Abstract
Gas cluster ion beam-secondary ion mass spectrometry (GCIB-SIMS) has shown the full potential of mapping intact lipids in biological systems with better than 10 μm lateral resolution. This study investigated further the capability of GCIB-SIMS in imaging high-mass signals from intact cardiolipin (CL) and gangliosides in normal brain and the effect of a controlled cortical impact model (CCI) of traumatic brain injury (TBI) on their distribution. A combination of enzymatic and chemical treatments was employed to suppress the signals from the most abundant phospholipids (phosphatidylcholine (PC) and phosphatidylethanolamine (PE)) and enhance the signals from the low-abundance CLs and gangliosides to allow their GCIB-SIMS detection at 8 and 16 μm spatial resolution. Brain CLs have not been observed previously using other contemporary imaging mass spectrometry techniques at better than 50 μm spatial resolution. High-resolution images of naive and injured brain tissue facilitated the comparison of CL species across three multicell layers in the CA1, CA3, and DG regions of the hippocampus. GCIB-SIMS also reliably mapped losses of oxidizable polyunsaturated CL species (but not the oxidation-resistant saturated and monounsaturated gangliosides) to regions including the CA1 and CA3 of the hippocampus after CCI. This work extends the detection range for SIMS measurements of intact lipids to above m/z 2000, bridging the mass range gap compared with MALDI. Further advances in high-resolution SIMS of CLs, with the potential for single cell or supra-cellular imaging, will be essential for the understanding of CL’s functional and structural organization in normal and injured brain.
Graphical abstract
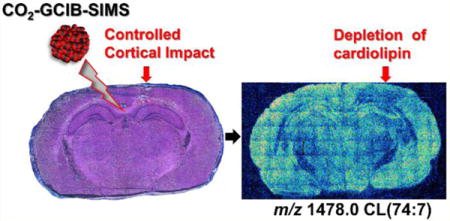
Cardiolipins (CLs) represent an important class of mitochondria-specific anionic phospholipids, which exhibit multiple structural and functional roles related to mitochondrial signaling, bioenergetics, and cellular death pathways.1–6 Overall, CLs exist in cells and tissues in low abundance compared to other phospholipids. The general structure of CL includes two phosphatidylglycerols connected by a glycerol backbone. This results in four fatty acyl chains for this unique phospholipid combining both the hydrophobic bulky body and two negatively charged phosphate groups at physiological pH.1–7 CLs that contain polyunsaturated fatty acyl chains readily undergo oxygenation via chemical (non-enzymatic) and biochemical (enzymatic) mechanisms.1–8 It has been shown that the latter represent an important source of numerous mitochondria-generated lipid signals, including many of the well characterized lipid mediators.8
CL diversity ranges from a few major CL species in tissues, such as heart, skeletal muscle, and liver, to a wide diversity of CL species in brain tissue.6 The low abundance of CL, which is distributed over multiple species, has contributed to the difficulty in imaging this particular anionic lipid species by mass spectrometry (MS), especially in brain tissue. While contemporary liquid chromatography–mass spectrometry (LC–MS) does allow us to identify, characterize, and quantify individual CLs, these types of methods are devoid of any tissue-specific spatial information. We have recently developed a protocol combining enzymatic and chemical modification of the samples aimed at the suppression of more abundant non-CL signals. We have successfully applied this protocol to image CL by MALDI mass spectrometry in naive brain tissue and studies involving traumatic brain injury in a controlled cortical impact (CCI) model.9,10 CL imaging of brain tissue has also been achieved by other groups using DESI-mass spectrometry imaging/liquid sample microjunction DESI-imaging11 as well as in other tissues using a silver nanoparticle technique.12 While the above CL imaging studies have allowed novel findings with regard to CL’s region-specific distribution in the brain and other tissues as well as changes in CL in response to traumatic brain injury (TBI), they are limited with regard to spatial resolution.
TOF-SIMS imaging has a lateral resolution which is unattainable (micrometer to sub-100 nm; 8 μm for CL in this article) with the other imaging mass spectrometry (IMS) techniques (e.g., MALDI, DESI, 50–630 μm for CL), making it possible to reveal the spatial distribution of CL on micrometer and submicrometer scales. This is important to further our understanding of the biological functionality of this low-abundance and highly diversified lipid species in brain tissue.13 However, the destructive nature of the energetic primary ion beam system can still pose problems for biological materials. The widely adapted atomic ion beams (e.g., Cs+, Bi+, Au+)14,15 only attain atomic ions or fragments with sufficient intensity for imaging but not molecular ions. Cluster ion beams (e.g., Bi3+, C60+, Aun+ (n=3−400))16,17 which have been developed to enhance the molecular ion information, can reduce the damage to some extent but still have very limited mass detection ranges, usually less than m/z 1000. There have been very few reports of the detection of CL utilizing these primary ion beams. Jerigova et al.18 detected fragments of CL in cardiac cells and salt adducts using Au3+ but was not able to chemically map CL molecular ions.
The recent development of gas cluster ion beams (GCIB), e.g., Arn+ (n=~10000),19 have provided new opportunities to probe higher mass species in biological samples since less chemical damage and less fragmentation is observed.20 The Ar-GCIB has been widely used as a sputtering source (i.e., nonanalytical surface shaving)21,22 due to the difficulties in focusing and generating sufficiently short pulses for analytical work with TOF instruments. The J105 3D Chemical Imager employs a continuous primary ion beam with a hybrid buncher-TOF, allowing high-quality mass spectra without beam pulsing.23 With this unique setup, oligopeptides have been studied by Ar1000+, and their molecular ions were observed with less fragmentation.24 Polymers with m/z above 1000 have been detected as intact protonated/deprotonated ions.25 The Ar-GCIB has also been successfully used for imaging of biological tissue with satisfactory lateral resolution.26 The advances in this technique have made lateral resolutions of a few micrometers possible for imaging a metal grid27 as well as biological samples.28 In addition, high mass species, such as phosphatidylcholine (PC) lipid dimers of approximately m/z 1545, were detected. The variation of the feeding gas has also improved the beam focus without the requirement of technical modification of the beam system. Resolutions of approximately 5 μm have been achieved when imaging ruberene coated metal grids with a 20 keV CO2-GCIB system.29 With the above advances, higher-mass lipid molecular ions such as CLs and ganglioside species may become excellent targets for GCIB-TOF-SIMS analyses.
The importance of CL with its multiple signaling and structural roles, especially in brain tissue, necessitates the development and application of high spatial resolution MSI protocols. However, the higher molecular masses (mostly m/z 1400–1550 in mammalian brain), high diversification, and low abundance of individual CL species limit successful imaging using TOF-SIMS. In this study, we developed new protocols that employ CO2-GCIB-TOF-SIMS imaging coupled with 1-ethyl-3-[3-(dimethylamino)propyl] carbodiimide hydrochloride and phospholipase C (EDC/PLC) tissue treatment to reveal CL species in naive and controlled cortical injured brain sections. The advantages of this approach over MALDI-TOF MSI are a higher lateral resolution (∼8 μm and ∼16 μm) combined with the relative simplicity of sample preparation. Here, we report on the feasibility of detection of diversified CL species in naive rat brain sections, including polyunsaturated fatty acid CL (PUFA-CL) species that are highly vulnerable to oxidative modification. In addition, we report that ganglioside molecular species are readily detectable under the same conditions utilized for CL imaging. We have also evaluated CL at 3 h after CCI resulting in the loss of this critical phospholipid in specific brain regions.
MATERIALS AND METHODS
Reagents
Water for tissue treatment was purified by a Milli-Q system (EMD Millipore, Billerica, MA). 2-(4-Morpholino)ethanesulfonic acid (MES) was BioReagent grade from Fisher Scientific (Pittsburgh PA). 1-Ethyl-3-[3-(dimethylamino)propyl] carbodiimide hydrochloride (EDC) was purchased from Thermo Scientific/Pierce Biotechnology (Rockford, IL). Phospholipase C from B. cerus and ammonium acetate were purchased from Sigma-Aldrich (St. Louis, MO). Harris Hematoxylin, Eosin Y, and Bluing Reagent for hematoxylin and eosin (H&E) staining were purchased from ThermoFisher Scientific (Waltham, MA). Histology grade ethanol and xylenes were also from ThermoFisher Scientific. Chloroform, water, and ethanol for cleaning the copper substrate were HPLC grade from OmniSolv EMD Millipore (Billerica, MA).
Animals and Tissue Preparation
All procedures were preapproved and performed according to the protocols approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh. CCI to induce TBI was performed on the left parietal cortex in one 17 day old male Sprague–Dawley rat (Harlan, Indianapolis, IN) as described previously.30 A metal pneumatically driven 6 mm impactor tip was used (impact velocity 4.0 ± 0.2 m/s, penetration depth 2.5 mm). One naive 17 day old male rat was used as a control. The brains were harvested 3 h after injury and immediately frozen in liquid nitrogen with neither fixation nor embedding and stored at −80°C until sectioning. Brain tissue was affixed to a cryotome block with minimal Tissue-Tek optimal cutting temperature (OCT) compound (Sakura FineTek USA, Torrance CA) to avoid the contamination of the region of interest. The blade and working surfaces of the cryomicrotome (Leica CM1950) were cleaned with 95% ethanol immediately prior and after to cutting, and at no time did the blade come into contact with the OCT. Copper plates at 800 μm thickness were used to receive the sections for GCIB-SIMS. Before use, the plates were polished using MetPrep (MetPrep 3 grinding and polishing machine/PH, Allied High Tech Products, Inc.) and sonicated in chloroform, water, and ethanol for 5 min each, respectively. Consecutive 6 μm coronal brain sections were cut at −21°C and placed on copper plates (for GCIB-SIMS with subsequent H&E histology) and plain glass slides (for conventional histology). Sections were stored for less than 2 days at −80°C until GCIB-SIMS analysis.
For CL imaging, the tissue was prepared as previously described.9 The frozen tissue sections were brought to room temperature while under vacuum. The tissue was first treated with freshly prepared 500 mM EDC in MES buffer (100 mM, pH 3.7) for 2 h at room temperature then washed three times with ice-cold 50 mM ammonium acetate buffer, pH 6.7 (200 μL per wash). The tissue was then treated with 200 μL of PLC solution (0.80 milli-units per μL) in ammonium acetate buffer (50 mM, pH 6.7) for 15 min at 37°C. The tissue was then washed again three times with ammonium acetate buffer as described above. After the chemical/enzymatic treatments, the tissue was very hydrophobic and was able to be dried with a gentle stream of dry N2.
Cluster SIMS Imaging
The dc beam buncher-TOF SIMS instrument, J105 3D Chemical Imager (Ionoptika, U.K.) was employed to image the naive and CCI brain sections with and without EDC/PLC treatment. The instrument is equipped with a 40 keV C60 source and a 20 keV gas cluster ion source (GCIB), described in detail elsewhere.23 For this study, GCIB with CO2 gas feeding is more beneficial for high mass species (e.g., CLs, gangliosides) and high spatial resolution. Specifically, a (CO2)3500+ beam was used for the study with a current of 9 pA. The beam was focused through a set of lenses down the beam column to either 8 or 16 μm for imaging. First, the SIMS spectra were acquired in both positive and negative mode from an area of 1100 × 1100 μm2 with an ion dose of 3.8 × 1011 ions cm−2 in the middle of the hippocampus from two serial naive brain sections with and without EDC/PLC treatment. Second, imaging of an entire tissue section was performed only in negative mode on naive and CCI tissue without/with EDC/PLC treatment at 16 μm lateral resolution using a tiled imaging mode. Each tile covers an area of 1100 × 1100 μm2 with 64 × 64 pixels and 200 shots per pixel, which gives an ion dose of 3.8 × 1011 ions cm−2. A higher lateral resolution, ∼8 μm, was employed to image the hippocampus and its immediately surrounding area. Each tile is set within an area of 1100 × 1100 μm2 with 128 × 128 pixels and 200 shots per pixel, making the total ion dose up to 1.5 × 1012 ions cm−2. The mass range for the spectra and imaging was m/z 200–3000 with a resolution of m/Δm = 6000. Charge neutralization was realized by using an electron beam (IOG-10, Ionoptika, U.K.) operating at 28 eV with approximately 10 μA current.
Histology
After GCIB-SIMS analysis, the tissue section (still on its copper plate) and a section serial or semiserial to it (on a glass slide) were stained with H&E without prior fixation. Coverslips were affixed with mounting media and dried overnight. Sections were scanned and montaged using a Nikon 90i upright microscope (Melville, NY) using Nikon Elements software with 4× and 10× dry plan apochromat objectives and a motorized stage.
Data Analysis
The chemical maps of ions of interest were generated using Ionoptika Image Analyzer (version 1.0.9015, Ionoptika, U.K.) and in-house software, ImagingSIMS (version 3.6, open access: https://raw.githubusercontent.com/jay5026/ImagingSIMS/master/deploy/pub/setup.exe). The mass for each ion was centered on the exact mass with a Δm/z of 0.6 Da.
RESULTS AND DISCUSSION
GCIB-SIMS Detects CL from Brain Tissue Only after EDC/PLC Treatment
Low relative abundance vs other phospholipids in cells preclude direct detection of CLs in biological tissues by TOF-SIMS. As shown in Figure 1b, negative ion mode GCIB-SIMS did not reveal any deprotonated CL molecular ions within the mass range of m/z 1300–1530 from the hippocampus of a naive rat brain tissue section. In contrast, after EDC/PLC treatment which partially removes highly abundant PC and aminophospholipids (e.g., PE), a diverse array of CL species was clearly seen from the same region of the hippocampus (Figure 1e). The prominent CL clusters contain 72 and 74 acyl carbons with 5–11 unsaturated carbon double bonds (CL 72:X and 74:X). The slightly less intense cluster is CL (70:X), followed by CL (68:X) and (76:X). Minor CL clusters include CL (66:X) and (78:X). A total of 34 identified CL species with reasonable intensities for imaging are listed in Table 1. This enhancement of CL signals from EDC/PLC tissue treatment is consistent with our previous results with MALDI imaging.9,10 Recently, Angerer et al. have shown that frozen hydrated tissue and dried tissue exposed to various reactive vapors aids in the analysis of brain lipids by TOF-SIMS. Enhanced high-mass species were observed, but no CL species were reported in that study.31,32
Figure 1.
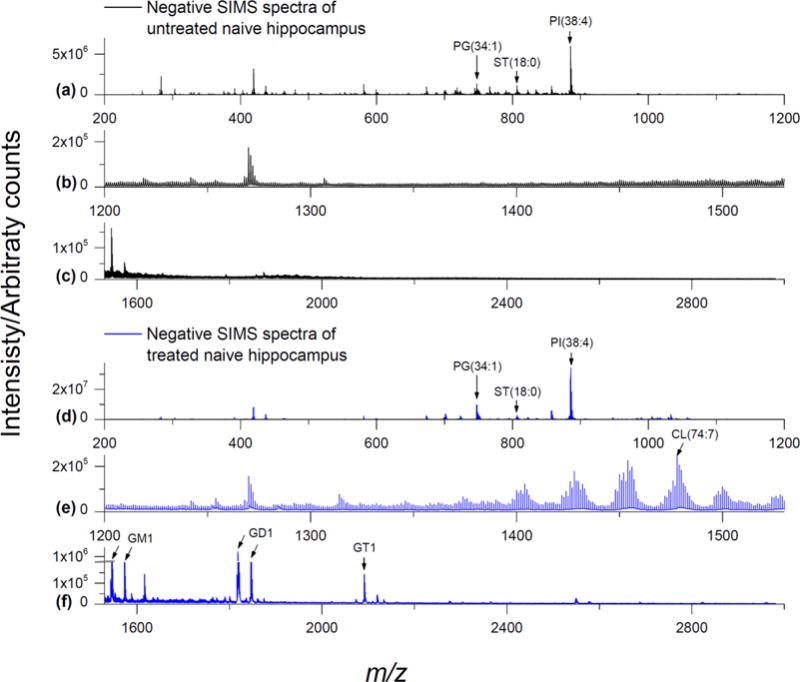
GCIB-SIMS negative mode spectra of naive rat brain tissue: SIMS negative ion mode spectra from the hippocampus of a naive rat brain coronal section (64 × 64 array, 16 μm spatial resolution). (a–c) Untreated tissue shows PI(38:4) at m/z 885.5, PG(34:1) at m/z 747.5, and several sulfatide species in the m/z 750–950 range including ST(18:0) at m/z 806.5 but very few lipid species above m/z 1000 are observed. (d–f) Tissue treated with EDC/PLC shows a remarkable increase in signals for various CLs (m/z 1370–1530) and gangliosides such as GM1, GD1, and GT1 (approximately m/z 1500–2700). See Table 1 for complete peak assignments.
Table 1.
High Mass Molecular Species in Negative Ion Modea
| observed m/z | assignment | accurate mass | Δppm |
|---|---|---|---|
| 747.4972 | PG(34:1) C40H76O10P | 747.5176 | −27.29 |
| 778.5093 | ST(16:0) C40H76SNO11 | 778.5139 | −5.91 |
| 806.5438 | ST(18:0) C42H80SNO11 | 806.5451 | −1.61 |
| 850.5656 | ST(h20:0) C44H84SNO12 | 850.5713 | −6.70 |
| 862.6032 | ST(22:0) C46H88SNO11 | 862.6078 | −5.33 |
| 878.5977 | ST(h22:0) C46H88SNO12 | 878.6027 | −5.69 |
| 888.6110 | ST(24:1) C48H90SNO11 | 888.6234 | −13.9 |
| 890.6326 | ST(24:0) C48H92SNO11 | 890.6391 | −7.30 |
| 904.6159 | ST(h24:1) C48H90SNO12 | 904.6183 | −2.65 |
| 906.6292 | ST(h24:0) C48H92SNO12 | 906.6340 | −5.29 |
| 857.5149 | PI(36:4) C45H78O13P | 857.5180 | −3.62 |
| 885.5437 | PI(38:4) C47H82O13P | 885.5493 | −6.32 |
| 1371.9568 | CL(66:4) C75H137O17P2 | 1371.9330 | 17.35 |
| 1373.9664 | CL(66:3) C75H139O17P2 | 1373.9487 | 12.88 |
| 1375.9658 | CL(66:2) C75H141O17P2 | 1375.9643 | 1.09 |
| 1395.9531 | CL(68:6) C77H137O17P2 | 1395.9330 | 14.40 |
| 1397.9720 | CL(68:5) C77H139O17P2 | 1397.9487 | 16.67 |
| 1399.9751 | CL(68:4) C77H141O17P2 | 1399.9643 | 7.71 |
| 1401.9920 | CL(68:3) C77H143O17P2 | 1401.9800 | 8.56 |
| 1403.9955 | CL(68:2) C77H145O17P2 | 1403.9956 | −0.07 |
| 1421.9591 | CL(70:7) C79H139O17P2 | 1421.9487 | 7.31 |
| 1423.9634 | CL(70:6) C79H141O17P2 | 1423.9643 | −0.63 |
| 1425.9839 | CL(70:5) C79H143O17P2 | 1425.9800 | 2.73 |
| 1427.9873 | CL(70:4) C79H145O17P2 | 1427.9956 | −5.81 |
| 1430.0229 | CL(70:3) C79H147O17P2 | 1430.0113 | 8.11 |
| 1445.9711 | CL(72:9) C81H139O17P2 | 1445.9784 | −5.05 |
| 1447.9623 | CL(72:8) C81H141O17P2 | 1447.9643 | −1.38 |
| 1449.9817 | CL(72:7) C81H143O17P2 | 1449.9800 | 1.17 |
| 1451.9982 | CL(72:6) C81H145O17P2 | 1451.9956 | 1.79 |
| 1454.0207 | CL(72:5) C81H147O17P2 | 1454.0113 | 6.46 |
| 1456.0308 | CL(72:4) C81H149O17P2 | 1456.0269 | 2.68 |
| 1458.0400 | CL(72:3) C81H151O17P2 | 1458.0425 | −1.71 |
| 1469.9427 | CL(74:11) C83H139O17P2 | 1469.9487 | −4.08 |
| 1471.9985 | CL(74:10) C83H141O17P2 | 1471.9643 | 23.23 |
| 1473.9535 | CL(74:9) C83H143O17P2 | 1473.9800 | −17.98 |
| 1476.0088 | CL(74:8) C83H145O17P2 | 1475.9956 | 8.94 |
| 1478.0367 | CL(74:7) C83H147O17P2 | 1478.0113 | 17.19 |
| 1480.0398 | CL(74:6) C83H149O17P2 | 1480.0269 | 8.72 |
| 1482.0535 | CL(74:5) C83H151O17P2 | 1482.0486 | 3.31 |
| 1495.9653 | CL(76:12) C85H141O17P2 | 1495.9643 | 0.67 |
| 1497.9636 | CL(76:11) C85H143O17P2 | 1497.9800 | −10.95 |
| 1500.0377 | CL(76:10) C85H145O17P2 | 1499.9956 | 28.07 |
| 1502.0540 | CL(76:9) C85H147O17P2 | 1502.0113 | 28.43 |
| 1504.0533 | CL(76:8) C85H149O17P2 | 1504.0269 | 17.55 |
| 1524.0355 | CL(78:12) C87H145O17P2 | 1523.9956 | 26.18 |
| 1526.0396 | CL(78:11) C87H147O17P2 | 1526.0113 | 18.55 |
| 1528.0405 | CL(78:10) C87H149O17P2 | 1528.0269 | 8.90 |
| 1544.8602 | GM1(d36:1) C73H130N3O31 | 1544.8688 | −5.57 |
| 1572.9107 | GM1(d38:1) C75H134N3O31 | 1572.9000 | 6.80 |
| 1817.9379 | GD1(d36:1) C84H145N4O38 | 1817.9536 | −8.64 |
| 1845.9744 | GD1(d38:1) C86H149N4O38 | 1845.9849 | −5.69 |
| 2091.0222 | GT1(d36:1) C95H160N5O45 | 2091.0384 | −7.75 |
| 2119.0435 | GT1(d38:1) C97H164N5O45 | 2119.0698 | −12.41 |
| 2364.0772 | GQ1(d36:1) C106H175N6O52 | 2364.1235 | −19.58 |
Lipids are assigned as [M − H]− except for gangliosides GD1, GT1, and GQ1 which are assigned as [M − H2O − H]−, [M − 2H2O − H]−, [M − 3H2O − H]−, respectively. Total ceramide carbons and double bonds are shown for gangliosides, while sulfatides are assigned to d18:1 sphingosines and only the fatty acyl chain composition is shown (h = hydroxyl fatty acyl).
Molecular ion signals for phosphatidylglycerol (PG), phosphatidylinositol (PI), and sulfatide (ST) were either unaffected or slightly enhanced by EDC/PLC treatment, especially for PG (34:1) (Figure 1d) vs untreated tissue (Figure 1a). The deprotonated ions from PG, PI, and ST are also listed in Table 1. All peaks are assigned according to literature values.
EDC/PLC Treatment of Tissue Greatly Reduces the PC Content
Next, the hippocampal regions of naive rat brain ± treatment with EDC/PLC were analyzed by GCIB-SIMS in positive ion mode (Figure 2). The m/z 700–850 region of untreated tissue exhibited numerous species of PC and sphingomyelin (SM) as mixtures of [M + H]+, [M + Na]+, and [M + K]+ (Figure 2a). This is similar to what was observed previously with Ar cluster SIMS of mouse brain.26 In the higher mass region, GCIB-SIMS detected PC as dimers (m/z 1450–1650) and trimers (m/z 2200–2500) of homomeric PC composition and heteromeric composition with phosphatidylethanolamine (PE) (Figure 2b,c). The detection of aggregates in samples with high concentrations of PC has also been reported with MALDI-TOF.33
Figure 2.
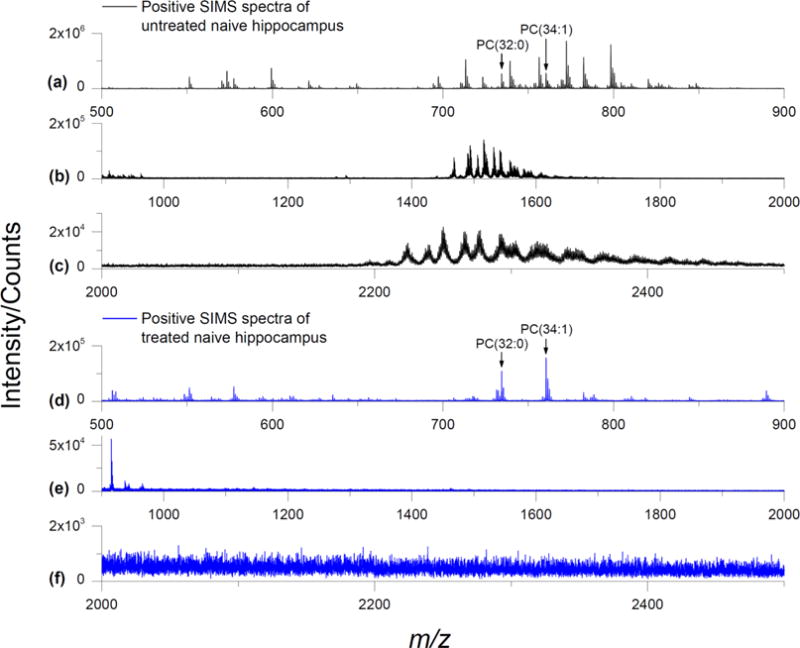
GCIB-SIMS positive mode spectra of naive rat brain tissue: SIMS positive ion mode spectra from the hippocampus of a naive rat brain coronal section (64 × 64 array, 16 μm spatial resolution). (a–c) Untreated tissue shows various species of PC in the m/z 700–900 range such as PC(32:0) [M + H]+ at m/z 734.5 and PC(34:1) at m/z 760.5. PC dimers and trimers are observed in the m/z 1400–1600 and m/z 2200–2400 ranges, respectively. (d–f) Tissue treated with EDC/PLC shows a sharp reduction in PC intensity and elimination of the dimers and trimers. Even though the PC [M + Na]+ and [M + K]+ ions have consolidated into [M + H]+ from the ammonium acetate washing, the overall PC intensity is approximately 10% of that from the untreated. Several species of sphingomyelin are still present after treatment. See Table 1 for complete peak assignments.
Tissue treatment with EDC/PLC greatly reduced the PC intensity as observed with GCIB-SIMS (Figure 2d). The remaining PC lipids were detected mainly as [M + H]+, e.g., m/z 734.6 for PC(32:0), m/z 760.6 for PC(34:1). This is due to removal of the extra salts by ammonium acetate washing during sample preparation. No PC aggregates were detected from treated tissue (Figure 2e,f).
Ganglioside GCIB-SIMS Signals Are Also Enhanced by EDC/PLC Treatment
The mass range of the larger gangliosides (m/z > 1500) are usually out of scope for TOF-SIMS detection. However, since our previous studies indicated that the tissue treatment protocol enhanced the signals for gangliosides in MALDI imaging,9,10 we examined the higher mass region for indications of them by GCIB-SIMS.9,10 Interestingly, signals for several brain ganglioside species were also enhanced by EDC/PLC treatment (Figure 1f). Without treatment, Ganglioside GM1 with putative 18:0 and 20:0 fatty acyl chains (d36:1 and d38:1, respectively) was hardly detectable by GCIB-SIMS as [M − H]− ions (m/z 1544.9 and 1572.9, respectively) and the spectrum was devoid of higher-order gangliosides (Figure 1c). EDC/PLC treatment not only resulted in the enhancement of the GM1 signal but also revealed signals for two higher-order gangliosides (GD1 and GT1) that were previously not detected by GCIB-SIMS.
Gangliosides GD1 and GT1 were primarily detected by GCIB-SIMS with losses of H2O dependent on their composition. While there was a faint signal for GD1(18:0) as [M − H]− at m/z 1836.0, the signal for it as [M − H2O − H]− at m/z 1818.0 was much stronger. Similarly, we observed a very weak GT1(18:0) signal as [M − H2O − H]− (m/z 2109.1) and a much stronger signal as [M − 2H2O − H]− (m/z 2091.0). To our knowledge, this is the first report of intact lipid species above m/z 2000 that has ever been detected using GCIB-TOF-SIMS. Our previous MALDI imaging studies also yielded this pattern, with enhanced ganglioside signals detected with losses of H2O corresponding to the number of internal sialic acid linkages.9,10
The structure of a ganglioside includes a ceramide attached to a chain of up to four central sugars, with one or more sialic acids attached to this central chain.34–36 Various mass spectrometric ionization methods are known to cause in-source glycosidic bond cleavages of gangliosides in addition to neutral losses of water and CO2.9,10,34,37–39 These issues have been circumvented to some degree by esterification of sialic acid residues with methyl/ethyl groups with various reagents including EDC.37–40 Our EDC tissue treatment protocol stabilizes the gangliosides by presumably forming a lactone ring between the carboxyl group of the alpha-2,3-linked sialic acid residue and the hydroxyl group of the neighboring saccharide residue.40 EDC has previously been used for stabilization of sialic acid containing glycans for MALDI-TOF analysis.41 Esterification of the sialic acids has also been shown to stabilize gangliosides during axial-MALDI-TOF analysis.37 However, complete esterification will remove all the sites for negative charging making it difficult to detect this species in negative ion mode. EDC treatment results in a mass loss of one water (18.02 u) per lactone ring and explains why the higher order gangliosides are preferentially detected by GCIB-SIMS with greater mass losses.
GCIB-SIMS Can Image CL in an Entire Coronal Brain Tissue Section at 16 μm
Knowing the capability of high-mass species detection of GCIB-SIMS, the CLs were mapped on an entire tissue section treated with EDC/PLC in negative ion mode at a spatial resolution of approximately 16 μm per pixel. The selected ion images for CL species at m/z 1456.0 and 1478.0 are shown in Figure 3. A total of 34 species of CL were detected in the EDC/PLC treated tissue (Table 1). In general, the more unsaturated species containing longer carbon chains tended to localize to the cortical region to a greater extent while those containing shorter carbon chains and fewer double bonds were observed more in the hippocampal region (Figure 3 and Supplemental Fig. S-1). This result is in accordance with our previous MALDI-TOF imaging work.9 However, in that study a pixel size of 200 μm was used, therefore only large regions of the brain were differentiated. Fine structures inside the hippocampus including the Dentate gyrus (DG), Cornu Ammonis 1 and 3 (CA1 and CA3), are now visible as shown in Figure 3 and match the features on the H&E stained image on the same section after SIMS analysis (Figure 3). Preparing tissue slices on glass coated with a transparent layer of indium–tin oxide (ITO) has been done for MALDI imaging and allows the actual tissue section to be subsequently examined after histological staining. The best signal for imaging CL by GCIB-SIMS was observed from tissue mounted on conductive copper plates. After SIMS imaging, H&E staining of the tissue was conducted directly on the copper plate and examined microscopically with incident light as opposed to transmitted light (Figure 3). Using this protocol, we still were able to generate acceptable optical images.
Figure 3.
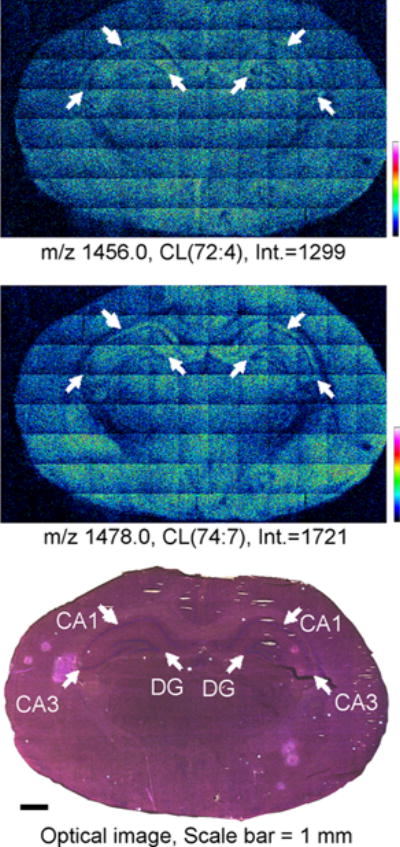
GCIB-SIMS images of CL in naive rat brain coronal tissue sections: SIMS negative ion mode images of CL [M − H]− species from an entire naive rat brain coronal section (spatial resolution, 16 μm) treated with EDC/PLC. Individual images for CL species are for CL(72:4) (top) and CL(74:7) (middle) indicating the number of carbons (72 or 74) and the total number of double bonds (:X). SIMS images are displayed as relative intensities with respect to the given ion species, with the absolute intensity of the top of the scale marked. The optical image of the H&E stained tissue section after GCIB-SIMS (scale bar 1 mm) is shown (bottom). White arrows indicate the CA1, CA3, and DG regions on the optical image with their corresponding locations on the SIMS images. See Table 1 for complete peak assignments.
GCIB-SIMS Can Image CL and Gangliosides for Regions of Interest at an 8 μm Pixel Size
With the CO2 cluster beam, it was possible to analyze the entire hippocampus and its immediate surrounding area with a more refined imaging resolution (8 μm) from a treated naive brain serial section (Figure 4). The total ion images of the hippocampus for CL species m/z 1456.0 and m/z 1478.0 are shown in Figure 4. The ion images match all of the important anatomical features as in the optical image of same tissue section. A representative CL spectrum generated from the 8 μm image from the center of the hippocampus is shown in Figure 4. Seven individual CL clusters are clearly detectable by GCIB-TOF-SIMS. Eight micrometer images for the additional CL species are displayed in Supplemental Fig. S-2. Because of the higher resolution, it was possible to compare the CL species in the CA1, CA3, and DG regions across three multicell layers. Most CL species are present at higher abundances in the oriens layer of the hippocampus, drop sharply in the pyramidal layer directly over the CA1 and CA3 regions, and increase moderately in the radial layer (Figure 4, Supplemental Figs. S-2 and S-3). While the increased imaging resolution at 8 μm approaches the individual cell level, it is difficult to ascertain if one or more cells are imaged in any given high-resolution pixel. In the adult rat, the thickness of the oriens layer varies from 75 to 125 μm; that of the pyramidal layer varies from 25 to 100 μm while the radial layer varies from 75 to 200 μm within different fields of the hippocampus.42 This is compounded by the fact that the regional differences in the thickness of the various subfields are related to the differences in the variety of efferent and afferent neuronal connections that run through the layers. This makes it difficult to determine if the signals are derived from a single cell body and/or multiple cellular processes.42
Figure 4.

GCIB-SIMS images of CL in naive rat brain hippocampus: SIMS negative ion mode images of CL [M − H]− species from the hippocampal region of a rat brain coronal tissue section (spatial resolution, 8 μm) treated with EDC/PLC. Individual images for CL species are for CL(72:4) (top left) and CL(74:7) (top middle) indicating the number of carbons (72 or 74) and the total number of double bonds (:X). SIMS images are displayed as relative intensities with respect to the given ion species, with the absolute intensity of the top of the scale marked. The optical image of the H&E stained tissue section after GCIB-SIMS (scale bar 250 μm) is shown (top right). A zoomed-in region of a representative spectrum from a single tile in the center of the hippocampus is shown (bottom). See Table 1 for complete peak assignments.
This refined imaging was also used for non-CL lipids such as the gangliosides GD1 (m/z 1818.0, 1846.0) and GT1 (m/z 2091.0) as well as PI(38:4) m/z 885.5 and ST(h24:1) m/z 904.6 (Figure 5). Sulfatides are normally most abundant in the white matter, and the mapping for ST(h24:1) confirms this. Gangliosides GM1, GD1, and GT1 exhibit either somewhat similar intensities in the cortex and hippocampus (for the putative 18:0 fatty acyl species) or stronger intensities in the hippocampus (for the putative 20:0 fatty acyl species).
Figure 5.

GCIB-SIMS images of non-CL lipids in naive hippocampus: SIMS negative ion mode images from the hippocampal region of a rat brain coronal tissue section (spatial resolution, 8 μm) treated with EDC/PLC. Individual images for PI, ST, and gangliosides are displayed. Ions are assigned as [M − H]− except for GD1 and GT1 which are [M − H2O − H]− and [M − 2H2O − H]−, respectively. The optical image of the H&E stained tissue section after GCIB-SIMS (scale bar 250 μm) is shown. SIMS images are displayed as relative intensities with respect to the given ion species, with the absolute intensity of the top of the scale marked. See Table 1 for complete peak assignments.
Imaging of CL with GCIB-SIMS in the Injured Brain
We applied our protocol to examine the effect of CCI on the intensity and spatial distribution of CL in the brain.9,30 Imaging was performed at a 16 μm pixel size for an entire brain tissue section (Figure 6, Supplemental Fig. S-4). As previously determined by MALDI-TOF imaging, CCI caused a significant loss of polyunsaturated CLs with the most dramatic CL changes being induced in the ipsilateral contusional cortex (blue arrow in Figure 6). Although losses in other lipids such as ganglioside GT1 were observed at the point of impact and areas immediately surrounding it, only CL exhibited additional selective losses in the CA3 region of the ipsilateral hippocampus and thalamus relative to the corresponding contralateral regions (Figure 6). The cortical regions far from the point of impact (Figure 6, Supplemental Fig. S-4) in ipsilateral and contralateral locations exhibited similar CL intensities. This is also typical of several of the more polyunsaturated/higher mass CL species (Supplemental Fig. S-4). The CA3 region is noted for its role in memory processes, seizure susceptibility, and neuro-degeneration.43 Therefore, specific losses of CL in this region may be responsible, at least in part, for the biochemical demise of mitochondria after TBI. Thus, CL loss may play a significant role in the poorer behavioral outcomes documented in our previous study,30 especially as it relates to cognitive functions. Roux et al. have reported changes in other lipids, from an entire brain tissue section (namely, ceramide, SM, cholesterol derivatives, and other glycerophospholipids, (50 μm resolution)) at later time points post-TBI using a combined sliver nanoparticle/ion mobility MALDI imaging technique.44
Figure 6.
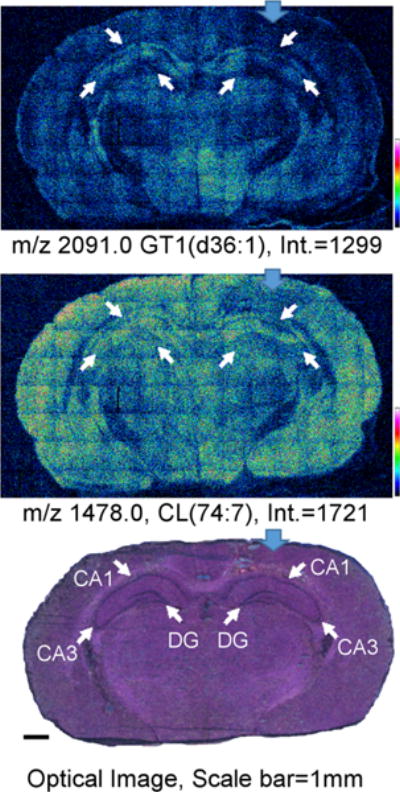
GCIB-SIMS images of CL lipids in CCI rat brain coronal tissue sections: SIMS negative ion mode images of ganglioside (top) and CL (middle) species from an entire CCI rat brain coronal section (spatial resolution, 16 μm) treated with EDC/PLC. Ions are assigned as [M − H]− (CL(74:7)) and [M − 2H2O − H]− (ganglioside GT1(d36:1)). The optical image of the H&E stained tissue section after GCIB-SIMS (scale bar 1 mm) is shown (bottom). SIMS images are displayed as relative intensities with respect to the given ion species, with the absolute intensity of the top of the scale marked. Blue arrows indicate point of impact on the ipsilateral hemisphere. White arrows indicate the CA1, CA3, and DG regions on the optical image with their corresponding locations on the SIMS images. See Table 1 for complete peak assignments.
CONCLUSION AND OUTLOOK
The present study demonstrates the feasibility and application of CO2-GCIB-SIMS technology for the assessment of larger lipid molecular ions, namely, cardiolipins and gangliosides, for successful imaging. The larger CO2 GCIB generates less damage to higher molecular mass lipid molecules. This combined with the EDC/PLC tissue treatment causing suppression of the signals from highly abundant phospholipids, like PC and PE, permits the mapping of individual CL molecular species despite their low abundance and high diversification in brain tissue. Moreover, GCIB-SIMS enhances the ionization of high mass lipid species, such as CLs, thus allowing for high-resolution imaging. On the basis of this, we were able to obtain more refined images of CLs and reliably establish the loss of CL polyunsaturated species in specific regions of the hippocampus and thalamus after traumatic brain injury. Thus, the combination of an appropriate gas cluster ion beam and method of tissue treatment can allow for successful imaging of larger (CL, gangliosides) and lower abundance (CL) lipid molecular ions by GCIB-TOF-SIMS.
As upgraded GCIB sources of SIMS with submicrometer focused ionization beams are becoming available, coupled with the latest development of dynamic reactive ionization of GCIB enhancing intact biomolecule signals up to 10-fold, the high spatial resolution imaging, particularly single cell imaging of low-abundance lipid molecules, like CL, becomes a readily achievable goal. Given the unique confinement of CLs to mitochondria, one can imagine that “biochemical microscopy” of it in intracellular organelles may become possible in the near future. Our current results, along with additional advances in GCIB-TOF-SIMS technology, will lay the groundwork for the imaging of lipids at submicrometer levels with higher resolution and less sample preparation.
Supplementary Material
Acknowledgments
This work was supported by NIH Grants 5R01GM113746-21, NS061817, NS076511, P01HL114453, and U19AI068021
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.7b00164.
GCIB-TOF-SIMS images of all 34 species of CL detected in naive brain tissue at both 8 and 16 μm resolution, injured brain tissue at 16 μm resolution, and intensities of CL species overlaid onto an optical image of hippocampal layers (PDF)
ORCID
Hua Tian: 0000-0002-3598-0219
Louis J. Sparvero: 0000-0001-6347-8448
Valerian E. Kagan: 0000-0002-7245-1885
Notes
The authors declare no competing financial interest.
References
- 1.Kagan VE, Bayir HA, Belikova NA, Kapralov O, Tyurina YY, Tyurin VA, Jiang JF, Stoyanovsky DA, Wipf P, Kochanek PM, Greenberger JS, Pitt B, Shvedova AA, Borisenko G. Free Radical Biol Med. 2009;46:1439–1453. doi: 10.1016/j.freeradbiomed.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoch FL. Biochim Biophys Acta, Rev Biomembr. 1992;1113:71–133. doi: 10.1016/0304-4157(92)90035-9. [DOI] [PubMed] [Google Scholar]
- 3.Schlame M. J Lipid Res. 2008;49:1607–1620. doi: 10.1194/jlr.R700018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claypool SM, Koehler CM. Trends Biochem Sci. 2012;37:32–41. doi: 10.1016/j.tibs.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osman C, Voelker DR, Langer T. J Cell Biol. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maguire JJ, Tyurina YY, Mohammadyani D, Kapralov AA, Anthonymuthu TS, Qu F, Amoscato AA, Sparvero LJ, Tyurin VA, Planas-Iglesias J, He R-R, Klein-Seetharaman J, Bayır H, Kagan VE. Biochim Biophys Acta, Mol Cell Biol Lipids. 2017;1862(1):8–24. doi: 10.1016/j.bbalip.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kooijman EE, Swim LA, Graber ZT, Tyurina YY, Bayır H, Kagan VE. Biochim Biophys Acta, Biomembr. 2017;1859:61–68. doi: 10.1016/j.bbamem.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tyurina YY, Poloyac SM, Tyurin VA, Kapralov AA, Jiang JF, Anthonymuthu TS, Kapralova VI, Vikulina AS, Jung MY, Epperly MW, Mohammadyani D, Klein-Seetharaman J, Jackson TC, Kochanek PM, Pitt BR, Greenberger JS, Vladimirov YA, Bayir H, Kagan VE. Nat Chem. 2014;6:542–552. doi: 10.1038/nchem.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sparvero LJ, Amoscato AA, Fink AB, Anthonymuthu T, New LA, Kochanek PM, Watkins S, Kagan VE, Bayır H. J Neurochem. 2016;139:659–675. doi: 10.1111/jnc.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amoscato AA, Sparvero LJ, He RR, Watkins S, Bayir H, Kagan VE. Anal Chem. 2014;86:6587–6595. doi: 10.1021/ac5011876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feider CL, Elizondo N, Eberlin LS. Anal Chem. 2016;88:11533. doi: 10.1021/acs.analchem.6b02798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang HYJ, Jackson SN, Woods AS. J Am Soc Mass Spectrom. 2007;18:567–577. doi: 10.1016/j.jasms.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vickerman J, Winograd N. Cluster Secondary Ion Mass Spectrometry. John Wiley & Sons, Inc.; Hoboken, NJ: 2013. pp. 269–312. [Google Scholar]
- 14.Blain MG, Della-Negra S, Joret H, Le Beyec Y, Schweikert EA. J Vac Sci Technol, A. 1990;8:2265–2268. [Google Scholar]
- 15.Davies N, Weibel DE, Blenkinsopp P, Lockyer N, Hill R, Vickerman JC. Appl Surf Sci. 2003;203–204:223–227. [Google Scholar]
- 16.Kollmer F. Appl Surf Sci. 2004;231:153–158. [Google Scholar]
- 17.Hill R, Blenkinsopp P, Barber A, Everest C. Appl Surf Sci. 2006;252:7304–7307. [Google Scholar]
- 18.Jerigova M, Biro C, Kirchnerova J, Chorvatova A, Chorvat D, Lorenc D, Velic D. Mol Imaging Biol. 2011;13:1067–1076. doi: 10.1007/s11307-010-0460-4. [DOI] [PubMed] [Google Scholar]
- 19.Yamada I, Matsuo J, Toyoda N, Kirkpatrick A. Mater Sci Eng, R. 2001;34:231–295. [Google Scholar]
- 20.Rabbani S, Barber AM, Fletcher JS, Lockyer NP, Vickerman JC. Anal Chem. 2011;83:3793–3800. doi: 10.1021/ac200288v. [DOI] [PubMed] [Google Scholar]
- 21.Ninomiya S, Ichiki K, Yamada H, Nakata Y, Seki T, Aoki T, Matsuo J. Rapid Commun Mass Spectrom. 2009;23:3264–3268. doi: 10.1002/rcm.4250. [DOI] [PubMed] [Google Scholar]
- 22.Ninomiya S, Ichiki K, Yamada H, Nakata Y, Seki T, Aoki T, Matsuo J. Rapid Commun Mass Spectrom. 2009;23:1601–1606. doi: 10.1002/rcm.4046. [DOI] [PubMed] [Google Scholar]
- 23.Hill R, Blenkinsopp P, Thompson S, Vickerman J, Fletcher JS. Surf Interface Anal. 2011;43:506–509. [Google Scholar]
- 24.Aoyagi S, Fletcher JS, Sheraz S, Kawashima T, Berrueta Razo I, Henderson A, Lockyer NP, Vickerman JC. Anal Bioanal Chem. 2013;405:6621–6628. doi: 10.1007/s00216-013-7139-z. [DOI] [PubMed] [Google Scholar]
- 25.Tian H, Wucher A, Winograd N. J Am Soc Mass Spectrom. 2016;27:2014–2024. doi: 10.1007/s13361-016-1492-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian H, Wucher A, Winograd N. Surf Interface Anal. 2014;46:115–117. doi: 10.1002/sia.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuo J, Torii S, Yamauchi K, Wakamoto K, Kusakari M, Nakagawa S, Fujii M, Aoki T, Seki T. Appl Phys Express. 2014;7:056602. [Google Scholar]
- 28.Angerer TB, Blenkinsopp P, Fletcher JS. Int J Mass Spectrom. 2015;377:591–598. [Google Scholar]
- 29.Tian H, Maciążek D, Postawa Z, Garrison BJ, Winograd N. J Am Soc Mass Spectrom. 2016;27:1476–1482. doi: 10.1007/s13361-016-1423-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji J, Kline AE, Amoscato A, Samhan-Arias AK, Sparvero LJ, Tyurin VA, Tyurina YY, Fink B, Manole MD, Puccio AM, Okonkwo DO, Cheng JP, Alexander H, Clark RSB, Kochanek PM, Wipf P, Kagan VE, Bayir H. Nat Neurosci. 2012;15:1407–1413. doi: 10.1038/nn.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angerer TB, Pour MD, Malmberg P, Fletcher JS. Anal Chem. 2015;87:4305–4313. doi: 10.1021/ac504774y. [DOI] [PubMed] [Google Scholar]
- 32.Angerer TB, Mohammadi AS, Fletcher JS. Biointerphases. 2016;11:02A319. doi: 10.1116/1.4941064. [DOI] [PubMed] [Google Scholar]
- 33.Eibisch M, Zellmer S, Gebhardt R, Suss R, Fuchs B, Schiller J. Rapid Commun Mass Spectrom. 2011;25:2619–2626. doi: 10.1002/rcm.5161. [DOI] [PubMed] [Google Scholar]
- 34.Colsch B, Woods AS. Glycobiology. 2010;20:661–667. doi: 10.1093/glycob/cwq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolter T. ISRN Biochem. 2012;2012:506160. doi: 10.5402/2012/506160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu RK, Tsai YT, Ariga T, Yanagisawa M. J Oleo Sci. 2011;60:537–544. doi: 10.5650/jos.60.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zarei M, Bindila L, Souady J, Dreisewerd K, Berkenkamp S, Müthing J, Peter-Katalinić J. J Mass Spectrom. 2008;43:716–725. doi: 10.1002/jms.1367. [DOI] [PubMed] [Google Scholar]
- 38.Jansen BC, Bondt A, Reiding KR, Lonardi E, de Jong CJ, Falck D, Kammeijer GSM, Dolhain RJEM, Rombouts Y, Wuhrer M. Sci Rep. 2016;6:23296. doi: 10.1038/srep23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steenackers A, Vanbeselaere J, Cazet A, Bobowski M, Rombouts Y, Colomb F, Bourhis XL, Guérardel Y, Delannoy P. Molecules. 2012;17:9559. doi: 10.3390/molecules17089559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiding KR, Blank D, Kuijper DM, Deelder AM, Wuhrer M. Anal Chem. 2014;86:5784–5793. doi: 10.1021/ac500335t. [DOI] [PubMed] [Google Scholar]
- 41.Bladergroen MR, Reiding KR, Ederveen ALH, Vreeker GCM, Clerc F, Holst S, Bondt A, Wuhrer M, van der Burgt YEM. J Proteome Res. 2015;14:4080–4086. doi: 10.1021/acs.jproteome.5b00538. [DOI] [PubMed] [Google Scholar]
- 42.Hussein AAW, George NT. J Fac Med Baghdad. 2009;51:323–327. [Google Scholar]
- 43.Cherubini E, Miles R. Front Cell Neurosci. 2015;9:19. doi: 10.3389/fncel.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roux A, Muller L, Jackson SN, Post J, Baldwin K, Hoffer B, Balaban CD, Barbacci D, Schultz JA, Gouty S, Cox BM, Woods AS. J Neurosci Methods. 2016;272:19–32. doi: 10.1016/j.jneumeth.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


