Abstract
Contractile forces eliminate cell contacts in many morphogenetic processes. However, mechanisms that balance contractile forces to promote subtler remodeling remain unknown. To address this gap, we investigated remodeling of Drosophila eye lattice cells (LCs), which preserve cell contacts as they narrow to form the edges of a multicellular hexagonal lattice. We found that during narrowing, LC-LC contacts dynamically constrict and expand. Similar to other systems, actomyosin-based contractile forces promote pulses of constriction. Conversely, we found that WAVE-dependent branched F-actin accumulates at LC-LC contacts during expansion and functions to expand the cell apical area, promote shape changes, and prevent elimination of LC-LC contacts. Finally, we found that small Rho GTPases regulate the balance of contractile and protrusive dynamics. These data suggest a mechanism by which WRC-based F-actin dynamics antagonize contractile forces to regulate cell shape and tissue topology during remodeling and thus contribute to the robustness and precision of the process.
Keywords: Epithelial morphogenesis; cell shape change; actomyosin contractility; non-muscle Myosin II; SCAR; Abi; WAVE regulatory complex; Arp2/3 complex; branched F-actin; small Rho GTPases; Rho1; Rac1; phosophoinositides; PI(3,4,5)P3
eTOC Blurb
During eye morphogenesis, lattice cells dramatically narrow, while maintaining contacts with neighboring cells. Here, Del Signore et al. demonstrate that localized pulsed actomyosin contractions promote narrowing, while pulsed assembly of WAVE-dependent branched F-actin transiently expands the narrowing cell to counter contractile forces and maintain cell-cell contacts during tissue remodeling.
Introduction
During development epithelial sheets undergo extensive remodeling to generate the intricate morphology of tissues and organs. Epithelial remodeling results from coordinated changes in cell shape, rearrangements of cell contacts, cell proliferation and cell death (Heisenberg and Bellaiche, 2013), and mechanical forces that coordinate these cell behaviors (Lecuit et al., 2011). To understand the underlying mechanisms we must determine the origin of the forces regulating these behaviors, and the mechanisms that regulate the generation and transmission of mechanical forces in space and time (Blanchard and Adams, 2011).
Epithelial remodeling is often powered by the assembly, activation and contraction of actomyosin networks (Levayer and Lecuit, 2012). During epithelial remodeling, MyoII structures assemble in pulses and are subsequently disassembled or remodeled (Gorfinkiel and Blanchard, 2011; Mason and Martin, 2011). Pulsed MyoII contractions drive a range of processes including the establishment of cell polarity (Munro et al., 2004), apical constriction (Azevedo et al., 2011; Blanchard et al., 2010; David et al., 2010; Martin et al., 2009; Solon et al., 2009), oocyte elongation (He et al., 2010) and tissue elongation (Fernandez-Gonzalez and Zallen, 2011; Kim and Davidson, 2011; Rauzi et al., 2010; Sawyer et al., 2011; Shindo and Wallingford, 2014; Skoglund et al., 2008). In many of these cases actomyosin contractility irreversibly eliminates cell-cell contacts, and it remains particularly unclear how cells might counterbalance contractile force to control more nuanced morphogenetic processes that depend primarily on cell shape changes.
Protrusive F-actin networks provide one potential mechanism to counteract contractile force during tissue remodeling (Betson et al., 2002; Nakagawa et al., 2001; Noren et al., 2001; Perez et al., 2008; Verma et al., 2004; Yamada and Nelson, 2007; Yamazaki et al., 2007). One candidate to regulate such force is the pentameric WAVE regulatory complex (WRC) (Pollitt and Insall, 2009; Takenawa and Suetsugu, 2007). Following activation by upstream signals (Chen et al., 2014; Eden et al., 2002; Ismail et al., 2009; Koronakis et al., 2011; Lebensohn and Kirschner, 2009; Mendoza, 2013; Oikawa et al., 2004), the WRC activates the Arp2/3 complex to generate a branched F-actin network that can push on and expand the cell surface (Pollard and Borisy, 2003). The WRC plays important roles in multiple developmental processes. During gastrulation in mouse, oocyte elongation in flies and ventral enclosure in worms the WRC localizes to the basal cell surface to promote collective cell migration (Barlan et al., 2017; Cetera et al., 2014; Patel et al., 2008; Rakeman and Anderson, 2006; Soto et al., 2002; Squarr et al., 2016). In brain development in mice and invagination of endodermal lineage cells in worms the WRC localizes to apical junctions to modulate cell adhesion and adherens junctions (AJs) remodeling (O’Leary et al., 2017; Sullivan-Brown et al., 2016). However, in each of these developmental processes, the molecular mechanisms by which the WRC remodels cell contacts remain poorly understood, and it is unknown how protrusive branched F-actin networks might interact with contractile actomyosin networks to promote tissue remodeling.
The apical epithelium of the fly eye is an established model with which to investigate the biochemical and physical basis of epithelial morphogenesis (Cagan, 2009; Carthew, 2007). It consists of ~800 photoreceptor clusters each capped by four cone cells, and surrounded by two large semi-circular 1° cells. Each of these units is surrounded by a single row of pigment cells known as lattice cells (LCs). Initially, the LCs arrange between ommatidia in a single row of largely isometric cells by cell intercalation (Bao and Cagan, 2005; Hayashi and Carthew, 2004). As development proceeds, excess LCs die and delaminate from the lattice, 2° LCs narrow and elongate to form the rectangular edges of the lattice and 3° LCs and three sensory bristles isometrically compact to form the vertices (Fig. 1A–B, Movie 1) (Cagan and Ready, 1989). Here, we investigated how 2° and 3° cells could undergo such elaborate shape changes while maintaining the precise topology required to form the hexagonal lattice. We provide evidence that pulses of MyoII accumulation promote specific shape changes of LCs. Conversely, we find that pulses of F-actin branching are required to limit contractile forces both to generate proper cell shape changes and to maintain cell-cell contacts and proper cell packing. Our data suggest that a balance between two dynamically opposing contractile and protrusive cytoskeletal networks affects the shape changes of the LCs and overall tissue topology.
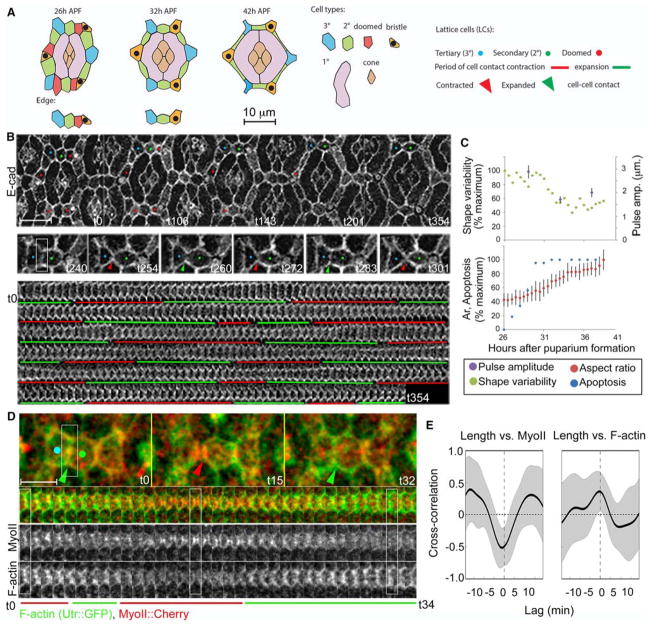
Figure 1. Fluctuations of LC-LC contact length correlate with shape changes and delamination of doomed LCs.
(A) Left: Schematic of an ommatidium (top) and a lattice edge (bottom) at 26, 32 and 42h APF. Cell types are color-coded. Right: Key to identifying LC types, periods of expansion and contractions of LC-LC contacts and contracted or expanded cell-cell contacts in this and subsequent Figures. (B) Upper panel: Snapshots from a time lapse of the apical epithelium of the fly eye marked with E-cad:: GFP, from 26h to 32h after puparium formation (APF) (t0=26h APF; Movie 1). Middle panel: A dynamic cell contact between 3° (left) and a 2° (right) LCs at ~30h APF that contracts (red arrowheads) and expands (green arrowheads) repeatedly. Lower panel: A kymograph of the LC-LC contacts shown above. Note repeating expansion (demarcated with green bars) and contraction (red bars) of the contact. (C) Quantification of the amplitude of contact fluctuations (purple), relative to cell shape variability (green), cell aspect ratio (red) and apoptosis (blue) in the lattice. (D) F-actin (green) and MyoII (red) dynamics. (E) Time-shifted Pearson correlation shows a negative correlation between contact length and MyoII levels (R=−.52, p<.001 from 0; time shift −1.0 +/− 2.3 min, p>.05 from 0) and positive correlation between contact length and F-actin (R=.36, p<.001 from 0; time shift −1.6 +/− 3.87 min, p>.05 from 0; n=21 contacts pooled from three eyes). In this and subsequent correlation plots: black lines mark the mean correlation, gray bands mark standard deviation. Scale bar = 10 μm in B, 5 μm in D.
Results
LC-LC contacts lengthen and shorten repeatedly during epithelial remodeling of the fly eye
From 26 to 42 hours after puparium formation (APF), the apical epithelium of the fly eye is remodeled primarily by cell shape changes and apoptosis of superfluous LCs (See schematics in Fig. 1A) (Cagan and Ready, 1989). To understand the underlying physical mechanisms we followed this process by live imaging of apical cell outlines labeled with GFP-tagged α-Catenin (α-Cat) or E-cadherin (E-cad) (Lye et al., 2014; Oda and Tsukita, 1999). We found that contacts connecting LCs contracted and expanded repeatedly as the LCs changed their shape (Fig. 1B, Movie 1). Each expansion or contraction of LC-LC contact lasted 14.2 +/− 3.0 min on average. The amplitudes of contact fluctuations were highest at 28–30 hours APF when the epithelium was most actively remodeling, as measured by a low average cell aspect ratio, and high cell shape variability and apoptosis (Fig. 1C). Contact pulse amplitudes decreased mildly at 32–34 and 36–38 hours APF as the cells progressively assumed their final shapes (19.7% over mean contact length at 28–30h, 14.9% at 32–34hr, 15.7% at 36–38h). Together, these data indicate that fluctuations of cell contact length correlate with the shape changes of LCs and lattice remodeling.
MyoII and F-actin accumulate dynamically along shortening and lengthening LC-LC contacts, respectively
To understand the forces that promote fluctuations in cell-cell contact length during lattice remodeling, we live imaged cytoskeletal dynamics using GFP-tagged myosin regulatory light chain (hereafter referred to as MyoII) (Martin et al., 2009) and F-actin, using lifeact::Ruby (Riedl et al., 2008) or a GFP-tagged actin-binding domain of Utrophin (Burkel et al., 2007; Rauzi et al., 2010). As expected, we found that MyoII levels increased along shortening contacts and decreased along lengthening contacts (Fig. 1D–E, S1A). Maximal accumulation of MyoII coincided with maximal shortening of the LC-LC contacts (Fig. 1E; R=−.52, p<.001 from 0; time shift −1.0 +/− 2.3 min, p>.05 from 0). Surprisingly, we found that F-actin levels increased along lengthening LC-LC contacts (Fig. 1D, S1A), and maximal accumulation of F-actin correlated with maximal lengthening of the LC-LC contacts (Fig. 1E; R=.36, p<.001 from 0; time shift −1.6 +/− 3.87 min, p>.05 from 0). We did not measure a significant correlation between F-actin and MyoII levels (not shown). However, we did observe that the contacts with the strongest correlation between F-actin or MyoII and contact length tended to have a stronger negative correlation between F-actin and MyoII (Fig. S1B). Together, these observations suggested opposite roles for MyoII and F-actin dynamics in this process, which we investigated below in further detail.
Rok and MyoII accumulate along shortening LC-LC contacts and myoII is required to preferentially shorten LC-LC contacts and compact the 3° LCs
We next analyzed the dynamics of the upstream MyoII regulator Rok relative to changes in levels of MyoII, F-actin, and cell contact length (Fig. 2A–F, S1C). Rok accumulated dynamically along shortening LC-LC contacts (Fig. 2A–C; R= −.50, p<.01), and positively correlated with MyoII levels (Fig. 2A–C; R=.79, p<.001). Consistent with the lack of direct relationship between F-actin and MyoII, we did not measure a significant correlation between Rok and F-actin accumulation (Fig. 2F). As in our previous experiment (Fig. 1E), F-actin levels correlated with contact length (R=.46, p<.001). Peak F-actin accumulation preceded maximal contact length by 2.5 +/− 3.12 min (p<.001), a shift which was not significantly different than that observed in Fig. 1E (p>.3). Neither Rok nor MyoII accumulation significantly preceded maximal contact contraction, reflecting the high variability in time delay.
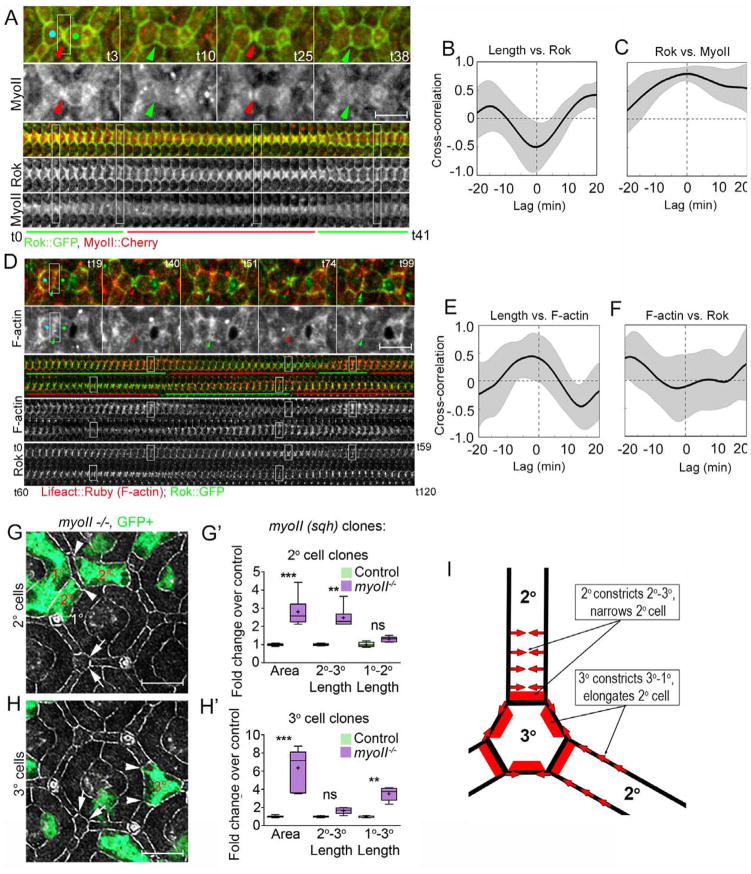
Figure 2.
MyoII accumulates with Rok along constricting LC-LC contacts and functions to preferentially constrict 2°-3° and 1°-3° cell contacts. (A, D) Dynamics and (B–C, E–F) cross correlation of Rok::GFP (green) relative to MyoII::Cherry (red, A–C) and F-actin, marked with Lifeact::mRuby, (red, D–F). (G–H′) Shape change analysis of single myoII mutant cells (green). (A–C) Rok accumulated along shortening LC-LC contacts (R=−.50, time shift .7+/− 2.6 min), while F-actin accumulated along lengthening contacts (R=.45, time shift −1.8 +/− 3.79 min; Movie 2; time correlations quantified in B–C, n=18 contacts from one representative eye). (D) Rok and MyoII accumulated with similar dynamics along constricting LC-LC contacts, and positively correlated with one another (E–F) Length vs. Rok (R=−.5, time shift= .7+/−2.62 min; Rok vs. Sqh R=.79, time shift= 0 +/− 4.47 min; Movie 2). (G, H) Shape changes and (G′, H′) shape analysis of genetically marked myoII mutant (G–G′) 2° and (H–H′) 3° cells. (G) Arrowheads demarcate expanded 2°-3° contact, arrows normal 2°-3° contact. (H) Arrowheads demarcate expanded 1°-3° contact, arrows normal 1°-3° contact. (F′, G′) Box-and-whisker plots in this and subsequent Figures: Median - horizontal band in the box; + indicates the mean; First and third quartile of data points - bottom and top of the boxes; Ends of the whiskers indicate 95% confidence interval; P-values: * p<.05, ** = p<.01, *** = p<.001, ns = not significant. (I) A model of the role of myoII in lattice remodeling. Red bars - sites of preferential accumulation and requirement for myoII for shortening cell-cell contacts; Red arrows - the direction of cell deformation. Scale bars = 5 μm
To determine whether polarized MyoII accumulation affects the shape of LCs, we created positively marked myoII mutant cells using the MARCM technique (Lee and Luo, 2001) and examined the changes in apical area and aspect ratio of mutant cells relative to wild type neighbors. The apical area of 1°, 2° and 3° myoII mutant cells expanded compared to wild type cells, as described previously (Warner and Longmore, 2009b). Consistent with the polarized accumulation of MyoII along LC-LC contacts, we further found that the expansion of the myoII mutant LCs was not uniform. Mutant 2° cells, preferentially expanded their LC-LC contacts relative to LC-1° contacts resulting in wider, more isometric 2° cells (Fig. 2G–G′). By contrast, myoII mutant 3° cells preferentially expanded their 1°-3° cell contacts, causing shortening of adjacent wild type 2° cells (Fig. 2H–H′). The polarized shape changes of myoII mutant LCs indicate that MyoII is required asymmetrically in LCs to control cell shape: MyoII in 3° cells contributes preferentially to the shortening of the 1°-3° contacts and non-autonomously to the lengthening of the 1°-2° contacts, while MyoII in 2° cells contributes preferentially to the shortening of the 2°-3° contacts (see model in Fig. 2I). As these contacts are shortened, but not eliminated, by actomyosin contractility, we next asked whether the pulses of F-actin might regulate contact maintenance during shape change of LCs.
Branched F-actin regulators accumulate along expanding LC-LC contacts
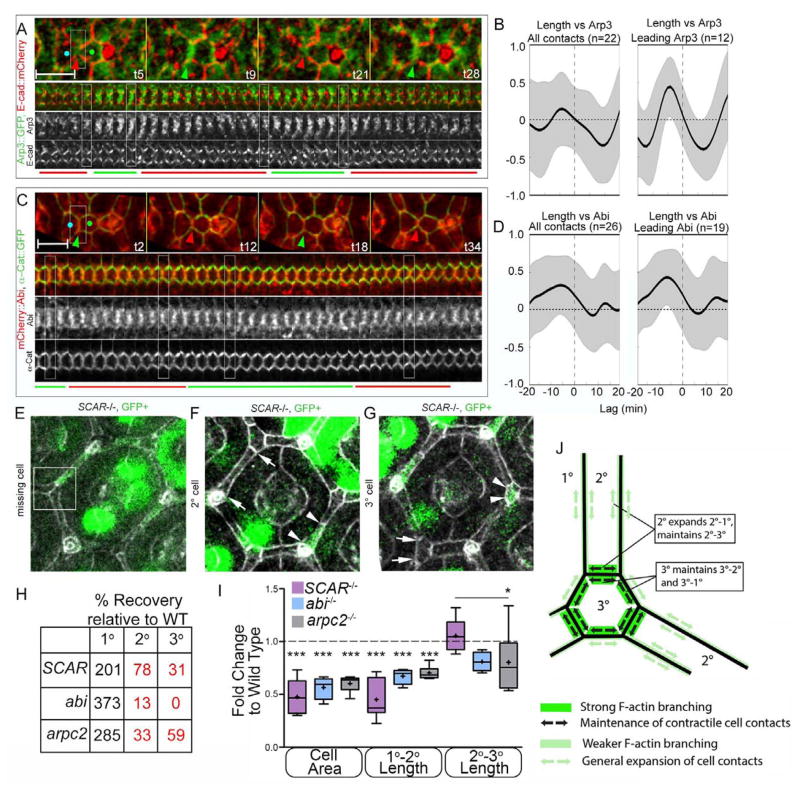
Branched F-actin networks can generate protrusive forces at the cell surface (Pollard and Borisy, 2003). To test whether F-actin pulses at expanding LC-LC contacts consisted of branched F-actin, we examined the localization and dynamics of GFP-tagged Arp3, a subunit of the Arp2/3 complex (Rajan et al., 2009). Arp3 localized to LC-LC contacts (Fig. S2A), and exhibited two distinct patterns of dynamics. Though overall there was no correlation between Arp3 and contact length, we detected two subpopulations of contacts: in one group (12/20), Arp3 accumulation preceded (time shift 5.0 +/− 2.3 min, p<.001 from 0) and correlated positively with contact expansion (Fig. 3A–B; R=.44, p<.001 from 0). In the remaining contacts (8/20), there was no significant relationship between contact length and Arp3 (Fig. S2D). Wasp and the WRC are the major Arp2/3 activators at the cell surface (Pollitt and Insall, 2009; Tajiri et al.; Takenawa and Suetsugu, 2007). To determine whether these proteins contribute to actin dynamics along LC-LC contacts, we examined their subcellular distribution. While Wasp localized in a diffuse pattern that did not coincide with cell-cell contacts (not shown), the WRC subunits SCAR and Abi localized with F-actin preferentially along LC-LC contacts (Fig. S2B–C). On average, Abi accumulation (Fig. 3C–D) preceded (−4.6+/−6.09 min, p<.001 from 0) and positively correlated with contact length (R=.33, p<.001 from 0). Similar to Arp3, we detected a subset of contacts (19/26) that preceded contact expansion with a stronger positive correlation (R=.47, p<.001 from 0; Fig. 3C–D), while the remainder exhibited no clear relationship (Fig. S2D). The long time delay between maximal Abi-Arp3 accumulation and contact expansion suggests that the targeting of these regulators to LC-LC contacts sets the stage for actin to assemble in branched networks. Overall, Abi and Arp3 localization and dynamics suggested that the pulsed accumulation of the WRC and the Arp2/3 complex at LC-LC contacts promotes assembly of branched F-actin that drives the lengthening of the contacts.
Figure 3.
Protrusive proteins accumulate preferentially along lengthening 2°–3° contacts and are required to promote the expansion of cells apical area and inhibit delamination of LCs. (A–D) Dynamics of branched F-actin regulators. (A) Arp3 (green) localizes to LC-LC contacts (AJ labeled with E-Cad:: mCherry). Overall, no correlation between Arp3 and contact length was measured (B, left). However, a subset of contacts in which Arp3 accumulation preceded contact expansion (12/20 contacts) exhibited a positive correlation (R=.44) with a time shift of 5.0 +/− 2.3 min. (C) mCherry::Abi (red) localizes to AJs marked with E-cad::GFP (green). (D) Abi levels positively correlated with contact length (D, left). Among the subset of contacts in which Abi preceded contact lengthening (19/26 contacts), Abi correlated with contact length (R=.48) with a time shift of 7.2 +/− 3.8 min. (E–I) Morphological analysis of SCAR, abi, and arpc2 mutant clones. (E–G) Eyes bearing positively marked SCAR mutant LCs exhibit multiple defects, including missing 2° and 3° cells (E, quantified in H), constricted and shortened 2° cells (F, quantified in I), and constricted tertiary cells (G). Comparative analysis of the (H) recovery and (I) shape of SCAR, abi and arpC2 mutant cells. (J) A model of the role of the WRC in lattice remodeling. Green bars indicate sites of preferential accumulation and requirement for SCAR and abi for lengthening cell contacts. Black arrows indicate the direction of cell deformation and role for SCAR and abi in contact maintenance. Green arrows indicate a role for SCAR, abi and arpC2 in contact lengthening (model also relies on Fig. 5, Movie 1). Scale bars = 5 μm
Branched F-actin regulators promote the expansion of cells apical perimeter and preferentially lengthen LC-1° contacts
To determine whether the WRC and the Arp2/3 complex regulate cell shape, we quantified cell shape of single SCAR, abi, and arpc2 mutant cell (Fig. 3E–G, I and Fig. S3). The recovery of mutant cells was lower compared to the recovery of wild type cells, and the distribution of clones was altered, with a greater percentage of 1° cells and a substantial loss of 2° and 3° cells (Fig. 3H). Consistent with this distribution, we occasionally found areas associated with SCAR clones in which tissue topology was disrupted, with 2° cells inappropriately meeting at a 3° cell niche (Fig. 3E, boxed area). Recovered mutant cells were significantly smaller and more isometric than nearby wild-type cells, driven primarily by shortening of the normally elongated 1°-2° cell contact (Fig. 3F–G, quantified in I). The length of LC-LC contacts was unaffected (Fig. F–G, 3I), suggesting that SCAR activity in adjacent wild type cells is sufficient to lengthen and maintain LC-LC contact in the mutant cell (See model in Fig. 3J). Importantly, we observed no changes in the levels of phospho-MyoII (p-MyoII, Fig. S3B) or adhesion molecules (Fig. S4), suggesting that constriction in SCAR clones is not due to altered MyoII activation or cell adhesion, respectively. Overall, these findings suggest that the WRC and F-actin branching are required to expand the apical cell area, regulate cell shape, and maintain tissue topology.
Branched F-actin regulators are required to maintain LC-LC contacts and inhibit formation of aberrant contacts between adjacent ommatidia
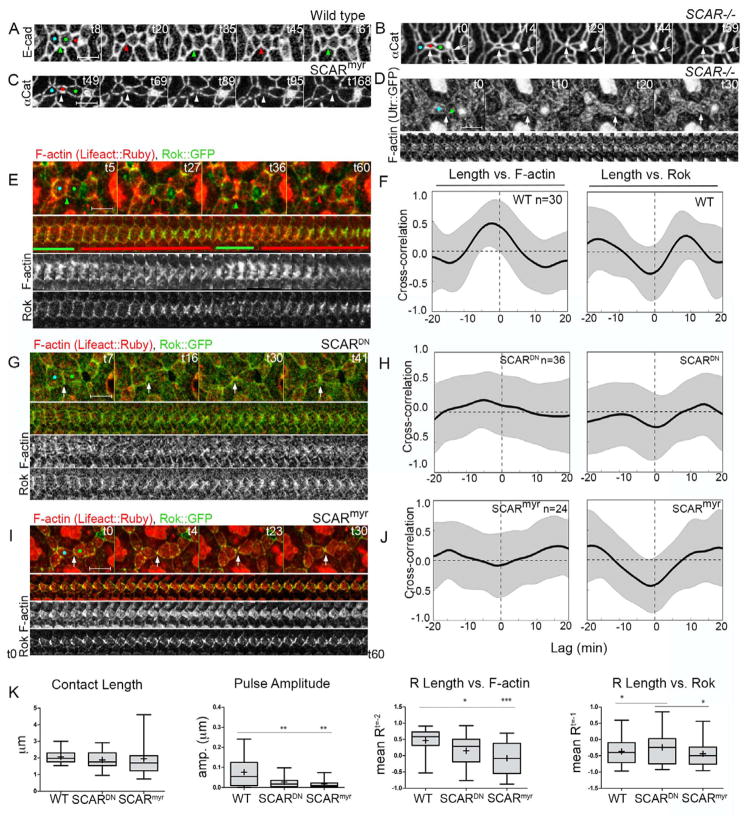
To determine how loss of branched F-actin regulators affects morphological dynamics, we generated eyes entirely mutant for SCAR, abi and arpC2 using the Eyeless-GAL4, UAS-FLP/FRT (EGUF) technique (Stowers and Schwarz, 1999). Adult mutant eyes were rough and small (Fig. S5A), suggesting epithelial remodeling defects. We followed epithelial remodeling in SCAR mutant eyes from 26 to 34h APF (Fig 4B, S5B–C, Movie 1). In wild type eyes by 26hr, LCs have intercalated to arrange in a single file around 1° cells, and excess cells have begun to delaminate. This process was delayed in SCAR mutant eyes (not shown), consistent with an earlier requirement for actin dynamics in cell intercalation (Johnson et al., 2011; Johnson et al., 2008). By 30h APF, intercalation was complete in SCAR mutant eyes, though LCs appeared constricted compared to wild type, and a subset of LC-LC contacts separated either transiently or permanently (Fig 4B, S5C, Movie 1). In contrast to wild-type eyes, in which cells smoothly re-establish cell contacts following delamination of excess cells, SCAR mutant LCs often failed to reestablish contacts properly, resulting in aberrant 1°-1° contacts or transient gaps in the contours of the AJs (Figs. 4B, S5C, arrowheads and arrows, respectively; Movie 1). Similar phenotypes, including defects in arrangement and shape of LCs and remodeling of LC-LC contacts were observed in arpC2 mutant eyes (Fig. S5D) and in eyes expressing dominant negative GFP-tagged SCAR protein (SCARDN, Fig. S5E) (Gildor et al., 2009). Interestingly, we found qualitatively similar phenotypes in eyes broadly expressing a myristoylated-SCAR (SCARmyr) protein (Fig. 4C) (Stephan et al., 2011). The SCARmyr protein localized broadly at both the apical cell surface and at AJs (Fig. S6A), and caused increased accumulation of F-actin at these sites (Fig. S6B), suggesting that polarized and/or dynamic activation of the WRC and Arp2/3 complex along LC-LC contacts is required for proper tissue remodeling.
Figure 4.
SCAR promotes protrusive F-actin dynamics to maintain LC-LC contacts. (A–C) Live imaging with either E-cad::GFP or α-Cat::Venus (Movie 1). Broad (B) loss of SCAR function (C) or expression of SCARmyr in the eye caused aberrant loss of LC-LC contacts (white arrowheads) following delamination of LCs (red). (D) Broad loss of SCAR caused decreased F-actin accumulation (Utr::GFP, Movie 3) at LC-LC contacts. (E–K) Comparison of F-actin (lifeact::Ruby, red) and Rok (Rok::GFP, green) dynamics following SCAR manipulation. (E–F) Wild type eyes exhibited a positive correlation between F-actin and contact length (R=.46; shift −2.5 +/− 3.1 min; n=30 contacts from 3 eyes). Broad expression of either (G–H, K) SCARDN or (I–J, K) SCARmyr abolished the correlation between F-actin and contact length (SCARDN: R=.15; shift=−1.5 +/− 6.6 min; n=36 contacts from 3 eyes; SCARmyr: R=−.09; shift=−1.3+/−5.9min; n=24 contacts from 3 eyes). Rok correlation was mildly affected only in SCARDN eyes. (K) Quantification of morphological and molecular dynamics in E–J.
Branched F-actin regulators promote pulsed F-actin accumulation and lengthening of LC-LC contacts
We hypothesized that the WRC and the Arp2/3 complex promote the assembly of protrusive branched F-actin along LC-LC contacts as a counterbalance to high tension to prevent the loss of LC-LC contacts. To examine the effects of SCAR on contractile and protrusive dynamics simultaneously, we examined F-actin and Rok dynamics in eyes expressing SCARDN or SCARmyr proteins (Fig. 4E–K, Movie 2). Each SCAR manipulation significantly decreased the amplitude of contact dynamics relative to control eyes, though the average contact length was unchanged (Fig. 4K). For this analysis, we selected only contacts that remained intact in order to identify changes in actin dynamics that precede contact separation, which likely underestimates any potential difference in contact length. Despite focusing on these more mildly affected contacts, we did detect changes in the relationship between F-actin accumulation and contact length. Expression of SCARDN abolished the normally positive correlation between F-actin and contact length (Fig. 4G–H, K), while SCARmyr caused it to become negative (Fig. 4I–J, K). By contrast, the normally negative correlation between Rok and contact length was somewhat diminished by SCARDN and unchanged by SCARmyr (Fig. 4G–H and I–J, respectively, quantification in K). Importantly, we also observed no change in the negative correlation between MyoII and contact length (Fig. S6F) with either manipulation, further suggesting that morphological defects are due to altered F-actin dynamics. Notably, the stronger imbalance of contractile and protrusive cytoskeletal dynamics in SCARmyr eyes relative to SCARDN correlated with more severe cell shape and topology phenotypes in SCARmyr eyes. Supporting these findings, we found decreased levels of F-actin (Fig. 4D, Movie 3), but no changes in MyoII (Fig. S6D) in SCAR mutant eyes compared to wild type (Fig. S6C). Finally, we observed similar changes in distribution of F-actin and MyoII in arpC2 mutant eyes (Fig. S6E). Overall, these data strongly suggest that SCAR promotes protrusive F-actin dynamics to actively lengthen LC-LC contacts to counterbalance the capacity of contractile forces to shorten and disassemble the LC-LC contact and alter epithelial cell shape and tissue topology.
Small Rho GTPases and phosphoinositide PI(3,4,5)3 accumulate dynamically along LC-LC contacts
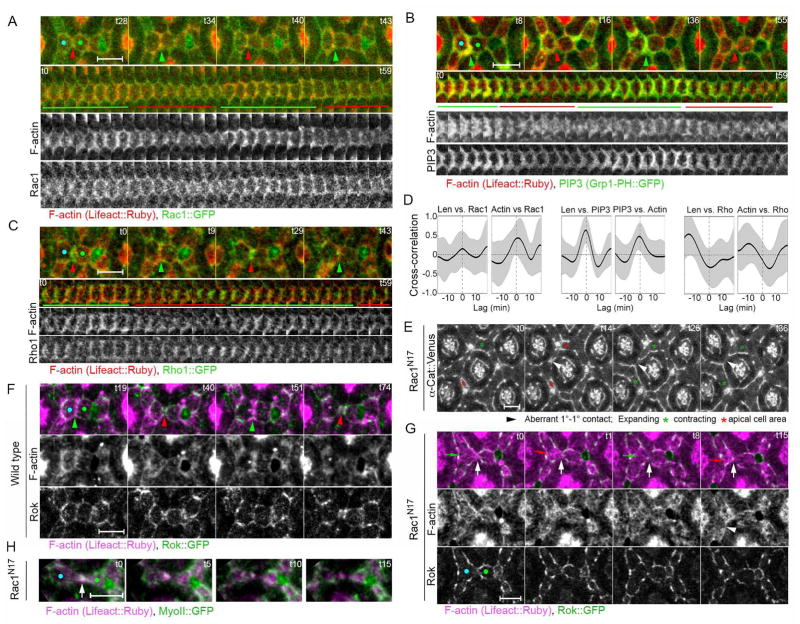
The small Rho GTPase Rac1 and the phosphoinositide PI(3,4,5)P3 can act synergistically to activate the WRC (Chen et al., 2014; Eden et al., 2002; Ismail et al., 2009; Koronakis et al., 2011; Lebensohn and Kirschner, 2009; Mendoza, 2013; Oikawa et al., 2004). To determine whether the same upstream signals can activate the WRC in vivo during lattice remodeling we examined Rac1 dynamics using GFP-tagged Rac1 and PI(3,4,5)3 dynamics using the PI(3,4,5)3 binding PH domain of GRP1 tagged with GFP relative to F-actin tagged with Lifeact::mRuby. We found that Rac1 and PI(3,4,5)3 both accumulate at LC-LC contacts, and their dynamics correlate strongly with F-actin accumulation (Fig. 5A–B, D; Movie 4). Further, we found that PI(3,4,5)P3 correlates positively with contact length (R=.62, p<.001 from 0), though Rac1 does not (R=.14, p>.05). We also examined the localization and dynamics of the small GTPase Rho1, which activates Rok to promote phosphorylation and activation of MyoII (Kimura et al., 1996; Mason et al., 2013). Similar to Rok, we found that Rho1 localizes to cell contacts and negatively correlates with contact lengthening (R=−.32, p<.05 from 0) and actin accumulation (Fig. 5C–D, Movie 4; R=−.35, p<.001 from 0). Taken together, these results raised the possibility that Rho GTPase and PI(3,4,5)P3 signaling may coordinate contractile and protrusive dynamics.
Figure 5.
Phosphoinositide PI(3,4,5)3 accumulate dynamically along LC-LC contacts, and small Rho GTPase signaling affects contractile and protrusive dynamics, cell contact length and lattice remodeling. (A–D) Live imaging of F-actin (Lifeact::Ruby, red) and upstream cytoskeletal regulators (green) in 28–30 hour APF eyes. (A) Rac1::GFP (green) and (B) GFP-tagged PI(3,4,5)3 binding PH domain of GRP1 accumulated along cell contacts and positively correlated with actin accumulation (red) (Rac1: R=.43; shift=45+/−210sec; n=8 contacts from 2 eyes; PI(3,4,5)P3: R=.47; shift=−1.4+/−3.1min n=18 contacts from 3 eyes). PI(3,4,5)P3, but not Rac1, positively correlated with contact length (R=.62; shift=−40+/−78sec). (C) Conversely, Rho correlated negatively with contact length and actin accumulation (length: R=−.32; shift=.5+/−3.7min; actin: R=−.35; shift=5.3+/−3.9min; n=12 contacts from 2 eyes). (D) Cross-correlation analysis of molecular dynamics in A–C. (E) Cell behavior in Rac1N17 expressing eyes. Stills from a time lapse of F-actin (Lifeact::Ruby, magenta) and Rok::GFP (green) in (F) control and (G) Rac1N17-expressing eyes (Movie 5). (F) Wild type eyes exhibit normal F-actin accumulation at expanded (green arrowheads) but not constricted (red arrowheads) contacts. (G) Rac1N17 expressing eyes exhibit loss of pulsed F-actin dynamics along LC-LC contact, as F-actin accumulated medioapically in constricting cells (red arrows), (H) MyoII accumulated with F-actin in separating LC-LC contacts in Rac1N17 eyes. Scale bars = 5 μm.
Small Rho GTPases signaling affects contractile and protrusive dynamics at LC-LC contacts
To determine whether small Rho GTPase signaling affects the balance between contractile and protrusive dynamics, we expressed dominant negative Rac1N17 in the eye. This manipulation has previously been shown to likely disrupt both Rac1 and Rho1 signaling in this tissue (Warner and Longmore, 2009a, b). Rac1N17 resulted in transient discontinuities in E-cad distribution at the cell surface during lattice remodeling as previously described (Fig. 5E)(Bruinsma et al., 2007), and high amplitude fluctuations of the apical cell perimeter of LCs. During this process a subset of LC-LC contacts abruptly separated (Fig. 5E, arrowhead points to separating LC-LC contact). To determine whether these defects correlated with changes in pulsed cytoskeletal dynamics, we examined F-actin and Rok dynamics in Rac1N17 expressing eyes (Fig. 5G compared to wild type in 5F, Movie 5). In Rac1N17 expressing eyes, F-actin no longer accumulated at lengthening LC-LC contacts (Fig. 5G, Movie 5). Instead, F-actin and MyoII frequently colocalized in intense foci at constricting and aberrantly eliminated LC-LC contacts (Fig. 5H). Moreover, Rok localization was mislocalized from LC-LC contacts to subregions along 1°-LC (Fig. 5G). These changes in cytoskeletal organization, cytoskeletal dynamics and cell behavior suggest important roles for Rho GTPase signaling in controlling the balance between contractile actomyosin dynamics and protrusive branched F-actin dynamics during lattice remodeling.
Discussion
Branched F-actin promotes active contact expansion during epithelial remodeling
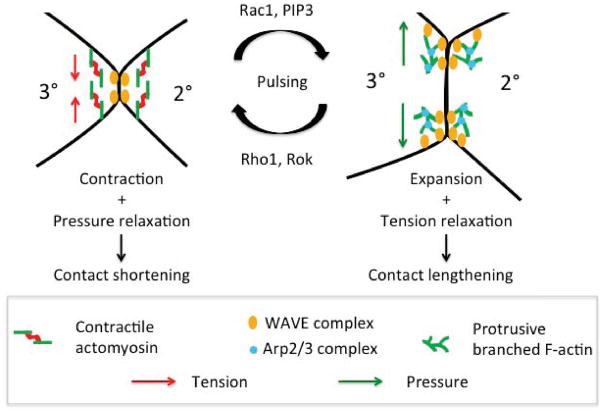
Cellular behaviors that mediate tissue morphogenesis involve both loss and gain of cell contacts and changes in length of cell contacts and apical cell perimeters. It had been generally assumed that following shortening or elimination of cell contacts by contractile force, the lengthening of new cell contacts results from passive relaxation of cell contacts in the direction of least resistance in the tissue. However, recent reports have identified mechanisms that actively promote expansion of cell contacts and apical area during epithelial tissue remodeling. During notum elongation, the phosphatase and tensin homolog PTEN decreases Rok levels and contractile force generation cell-autonomously to promote the lengthening of newly formed cell contacts (Bardet et al., 2013). During germ band extension, a decrease in MyoII levels along lengthening contacts coordinates with a dynamic increase in MyoII levels in neighboring cells to lengthen newly formed contacts (Collinet et al., 2015; Hara et al., 2016; Yu and Fernandez-Gonzalez, 2016). During radial cell intercalation in Xenopus, decreased junctional tension and Formin1-mediated F-actin assembly promote apical expansion (Sedzinski et al., 2016). Here, we identified a distinct actin-based mechanism by which cells lengthen cell contacts and counterbalance contractile forces to control cell shape (See model in Fig. 6). We found that actomyosin contractility generally constricts the apical cell area and preferentially the 2°-3° and 3°-1° contacts to control cell shape (See model in Fig. 2I). In turn, protrusive branched F-actin expands the apical cell perimeter and both the 2° and 3° cells cooperate to maintain the LC-LC contacts (See model in Fig. 3J). This interplay between actomyosin contractility and protrusive branched F-actin controls the shape of the cells, maintains cell connectivity and contributes to the resilience and precision of the process.
Figure 6.
Graphical summary of the control of LC-LC contact length during lattice remodeling. Pulsed MyoII accumulation along LC-LC contacts generates contractile force that drives polarized narrowing of 2° cells. Regulators of actomyosin contractility Rho1 and Rok accumulate dynamically with MyoII along shortening contacts suggesting that they integrate mechanical inputs to regulate actomyosin contractility. Opposing these contractile pulses, dynamic assembly of a branched F-actin network transiently lengthens LC-LC contacts to prevent loss and maintain topology of LC-LC contacts. WRC regulators Rac1 and PIP3 accumulate dynamically with F-actin suggesting that mechanisms exist to dynamically recruit and activate the WRC at LC-LC contacts.
Polarized contractile and protrusive forces contribute to shape changes of LCs
Previous experimental analyses (Bao and Cagan, 2005; Hayashi and Carthew, 2004) and biophysical modeling (Hilgenfeldt et al., 2008; Kafer et al., 2007) of epithelial morphogenesis in the fly eye suggested that cortical tension in the cells is uniform and the shape of the cells is regulated primarily by maximizing adhesion between cell types to produce more thermodynamically stable epithelial topology. Here, we find that contractile and protrusive cytoskeletal proteins accumulate in a polarized pattern preferentially along LC-LC contacts and act both autonomously and non-autonomously to regulate cell shape and epithelial morphology. It is yet unclear what cues organize these cytoskeletal dynamics in space. Several adhesion molecules, such as Hibris and Roughest, localize to a subset of the contacts and not others (Bao and Cagan, 2005). One possibility is that adhesion molecules that localize preferentially to LC-LC contacts could recruit the WRC or regulators of Rho family GTPases and/or PI(3,4,5)P3 production to locally modulate contractile and protrusive proteins along the LC-LC contacts.
Coordination of pulsatile contractile and protrusive dynamics in eye morphogenesis
The finding that polarized MyoII dynamics drive LC shape change is generally consistent with the role of MyoII dynamics in other systems, where pulsed accumulation of MyoII constricts the apical cell perimeter or cell-cell contacts to drive tissue level behaviors such as apical constriction and cell intercalation, respectively (Gorfinkiel and Blanchard, 2011; Mason and Martin, 2011). Interestingly, in these systems the pulse duration is in the range of 1–2 minutes, whereas we observed pulse durations greater than 10 minutes, with very weak periodicity. This is similar to the prolonged pulsed accumulation of basal MyoII in follicle cells during oocyte elongation (He et al., 2010). Importantly, these processes, eye development and oocyte elongation, differ from apical constriction and germ-band extension in that contractile contacts are preserved, instead of eliminated, suggesting different underlying mechanisms. Consistent with this, we also detect longer lags and high variability among and between molecular and morphological dynamics. These could reflect ‘competition’ between cytoskeletal architectures (Burke et al., 2014; Lomakin et al., 2015), and/or that multiple intrinsic pathways and external forces such as tension and compression may collaboratively regulate both the molecular and morphological dynamics described here.
This dynamic interplay may contribute to normal epithelial development in several ways. Robust contact dynamics could allow cells to coordinate their behavior with immediate neighbors to prevent local topological rearrangements and/or tearing (Jodoin et al., 2015). More broadly, pulsed dynamics might enable larger cohorts of cells to coordinately explore a range of energy landscapes in search of an optimal one. Prevailing models of epithelial remodeling suggest that the final topology represents a minimal interfacial energy state (Bao and Cagan, 2005; Hayashi and Carthew, 2004; Hilgenfeldt et al., 2008; Kafer et al., 2007). However, the acquisition of that topology must pass through energetically unfavorable states, and fluctuations of cell contact length could allow individual cell contacts to sense and respond to the overall tension landscape to achieve an energy minimum that is compatible with the molecular patterning of the tissue. Both AJs (Borghi et al., 2012; Dufour et al., 2013; le Duc et al., 2010; Twiss and de Rooij, 2013) and MyoII (Clark et al., 2007; Fernandez-Gonzalez et al., 2009) as well as the WRC as suggested in this study, are responsive to tension, supporting a mechanism for such coordination. The engagement of both contractile and protrusive proteins could enable epithelial cells to more robustly search for stable shapes to form the crystalline lattice of the fly eye.
STAR Methods
Contact for reagent and resource sharing
Further information and requests for reagents should be directed to and will be fulfilled by the Lead Contact, Victor Hatini (victor.hatini@tufts.edu).
Experimental model and subject details
Fly strains
Fly lines from the Bloomington Drosophila Stock Center were: UAS-Lifeact::Ruby, UAS-Arp3::GFP, ubi-Abi::mCherry, sqh-GFP::Rok, sqhAX3, sqh-Sqh::GFP, SCARΔ37 FRT40A, sqhAX3 FRT19A, GMR-GAL4, A>y>GAL4, UAS-GFP, Shg::mCherry, tGPH (tub-GRP1-PH::GFP), Rac1::GFP, UAS-Rac1N17, Rho1::GFP. Fly lines from the Kyoto stock center were α–Cat::Venus (CPTI002596), arp383F FRT40A, arp3515FCFRT40A and arpc2KG04658FRT40A. Only arpc2KG04658 mutant cells generated by the MARCM technique were recovered 2 days after clone induction. Additional stocks used were: sqh-sqh::mCherry and sqh-Utr::GFP, sqh-Sqh::mCherry (gift of A. Martin), abiD20 and UAS-myr-WAVE (gift of S. Bogdan), UAS-SCAR:: GFP (gift of E. Schejter), GMR-GAL4, UAS-α–Cat::GFP (gift of R. Cagan). The following stocks were created in this study: (1) GMR-GAL4, α–Cat::Venus, (2) UAS-Lifeact::Ruby; GMR-GAL4, (3) UAS-Lifeact:: GFP; sqh-Sqh::mCherry, GMR-GAL4, (4) UAS-Lifeact::Ruby; sqh-Rok::GFP, GMR-GAL4, (5) arpC2 KG04658 40AFRT; αCat::Venus, (6) SCARΔ37 FRT40A; αCat::Venus, (7) SCARΔ37 FRT40A; Utr::GFP (8) sqh-UtrABD::GFP; ubi-Abi::mCherry, (9) GMR-GAL4, UAS-αCat.GFP; Abi::mCherry, (10) Shg::Cherry; Arp3::GFP, (11) UAS-Lifeact::Ruby; tGPH, GMR-GAL4
Genetic analyses
Transgenes were expressed broadly in the eye with GMR-GAL4. The FLP-out/GAL4 technique was used to generate genetically marked clones that over express desired UAS transgenes (Ito et al., 1997). The FLP/FRT and MARCM techniques were used to generate genetically marked clones by mitotic recombination (Lee and Luo, 2001). The Hid/EGUF technique was used to generate eyes composed entirely of SCARD37, abiD20 or arpc2 KG04658 mutant cells (Stowers and Schwarz, 1999). Mitotic recombination in retinal progenitors rescued these eyes from Hid-mediated apoptosis by the loss of the GMR-Hid transgens from mutant cells. hsFLP, tub-GAL80 w 19AFRT; UAS-mCD8::GFP (Bloomington Stock Center) was used to generate sqh mutant clones by the MARCM technique; ywhsFLP, UAS-GFPnls, tub-GAL4; tub-GAL80 FRT40A (gift of G. Struhl) to generate SCAR and arpC2 KG04658 mutant clones, and y w hsFLP, UAS-GFPnls, tub-GAL4; 82BFRT tub-GAL80 (gift of G. Struhl) to generate abi mutant clones. GMR-Hid 40AFRT; ey-GAL4, UAS-Flp was used to generate SCARΔ37or arpC2KG0465 mutant eyes, and ey-GAL4 UAS-Flp; FRT82B GMR-Hid was used to generate abiD20 mutant eyes using the Hid/EGUF technique. Mitotic clones generated by the MARCM technique were induced by a heat shock for 1 hour at 37° during late third instar (120–144 hours). Flp-out clones were induced by a heat shock for 30 minutes at 34°C. There are no known differences in eye morphogenesis between males and females. Therefore, sex has not been factored in as a biological variable into the experiments.
Method detail
Immunofluorescence
White prepupae were selected and aged on glass slides in a humidifying chamber at 25°C. Pupal eyes were dissected in phosphate buffered saline, fixed for 35 minutes in 4% paraformaldehyde in PBS and stained with antibodies in PBS with 3% BSA, 0.3% Triton X-100 and 0.01% Sodium Azide. Primary antibodies used were rat anti-E-cad (DSHB #DCAD2, 1:100), mouse anti-SCAR (DSHB #P1C1, 1:100), mouse anti-Wasp (DSHB, P5E1, 1:100), mouse anti-Dlg (4F3), mouse anti-FasIII (7G10), guinea pig anti-Sqh1P (gift of R. Ward; 1:100), rabbit anti-GFP (Molecular Probes, # A11122), rabbit anti-dsRed (Clontech, # 632496; cross-reacts with mCherry). Alexa488, Alexa647 (Molecular Probes) and Cy3 (Jackson ImmunoResearch) conjugated secondary antibodies were used at 1:100. Actin was stained with Acti-stain 488 palloidin and Actin-stain 555 phalloidin (Cytoskeleton, Inc. # PHDG1 and #PHDR1, respectively at 1:150).
Sample preparation for live imaging
Image data were collected on a LSM800 laser scanning confocal microscope. Live imaging of pupal eyes was conducted according to a previously published protocol (Larson et al., 2008). Briefly, prior to imaging, the operculum and surrounding pupal case were peeled carefully to expose the eyes. Intact pupae were inserted in a slit created in an agarose block with eyes facing the coverslip. The agarose block was fitted with a custom-made rectangular SealGuard gasket and capped with custom-built humidified chamber.
Confocal imaging
For analysis of epithelial remodeling and protein dynamics, an image stack was obtained every 1 minute unless otherwise noted with optimal pinhole using a 63X, 1.4 NA, plan Apochromat oil immersion objective, 0.8 micron per optical section with a 50 % overlap between sections, at a scan speed of 7, averaging of 1 with an overall pixel dwell time of ~1 sec.
Quantification and Statistical Analysis
Image Processing and Analysis
All image processing was performed in Fiji. Time-lapse data were bleach corrected with an exponential fit. Z-projections were created by sum intensity projections of three optical sections encompassing the AJs.
Contact dynamics were analyzed by manual tracing with a line segment selection (width=5px). Contact length, pixel mean and total intensity for each channel at each time point, and background were collected using a custom Fiji macro. Time-resolved Pearson cross-correlations (time windows from +/−19 minutes) were compared between the mean fluorescent marker intensities and contact length using 40 min long movies with an applied Gaussian filter of sigma=3 and calculated using a custom Python script along with packages NumPy and ScipPy.
Data presented are the average Pearson cross correlations of individual contacts as noted in figure legends, and are presented as the mean R +/− SD. To calculate whether average Pearson R values and time shifts were significantly different than zero, we utilized a one-sample T-test. For comparisons between groups, ANOVA with post-hoc Bonferroni corrected t-tests were used to analyze normally distributed data for pulse amplitude in Fig. 4K. Kruskal-Wallace with post-hoc Dunn’s test was used to analyze non-normally distributed data for contact length and cross correlations in Fig. 4K.
Measurements of cell area and shape
Apical cell area and cell contact lengths for sqhAX3, SCARD37, abiD20 and arpC KG04658 mutant cells were measured manually in Fiji. For analysis of apical cell area, data are presented as the ratio of the apical area of a mutant cell to the mean of two wild-type 2° in an adjacent ommatidium, which were not in contact with a mutant clone. Morphological analyses presented in Fig 1C were performed using Fiji: aspect ratio is measured as the major/minor axis of an ellipse fitted to each cell outline in a field at a given time point, while shape variability is the standard deviation of these aspect ratios. ANOVA with post-hoc Bonferroni corrected t-tests were used to analyze normally distributed data for myoII and SCAR clonal analyses and Chi squared was used to analyze clone distribution.
KEY RESOURCES TABLE
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal α-DE-cadherin (clone DCAD2) | Developmental Studies Hybridoma bank (DSHB) | RRID: AB_528120 |
| Mouse monoclonal α-SCAR (clone P1C1) | Developmental Studies Hybridoma bank (DSHB) | RRID: AB_2618386 |
| Mouse monoclonal α-WASP (clone P5E1) | Developmental Studies Hybridoma bank (DSHB) | RRID: AB_2618392 |
| Mouse monoclonal α-Dlg (4F3) | Developmental Studies Hybridoma bank (DSHB) | RRID:AB_528203 |
| Mouse monoclonal α-FasIII (7G10) | Developmental Studies Hybridoma bank (DSHB) | RRID:AB_528238 |
| Guinea Pig α-Sqh1P | (Zhang & Ward, 2012) | NA |
| Rabbit polyclonal α-GFP | Molecular Probes - Thermo Fisher Scientific | A11122; RRID: AB_221569 |
| Rabbit polyclonal α-DsRed | Clonetech - Takara | Cat. #: 632496 |
| Goat anti-mouse Alexa Flour 488 | Molecular Probes - Thermo Fisher Scientific | Cat.#: A-11029; RRID: AB_2534088 |
| Cy3 AffiniPure Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | Code: 715-165-150 |
| Cy3 AffiniPure Donkey Anti-Guinea Pig IgG (H+L) | Jackson ImmunoResearch | Code: 716-165-148 |
| Goat anti-rat Alexa Flour 647 | Molecular Probes – Thermo Fisher Scientific | Cat. #: A-21247; RRID: AB_141778 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Acti-stain 555 Fluorescent Phalloidin | Cytoskeleton Inc. | Cat. # PHDH1 (Lot 023) |
| Acti-stain 488 Fluorescent Phalloidin | Cytoskeleton Inc. | Cat. # PHDG1 (Lot 027) |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: UAS-Lifeact::Ruby | Bloomington Drosophila Stock Center | RRID:BDSC_35544 |
| D. melanogaster: UAS-Lifeact::GFP | Bloomington Drosophila Stock Center | RRID:BDSC_35544 |
| D. melanogaster: UAS-Arp3::GFP | Bloomington Drosophila Stock Center | RRID:BDSC_39723 |
| D. melanogaster: ubi-mCherry::Abi | Bloomington Drosophila Stock Center | RRID:BDSC_58729 |
| D. melanogaster: sqh-GFP::Rok | Bloomington Drosophila Stock Center | RRID:BDSC_52289 |
| D. melanogaster: sqhAX3, sqh-Sqh::GFP | Bloomington Drosophila Stock Center | RRID:BDSC_57144 |
| D. melanogaster: SCARΔ37 FRT40A | Bloomington Drosophila Stock Center | RRID:BDSC_8754 |
| D. melanogaster: sqhAX3 FRT19A | Bloomington Drosophila Stock Center | RRID:BDSC_25712 |
| D. melanogaster: GMR-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_8121 |
| D. melanogaster: GMR-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_8605 |
| D. melanogaster: Act>y>GAL4, UAS-GFP | Bloomington Drosophila Stock Center | RRID:BDSC_4411 |
| D. melanogaster: ubi-E-cad::GFP | Kyoto Stock Center | DGRC #: 109007 |
| D. melanogaster: E-cad::mCherry | Bloomington Drosophila Stock Center | RRID:BDSC_59014 |
| D. melanogaster : tGPH (tub-GRP1-PH::GFP) | Bloomington Drosophila Stock Center | RRID:BDSC_8164 |
| D. melanogaster : Rac1::GFP | Bloomington Drosophila Stock Center | RRID:BDSC_52284 |
| D. melanogaster : UAS-Rac1N17 | Bloomington Drosophila Stock Center | RRID:BDSC_6292 |
| D. melanogaster: Rho1::GFP | Bloomington Drosophila Stock Center | RRID:BDSC_9528 |
| D. melanogaster: α–Cat::Venus (CPTI002596) | Kyoto Stock Center | NA |
| D. melanogaster: arp383F FRT40A | Bloomington Drosophila Stock Center | RRID:BDSC_39726 |
| D. melanogaster: arp3515FCFRT40A | Bloomington Drosophila Stock Center | RRID:BDSC_39727 |
| D. melanogaster: arpc2KG04658FRT40A | Kyoto Stock Center | Stock #: 111578 |
| D. melanogaster: sqh-sqh::mCherry | (Martin et al., 2009) | NA |
| D. melanogaster: sqh-UtrABD::GFP | (Rauzi et al., 2010) | NA |
| D. melanogaster: abiD20 | (Chen et al., 2014b) | NA |
| D. melanogaster: UAS-myr-SCAR | (Stephan et al., 2011) | NA |
| D. melanogaster: UAS-GFP::SCAR | (Gildor et. al., 2009) | NA |
| D. melanogaster: GMR-GAL4, UAS-α–Cat::GFP | (Larson et al., 2008) | NA |
| D. melanogaster: hsFLP, tub-GAL80 w 19AFRT; UAS-mCD8::GFP | Bloomington Drosophila Stock Center | RRID:BDSC_5134 |
| D. melanogaster: y w hsFLP, UAS-GFPnls, tub-GAL4 | (Lawrence et al., 2004) | NA |
| D. melanogaster: y w hsFLP, UAS-GFPnls, tub-GAL4; tub-GAL80 FRT40A | This study | NA |
| D. melanogaster: y w hsFLP, UAS-GFPnls, tub-GAL4; 82BFRT tub-GAL80 | This study | NA |
| D. melanogaster: GMR-Hid 40AFRT; ey-GAL4, UAS-Flp | (Stowers and Schwarz, 1999) | NA |
| D. melanogaster: ey-GAL4 UAS-Flp; FRT82B GMR-Hid | Bloomington Drosophila Stock Center | RRID:BDSC_43658 |
| D. melanogaster : GMR-GAL4, α–Cat::Venus | This paper | NA |
| D. melanogaster: UAS-Lifeact::Ruby; GMR-GAL4 | This paper | NA |
| D. melanogaster : UAS-Lifeact::GFP; sqh-Sqh::mCherry, GMR-GAL4 | This paper | NA |
| D. melanogaster: UAS-Lifeact::Ruby; sqh-Rok::GFP, GMR-GAL4 | This paper | NA |
| D. melanogaster: arpC2 KG04658 40AFRT; α–Cat::Venus | This paper | NA |
| D. melanogaster: SCARΔ37 FRT40A; α–Cat::Venus | This paper | NA |
| D. melanogaster: SCARΔ37 FRT40A; UtrABD::GFP | This paper | NA |
| D. melanogaster: FRT82B abiD20 | This paper | NA |
| D. melanogaster: sqh-UtrABD::GFP; ubi-Abi::mCherry | This paper | NA |
| D. melanogaster: GMR-GAL4, UAS-α–Cat.GFP; Abi::mCherry | This paper | NA |
| D. melanogaster: Shg::Cherry; UAS-Arp3::GFP | This paper | NA |
| D. melanogaster: UAS-Lifeact::Ruby; tGPH, GMR-GAL4 | This paper | NA |
| Oligonucleotides | ||
| Recombinant DNA | ||
| Software and Algorithms | ||
| Zen Blue | Carl Zeiss, Inc. | https://www.zeiss.com/microscopy/us/downloads.html |
| Fiji | Fiji | https://imagej.net/Fiji/Downloads |
| Python (x,y) | https://github.com/python-xy | |
| Graphpad | Prism | |
| Other | ||
Supplementary Material
Relates to Figures 1 and 4. Lengthening of LC-LC contacts decreased while the separation of these contacts increased in SCAR mutant eyes and eyes expressing SCARmyr compared to wild type. The movies follow cell behavior in a SCAR mutant eye and a SCARmyr expressing eye compared to wild type. Boxes in top panels demarcate edges, arrows in bottom panels point to separating LC-LC contact in SCAR mutant and SCARmyr eyes compared to a dynamic LC-LC contact in wild type
Relates to Figure 4. F-actin dynamics diminished while Rok persisted at LC-LC contacts in eyes expressing SCARDN or SCARmyr. The movies show the inverse pulse dynamics of F-actin and Rok in wild type and the decrease in F-actin dynamics and loss of coordination between F-actin and Rok dynamics in eyes expressing SCARDN and SCARmyr compared to wild type. Boxes surround LC-LC contacts between a 3° LC (left) and a 2° LC (right).
Relates to Figure 4. Pulsed dynamics of F-actin diminished along LC-LC contacts in SCAR mutant eyes. It shows persistent medioapical F-actin dynamics and a severe loss of pulsed F-actin dynamics along LC-LC contacts in SCAR mutant eyes compared to wild type. Boxes surround LC-LC contacts between a 3° LC (left) and a 2° LC (right).
Relates to Figure 5. Rac1, PIP3 and Rho1 accumulate dynamically along LC-LC contacts. The movies show that Rac1, PIP3 and Rho1 accumulate generally along cell contacts. Additionally, Rac1 and PIP3 accumulate along lengthening LC-LC contacts and both Rac1 and PIP3 accumulate in F-actin protrusions, while Rho1 accumulates preferentially along shortening LC-LC contacts. Boxes surround LC-LC contacts between a 3° LC (left) and a 2° LC (right).
Relates to Figure 5. F-actin dynamics diminished along LC-LC contacts in Rac1N17 expressing eyes, while Rok redistributed to subregions along 1°-LC contacts. It shows that the dynamics of F-actin and Rok along LC-LC contacts diminished, that Rok redistributed from LC-LC contacts to 1°-LC contacts and that the dynamics of the medioapical pool of F-actin coincided with expansion and constriction of the apical cell perimeter of LCs. The movies were taken at a 5 second acquisition rate. All other movies were taken at a 1 minute acquisition rate. Arrows in top panels point to LC-LC contacts.
Highlights.
Cell contacts transiently constrict and expand during shape changes
Myosin II assembles at and constricts lattice cell contacts to drive shape change
Branched F-actin assembles at and expands cell contacts to balance constriction
WAVE is required for normal shape changes and to preserve tissue topology
Acknowledgments
We thank S. Bogdan, R. Cagan, A. Martin and E. Schejter for generous gifts of flies, R. Ward for a generous gift of antibodies, the Bloomington Stock Center, the Vienna Drosophila Research Center, and the Kyoto Stock Center for flies, the Developmental Studies Hybridoma Bank for antibodies. Special thanks to A. Martin and J. Treisman for discussions, advise and comments on the manuscript and anonymous reviewers for helpful suggestions. This work was supported by a grant from the NIH to V.H. (R01GM06806).
Footnotes
Authors Contributions
V.H. and S.D. designed and performed the experiments, analyzed data and wrote the paper. RC processed several movies and contributed to data analyses. All authors have read, suggested revisions, and approved the final version of the manuscript.
Declaration of Interests
The authors declare no competing interests
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azevedo D, Antunes M, Prag S, Ma X, Hacker U, Brodland GW, Hutson MS, Solon J, Jacinto A. DRhoGEF2 regulates cellular tension and cell pulsations in the Amnioserosa during Drosophila dorsal closure. PLoS One. 2011;6:e23964. doi: 10.1371/journal.pone.0023964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Cagan R. Preferential adhesion mediated by Hibris and Roughest regulates morphogenesis and patterning in the Drosophila eye. Dev Cell. 2005;8:925–935. doi: 10.1016/j.devcel.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Bardet PL, Guirao B, Paoletti C, Serman F, Leopold V, Bosveld F, Goya Y, Mirouse V, Graner F, Bellaiche Y. PTEN controls junction lengthening and stability during cell rearrangement in epithelial tissue. Dev Cell. 2013;25:534–546. doi: 10.1016/j.devcel.2013.04.020. [DOI] [PubMed] [Google Scholar]
- Barlan K, Cetera M, Horne-Badovinac S. Fat2 and Lar Define a Basally Localized Planar Signaling System Controlling Collective Cell Migration. Dev Cell. 2017;40:467–477. e465. doi: 10.1016/j.devcel.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betson M, Lozano E, Zhang J, Braga VM. Rac activation upon cell-cell contact formation is dependent on signaling from the epidermal growth factor receptor. J Biol Chem. 2002;277:36962–36969. doi: 10.1074/jbc.M207358200. [DOI] [PubMed] [Google Scholar]
- Blanchard GB, Adams RJ. Measuring the multi-scale integration of mechanical forces during morphogenesis. Curr Opin Genet Dev. 2011;21:653–663. doi: 10.1016/j.gde.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Blanchard GB, Murugesu S, Adams RJ, Martinez-Arias A, Gorfinkiel N. Cytoskeletal dynamics and supracellular organisation of cell shape fluctuations during dorsal closure. Development. 2010;137:2743–2752. doi: 10.1242/dev.045872. [DOI] [PubMed] [Google Scholar]
- Borghi N, Sorokina M, Shcherbakova OG, Weis WI, Pruitt BL, Nelson WJ, Dunn AR. E-cadherin is under constitutive actomyosin-generated tension that is increased at cell-cell contacts upon externally applied stretch. Proc Natl Acad Sci U S A. 2012;109:12568–12573. doi: 10.1073/pnas.1204390109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma SP, Cagan RL, Baranski TJ. Chimaerin and Rac regulate cell number, adherens junctions, and ERK MAP kinase signaling in the Drosophila eye. Proc Natl Acad Sci U S A. 2007;104:7098–7103. doi: 10.1073/pnas.0701686104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TA, Christensen JR, Barone E, Suarez C, Sirotkin V, Kovar DR. Homeostatic actin cytoskeleton networks are regulated by assembly factor competition for monomers. Curr Biol. 2014;24:579–585. doi: 10.1016/j.cub.2014.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil Cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan R. Principles of Drosophila eye differentiation. Curr Top Dev Biol. 2009;89:115–135. doi: 10.1016/S0070-2153(09)89005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagan RL, Ready DF. The emergence of order in the Drosophila pupal retina. Dev Biol. 1989;136:346–362. doi: 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- Carthew RW. Pattern formation in the Drosophila eye. Curr Opin Genet Dev. 2007;17:309–313. doi: 10.1016/j.gde.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera M, Ramirez-San Juan GR, Oakes PW, Lewellyn L, Fairchild MJ, Tanentzapf G, Gardel ML, Horne-Badovinac S. Epithelial rotation promotes the global alignment of contractile actin bundles during Drosophila egg chamber elongation. Nat Commun. 2014;5:5511. doi: 10.1038/ncomms6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Squarr AJ, Stephan R, Chen B, Higgins TE, Barry DJ, Martin MC, Rosen MK, Bogdan S, Way M. Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev Cell. 2014;30:569–584. doi: 10.1016/j.devcel.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Langeslag M, Figdor CG, van Leeuwen FN. Myosin II and mechanotransduction: a balancing act. Trends Cell Biol. 2007;17:178–186. doi: 10.1016/j.tcb.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Collinet C, Rauzi M, Lenne PF, Lecuit T. Local and tissue-scale forces drive oriented junction growth during tissue extension. Nat Cell Biol. 2015;17:1247–1258. doi: 10.1038/ncb3226. [DOI] [PubMed] [Google Scholar]
- David DJ, Tishkina A, Harris TJ. The PAR complex regulates pulsed actomyosin contractions during amnioserosa apical constriction in Drosophila. Development. 2010;137:1645–1655. doi: 10.1242/dev.044107. [DOI] [PubMed] [Google Scholar]
- Dufour S, Mege RM, Thiery JP. alpha-catenin, vinculin, and F-actin in strengthening E-cadherin cell-cell adhesions and mechanosensing. Cell Adh Migr. 2013;7:345–350. doi: 10.4161/cam.25139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature. 2002;418:790–793. doi: 10.1038/nature00859. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, de Simoes SM, Roper JC, Eaton S, Zallen JA. Myosin II dynamics are regulated by tension in intercalating cells. Dev Cell. 2009;17:736–743. doi: 10.1016/j.devcel.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Zallen JA. Oscillatory behaviors and hierarchical assembly of contractile structures in intercalating cells. Phys Biol. 2011;8:045005. doi: 10.1088/1478-3975/8/4/045005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildor B, Massarwa R, Shilo BZ, Schejter ED. The SCAR and WASp nucleation-promoting factors act sequentially to mediate Drosophila myoblast fusion. EMBO Rep. 2009;10:1043–1050. doi: 10.1038/embor.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorfinkiel N, Blanchard GB. Dynamics of actomyosin contractile activity during epithelial morphogenesis. Curr Opin Cell Biol. 2011;23:531–539. doi: 10.1016/j.ceb.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Hara Y, Shagirov M, Toyama Y. Cell Boundary Elongation by Non-autonomous Contractility in Cell Oscillation. Curr Biol. 2016;26:2388–2396. doi: 10.1016/j.cub.2016.07.003. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Carthew RW. Surface mechanics mediate pattern formation in the developing retina. Nature. 2004;431:647–652. doi: 10.1038/nature02952. [DOI] [PubMed] [Google Scholar]
- He L, Wang X, Tang HL, Montell DJ. Tissue elongation requires oscillating contractions of a basal actomyosin network. Nat Cell Biol. 2010;12:1133–1142. doi: 10.1038/ncb2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisenberg CP, Bellaiche Y. Forces in tissue morphogenesis and patterning. Cell. 2013;153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- Hilgenfeldt S, Erisken S, Carthew RW. Physical modeling of cell geometric order in an epithelial tissue. Proc Natl Acad Sci U S A. 2008;105:907–911. doi: 10.1073/pnas.0711077105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK. The WAVE regulatory complex is inhibited. Nat Struct Mol Biol. 2009;16:561–563. doi: 10.1038/nsmb.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124:761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Jodoin JN, Coravos JS, Chanet S, Vasquez CG, Tworoger M, Kingston ER, Perkins LA, Perrimon N, Martin AC. Stable Force Balance between Epithelial Cells Arises from F-Actin Turnover. Dev Cell. 2015;35:685–697. doi: 10.1016/j.devcel.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RI, Sedgwick A, D’Souza-Schorey C, Cagan RL. Role for a Cindr-Arf6 axis in patterning emerging epithelia. Mol Biol Cell. 2011;22:4513–4526. doi: 10.1091/mbc.E11-04-0305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RI, Seppa MJ, Cagan RL. The Drosophila CD2AP/CIN85 orthologue Cindr regulates junctions and cytoskeleton dynamics during tissue patterning. J Cell Biol. 2008;180:1191–1204. doi: 10.1083/jcb.200706108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafer J, Hayashi T, Maree AF, Carthew RW, Graner F. Cell adhesion and cortex contractility determine cell patterning in the Drosophila retina. Proc Natl Acad Sci U S A. 2007;104:18549–18554. doi: 10.1073/pnas.0704235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY, Davidson LA. Punctuated actin contractions during convergent extension and their permissive regulation by the non-canonical Wnt-signaling pathway. J Cell Sci. 2011;124:635–646. doi: 10.1242/jcs.067579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V, Hume PJ, Humphreys D, Liu T, Horning O, Jensen ON, McGhie EJ. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc Natl Acad Sci U S A. 2011;108:14449–14454. doi: 10.1073/pnas.1107666108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DE, Liberman Z, Cagan RL. Cellular behavior in the developing Drosophila pupal retina. Mech Dev. 2008;125:223–232. doi: 10.1016/j.mod.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T, Lenne PF, Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu Rev Cell Dev Biol. 2011;27:157–184. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Levayer R, Lecuit T. Biomechanical regulation of contractility: spatial control and dynamics. Trends Cell Biol. 2012;22:61–81. doi: 10.1016/j.tcb.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Lomakin AJ, Lee KC, Han SJ, Bui DA, Davidson M, Mogilner A, Danuser G. Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat Cell Biol. 2015;17:1435–1445. doi: 10.1038/ncb3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye CM, Naylor HW, Sanson B. Subcellular localisations of the CPTI collection of YFP-tagged proteins in Drosophila embryos. Development. 2014;141:4006–4017. doi: 10.1242/dev.111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AC, Kaschube M, Wieschaus EF. Pulsed contractions of an actin-myosin network drive apical constriction. Nature. 2009;457:495–499. doi: 10.1038/nature07522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason FM, Martin AC. Tuning cell shape change with contractile ratchets. Curr Opin Genet Dev. 2011;21:671–679. doi: 10.1016/j.gde.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza MC. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin Cell Dev Biol. 2013;24:272–279. doi: 10.1016/j.semcdb.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro E, Nance J, Priess JR. Cortical flows powered by asymmetrical contraction transport PAR proteins to establish and maintain anterior-posterior polarity in the early C. elegans embryo. Dev Cell. 2004;7:413–424. doi: 10.1016/j.devcel.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Fukata M, Yamaga M, Itoh N, Kaibuchi K. Recruitment and activation of Rac1 by the formation of E-cadherin-mediated cell-cell adhesion sites. J Cell Sci. 2001;114:1829–1838. doi: 10.1242/jcs.114.10.1829. [DOI] [PubMed] [Google Scholar]
- Noren NK, Niessen CM, Gumbiner BM, Burridge K. Cadherin engagement regulates Rho family GTPases. J Biol Chem. 2001;276:33305–33308. doi: 10.1074/jbc.C100306200. [DOI] [PubMed] [Google Scholar]
- O’Leary CJ, Nourse CC, Lee NK, White A, Langford M, Sempert K, Cole SJ, Cooper HM. Neogenin Recruitment of the WAVE Regulatory Complex to Ependymal and Radial Progenitor Adherens Junctions Prevents Hydrocephalus. Cell Rep. 2017;20:370–383. doi: 10.1016/j.celrep.2017.06.051. [DOI] [PubMed] [Google Scholar]
- Oda H, Tsukita S. Dynamic features of adherens junctions during Drosophila embryonic epithelial morphogenesis revealed by a Dalpha-catenin-GFP fusion protein. Dev Genes Evol. 1999;209:218–225. doi: 10.1007/s004270050246. [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, Suetsugu S, Takenawa T. PtdIns(3,4,5)P3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat Cell Biol. 2004;6:420–426. doi: 10.1038/ncb1125. [DOI] [PubMed] [Google Scholar]
- Patel FB, Bernadskaya YY, Chen E, Jobanputra A, Pooladi Z, Freeman KL, Gally C, Mohler WA, Soto MC. The WAVE/SCAR complex promotes polarized cell movements and actin enrichment in epithelia during C. elegans embryogenesis. Dev Biol. 2008;324:297–309. doi: 10.1016/j.ydbio.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez TD, Tamada M, Sheetz MP, Nelson WJ. Immediate-early signaling induced by E-cadherin engagement and adhesion. J Biol Chem. 2008;283:5014–5022. doi: 10.1074/jbc.M705209200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Pollitt AY, Insall RH. WASP and SCAR/WAVE proteins: the drivers of actin assembly. J Cell Sci. 2009;122:2575–2578. doi: 10.1242/jcs.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan A, Tien AC, Haueter CM, Schulze KL, Bellen HJ. The Arp2/3 complex and WASp are required for apical trafficking of Delta into microvilli during cell fate specification of sensory organ precursors. Nat Cell Biol. 2009;11:815–824. doi: 10.1038/ncb1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakeman AS, Anderson KV. Axis specification and morphogenesis in the mouse embryo require Nap1, a regulator of WAVE-mediated actin branching. Development. 2006;133:3075–3083. doi: 10.1242/dev.02473. [DOI] [PubMed] [Google Scholar]
- Rauzi M, Lenne PF, Lecuit T. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature. 2010;468:1110–1114. doi: 10.1038/nature09566. [DOI] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versatile marker to visualize F-actin. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JK, Choi W, Jung KC, He L, Harris NJ, Peifer M. A contractile actomyosin network linked to adherens junctions by Canoe/afadin helps drive convergent extension. Mol Biol Cell. 2011;22:2491–2508. doi: 10.1091/mbc.E11-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J, Hannezo E, Tu F, Biro M, Wallingford JB. Emergence of an Apical Epithelial Cell Surface In Vivo. Dev Cell. 2016;36:24–35. doi: 10.1016/j.devcel.2015.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo A, Wallingford JB. PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science. 2014;343:649–652. doi: 10.1126/science.1243126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P, Rolo A, Chen X, Gumbiner BM, Keller R. Convergence and extension at gastrulation require a myosin IIB-dependent cortical actin network. Development. 2008;135:2435–2444. doi: 10.1242/dev.014704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon J, Kaya-Copur A, Colombelli J, Brunner D. Pulsed forces timed by a ratchet-like mechanism drive directed tissue movement during dorsal closure. Cell. 2009;137:1331–1342. doi: 10.1016/j.cell.2009.03.050. [DOI] [PubMed] [Google Scholar]
- Soto MC, Qadota H, Kasuya K, Inoue M, Tsuboi D, Mello CC, Kaibuchi K. The GEX-2 and GEX-3 proteins are required for tissue morphogenesis and cell migrations in C. elegans. Genes Dev. 2002;16:620–632. doi: 10.1101/gad.955702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarr AJ, Brinkmann K, Chen B, Steinbacher T, Ebnet K, Rosen MK, Bogdan S. Fat2 acts through the WAVE regulatory complex to drive collective cell migration during tissue rotation. J Cell Biol. 2016;212:591–603. doi: 10.1083/jcb.201508081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan R, Gohl C, Fleige A, Klambt C, Bogdan S. Membrane-targeted WAVE mediates photoreceptor axon targeting in the absence of the WAVE complex in Drosophila. Mol Biol Cell. 2011;22:4079–4092. doi: 10.1091/mbc.E11-02-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers RS, Schwarz TL. A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics. 1999;152:1631–1639. doi: 10.1093/genetics/152.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan-Brown JL, Tandon P, Bird KE, Dickinson DJ, Tintori SC, Heppert JK, Meserve JH, Trogden KP, Orlowski SK, Conlon FL, et al. Identifying Regulators of Morphogenesis Common to Vertebrate Neural Tube Closure and Caenorhabditis elegans Gastrulation. Genetics. 2016;202:123–139. doi: 10.1534/genetics.115.183137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajiri R, Misaki K, Yonemura S, Hayashi S. Joint morphology in the insect leg: evolutionary history inferred from Notch loss-of-function phenotypes in Drosophila. Development. 138:4621–4626. doi: 10.1242/dev.067330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S. The WASP-WAVE protein network: connecting the membrane to the cytoskeleton. Nat Rev Mol Cell Biol. 2007;8:37–48. doi: 10.1038/nrm2069. [DOI] [PubMed] [Google Scholar]
- Twiss F, de Rooij J. Cadherin mechanotransduction in tissue remodeling. Cell Mol Life Sci. 2013;70:4101–4116. doi: 10.1007/s00018-013-1329-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma S, Shewan AM, Scott JA, Helwani FM, den Elzen NR, Miki H, Takenawa T, Yap AS. Arp2/3 activity is necessary for efficient formation of E-cadherin adhesive contacts. J Biol Chem. 2004;279:34062–34070. doi: 10.1074/jbc.M404814200. [DOI] [PubMed] [Google Scholar]
- Warner SJ, Longmore GD. Cdc42 antagonizes Rho1 activity at adherens junctions to limit epithelial cell apical tension. J Cell Biol. 2009a;187:119–133. doi: 10.1083/jcb.200906047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner SJ, Longmore GD. Distinct functions for Rho1 in maintaining adherens junctions and apical tension in remodeling epithelia. J Cell Biol. 2009b;185:1111–1125. doi: 10.1083/jcb.200901029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Nelson WJ. Localized zones of Rho and Rac activities drive initiation and expansion of epithelial cell-cell adhesion. J Cell Biol. 2007;178:517–527. doi: 10.1083/jcb.200701058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Oikawa T, Takenawa T. Rac-WAVE-mediated actin reorganization is required for organization and maintenance of cell-cell adhesion. J Cell Sci. 2007;120:86–100. doi: 10.1242/jcs.03311. [DOI] [PubMed] [Google Scholar]
- Yu JC, Fernandez-Gonzalez R. Local mechanical forces promote polarized junctional assembly and axis elongation in Drosophila. Elife. 2016;5 doi: 10.7554/eLife.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relates to Figures 1 and 4. Lengthening of LC-LC contacts decreased while the separation of these contacts increased in SCAR mutant eyes and eyes expressing SCARmyr compared to wild type. The movies follow cell behavior in a SCAR mutant eye and a SCARmyr expressing eye compared to wild type. Boxes in top panels demarcate edges, arrows in bottom panels point to separating LC-LC contact in SCAR mutant and SCARmyr eyes compared to a dynamic LC-LC contact in wild type
Relates to Figure 4. F-actin dynamics diminished while Rok persisted at LC-LC contacts in eyes expressing SCARDN or SCARmyr. The movies show the inverse pulse dynamics of F-actin and Rok in wild type and the decrease in F-actin dynamics and loss of coordination between F-actin and Rok dynamics in eyes expressing SCARDN and SCARmyr compared to wild type. Boxes surround LC-LC contacts between a 3° LC (left) and a 2° LC (right).
Relates to Figure 4. Pulsed dynamics of F-actin diminished along LC-LC contacts in SCAR mutant eyes. It shows persistent medioapical F-actin dynamics and a severe loss of pulsed F-actin dynamics along LC-LC contacts in SCAR mutant eyes compared to wild type. Boxes surround LC-LC contacts between a 3° LC (left) and a 2° LC (right).
Relates to Figure 5. Rac1, PIP3 and Rho1 accumulate dynamically along LC-LC contacts. The movies show that Rac1, PIP3 and Rho1 accumulate generally along cell contacts. Additionally, Rac1 and PIP3 accumulate along lengthening LC-LC contacts and both Rac1 and PIP3 accumulate in F-actin protrusions, while Rho1 accumulates preferentially along shortening LC-LC contacts. Boxes surround LC-LC contacts between a 3° LC (left) and a 2° LC (right).
Relates to Figure 5. F-actin dynamics diminished along LC-LC contacts in Rac1N17 expressing eyes, while Rok redistributed to subregions along 1°-LC contacts. It shows that the dynamics of F-actin and Rok along LC-LC contacts diminished, that Rok redistributed from LC-LC contacts to 1°-LC contacts and that the dynamics of the medioapical pool of F-actin coincided with expansion and constriction of the apical cell perimeter of LCs. The movies were taken at a 5 second acquisition rate. All other movies were taken at a 1 minute acquisition rate. Arrows in top panels point to LC-LC contacts.