Abstract
The most common primary tumors of the human brain are thought to be of glial cell origin. However, glial cell neoplasms cannot be fully classified by cellular morphology or with conventional markers for astrocytes, oligodendrocytes, or their progenitors. Recent insights into central nervous system tumorigenesis suggest that novel molecular markers might be found among factors that have roles in glial development. Oligodendrocyte lineage genes (Olig1/2) encode basic helix–loop–helix transcription factors. In the rodent central nervous system, they are expressed exclusively in oligodendrocytes and oligodendrocyte progenitors, and Olig1 can promote formation of an chondroitin sulfate proteoglycon-positive glial progenitor. Here we show that human OLIG genes are expressed strongly in oligodendroglioma, contrasting absent or low expression in astrocytoma. Our data provide evidence that neoplastic cells of oligodendroglioma resemble oligodendrocytes or their progenitor cells and may derive from cells of this lineage. They further suggest the diagnostic potential of OLIG markers to augment identification of oligodendroglial tumors.
Glial neoplasms of the central nervous system (CNS) account for less than 1.5% of all new cancer cases reported in the United States each year. Nevertheless, these tumors are the fourth leading cause of cancer-related deaths in the United States (1). The high mortality rate of these infrequent cancers reflects the fact that two of the major subclasses—anaplastic astrocytoma and glioblastoma (GBM)—are refractory to conventional modalities of surgical resection, radiotherapy, and chemotherapy. On the other hand, selected patients diagnosed with anaplastic oligodendroglioma as well as low-grade oligodendroglioma can respond dramatically to the chemotherapeutic regimen of procarbazine, lomustine (CCNU), and vincristine (2–4). The presence of oligodendroglial features in malignant astrocytoma also appears to connote a better prognosis (5, 6). Thus there is prognostic and therapeutic value in the accurate diagnosis of oligodendroglial features within human brain tumors.
Although the biological distinction of oligodendroglial tumors from other gliomas holds great significance, the histopathological diagnosis of oligodendroglioma can be difficult, because there are currently no available molecular markers that reliably distinguish oligodendroglial tumors from astrocytomas. Cell type-specific marker proteins for mature oligodendroglial cells, such as myelin basic protein, myelin-associated glycoprotein, myelin proteolipid protein, 2′,3′-cyclic nucleotide-3′-phosphodiesterase, and galactolipids (GalC, O1, O4), are not expressed at detectable levels in oligodendrogliomas (7–10) (E.N. and P.M.B., unpublished observations). Likewise, the mRNAs that encode mature oligodendrocyte marker proteins do not correlate with the diagnosis of oligodendrogliomas by morphological classification (11). Astrocytic glial cell markers such as glial fibrillary acidic protein (GFAP) and S-100 β are expressed in both astrocytomas and oligodendrogliomas at various levels. They may be very useful for the diagnosis of astrocytomas but not for oligodendrogliomas (7, 12, 13).
The absence of lineage-specific marker proteins for oligodendroglioma raises another question. Does oligodendroglioma actually arise from oligodendrocytes or their progenitors? The term “oligodendroglioma” is empirical and reflects similarities between the cellular morphology of well-differentiated tumors and nonneoplastic oligodendrocytes. Certain glial cell neoplasms do express genes that are associated with undifferentiated oligodendrocyte precursor cells (14). For instance, immature glial markers, chondroitin sulfate proteoglycon (NG2) and α receptor of platelet-derived growth factor (PDGFαR), are detected in oligodendroglial tumors and pilocytic astrocytoma (PA) by immunohistochemical methods (15). However, NG2 chondroitin sulfate proteoglycan antibody is known to label the O-2A progenitor (16), which can give rise both to oligodendrocytes and type 2 astrocytes in vitro as well as other tissue types (e.g., capillaries) that might be found as components of brain tumors. PDGFαR is likewise expressed in other CNS cell types. Thus, neither of these markers is clearly specific to oligodendrocyte progenitor cells. Indeed, the paucity of molecular markers for early stages of normal glial cell development has made it difficult to establish the cellular origins of oligodendroglioma.
Recently rodent Olig1 and Olig2 genes were identified as oligodendrocyte lineage genes, encoding basic helix–loop–helix factors (17, 18). The Olig genes are expressed in rodent CNS specifically in cells of the oligodendrocyte lineage (17–19). Strikingly, Olig genes are expressed in embryonic oligodendrocyte precursors of the neural tube as well as in mature oligodendrocytes. Thus, oligodendrocytes in the human CNS might be expected to express human OLIG genes regardless of their degree of differentiation. This observation raised the question of whether human OLIG genes might serve as useful markers for identification of oligodendrogliomas or brain tumors derived from oligodendroglial cells. Here we report that OLIG genes are expressed in human oligodendrocytes and show strong expression in oligodendroglioma but not in other glial cell tumors. These results suggest the diagnostic potential of OLIG markers to augment identification of oligodendroglial tumors and shed light on the biological nature of these neoplasms.
Materials and Methods
Human Brain Tissue Collection, Processing, and Histological Analysis.
In accordance with institutional review board-approved human tissue collection protocols, brain tumor samples were obtained via open neurosurgical resection of masses from 23 patients; two normal brain samples were collected during temporal lobe resection for intractable epilepsy. In all cases, parts of the specimen were fixed and paraffin-embedded for histological analysis, and parts were snap-frozen in liquid nitrogen for analysis by in situ hybridization and/or immunostaining. An independent neuropathologist (D.N.L) reconfirmed clinical neuropathological diagnoses on frozen sections. Hematoxylin/eosin staining was carried out according to standard procedures. For methyl green staining, frozen tissue sections were immersed in 1% methyl green (Vector Laboratories) in 20 mM sodium acetate and sodium barbital (pH 4.0) for 1 min, then washed with the phosphate buffer to maintain green nuclear staining.
In Situ Hybridization Analysis of OLIG Expression in Human Tumors.
After storage in liquid nitrogen, brain tissues were drop-fixed in fresh 4% paraformaldehyde for 12–16 h. They were then equilibrated in 20% glucose/PBS (pH 7.4) at 4°C, embedded in optimal cutting temperature compound, and sectioned at 15 μm on a cryostat (Bright, Huntington, U.K.). In situ hybridization was performed as previously described (17). A detailed protocol is available on request. The following cRNA probes were used: human OLIG1, OLIG2 (cloned on the basis of GenBank published sequence AP000109; ref. 16), and GFAP (11). All analyses of expression level were performed at low-power magnification, and the entire tumor on the slide was evaluated, rather than specific foci or selected high-power fields. OLIG gene expression levels were scored from low to strongly positive (+/− to +++++) on a six-grade arbitrary scale, depending on the percentage and intensity of OLIG-positive cells in tumors, as indicated in the legend to Table 1. These subjective measures were scored by two independent investigators, and the average values of classification are shown in Table 1. To ensure consistency, we used an identical preparation of digoxigenin–OLIG1 probe on all tumors, and incubation with chromogenic substrate (nitro blue tetrazolium (NBT)/5-bromo-4-chloro-3-inodolyl-phosphate (BCIP) was only for 24 h. The higher intensity of OLIG expression reflects the fact that less time is required to develop the purple color formation of the substrate. Furthermore, as a measure of the quality of the mRNA species in the astrocytic tumors, testing for GFAP expression was done in cases where OLIG expression levels were less than “+/−.” For double in situ hybridization, we used digoxigenin-labeled cRNA for OLIG1 and fluorescein-labeled cRNA for GFAP, respectively. The different labeled cRNAs were recognized with anti-digoxigenin–alkaline phosphatase (AP) Fab fragment or anti-fluorescein–AP Fab fragment and developed with corresponding substrates NBT/BCIP (blue purple) or 1-(4-iodophenyl)-5-(4-nitrophenyl)-3-phenyltetrazolium chloride/BCIP (dark brown) sequentially.
Table 1.
OLIG expression in normal and neoplastic brain
| Clinical diagnosis | mRNA expression level
|
|
|---|---|---|
| OLIG1 | OLIG2 | |
| Nonneoplastic brain | ||
| Cerebral cortex | +/− | +/− |
| White matter | + | + |
| Brain tumors | ||
| Oligodendrogliomas | ||
| Oligodendroglioma (grade II) | +++++ | +++++ |
| Oligodendroglioma (grade II) | ++++ | ++++ |
| Oligodendroglioma (grade II) | ++++ | ++++ |
| Anaplastic oligodendroglioma (grade III) | +++++ | +++++ |
| Anaplastic oligodendroglioma (grade III) | ++++ | ++++ |
| Anaplastic oligodendroglioma (grade III) | +++ | +++ |
| Astrocytomas | ||
| Pilocytic astrocytoma (grade I) | +/− | +/− |
| Pilocytic astrocytoma (grade I) | +/− | +/− |
| Astrocytoma (grade II) | +/− | +/− |
| Astrocytoma (grade II) | +/− | +/− |
| Astrocytoma (grade II) | +/− | +/− |
| Astrocytoma (grade II) | +/− | +/− |
| Astrocytoma (grade II) | + | + |
| Anaplastic astrocytoma (grade III) | +/− | +/− |
| Anaplastic astrocytoma (grade III) | +/− | +/− |
| Glioblastomas | ||
| Glioblastoma (grade IV) | +/− | +/− |
| Glioblastoma (grade IV) | +/− | +/− |
| Glioblastoma (grade IV) | + | + |
| Glioblastoma (grade IV) | + | + |
| Glioblastoma (grade IV) | + | + |
| Glioblastoma (grade IV) | + | + |
| Glioblastoma (grade IV) | ++ | ++ |
| Glioblastoma (grade IV) | ++ | ++ |
Immunohistochemistry.
After in situ hybridization, certain sections were pretreated with 3% H2O2 in PBS for 30 min at room temperature and then immunolabeled with rabbit polyclonal antibody against myelin basic protein [1:200 dilution (Biomedia, Foster City, CA)], and subsequent visualization with avidin–biotin complex (Vector) in the presence of diaminobenzidine, per manufacturers' protocols.
Results
Specific Expression of OLIG1 in Human Oligodendrocytes.
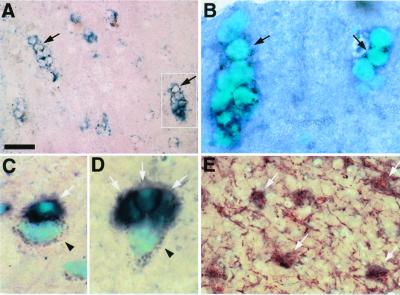
To determine the specificity of human OLIG gene expression in oligodendrocytes, we analyzed nonneoplastic cerebral cortex and temporal lobe tissues by in situ hybridization with an OLIG1 cRNA probe. As shown in Fig. 1 A and B, OLIG1-expressing cells were small with round nuclei and scant cytoplasm and formed interfascicular linear arrays, features typical of oligodendroglial cells. OLIG1-expressing cells were often detected adjacent to neurons, identified morphologically by the presence of large nuclei and dark brown lipofuscin granules (Fig. 1 C and D; filled arrowhead). Indeed, although most regions of the brain showed single oligodendrocytes in association with neuron cell bodies (Fig. 1C, white arrow), a characteristic finding in the temporal lobe was the presence of perineuronal oligodendroglial satellites (Fig. 1D, white arrows). Moreover, we observed colabeling of OLIG1-expressing cells with the mature oligodendrocyte marker, myelin basic protein (Fig. 1E, white arrows). Together, these results indicate that OLIG1 specifically marks oligodendrocytes in human brain and are consistent with previous observations in the rodent CNS (17–19).
Figure 1.
OLIG1 expression specifically identifies human oligodendrocytes. Frozen sections of temporal lobe tissue, resected from a patient with epilepsy, were analyzed by in situ hybridization with an antisense human OLIG1 cRNA probe and immunolabeling with the mature oligodendrocyte marker, myelin basic protein (MBP). (A, B) OLIG1 expression was detected in cells with oligodendrocyte morphological characteristics including, small round nuclei that counterstain with methyl green (B), scarce cytoplasm, and formation interfascicular cellular arrays (A and B, filled arrows). The white boxed area in A is shown at higher power in B. (C, D) OLIG1 expression in cells adjacent to neuron cell bodies, which contain large nuclei and dark brown lipofuscin granules (arrowheads). Single OLIG1-positive cells (indicated by white arrows) were observed in association with neurons (C) and as perineuronal oligodendrocyte satellites (D). (E) In situ hybridization of OLIG1 mRNA transcripts in conjunction with MBP immunolabeling, which identified cell body and myelinated fibers of oligodendrocytes, indicating that OLIG1-expressing cells are oligodendrocytes (white arrows). [Bars = 50 μm (A); 15 μm (B–D); 25 μm (E).]
Oligodendrogliomas Express High Levels of OLIG mRNA Transcripts.
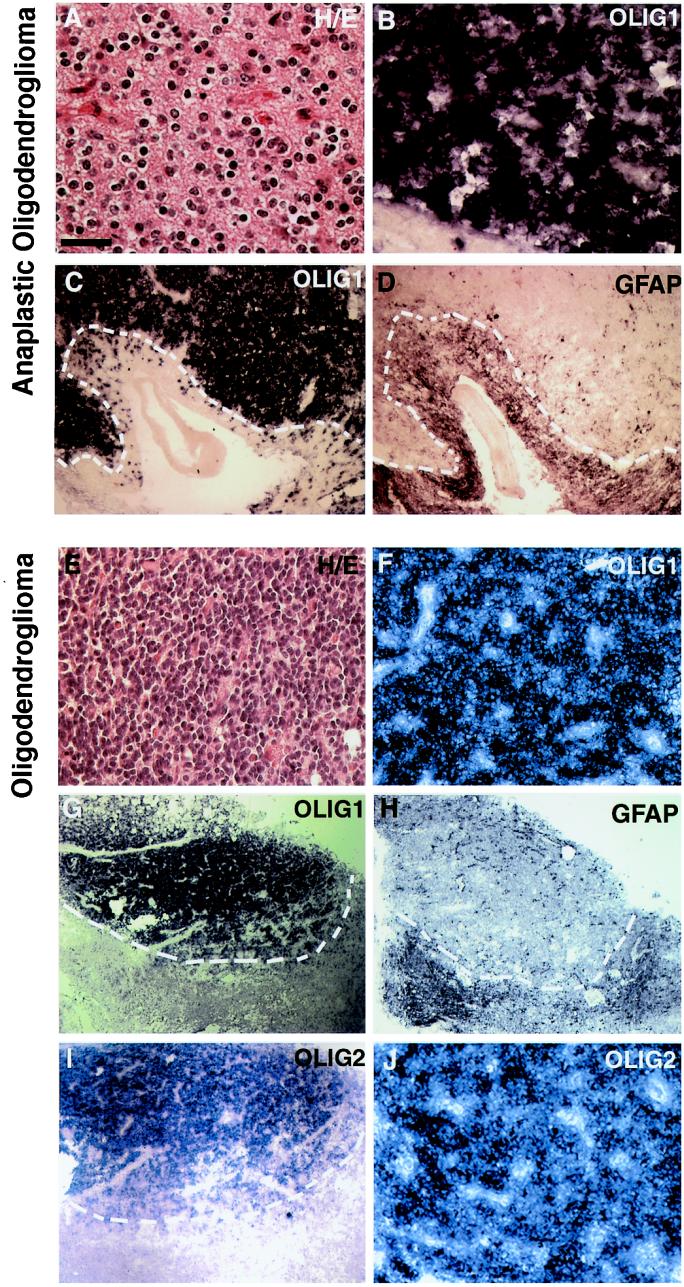
To determine whether OLIG genes were expressed in human glial tumors, we screened a panel of 23 glial tumors with independently reconfirmed diagnoses (see Materials and Methods) by in situ hybridization. All tumors were also tested for expression of GFAP, which served both as an independent measure of tissue quality and an indicator of certain tumor characteristics (e.g., reactive astrocytosis). As indicated in Table 1, all six of the oligodendrogliomas analyzed showed high levels of OLIG expression, and two representative cases are shown in Fig. 2. Histological analysis of anaplastic (Fig. 2A) and low-grade (grade II; Fig. 2E) oligodendroglioma at high magnification shows moderately to densely cellular tumors composed of cells with rounded and homogeneous nuclei as well as perinuclear halos (Fig. 2A)—all characteristic features of oligodendrogliomas. The OLIG1 gene was strongly expressed in the neoplastic cells of these oligodendrogliomas, labeling densely packed cells (Figs. 2 B and F). Low- magnification images are shown of these tumors to illustrate the interface between neoplastic areas with high OLIG1 expression and adjacent nonneoplastic tissue with minimal OLIG1 expression (Fig. 2 C and G). Note that the intense OLIG1-expressing cellular foci are devoid of expression of the astrocytic marker, GFAP (Fig. 2 D and H). The GFAP-positive cells found outside of the tumor were judged to be reactive astrocytes surrounding the neoplastic tissue, given their location and characteristic stellate appearance. Similar to OLIG1 characteristics, OLIG2 was expressed at high levels in the oligodendroglial tumors (Fig. 2 I and J). Our results demonstrate that neoplastic cells of oligodendrogliomas strongly express OLIG genes.
Figure 2.
Neoplastic oligodendroglioma cells express high levels of OLIG mRNA transcripts. (A, E) Histological analysis and (B–D, F–J) in situ hybridization for expression of human OLIG1, OLIG2, and GFAP were performed on samples of (A–D) anaplastic oligodendroglioma and (E–J) oligodendroglioma (grade II), as described in Materials and Methods. (A, E) In both cases, histological analysis with hematoxylin/eosin staining reveals densely packed cells with round nuclei. Perinuclear halos in paraffin process section are also observed (A, high magnification). (B, F, J) Strong OLIG expression was found in densely packed homogeneous-appearing neoplastic cells of oligodendrogliomas; OLIG1 (B, F) and OLIG2 (J). (C, D, G–I) Low-magnification images of tumors showing expression of OLIG1 (C, G), OLIG2 (I), or GFAP (D, H). Note apparent segregation of GFAP mRNA transcripts from neoplastic cells of oligodendroglioma with strong OLIG expression. The border of tumor and the surrounding nonneoplastic tissue is indicated by the white dotted line. This indicates that the GFAP+-reactive astrocytes are found in the tissue surrounding tumor neoplasm. [Bars = 50 μm (A and B); 200 μm (C and D); 100 μm (E, F, and J); 400 μm (G–I).]
Characterization of OLIG Expression in Other Gliomas.
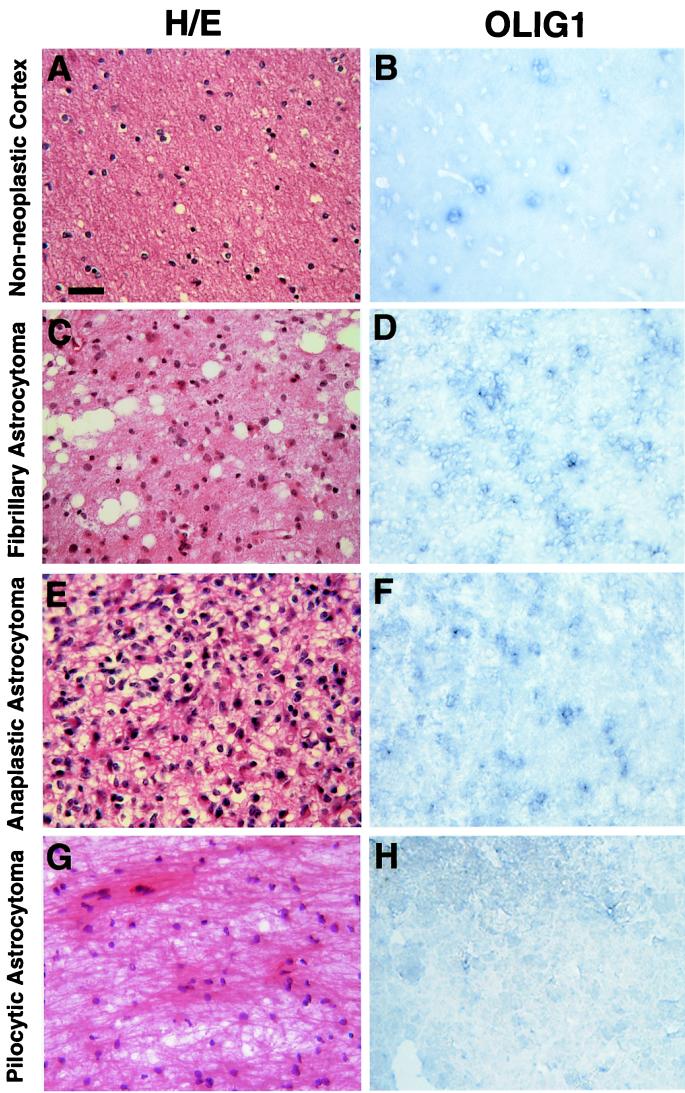
To test whether high levels of OLIG gene expression could distinguish oligodendrogliomas from astrocytomas, we examined OLIG1 expression in other glial tumor samples. As shown in Fig. 3 A and B, the nonneoplastic human cerebral cortex contains only scattered OLIG1-positive cells. In contrast to the high levels of OLIG expression in oligodendroglioma (Fig. 2), we found only weak or moderate expression in astrocytomas such as fibrillary astrocytoma (Fig. 3 C and D), anaplastic astrocytoma (Fig. 3 E and F), and PA (Fig. 3 I and J). Similar patterns and levels of mRNA expression were also observed with human OLIG2 in sections of astrocytomas (Table 1).
Figure 3.
OLIG1 is expressed at low levels in most astroglial tumors. (A, C, E, G) Histological analysis and (B, D, F, H) in situ hybridization by using an OLIG1 cRNA probe were performed on samples of: (A, B) nonneoplastic human cortex, (C, D) fibrillary astrocytoma, (E, F) anaplastic astrocytoma, and (G, H) PA. Note astrocytic cells in astrocytomas with pleomorphic nuclei, large amounts of cytoplasm, and dense fiber bundles. Expression of OLIG1 in these tumors was low and contrasted strong expression in oligodendroglioma (see Fig. 2). (Bar = 50 μm.)
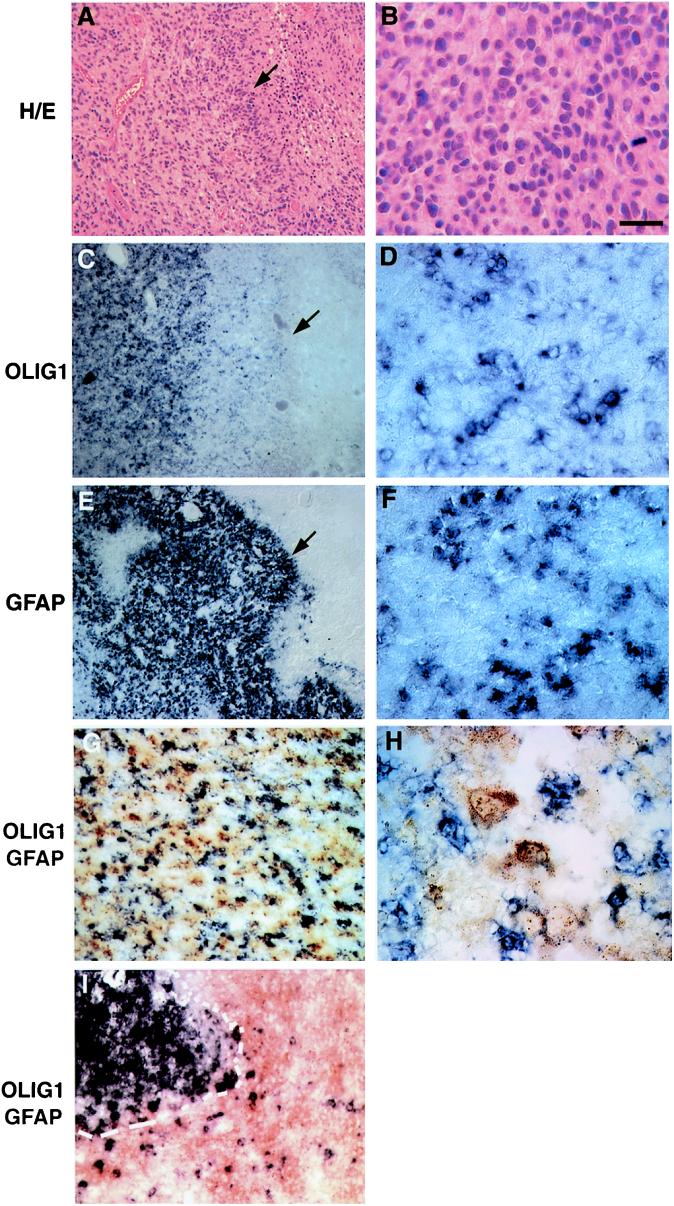
GBM is a highly anaplastic brain tumor composed of poorly differentiated neoplastic cells and pleomorphic cells displaying astrocytic differentiation, mitoses, microvascular endothelial proliferation, and pseudopalisading necrosis (20). As indicated in Table 1, we found OLIG1 gene expression in GBM tumor sections to be highly variable. One case of GBM with a moderate level (++) of OLIG1 expression is shown in Fig. 4. In contrast to results obtained with all oligodendroglial tumors tested, both GFAP (Fig. 4E) and OLIG1 (Fig. 4C) were expressed in this GBM sample in the solid tumor region and the region adjacent to necrosis (peripheral pseudopalisading tumor cells indicated by arrows in Fig. 4 A, C, E). OLIG1-expressing cells in GBM were not densely packed, and many intervening OLIG1-negative cells were present (Fig. 4D). GFAP- and OLIG1-positive cells appeared to be intermingled (compare Fig. 4 D and F). The double-labeling images of OLIG1 and GFAP clearly show that OLIG and GFAP do not mark the same cells (Fig. 4 G and H). In contrast, most—if not all—neoplastic cells in oligodendroglioma expressed high levels of OLIG. Essentially no GFAP expression was detected within oligodendrogliomas, and GFAP+-reactive astrocytes were found only surrounding the neoplasm (Fig. 4I).
Figure 4.
OLIG1 expression in a case of GBM. (A, B) Histological analysis and (C–F) in situ hybridization by using (C, D) OLIG1 or (E, F) GFAP cRNA probes were performed on a case of GBM. (A, C, E) Low-magnification images of this GBM. Note areas of necrosis (Right) and peripheral pseudopalisading tumor cells in the sections (filled arrows). (B, D, F) At high magnification, histological analysis in conjunction with in situ hybridization revealed expression of both OLIG1 (D) and GFAP (F) in areas of solid tumor. OLIG1- (purple) and GFAP- (brown) expressing cells are intermingled but distinct from each other shown in low (G) and high (H) magnification, in contrast to oligodendroglioma, wherein GFAP+ cells surround the neoplasm (I). [Bars = 200 μm (A, C, and E); 150 μm (G, I); 50 μm (B, D, F); 30 μm (H).]
In summary, OLIG expression per se does not distinguish oligodendroglioma from GBM. However, GBM is a very distinctive tumor that is easily diagnosed by other histopathologic criteria. High-grade gliomas typically infiltrate into normal adjacent white matter that is rich in oligodendrocytes. Thus it is expected that some background cells within high-grade gliomas will express OLIG genes. In addition, high-grade gliomas such as GBMs are genetically unstable, and the neoplastic cells of these tumors are notorious for expressing a wide range of marker genes for different neural lineages.
Discussion
Molecular analysis of mechanisms underlying CNS development has yielded significant insight into tumorigenesis. For example, activity of the Sonic hedgehog pathway, essential for proliferation of granule neuron precursors during development of the cerebellum (21–23), is etiologic in perhaps 10–20% of cases of the human cerebellar tumor, medulloblastoma (24, 25). Although parallels have been drawn between glial tumors and immature glial precursors (14), such characterization has been limited by the paucity of molecular markers for early stages of glial development. Our results indicate strong OLIG gene expression in oligodendroglioma and highlight the diagnostic potential of OLIG markers to distinguish between types of human glial tumors. Together with previous observations, these data further suggest that neoplastic cells of oligodendroglioma bear resemblance to developing oligodendrocytes.
Currently, the identification of tumor-specific molecules remains the major challenge for precise diagnosis of brain tumor subtypes. Oligodendrogliomas manifest weak or absent immunoreactivity to conventional markers for mature oligodendrocytes (8, 9, 26), and in situ hybridization for mRNA transcripts of myelin-specific proteins was essentially negative in oligodendroglioma (11). These findings have led investigators to question whether oligodendroglioma is indeed a derivative of oligodendroglial cells or represents some other cell of origin (27).
To further investigate the biological nature of oligodendroglioma, we used homologues of the recently cloned oligodendrocyte lineage gene markers, Olig1 and Olig2, that encode basis helix–loop–helix proteins (17, 18). During rodent CNS development, Olig genes are expressed in oligodendrocyte precursors and differentiated oligodendrocytes (17, 18). In keeping with these observations, our results indicate that human OLIG1 expression is specific for oligodendrocytes. In addition, of 23 glial tumors screened, neoplasms expressing high levels of OLIG genes were observed only in oligodendroglial tumors but not in other tumor types. Interestingly, multiple GenBank entries show high representation of human OLIG1 expression in expressed sequence tag libraries derived from oligodendrogliomas. It is therefore possible that high levels of OLIG expression are a general characteristic of oligodendroglial tumors. Indeed, that OLIG genes are consistently expressed in oligodendroglial tumors, coupled with the absence of differentiated markers for oligodendrocytes (11), strongly argues for an immature character, similar to oligodendrocyte progenitor cells of the developing CNS. These findings extend those of Shoshan et al. (15), who determined that the immature glial marker, NG2, was expressed in oligodendroglial tumors. Although NG2 expression is a marker for oligodendrocyte precursors, it has also been observed in bipotential glial precursors. Thus, the additional finding of OLIG expression strongly argues for the oligodendroglial nature of this tumor. Ectopic expression of OLIG1 has been shown to promote the formation of NG2+ cells in vitro (17). Thus, an intriguing possibility is that OLIG expression in oligodendroglioma itself may be mechanistically linked to initiation and/or maintenance of expression of early markers of oligodendrocyte development.
It is also significant that astroglial tumor types showed low or absent levels of OLIG gene expression. In particular, we found consistently low levels of OLIG expression in low-grade astrocytomas. The scattered pattern of OLIG+ cells in the astrocytoma samples, moreover, could represent the native oligodendrocytes in tissue that has been infiltrated by the neoplastic astrocytes (11), as OLIG expression marks mature oligodendrocytes as well as precursors (17). In contrast to results of Shoshan et al. (15), who detected NG2 labeling in PA, we found PA to be OLIG-negative. Although our numbers were small, strong GFAP expression in our PA samples (data not shown) rules out the possibility that absence of OLIG expression was because of poor tissue quality. Thus, although these results will have to be confirmed on larger numbers of PAs, they raise the possibility that OLIG marker expression may help distinguish oligodendroglioma from PA.
The diagnosis of GBM is made in tumors that are thought to be astrocytic in nature with the malignant features of marked cytological atypia, mitoses, neovascular proliferation, and necrosis. Additionally, GBM may show significant cytological and morphological heterogeneity and variability even within the same tumor, making it difficult to distinguish between GBM and some anaplastic oligodendrogliomas, which can also show pleomorphism, necrosis, and vascular proliferation. This issue currently represents a diagnostic dilemma in the field of neuropathology and neurooncology, as there is evidence that anaplastic oligodendrogliomas and tumors with oligodendroglial components may respond better to therapy and have a more favorable prognosis (5, 6).
Interestingly, it has been proposed that the so-called oligodendrocyte precursor cells (also known as O-2A cells) may be the cellular origin of a subset of GBMs (14). Some of the GBMs examined in this study expressed relatively high levels of OLIG mRNA. Thus, our results are consistent with the idea that different components of certain anaplastic oligoastrocytomas may arise from a common precursor cell. However, it should be pointed out that high-grade gliomas such as GBMs often show aberrant expression patterns because of progressive genetic instability. Thus, an alternative possibility is that OLIG expression in GBM might reflect dysregulation. These observations raise further questions about the origins of complex human glial tumors and are compelling reasons to investigate further the origins of glial tumors by using animal models. It was reported that ectopic expression of polyoma middle T-antigen driven by GFAP regulatory sequences produced mixed astrocytoma–oligodendroglioma (28). Although GFAP is known to mark differentiated astrocytes, labeling has recently been described in association with putative multipotent neural stem cells (29). Thus, it is possible that the mixed gliomas arising in this model may derive from oncogenic transformation of multipotential precursor. In any case, our observations suggest that OLIG genes will be useful markers in the characterization of early oligodendroglial tumor development and determination of their ontogeny.
In terms of clinical implications, we found OLIG expression to be a molecular marker whose expression characteristics varied between different glial tumor types. It may be that use of OLIG expression, in conjunction with standard neuropathological approaches, will eventually facilitate more accurate diagnoses of glial tumors. This is particularly important for the diagnosis of oligodendrogliomas, a relatively rare yet chemosensitive tumor (2, 3, 30). Recent data suggest that molecular genetic analyses such as loss of heterozygosity (LOH) may be a predictive means of determining which oligodendroglial tumors will be chemosensitive (2–4). OLIG gene expression could be used to triage samples for LOH analysis, because this process is too expensive and labor-intensive to be performed as a matter of routine on all brain tumors. Furthermore, OLIG levels will need to be measured against other clinical parameters such as chemosensitivity and survival (2, 3), as well as predictive diagnostic tests (31, 32). Finally, it bears mentioning that ectopic expression of OLIG2 has been reported in one case of T-cell leukemia (33). Indeed, misexpression of basis helix–loop–helix-encoding genes (e.g., SCL/Tal) is commonly associated with dysregulation of the cell cycle. This raises the possibility that OLIG expression itself may play a role in the initiation or maintenance of the glial tumor phenotype. In any case, the preponderance of available evidence in conjunction with results of this study strongly supports the characterization of oligodendroglial tumors as bearing strong similarity to immature oligodendrocytes. If so, a better understanding of factors regulating cell death and differentiation in oligodendrocyte precursors may facilitate development of novel adjuvants to current antioligodendroglial tumor therapies.
Acknowledgments
We thank Dr. Umberto De Girolami for review of tumor slides and Drs. Patricia Dahia and Keith Ligon for stimulating discussions. We are grateful to Dr. Luning Hao for technical assistance. Q.R.L. is a recipient of a postdoctoral fellowship award from the National Institutes of Health and the Charles A. King Trust Medical Foundation. D.H.R. is a Claudia Adams Barr Investigator, a Kimmel Scholar, and a Weaver Neuroscience Scholar of the United States National Multiple Sclerosis Society. This study was funded by grants from the National Institute of Child Health and Human Development (HD24296 to C.D.S.), the National Institute of Neurological Disorders and Stroke (NS41764 to D.H.R.), the National Cancer Institute (CA 57683 to D.N.L.), and the Mahoney Center for Neuro-oncology at the Dana–Farber Cancer Institute.
Abbreviations
- CNS
central nervous system
- GFAP
glial fibrillary acidic protein
- GBM
glioblastoma
- PA
pilocytic astrocytoma
- NG2
chondroitin sulfate proteoglycon
Note
During the course of this work, we have learned that the high level of OLIG2 expression in oligodendroglial tumor cells was also observed by Dr. Boris Zalc and his colleagues (personal communication). We thank Dr. Zalc for sharing the unpublished information.
References
- 1.DeAngelis L M. N Engl J Med. 2001;344:114–123. doi: 10.1056/NEJM200101113440207. [DOI] [PubMed] [Google Scholar]
- 2.Cairncross J G, Macdonald D R. Ann Neurol. 1988;23:360–364. doi: 10.1002/ana.410230408. [DOI] [PubMed] [Google Scholar]
- 3.Macdonald D R, Gaspar L E, Cairncross J G. Ann Neurol. 1990;27:573–574. doi: 10.1002/ana.410270519. [DOI] [PubMed] [Google Scholar]
- 4.Fortin D, Cairncross G J, Hammond R R. Neurosurgery. 1999;45:1279–1291. doi: 10.1097/00006123-199912000-00001. [DOI] [PubMed] [Google Scholar]
- 5.Donahue B, Scott C B, Nelson J S, Rotman M, Murray K J, Nelson D F, Banker F L, Earle J D, Fischbach J A, Asbell S O, et al. Int J Radiat Oncol Biol Phys. 1997;38:911–914. doi: 10.1016/s0360-3016(97)00126-0. [DOI] [PubMed] [Google Scholar]
- 6.Ino Y, Zlatescu M C, Sasaki H, Macdonald D R, Stemmer-Rachamimov A O, Jhung S, Ramsay D A, von Deimling A, Louis D N, Cairncross J G. J Neurosurg. 2000;92:983–990. doi: 10.3171/jns.2000.92.6.0983. [DOI] [PubMed] [Google Scholar]
- 7.Nakagawa Y, Perentes E, Rubinstein L J. Acta Neuropathol (Berlin) 1986;72:15–22. doi: 10.1007/BF00687942. [DOI] [PubMed] [Google Scholar]
- 8.Sung C C, Collins R, Li J, Pearl D K, Coons S W, Scheithauer B W, Johnson P C, Yates A J. Glycoconj J. 1996;13:433–443. doi: 10.1007/BF00731476. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy P G, Watkins B A, Thomas D G, Noble M D. Neuropathol Appl Neurobiol. 1987;13:327–347. doi: 10.1111/j.1365-2990.1987.tb00190.x. [DOI] [PubMed] [Google Scholar]
- 10.Szymas J, Wajgt A. Neuropatol Polska. 1985;23:239–246. [PubMed] [Google Scholar]
- 11.Landry C F, Verity M A, Cherman L, Kashima T, Black K, Yates A, Campagnoni A T. Cancer Res. 1997;57:4098–4104. [PubMed] [Google Scholar]
- 12.Reifenberger G, Szymas J, Wechsler W. Acta Neuropathol (Berlin) 1987;74:105–123. doi: 10.1007/BF00692841. [DOI] [PubMed] [Google Scholar]
- 13.Kurpad S N, Zhao X G, Wikstrand C J, Batra S K, McLendon R E, Bigner D D. Glia. 1995;15:244–256. doi: 10.1002/glia.440150306. [DOI] [PubMed] [Google Scholar]
- 14.Noble M, Gutowski N, Bevan K, Engel U, Linskey M, Urenjak J, Bhakoo K, Williams S. Glia. 1995;15:222–230. doi: 10.1002/glia.440150304. [DOI] [PubMed] [Google Scholar]
- 15.Shoshan Y, Nishiyama A, Chang A, Mork S, Barnett G H, Cowell J K, Trapp B D, Staugaitis S M. Proc Natl Acad Sci USA. 1999;96:10361–10366. doi: 10.1073/pnas.96.18.10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raff M C, Miller R H, Noble M. Nature (London) 1983;303:390–396. doi: 10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- 17.Lu Q R, Yuk D, Alberta J A, Zhu Z, Pawlitzky I, Chan J, McMahon A P, Stiles C D, Rowitch D H. Neuron. 2000;25:317–329. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- 18.Zhou Q, Wang S, Anderson D J. Neuron. 2000;25:331–343. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- 19.Takebayashi H, Yoshida S, Sugimori M, Kosako H, Kominami R, Nakafuku M, Nabeshima Y. Mech Dev. 2000;99:143–148. doi: 10.1016/s0925-4773(00)00466-4. [DOI] [PubMed] [Google Scholar]
- 20.Kleihues P, Burger P C, Collins V P, Newcomb E W, Ohgaki H, Cavenee W K. In: World Health Organization Classification of Tumors of the Central Nervous System. Kleihues P, Cavenee W K, editors. Lyon, France: International Agency for Research on Cancer (IARC))/World Health Organization (WHO); 2000. pp. 29–39. [Google Scholar]
- 21.Wechsler-Reya R J, Scott M P. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 22.Wallace V A. Curr Biol. 1999;9:445–448. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- 23.Dahmane N, Ruiz-i-Altaba A. Development (Cambridge, UK) 1999;126:3089–3100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- 24.Hahn H, Wojnowski L, Specht K, Kappler R, Calzada-Wack J, Potter D, Zimmer A, Muller U, Samson E, Quintanilla-Martinez L. J Biol Chem. 2000;275:28341–28344. doi: 10.1074/jbc.C000352200. [DOI] [PubMed] [Google Scholar]
- 25.Raffel C, Jenkins R B, Frederick L, Hebrink D, Alderete B, Fults D W, James C D. Cancer Res. 1997;57:842–845. [PubMed] [Google Scholar]
- 26.Kamitani H, Masuzawa H, Sato J, Kanazawa I. J Neurol, Sci. 1988;83:219–225. doi: 10.1016/0022-510x(88)90070-6. [DOI] [PubMed] [Google Scholar]
- 27.Reifenberger G, Kros J M, Berger P C, Louis D N, Collins V P. In: World Health Organization Classification of Tumors of the Central Nervous System. Kleihues P, Cavenee W K, editors. Lyon, France: International Agency for Research on Cancer (IARC)/World Health Organization (WHO); 2000. , pp. 56–67. [Google Scholar]
- 28.Holland E C, Li Y, Celestino J, Dai C, Schaefer L, Sawaya R A, Fuller G N. Am J Pathol. 2000;157:1031–1037. doi: 10.1016/S0002-9440(10)64615-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doetsch F, Caille I, Lim D A, Garcia-Verdugo J M, Alvarez-Buylla A. Cell. 1999;97:703–716. doi: 10.1016/s0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 30.Coons S W, Johnson P C, Scheithauer B W, Yates A J, Pearl D K. Cancer. 1997;79:1381–1393. doi: 10.1002/(sici)1097-0142(19970401)79:7<1381::aid-cncr16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 31.Cairncross J G, Ueki K, Zlatescu M C, Lisle D K, Finkelstein D M, Hammond R R, Silver J S, Stark P C, Macdonald D R, Ino Y, et al. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- 32.Ino Y, Betensky R A, Zlatescu M C, Sasaki H, Macdonald D R, Stemmer-Rachamimov A O, Ramsay D A, Cairncross J G, Louis D N. Clin. Cancer Res. 2001. [PubMed] [Google Scholar]
- 33.Wang J, Jani-Sait S N, Escalon E A, Carroll A J, de Jong P J, Kirsch I R, Aplan P D. Proc Natl Acad Sci USA. 2000;97:3497–3502. doi: 10.1073/pnas.97.7.3497. [DOI] [PMC free article] [PubMed] [Google Scholar]