Abstract
Precision medicine has a goal of customizing disease prevention and treatment strategies. Under the precision medicine paradigm, each patient has unique pathologic processes resulting from cellular genomic, epigenomic, proteomic, and metabolomic alterations, which are influenced by pharmacological, environmental, microbial, dietary, and lifestyle factors. Hence, to realize the promise of precision medicine, multi-level research methods that can comprehensively analyze many of these variables are needed. In order to address this gap, the integrative field of molecular pathology and population data science (i.e., molecular pathological epidemiology) has been developed to enable such multi-level analyses, especially in gastrointestinal cancer research. Further integration of pharmacology can improve our understanding of drug effects, and inform decision-making of drug use at both the individual and population levels. Such integrative research demonstrated potential benefits of aspirin in colorectal carcinoma with PIK3CA mutations, providing the basis for new clinical trials. Evidence also suggests that HPGD (15-PDGH) expression levels in normal colon and the germline rs6983267 polymorphism that relates to tumor CTNNB1 (β-catenin)/WNT signaling status may predict the efficacy of aspirin for cancer chemoprevention. As immune checkpoint blockade targeting the CD274 (PD-L1)/PDCD1 (PD-1) pathway for microsatellite instability-high (or mismatch repair-deficient) metastatic gastrointestinal or other tumors has become standard of care, potential modifying effects of dietary, lifestyle, microbial, and environmental factors on immunotherapy need to be studied to further optimize treatment strategies. With its broad applicability, our integrative approach can provide insights into the interactive role of medications, exposures, and molecular pathology, and guide the development of precision medicine.
Introduction–emergence of precision medicine
Science is composed of specific fields of research, which are structured to organize education, training, and scientists themselves. By its nature, science never stops evolving. The medical and health sciences are no exception. As science continuously progresses with the development of concepts, methods, and discoveries, new scientific paradigms and fields emerge, which may augment or replace existing ones.
A good example of this phenomenon is the recent development of precision medicine, which has attracted heated attention, particularly in the field of oncology, with the hope of individualized patient treatment and care. In addition to precision treatment, precision prevention is a part of precision medicine.1–4 However, to realize the promise of precision medicine, the development of supporting method-based disciplines is needed. There have been growing concerns about the validity of published study findings5,6 and, unless we take action, this problem will only be exacerbated in the era of big data and omics research. Rigorous and standardized research methods are essential for generating reproducible data and generalizable knowledge, to facilitate the realization of precision medicine. Furthermore, a better understanding of research methods can improve not only the quality of research publications but also peer-review processes and evaluation of evidence in the literature. Therefore, we cannot over-emphasize the importance of the development of method-based science.
Pathology is a discipline that concerns pathogenic mechanisms, as well as methods to analyze tissues, cells, and molecules in disease processes. In recent decades, molecular pathology has become dominant, and large amounts of molecular pathology data have accumulated worldwide.7,8 The advancement of molecular pathology is not only transforming disease classification schemes, but is also generating opportunities for improved prevention and treatment.7,8 However, to optimally utilize molecular pathologic data, there is a pressing need to integrate data science into pathology.
Epidemiology is a method-based discipline that concerns not only examining determinants of disease and health outcomes but also the development of data analysis methods. Epidemiologic research provides data to inform evidence-based clinical practice and health policy-making. Essentially every medical study explicitly or implicitly uses epidemiologic principles to produce generalizable scientific knowledge. These facts attest to the importance of epidemiology as a core method-based discipline. Epidemiology has been applied in not only various disease-based fields (to generate subfields such as cancer epidemiology) but also research areas that focus on specific endogenous or exogenous health-related factors (to generate subfields such as pharmacoepidemiology) (Fig. 1). As various subfields emerge and evolve, the field of epidemiology constantly transforms to generate new concepts and address ever-changing practical and educational needs.
Fig. 1.
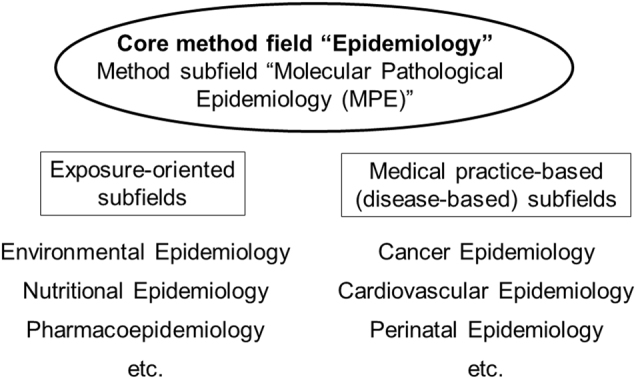
Structure of epidemiology and its subfields. Since the formation of the field of epidemiology, a number of subfields have emerged to specialize into particular subject matters, including detailed analyses of exposure factors (depicted on the left) and detailed analyses of health outcomes or diseases (depicted on the right). Six such subfields among many are shown. In addition, a method subfield "molecular pathological epidemiology (MPE)" has been developed under the core method field of epidemiology. MPE can be applied to any exposure and disease settings, and can be integrated with any other subfield of epidemiology
In this article, we discuss the value and implications of integrating pharmacology, molecular pathology, and epidemiology, utilizing gastrointestinal cancers as a disease model. Our intent is to illustrate the clinical and public health implications of this integrative concept that can be instrumental in advancing the field of cancer precision medicine.
Molecular pathological epidemiology
Although molecular pathology was incorporated into the broad umbrella of molecular epidemiology in the 1990s, a substantial gap between molecular pathology and epidemiology resulted in limited interest in developing integrated approaches.9 To address this, a single unified field of molecular pathological epidemiology (MPE), which integrates research approaches in molecular pathology and epidemiology, has emerged.10–12 The basic principle of MPE resides in the fact that many endogenous and exogenous factors (commonly referred to as "exposures"), in combination, modify phenotypes of diseases including cancer (Fig. 2); hence, better understanding of cancer requires comprehensive analyses of exposures and tumor phenotypes. In MPE research, an exposure or risk factor can be connected to specific pathogenic signatures.12 As a more homogeneous group of patients (i.e., disease subtype based on common pathogenic features) have similar etiologies, the MPE approach can yield a more accurate risk measure.11,12 This new research framework of MPE drives the development of epidemiological and statistical methods.13–18 Although challenges exist in MPE,11 the paradigm of MPE has been widely accepted,19–29 and discussed at various international meetings.30–33 MPE can also provide evidence linking drugs and disease outcomes,34–37 thereby supporting the precision medicine initiative. As a versatile method-based discipline, MPE can be applied to any disease setting, and further integrated with other investigative fields. To date, the MPE approach has been most commonly used in research of cancers, especially colorectal cancer.11,38
Fig. 2.
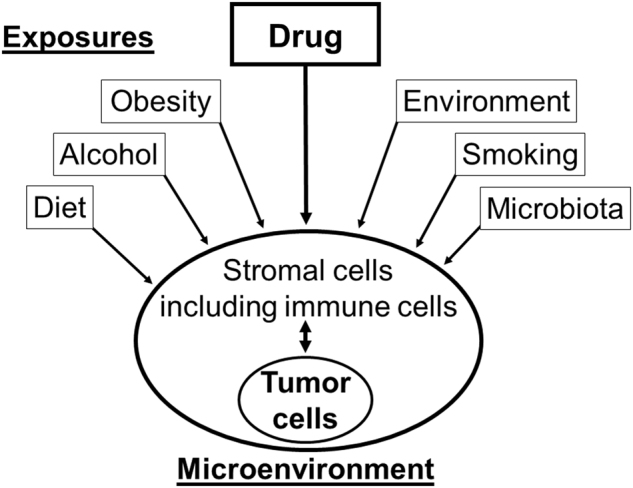
Influences of various exposures on pathogenic process. A wide variety of endogenous and exogenous factors (including drugs), individually or in combination, can modify phenotypes of cancer, leading to interpersonal heterogeneity. The molecular pathological epidemiology (MPE) approach utilizes integrated analyses of these exposures and tumor phenotypes to improve our understanding of tumor development and progression. Of note, for the sake of simplicity, this illustration does not depict complex interactions between the exposure factors
Pharmacoepidemiology
Pharmacoepidemiology is the study of the use and effects of drugs and other medical devices in populations.39 In the 1980s, the field of pharmacoepidemiology arose as the integration of pharmacology and epidemiology (Fig. 3), primarily to improve drug surveillance programs for ensuring safe and effective medication use.39 Hence, drug surveillance and effectiveness research has been a major focus of pharmacoepidemiology. With the precision medicine initiative,40 molecular diagnostics has become an important part of clinical practice and decision making, especially to guide medication use. Therefore, there are ample opportunities for pharmacoepidemiology to expand and incorporate advances of molecular pathology.
Fig. 3.
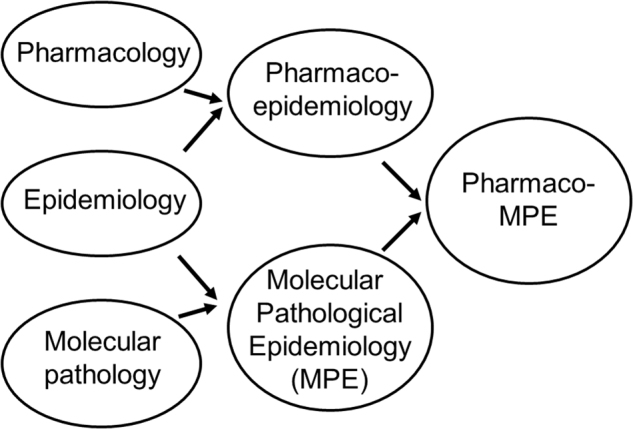
Trans-multidisciplinary integration of pharmacology, epidemiology, and molecular pathology. The integration of pharmacology and epidemiology has generated pharmacoepidemiology, while the integration of molecular pathology and epidemiology has generated molecular pathological epidemiology (MPE). We propose the integration of pharmacoepidemiology and MPE to generate pharmaco-MPE
Value of integrating pharmacoepidemiology and MPE
When we study pharmacologic factors and molecular pathology using the epidemiologic principles, the integration of pharmacoepidemiology and MPE (herein referred to as pharmaco-MPE) can be considered (Fig. 3). What is the value of this integration? Taking advantage of the complementary, synergistic strengths of pharmacoepidemiology and MPE (as illustrated in Fig. 4), the integrative approach can provide new insights into interactions of drug, environmental, and host factors in pathologic processes, thereby helping personalize therapies and advance precision medicine.
Fig. 4.
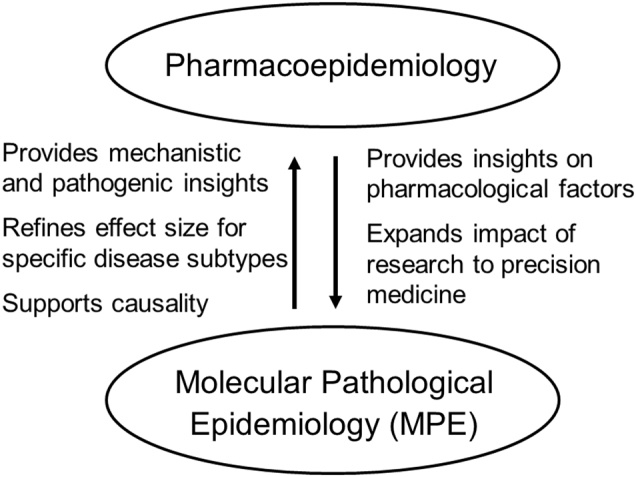
Collaborative relationship between pharmacoepidemiology and molecular pathological epidemiology (MPE). Both are subfields of epidemiology, and cover the entire spectrum of human diseases. Both fields can be synergized to create an integrative field of pharmaco-MPE, which can further enhance research and education for precision medicine
For the past decades, research on inter-individual differences in effects of drugs has become increasingly relevant. In precision medicine, it is critical to understand the effects of drugs in relation to the spectrum of molecular pathological characteristics.41 In fact, the integrative MPE approach has been useful in investigating effects of exposures, including drugs, and their interactive effects with disease molecular pathology (as explained in the next section).38
With the precise definition of epidemiology, any clinical investigation (regardless of its design) explicitly or implicitly uses epidemiologic principles with a goal of obtaining generalizable knowledge, and therefore can be regarded as epidemiologic research. Despite this fact, many investigators do not have adequate knowledge of epidemiologic principles, which has led to the situation that "most published research findings are false".5 Thus, epidemiologic principles that can enhance scientific rigor are of paramount importance. Essentially all clinical studies on drugs (including clinical trials) utilize epidemiologic principles, and can be regarded as pharmacoepidemiologic research. By these definitions, clinical studies that examine both drugs and molecular pathology can be regarded as pharmaco-MPE research. With this "pharmaco-MPE" designation, any investigators who examine drugs, molecular pathology, and clinical outcomes should be aware that they must utilize appropriate multidisciplinary expertise, and follow research standards of all of the component fields, i.e., pharmacology, molecular pathology, and epidemiology. While invalid results from poorly-designed studies confuse both researchers and practitioners, evidence from high quality pharmaco-MPE research can potentially improve clinical practice and guide future research.
Integrative research model on aspirin and colorectal cancer
In this section, we discuss studies on aspirin and colorectal cancer as a pharmaco-MPE research model, to illustrate its potential for impacting clinical practice. Colorectal cancer remains a major cause of morbidity and mortality. Colorectal carcinoma represents a group of molecularly heterogeneous tumors with varying combinations of somatic genetic and epigenetic changes,42–47 as well as varying degrees of immune response to tumor.48–53 As the colorectum is a long organ with differing contents and microbiota along its length, tumor characteristics vary according to its subsites.54–58
Accumulating evidence supports the anti-cancer effect of aspirin, one of the non-steroidal anti-inflammatory drugs (NSAIDs).59–61 Aspirin is a modulator of immunity and inflammation.49,62,63 A recent study has shown a stronger cancer-preventive effect of aspirin for colorectal cancer compared to other major cancer types.64 In addition, aspirin may be used for colorectal cancer therapy.65,66 As it is conceivable that the effect of aspirin differs by tumor subtype, the pharmaco-MPE approach could be useful to determine which individuals would gain more benefits from aspirin therapy. Previous pharmaco-MPE studies have utilized population-based datasets to examine potential influences of common medications (including aspirin) on incidence and progression of colorectal cancer subtypes as classified by tissue biomarkers including PTGS2 (cyclooxygenase 2) expression.34–37,67–72 Notably, a pharmaco-MPE study provided evidence for a strong beneficial effect of aspirin for PIK3CA-mutated colorectal cancer but not for PIK3CA-wild-type cancer.71 Hence, PIK3CA mutation in colorectal cancer may be a predictive biomarker for response to aspirin, while its prognostic role may not be significant.73–79 The findings have been subsequently tested in independent datasets,80–83 systematic review and meta-analyses,73,84 and in vitro experiments on colon and breast cancer cells.85,86 Although most of these follow-up studies support the original observations in the human population, new prospective trials on aspirin are needed to validate the findings in addition to the existing trials on aspirin.63,65,87 Pharmaco-MPE studies on the association of postdiagnosis aspirin use with colorectal cancer survival according to other tumor molecular markers are also warranted. For example, alterations in the TGFB1 (transforming growth factor beta 1, TGF-beta) signaling have been implicated in colorectal carcinogenesis,88 and loss-of-function mutations in TGFBR2 (TGFB1 receptor 2) may serve as prognostic biomarkers for colorectal cancer patients. Interestingly, this signaling pathway interacts with the PI3K-AKT-MTOR pathway,88 which may modify the anti-tumor effects of aspirin, as described.
With regard to primary prevention, MPE research may help identify a biomarker to predict efficacy of aspirin. Studies provide evidence to support varying cancer-preventive effect of aspirin for different tumor subtypes.34–36,67 Notably, aspirin has been associated with lower risk for colorectal cancer with higher expression of HPGD (hydroxyprostaglandin dehydrogenase 15-(NAD), 15-PGDH) in normal adjacent colon mucosa.72 HPGD, a prostaglandin-degrading enzyme, has been shown to down-regulate the proinflammatory reaction.89,90 HPGD in normal colon in cancer-free individuals may be a candidate biomarker to predict the efficacy of aspirin for primary cancer prevention.72 In addition, in vivo imaging technologies (such as low-coherence enhanced backscattering spectroscopy91) can be utilized to evaluate cellular molecular changes associated with use of aspirin or other drugs.92 Another candidate predictor of aspirin efficacy is the rs6983267 single nucleotide polymorphism in 8q24. A study has shown an association of aspirin with lower colorectal cancer risk only in individuals carrying the minor T allele (rs6983267), and a specificity of this association for the CTNNB1 (beta-catenin)-positive WNT signaling activated tumor subtype.67 Together with the gene-environment interaction analysis,93 integrated analyses of drugs, germline genetics, and somatic molecular alterations will further shed lights on inter-individual differences in drug effects.
As exemplified by pharmaco-MPE research on aspirin, integration of existing data on medications, molecular pathology, and clinical outcomes can help discover a new biomarker-drug combination to predict drug efficacy in specific populations stratified by the biomarker. Pharmaco-MPE can be applied for other medications including statins, metformins, and bisphosphonates. For example, statins, 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase inhibitors, can suppress the RAS protein and the Rho family of proteins,94 which are strongly implicated in carcinogenesis. Therefore, pharmaco-MPE studies on the association of statin use with colorectal cancer incidence and survival according to KRAS mutation status have been reasonably conducted.69,95 Because molecular pathology tests have become part of routine clinical practice to guide decision making, molecular pathology data will rapidly accumulate among various populations, and can be utilized in pharmaco-MPE research.41 Findings from pharmaco-MPE research can guide not only drug repurposing96 for efficient utilization of common medications, but also tailored drug abstention or dosing adjustment to avoid potential side effects that can be predicted from individual’s characteristics.
Study designs
Better understanding of study designs, including their strengths and weaknesses, is a prerequisite for rigor and reproducibility in science. There are four main study designs in pharmaco-MPE research (Fig. 5). Characteristics of the four designs are compared in Table 1. An observational study on a subject sample drawn from one or more institutions is herein referred to as the "observational hospital-based design" (Fig. 5a). Compared to the three other designs, internal, and external validities (i.e., generalizability) of this study design's findings may be relatively limited.
Fig. 5.
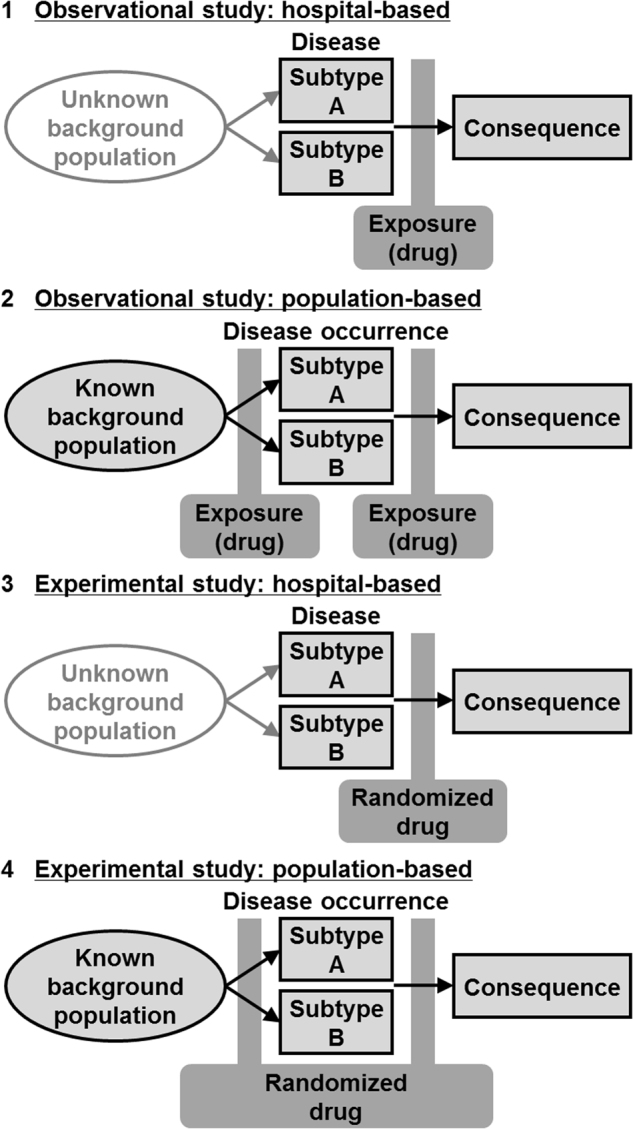
Study Designs in pharmaco-molecular pathological epidemiology (MPE) research. Arrows indicate time sequence. Here, the disease of interest is sub-classified based on pathogenic signatures into binary subtypes A and B for simplicity. Note that multiple subtypes can be evaluated in pharmaco-MPE research. Analyses can be conducted to assess effects of a drug of interest on the occurrence and/or consequential event (such as death) of a specific disease subtype. In the MPE research framework, a difference in the associations between disease subtypes is assessed. Panels indicate specific designs (with the corresponding column number in Table 1) as follows: 1, observational hospital-based design; 2, observational population-based design; 3, experimental hospital-based trial; and 4, experimental population-based trial
Table 1.
Comparisons of study designs in pharmaco-molecular pathological epidemiology (MPE)
| Study design (panel number in Fig. 5) | Observational study: hospital-baseda (1) | Observational study: population-baseda (2) | Experimental trial study: hospital-basedb (3) | Experimental trial study: population-basedb (4) |
|---|---|---|---|---|
| Typical study base population | Patients with a certain disease in question from a single to several hospitals (or institutions) | General population (broadly selected population) | Patients with a certain disease in question from a single to several hospitals (or institutions) | General population. Study population may be a highly selected group of individuals, to conduct a trial study |
| Typical study outcome | Consequence of the disease | Disease occurrence or consequence in a prospective cohort design. Disease prevalence in a case-control design. A prospective design enables assessment of occurrences of a number of diseases | Consequence of the disease | Disease occurrence or consequence. This design enables assessment of occurrences of a number of diseases |
| Use of a certain hospital or healthcare system in selection of study subjects | Yes | Usually no (but can be yes) | Yes | Usually yes, to conduct a trial study |
| Issues to consider for potential sources of sample selection bias | • The source population which has given rise to cases is unaccounted and inexplicable | • The source population which will give (or has given) rise to cases can be characterized | • The source population which has given rise to cases is unaccounted and inexplicable | • The source population which will give (or has given) rise to cases can be characterized |
| • Selection bias may arise based on study inclusion criteria, healthcare coverage, geographic restriction, initial recruitment rate, and/or stability of follow-up | • Selection bias may arise based on study inclusion criteria, initial recruitment rate, and/ or stability of follow-up | • Selection bias may arise based on study inclusion criteria, incentives to enroll (or not to enroll), healthcare coverage, geographic restriction, initial recruitment rate, and/or stability of follow-up | • Selection bias may arise based on study inclusion criteria, incentives to enroll (or not to enroll), initial recruitment rate, and/or stability of follow-up | |
| Sample size | Usually small; can be large | Usually large | Usually small; can be large | Usually large |
| Molecular pathologic analyses of cases | Maybe not difficult in a study with one to a few hospitals that have protocols in place for collection of molecular pathology data | Usually difficult because cases are typically seen in many different hospitals | Maybe not difficult in a study with one to a few hospitals that have protocols in place for collection of molecular pathology data | Usually difficult because cases are typically seen in many different hospitals |
| Medication data | Available through hospital records and/or recall of participants | Available through hospital records, and/or recall of participants. | Randomized medication data are always available. Other medication data are usually available through hospital records and/or recall of participants. | Randomized medication data are always available. Other medication data are usually available through hospital records and/or recall of participants. |
| Typical cost to establish a base study cohort and perform a pharmaco-MPE study | Least expensive | Very expensive (less expensive in a case-control design than a prospective cohort design) | Expensive | Most expensive |
| Issues that may affect internal and external validities | • Internal validity may be limited by residual and unmeasured confounding | • Internal validity may be limited by residual and unmeasured confounding, and/or recall bias | • External validity may be limited by subject selection bias, and/or small sample size | • External validity may be limited by subject selection bias |
| • External validity may be limited by subject selection bias, and/ or small sample size | • External validity may be limited by subject selection bias. | • Typically, higher generalizability compared to the other designs | ||
| • Typically, lower generalizability compared to the other designs |
a The distinction between these two observational designs can be ambiguous
b The distinction between these two experimental trial designs can be ambiguous
A second study design, herein referred to as "observational population-based design" (Fig. 5b), is intended to analyze a sample of individuals based on large populations (but not on a small number of institutions), so that findings will be likely generalizable. The sample sizes of these prospective cohort studies are considerably large, to have a sufficient number of occurrences of the disease of interest. As an alternative to the prospective design, investigators can design a case-control study where they intend to sample representative cases and controls from the same background population.
A third design, an experimental trial design using patients in one or more institutions (Fig. 5c), has the main purpose of removing confounding in the assessment of drug effects with randomized assignment to the drug. Occasionally, dozens of institutions can participate and provide cases to increase the sample size; examples are clinical trials of cooperative groups such as the Alliance in Clinical Trials in Oncology, Pan-European Trials in Alimentary Tract Cancers (PETACC), and the National Surgical Adjuvant Breast and Bowel Project (NSABP). Because of the challenge in securing a large sample in a trial setting, most ongoing trials have sample sizes sufficient to draw reasonable conclusions for the main aim of determining drug effects in the entire trial sample. As molecular subtyping classifies patients into smaller subgroups, statistical inference may be an important challenge in pharmaco-MPE research.
Rarely, an experimental trial is performed on a large sample of individuals who have not had the disease condition in question (Fig. 5d). Its main purpose is to analyze a large subject sample that is not based on a small number of institutions, at the same time removing confounding in the assessment of drug effects. Hence, the generalizability of findings is considered higher than the other designs. A substantial challenge is its cost and labor-intensiveness.
Challenges and opportunities in pharmaco-MPE
As a subfield of epidemiology, pharmaco-MPE shares many strengths and pitfalls with epidemiology. In addition, the current field of MPE has its own pitfalls, as described above. Major challenges include the paucity of transdisciplinary experts and education programs. One purpose of this article is to promote awareness of new opportunities in this emerging field of pharmaco-MPE, as more investigators with appropriate cross-disciplinary training are needed.
New opportunities in MPE are emerging in the context of a synergistic integration of pharmacoepidemiology and MPE methods, as each subfield has its own unique strengths (Fig. 4). The field of MPE can provide molecular pathologic insights into drug surveillance and safety research where traditional pharmacoepidemiology has a considerable strength. MPE methods also enable investigators to evaluate the effect of a drug in specific disease subtypes, which can improve causal inference. On the other hand, pharmacoepidemiology, which has been developed in close relationships with industries and regulatory agencies (e.g., the U.S. Food and Drug Administration, FDA), can provide not only methods to examine utilization of drugs in populations, but also provide opportunities to extend findings to improve public health policies. Implementation science97 has been one of major focuses of pharmacoepidemiology. Hence, the strengths of pharmacoepidemiology and those of MPE are not only complementary but also synergistic in the pharmaco-MPE approach. Once a population-based pharmaco-MPE database is established, a multi-level approach can be possible to combine with germline genetics and other blood biomarkers. Figure 6 illustrates a roadmap of integrative pharmaco-MPE research to conduct rigorous studies, realize precision medicine, and exert clinical impact.
Fig. 6.
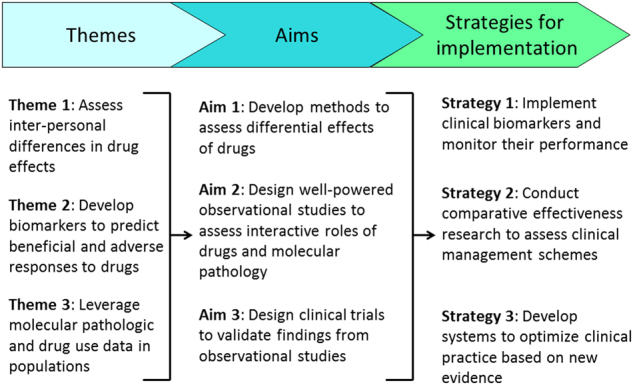
Roadmap for implementing molecular pathologic biomarkers for precision medicine. Three themes are set to launch integrated pharmaco-molecular pathological epidemiology (MPE) research and achieve three specific aims. Based on data obtained by research for the specific aims, Strategies 1 through 3 will help implement, monitor, and optimize tumor biomarker testing for clinical impact
In addition, together with diet and nutrients,98 drugs can be used to modulate the microbiota (and the relationship of host immunity to the microbiota), while the microbiota can also modulate drug effects.99 Regular use of antibiotics has been associated with a higher risk of colorectal adenoma likely due to pertubation of the gut microbiota.100 Investigations into the complex inter-relationship between drugs, the microbiota, immunity, and disease outcomes are important, because changes in the microbiota and immunity can cause not only benefits but also harms to individuals.
Immunotherapeutic strategies have been revolutionizing oncology practice. Recent evidence indicates that mismatch repair (MMR) deficiency, manifested as the microsatellite instability (MSI)-high phenotype, can predict response to immune checkpoint blockade targeting the CD274 (PD-L1)/PDCD1 (PD-1) pathway in a wide variety of cancer types including metastatic gastrointestinal cancers.101 Hence, the FDA has approved use of the anti-PDCD1 (PD-1) antibody pembrolizumab for the treatment of mismatch repair-deficient or MSI-high tumors irrespective of primary organ site (https://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm560040.htm; last visited on August 3, 2017). This is the first FDA approval of a drug use based on MMR or MSI test results, attesting to the importance of tumor molecular characteristics in determining response to therapy. There has been ample evidence indicating the important influence of diet and the environment on tumor-immune interactions.98 To further optimize treatment strategies and improve clinical outcomes, potential modifying effects of diet, lifestyle, microbial, and environmental factors on immunotherapy should be studied, as new data could inform patient management.
Conclusion
Given the importance of drugs not only for patient care and public health but also for pharmaceutical industries and the economy at large, the fields of pharmacoepidemiology and pharmaco-MPE can be expected to grow. The process of integrating pharmaco-MPE has been somewhat slow due to disciplinary barriers and the complex nature of the molecular pathology of disease. Nonetheless, research efforts integrating pharmacoepidemiology and MPE are underway. To further advance the field, additional efforts are needed to establish integrated education and training systems. Funding mechanisms must be adapted to ensure fair peer-review for trans- and inter-disciplinary science. Although transdisciplinary research has the potential to make a stronger impact,102 its funding success has been lower than traditional scientific approaches.103 We anticipate that the value of the integrative science of pharmaco-MPE will become increasingly recognized in the age of precision medicine, as improved understanding of drugs within the context of molecular pathology, and vice versa, can inform the development of customized preventive and therapeutic strategies.
Use of standardized official symbols
We use HUGO (Human Genome Organisation)-approved official symbols and root symbols for genes, gene products, and gene family, including AKT, CD274, CTNNB1, CYP2D6, HPGD, KRAS, MTOR, PDCD1, PIK3CA, PTGS2, TGFB1, TGFBR2, and WNT; all of which are described in detail at www.genenames.org. Symbols of genes and gene products are italicized, to differentiate from colloquial names that are used only in parenthesis with non-italicized forms. This format enables readers to familiarize the official gene symbols together with common colloquial names.
Acknowledgements
This work was supported in part by grants from the USA National Institutes of Health [R35 CA197735 (to SO), K07 CA190673 (to RN), R01 CA138962 (to JEM), R01 CA137178 (to ATC), K24 DK098311 (to ATC)], and Nodal Award (to SO) from the Dana-Farber Harvard Cancer Center.
Author contributions
Conceptualization: S.O., Funding acquisition: S.O., R.N., J.E.M., A.T.C., writing of the original draft: S.O., I.J., T.R.S., T.H., Y.C., Reviewing and editing: S.O., I.J., D.A.M., T.R.S., T.H., L.L., R.N., M.G., Y.C., J.E.M., J.A.N., A.T.C.
Competing interests
JoAnn E. Manson and colleagues at Brigham and Women’s Hospital, Harvard Medical School, are recipients of funding from the National Institutes of Health to conduct the VITamin D and OmegA-3 TriaL (VITAL), a large-scale randomized trial of vitamin D and omega-3 fatty acids in the prevention of cancer and cardiovascular disease. Andrew T. Chan was a consultant of Bayer Healthcare, andPfizer Inc. This work was not funded by Bayer Healthcare, or Pfizer Inc. The remaining authors declare no competing financial interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Yin Cao, JoAnn E. Manson, Jonathan A. Nowak and Andrew T. Chan contributed equally to this work.
Contributor Information
Shuji Ogino, Phone: +617-632-1972, Email: shuji_ogino@dfci.harvard.edu.
Andrew T. Chan, Email: achan@mgh.harvard.edu
References
- 1.Spira A, et al. Precancer atlas to drive precision prevention trials. Cancer Res. 2017;77:1510–1541. doi: 10.1158/0008-5472.CAN-16-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyskens FL, Jr., et al. Cancer prevention: obstacles, challenges and the road ahead. J. Natl Cancer Inst. 2016;108:djv309. doi: 10.1093/jnci/djv309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart BW, et al. Cancer prevention as part of precision medicine: 'plenty to be done'. Carcinogenesis. 2016;37:2–9. doi: 10.1093/carcin/bgv166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kensler TW, et al. Transforming cancer prevention through precision medicine and immune-oncology. Cancer Prev. Res. 2016;9:2–10. doi: 10.1158/1940-6207.CAPR-15-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ioannidis JP. Why most published research findings are false. PLoS Med. 2005;2:e124. doi: 10.1371/journal.pmed.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ioannidis JP. How to make more published research true. PLoS Med. 2014;11:e1001747. doi: 10.1371/journal.pmed.1001747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogino S, et al. Molecular pathological epidemiology of epigenetics: Emerging integrative science to analyze environment, host, and disease. Mod. Pathol. 2013;26:465–484. doi: 10.1038/modpathol.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishi A, et al. Integration of molecular pathology, epidemiology, and social science for global precision medicine. Expert Rev. Mol. Diagn. 2016;16:11–23. doi: 10.1586/14737159.2016.1115346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogino S, et al. Interdisciplinary education to integrate pathology and epidemiology: towards molecular and population-level health science. Am. J. Epidemiol. 2012;176:659–667. doi: 10.1093/aje/kws226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogino S, Stampfer M. Lifestyle factors and microsatellite instability in colorectal cancer: The evolving field of molecular pathological epidemiology. J .Natl Cancer. Inst. 2010;102:365–367. doi: 10.1093/jnci/djq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60:397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ogino S, et al. The role of molecular pathological epidemiology in the study of neoplastic and non-neoplastic diseases in the era of precision medicine. Epidemiology. 2016;27:602–611. doi: 10.1097/EDE.0000000000000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.VanderWeele TJ, Knol MJ. Interactions and complexity: goals and limitations. Epidemiol. Meth. 2014;3:79–81. [Google Scholar]
- 14.Begg CB, et al. Identifying etiologically distinct sub-types of cancer: a demonstration project involving breast cancer. Cancer Med. 2015;4:1432–1439. doi: 10.1002/cam4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang M, et al. Statistical methods for studying disease subtype heterogeneity. Stat. Med. 2016;35:782–800. doi: 10.1002/sim.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Kuchiba A, Ogino S. A meta-regression method for studying etiologic heterogeneity across disease subtypes classified by multiple biomarkers. Am. J. Epidemiol. 2015;182:263–270. doi: 10.1093/aje/kwv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richiardi L, Barone-Adesi F, Pearce N. Cancer subtypes in aetiological research. Eur. J. Epidemiol. 2017;32:353–361. doi: 10.1007/s10654-017-0253-z. [DOI] [PubMed] [Google Scholar]
- 18.Zabor, E. C. & Begg, C. B. A comparison of statistical methods for the study of etiologic heterogeneity. Stat. Med. 36, 4050–4060 (2017). [DOI] [PMC free article] [PubMed]
- 19.Rescigno T, Micolucci L, Tecce MF, Capasso A. Bioactive nutrients and nutrigenomics in age-related diseases. Molecules. 2017;22:105. doi: 10.3390/molecules22010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Pathol. Res. Int. 2011;2011:902674. doi: 10.4061/2011/902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bishehsari F, Mahdavinia M, Vacca M, Malekzadeh R, Mariani-Costantini R. Epidemiological transition of colorectal cancer in developing countries: Environmental factors, molecular pathways, and opportunities for prevention. World J. Gastroenterol. 2014;20:6055–6072. doi: 10.3748/wjg.v20.i20.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang M, Dai J, Gu D, Huang Q, Tian L. Aspirin in pancreatic cancer: chemopreventive effects and therapeutic potentials. BBA Rev. Cancer. 2017;1866:163–176. doi: 10.1016/j.bbcan.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Useros J, Garcia-Foncillas J. Obesity and colorectal cancer: molecular features of adipose tissue. J. Transl. Med. 2016;14:21. doi: 10.1186/s12967-016-0772-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campbell PT, et al. Association between body mass index and mortality for colorectal cancer survivors: overall and by tumor molecular phenotype. Cancer Epidemiol. Biomarkers Prev. 2015;24:1229–1238. doi: 10.1158/1055-9965.EPI-15-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Serafino A, et al. Developing drugs that target the Wnt pathway: recent approaches in cancer and neurodegenerative diseases. Expert Opin. Drug Discov. 2017;12:169–186. doi: 10.1080/17460441.2017.1271321. [DOI] [PubMed] [Google Scholar]
- 26.Patil H, et al. Chasing the personalized medicine dream through biomarker validation in colorectal cancer. Drug Discov. Today. 2017;22:111–119. doi: 10.1016/j.drudis.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 27.Kuroiwa-Trzmielina J, et al. SNP rs16906252C>T is an expression and methylation quantitative trait locus associated with an increased risk of developing MGMT-methylated colorectal cancer. Clin. Cancer Res. 2016;22:6266–6277. doi: 10.1158/1078-0432.CCR-15-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slattery ML, et al. Infrequently expressed miRNAs in colorectal cancer tissue and tumor molecular phenotype. Mod. Pathol. 2017;30:1152–1169. doi: 10.1038/modpathol.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alnabulsi, A. & Murray, G. I. Integrative analysis of the colorectal cancer proteome: potential clinical impact. Expert Rev. Proteomics (2017). [DOI] [PubMed]
- 30.Kuller LH, Bracken MB, Ogino S, Prentice RL, Tracy RP. The role of epidemiology in the era of molecular epidemiology and genomics: Summary of the 2013 AJE-sponsored society of epidemiologic research symposium. Am. J. Epidemiol. 2013;178:1350–1354. doi: 10.1093/aje/kwt239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Epplein M, et al. Challenges and opportunities in international molecular cancer prevention research: An ASPO molecular epidemiology and the environment and international cancer prevention interest groups report. Cancer Epidemiol. Biomarkers Prev. 2014;23:2613–2617. doi: 10.1158/1055-9965.EPI-14-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogino S, et al. Proceedings of the second international molecular pathological epidemiology (MPE) meeting. Cancer Causes Control. 2015;26:959–972. doi: 10.1007/s10552-015-0596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Campbell PT, et al. Proceedings of the third international molecular pathological epidemiology (MPE) meeting. Cancer Causes Control. 2017;28:167–176. doi: 10.1007/s10552-016-0845-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan AT, Ogino S, Fuchs CS. Aspirin and the risk of colorectal cancer in relation to the expression of COX-2. N. Engl J. Med. 2007;356:2131–2142. doi: 10.1056/NEJMoa067208. [DOI] [PubMed] [Google Scholar]
- 35.Nishihara R, et al. Aspirin use and risk of colorectal cancer according to BRAF mutation status. JAMA. 2013;309:2563–2571. doi: 10.1001/jama.2013.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao Y, et al. Regular aspirin use associates with lower risk of colorectal cancers with low numbers of tumor-infiltrating lymphocytes. Gastroenterology. 2016;151:879–892. doi: 10.1053/j.gastro.2016.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamada T, et al. Aspirin use and colorectal cancer survival according to tumor CD274 (PD-L1) expression status. J. Clin. Oncol. 2017;35:1836–1844. doi: 10.1200/JCO.2016.70.7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamada T, Keum N, Nishihara R, Ogino S. Molecular pathological epidemiology: new developing frontiers of big data science to study etiologies and pathogenesis. J. Gastroenterol. 2017;52:265–275. doi: 10.1007/s00535-016-1272-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strom, B. L. What is Pharmacoepidemiology? in Textbook of Pharmacoepidemiology (eds Strom, B.L., Kimmel, S.E. & Hennessy, S.) (John Wiley & Sons Ltd, 2013).
- 40.Collins FS, Varmus H. A new initiative on precision medicine. N. Engl J. Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ogino S, et al. Discovery of colorectal cancer PIK3CA mutation as potential predictive biomarker: power and promise of molecular pathological epidemiology. Oncogene. 2014;33:2949–2955. doi: 10.1038/onc.2013.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colussi D, Brandi G, Bazzoli F, Ricciardiello L. Molecular pathways involved in colorectal cancer: implications for disease behavior and prevention. Int. J. Mol. Sci. 2013;14:16365–16385. doi: 10.3390/ijms140816365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kocarnik JM, Shiovitz S, Phipps AI. Molecular phenotypes of colorectal cancer and potential clinical applications. Gastroenterol. Rep. 2015;3:269–276. doi: 10.1093/gastro/gov046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kudryavtseva AV, et al. Important molecular genetic markers of colorectal cancer. Oncotarget. 2016;7:53959–53983. doi: 10.18632/oncotarget.9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dienstmann R, Salazar R, Tabernero J. Personalizing colon cancer adjuvant therapy: selecting optimal treatments for individual patients. J. Clin. Oncol. 2015;33:1787–1796. doi: 10.1200/JCO.2014.60.0213. [DOI] [PubMed] [Google Scholar]
- 46.Dienstmann R, et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer. 2017;17:79–92. doi: 10.1038/nrc.2016.126. [DOI] [PubMed] [Google Scholar]
- 47.Phipps AI, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology. 2015;148:77–87.e72. doi: 10.1053/j.gastro.2014.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nosho K, et al. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer and prognosis: cohort study and literature review. J. Pathol. 2010;222:350–366. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Basile D, et al. Immunotherapy for colorectal cancer: where are we heading? Expert Opin. Biol. Ther. 2017;17:709–721. doi: 10.1080/14712598.2017.1315405. [DOI] [PubMed] [Google Scholar]
- 50.Galon J, et al. Towards the introduction of the Immunoscore in the classification of malignant tumors. J. Pathol. 2014;232:199–209. doi: 10.1002/path.4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Di Caro G, Marchesi F, Laghi L, Grizzi F. Immune cells: plastic players along colorectal cancer progression. J. Cell Mol. Med. 2013;17:1088–1095. doi: 10.1111/jcmm.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mlecnik B, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44:698–711. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 53.Rozek LS, et al. Tumor-Infiltrating lymphocytes, Crohn's-Like lymphoid reaction, and survival from colorectal cancer. J. Natl Cancer Inst. 2016;108:djw027. doi: 10.1093/jnci/djw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yamauchi M, et al. Assessment of colorectal cancer molecular features along bowel subsites challenges the conception of distinct dichotomy of proximal versus distal colorectum. Gut. 2012;61:847–854. doi: 10.1136/gutjnl-2011-300865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamauchi M, et al. Colorectal cancer: a tale of two sides or a continuum? Gut. 2012;61:794–797. doi: 10.1136/gutjnl-2012-302014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosty C, et al. PIK3CA activating mutation in colorectal carcinoma: Associations with molecular features and survival. PLoS One. 2013;8:e65479. doi: 10.1371/journal.pone.0065479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bae JM, Kim JH, Cho NY, Kim TY, Kang GH. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br. J. Cancer. 2013;109:1004–1012. doi: 10.1038/bjc.2013.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mima K, et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin. Transl. Gastroenterol. 2016;7:e200. doi: 10.1038/ctg.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Drew DA, Cao Y, Chan AT. Aspirin and colorectal cancer: the promise of precision chemoprevention. Nat. Rev. Cancer. 2016;16:173–186. doi: 10.1038/nrc.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li P, et al. Aspirin use after diagnosis but not prediagnosis improves established colorectal cancer survival: a meta-analysis. Gut. 2015;64:1419–1425. doi: 10.1136/gutjnl-2014-308260. [DOI] [PubMed] [Google Scholar]
- 61.Herbert K, Kerr R, Kerr DJ, Church DN. Are NSAIDs coming back to colorectal cancer therapy or not? Curr. Colorectal. Cancer Rep. 2014;10:363–371. doi: 10.1007/s11888-014-0247-0. [DOI] [Google Scholar]
- 62.Umar A, Steele VE, Menter D, Hawk ET. Mechanisms of non-steroidal anti-inflammatory drugs (NSAIDs) in cancer prevention. Semin. Oncol. 2016;43:67–77. doi: 10.1053/j.seminoncol.2015.09.010. [DOI] [PubMed] [Google Scholar]
- 63.Reimers MS, Zeestraten EC, Kuppen PJ, Liefers GJ, van de Velde JH. Biomarkers in precision therapy in colorectal cancer. Gastroenterol. Rep. 2013;1:166–183. doi: 10.1093/gastro/got022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao Y, et al. Population-wide impact of long-term use of aspirin and the risk for cancer. JAMA Oncol. 2016;2:762–769. doi: 10.1001/jamaoncol.2015.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chia WK, Ali R, Toh HC. Aspirin as adjuvant therapy for colorectal cancer-reinterpreting paradigms. Nat. Rev. Clin. Oncol. 2012;9:561–570. doi: 10.1038/nrclinonc.2012.137. [DOI] [PubMed] [Google Scholar]
- 66.Tougeron D, Sha D, Manthravadi S, Sinicrope FA. Aspirin and colorectal cancer: Back to the future. Clin Cancer Res. 2014;20:1087–1094. doi: 10.1158/1078-0432.CCR-13-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nan H, et al. Aspirin use, 8q24 single nucleotide polymorphism rs6983267, and colorectal cancer according to CTNNB1 alterations. J. Natl Cancer. Inst. 2013;105:1852–1861. doi: 10.1093/jnci/djt331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan AT, Ogino S, Fuchs CS. Aspirin use and survival after diagnosis of colorectal cancer. JAMA. 2009;302:649–658. doi: 10.1001/jama.2009.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee JE, et al. Statin use and colorectal cancer risk according to molecular subtypes in two large prospective cohort studies. Cancer Prev. Res. 2011;4:1808–1815. doi: 10.1158/1940-6207.CAPR-11-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khalili H, Huang ES, Ogino S, Fuchs CS, Chan AT. A prospective study of bisphosphonate use and risk of colorectal cancer. J. Clin. Oncol. 2012;30:3229–3233. doi: 10.1200/JCO.2011.39.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liao X, et al. Aspirin use, tumor PIK3CA mutation status, and colorectal cancer survival. N. Engl. J. Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fink SP, et al. Aspirin and the risk of colorectal cancer in relation to the expression of 15-hydroxyprostaglandin dehydrogenase (HPGD) Sci. Transl. Med. 2014;6:233re232. doi: 10.1126/scitranslmed.3008481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mei Z, Duan C, Li C, Cui L, Ogino S. Prognostic role of tumor PIK3CA mutation in colorectal cancer: a systematic review and meta-analysis. Ann. Oncol. 2016;27:1836–1848. doi: 10.1093/annonc/mdw264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.De Roock W, Vriendt VD, Normanno N, Ciardiello F, Tejpar S. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol. 2011;12:594–603. doi: 10.1016/S1470-2045(10)70209-6. [DOI] [PubMed] [Google Scholar]
- 75.Liao X, et al. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin. Cancer Res. 2012;18:2257–2268. doi: 10.1158/1078-0432.CCR-11-2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eklof V, et al. The prognostic role of KRAS, BRAF, PIK3CA and PTEN in colorectal cancer. Br. J. Cancer. 2013;108:2153–2163. doi: 10.1038/bjc.2013.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Manceau G, et al. PIK3CA mutations predict recurrence in localized microsatellite stable colon cancer. Cancer Med. 2015;4:371–382. doi: 10.1002/cam4.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogelaar F, et al. The prognostic value of microsatellite Instability, KRAS, BRAF and PIK3CA mutations in stage II colon cancer patients. Mol. Med. 2015;21:1038–1046. doi: 10.2119/molmed.2015.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ogino S, et al. Predictive and prognostic analysis of PIK3CA mutation in stage III colon cancer intergroup trial. J. Natl Cancer Inst. 2013;105:1789–1798. doi: 10.1093/jnci/djt298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Domingo E, et al. Evaluation of PIK3CA mutation as a predictor of benefit from NSAID therapy in colorectal cancer. J. Clin. Oncol. 2013;31:4297–4305. doi: 10.1200/JCO.2013.50.0322. [DOI] [PubMed] [Google Scholar]
- 81.Reimers MS, et al. Expression of HLA Class I antigen, aspirin use, and survival after a diagnosis of colon cancer. JAMA Intern. Med. 2014;174:732–739. doi: 10.1001/jamainternmed.2014.511. [DOI] [PubMed] [Google Scholar]
- 82.Kothari N, et al. Impact of regular aspirin use on overall and cancer-specific survival in patients with colorectal cancer harboring a PIK3CA mutation. Acta Oncol. 2015;54:487–492. doi: 10.3109/0284186X.2014.990158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gray RT, et al. Evaluation of PTGS2 expression, PIK3CA mutation, aspirin use and colon cancer survival in a population-based cohort study. Clin. Transl. Gastroenterol. 2017;8:e91. doi: 10.1038/ctg.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paleari L, et al. PIK3CA mutation, aspirin use after diagnosis and survival of colorectal cancer. A systematic review and meta-analysis of epidemiological studies. Clin. Oncol. 2016;28:317–326. doi: 10.1016/j.clon.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 85.Zumwalt TJ, et al. Aspirin-induced chemoprevention and response kinetics are enhanced by PIK3CA mutations in colorectal cancer cells. Cancer Prev. Res. 2017;10:208–218. doi: 10.1158/1940-6207.CAPR-16-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Turturro SB, Najor MS, Ruby CE, Cobleigh MA, Abukhdeir AM. Mutations in PIK3CA sensitize breast cancer cells to physiologic levels of aspirin. Breast Cancer Res. Treat. 2016;156:33–43. doi: 10.1007/s10549-016-3729-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ali R, Toh HC, Chia WK. The utility of aspirin in Dukes C and high risk Dukes B colorectal cancer--the ASCOLT study: study protocol for a randomized controlled trial. Trials. 2011;12:261. doi: 10.1186/1745-6215-12-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jung B, Staudacher JJ, Beauchamp D. Transforming growth factor beta superfamily signaling in development of colorectal cancer. Gastroenterology. 2017;152:36–52. doi: 10.1053/j.gastro.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan M, et al. 15-Hydroxyprostaglandin dehydrogenase, a COX-2 oncogene antagonist, is a TGF-beta-induced suppressor of human gastrointestinal cancers. Proc. Natl Acad. Sci. USA. 2004;101:17468–17473. doi: 10.1073/pnas.0406142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Myung SJ, et al. 15-Hydroxyprostaglandin dehydrogenase is an in vivo suppressor of colon tumorigenesis. Proc. Natl Acad. Sci. USA. 2006;103:12098–12102. doi: 10.1073/pnas.0603235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Roy HK, et al. Spectral biomarkers for chemoprevention of colonic neoplasia: a placebo-controlled double-blinded trial with aspirin. Gut. 2017;66:285–292. doi: 10.1136/gutjnl-2015-309996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.O'Connor JP, et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 2017;14:169–186. doi: 10.1038/nrclinonc.2016.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nan H, et al. Association of aspirin and NSAID use with risk of colorectal cancer according to genetic variants. JAMA. 2015;313:1133–1142. doi: 10.1001/jama.2015.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gronich N, Rennert G. Beyond aspirin-cancer prevention with statins, metformin and bisphosphonates. Nat. Rev. Clin. Oncol. 2013;10:625–642. doi: 10.1038/nrclinonc.2013.169. [DOI] [PubMed] [Google Scholar]
- 95.Ng K, et al. Relationship between statin use and colon cancer recurrence and survival: results from CALGB 89803. J. Natl Cancer Inst. 2011;103:1540–1551. doi: 10.1093/jnci/djr307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bertolini F, Sukhatme VP, Bouche G. Drug repurposing in oncology--patient and health systems opportunities. Nat. Rev. Clin. Oncol. 2015;12:732–742. doi: 10.1038/nrclinonc.2015.169. [DOI] [PubMed] [Google Scholar]
- 97.Emmons KM, Colditz GA. Realizing the potential of cancer prevention-the role of implementation science. N. Engl J. Med. 2017;376:986–990. doi: 10.1056/NEJMsb1609101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zitvogel L, Pietrocola F, Kroemer G. Nutrition, inflammation and cancer. Nat. Immunol. 2017;18:843–850. doi: 10.1038/ni.3754. [DOI] [PubMed] [Google Scholar]
- 99.Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 2016;14:273–287. doi: 10.1038/nrmicro.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cao, Y. et al. Long-term use of antibiotics and risk of colorectal adenoma. Gut (2018). [DOI] [PMC free article] [PubMed]
- 101.Le DT, et al. Mismatch-repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wuchty S, Jones BF, Uzzi B. The increasing dominance of teams in production of knowledge. Science. 2007;316:1036–1039. doi: 10.1126/science.1136099. [DOI] [PubMed] [Google Scholar]
- 103.Bromham L, Dinnage R, Hua X. Interdisciplinary research has consistently lower funding success. Nature. 2016;534:684–687. doi: 10.1038/nature18315. [DOI] [PubMed] [Google Scholar]


