Abstract
The ability to enter monocyte-derived macrophage (MDM) in vitro is commonly used to define macrophage-tropic HIV-1 despite the fact that viruses vary continuously in their ability to enter MDMs in vitro, and MDMs vary in their ability to support HIV-1 entry (Joseph et al., 2014; Peters et al., 2006). This makes it difficult to distinguish viruses that are adapted to replicating in macrophage from those that are adapted to replicating in T cells. We use the Affinofile cell line (Johnston et al., 2009) to assay for macrophage tropism by capitalizing on the fact that macrophage-tropic HIV-1 has an enhanced ability to enter cells expressing low levels of CD4 (Joseph et al., 2014; Peters et al., 2006; Duenas-Decamp et al., 2009; Dunfee et al., 2006; Gorry et al., 2002; Martin-Garcia et al., 2006; Peters et al., 2004) and Affinofile cells can be induced to express a wide range of CD4 densities (Johnston et al., 2009). We induce Affinofile cells to express either high or low CD4, infect those cells with pseudotyped reporter virus, and quantify percent infectivity at low CD4 relative to infectivity at high CD4. Macrophage-tropic viruses have an enhanced ability to infect at low CD4. Using this approach we have found that macrophage-tropic strains of HIV-1 are relatively rare and that most HIV-1 variants require high levels of CD4 to enter cells, a phenotype we have referred to as R5 T cell-tropic.
Materials and Reagents
Affinofile cells (Johnston et al., 2009)
Luciferase Assay System (Promega Corporation, catalog number: E1501)
5× Reporter Lysis Buffer (Promega Corporation, catalog number: E397A)
PE-conjugated anti-CD4 antibody (clone RPA-T4) (BD, catalog number: 555347)
PE-conjugated anti-CCR5 antibody (clone 2D7) (BD, catalog number: 555993)
Aqua live/dead stain (Life Technologies, catalog number: L34957)
Env-pseudotyped luciferase reporter virus stock (frozen and tittered)
Blasticidin (Life Technologies, catalog number: A11139-03)
FBS (Atlanta Biologicals, catalog number: S12850)
DMEM-H (Cellgro, catalog number: 10-013-CV)
10% Buffered Formalin (Thermo Fisher Scientific, catalog number: SF100)
Poly-L-Lysine (Sigma-Aldrich, catalog number: P4707) (see Recipes)
Ponasterone A (Life Technologies, catalog number: 45-0478) (see Recipes)
Doxycycline (Sigma-Aldrich, catalog number: D9891) (see Recipes)
DMEM-F10/B (see Recipes)
1× staining solution (see Recipes)
Fixing solution (see Recipes)
Equipment
96-well plate (sterile, black) (Corning, Costar®, catalog number: 3916)
24 well plate
37 °C, 5% CO2 cell culture incubator (BSL-2)
Hemocytometer
Light microscope
PE-conjugated QuantiBRITE beads (BD, catalog number: 340495)
Incubated plate centrifuge (BSL-2)
Laminar flow biosafety cabinet (BSL-2)
Plate-reading luminometer
Procedure
A. For high versus low CD4 comparison
Day 1
Add 50 µl of diluted Poly-L-Lysine to sterile, black 96 well plates and incubate at 37 °C for 20 min. Remove Poly-L-Lysine.
Treat Affinofile cells with trypsin-EDTA and harvest in DMEM-F10/B.
Count cells and dilute to a concentration of 1.8 × 105 cells/ml in DMEM-F10/B.
Plate 100 µl of cells/well in 96 well plates.
Plate 0.75 ml of cells/well in 24 well plate for flow cytometry. Each well will be stained for flow cytometry. Plate enough cells to perform the necessary isotype and compensation controls.
Incubate cells at 37 °C overnight.
Day 2
- Prepare media for high and low CD4 induction.
- Dilute Doxycycline (Doxy) stock to 4× in DMEM-F10/B. A concentration of 24 ng/ml corresponds to 4× the concentration necessary for maximum CD4 induction.
- Dilute Ponasterone A (Pon A) stock to 4× in DMEM-F10/B. A concentration of 20 µM corresponds to 4× the concentration necessary for maximum CCR5 induction.
- Low CD4 induction medium (Low CD4/High CCR5): Mix equal volumes of 4× Pon A and DMEM-F10/B (=2× Pon A and 0× Doxy).
- High CD4 induction medium (High CD4/High CCR5): Mix equal volumes of 4× Pon A and 4× Doxy (=2× Pon A and 2× Doxy).
- Induce cells for infection.
- Remove 50 µl of medium from each well of the 96-well plate.
- For each virus being tested, add 50 µl of low CD4 induction medium to 3 wells (=1× Pon A and 0× Doxy) and 50 µl of High CD4 induction medium to 3 wells (=1× Pon A and 1× Doxy).
- Incubate cells at 37 °C.
- Induce cells for flow cytometry.
- Remove all medium from the 24 well plate, replace with 750 µl of warm DMEM-F10/B and 250 µl of either 4× Doxy or 4× Pon A (=1× desired amount of Pon A or Doxy).
- Incubate cells at 37 °C.
Day 3
- Infect cells 18–24 h after adding drugs (Pon A and Doxy).
- Remove induction medium from wells and immediately replace with 50 µl of warm DMEM-D10/B.
- Warm plate centrifuge to 37 °C.
- Thaw pseudovirus stock and dilute in DMEM-D10/B so that 50 µl of diluted virus produces a desired signal when titered on Affinofile cells expressing maximum CD4 and CCR5. This is determined by pre-titering the virus to ensure the amount of virus added is within the linear dose-response curve for the readout (in this case luciferase) and many fold above the assay background. As an additional control, include a dilution curve of a high titer virus to ensure all signals are within the linear range.
- Add 50 µl of diluted virus to each well of a 96-well plate (3 High CD4 wells and 3 Low CD4 wells per virus).
- Centrifuge plates at 2,000 rpm (849 × g) for 2 h at 37 °C (spinoculation; O'Doherty et al., 2000).
- Incubate cells at 37 °C for 48 h.
- Prepare cells for flow cytometry.
- Remove medium from a well of a 24 well plate, then use 1 ml of cold PBS to harvest the cells from that well and store the cells in eppendorf tubes on ice until all wells are harvested.
- Centrifuge the harvested cells in a table top microcentrifuge at 4 °C at 2,600 rpm for 10 min.
- While centrifuging, make staining solutions (Recipe 5)..
- Resuspend the cells in 50 µl of staining solution and stain at room temperature, in the dark, for 60 min.
- Add, with gentle mixing, 1 ml of fresh fixing solution (Recipe 6).
- Centrifuge in cold room at 2,600 rpm for 10 min, dispose of supernatant.
- Resuspend cells in 1 ml PBS.
- Centrifuge in cold room at 2,600 rpm for 10 min.
- Resuspend cells in 300 µl PBS.
- Store in the dark at 4 °C until ready to analyze.
- Analyze cells and QuantiBRITE beads using flow cytometer equipped with a 405 nm laser and a filter that measures emissions at approximately 526 nm and a 488 nm laser with a filter that measures emissions at approximately 578 nm.
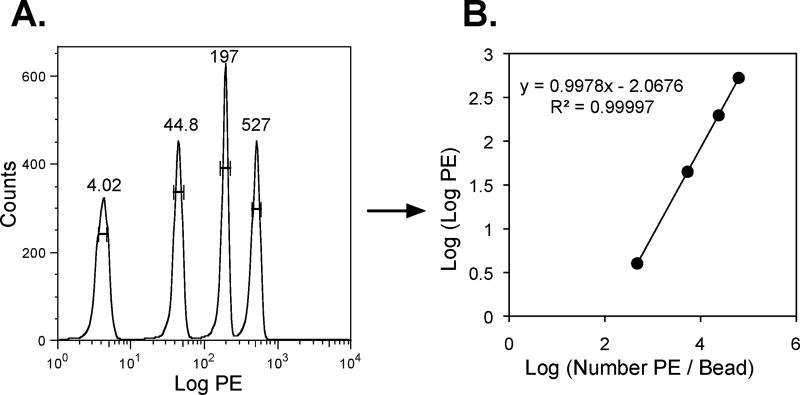
- Use Quantibrite beads, with their four known levels of PE, to determine the relationship between mean fluorescence and number of PE molecules (Figure 1). This relationship can be used to translate the PE fluorescence of stained cells into mean number of CD4 or CCR5 antibody binding sites (ABS) per cell. This method requires that cells be stained with saturating levels of antibody and assumes that each antibody is conjugated to a single PE molecule.
Figure 1. QuantiBRITE beads are used to quantify the relationship between PE fluorescence and number of PE molecules attached to a bead/cell.
A) QuantiBRITE beads are analyzed by flow cytometry to calculate the mean, log PE fluorescence of the four bead populations, each conjugated to a different, known number of PE molecules. B) The relationship between log (PE molecules/bead) and log (log PE) is then determined. This relationship can then be used to translate the mean PE fluorescence of stained cells into the mean number of CD4 or CCR5 antibody binding sites (ABS) per cell.
Day 5
- Harvest infected cells 48 h after infection.
- Remove medium from wells.
- Gently rinse 2× with warm PBS, discard PBS after each wash.
- Add 50 µl of 1× Reporter Lysis Buffer and incubate at room temperature for 15 min.
- Seal plates with aluminum foil covers and freeze at −80 °C for at least 2 h.
- Thaw plates and Luciferase Assay System reagent at room temperature.
- Prepare Luciferase Assay reagents following the manufacturer’s protocol, and read plate using a plate-reading luminometer.
- Calculate percent infectivity at low CD4 relative to infectivity at high CD4 for each individual virus.
B. For continuous CD4 comparison
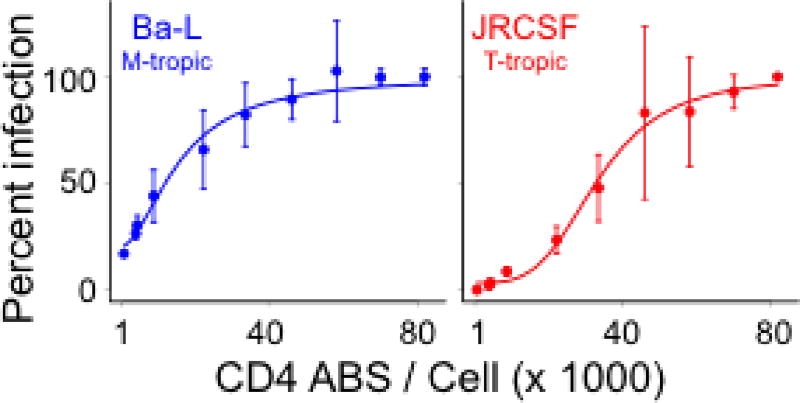
Macrophage-tropism can also be assessed by infecting Affinofile cells expressing a wide range of CD4 densities (Joseph et al., 2014). CD4 usage curves reveal that, relative to T cell-tropic viruses, macrophage-tropic viruses are significantly better at infecting cells expressing low levels of CD4, reach 50% infectivity at lower CD4 levels (CD4 ED50) and often approach a plateau level of infectivity at a lower CD4 density (Figure 2; Joseph et al., 2014). This procedure is identical to the ‘Procedure for High Versus Low CD4 Comparison’ (part A above) except that cells are induced to express 10 levels of CD4 and 30 wells of a 96-well plate are needed to test each virus (10 CD4 levels × 3 replicates per level).
Figure 2. Dose-response curves for infectivity of a macrophage-tropic virus (Ba-L) and a T cell-tropic viruses (JRCSF) to Affinofile cells expressing various densities of CD4.
The sensitivity of pseudotyped viruses to CD4 levels was assessed by measuring their ability to infect Affinofile cells expressing 10 CD4 levels (1,425, 4,590, 4,981, 9,374, 22,667, 33,842, 46,204, 58,153, 69,897, and 81,649 CD4 ABS per cell), with CCR5 fully induced. In this assay, the vast majority of blood-derived viruses have an infectivity profile similar to that of JRCSF (Ping et al., 2013).
Day 2
- Prepare media for continuous CD4 comparison
- Dilute Doxycycline stock to 24 ng/ml in DMEM-F10/B.
- Serially dilute Doxycycline to the following concentrations: 6, 4, 2.8, 2, 1.2, 0.8, 0.4, 0.28 ng/ml.
- Dilute Ponasterone A stock to 20 µM in DMEM-F10/B.
- Induction medium (10 levels of Doxycycline and 1 high level of Pon A): Mix equal volumes of 4× Doxycycline and 4× Ponasterone A (Table 1).
- Induce cells for infection.
- Remove 50 µl of media from each well of the 96-well plate.
- For each virus being tested, at each Doxycycline level, add 50 µl of each induction medium to 3 wells (10 induction levels × 3 replicates = 30 wells / virus).
- Incubate cells at 37 °C.
- Induce cells for flow cytometry
- Remove all medium from the 24 well plate, replace with 750 µl of warm DMEM-F10/B and 250 µl of either 4× Doxy or 4× Pon A (=1× desired amount of Pon A or Doxy).
- Incubate cells at 37 °C.
Table 1.
Dilutions of Doxycycline and Ponasterone A to induce ten levels of CD4 expression and a single, high level of CCR5 expression
| 4× Doxycycline (ng/ml) |
4× Ponasterone A (µM) |
1× Final Doxycycline concentration |
1× Final Ponasterone A concentration |
|---|---|---|---|
| 24 | 20 | 6 | 5 |
| 6 | 20 | 1.5 | 5 |
| 4 | 20 | 1 | 5 |
| 2.8 | 20 | 0.7 | 5 |
| 2 | 20 | 0.5 | 5 |
| 1.2 | 20 | 0.3 | 5 |
| 0.8 | 20 | 0.2 | 5 |
| 0.4 | 20 | 0.1 | 5 |
| 0.28 | 20 | 0.07 | 5 |
| 0 | 20 | 0 | 5 |
Days 3 and 5 are the same as described in ‘Procedure for High Versus Low CD4 Comparison’ (part A above).
Recipes
-
Poly-L-Lysine (0.1 mg/ml)
Diluted 10−1 in sterile PBS (10 µg/ml) and stored at 4 °C
-
Ponasterone A
Diluted in 50% ethanol to 1 mM and stored in 100 µl aliquots at −20 °C
-
Doxycycline
Diluted in sterile water to 10 mg/ml and frozen in 20 µl aliquots at −20 °C (do not freeze-thaw)
-
DMEM-F10/B
450 ml Cellgro DMEM-H
2.5 ml of blasticidin (final conc = 50 mg/ml, do not freeze/thaw blasticidin)
50 ml of dialyzed FBS (final = 10%)
Stored at 4 °C
-
1× staining solution
3 µl of antibody (anti-CD4, anti-CCR5 or isotype controls)
2 µl of aqua live/dead stain
45 µl of PBS
1% FBS
-
Fixing solution
200 µl of 10% formalin
800 µl H2O
Acknowledgments
We would like to thank Kathryn T. Arrildt for helpful comments that improved this protocol. This work was funded by the National Institutes of Health grant R37 AI44667 to RS.
References
- 1.Dunfee RL, Thomas ER, Gorry PR, Wang J, Taylor J, Kunstman K, Wolinsky SM, Gabuzda D. The HIV Env variant N283 enhances macrophage tropism and is associated with brain infection and dementia. Proc Natl Acad Sci U S A. 2006;103(41):15160–15165. doi: 10.1073/pnas.0605513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duenas-Decamp MJ, Peters PJ, Burton D, Clapham PR. Determinants flanking the CD4 binding loop modulate macrophage tropism of human immunodeficiency virus type 1 R5 envelopes. J Virol. 2009;83(6):2575–2583. doi: 10.1128/JVI.02133-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorry PR, Taylor J, Holm GH, Mehle A, Morgan T, Cayabyab M, Farzan M, Wang H, Bell JE, Kunstman K, Moore JP, Wolinsky SM, Gabuzda D. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J Virol. 2002;76(12):6277–6292. doi: 10.1128/JVI.76.12.6277-6292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston SH, Lobritz MA, Nguyen S, Lassen K, Delair S, Posta F, Bryson YJ, Arts EJ, Chou T, Lee B. A quantitative affinity-profiling system that reveals distinct CD4/CCR5 usage patterns among human immunodeficiency virus type 1 and simian immunodeficiency virus strains. J Virol. 2009;83(21):11016–11026. doi: 10.1128/JVI.01242-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joseph SB, Arrildt KT, Swanstrom AE, Schnell G, Lee B, Hoxie JA, Swanstrom R. Quantification of Entry Phenotypes of Macrophage-Tropic HIV-1 across a Wide Range of CD4 Densities. J Virol. 2014;88(4):1858–1869. doi: 10.1128/JVI.02477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O'Doherty U, Swiggard WJ, Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74(21):10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peters PJ, Bhattacharya J, Hibbitts S, Dittmar MT, Simmons G, Bell J, Simmonds P, Clapham PR. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J Virol. 2004;78(13):6915–6926. doi: 10.1128/JVI.78.13.6915-6926.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peters PJ, Sullivan WM, Duenas-Decamp MJ, Bhattacharya J, Ankghuambom C, Brown R, Luzuriaga K, Bell J, Simmonds P, Ball J, Clapham PR. Non-macrophage-tropic human immunodeficiency virus type 1 R5 envelopes predominate in blood, lymph nodes, and semen: implications for transmission and pathogenesis. J Virol. 2006;80(13):6324–6332. doi: 10.1128/JVI.02328-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ping LH, Joseph SB, Anderson JA, Abrahams MR, Salazar-Gonzalez JF, Kincer LP, Treurnicht FK, Arney L, Ojeda S, Zhang M, Keys J, Potter EL, Chu H, Moore P, Salazar MG, Iyer S, Jabara C, Kirchherr J, Mapanje C, Ngandu N, Seoighe C, Hoffman I, Gao F, Tang Y, Labranche C, Lee B, Saville A, Vermeulen M, Fiscus S, Morris L, Karim SA, Haynes BF, Shaw GM, Korber BT, Hahn BH, Cohen MS, Montefiori D, Williamson C, Swanstrom R, Study C, A. I. the Center for, H. I. V. A. V. I. C Comparison of viral Env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol. 2013;87(13):7218–7233. doi: 10.1128/JVI.03577-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Garcia J, Cao W, Varela-Rohena A, Plassmeyer ML, Gonzalez-Scarano F. HIV-1 tropism for the central nervous system: Brain-derived envelope glycoproteins with lower CD4 dependence and reduced sensitivity to a fusion inhibitor. Virology. 2006;346(1):169–179. doi: 10.1016/j.virol.2005.10.031. [DOI] [PubMed] [Google Scholar]