value estimates for cell-free noninvasive prenatal screening from data of a large referral genetic diagnostic laboratory. American journal of obstetrics and gynecology. 2017;217:691, e1–e6. doi: 10.1016/j.ajog.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Martin K, Iyengar S, Kalyan A, Lan C, Simon AL, Stosic M, et al. Clinical experience with a single-nucleotide polymorphism-based non-invasive prenatal test for five clinically significant microdeletions. Clin Genet. 2017 doi: 10.1111/cge.13098. [DOI] [PubMed] [Google Scholar]
- 59.Armengol L, Nevado J, Serra-Juhe C, Plaja A, Mediano C, Garcia-Santiago FA, et al. Clinical utility of chromosomal microarray analysis in invasive prenatal diagnosis. Human genetics. 2012;131:513–23. doi: 10.1007/s00439-011-1095-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fiorentino F, Caiazzo F, Napolitano S, Spizzichino L, Bono S, Sessa M, et al. Introducing array comparative genomic hybridization into routine prenatal diagnosis practice: a prospective study on over 1000 consecutive clinical cases. Prenatal diagnosis. 2011;31:1270–82. doi: 10.1002/pd.2884. [DOI] [PubMed] [Google Scholar]
- 61.Lee CN, Lin SY, Lin CH, Shih JC, Lin TH, Su YN. Clinical utility of array comparative genomic hybridisation for prenatal diagnosis: a cohort study of 3171 pregnancies. BJOG : an international journal of obstetrics and gynaecology. 2012;119:614–25. doi: 10.1111/j.1471-0528.2012.03279.x. [DOI] [PubMed] [Google Scholar]
- 62.Dondorp W, de Wert G, Bombard Y, Bianchi DW, Bergmann C, Borry P, et al. Non-invasive prenatal testing for aneuploidy and beyond: challenges of responsible innovation in prenatal screening. Eur J Hum Genet. 2015;23:1592. doi: 10.1038/ejhg.2015.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genetics in medicine : official journal of the American College of Medical Genetics. 2016;18:1056–65. doi: 10.1038/gim.2016.97. [DOI] [PubMed] [Google Scholar]
- 64.Pertile MD, Halks-Miller M, Flowers N, Barbacioru C, Kinnings SL, Vavrek D, et al. Rare autosomal trisomies, revealed by maternal plasma DNA sequencing, suggest increased risk of feto-placental disease. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aan1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malvestiti F, Agrati C, Grimi B, Pompilii E, Izzi C, Martinoni L, et al. Interpreting mosaicism in chorionic villi: results of a monocentric series of 1001 mosaics in chorionic villi with follow-up amniocentesis. Prenatal diagnosis. 2015;35:1117–27. doi: 10.1002/pd.4656. [DOI] [PubMed] [Google Scholar]
- 66.Amant F, Verheecke M, Wlodarska I, Dehaspe L, Brady P, Brison N, et al. Presymptomatic Identification of Cancers in Pregnant Women During Noninvasive Prenatal Testing. JAMA Oncol. 2015;1:814–9. doi: 10.1001/jamaoncol.2015.1883. [DOI] [PubMed] [Google Scholar]
- 67.Bianchi DW, Chudova D, Sehnert AJ, Bhatt S, Murray K, Prosen TL, et al. Noninvasive Prenatal Testing and Incidental Detection of Occult Maternal Malignancies. JAMA : the journal of the American Medical Association. 2015;314:162–9. doi: 10.1001/jama.2015.7120. [DOI] [PubMed] [Google Scholar]
- 68.Dharajiya NG, Namba A, Horiuchi I, Miyai S, Farkas DH, Almasri E, et al. Uterine leiomyoma confounding a noninvasive prenatal test result. Prenatal diagnosis. 2015;35:990–3. doi: 10.1002/pd.4629. [DOI] [PubMed] [Google Scholar]
- 69.Osborne CM, Hardisty E, Devers P, Kaiser-Rogers K, Hayden MA, Goodnight W, et al. Discordant noninvasive prenatal testing results in a patient subsequently diagnosed with metastatic disease. Prenatal diagnosis. 2013;33:609–11. doi: 10.1002/pd.4100. [DOI] [PubMed] [Google Scholar]
- 70.Chan YM, Leung WC, Chan WP, Leung TY, Cheng YK, Sahota DS. Women's uptake of non-invasive DNA testing following a high-risk screening test for trisomy 21 within a publicly funded healthcare system: findings from a retrospective review. Prenatal diagnosis. 2015;35:342–7. doi: 10.1002/pd.4544. [DOI] [PubMed] [Google Scholar]
- 71.Chetty S, Garabedian MJ, Norton ME. Uptake of noninvasive prenatal testing (NIPT) in women following positive aneuploidy screening. Prenatal diagnosis. 2013;33:542–6. doi: 10.1002/pd.4125. [DOI] [PubMed] [Google Scholar]
- 72.Williams J, 3rd, Rad S, Beauchamp S, Ratousi D, Subramaniam V, Farivar S, et al. Utilization of noninvasive prenatal testing: impact on referrals for diagnostic testing. American journal of obstetrics and gynecology. 2015;213:102, e1–6. doi: 10.1016/j.ajog.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 73.Akolekar R, Beta J, Picciarelli G, Ogilvie C, D'Antonio F. Procedure-related risk of miscarriage following amniocentesis and chorionic villus sampling: a systematic review and meta-analysis. Ultrasound in obstetrics & gynecology : the official journal of the International Society of Ultrasound in Obstetrics and Gynecology. 2015;45:16–26. doi: 10.1002/uog.14636. [DOI] [PubMed] [Google Scholar]
- 74.Bayefsky MJ, White A, Wakim P, Hull SC, Wasserman D, Chen S, et al. Views of American OB/GYNs on the ethics of prenatal whole-genome sequencing. Prenatal diagnosis. 2016;36:1250–6. doi: 10.1002/pd.4968. [DOI] [PubMed] [Google Scholar]
- 75.Farrell RM, Agatisa PK, Mercer MB, Mitchum AG, Coleridge MB. The use of noninvasive prenatal testing in obstetric care: educational resources, practice patterns, and barriers reported by a national sample of clinicians. Prenatal diagnosis. 2016;36:499–506. doi: 10.1002/pd.4812. [DOI] [PubMed] [Google Scholar]
- 76.American College of O, Gynecologists Committee on G. Committee Opinion No. 581: the use of chromosomal microarray analysis in prenatal diagnosis. Obstetrics and gynecology. 2013;122:1374–7. doi: 10.1097/01.AOG.0000438962.16108.d1. [DOI] [PubMed] [Google Scholar]
- 77.American College of O, Gynecologists' Committee on Practice B-O, Committee on G, Society for Maternal-Fetal M. Practice Bulletin No. 162: Prenatal Diagnostic Testing for Genetic Disorders. Obstetrics and gynecology. 2016;127:e108–22. doi: 10.1097/AOG.0000000000001405. [DOI] [PubMed] [Google Scholar]
- 78.American College of OG. ACOG Practice Bulletin No. 88, December 2007. Invasive prenatal testing for aneuploidy. Obstetrics and gynecology. 2007;110:1459–67. doi: 10.1097/01.AOG.0000291570.63450.44. [DOI] [PubMed] [Google Scholar]
- 79.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in medicine : official journal of the American College of Medical Genetics. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST Working Group of the American College of Medical Genetics Laboratory Quality Assurance C. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genetics in medicine : official journal of the American College of Medical Genetics. 2011;13:680–5. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 81.Bernhardt BA, Soucier D, Hanson K, Savage MS, Jackson L, Wapner RJ. Women's experiences receiving abnormal prenatal chromosomal microarray testing results. Genetics in medicine : official journal of the American College of Medical Genetics. 2013;15:139–45. doi: 10.1038/gim.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang YW, Chang CM, Sung PL, Yang MJ, Li WH, Li HY, et al. An overview of a 30-year experience with amniocentesis in a single tertiary medical center in Taiwan. Taiwan J Obstet Gynecol. 2012;51:206–11. doi: 10.1016/j.tjog.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 83.Hanson E, Bernier R, Porche K, Jackson FI, Goin-Kochel RP, Snyder LG, et al. The cognitive and behavioral phenotype of the 16p11.2 deletion in a clinically ascertained population. Biol Psychiatry. 2015;77:785–93. doi: 10.1016/j.biopsych.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]