Abstract
Cancer is one of the major and leading causes of death worldwide. Two of the greatest challenges infighting cancer are early detection and effective treatments with no or minimum side effects. Widespread use of targeted therapies and molecular imaging in clinics requires high affinity, tumor-specific agents as effective targeting vehicles to deliver therapeutics and imaging probes to the primary or metastatic tumor sites. Combinatorial libraries such as phage-display and one-bead one-compound (OBOC) peptide libraries are powerful approaches in discovering tumor-targeting peptides. This review gives an overview of different combinatorial library technologies that have been used for the discovery of tumor-targeting peptides. Examples of tumor-targeting peptides identified from each combinatorial library method will be discussed. Published tumor-targeting peptide ligands and their applications will also be summarized by the combinatorial library methods and their corresponding binding receptors.
Keywords: Tumor-targeting peptide, Biological library, Phage-display peptide library, One-bead one-compound peptide library, High throughput screening, Cell surface receptor
1. Introduction
Breakthrough advances have been achieved in cancer diagnosis and treatment in the last decade including the recently FDA-approved immunotherapeutic agents, which have provided patients with new hope. However, cancer continues to be the second cause of death in the US (584,881 vs 611,105 in heart disease from the 2015 Fast Stats provided by CDC), and it is expected to surpass heart disease to become the No. 1 killer by 2030. Conventional chemotherapies have low specificity towards cancer cells and therefore exhibit serious toxic side effects. Target-specific delivery of chemotherapeutic drugs to the tumor cells can help improve the outcome of existing anti-cancer drugs. Widespread use of targeted therapies and molecular imaging in the clinic requires high affinity, tumor-specific agents as effective targeting vehicles to deliver therapeutics and imaging probes to the tumor sites. Tumor-targeting agents can be antibodies, proteins, peptides, peptidomimetics, glycopeptides, peptoids, aptamers or small molecules. Several cell surface-targeting antibodies have been approved by the FDA as vehicles to deliver radionuclides (e.g. Zevalin or Bexxar, anti-CD20 antibodies loaded with 90Y or 131I, respectively), toxins (e.g. Adcetris, an anti-CD30 antibody-MMAE conjugate directed against systemic anaplastic large cell lymphoma and Hodgkin's lymphoma), or cytotoxic chemotherapeutic agents (e.g., Trastuzumab emtansine) to the cancer cells. Cancer-targeting antibodies have proven success in the clinic, but they also suffer some limitations because (i) the Fc region of the antibodies binds to the reticuloendothelial system resulting in significant toxicities to liver, bone marrow, and spleen; (ii) antibodies against the cancer cells have difficulty in infiltrating the entire tumor mass due to their large size (M.W. ∼160,000 Da); (iii) they are difficult to manufacture in large-scale; therefore, they are expensive. Tumor-targeting peptides are efficient alternative vehicles for selective delivery of high dose of chemotherapeutic drugs or diagnostic agents to tumor sites while sparing normal tissues. Several peptide hormones have already been used for tumor targeting. For example, octreotide, a cyclic octapeptide analogue of somatostatin, has been used for radiotargeting of neuroendocrine tumor [1]. AN-152, a linear peptide analogue of LHRH, has also been used to target LHRH receptor of ovarian cancer, breast cancer and prostate cancer [2]. Peptides consisting of only eukaryotic amino acids in general are not stable in vivo, but their stability against proteolysis can be significantly improved if they contain D-amino acids and unnatural amino acids, are cyclized, and/or are N-and C-terminally blocked. Advantages of peptides over currently-used biomolecules such as antibodies are their rapid blood clearance, increased diffusion and tissue penetration, chemical stability, and ease of synthesis in large scale. In addition, they can be readily conjugated to cytotoxic drugs, radionuclides, or toxins in a chemically defined manner.
The subject of using combinatorial libraries to discover tumor-targeting ligands has been reviewed by us and other investigators in the past [3–9].Inthis review, we attempt to give an update on combinatorial library technologies and newly identified tumor-targeting peptides. Additionally, we categorize the peptide ligands according to their interacting receptor(s) and library screening.
2. Overview of approaches to discover tumor-targeting peptides
Tumor-targeting ligands generally target one of the following three sites: (i) cancer cell surface receptors, (ii) the tumor's extracellular matrix, and (iii) tumor vessel endothelial cell surface receptors. Many of these targeting molecules can be developed through (i) the use of native ligands or their analogues such as octreotide against somatostatin receptors, bradykinin analogues against bradykinin receptors, AN-152 against LHRH receptors, and folic acid against the folate receptor; (ii) computer-aided design if the X-ray crystallographic structure of the cancer-associated receptor or a related receptor is known, in combination with medicinal chemistry, and/or (iii) screening combinatorial libraries. The combinatorial library method allows rapid identification of tumor-targeting ligands from a large number of diverse compounds. In this review, we shall focus on different library technologies and their use for discovery of tumor-targeting peptide ligands. The phage-display peptide library [10] and the OBOC combinatorial peptide library [11] are the two most popular approaches that have been successfully applied to discover tumor-targeting peptides. Other biological-display peptide libraries such as yeast-display, bacteria-display, ribosome-display, and mRNA-display peptide libraries have also been used but are less common. In addition to OBOC library, peptide nucleic acid (PNA)-encoded solution phase peptide library and peptide microarrays are two other synthetic chemical libraries that have been used for the discovery of tumor-targeting ligands. The former takes advantage of its ease in decoding (see below). Peptide microarrays have been mainly used for ligand optimization and structure–activity relationship studies. Other synthetic combinatorial library methods such as positional-scanning peptide library have also been used to discover cancer targeting pep-tides but are much less often applied. A summary of the advantages and disadvantages of different library approaches is shown in Table 1. Live cancer cells or cancer-related proteins are commonly used as probes to screen combinatorial libraries. The advantages of using live cells for library screening include (i) the target receptors on cell surface are in the native state; (ii) cloning, expression, and purification of cell surface targets are not needed; (iii) the cell surface target can be an unknown receptor; (iv) targeting ligands with cell-penetrating ligands can be identified; and (v) functional ligands that trigger downstream signaling can be readily identified, particularly with the OBOC method. Therefore, most cancer-targeting peptides target cancer cell surface and have been discovered from cell-binding assays. Soluble cancer-associated proteins, if available, can also be used as probes for identification of ligands, although their specificity against other proteins needs to be verified. The technology of biological-display, OBOC, PNA-encoded solution-phase peptide libraries and peptide microarray, as well as their applications in the discovery of cancer-targeting peptides will be discussed below.
Table 1.
Comparison of different combinatorial peptide library methods.
| Combinatorial library methods | Advantages | Disadvantages | |
|---|---|---|---|
| Biological library | Phage-display |
|
|
| Yeast-display |
|
|
|
| Bacteria-display |
|
|
|
| Ribosome-or mRNA-display |
|
|
|
| Chemical library | OBOC |
|
|
| PNA- encoded solution phase peptide library |
|
|
|
| Peptide microarray |
|
|
|
3. Biological library methods
The surface profile difference between cancer cells and their nonma-lignant counterparts can serve as an excellent molecular address for targeted delivery of therapeutic agents, diagnostic agents or both to cancer cells. The biological-display system is an efficient tool in discovering novel tumor-targeting peptides via high-throughput screening. The peptide-displaying microbes can be considered as peptide-covered micro-particles. The peptides displayed on the surface of the microbes can be directly used to screen live cancer cells and/or purified receptor proteins. The sequence of the peptide hits can be easily determined by DNA sequencing of the microbial plasmid. The biological-display system includes phage, yeast, bacteria, ribosome, mRNA, and mammalian cell-display [12]. Phage-display peptide library remains the most commonly used combinatorial library technique to discover tumor-targeting peptides. To the best of our knowledge, mammalian cell-display has not been applied to the discovery of tumor-targeting peptides so far; therefore, it will not be discussed in this review.
3.1. Phage-display peptide library method
3.1.1. Methodology
A phage-display peptide library is comprised of a heterogeneous mixture of billions of bacteriophage clones, each carrying a different foreign DNA insert and therefore displaying a different peptide on its surface [10]. Peptides are typically displayed at the N-terminus of the pIII or pVIII proteins [4,13]. Inverted pVIII proteins have been used to display peptide with free carboxy terminus [14]. The pVIII “landscape” (all pVIII proteins express peptides) has also been used as peptide-displaying coat proteins [4]. Techniques for generating chimera phage with both peptide-displaying coat proteins and normal wild-type coat protein have been developed. The M13 filamentous phage is the most widely used phage-display system due to its high capacity for replication and ability to accommodate large foreign DNA. The fd filamentous phage, which is closely related M13 phage, is also commonly used for pIII and pVIII display [10,15]. The less commonly used T7 lytic phages that lyse their bacterial hosts have also been used for peptide library display. In T7 phage, peptide sequences are typically displayed as C-terminal fusions of the 10B capsid protein. T7 libraries exhibit less bias than filamentous phage libraries and longer peptide libraries (12-20mer) are most often used for the discovery of cell binding peptides. In contrast, short peptide libraries (7-12mer) are more popular in M13 phage display.
The phage-display library method has several advantages: (i) it can display combinatorial peptide libraries with huge permutations (109); (ii) the size of the grafted peptide is not limited by the constraints as in the case of a synthetic peptide library; (iii) it can take advantage of known protein folds (e.g. zinc-finger fold, conotoxin fold, immunoglobulin fold, or cystine-knot) by grafting random oligopeptides on such tertiary folds; (iv) the method is highly efficient, inexpensive, and amenable to both short and long peptides, linear and simple cyclic peptides (disulfide formation with two l-cysteines), and can be carried out in most molecular biology laboratories; (v) phage-display peptide libraries are commercially available, e.g., Ph.D.-7, Ph.D.-12 and Ph.D.-C7C (cyclic with a disulfide bond) libraries can be purchased from New England Biolabs (Ipswich, MA, USA); (vi) phage-displayed libraries can be screened with in vivo selection techniques in xenograft models; (vii) the standard library can be easily generated by simply growing the microorganisms; (viii) usage of this method may identify novel ligands even without prior knowledge about the identity of the target receptor on cell surface; and (ix) recent advances in chemical modification of phage-display libraries with thiol-reactive compounds or crosslinkers prior to screening enable introduction of limited cyclization and derivatization to the displayed peptides [16-18].
However, despite the many advantages, this biological library approach suffers some important limitations: (i) only the natural l-amino acid peptide libraries (comprised of 20 eukaryotic amino acids) can be incorporated into the phages. Such peptides are generally susceptible to proteolysis particularly if the N- and C-termini are not blocked; (ii) because of bias in genetic codons, the peptide library is not totally random; (iii) screening assays of the phage-display libraries are generally limited to binding and a few functional assays such as protease substrate determination; (iv) complicated bicyclic, compact scaffolding, branched structures, or molecules with special chemistry of cyclization are more difficult with this method; and (v) optimizing a phage-display peptide into a proteolytic stable molecule while still retaining a high binding activity and specificity to the cell surface receptor is not a trivial matter.
Fig. 1 illustrated the process of selecting phage clones that bind a specific target, which is called “biopanning” including in vitro, in vivo, and ex vivo biopannings. Kay et al. have published a step-by-step screening protocol which facilitates new investigators to learn this technology [19]. The following factors need to be taken into account because they may affect the quality of hits identified from screening phage-display libraries: the number of phages used, stringency of selection process, the number of selection rounds, competitive selection and subtractive panning. Panning of a phage-display library with living cells provides the option to select phages that bind and internalize inside the cells, which can be achieved by optimizing the washing techniques [20]. Decoding of peptide ligands identified from phage-display libraries can be achieved with DNA sequencing, which is simple and inexpensive.
Fig. 1.
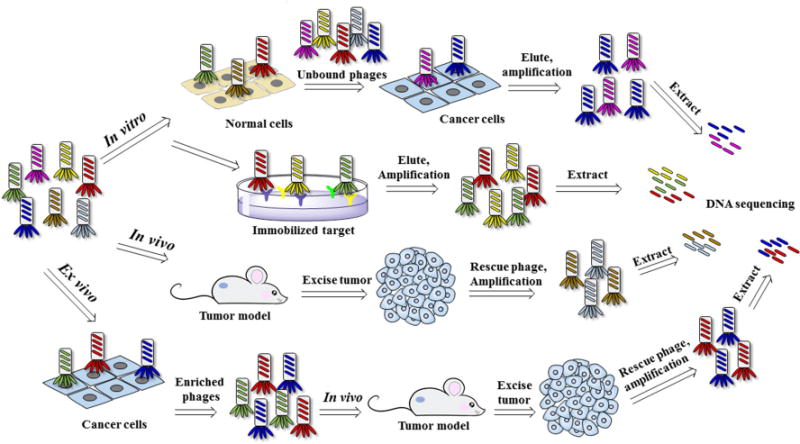
Biopanning of phage-display peptide library.
3.1.2. Discovery of tumor-targeting peptides from phage-display peptide library
Tumor-targeting peptides can be discovered from phage-display libraries via one of the three screening approaches: in vitro, in vivo or ex vivo selections (Fig. 1). Cancer-associated proteins, specific cancer cell lines, patient tissues, and tumor xenograft mouse models have been successfully used as screening probes. In vivo selection of phages has recently shifted from using xenograft mouse models to transgenic and clinically relevant patient-derived xenograft (PDX) models.
Unbiased bio-panning of phage-displayed peptide libraries has yielded a number of peptides that bind cancer cells and cancer-associated antigens presented on cancer cell surface, tumor vasculature or tumor lymphatic vessels. Different peptide ligands have been identified for a variety of tumor types including breast, lung, thyroid, head and neck, liver, prostate, bladder, colon and gastric cancers, osteo-sarcomas, as well as pancreatic ductal adenocarcinomas and squamous cell carcinoma. Table 2 lists some tumor-targeting peptides that were identified from in vitro selection of phage-display peptide library using purified receptor as a screening probe. Examples of tumor-targeting peptides that target specific receptor(s) using live cells, tissues, or tumors as screening probes are listed in Table 3. Phage-display library screening also yields many tumor-targeting peptides of which the binding partners have not yet been identified. This latter group of pep-tides will not be covered here but can be found in other reviews [3–7].
Table 2.
Tumor-targeting peptides identified from phage-display peptide libraries by in vitro biopanning against purified receptors.
| Receptor | Sequence* | Ref | Library | Application | |
|---|---|---|---|---|---|
|
|
|||||
| Imaging | Therapy | ||||
| GPC3 | DHLASLWWGTEL (TJ12P1) | [174] | Ph.D.-12 | [174] | – |
| PD-L1 | NYSKPTDRQYHF** (D APP1) | [175] | Ph.D.-12 | [175] | [175] |
| β-Catenin | AC#AQKLDGC#SYISWSC#G (BC1) AC#SGWWPKC#QGYIPGC#G (BC2) | [176] | ACX6CX6CG | – | – |
| AC‡APGVYRC‡NQNFIWC‡G (BC3) | |||||
| PDGFRβ | IPLPPPSRPFFK | [55] | Ph.D.-12 | [55] | – |
| PKCδ | LMNPNNHPRTPR (PKC-bp) | [177] | Ph.D.-12 | – | – |
| PTPRJ | CHHNLTHAC (PTPRJ-pep19) CLHHYHGSC (PTPRJ-pep24) | [59] | Ph.D.-C7C | ||
| CHHALTHAC (PTPRJ-pep19.4) | [60] | Ph.D.-C7C | – | – | |
| TfR 1 | SPRPRHTLRLSL (B18) | [178] | Ph.D.-12 | – | – |
| Tie 2 | TMGFTAPRFPHY | [179] | Ph.D.-12 | – | [179] |
| CD-21 | RMWPSSTVNLSAGRR (P1) | [180] | 15mer | – | – |
| VEGFRI (Flt-1) | NGYEIEWYSWVTHGMY (SP5.2) | [181] | 16mer | – | – |
| IL-10 RA | FRSFESCLAKSH | [182] | PH.D.-12 | – | – |
| EGFR | YHWYGYTPQNVI (GE11) | [21] | Ph.D.-12 | [183,184] | [21,183,185–189] |
| QHYNIVNTQSRVa | [190] | Ph.D.-12 | – | – | |
| QRHKPRE | [191] | Ph.D.-7 | [191] | – | |
| FGF8b | HSQAAVP (P12) | [192] | Ph.D.-7 | [192] | – |
| aFGF | AGNWTPI (AP8) | [193] | Ph.D.-7 | – | – |
| bFGF | PLLQATL (P7) | [194] | Ph.D.-7 | [194] | – |
| IL-6Rα | LSLITRL (S7) | [195] | Ph.D.-7 | – | [195] |
| α5β1 | CRGDCL | [196] | 6mer | – | – |
| GACRGDCLGA (synthetic peptide) | |||||
| CRRETAWAC | [197] | CX7C | – | – | |
| GACRRETAWACGA (synthetic peptide) | |||||
| α6β1 | VSWFSRHRYSPFAVS (P3) | [198] | 15mer | – | – |
| αvβ3/αvβ5 | CDCRGDCFC (RGD-4C) | [40,199] | CX9 | [200–202] | [199,201–204] |
| αvβ6 | RTDLDSLRTYTL | [27] | Ph.D.-12 | – | [205] |
| MMP-9 | CTTHWGFTLC (CTT) | [206] | CX C | [206–211] | [206,212–216] |
| CD133 | APSPMIW, LQNAPRS | [217] | Ph.D.-7 | – | – |
| N-cadherin | SWTLYTPSGQSK | [218] | Ph.D.-12 | – | – |
| E-cadherin | SWELYYPLRANL | [219] | Ph.D.-12 | – | – |
| PSMA | WQPDTAHHWATL | [220] | Ph.D.-12 | – | – |
| VEGFR-3 | CSDSWHYWC (P1) | [53] | Ph.D.-C7C | – | – |
| WHWLPNLRHYAS (peptide III) | [221] | Ph.D.-12 | [221] | – | |
| EGFRvIII/EGFR | WHTEILKSYPHEb, LPAFFVTNQTQDb | [222] | PH.D.-12 | – | – |
| Carbonic anhydrase IX | YNTNHVPLSPKY (CAIX-P1) | [223] | Ph.D.-12 | [223] | – |
| EphA2 | YSAYPDSVPMMS (YSA) | [224] | Ph.D.-12 | [225,226] | [227–229] |
| EphB4 | TNYLFSPNGPIA (TNYL) | [230] | Ph.D.-12 | [231] | [232] |
| PS | CLSYYPSYC | [233] | CX7C | [233] | – |
| HER2 | AC#SLQDPNC#DWWGHYC#G (H8)c | [234] | ACX6CX6CG | – | – |
| ACGLQGYGCWGMYGKCG (H30)c | |||||
| CVGVLPSQDAIGIC (L-26-19)d | [235] | Ph.D.-12 | |||
| CGPLPVDWYWC (L-26-24)d | |||||
| CEWKFDPGLGQARC (N-12-1)e | |||||
| CDYMTDGRAASKIC (N-12-2)e | |||||
| KCCYSL (p6.1) | [236] | 6mer | [237–239] | – | |
| MARSGL, MARAKE, MSRTMS | [240] | 6mer | – | – | |
| WTGWCLNPEESTWGFCTGSF (EC-1) | [241] | 20mer | – | – | |
| MCGVCLSAQRWT, SGLWWLGVDILG | [242] | Ph.D.-12 | – | – | |
| TGA-72 | NPGTCKDKWIECLLNG (A3-10) | [243] | 16mer | [243,244] | – |
| DPRHCQKRVLPCPAWL FRERCDKHPQKCTKFL | [245] | 16mer | [245] | – | |
| GGVSCMQTSPVCENNL (A2-6) | [246] | 16mer | – | – | |
| Galectin-3 | ANTPCGPYTHDCPVKR (G3-C12) PQNSKIPGPTFLDPH (G3-A9) | [247] | 16mer | [248,249] | – |
| T antigen | IVWHRWYAWSPASRI (P30-1) | [250,251] | 15mer | [252] | – |
| HGRFILPWWYAFSPS (P-30) | [253] | 15mer | – | – | |
| Fibrin-fibronectin complexes | CGLIIQKNEC (CTL1)f | [254] | CX8C | [254] | – |
| CNAGESSKNC (CTL2)f | |||||
| FGFR | AESGDDYCVLVFTDSAWTKICDWSHFRN (C19) | [255] | 26mer | – | – |
| MQLPLAT | [256] | Ph.D.-7 | – | – | |
| CRALLRGAPFHLAEC | [257] | 15mer | – | – | |
| E-selectin | IELLQAR | [258] | Ph.D.-7 | – | [258] |
| MMP2-processed collagen IV | TLTYTWS | [259] | Ph.D.-7 | – | – |
| PSA | CVAYCIEHHCWTC (C-4) | [260] | CX3CX4CX2C | – | – |
| Notch1 NRR | AC#ERYQGC#FSVGGYC#G (NRR17) | [261] | ACX6CX6CG | – | – |
| CD44 | THENWPA (CV-1) | [262] | Ph.D.-7 | – | – |
| WHPWSYLWTQQA (RP-1) | [263] | Ph.D.-12 | – | – | |
| FGF3 | VLWLKNR (FP16) | [264] | Ph.D.-7 | – | – |
| Extradomain-B fibronectin | CTVRTSADC (ZD2) | [265] | Ph.D. C7C | [265] | – |
| APRIL | AAAPLAQPHMWA (sAPRIL-BP1) | [266] | Ph.D.-12 | – | [266] |
| p16 | SHSLLSS | [267] | Ph.D.-7 | – | – |
| pre-miR-21 | ALWPPNLHAWVP | [268] | Ph.D.-12 | – | – |
Cysteine residues that form disulfide bonds are indicated in bold and italic
Mirror-image phage display strategy was used. Synthetic D-version of the IgV domain of PD-L1 was used as screening target D-version of this peptide targets PD-L1.
Bicyclic peptide cyclized by1,3,5-tris(bromomethyl)benzene.
Bicyclic peptide cyclized by1,3,5-triacryloyl-1,3,5-triazinane.
ICR-62 mAb was used as a panning probe;
12H23 mAb was used as a panning probe.
Bicyclic peptides binding to the ECD of Her2.
L-26 mAb was used as a panning probe;
N-12 mAb was used as a panning probe;
human plasma clots were used for panning probes; GPC3: Glypican-3;T antigen: Thomsen-Friedenreich glycoantigen; PS: Phosphatidyl serine; IL-10 RA: human interleukin-10 receptor alpha; APN: Aminopeptidase N; PPP2R1A: phosphatase 2 regulatory subunit A, α-isoform; CXCR2: CXC chemokine receptor-2; TfR1: human transferrin receptor 1; IL-6Rα: Interleukin 6 receptor chain α; Notch1 NRR, negative regulatory region in Notch 1. p16: Protein p (16INK4a).
Table 3.
Tumor-targeting peptides identified from phage-display peptide libraries by screening cancer cells or tumors.
| Receptor | Sequencea | Ref | Library | Selection | Cells or tumors | Cancer type | Imaging | Therapy |
|---|---|---|---|---|---|---|---|---|
| HER2 | LTVSPWY | [269,270] | Ph.D.-7 | In vitro | SKBR3 | Breast cancer | [271,272] | [269,273] |
| α-Enolase | SSMDIVLRAPLM (pHCT74) | [274] | Ph.D.-12 | In vitro | HCT116 | Colorectal cancer | – | [274] |
| EGFR | FPMFNHWEQWPP | [275] | Ph.D.-12 | In vitro | U-87 MG | Glioblastoma | [275] | – |
| SYPIPDT (P1) | [22] | Ph.D.-7 | In vitro | A431 | Epidermoid | – | – | |
| HTSDQTN (P2) | carcinoma | |||||||
| MUC18 | CLFMRLAWC | [276] | CX7C | In vitro | B16 cells cocultured with B-1 lymphocytes | Melanoma | – | – |
| Nucleolin | DMPGTVLP | [277,278] | Landscape phage- | In vitro | MCF-7 | Breast cancer | – | [277,279] |
| DWRGDSMDS | display libraries | |||||||
| VPTDTDYS | f8/8 and f8/9 | |||||||
| VEEGGYIAA | ||||||||
| GP130b | VTWTPQAWFQWV (VTW) | [280] | PH.D.-12 | In vitro | U-87 MG | Glioblastoma | – | [281,282] |
| Nestin | AQYLNPS | [283] | Ph.D.-7 | In vitro | Glioma stem cells | Glioblastoma | – | [283] |
| Cadherins | CSSRTMHHC | [284] | CX7C | In vitro | B16-F10-Nex2 | Melanoma | – | [284] |
| α4β1 | CPLDIDFYC | CX7C | In vitro | Kasumi-1 | Leukemia | – | – | |
| α5β1 | CPIEDRPMC (RPMrel) | [285,286] | CX7C | In vitro | HT29 | Colon cancer | [285] | [286] |
| αvβ6 | RGDLATLRQLAQEDGVVG-VR | [24,25] | Ph.D.-20 | In vitro | H2009 | NSCLC | [287] | [288–293] |
| (H2009.1) SPRGDLAVLGHK (HBP) | [26] | PH.D.-12 | In vitro | HNO223 | Head and neck cancer | [26] | – | |
| SPRGDLAVLGHKY (HBP-1) | ||||||||
| αvβ3 (RMS-I) | CQQSNRGDRKRC (RMS-I) | [41] | CX7–10C | In vitro | RD | Rhabdo-myosarcoma | [41] | – |
| IL-13Rα2 | CMGNKCRSAKRP (RMS-II) CGEMGWVRC | [50] | Ph.D-C7C | In vitro | G26-H2 and SnB19-pcDNA cells | GBM | [50] | – |
| VPAC1 | GFRFGALHEYNS (VP2) | [294] | Ph.D.-12 | In vitro | CHO-K1 cell transfected wi VPAC1 | th Colorectal cancer | – | – |
| IGHC | CTLPHLKMC | [295] | CX C | In vitro | Raji | Lymphoma | – | – |
| HSPGb | ASGALSPSRLDT (OSP-1) | [296] | Ph.D.-12 | In vitro | 143B | Osteosarcoma | [296] | – |
| Adenoviral receptor | SWDIAWPPLKVP | [297] | PhD-12 | In vitro | A172 | Glioblastoma | – | – |
| GRP78 | CTVALPGGYVRVC (Pep42) | [298] | CX3-12C | In vitro | Me6652/4 | Melanoma | – | [298,299] |
| GRP78 | ETAPLSTMLSPY (GMBP1) | [300] | Ph.D.-12 | In vitro | SGC7901 | Gastric cancer | – | [300] |
| GRP78 | GIRLRG | [301] | Ph.D.-7 | In vivo | Irradiated GL261 gliomas i mice | n Murine glioma | [301] | [301] |
| APP | CPGPEGAGC | [302] | CX C | In vivo | Mammary tissue | Breast cancer | – | – |
| IL-11Rα | CGRRAGGSC | [303] | CX7C | In vivo | Prostate from a human patient | Waldenström macro-globulinemia | [304] | [305] |
| PDGFRβb | CRGRRST (RGR) | [306] | CX7C | In vivo | angiogenic islets in RIP1-Tag2 mice | Pancreatic istets | – | [307] |
| APN (CD13) | CNGRCVSGCAGRC (NGR) | [199,308] | CX9 | In vivo | MDA-MB-435 xenografts | Breast cancer | – | [199,309–317] |
| p32/gC1qR | CGNKRTRGC (LyP-1) | [318,319] | CX7C | Ex vivo | MDA-MB-435 xenograft | Breast cancer | [211] | [210,320–323] |
| TIP-1 | HVGGSSV | [324,325] | Ph.D.-7 | In vivo In vivo | Irradiated and SU11248- | LLC, murine | [325–328] | [325,327] |
| α2bβ3 | RGDGSSV | [329] | Ph.D.-7 | In vivo | treated LLC and GL261 tumo irradiated LLC and GL261 tumors | rs glioblastoma LLC, murine glioblastoma | [329] | [329] |
| α3β1b | SWKLPPS | [330] | Ph.D.-7 | In vivo | AZ-P7a tumor | Gastric cancer | – | [330] |
| αvβ3 αvβ5 | CRGDKRGPDC (iRGD) | [42] | CX7C | In vivo | PC-3 xenograft | Prostate cancer | [331] | [331,332] |
| NRP-1 | GGKRPAR (P4) | [38] | Ph.D.-7 | In vitro | PC-1 | Prostate cancer | – | – |
| RIGRPLR (P7) | ||||||||
| CGFYWLRSC | [35] | CX7C | In vitro | Molt-4 | Lymphoma | – | [35] | |
| RPARPAR | [37] | Ph.D.-7 | Ex vivo | PPC-1 human prostate | Prostate cancer | [333] | [333] | |
| In vivo | carcinoma xenograft | |||||||
| MMP2-processe collagen IV | d TLTYTWS | [259] | Ph.D.-7 | In vivo In vitro | In vivo LLC tumor-bearing mouse, then in vitro | Lewis lung carcinoma | – | [259] |
| VAV3 | SSQPFWS | [61] | Ph.D.-7 | In vivo | MMP2-processed collagen glioblastoma xenograft | IV xxx | [61] | – |
| CRKL | YRCTLNSPFFWEDMTHEC-HA | [334] | 20mer | In vivo | DU145 xenograft | Prostate cancer | – | [334] |
| Plectin-1 | KTLLPTP (PTP) | [335] | Ph.D.-7 | Ex vivo | PDAC cells from the Kras/p53 mouse | PDAC | [335,336] | – |
IGHC: immuno-globulin heavy chain; HSPG: heparin sulfate proteoglycans; PDAC: Pancreatic ductal adenocarcinoma; IL-13Rα2: Interleukin 13 receptor α2; GBM: glioblastoma multiforme; LLC: Lewis lung carcinoma; GRP78: glucose-regulated protein 78; APP: aminopeptidase P; NRP-1: neuropilin-1; APN: aminopeptidase N.
Cysteine residues that form disulfide bonds are indicated in bold and italic.
Candidate cellular receptor.
3.1.2.1. Epidermal growth factor receptor (EGFR)-binding peptides
EGFR is a cell surface protein that binds to epidermal growth factors and is over expressed in a variety of human cancer cells, thus making it an excellent target for drug delivery. Li et al. screened a phage-display peptide library and identified a peptide named GE11 (YHWYGYTPQNVI) that binds to EGFR specifically and efficiently with a dissociation constant (KD) of approximately 22 nM [21]. GE11 has a much lower mitogenic activity towards EGF. GE11 is internalized preferentially into cancer cells with high expression level of EGFR and is furthermore accumulated in EGFR over expressing tumor xenografts in vivo after i.v. administration. In gene delivery studies, GE11-conjugated polyethylenimine (PEI) was found to deliver genes efficiently into EGFR over expressing cells and tumor xenografts. Taken together, GE11 could be potentially used as a safe and efficient tumor-targeting vehicle for selective drug delivery mediated through EGFR.
Hamzeh-Mivehroud et al. recently identified two short peptide ligands (SYPIPDT and HTSDQTN) against EGFR from screening of a phage-display peptide library Ph.D.-7 [22]. EGFR expressing A-431 cells were used as the matrix in a cell-based subtractive biopanning approach. The identified peptides were able to inhibit the EGF-induced phosphorylation of EGFR in a concentration-dependent manner. The results of affinity binding experiments showed that EGF was able to inhibit competitively the binding of peptide-bearing phage to A-431 cells.
3.1.2.2. ανβ6 integrin-binding peptides
Integrin ανβ6 is highly expressed in many malignancies but is usually expressed at low or undetectable levels in normal adult tissues. It supports epithelial cell proliferation during inflammation, fibrosis, wound healing and carcinoma progression [23]. Overexpression of integrin ανβ6 usually correlates with malignant potential and poor prognosis [24]. Integrin ανβ6 can also increase cancer metastasis by promoting cancer cell invasion and migration. Because the expression pattern of integrin ανβ6 is restricted to tumors and other pathological tissues, it has become a promising diagnostic and therapeutic target.
A peptide specifically targeting lung cancer cells, named H2009.1 (RGDLATLRQLAQEDGVVGVR), was isolated by Oyama et al [25] through panning of a Ph.D.-20 peptide library. However, its cellular receptor target was not identified at that time. Later, Elayadi et al. found out that integrin ανβ6 is the binding receptor of this peptide [24]. H2009.1 is able to mediate cell-specific uptake of a fluorescent nanoparticle via this receptor. Nothelfer et al. identified a shorter ανβ6-targeting peptide, namely, HBP (SPRGDLAVLGHK), through screening a Ph.D.-12 phage-display peptide library against HNO223 head and neck squamous cell carcinoma cell (HNSCC) line [26]. HBP-1 (SPRGDLAVLGHKY) with a tyrosine added to the C-terminal of HBP for easy radiolabeling with 131I showing preferential binding to ανβ6 over ανβ3 integrin. 125I-labeled HBP-1 showed binding to 5 different HNSCC cell lines. [131I]-Labeled HBP-1 accumulated rapidly in 2 different HNSCC tumor xenografts, with stable uptake until 45 min after intravenous administration. Peptide histochemistry by HBP-1 was positive for HNSCC tissue sections but negative for normal tissues. Goodman and co-workers reported the discovery of an unexpected non-RGD recognition motif for integrin ανβ6 [27]. They compared the recognition profiles of recombinant ανβ6 and ανβ3 integrins by screening Ph.D.-7 and Ph.D.-12 peptide libraries. As predicted, phages binding strongly to ανβ3 were found to contain ubiquitous RGD sequences. However, in addition to RGD-containing phages, one-quarter of the phages isolated from the Ph.D.-12 library contained the distinctive consensus motif DLXXL for ανβ6. A synthetic DLXXL peptide, RTDLDSLRTYTL, was determined to be a selective inhibitor of RGD-dependent ligand binding to ανβ6 in an isolated receptor assay (IC50 = 20 nM), and in a cell adhesion assay (IC50 = 50 μM). DLXXL peptides were specific inhibitors of ανβ6-fibronectin interaction since synthetic scrambled or reversed DLXXL peptides were inactive. Further studies revealed that a 7-mer peptide sequence RG/TDLXXL is the minimum motif required for high affinity and selectivity towards ανβ6 and additional flanking amino acids resulted in further improvement in binding affinity and specificity.
Because linear peptides may suffer from in vivo instability and rapid in vivo clearance, several peptides cyclized by disulfide bond have been recently developed that exhibit high affinity for ανβ6 [28]. Several ανβ6-targeting ligands have been labeled with radionuclides for in vivo PET or SPECT imaging in animal models of cancer and other diseases. For a review, please refer to ref. [29].
3.1.2.3. Neuropilin-1 (NRP-1)-bindingpeptides
NRP-1 is a membrane-bound receptor and plays an essential role in normal neuronal and vascular development [30,31]. NRP-1 has also been implicated as a novel mediator of the primary immune response [32]. The expression of NRP-1 can be stimulated in response to tissue injury or hypoxic conditions [33]. NRP-1 was found to be highly expressed in a variety of solid tumors, such as prostate, breast, pancreatic, lung, ovarian, and gastrointestinal carcinomas [34]. In addition, the expression of NRP-1 is elevated in acute lymphoblastic leukemia (ALL) and acute myelogenous leukemia (AML) [35], and is correlated with poor survival of AML patients [36]. NRP-1 expression was found increased in the bone marrow of ALL and AML patients compared with normal bone marrow. Ligands that bind to NRP-1 should potentially exhibit an increased homing specificity towards leukemic bone marrow. Increased expression of NRP-1 was found correlated with tumor growth and vascularization in vivo and with invasiveness in human cancer. Therefore, NRP-1 could serve as an excellent target for effective receptor-mediated drug delivery. Teesalu et al. reported discovery peptide RPARPAR from ex vivo screening with T7 phage. NRP-1 was found to be the cellular receptor of this peptide [37]. Wang and colleagues isolated two peptide sequences GGKRPAR (P4) and RIGRPLR (P7) with superior binding affinity and specificity relative to the RPARPAR peptide by using a microfluidic phage selection (MiPS) system [38]. Compared to conventional biopanning methods, the MiPS is a more efficient system to identify peptides with higher affinity and specificity by applying stringent selection conditions against minimal amount of target cells [38]. By screening of human T-cell lymphoma Molt-4 cells with a phage-display peptide library CX7C, Karjalainen et al. discovered a motif F(F/Y)XLRS (X = any amino acids) targeting NRP-1 [35]. A cyclic peptide CGFYWLRSC, when linked to a pro-apoptotic peptide (klaklak)2 (wherein small letters stand for D-amino acids) decreased cell viability at a relatively low μM level in human leukemia and lymphoma cell lines, while the control, a mixture of peptides CGFYWLRSC and (klaklak)2, did not show cell killing effect at the same concentration.
3.1.2.4. ανβ3 integrin-binding peptides
The ανβ3 integrin is a receptor expressed on the surface of various normal and cancer cells, and binds to extracellular matrix proteins displaying the RGD sequence [39]. It plays a key role in angiogenesis and metastasis of human tumors. Peptide ligands targeting ανβ3 integrin have great potential for developing cancer targeting therapy and imaging. Phage-display peptide library screening has successfully been used to identify peptide ligands targeting ανβ3 integrin. By screening a phage-display library, a nonapeptide named RGD-4C (CDCRGDCFC) was identified to be highly selective against αν integrins [40]. Witt et al. screened phage-display peptide libraries with a human rhabdomyosarcoma (RMS) cell line RD, and discovered RMS-I (CQQSNRGDRKRC) targeting ανβ3 integrin expressed on the RD cells [41]. This peptide binds to RMS and other tumor cell lines, but not to normal skeletal muscle cells and fibroblasts. Sugahara et al. screened a cyclic CX7C peptide library displayed on T7 phage against tumor blood vessels in metastasis mouse models of human prostate cancer [42]. After three rounds of ex vivo selection with cell suspensions from bone tumors, followed by one in vivo selection for homing to the bone tumors, a tumor-homing peptide named iRGD (internalizing RGD) with sequence CRGDKRGPDC was discovered. It has been proposed that iRGD homes to tumors involving three steps: first iRGD bound to αν integrins on tumor endothelium via RGD motif, and was then cleaved by proteolysis to expose a binding motif for neuropilin-1 (NRP-1), which mediates penetration into tissue and cells [42]. MRI imaging with iRGD peptide-decorated superparamagnetic iron oxide nanoworms in mice bearing 22Rv1 orthotopic xenograft has shown significantly improved sensitivity in tumor imaging. In vivo anti-tumor efficacy study with iRGD-coated Abraxane demonstrated that 8-fold more Abraxane was found accumulatedinthe tumors than that of non-targeted-Abraxane. iRGD was able to bind to tumor vessels and then spread into the extravascular tumor parenchyma, whereas conventional RGD peptides only delivered the cargo to the blood vessels.
Optimization of RGD cyclopeptides through changing the ring size and amino acid chirality, and introducing constrained building blocks such as N-methylated amino acids led to the discovery of Cilengitide, a head-tail cyclic peptide with sequence [RGDf-N(Me)V-] [43–45]. Cilengitide binds strongly and relatively selectively to αvβ3 integrin. Preclinical studies of Cilengitide in mice demonstrated efficacious tumor regression [46]. Phase II studies also demonstrated that Cilengitide is a potential monotherapy in patients with recurrent glioblastoma [47]. However, in a phase III clinical trial investigating Cilengitideas anti-tumor therapyin combination with standard chemo-radiotherapy, it did not improve overall survival in patients with newly diagnosed glioblastoma [48]. Nevertheless, αvβ3 integrin remains a potential therapeutic and imaging target for cancer. RGD peptides could still be developed as vehicles to deliver drug payload or imaging probes to tumor sites. Cilengitide, however, is a head-to-tail cyclic peptide without any handle for attachment. One residue will need to be modified to introduce a handle.
3.1.2.5. Interleukin 13 receptor α2 (IL-13Rα2)-binding peptides
IL-13Rα2 is a plasma membrane receptor that is over-expressed in a majority of patients with glioblastoma multiforme (GBM) but not found in normal brain [49]. Debinski's group screened a disulfide-constrained heptapeptide phages display library, and identified several peptide ligands; one of these peptides, CGEMGWVRC, was found to bind to IL-13Rα2 with the highest specificity [50]. Surprisingly, the linear form of this peptide bound to IL-13Rα2 even more avidly than the disulfide-cyclized form. Furthermore, they found this linear peptide is capable of homing to both subcutaneous and orthotopic human GBM xenografts expressing IL-13Rα2 when administered intravenously [50].
3.1.2.6. Vascular endothelial growth factor receptor 3 (VEGFR-3)-binding peptides
VEGFR-3 belongs to class III receptor tyrosine kinase family and is initially expressed in all embryonic endothelia, but its expression in the blood vessel endothelium decreases during development. Human VEGFR-3 is up-regulated in a variety of human cancers. Inhibiting the signal pathway of VEGFR-3 could prevent the growth and metastasis of tumor [51,52]. To identify novel ligands with specific binding capabilities to VEGFR-3, Qin and coworkers screened a phage-display peptide library against VEGFR-3 and discovered a consensus peptide motif CSDXXHXWC (X is any amino acid). Peptide P1, CSDSWHYWC, exhibited the highest affinity to VEGFR-3 in phage ELISA. Chemically synthesized P1 can bind to VEGFR-3 positive carcinoma cells with specificity [53].
3.1.2.7. Platelet-derived growth factor receptor β (PDGFRβ)-binding peptides
The PDGFRβ is a transmembrane glycoprotein in the receptor tyrosine kinase family. It is an important factor for regulating cell proliferation, cellular differentiation, cell growth and development. PDGFRβ is implicated in tumor growth through angiogenesis activation and in early stages of fibrosis. PDGFRβ is upregulated in various solid tumors, and its signaling plays a key role in the regulation of tumor interstitial fluid pressure [54]. This receptor represents an attractive and potentially valuable target for treatment and molecular imaging in oncologic and fibrotic diseases.
Peptide IPLPPPSRPFFK targeting PDGFRβ has been identified through biopanning with a Ph.D.-12 library against the recombinant extracellular domain of PDGFRβ [55]. PDGFR-P1 (IPLPPPSRPFFKY-NH2) is a synthetic peptide with a tyrosine was added to the C-terminal of the identified peptide for radiolabeling = with 125I or 131I. In vitro studies demonstrated a higher binding to PDGFRβ-expressing BxPC3 and MCF7 cells as well as PDGFRβ-transfected-HEK cells in comparison to negative control wtHEK293 and CaIX-transfected HEK cells. Binding was inhibited up to 90% by the unlabeled PDGFR-P1 peptide. In vivo biodistribution experiments were performed in subcutaneous BxPC3 tumor mouse model in Balb/c nude mice. Ex vivo distribution studies revealed a higher accumulation in BxPC3 tumors than in most normal organs. Therefore, PDGFR-P1 is a promising candidate for targeting human PDGFRβ.
3.1.2.8. Protein tyrosine phosphatase receptor type J (PTPRJ)-binding peptides
Mutations or overexpression of protein tyrosine kinases (PTKs) often result in cell malignant transformation [56]. Protein tyro-sine phosphatases (PTPs) that antagonize the oncogenic PTK signaling are considered potential tumor suppressors and, consequently, potential targets for novel anticancer therapies [57]. PTPRJ is a receptor protein tyrosine phosphatase whose expression is strongly reduced in many cancer cell lines and tumor specimens [58]. Using PTPRJ-His-6 recombinant protein as a probe to screen a phage display library Ph.D.-C7C, Paduano et al. isolated two peptide ligands PTPRJ-pep19 (CHHNLTHAC) and PTPRJ-pep24 (CLHHYHGSC), which could induce mitogen-activated protein kinase (MAPK) dephosphorylation and inhibit cell growth of HeLa and human umbilical vein endothelial cell (HUVEC) cells [59]. In a subsequent work, they developed a panel of nonapeptide analogues based on the sequence of PTPRJ-pep19; one of this PTPRJ-19.4 (CHHALTHAC) was able to dramatically reduce cell proliferation and effectively trigger apoptosis of both HeLa and HUVEC cells, as well as inhibiting in vitro tube formation on Matrigel [60].
3.1.2.9. VAV3-binding peptides
VAV3 is a guanine nucleotide exchange factor and activates the Rho GTPase pathway to promote invasion, proliferation, and survival. Expression level of VAV3 in recurrent glioblastoma is higher than that of primary glioblastoma [61]. Elevated VAV3 expression correlates with higher grade tumor and poor prognosis. Self-renewing glioma-initiating cells (GICs) in glioblastoma were found to express high level of VAV3, at the apex. Targeting VAV3 by ribonucleic acid interference decreased GIC growth, migration, invasion and in vivo tumorigenesis. A VAV3-targeting peptide SSQPFWS was identified by the Rich group through in vivo biopanning of the Ph.D.-7 library in mice implanted subcutaneously with patient-derived glioblastoma xenografts [61]. This peptide specifically internalized into GICs. When labeled with a fluorescent dye, this peptide was found to be able to identify and sort functional GIC cells from bulk and unsorted glioma culture using FACS.
3.2. Yeast-display peptide library
3.2.1. Methodology
Yeast-display technique was first published by the Wittrup lab [62]. Like other directed evolution display technologies, yeast-display relies on an intimate linkage between genotype (plasmid encoding the gene) and phenotype (protein scaffold expressed on the cell surface) [63]. Among different yeast strains and many cell wall anchors that have been reported inyeast-display, Aga2 subunit of the mating protein a-agglutinin in Saccharomyces cerevisiae is most commonly used. In this system, mutant proteins or library peptides are displayed as fusion protein with Aga2p, which in turn are covalently linked via two disulfide bonds to Aga1p subunit that anchors on the yeast cell wall. Incorporation of a hemagglutinin (HA) and/or c-myc tag adjacent to the displayed protein/peptide enables easy detection with immunoflu-orescence (see Fig. 2). In a yeast-display library, each yeast cell displays ∼50,000 copies of the mutant proteins or peptides; in some cases, the expression level may be lower.
Fig. 2.
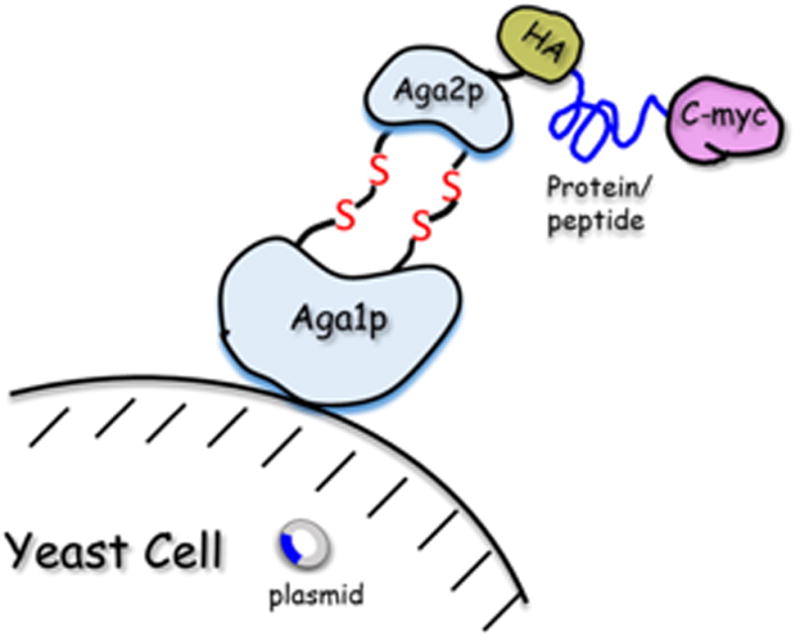
Display surface of the yeast cell.
Library screening in general involves incubation of the library with target-coated magnetic beads or fluorescently labeled soluble target, followed by isolation of the positive yeast through magnetic separation or fluorescence activated cell sorting (FACS) [64] (Fig. 3).
Fig. 3.
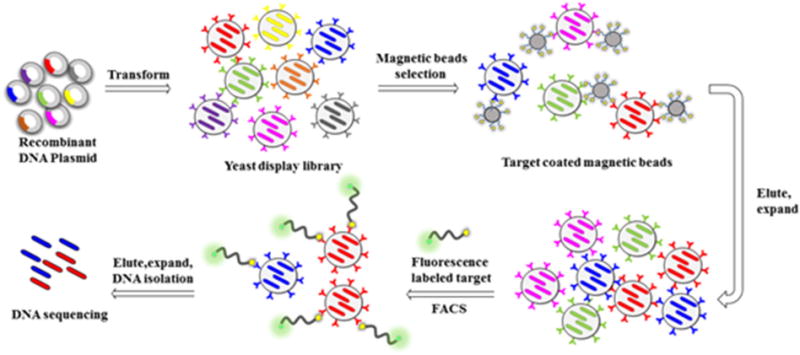
Construction and screening of yeast-display peptide library.
3.2.2. Discovery of tumor-targeting peptides from yeast-display peptide library
3.2.2.1. ανβ3 integrin-binding peptides
Silverman et al. replaced a six amino acid loop in agouti-related protein (AgRP) with a nine amino acid loop containing an RGD motif, created a yeast surface display library by randomizing the residues flanking the RGD sequence, and identified six clones with high affinity for ανβ3 integrins [65]. Binding data showed that the engineered AgRP peptides bound to cells expressing ανβ3 integrins with affinities ranging from 15 nM to 780 pM and were highly specific for ανβ3 over other integrins [65].
3.2.2.2. Cytotoxic T lymphocyte-associated antigen 4 (CTLA-4)-binding peptides
CTLA-4, an inhibitory receptor expressed by T lymphocytes, has emerged as a target for the treatment of metastatic melanoma. In a recent research, Maaβ et al. isolated ten different cystine-knot peptides that bind to the extracellular domain of CTLA-4 from knowledge-based combinatorial library based on the cystine-knot peptide McoTI-II. Cystine-knots (also known as knottins) are small, compact peptides (typically 20-60 amino acids) that consist of a core of at least three disulfide bonds that are interwoven into a “knot” conformation [66]. The most potent peptide MC-CT-010 (SPRCKYSHVPCRRDSDCPGKCICRGNGYCG) containing four cyclic loops was conjugated with neutravidin, and fused to antibody Fc domain or the oligomerization domain of C4b binding protein, resulting in enhanced dissociation constants in the nM range [67].
The tumor-targeting peptides identified by the yeast-display library method are summarized in Table 4.
Table 4.
Tumor-targeting peptides identified from yeast-display peptide library.
| Receptor | Sequence | Ref | Library type |
|---|---|---|---|
| Mcl-1 | RPEIWMTQGLRRLGDEINAYYAR (MS1) RPEIWLTQSLQRLGDEINAYYAR (MS2) | [337] | Bim-BH3 variants |
| RPEIWLTQHLQRLGDEINAYYAR (MS3) | |||
| RPEIWIAQEIDRIGDEVNAYYAR (MB1) | [338] | Bim-BH3 | |
| CTLA-4 | SPR3CKYSHVP2CRRDSD1CPGK3CI2CRGNGY1CG (MC-CT010)a | [67] | oMCoTI-II mutant Saccharomyces cerevisiae |
| αvβ3 Integrin | G4CVRLHES3CLGQQVP1C4CDPAAT3CY2CTGRGDEKLR2CY1CR (6C)α | [65] | AgRP mutants Pichia pastoris |
| αvβ3, αvβ5, and α5β1 Integrin | G3CPRPRGDNPPLT2CKQDSD1CLAG3CV2CGPNGF1CG (EETI2.5F)α | [339] | EETI-II mutant S. cerevisiae |
| Bcl-XL | RPEIWVAQELKRNGDEFNAYYAR (BCL-XL) | [338] | Bim-BH3 |
Disulfide bonds formed between 1C and 1C, 2C and 2C, 3C and 3C, 4C and 4C, respectively. Mcl-1: myeloid cell leukemia 1. CTLA-4: cytotoxic T lymphocyte-associated antigen 4.
3.3. Bacteria-display peptide library
3.3.1. Methodology
Display of heterologous peptides on the surface of bacterial cells was reported as early as 1986 [68]. Bacteria-display library (up to 1011 different peptides) utilizing membrane flagella to display random pep-tides enables bound clones to be readily eluted from the immobilized target by mechanical shearing of the flagella even if the peptide-target interaction is very strong. In contrast, elution of phages can only be achieved by breaking the peptide-target interaction, which could be problematic if the affinity is extremely high. Furthermore, bacteria are easier to cultivate with a selection marker that helps to prevent library contamination [69]. In bacteria-display peptide libraries, peptides are typically fused to the C- or N-terminus, or inserted into the middle of one of the four bacterial proteins (FliTrx, CPX, OmpA, or invasion). Bacterial surface display is a well-established methodology for the discovery and optimization of peptides with desired activities such as binding to cancer cells [70]. The subject of bacteria-display library has previously been reviewed [71–73]. A step-by-step protocol of the design and screening of bacteria-display peptide library has been published by the Daugherty group [74]. Bacteria-display combines the advantages of phage- and yeast-display since large libraries can be constructed efficiently in Escherichia coli [75]. E. coli is ideal for display libraries because it grows quickly and is easy to manipulate. Similar to yeast-display, bacteria that display target-binding peptides can be enriched from large libraries using sequential magnetic-activated cell sorting (MACS) and FACS [76]. A screening process of bacteria-display peptide is shown in Fig. 4.
Fig. 4.
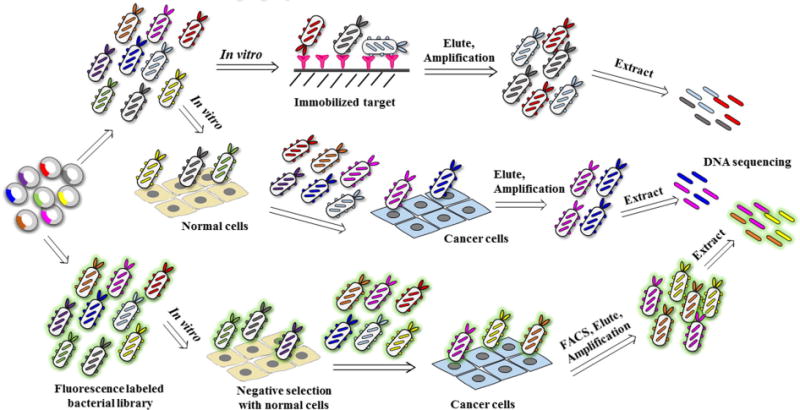
Construction and screening of bacteria-display peptide library.
3.3.2. Discovery of cancer targeting peptides from bacteria-display peptide library
3.3.2.1. Vascular endothelial growth factor (VEGF)-binding peptides
VEGF is a potent angiogenic factor and an essential growth factor for vascular endothelial cells [77]. VEGF is up-regulated in many tumors and plays important role in tumor angiogenesis and vascular permeability. VEGF-mediated signaling occurs in tumor cells, and this signaling contributes to key aspects of tumorigenesis, including the function of cancer stem cells and tumor initiation. Combination therapies using anti-VEGF therapies with chemotherapy and/or radiotherapy are effective against many types of tumor. To identify peptide ligands specific for VEGF, Kenrick and Daugherty constructed and screened bacteria-display peptide libraries, which were displayed on E. coli strain MC1061 on the N-terminus of circularly permuted OmpX (CPX) [76], by one round of Magnetic selection (MACS) followed by four rounds of FACS. They discovered a core motif of WE/DWE/D that conferred binding to VEGF from a random 15mer peptide library. A focused library X6W(E/D)W(E/D)X9 was constructed and screened. The resulting pep-tides from screening exhibited a consensus of CSR(F/L)(V/L)MWEWECF (account for 12 of 19 amino acid positions). A third library was constructed based on the consensus from the second generation of the form X4CX4(M/I)W(E/D)W(E/D)C(I/L/M/F)X3. A potent peptide named 3.30 (WPVRCSRFVMWEWECFLRA, KD = 470 nM) was identified. This peptide has a comparable binding affinity to peptide v114 (VEPNCDIHVMWEWECFERL, KD = 230 nM), which was affinity-matured using phage-display library approach. These two peptides share a high consensus motif CXXXVMWEWECFXR(A/L).
The tumor-targeting peptides identified by the bacteria-display library method are summarized in Table 5.
Table 5.
Tumor-targeting peptides identified from bacteria-display peptide library.
| Receptor | Peptide sequencea | Ref | Cancer type | Cell line used for selection | Library type |
|---|---|---|---|---|---|
| VEGF | GPGPCSRLVMWEWECFAAL WPVRCSRFVMWEWECFLRA | [76] | N/A | N/A | CPX |
| NS | IAVAPGWLWEEE (Hep1) | [340] | Liver cancer | HepG2 | FliTrx |
| KELCELDSLLRI (Hep2) | |||||
| IRELYSYDDDFG (Hep3) | |||||
| NS | CPGDRGQRRLFSKIEGPC (MM-2) | [341] | Prostate Cancer | PC-3 | FliTrx |
| NS | NVVRQ (TMTP1) | [342] | Prostate Cancer | PC-3M-1E8 | FliTrx |
| NS | VECYLIRDNLCIY | [343] | Breast cancer | ZR-75-1 | CPX |
| NS | EWCGIVRVGYCLGGGKK (PepC3) | [344] | Breast tumor | MDA-MB-231, MCF-7, and T47-D | CPX |
| NS | CGGRRLGGC | [345] | Murine squamous carcinoma | SCC VII | FliTrx |
| NS | WFCSWYGGDTCVQ | [346] | Lung cancer | A549 | CPX fluorescence-library |
Cysteine residues that form disulfide bonds are indicated in bold and italic. NS: not specified. CPX: circularly permuted outer membrane protein OmpX.
3.4. Ribosome-display and mRNA-display peptide libraries
3.4.1. Methodology
Ribosome-display is an in vitro selection and evolution technology for large libraries of proteins and peptides [78]. An important feature of this technology is that the RNA encoding the peptide library is translated in vitro while peptides and their corresponding RNA remain associated with the ribosome. The sequence information for the peptide of interest can be selected by affinity purification of the resulting peptide–ribosome–mRNA (PRM) complex [79]. Kawasaki first reported this technology in a patent application in 1991 [80]. In 1994, Mattheakis et al. used it to select peptide ligands against an antibody [81]. Heyduk et al. recently reported the improvement of ribosome-display method by using next generation sequencing (NGS) for decoding of the isolated PRMs [82]. A single NGS analysis can reveal the identity of millions of sequences in complex nucleic acid mixtures. Read count of a given pep-tide sequence is proportional to the relative abundance of a sequence in thesample. This method greatly reduces the number of selection rounds that were required to identify specific ligands. Since ribosome-display procedures are performed entirely in vitro, there are two main advantages over other biological-display technologies. First, the diversity of the peptide library is not limited by the transformation efficiency of bacterial cells but only by the number of ribosomes and different mRNA molecules present in the test tube. Second, random mutations can be introduced easily after each selection round. Fig. 5 demonstrates generation and screening of ribosome-display peptide library.
Fig. 5.
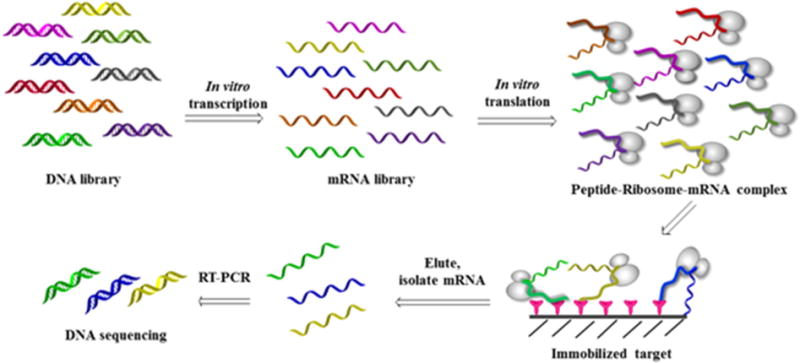
Construction and screening of ribosome-display library.
In mRNA-display library method, the mRNA is first ligated to a PEG linker connected to puromycin and translated in vitro [83]. The ribosome stalls at the junction between RNA and DNA. Puromycin then binds to the A-site of ribosome and attacks the peptidyl-tRNA at the P-site. The nascent peptide is thereby transferred to puromycin. The resulting covalently linked mRNA–peptide complex has the puromycin-linker-mRNA on one side of the tunnel, and the peptide on the other side of the tunnel. It is then reverse transcribed and used for selection. The DNA strandisrecovered from target-bound complexes by hydrolyzing the complementary mRNA at high pH, and then amplified by PCR [83]. Since all the steps of mRNA-display are in vitro and cell-free, library size is not limited by the need to transform bacteria and in principle can reach as large as ∼1013. Furthermore, using synthetic tRNAs pre-charged with desired unnatural amino acids, the PURE (protein synthesis using recombinant elements) system [84] enables the incorporation of D-, α-hydroxyl and N-methyl amino acids, N-substituted glycines (peptoid building blocks), and amino acids with reactive side chains for library cyclization into the macrocyclic or linear peptide libraries. Fig. 6 demonstrates generation and screening of ribosome-display peptide library. Since the peptide and mRNA are not topographically segregated, the mRNA can potentially interfere with the peptide.
Fig. 6.
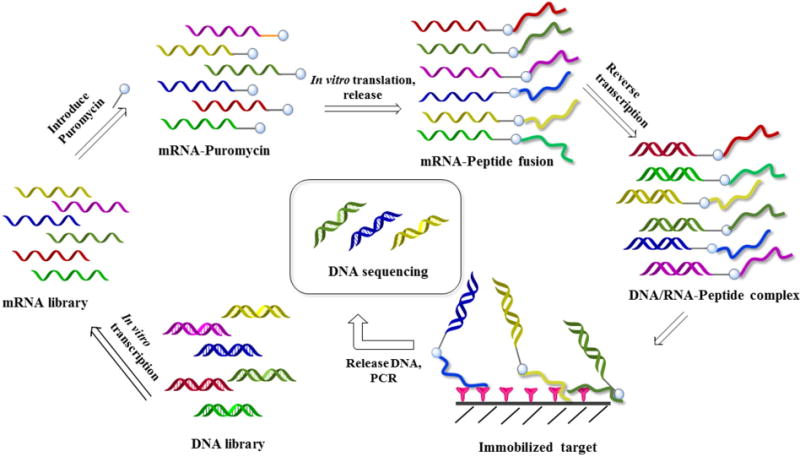
Construction and screening of mRNA-display library.
3.4.2. Discovery of tumor-targeting peptides from ribosome-display peptide library
Rong and Wen reported a ribosome-display system to isolate specific anti-tumor peptides from a designed random DNA library with affinity to membrane model [85]. Pore-forming peptides Mast21 and MastoparanX were chosen as positive control peptides to optimize the artificially synthesized tumor liposome cell membrane model. After 6 rounds of successive selection with improved membrane model (PE/PS, 3:7) and ribosome-display system, several peptides were obtained. Anti-tumor effects of the isolated peptides on non-small cell lung cancer cell line NCI-H460 were analyzed with MTT assay. In vitro experiment confirmed that two peptides SeqA3 (MKYDWEGVRDMFRRCLWISLRSWCVH) and SeqB3 (MKYDWRCLAGHAIKGWALRSHLAVYD) showed anti-tumor effects against NCI-H460 cells, with IC50 of 22.5 μM and 11.3 μM, respectively, compared to that of normal human fibroblast cell line CCD-27SK at a concentration greater than 100 μM.
3.4.3. Discovery of tumor-targeting peptides from mRNA-display peptide library
3.4.3.1. Interleukin-6 (IL-6)-binding peptides
IL-6 is a multifunctional cytokine and plays an important role in the host immune defense and the modulation of growth/differentiation in various malignancies [86–88]. Clinical studies have revealed that increased serum IL-6 concentrations in patients are associated with advanced tumor stages and short survival in patients [86]. Therefore, blocking IL-6 signaling is a potential targeted therapeutic strategy for cancer (i.e., anti-IL-6 therapy). Kobayashi et al. isolated a novel peptide inhibitor against IL-6 by in vitro selection using mRNA-display technology. Of a total of 39 clones analyzed, 12 had an identical sequence of NQQLIEEIIQILHKIFEIL, which was named CA11. After the amino acid sequence of CA11 was partially randomized and submitted for further selection, a new peptide RA07 (INTLLSEINSILLDIISLL) was obtained. RA07 specifically interacted with IL-6 and prevented the IL-6/IL-6R complex from binding to gp130 [89].
3.4.3.2. Vascular endothelial growth factor receptor 2 (VEGFR2)-binding peptides
Suga's group recently selected inhibitors of VEGFR2 from libraries produced in a PURE translation system with 16 natural amino acids and four backbone-modified unnatural amino acids (cycloleucine, D-phenylalanine, D-tyrosine, N-methyl-histidine, and N-methyl-phenyl-alanine) [90]. The most potent compound L1 is a cyclic head-to-tail thioether macrocyclic peptide, which showed relatively high serum stability and was much more potent than previously reported small molecule (“monomer”) antagonists obtained from combinatorial libraries [91-93]. It blocked VEGF-induced HUVEC proliferation with an IC50 of 60 nM and inhibited angiogenesis as measured by the HUVEC tube formation assay.
The tumor-targeting peptides identified by the ribosome- or mRNA-display library method are summarized in Table 6.
Table 6.
Tumor-targeting peptides identified from mRNA-display peptide library.
| Receptor | Sequence | Ref | Note |
|---|---|---|---|
| AKT2 | *YILVRNRLLRVD *CG (Pakti-L1) | [347] | Inhibited Akt2 with IC50 of 100 nM, and exhibited 10- and 25-fold selectivity over Akt1 and Akt3, respectively |
| IL-6 | NQQLIEEIIQILHKIFEIL (CA11) | [89] | RA07 prevented the L-6/IL-6R complex from binding to gp130 |
| INTLLSEINSILLDIISLL (RA07) | |||
| VEGFR | *FVVVSTDPWVNGLYID C (L1) | [90] | Inhibited HUVEC tube formation. Inhibit VEGF-induced HUVEC growth with IC50 of 60 nM |
| SIRT2 | *YSNFRIK(Tfa)RYSNSS *C (S2iL8) | [348] | Bound and inhibited SIRT2 with IC50 of 3.8 nM and exhibited 10- and 100-fold selectivity over SIRT1 and SIRT3, respectively |
An N-terminal chloroacetyl-l-tyrosine or chloroacetyl-,l-phenylalanine and a cysteine are cyclized with a thioether bond. Tfa: trifluoroacetyl.
4. OBOC peptide library
4.1. Design, synthesis and decoding of OBOC peptide library
Unlike phage-display technology, which limits the peptide library to l-amino acids and simple configuration structure, the OBOC technology offers a lot more structural possibilities, e.g. linear, cyclic, branch and macrocyclic peptide libraries, as well as peptide libraries comprised of both natural and unnatural amino acids (l-/d-, α-/β-/-γ-amino acids and amino acids with posttranslational modifications such as phosphorylation and glycosylation). Fig. 7 shows examples of different peptide libraries that can be constructed and encoded using OBOC library approach. OBOC library is synthesized on solid phase such as TentaGel resin beads from Rapp Polymere (Tübingen, Germany). Standard solid phase peptide synthesis employing Fmoc-chemistry and split-mix strategy are commonly used for the synthesis of OBOC peptide libraries (Fig. 8) [11,94]. Each 80-100 μm bead displays only one chemical entity and contains approximately 100 pmol of the same compound.
Fig. 7.
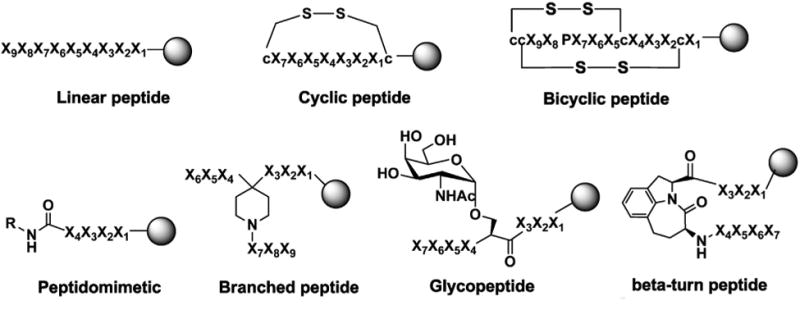
Examples of synthetic OBOC peptide and peptidomimetic libraries for discovery of tumor-targeting ligands. X1–X9 stand for natural and unnatural amino acids, l- or d-amino acids. c represents d-cysteine, K stands for l-lysine. R: alkyl or aryl.
Fig. 8.
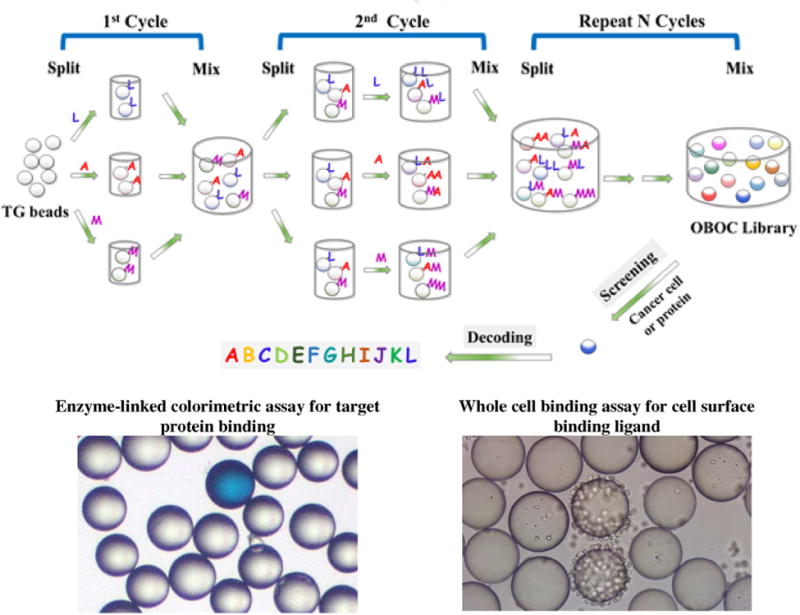
Synthesis and screening of an OBOC combinatorial peptide library. Eukaryotic amino acids use standard single-letter codes. B and J represent unnatural amino acids.
To successfully apply OBOC peptide library approach to the discovery of cancer-targeting peptides, good library design is important. Since the peptide concentration on bead surface is very high that may lead to identification of low-affinity ligands, a down-substitution method [95,96] employing topologically segregated bi-layer beads [97] has been developed to reduce loading only on bead surface while keeping full loading in the bead interior for decoding. Cell surface is generally negatively charged due to presence of the sialic acids and phospholipid head groups. To avoid non-specific anionic–cationic interactions between the cells and the peptide–bead surface, the amount of basic amino acids (i.e. lysine and arginine) in the library construction can be lowered, but not eliminated because basic residues may be required for binding or internalization. This is certainly true for the RGD motif of αvβ3, αvβ5 or α5β1 integrin and many cell-penetrating peptides. To reduce the rate of false positive beads, Kodadek's group introduced the concept of redundant OBOC libraries, in which more than one bead displays an identical compound [98]. If compounds isolated from the screening of a redundant library are present more than once, they are likely to be high quality ligands. While useful, redundant libraries limit the number of unique compounds that can be synthesized and screened at one time.
Tumor-targeting ligands can be discovered through screening OBOC libraries with live cancer cells (see below) or with an extracellular domain of cell surface receptors. In general, fully random peptide libraries (i.e., each amino acid has an equal chance to present in the library) are screened for the discovery of initial hits. Subsequent lead optimization can be achieved through screening-focused libraries, e.g. using motifs of the initial hits. Positive beads are picked up manually under a stereo-microscope or by automated sorting with a COPAS instrument [99]. If the target is a protein, it can be used to decorate magnetic nanoparticles, which in turn can be used as probes to detect and isolate positive beads with magnetic bead sorting [100,101]. After positive beads are isolated, chemical decoding can be performed directly with Edman microsequencing if coding tag consists of α-amino acids [102]. Alternatively, coding tag can be released from a bead for decoding via mass spectrometry (MS) [103,104]. A third method is partial Edman degradation-mass spectrometry (PED-MS) method developed by Pei et al. [104], in which partial Edman chemistry is performed on beads prior to release of “ladder sequences” for MS analysis. For OBOC peptide library comprising long sequence (e.g., >15mer), huge permutation or complex structure, decoding by MS remains a significant challenge. In that case, traditional automatic Edman microsequencing may be preferred, but it is slower and more expensive.
The main advantages of the OBOC method are (i) a large number (106-108) of compounds which can be rapidly synthesized and screened concurrently without the need for any special equipment, and therefore can be employed in any chemical or biochemical laboratory; (ii) OBOC library can be screened using either one or a combination of both on-bead binding and solution phase functional assays (via a cleavable linker) [105]; (iii) both binding and functional ligands (e.g., pro-apoptotic agents) [106,107] can be discovered. OBOC library can also be used for discovery of cell penetrating peptide ligands, but a cleavable linker and a reporting probe are needed in between the library compound and beads; (iv) OBOC combinatorial libraries are based on synthetic chemistry; therefore, it enables incorporation of D-amino acids, unnatural amino acids, many other organic building blocks, and investigation of cyclic, turned or branched peptides and secondary structures which can confer enhanced resistance to proteolytic degradation, a key requirement for clinical applications; (v) tumor-targeting ligands identified by OBOC library approach are not limited to peptides, but also N-methylated peptides [108], glycopeptides [109-111], peptide tertiary amides [112,113], peptidomimetics [114,115], and peptoids [116]; and (vi) the OBOC method can be used for rapid optimization of the initial lead compounds, whether they are native or identified from biological or synthetic peptide libraries. In OBOC method, each library compound is tethered to the solid support via a linker such as polyethylene glycol. In tumor imaging and drug delivery applications, the linker could be used as a convenient handle to connect the cancer targeting ligand with the therapeutic or diagnostic payload. However, unlike phage-displayed libraries, it is technically difficult to screen tumor-targeting ligand in live animals with OBOC technology.
4.2. Screening of OBOC library
4.2.1. Screening of OBOC library with live cancer cells
To identify ligands that bind to the surface of cancer cells, the direct way is to incubate OBOC library beads with live cancer cells and inspect under a microscope for cell-bound beads (Fig. 8). The screening process includes 5 steps: extensive washing of the library beads, incubating beads with cells, isolating positive beads, stripping of the cells from beads and decoding the peptide sequence of each positive bead. Beads with the appropriate ligands that bind to the cell surface receptors will be coated by a monolayer of cells and considered as “hits”. In order to eliminate the false “hits” due to non-specific binding, Lam and co-workers have developed two “subtraction screening” methods [117]. In the first method, the OBOC library beads are first screened with cancer cells, the positive beads coated with cancer cells are picked and recycled by stripping the cells off with 8 M guanidine hydrochloride followed by thorough washing. The recycled beads are then incubated with “normal” cells. Those beads that bind to both cancer and normal cell types are considered false positive. The second method is termed “dual-color screening method” which involves labeling the cancer cells with a fluorochrome (e.g. calcein AM) and mixing them with unlabeled “control” cells prior to screening the bead-library. Those beads that only bind to the fluorescent cells are considered “true positive” beads. In addition to screening for cell binding ligands, OBOC libraries can screen for both cell attachment and cell functions (e.g., cell signaling or apoptosis). One may use a green fluorescent protein (GFP) transfected cell line in which GFP will be expressed upon activation of a specific cell-signaling pathway. For the discovery of pro-apoptotic peptides, caspase-3 fluorescent substrate may be used to identify beads that are coated with cells undergoing apoptosis. This OBOC library method can be applied to both suspension and adherent cells, as well as fresh cancer cells isolated from patient blood, pleural fluid, ascites, or biopsy specimens.
Traditionally, positive beads identified from screening of OBOC library are picked up manually with a micropipette. However, it could be tedious and time-consuming if there were too many beads to pick. To speed up the screening process, fluorescence-based sorting platform such as the Complex Object Parametric Analyzer and Sorter (COPAS, from Union Biometrica) has been developed. Lewis group reported high-throughput screening of OBOC peptide libraries using intact cells. They evaluated a COPAS large particle biosorter for high-throughput sorting of beads that were bound by MDA-MB-435 GFP breast cancer cells [118]. When an OBOC library of GRGDXX was screened against human cancer cells that express αvβ3 integrin, cells bound to beads were rapidly dissociated when sorted through the COPAS instrument. However, after the bound cells were reversibly cross-linked onto the beads with 3% formaldehyde, they have successfully sorted positive beads together with cells. This approach should facilitate screening of OBOC library with live cells to identify novel peptide ligands against cell surface targets in their native state. Major drawbacks of COPAS screening are that the equipment is expensive and generally not available in most laboratories, and formaldehyde fixation may preclude sequential screening with other cells. Furthermore, overfixation can potentially destroy the coding tags.
4.2.2. Screening of OBOC library with cancer-associated protein
Cancer-associated soluble proteins can be screened for binding to OBOC peptide libraries. For target protein that cannot be visualized directly through a microscope, a reporter system such as an enzyme, fluorophore, colorimetric dye or radionuclide conjugated to the target protein is needed to identify the interacting beads. A general approach is to use biotinylated protein and probe with streptavidin-alkaline phosphatase (AP) employing an enzyme-linked colorimetric assay. The selection process is relatively simple and involves the catalysis of the colorimetric substrate bromochloro-indolyl phosphate (BCIP) by the bead-bound AP, turning the positive beads turquoise (Fig. 8). A two-step subtraction approach is used to eliminate false positive beads that bind to streptavidin [117]. If an antibody to the target protein is readily available, a secondary antibody conjugated to AP can be used as the reporter system. The success of OBOC library screening is dependent on the stringency of the screening conditions and effective negative control. COPAS bead sorting can be used for screening of OBOC library with soluble proteins [119]. Alternatively, bead sorting can be achieved with the aid of streptavidin-coated magnetic nano-particles. Bulk magnetic separation can be performed on the bench of any lab at low cost without the need for expensive specialized equipment.
4.3. Discovery of tumor-targeting peptides from OBOC peptide library
We and other researchers have reported the use of the whole-cell bead binding assay to identify cell surface ligands against a number of different cancer cell lines, including both adherent and non-adherent cells.We have also performed experiments on fresh cancer cells isolated from patients. A variety of peptide libraries have been designed, synthesized and screened including libraries consisting of all l-amino acids or d-, both l- and d-amino acids. Some of these libraries are linear; others are cyclic (Fig. 7). Upon further optimization with focused libraries, we have found that unnatural amino acid substitution and/or addition of organic moieties often lead to the development of ligands with higher affinity and specificity. Tumor-targeting ligands identified from OBOC peptide library are summarized in Table 7.
Table 7.
Tumor-targeting peptides identified by OBOC combinatorial library methods.
| Receptor | Sequencea | Ref | Cancer type | Cancer cell line used for screening | In vivo Imaging | Therapy |
|---|---|---|---|---|---|---|
| α3β1 integrin | cdG-Phe(3,5-diF)-G-Hyp-NcR (LXY30) | [134] | Glioblastoma | U-87 MG | [134] | – |
| cNGQGEQc (pA) | [349] | NSCLC | A549 | – | – | |
| cDGLGDDc | [350] | Ovarian cancer | CaOV-3, ES-2, SKOV-3, OVCAR-3 | – | – | |
| cdG-HCit-GPQc (OA02) | [131] | Ovarian cancer | CaOV-3, ES-2, SKOV-3 | [3,131] | [121] | |
| cNGRGEQc | [351] | NSCLC | A549 | – | – | |
| cdGLG-Hyp-Nc (LXY1) | [132] | Glioblastoma | U-87 MG | [132] | – | |
| cdG-Tyr(3-NO2)-G-Hyp-Nc (LXY3) | [133] | Breast cancer | MDA-MB-231 | [133] | – | |
| kmviywkag (RZ-3) | [352] | Prostate cancer | DU145 | – | – | |
| α3β1/α6β1 integrin | kikmviswkg (HYD-1) | [352] | Prostate cancer | DU145 | – | [353] |
| α6β1 integrin | LNIVSVNGRH (RU-1)b | [354] | Prostate cancer | DU145 | – | – |
| NS | QMARIPKRLARH | [355] | Prostate cancer | LNCap | – | – |
| IgM kappa | wGeyvmvnG | [356] | Murine lymphoma | WEHI-231 | – | – |
| APN | YVEYHLC (AP-1) | [162] | Liver cancer | HepG2 | [162] | – |
| α4β1 integrin | c-Nle-D-Nle-T-Hyp-rc (pM2) | [357] | NSCLC | H1650 | – | – |
| LTGpLDI | [145] | Leukemia | Jurkat | – | – | |
| cLDYWDc, cWDLDHHc, sppLDIn, eapLDId, | [3] | Lymphoma | Raji | – | – | |
| fypLDFf, FSIpLDI, QSYpLDF | ||||||
| “LLP2A” peptidomimetic and a series | [115,358] | Leukemia | Jurkat Molt-4, Raji | [115,149–151,153] | – | |
| of analogues [115,358] | Lymphoma | |||||
| c-Nle-DWEEc, c-Nle-DVDEc, c-Nle-D-Chg-YMc, | [3,350] | Ovarian cancer | ES-2 | – | – | |
| cSD-Nle-D-Chg-c | ||||||
| yminp-Nle-DIdnhh | ||||||
| vswap-Nle-DIgspd | ||||||
| vqgp-Nle-DIafvl | ||||||
| vgnvp-Nle-DIgqea | ||||||
| wdinp-Nle-DIgsfn | ||||||
| wsrip-Nle-DIqeps | ||||||
| cLDI-Chg-Hyp-Yc | [3,350] | Ovarian cancer | SKOV-3 | – | – | |
| c-Nle-D-Chg-NDFc | ||||||
| c-Nle-D-Nle-PhgDc | ||||||
| cDEL-Nle-EWc | ||||||
| NS | cQDGRMGFc (PLZ4) | [359] | Bladder cancer | 5637 | [359,360] | [361] |
| αvβ3 integrin | cGRGDdvc (LXW7) | [155] | Glioblastoma | U-87 MG | [155] | – |
| cGRGDdvc (LXW7) | [155] | Melanoma | A375M | [155] | – | |
| cGRGDd-nal1-c (LXW64) | [108] | Glioblastoma | U-87 MG | [108] | – | |
| CD21 | YILIHRN (B1), PTLDPLP (B2), LVLLTRE (B3) | [163] | – | – | – | – |
NS: not specified. NSCLC: non-small cell lung cancer.
D-Cysteine residues that form disulfide bonds are indicated in bold and italic.
Original sequence has an uncertain amino acid at the c-terminal of RU-1. When omitted, the peptide retained binding to DU 145 cells in FACS and plate adhesion assay.
4.3.1. α3β1 integrin-binding peptides
Overexpressionofα3β1 integrin has been reportedinseveral cancer types, such as glioblastoma [120], ovarian cancer [121], breast cancer [122–124], lung cancer [125], and melanoma [126], and has been associated with poor prognosis, tumorigenesis, tumor metastasis, invasion, and resistance to cancer treatment [127–130]. Thus, α3β1 integrin has been investigated as a promising cancer-specific biomarker and therapeutic target.
Through screening of random OBOC libraries, we previously discovered a cyclic motif cDGXGXXc (c stands for D-cysteine) that binds to the α3 sub-unit of α3β1 integrin on ovarian adenocarcinoma cell lines ES-2, SKOV-3, and CaOV-3. We subsequently synthesized and screened two secondary libraries based on this motif and identified several new peptides with higher affinity towards these cell lines including a cyclic peptide named OA02 with the sequence cdG-HoCit-GPQc (wherein HoCit is l-homocitrulline) [131]. OA02-decorated paclitaxel-loaded nanomicelles showed improved tumor-targeting in the SKOV-3 mouse model compared with drug-loaded nanomicelles without OA02 [121]. We subsequently synthesized four OBOC peptide libraries based on cdGXGXXc and screened against MDA-MB-231 breast cancer cells. LXY1 with sequence cdGLG-Hyp-Nc (wherein Hyp is l-hydroxyproline) was identified with high binding affinity (KD = 0.4 μM) and specificity to α3 integrin [132]. Based on the established SAR information, two highly focused OBOC cyclic peptide libraries were further designed, synthesized, and screened against MDA-MB-231 breast cancer cells under stringent conditions. A novel cyclic peptide LXY3 (cyclic cdG-Tyr(3-NO2)-G-Hyp-Nc) with a high binding affinity (IC50 = 57 nM) was identified [133]. Moreover, the targeting efficiency and specificity of LXY3 to the breast adenocarcinoma tumors in mouse xenografts were further confirmed by in vivo andexvivo near-infraredfluorescence optical imaging. Using OBOC library approach followed by optimization with medicinal chemistry, we have very recently developed a cyclic nonapeptide ligand named LXY30 (cyclic cdG-Phe(3,5-diF)-G-Hyp-GcR), which showed further improved in vivo tumor targeting property [134]. Chemical structures of OA02 and LXY30 are shown in Fig. 9. Tumor-targeting of LXY30 has been verified by in vitro binding to SKOV3 cells (Fig. 10A) and clinical ovarian tumor tissue (Fig. 10B), as well as in vivo tumor uptake in SKOV3 xenograft mouse model (in vivo and ex vivo optical images are shown in Fig. 10C and D, respectively).
Fig. 9.
Chemical structuresof selected tumor-targeting ligands identified by the OBOC library approach. OA02 and LXY30 are peptide ligands of α3β1 integrin. LXW64 and LLP2A are ligand of αvβ3 and α4β1 integrin, respectively.
Fig. 10.
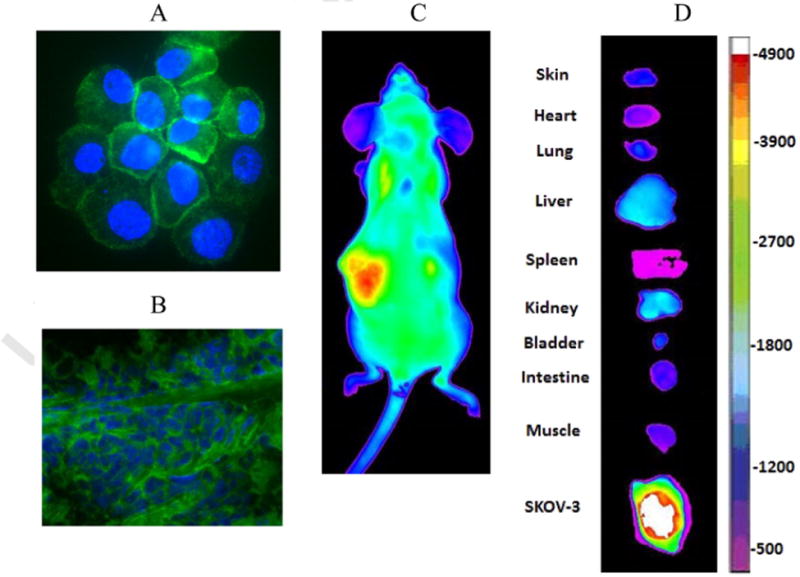
Applications of tumor-targeting peptides identified by OBOC library approach (examples). A: in vitro staining of SKOV3 cells with LXY30-Biotin-SA-PE; B: Staining of human ovarian tumor tissue with LXY30-Biotin-SA-Alexa488 (1 μM of LXY30-Biotin). Green fluorescence: Positive staining with Alexa488. Blue fluorescence: Nuclei counter-stained with DAPI; C: In vivo optical imaging of SKOV3 subcutaneous tumor with LXY30-SA-Cy5.5; D: Ex vivo optical imaging of SKOV3 subcutaneous tumor and normal organ.
4.3.2. α4β1 integrin-binding ligands
α4β1 integrin is a non-covalent heterodimeric transmembrane receptor, recognizing the QIDS and ILDV sequences in the vascular cell adhesion molecule-1 (VCAM-1) and fibronectin, respectively [135, 136]. Activated α4β1 integrin is expressed in lymphomas, leukemias, sarcomas, and melanomas [137]. It strengthens tumor cell adhesion to vasculature endothelium and facilitates tumor cell extravasation, thus promoting dissemination of tumor cells to distal organs [137–139]. α4β1 integrin prevents the apoptosis of malignant chronic lymphocytic leukemia (CLL) cells [140] and plays important roles in the drug resistance of both multiple myeloma [141] and acute myelogenous leukemia [142]. Anti-α4 integrin antibody has been found to inhibit multiple myeloma growth in a murine model [143]. Furthermore, α4β1 is expressed on proliferating but not on quiescent endothelial cells in angiogenesis during tumor development [144]. Therefore, α4β1 integrin is an attractive drug target against cancers, especially lymphoid malignancies. High-affinity and high-specificity ligands of α4β1 integrin can be used to noninvasively image α4β1 integrin-expressing tumors and to develop α4β1-targeting anti-cancer therapy.
Through screening random OBOC peptide libraries against live Jurkat T-lymphoid leukemia cells, we have identified peptide ligands which contain LD(V/I/F) motif against α4β1 integrin [145]. However, these ligands from random peptide libraries have relatively low affinity, and the binding is barely detectable on α4β1-expressing cells using radiolabeled or fluorescence-labeled ligands. Using a highly focused OBOC combinatorial peptidomimetic library in conjunction with a high stringency screening method, we have successfully identified a high-affinity (IC50 = 2 pM) and high-specificity tri-peptide derived molecule LLP2A (Fig. 9) that bind to activated α4β1 integrin of lym-phoid cancers [115]. LLP2A provides an important tool to noninvasively monitor α4β1 expression and activity during tumor progression and has been used successfully to detect α4β1-expressing tumors in vivo [115,146–154].
4.3.3. ανβ3 integrin-binding peptides
Lam and colleagues have designed several cyclic RGD-based OBOC peptide libraries and screened against ανβ3 integrin receptor transfected K562 myeloid leukemia cells using whole cell on-bead screening. A new RGD peptide, cGRGDdvc (LXW7) [155], cyclized by a disulfide bond and with a built-in handle at the carboxyl terminus, was identified. LXW7 shows high specificity against ανβ3 integrin and comparable binding affinity (IC50 = 0.68 μM) to some of the well-known RGD “head-to-tail” cyclic pentapeptide ligands reported in the literatures. It binds to both U-87MG glioblastoma and A375M melanoma cell lines, both of which express high levels of ανβ3 integrin. Optical in vivo imaging study with LXW7-biotin/streptavidin-Cy5.5 complex showed higher tumor uptake in U-87MG glioblastoma and A375M melanoma xenografts, and lower uptake by liver when compared to biotinylated RGD cyclo-pentapeptide ligands. Further optimization of LXW7 led to development of LXW64 with sequence cGRGDd-nal1-c (where nal1 stands for D-1-naphthylalanine). LXW64 exhibits 6.6-fold more potent binding affinity against ανβ3-expressing U-87MG cell in vitro and better in vivo tumor targeting compared to LXW7 [108].
RGD-based peptides may affect the biology of tumors resulting in increased tumor aggressiveness, invasiveness [156] and micrometastases [157]. Cancer cells treated with RGD-labeled iron oxide nanoparticles lose intercellular contacts and have decreased cell adhesion [158]. Low concentrations of RGD peptide can enhance tumor growth in vivo by promoting VEGF mediated angiogenesis [159]. Therefore, there is a great need to identify new non-RGD peptides that do not negatively affect tumor biology yet potently bind to ανβ3 integrin. The Lewis group recently identified several non-RGD peptides against ανβ3 integrin containing a (K/H)_(K/H) motif, through screening an OBOC octapeptide library via a multiplex “beads on a bead” library screening approach [160]. In this method, ανβ3 integrin was first biotinylated incompletely to average one biotin per molecule. The biotinylated integrin was then immobilized to 2 μm streptavidin-coated beads that are both magnetic (from iron oxide core) and fluorescent (from rhoda- mine), followed by incubating with OBOC peptide library. The positive beads, with the highest affinity peptides for ανβ3, were isolated by magnetic sorting and flow-based fluorescence separation. The positive beads were further validated with binding to ανβ3 integrin-expressing live cells, prior to MS decoding. The two peptides with the highest affinity, LCE62 (MAFKHKAH) and LCE64 (KTKKVHSQ), have KD value of 10.8 ± 1.2 and 4.7 ± 0.3 nM, respectively, against MDA-MB-435 breast cancer cells that express ανβ3 integrin. No significant uptake of FITC- LCE62 and FITC-LCE64 was observed in ανβ3-knockdown MDA-MB- 435 cells as well as ανβ3-low expression HT-1080 fibrosarcoma cells. Interestingly, peptides LCE62 and LCE64 do not appreciably alter the morphology, cell adhesion, and cell spreading of MDA-MB-435 cells, nor do they affect the tube formation of HUVEC cells in vitro. Taken together, non-RGD peptides LCE62 and LCE64 could be utilized as vehicles that effectively deliver imaging and therapeutic agents to ανβ3-expressing tumor cells without negatively affecting the biology of tumor cells.
4.3.4. Aminopeptidase N (APN)-binding peptides
APN is a cell membrane protein that plays a key role in tumor angio- genesis [161]. Wang et al. reported a continuous-flow microfluidic method for OBOC combinatorial peptide library screening [162]. They used an integrated screening strategy based on a lab-on-chip system that included high-throughput positive peptide isolation, magnetic bead sorting, single bead trapping, and in situ MS decoding. A 7mer OBOC peptide library containing ∼2 × 105 peptide beads was screened within 4 h, and 140 peptides targeting APN with a conserved YXXY sequence at the N-terminal were discovered. Peptide AP-1 (YVEYHLC) has high affinity (KD = 37.5 nM) and high specificity for APN. Ex vivo and in vivo optical imaging demonstrated high tumor uptake in a HepG2 liver cancer xenograft mouse model.
4.3.5. CD21-binding peptides
CD21 receptor is a cell surface marker of malignant B cell lymphoma. The Kopecek group has identified four heptapeptide ligands of CD21 through screening a 7mer peptide OBOC library with a two-step fluorescence screening method [163]. The binding affinities of selected peptides, YILIHRN (B1), PTLDPLP (B2), and LVLLTRE (B3), were in the μM range as determined by a fluorescence quenching assay. Peptide B1 was conjugated to N-(2-hydroxypropyl)-methacrylamide (HPMA) copolymer via linkers with different lengths containing one to four repeats of the 8-amino-3,6-dioxaoctanoic acid. The HPMA copolymer-B1 conjugates with three repeats of linker in the spacer showed optimal bio-recognition by the CD21 receptor.
5. PNA-encoded solution phase peptide library
5.1. Methodology
In 2004, Winssinger et al. reported a self-assembled PNA-encoded peptide library method [164]. In this method, peptides were first prepared by the “split-mix” synthesis method and then cleaved from the resin to form a PNA-encoded solution phase peptide library such that each peptide was linked to a PNA coding tag via a hydrophilic linker. The peptide library was then mixed with the protein of interest followed by exposure to a planar oligonucleotide microarray of predetermined sequences [165]. Alternatively, the PNA-encoded soluble peptide library can be first hybridized to the oligonucleotide microarrays and then mixed with the target protein. Decoding can be achieved via direct read-out from the oligonucleotide microarray. Svensen et al. recently reported a related PNA-encoded peptide library, which allows the identification of versatile cell-penetrating homing peptides and/or cell surface receptor ligands [166]. The PNA-tagged peptide library was first incubated with cells, which were then lysed, PNA extracted and hybridized with random DNAs, followed by amplification of the hybridized DNA for decoding via microarray hybridization readout. The advantages of this method include easy decoding and only a tiny amount of tags are needed for decoding. The disadvantage for this method is that since the peptide and PNA code are not topographically segregated, interference by the PNA coding tag could be problematic.
5.2. Discovery of tumor-targeting peptides from PNA-encoded solution phase peptide library
5.2.1. ανβ5/ανβ3 integrins- and CCR6-binding peptides
Svensen and Bradley reported discovery of peptide ligands targeting ανβ5/ανβ3 integrins and receptor CCR6 by screening a PNA-encoded peptide library (Library-1) with human D54 cells (overexpressing ανβ5 and ανβ3 integrins) and HEK293T-CCR6 (over-expressing CCR6) [166,167], respectively. Library-1, Ac-FQX4X3YX2X1IK-PNA17-fluorescein, consisted of 10,000 nonapeptide-PNA conjugates with each variable amino acid (X = 10 l- or D-amino acids) encoded by a PNA-triplet. Library screening identified two peptides with sequences of FQSIYPpIK [166] and FQpIYIIIK [167] bound to D54, and one peptide with sequence of FQIPYIIIK [166] bound to HEK293T-CCR6 cells, respectively. Their binding to specific cells was confirmed with flow cytometry analysis when cells were incubated with the FAM-labeled peptides or with CCR6 antibodies. These three peptides did not show cytotoxicity at 100 μM as assessed with standard MTT assays.
6. Peptide microarray
6.1. Methodology
Peptide microarrays are usually prepared by printing peptides on a solid surface (e.g. glass slides, cellulose sheets, polymer-based membranes, microchip, etc) through in situ synthesis (e.g., SPOT synthesis), in situ immobilization, or chemical ligation. Details on this technology can be found in the previous reviews [168,169]. Peptide microarrays enable investigators to rapidly analyze, in parallel, molecular interactions between immobilized molecules and complex biological mixtures. Screening is accomplished by a binding assay with cells or fluorescent-labeled protein. The peptide sequences of immobilized peptides on microarrays are known; therefore, decoding is not required. Another advantage of peptide microarray is peptides with different lengths or structures can be included in the same library to achieve maximal structure-activity relationship (SAR) information. Peptide microarrays enable rapid identification of critical amino acid residues for target binding and determination of the minimal binding sequence. Although there is no report so far, microfluidic technology holds the promise to prepare peptide microarray more efficiently. Work is currently underway in our laboratory to develop microfluidic print-head [170,171] that allows solid phase peptide synthesis on solid planar substrates using standard Fmoc-chemistry.
One main drawback of peptide microarray is that only a relatively small number of peptides can be generated and screened, therefore this technology is a medium throughput approach and mainly used for ligand optimization and SAR studies. Immobilization of peptides on a planar surface might influence their conformation, resulting in a decrease of their target affinities. The results provided by an array might be biased by unequal peptide quantity (concentration) or surface immobilization.
6.2. Discovery of tumor-targeting peptides from peptide microarray
6.2.1. PDGFRfi-binding peptides
Peptide microarrays have been successfully applied for optimization of the PDGFRp binding peptide PDGFR-P1 (IPLPPPSRPFFKY), which was originally identified from a phage-display peptide library [172]. A peptide array randomizing PDGFR-P1 by replacement of each amino acid with 20 natural amino acids was constructed via spotting. Fc-tagged PDGFRp and fibroblast growth factor receptor (FGFR, as a negative control target) were incubated with the peptide arrays. After washing, spot intensity was determined using an Fc-specific antibody conjugated to horseradish peroxidase. Selected derivatives and fragments of PDGFR-P1 were chemically synthesized, radiolabeled, and evaluated in cell-based assays, using PDGFRβ-overexpressing BxPC3 (human pancreatic carcinoma cells) and control MCF7 cells (human breast cancer expressing FGFR). Through addition of a D-Tyr to the N-terminus and replacement of Ser at residue 7 with Arg, the binding affinity of the resulting peptide yG2 (yIPLPPPRRPFFK, wherein y = 125I-tyrosine) against BxPC3 cells was increased (KD = 0.57 μM for yG2 vs 3.43 μM for PDGFR-P1). Serum stability of yG2 was found to improve 20-fold (t1/2 = 80 min for yG2 and 4 min for PDGFR-P1).
7. Perspectives
Cancer cell-targeting peptides identified from combinatorial libraries are excellent vehicles for delivery of therapeutic agents or imaging probes to the primary or metastatic tumor sites. Therapeutic payload includes radionuclides, potent cytotoxic peptides such as MMAE, therapeutic oligonucleotides such as siRNA and microRNA, biologics such as IL-2, nanocarriers encapsulating chemotherapeutic agents, and therapeutic check-point blockade antibodies. To be effective clinically, the cancer cell-targeting peptides need to have a high affinity and a high specificity to their target tissue(s). Non-specific targeting of these peptides to circulating blood cells, plasma proteins, or normal organs should be low. They should be stable in circulation and tissue proteases. For tumor imaging, uptake of the imaging probes into the cancer cells may not be needed. However, for many of the aforementioned therapeutic payloads, cancer cell uptake is essential. Although nanotherapeutics without targeting ligands can be passively taken up by the tumors via the “enhanced permeability retention (EPR)” effect, in vivo xenograft studies demonstrated that the addition of targeting ligands can enhance intra-tumoral distribution and cellular uptake of the nanotherapeutics into the tumor cells, resulting in better tumor response [121].
Drug resistance often develops with cancer treatment. This could be due to the lack of targeting to the “tumor stem cells”, emergence of mutant tumor cells that evade the targeting ligands or therapeutic payloads, or regional inaccessibility of some parts of the tumor to the therapeutic agent. Some of the solutions to this problem include (i) development of ligands that target all tumor cells including “tumor stem cells”, (ii) the use of therapeutic radionuclides that can exert “bystander effects”, (iii) the use of multiple different therapeutic pay-loads, and (iv) the addition of immunotherapy, such as checkpoint blockade, resulting in active targeting by the host's immune cells.
It is now clear that the tumor microenvironment plays a very important role in oncogenesis and tumor progression. Thus, there is a need to develop peptides that target specific cellular or acellular components within the tumor microenvironment. Therapeutic payloads against these targets are probably very different from those used in cancer cell targeting.
There is an increasing interest in developing therapeutic peptides or proteins that block intracellular protein-protein interactions. Peptide libraries have been used to discover such blocking agents. However, in vivo tumor-specific delivery of these peptides inside the target tumor cells could prove to be challenging. Cell-penetration peptides (CPP) have been used as the delivery agent, but CPPs are often nonspecific and they, together with the therapeutic payloads, will likely be taken up by a large number of circulating blood cells before they ever reach the tumor sites. One approach to overcome this hurdle is to use nanocarriers to protect the CPP-drug conjugates during the circulation and release them at the tumor sites. Another approach is to create “pro-CPP” that does not bind and enter circulating blood cells, but can be enzymatically activated at the tumor sites to become CPP. To further enhance tumor cell delivery of such drugs, one may incorporate both pro-CPP and tumor-targeting peptides to the final drug conjugate. Development of trifunctional peptides equipped with (i) tumor-targeting, (ii) CPP or pro-CPP, and (iii) protein-protein blocking functions could be very challenging. One approach is to incorporate one or more of these functional motifs (e.g. tumor-targeting and CPP functions) in the OBOC library design and screen for ligands that bind strongly to the target protein. Ligands identified from such libraries are expected to be able to target and penetrate target tumor cells in vivo as well as block specific protein-protein interaction inside the tumor cells.
Although many short peptides (e.g. <20mer) identified from combinatorial library screening are foreign to the host, they probably are not immunogenic unless they are conjugated to a carrier protein and administered together with immune adjuvants.
There is no set rule for optimal length and structure of peptides for receptor binding; it all depends on the nature of the receptors and the corresponding ligands. For example, integrins often bind to relatively short peptide motifs, such as -RGD-, which is the minimal binding motif for ανβ3, ανβ5, ανβ6, ανβ3, and ανβ1 integrin. Binding specificity and affinity of this tripeptide motif to specific integrin(s) depend greatly on the nature and chirality of the residues flanking the RGD sequence as well as the overall size of cyclization. Many peptides with RGD motifs have been discovered by both OBOC and phage-display library methods (Tables 2, 3, 4, 7).
Although structural information of the receptor is not needed in combinatorial library approaches, it could be very helpful. Many related peptides are often discovered from combinatorial library screening. If structural information of the receptor and binding affinity of the peptides are available, computational chemistry can guide rational design of peptides and focused peptide libraries; this can greatly facilitate the discovery process.
There have been great advances in the drug delivery field in recent years. Peptidic and non-peptidic ligands that bind to target cells continue to play a very important role in this field. In addition to cancer targeting, there are needsin other disease areas. For example, there is an urgent need for targeted-delivery of antibiotics to bacterial biofilms or pathogens hiding inside host cells. As stem cell biology advances, there is a great need for efficient delivery of therapeutic stem cells to target organ sites [173]. Undoubtedly, combinatorial technologies discussed in this review will play a significant role in the development of efficacious vehicles for drug, nucleic acid, and cell deliveries in these therapeutic areas.
Acknowledgments
The authors would like to thank the funding support from the NIH (R21 CA135345 for Liu and R01CA115483 for Lam). The authors wish to thank Tuong V. Pham and Jonathan S. Huynh for proofreading the manuscript.
Footnotes
This review is part of the Advanced Drug Delivery Reviews theme issue on “Peptides and Peptide Conjugates in Medicine”.
References
- 1.Lopci E, Nanni C, Rampin L, Rubello D, Fanti S. Clinical applications of 68Ga-DOTANOC in neuroendocrine tumours. Minerva Endocrinol. 2008;33:277–281. [PubMed] [Google Scholar]
- 2.Emons G, Sindermann H, Engel J, Schally AV, Grundker C. Luteinizing hormone-releasing hormone receptor-targeted chemotherapy using AN-152. Neuroendocrinology. 2009;90:15–18. doi: 10.1159/000225410. [DOI] [PubMed] [Google Scholar]
- 3.Aina OH, Liu R, Sutcliffe JL, Marik J, Pan CX, Lam KS. From combinatorial chemistry to cancer-targeting peptides. Mol Pharm. 2007;4:631–651. doi: 10.1021/mp700073y. [DOI] [PubMed] [Google Scholar]
- 4.Gray BP, Brown KC. Combinatorial peptide libraries: mining for cell-binding pep-tides. Chem Rev. 2014;114:1020–1081. doi: 10.1021/cr400166n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gautam A, Kapoor P, Chaudhary K, Kumar R. C. Open Source Drug Discovery, G.P. Raghava, Tumor homing peptides as molecular probes for cancer therapeutics, diagnostics and theranostics. Curr Med Chem. 2014;21:2367–2391. doi: 10.2174/0929867321666140217122100. [DOI] [PubMed] [Google Scholar]
- 6.Ullman CG, Frigotto L, Cooley RN. In vitro methods for peptide display and their applications. Brief Funct Genomics. 2011;10:125–134. doi: 10.1093/bfgp/elr010. [DOI] [PubMed] [Google Scholar]
- 7.Williams RM, Sooter LJ. In vitro selection of cancer cell-specific molecular recognition elements from amino acid libraries. J Immunol Res. 2015;2015:186586. doi: 10.1155/2015/186586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheetham AG, Keith D, Zhang P, Lin R, Su H, Cui H. Targeting tumors with small molecule peptides. Curr Cancer Drug Targets. 2015;16 doi: 10.2174/1568009616666151130214646. http://dx.doi.org/10.2174/1568009616666151130214646 Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soudy R, Byeon N, Raghuwanshi Y, Ahmed S, Lavasanifar A, Kaur K. Engineered peptides for applications in cancer targeted drug delivery and tumor detection. Mini Rev Med Chem. 2016;16 doi: 10.2174/1389557516666160219121836. http://dx.doi.org/10.2174/1389557516666160219121836 Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 10.Scott JK, Smith GP. Searching for peptide ligands with an epitope library. Science. 1990;249:386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- 11.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 12.Wolkowicz R, Jager GC, Nolan GP. A random peptide library fused to CCR5 for selection of mimetopes expressed on the mammalian cell surface via retroviral vectors. J Biol Chem. 2005;280:15195–15201. doi: 10.1074/jbc.M500254200. [DOI] [PubMed] [Google Scholar]
- 13.Kehoe JW, Kay BK. Filamentous phage display in the new millennium. Chem Rev. 2005;105:4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 14.McLaughlin ME, Sidhu SS. Engineering and analysis of peptide-recognition domain specificities by phage display and deep sequencing. Methods Enzymol. 2013;523:327–349. doi: 10.1016/B978-0-12-394292-0.00015-1. [DOI] [PubMed] [Google Scholar]
- 15.Kuzmicheva GA, Jayanna PK, Sorokulova IB, Petrenko VA. Diversity and censoring of landscape phage libraries. Protein Eng Des Sel. 2009;22:9–18. doi: 10.1093/protein/gzn060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jafari MR, Deng L, Kitov PI, Ng S, Matochko WL, Tjhung KF, Zeberoff A, Elias A, Klassen JS, Derda R. Discovery of light-responsive ligands through screening of a light-responsive genetically encoded library. ACS Chem Biol. 2014;9:443–450. doi: 10.1021/cb4006722. [DOI] [PubMed] [Google Scholar]
- 17.Chen S, Touati J, Heinis C. Tracking chemical reactions on the surface of filamentous phage using mass spectrometry. Chem Commun (Camb) 2014;50:5267–5269. doi: 10.1039/c3cc47496h. [DOI] [PubMed] [Google Scholar]
- 18.Heinis C, Rutherford T, Freund S, Winter G. Phage-encoded combinatorial chemical libraries based on bicyclic peptides. Nat Chem Biol. 2009;5:502–507. doi: 10.1038/nchembio.184. [DOI] [PubMed] [Google Scholar]
- 19.Kay BK, Kasanov J, Yamabhai M. Screening phage-displayed combinatorial peptide libraries. Methods. 2001;24:240–246. doi: 10.1006/meth.2001.1185. [DOI] [PubMed] [Google Scholar]
- 20.McGuire MJ, Samli KN, Chang YC, Brown KC. Novel ligands for cancer diagnosis: selection of peptide ligands for identification and isolation of B-cell lymphomas. Exp Hematol. 2006;34:443–452. doi: 10.1016/j.exphem.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Zhao R, Wu X, Sun Y, Yao M, Li J, Xu Y, Gu J. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J. 2005;19:1978–1985. doi: 10.1096/fj.05-4058com. [DOI] [PubMed] [Google Scholar]
- 22.Hamzeh-Mivehroud M, Mahmoudpour A, Dastmalchi S. Identification of new peptide ligands for epidermal growth factor receptor using phage display and computationally modeling their mode of binding. Chem Biol Drug Des. 2012;79:246–259. doi: 10.1111/j.1747-0285.2011.01282.x. [DOI] [PubMed] [Google Scholar]
- 23.Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elayadi AN, Samli KN, Prudkin L, Liu YH, Bian A, Xie XJ, Wistuba II, Roth JA, McGuire MJ, Brown KC. A peptide selected by biopanning identifies the integrin alphavbeta6 as a prognostic biomarker for nonsmall cell lung cancer. Cancer Res. 2007;67:5889–5895. doi: 10.1158/0008-5472.CAN-07-0245. [DOI] [PubMed] [Google Scholar]
- 25.Oyama T, Sykes KF, Samli KN, Minna JD, Johnston SA, Brown KC. Isolation of lung tumor specific peptides from a random peptide library: generation of diagnostic and cell-targeting reagents. Cancer Lett. 2003;202:219–230. doi: 10.1016/j.canlet.2003.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Nothelfer EM, Zitzmann-Kolbe S, Garcia-Boy R, Kramer S, Herold-Mende C, Altmann A, Eisenhut M, Mier W, Haberkorn U. Identification and characterization of a peptide with affinity to head and neck cancer. J Nucl Med. 2009;50:426–434. doi: 10.2967/jnumed.108.058123. [DOI] [PubMed] [Google Scholar]
- 27.Kraft S, Diefenbach B, Mehta R, Jonczyk A, Luckenbach GA, Goodman SL. Definition of an unexpected ligand recognition motif for alphav beta6 integrin. J Biol Chem. 1999;274:1979–1985. doi: 10.1074/jbc.274.4.1979. [DOI] [PubMed] [Google Scholar]
- 28.Kimura RH, Teed R, Hackel BJ, Pysz MA, Chuang CZ, Sathirachinda A, Willmann JK, Gambhir SS. Pharmacokinetically stabilized cystine knot peptides that bind alpha-v-beta-6 integrin with single-digit nanomolar affinities for detection of pancreatic cancer. Clin Cancer Res. 2012;18:839–849. doi: 10.1158/1078-0432.CCR-11-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Wu Y, Wang F, Liu Z. Molecular imaging of integrin alphavbeta6 expression in living subjects. Am J Nucl Med Mol Imaging. 2014;4:333–345. [PMC free article] [PubMed] [Google Scholar]
- 30.Gu C, Rodriguez ER, Reimert DV, Shu T, Fritzsch B, Richards LJ, Kolodkin AL, Ginty DD. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev Cell. 2003;5:45–57. doi: 10.1016/s1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herzog Y, Kalcheim C, Kahane N, Reshef R, Neufeld G. Differential expression of neuropilin-1 and neuropilin-2 in arteries and veins. Mech Dev. 2001;109:115–119. doi: 10.1016/s0925-4773(01)00518-4. [DOI] [PubMed] [Google Scholar]
- 32.Tordjman R, Lepelletier Y, Lemarchandel V, Cambot M, Gaulard P, Hermine O, Romeo PH. A neuronal receptor, neuropilin-1, is essential for the initiation of the primary immune response. Nat Immunol. 2002;3:477–482. doi: 10.1038/ni789. [DOI] [PubMed] [Google Scholar]
- 33.Matthies AM, Low QE, Lingen MW, DiPietro LA. Neuropilin-1 participates in wound angiogenesis. Am J Pathol. 2002;160:289–296. doi: 10.1016/S0002-9440(10)64372-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klagsbrun M, Takashima S, Mamluk R. The role of neuropilin in vascular and tumor biology. Adv Exp Med Biol. 2002;515:33–48. doi: 10.1007/978-1-4615-0119-0_3. [DOI] [PubMed] [Google Scholar]
- 35.Karjalainen K, Jaalouk DE, Bueso-Ramos CE, Zurita AJ, Kuniyasu A, Eckhardt BL, Marini FC, Lichtiger B, O'Brien S, Kantarjian HM, Cortes JE, Koivunen E, Arap W, Pasqualini R. Targeting neuropilin-1 in human leukemia and lymphoma, Blood. 2011;117:920–927. doi: 10.1182/blood-2010-05-282921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kreuter M, Woelke K, Bieker R, Schliemann C, Steins M, Buechner T, Berdel WE, Mesters RM. Correlation of neuropilin-1 overexpression to survival in acute myeloid leukemia. Leukemia. 2006;20:1950–1954. doi: 10.1038/sj.leu.2404384. [DOI] [PubMed] [Google Scholar]
- 37.Teesalu T, Sugahara KN, Kotamraju VR, Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc Natl Acad Sci U S A. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Liu Y, Teesalu T, Sugahara KN, Kotamrajua VR, Adams JD, Ferguson BS, Gong Q, Oh SS, Csordas AT, Cho M, Ruoslahti E, Xiao Y, Soh HT. Selection of phage-displayed peptides on live adherent cells in microfluidic channels. Proc Natl Acad Sci U S A. 2011;108:6909–6914. doi: 10.1073/pnas.1014753108. doi:S6909/6901-S6909/6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin H, Varner J. Integrins: roles in cancer development and as treatment targets. Br J Cancer. 2004;90:561–565. doi: 10.1038/sj.bjc.6601576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koivunen E, Wang B, Ruoslahti E. Phage libraries displaying cyclic peptides with different ring sizes: ligand specificities of the RGD-directed integrins. Biotechnology (N Y) 1995;13:265–270. doi: 10.1038/nbt0395-265. [DOI] [PubMed] [Google Scholar]
- 41.Witt H, Hajdin K, Iljin K, Greiner O, Niggli FK, Schafer BW, Bernasconi M. Identification of a rhabdomyosarcoma targeting peptide by phage display with sequence similarities to the tumour lymphatic-homing peptide LyP-1. Int J Cancer. 2009;124:2026–2032. doi: 10.1002/ijc.24170. [DOI] [PubMed] [Google Scholar]
- 42.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell. 2009;16:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kessler RGH, Hessler G, Gurrath M, Muller G. Conformation of cyclic peptides. Principle concepts and the design of selectivity and superactivity in bioactive sequences by spatial screening'. Pure Appl Chem. 1996;68:5. [Google Scholar]
- 44.Haubner R, Gratias R, Diefenbach B, Goodman SL, Jonczyk A, Kessler H. Structural and functional aspects of RGD-containing cyclic pentapeptides as highly potent and selective integrin αvβ3 antagonists. J Am Chem Soc. 1996;118:13. [Google Scholar]
- 45.Dechantsreiter MA, Planker E, Matha B, Lohof E, Holzemann G, Jonczyk A, Goodman SL, Kessler H. N-methylated cyclic RGD peptides as highly active and selective alpha(V)beta(3) integrin antagonists. J Med Chem. 1999;42:3033–3040. doi: 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- 46.Yamada S, Bu XY, Khankaldyyan V, Gonzales-Gomez I, McComb JG, Laug WE. Effect of the angiogenesis inhibitor Cilengitide (EMD 121974) on glioblastoma growth in nude mice. Neurosurgery. 2006;59:1304–1312. doi: 10.1227/01.NEU.0000245622.70344.BE. discussion 1312. [DOI] [PubMed] [Google Scholar]
- 47.Reardon DA, Fink KL, Mikkelsen T, Cloughesy TF, O'Neill A, Plotkin S, Glantz M, Ravin P, Raizer JJ, Rich KM, Schiff D, Shapiro WR, Burdette-Radoux S, Dropcho EJ, Wittemer SM, Nippgen J, Picard M, Nabors LB. Randomized phase II study of cilengitide, an integrin-targeting arginine-glycine-aspartic acid peptide, in recurrent glioblastoma multiforme, <st>J. Clin. Oncol. 2008;26:5610–5617. doi: 10.1200/JCO.2008.16.7510. [DOI] [PubMed] [Google Scholar]
- 48.Stupp R, Hegi ME, Gorlia T, Erridge SC, Perry J, Hong YK, Aldape KD, Lhermitte B, Pietsch T, Grujicic D, Steinbach JP, Wick W, Tarnawski R, Nam DH, Hau P, Weyerbrock A, Taphoorn MJ, Shen CC, Rao N, Thurzo L, Herrlinger U, Gupta T, Kortmann RD, Adamska K, McBain C, Brandes AA, Tonn JC, Schnell O, Wiegel T, Kim CY, Nabors LB, Reardon DA, van den Bent MJ, Hicking C, Markivskyy A, Picard M, Weller M. R. European Organisation for, C. Treatment of, C. Canadian Brain Tumor, C.s. team, Cilengitide combined with standard treatment for patients with newly diagnosed glioblastoma with methylated MGMT promoter(CENTRIC EORTC26071-22072 study):amulticentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2014;15:1100–1108. doi: 10.1016/S1470-2045(14)70379-1. [DOI] [PubMed] [Google Scholar]
- 49.Wykosky J, Gibo DM, Stanton C, Debinski W. Interleukin-13 receptor alpha 2, EphA2, and Fos-related antigen 1 as molecular denominators of high-grade astro-cytomas and specific targets for combinatorial therapy. Clin Cancer Res. 2008;14:199–208. doi: 10.1158/1078-0432.CCR-07-1990. [DOI] [PubMed] [Google Scholar]
- 50.Pandya H, Gibo DM, Garg S, Kridel S, Debinski W. An interleukin 13 receptor α 2-specific peptide homes to human glioblastoma multiforme xenografts. Neuro-Oncology. 2012;14:6–18. doi: 10.1093/neuonc/nor141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kubo H, Cao R, Brakenhielm E, Makinen T, Cao Y, Alitalo K. Blockade of vascular endothelial growth factor receptor-3 signaling inhibits fibroblast growth factor-2-induced lymphangiogenesis in mouse cornea. Proc Natl Acad Sci U S A. 2002;99:8868–8873. doi: 10.1073/pnas.062040199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Laakkonen P, Waltari M, Holopainen T, Takahashi T, Pytowski B, Steiner P, Hicklin D, Persaud K, Tonra JR, Witte L, Alitalo K. Vascular endothelial growth factor receptor 3 is involved in tumor angiogenesis and growth. Cancer Res. 2007;67:593–599. doi: 10.1158/0008-5472.CAN-06-3567. [DOI] [PubMed] [Google Scholar]
- 53.Qin X, Wan Y, Li M, Xue X, Wu S, Zhang C, You Y, Wang W, Jiang C, Liu Y, Zhu W, Ran Y, Zhang Z, Han W, Zhang Y. Identification of a novel peptide ligand of human vascular endothelia growth factor receptor 3 for targeted tumor diagnosis and therapy. J Biochem. 2007;142:79–85. doi: 10.1093/jb/mvm109. [DOI] [PubMed] [Google Scholar]
- 54.Pietras K, Rubin K, Sjoblom T, Buchdunger E, Sjoquist M, Heldin CH, Ostman A. Inhibition of PDGF receptor signaling in tumor stroma enhances antitumor effect of chemotherapy. Cancer Res. 2002;62:5476–5484. [PubMed] [Google Scholar]
- 55.Askoxylakis V, Marr A, Altmann A, Markert A, Mier W, Debus J, Huber PE, Haberkorn U. Peptide-based targeting of the platelet-derived growth factor receptor beta. Mol Imaging Biol. 2013;15:212–221. doi: 10.1007/s11307-012-0578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 57.Zhang JM, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PubMed] [Google Scholar]
- 58.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 59.Paduano F, Ortuso F, Campiglia P, Raso C, Iaccino E, Gaspari M, Gaudio E, Mangone G, Carotenuto A, Bilotta A, Narciso D, Palmieri C, Agosti V, Artese A, Gomez-Monterrey I, Sala M, Cuda G, Iuliano R, Perrotti N, Scala G, Viglietto G, Alcaro S, Croce CM, Novellino E, Fusco A, Trapasso F. Isolation and functional characterization of peptide agonists of PTPRJ, a tyrosine phosphatase receptor endowed with tumor suppressor activity. ACS Chem Biol. 2012;7:1666–1676. doi: 10.1021/cb300281t. [DOI] [PubMed] [Google Scholar]
- 60.Ortuso F, Paduano F, Carotenuto A, Gomez-Monterrey I, Bilotta A, Gaudio E, Sala M, Artese A, Vernieri E, Dattilo V, Iuliano R, Brancaccio D, Bertamino A, Musella S, Alcaro S, Grieco P, Perrotti N, Croce CM, Novellino E, Fusco A, Campiglia P, Trapasso F. Discovery of PTPRJ agonist peptides that effectively inhibit in vitro cancer cell proliferation and tube formation. ACS Chem Biol. 2013;8:1497–1506. doi: 10.1021/cb3007192. [DOI] [PubMed] [Google Scholar]
- 61.Liu JK, Lubelski D, Schonberg DL, Wu Q, Hale JS, Flavahan WA, Mulkearns-Hubert EE, Man J, Hjelmeland AB, Yu J, Lathia JD, Rich JN. Phage display discovery of novel molecular targets in glioblastoma-initiating cells. Cell Death Differ. 2014;21:1325–1339. doi: 10.1038/cdd.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boder ET, Wittrup KD. Yeast surface display for screening combinatorial polypep-tide libraries. Nat Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- 63.Angelini A, Chen TF, de Picciotto S, Yang NJ, Tzeng A, Santos MS, Van Deventer JA, Traxlmayr MW, Wittrup KD. Protein engineering and selection using yeast surface display. Methods Mol Biol. 2015;1319:3–36. doi: 10.1007/978-1-4939-2748-7_1. [DOI] [PubMed] [Google Scholar]
- 64.Gera N, Hussain M, Rao BM. Protein selection using yeast surface display. Methods. 2013;60:15–26. doi: 10.1016/j.ymeth.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 65.Silverman AP, Levin AM, Lahti JL, Cochran JR. Engineered cystine-knot peptides that bind alpha(v)beta(3) integrin with antibody-like affinities. J Mol Biol. 2009;385:1064–1075. doi: 10.1016/j.jmb.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kolmar H. Alternative binding proteins: biological activity and therapeutic potential of cystine-knot miniproteins. FEBS J. 2008;275:2684–2690. doi: 10.1111/j.1742-4658.2008.06440.x. [DOI] [PubMed] [Google Scholar]
- 67.Maass F, Wustehube-Lausch J, Dickgiessr S, Valldorf B, Reinwarth M, Schmoldt HU, Daneschdar M, Avrutina O, Sahin U, Kolmar H. Cystine-knot peptides targeting cancer-relevant human cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) J Pept Sci. 2015;21:651–660. doi: 10.1002/psc.2782. [DOI] [PubMed] [Google Scholar]
- 68.Freudl R, MacIntyre S, Degen M, Henning U. Cell surface exposure of the outer membrane protein OmpA of Escherichia coli K-12. J Mol Biol. 1986;188:491–494. doi: 10.1016/0022-2836(86)90171-3. [DOI] [PubMed] [Google Scholar]
- 69.Lunder M, Bratkovic T, Doljak B, Kreft S, Urleb U, Strukelj B, Plazar N. Comparison of bacterial and phage display peptide libraries in search of target-binding motif. Appl Biochem Biotechnol. 2005;127:125–131. doi: 10.1385/abab:127:2:125. [DOI] [PubMed] [Google Scholar]
- 70.Getz JA, Schoep TD, Daugherty PS. Peptide Discovery Using Bacterial Display and Flow Cytometry. 2012 doi: 10.1016/B978-0-12-396962-0.00004-5. [DOI] [PubMed] [Google Scholar]
- 71.Daugherty PS. Protein engineering with bacterial display. Curr Opin Struct Biol. 2007;17:474–480. doi: 10.1016/j.sbi.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 72.Lee SY, Choi JH, Xu Z. Microbial cell-surface display. Trends Biotechnol. 2003;21:45–52. doi: 10.1016/s0167-7799(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 73.Jostock T, Dubel S. Screening of molecular repertoires by microbial surface display. Comb Chem High Throughput Screen. 2005;8:127–133. doi: 10.2174/1386207053258479. [DOI] [PubMed] [Google Scholar]
- 74.Getz JA, Schoep TD, Daugherty PS. Peptide discovery using bacterial display and flow cytometry. Methods Enzymol. 2012;503:75–97. doi: 10.1016/B978-0-12-396962-0.00004-5. [DOI] [PubMed] [Google Scholar]
- 75.Bessette PH, Rice JJ, Daugherty PS. Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng Des Sel. 2004;17:731–739. doi: 10.1093/protein/gzh084. [DOI] [PubMed] [Google Scholar]
- 76.Kenrick SA, Daugherty PS. Bacterial display enables efficient and quantitative peptide affinity maturation. Protein Eng Des Sel. 2010;23:9–17. doi: 10.1093/protein/gzp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Goel HL, Mercurio AM. VEGF targets the tumour cell. Nat Rev Cancer. 2013;13:871–882. doi: 10.1038/nrc3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zahnd C, Amstutz P, Pluckthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat Methods. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 79.Hara S, Liu MZ, Wang W, Xu MY, Li Z, Ito Y. Stabilized ribosome display for in vitro selection. Methods Mol Biol. 2012;805:59–73. doi: 10.1007/978-1-61779-379-0_4. [DOI] [PubMed] [Google Scholar]
- 80.Kawasaki G. Screening Randomized Peptides and Proteins With Polysomes. PCT Int Appl WO. 1991 91/05058. [Google Scholar]
- 81.Mattheakis LC, Bhatt RR, Dower WJ. An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc Natl Acad Sci U S A. 1994;91:9022–9026. doi: 10.1073/pnas.91.19.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Heyduk E, Heyduk T. Ribosome display enhanced by next generation sequencing: a tool to identify antibody-specific peptide ligands. Anal Biochem. 2014;464:73–82. doi: 10.1016/j.ab.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 83.Pluckthun A. Ribosome display: a perspective. Methods Mol Biol. 2012;805:3–28. doi: 10.1007/978-1-61779-379-0_1. [DOI] [PubMed] [Google Scholar]
- 84.Josephson K, Ricardo A, Szostak JW. mRNA display: from basic principles to macrocycle drug discovery. Drug Discov Today. 2014;19:388–399. doi: 10.1016/j.drudis.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 85.Wen ZS, Rong TH. Establishment of tumor cell membrane model and screening for specific anti-tumor peptides. Ai Zheng. 2004;23:1660–1665. [PubMed] [Google Scholar]
- 86.Guo Y, Xu F, Lu T, Duan Z, Zhang Z. Interleukin-6 signaling pathway in targeted therapy for cancer. Cancer Treat Rev. 2012;38:904–910. doi: 10.1016/j.ctrv.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 87.Waldner MJ, Foersch S, Neurath MF. Interleukin-6—a key regulator of colorectal cancer development. Int J Biol Sci. 2012;8:1248–1253. doi: 10.7150/ijbs.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. J Clin Invest. 2007;117:3660–3663. doi: 10.1172/JCI34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kobayashi T, Kakui M, Shibui T, Kitano Y. In vitro selection of a peptide inhibitor of human IL-6 using mRNA display. Mol Biotechnol. 2011;48:147–155. doi: 10.1007/s12033-010-9355-5. [DOI] [PubMed] [Google Scholar]
- 90.Kawakami T, Ishizawa T, Fujino T, Reid PC, Suga H, Murakami H. In vitro selection of multiple libraries created by genetic code reprogramming to discover mac-rocyclic peptides that antagonize VEGFR2 activity in living cells. ACS Chem Biol. 2013;8:1205–1214. doi: 10.1021/cb300697h. [DOI] [PubMed] [Google Scholar]
- 91.Udugamasooriya DG, Dunham G, Ritchie C, Brekken RA, Kodadek T. The pharmacophore of a peptoid VEGF receptor 2 antagonist includes both side chain and main chain residues. Bioorg Med Chem Lett. 2008;18:5892–5894. doi: 10.1016/j.bmcl.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Udugamasooriya DG, Ritchie C, Brekken RA, Kodadek T. A peptoid antagonist of VEGF receptor 2 recognizes a ‘hotspot in the extracellular domain distinct from the hormone-binding site. Bioorg Med Chem. 2008;16:6338–6343. doi: 10.1016/j.bmc.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shrivastava A, von Wronski MA, Sato AK, Dransfield DT, Sexton D, Bogdan N, Pillai R, Nanjappan P, Song B, Marinelli E, DeOliveira D, Luneau C, Devlin M, Muruganandam A, Abujoub A, Connelly G, Wu QL, Conley G, Chang Q, Tweedle MF, Ladner RC, Swenson RE, Nunn AD. A distinct strategy to generate high-affinity peptide binders to receptor tyrosine kinases. Protein Eng Des Sel. 2005;18:417–424. doi: 10.1093/protein/gzi049. [DOI] [PubMed] [Google Scholar]
- 94.Lam KS, Lehman AL, Song A, Doan N, Enstrom AM, Maxwell J, Liu R. Synthesis and screening of “one-bead one-compound” combinatorial peptide libraries. Methods Enzymol. 2003;369:298–322. doi: 10.1016/S0076-6879(03)69017-8. [DOI] [PubMed] [Google Scholar]
- 95.Wang X, Peng L, Liu R, Xu B, Lam KS. Applications of topologically segregated bi-layer beads in ‘one-bead one-compound’ combinatorial libraries. J Pept Res. 2005;65:130–138. doi: 10.1111/j.1399-3011.2005.00192.x. [DOI] [PubMed] [Google Scholar]
- 96.Chen X, Tan PH, Zhang Y, Pei D. On-bead screening of combinatorial libraries: reduction of nonspecific binding by decreasing surface ligand density. J Comb Chem. 2009;11:604–611. doi: 10.1021/cc9000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Liu R, Marik J, Lam KS. A novel peptide-based encoding system for “one-bead one-compound” peptidomimetic and small molecule combinatorial libraries. J Am Chem Soc. 2002;124:7678–7680. doi: 10.1021/ja026421t. [DOI] [PubMed] [Google Scholar]
- 98.Doran TM, Gao Y, Mendes K, Dean S, Simanski S, Kodadek T. Utility of redundant combinatorial libraries in distinguishing high and low quality screening hits. ACS Comb Sci. 2014;16:259–270. doi: 10.1021/co500030f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hintersteiner M, Kimmerlin T, Kalthoff F, Stoeckli M, Garavel G, Seifert JM, Meisner NC, Uhl V, Buehler C, Weidemann T, Auer M. Single bead labeling method for combining confocal fluorescence on-bead screening and solution validation of tagged one-bead one-compound libraries. Chem Biol. 2009;16:724–735. doi: 10.1016/j.chembiol.2009.06.011. [DOI] [PubMed] [Google Scholar]
- 100.Astle JM, Simpson LS, Huang Y, Reddy MM, Wilson R, Connell S, Wilson J, Kodadek T. Seamless bead to microarray screening: rapid identification of the highest affinity protein ligands from large combinatorial libraries. Chem Biol. 2010;17:38–45. doi: 10.1016/j.chembiol.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mendes K, Ndungu JM, Clark LF, Kodadek T. Optimization of the magnetic recovery of hits from one-bead-one-compound library screens. ACS Comb Sci. 2015;17:506–517. doi: 10.1021/acscombsci.5b00090. [DOI] [PubMed] [Google Scholar]
- 102.Liu R, Lam KS. Automatic Edman microsequencing of peptides containing multiple unnatural amino acids. Anal Biochem. 2001;295:9–16. doi: 10.1006/abio.2001.5172. [DOI] [PubMed] [Google Scholar]
- 103.Paulick MG, Hart KM, Brinner KM, Tjandra M, Charych DH, Zuckermann RN. Cleavable hydrophilic linker for one-bead-one-compound sequencing of oligomer libraries by tandem mass spectrometry. J Comb Chem. 2006;8:417–426. doi: 10.1021/cc0501460. [DOI] [PubMed] [Google Scholar]
- 104.Joo SH, Xiao Q, Ling Y, Gopishetty B, Pei D. High-throughput sequence determination of cyclic peptide library members by partial Edman degradation/mass spectrometry. J Am Chem Soc. 2006;128:13000–13009. doi: 10.1021/ja063722k. [DOI] [PubMed] [Google Scholar]
- 105.Townsend JB, Shaheen F, Liu R, Lam KS. Jeffamine derivatized TentaGel beads and poly(dimethylsiloxane) microbead cassettes for ultrahigh-throughput in situ releasable solution-phase cell-based screening of one-bead-one-compound combinatorial small molecule libraries. J Comb Chem. 2010;12:700–712. doi: 10.1021/cc100083f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kumaresan PR, Wang Y, Saunders M, Maeda Y, Liu R, Wang X, Lam KS. Rapid discovery of death ligands with one-bead-two-compound combinatorial library methods. ACS Comb Sci. 2011;13:259–264. doi: 10.1021/co100069t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu R, Shih TC, Deng X, Anwar L, Ahadi S, Kumaresan P, Lam KS. Design, synthesis, and application of OB2C combinatorial peptide and peptidomimetic libraries. Methods Mol Biol. 2015;1248:3–22. doi: 10.1007/978-1-4939-2020-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang Y, Xiao W, Zhang Y, Meza L, Tseng H, Takada Y, Ames JB, Lam KS. Optimization of RGD containing cyclic peptides against alphavbeta3 integrin. Mol Cancer Ther. 2016;15:232–240. doi: 10.1158/1535-7163.MCT-15-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kracun SK, Clo E, Clausen H, Levery SB, Jensen KJ, Blixt O. Random glycopeptide bead libraries for seromic biomarker discovery. J Proteome Res. 2010;9:6705–6714. doi: 10.1021/pr1008477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Clo E, Kracun SK, Nudelman AS, Jensen KJ, Liljeqvist JA, Olofsson S, Bergstrom T, Blixt O. Characterization of the viral O-glycopeptidome: a novel tool of relevance for vaccine design and serodiagnosis. J Virol. 2012;86:6268–6278. doi: 10.1128/JVI.00392-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ying L, Liu R, Zhang J, Lam K, Lebrilla CB, Gervay-Hague J. A topologically segregated one-bead-one-compound combinatorial glycopeptide library for identification of lectin ligands. J Comb Chem. 2005;7:372–384. doi: 10.1021/cc049836e. [DOI] [PubMed] [Google Scholar]
- 112.Gao Y, Kodadek T. Split-and-pool synthesis and characterization of peptide tertiary amide library. J Vis Exp. 2014:e51299. doi: 10.3791/51299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Pels K, Kodadek T. Solid-phase synthesis of diverse peptide tertiary amides by reductive amination. ACS Comb Sci. 2015;17:152–155. doi: 10.1021/acscombsci.5b00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Teng P, Zhang X, Wu H, Qiao Q, Sebti SM, Cai J. Identification of novel inhibitors that disrupt STAT3-DNA interaction from a gamma-AApeptide OBOC combinatorial library. Chem Commun (Camb) 2014;50:8739–8742. doi: 10.1039/c4cc03909b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peng L, Liu R, Marik J, Wang X, Takada Y, Lam KS. Combinatorial chemistry identifies high-affinity peptidomimetics against alpha4beta1 integrin for in vivo tumor imaging. Nat Chem Biol. 2006;2:381–389. doi: 10.1038/nchembio798. [DOI] [PubMed] [Google Scholar]
- 116.Qi X, Astle J, Kodadek T. Rapid identification of orexin receptor binding ligands using cell-based screening accelerated with magnetic beads. Mol BioSyst. 2010;6:102–107. doi: 10.1039/b915611a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lehman A, Gholami S, Hahn M, Lam KS. Image subtraction approach to screening one-bead-one-compound combinatorial libraries with complex protein mixtures. J Comb Chem. 2006;8:562–570. doi: 10.1021/cc0600268. [DOI] [PubMed] [Google Scholar]
- 118.Cho CF, Behnam Azad B, Luyt LG, Lewis JD. High-throughput screening of one-bead-one-compound peptide libraries using intact cells. ACS Comb Sci. 2013;15:393–400. doi: 10.1021/co4000584. [DOI] [PubMed] [Google Scholar]
- 119.Townsend J, Do A, Lehman A, Dixon S, Sanii B, Lam KS. 3-Nitro-tyrosine as an internal quencher of autofluorescence enhances the compatibility of fluorescence based screening of OBOC combinatorial libraries. Comb Chem High Throughput Screen. 2010;13:422–429. doi: 10.2174/138620710791292994. [DOI] [PubMed] [Google Scholar]
- 120.Paulus W, Baur I, Schuppan D, Roggendorf W. Characterization of integrin receptors in normal and neoplastic human brain. Am J Pathol. 1993;143:154–163. [PMC free article] [PubMed] [Google Scholar]
- 121.Xiao K, Li Y, Lee JS, Gonik AM, Dong T, Fung G, Sanchez E, Xing L, Cheng HR, Luo J, Lam KS. “OA02” peptide facilitates the precise targeting of paclitaxel-loaded micellar nanoparticles to ovarian cancer in vivo”. Cancer Res. 2012;72:2100–2110. doi: 10.1158/0008-5472.CAN-11-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Coopman PJ, Thomas DM, Gehlsen KR, Mueller SC. Integrin alpha 3 beta 1 participates in the phagocytosis of extracellular matrix molecules by human breast cancer cells. Mol Biol Cell. 1996;7:1789–1804. doi: 10.1091/mbc.7.11.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Patriarca C, Ivanyi D, Fles D, de Melker A, van Doornewaard G, Oomen L, Alfano RM, Coggi G, Sonnenberg A. Distribution of extracellular and cytoplasmic domains of the alpha 3 and alpha 6 integrin subunits in solid tumors. Int J Cancer. 1995;63:182–189. doi: 10.1002/ijc.2910630206. [DOI] [PubMed] [Google Scholar]
- 124.Morini M, Mottolese M, Ferrari N, Ghiorzo F, Buglioni S, Mortarini R, Noonan DM, Natali PG, Albini A. The alpha 3 beta 1 integrin is associated with mammary carcinoma cell metastasis, invasion, and gelatinase B (MMP-9) activity. Int J Cancer. 2000;87:336–342. [PubMed] [Google Scholar]
- 125.Gogali A, Charalabopoulos K, Constantopoulos S. Integrin receptors in primary lung cancer. Exp Oncol. 2004;26:106–110. [PubMed] [Google Scholar]
- 126.Mizejewski GJ. Role of integrins in cancer: survey of expression patterns. Proc Soc Exp Biol Med. 1999;222:124–138. doi: 10.1177/153537029922200203. [DOI] [PubMed] [Google Scholar]
- 127.Takenaka K, Shibuya M, Takeda Y, Hibino S, Gemma A, Ono Y, Kudoh S. Altered expression and function of beta1 integrins in a highly metastatic human lung adenocarcinoma cell line. Int J Oncol. 2000;17:1187–1194. doi: 10.3892/ijo.17.6.1187. [DOI] [PubMed] [Google Scholar]
- 128.Varzavand A, Drake JM, Svensson RU, Herndon ME, Zhou B, Henry MD, Stipp CS. Integrin alpha3beta1 regulates tumor cell responses to stromal cells and can function to suppress prostate cancer metastatic colonization. Clin Exp Metastasis. 2013;30:541–552. doi: 10.1007/s10585-012-9558-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yoshimasu T, Sakurai T, Oura S, Hirai I, Tanino H, Kokawa Y, Naito Y, Okamura Y, Ota I, Tani N, Matsuura N. Increased expression of integrin alpha3beta1 in highly brain metastatic subclone of a human non-small cell lung cancer cell line. Cancer Sci. 2004;95:142–148. doi: 10.1111/j.1349-7006.2004.tb03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhou B, Gibson-Corley KN, Herndon ME, Sun Y, Gustafson-Wagner E, Teoh-Fitzgerald M, Domann FE, Henry MD, Stipp CS. Integrin alpha3beta1 can function to promote spontaneous metastasis and lung colonization of invasive breast carcinoma. Mol Cancer Res. 2014;12:143–154. doi: 10.1158/1541-7786.MCR-13-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Aina OH, Marik J, Gandour-Edwards R, Lam KS. Near-infrared optical imaging of ovarian cancer xenografts with novel alpha 3-integrin binding peptide “OA02”. Mol Imaging. 2005;4:439–447. doi: 10.2310/7290.2005.05169. [DOI] [PubMed] [Google Scholar]
- 132.Xiao W, Yao N, Peng L, Liu R, Lam KS. Near-infrared optical imaging in glioblastoma xenograft with ligand-targeting alpha 3 integrin. Eur J Nucl Med Mol Imaging. 2009;36:94–103. doi: 10.1007/s00259-008-0920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yao N, Xiao W, Wang X, Marik J, Park SH, Takada Y, Lam KS. Discovery of targeting ligands for breast cancer cells using the one-bead one-compound combinatorial method. J Med Chem. 2009;52:126–133. doi: 10.1021/jm801062d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xiao W, Li T, Bononi F, Lac D, Kekessie I, Liu Y, Sanchez E, Mazloom A, Ma AH, Lin J, Tran J, Yang K, Lam K, Liu R. Discovery and characterization of a high affinity and specificity peptide ligand LXY30 for in vivo targeting of α3 integrin-expressing human tumors. EJNMMI Res. 2016;6:18. doi: 10.1186/s13550-016-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Osborn L, Vassallo C, Browning BG, Tizard R, Haskard DO, Benjamin CD, Dougas I, Kirchhausen T. Arrangement of domains, and amino acid residues required for binding of vascular cell adhesion molecule-1 to its counter-receptor VLA-4 (alpha 4 beta 1) J Cell Biol. 1994;124:601–608. doi: 10.1083/jcb.124.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Komoriya A, Green LJ, Mervic M, Yamada SS, Yamada KM, Humphries MJ. The minimal essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J Biol Chem. 1991;266:15075–15079. [PubMed] [Google Scholar]
- 137.Holzmann B, Gosslar U, Bittner M. Alpha 4 integrins and tumor metastasis. Curr Top Microbiol Immunol. 1998;231:125–141. doi: 10.1007/978-3-642-71987-5_8. [DOI] [PubMed] [Google Scholar]
- 138.Till KJ, Lin K, Zuzel M, Cawley JC. The chemokine receptor CCR7 and alpha4 integrin are important for migration of chronic lymphocytic leukemia cells into lymph nodes. Blood. 2002;99:2977–2984. doi: 10.1182/blood.v99.8.2977. [DOI] [PubMed] [Google Scholar]
- 139.Vincent AM, Cawley JC, Burthem J. Integrin function in chronic lymphocytic leukemia. Blood. 1996;87:4780–4788. [PubMed] [Google Scholar]
- 140.de la Fuente MT, Casanova B, Garcia-Gila M, Silva A, Garcia-Pardo A. Fibronectin interaction with alpha4beta1 integrin prevents apoptosis in B cell chronic lymphocytic leukemia: correlation with Bcl-2 and Bax. Leukemia. 1999;13:266–274. doi: 10.1038/sj.leu.2401275. [DOI] [PubMed] [Google Scholar]
- 141.Damiano JS, Dalton WS. Integrin-mediated drug resistance in multiple myeloma. Leuk Lymphoma. 2000;38:71–81. doi: 10.3109/10428190009060320. [DOI] [PubMed] [Google Scholar]
- 142.Matsunaga T, Takemoto N, Sato T, Takimoto R, Tanaka I, Fujimi A, Akiyama T, Kuroda H, Kawano Y, Kobune M, Kato J, Hirayama Y, Sakamaki S, Kohda K, Miyake K, Niitsu Y. Interaction between leukemic-cell VLA-4 and stromal fibronectin is a decisive factor for minimal residual disease of acute myelogenous leukemia. Nat Med. 2003;9:1158–1165. doi: 10.1038/nm909. [DOI] [PubMed] [Google Scholar]
- 143.Olson DL, Burkly LC, Leone DR, Dolinski BM, Lobb RR. Anti-alpha4 integrin monoclonal antibody inhibits multiple myeloma growth in a murine model. Mol Cancer Ther. 2005;4:91–99. [PubMed] [Google Scholar]
- 144.Garmy-Susini B, Jin H, Zhu Y, Sung RJ, Hwang R, Varner J. Integrin alpha4beta1-VCAM-1-mediated adhesion between endothelial and mural cells is required for blood vessel maturation. J Clin Invest. 2005;115:1542–1551. doi: 10.1172/JCI23445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Park SI, Manat R, Vikstrom B, Amro N, Song LW, Lam KS. The use of one-bead one-compound combinatorial library method to identify peptide ligands for alpha-4-beta-1 integrin receptor in non-Hodgkin's lymphoma. Lett Pept Sci. 2002;8:171–178. [Google Scholar]
- 146.Soodgupta D, Zhou H, Beaino W, Lu L, Snee M, Skeath J, DiPersio JF, Akers WJ, Laforest R, Anderson CJ, Tomasson MH, Shokeen M. Ex vivo and in vivo evaluation of over-expressed VLA-4 in multiple myeloma using LLP2A imaging agents. J Nucl Med. 2016 doi: 10.2967/jnumed.115.164624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Beaino W, Anderson CJ. PET imaging of very late antigen-4 in melanoma: comparison of 68Ga- and 64Cu-labeled NODAGA and CB-TE1A1P-LLP2A conjugates. J Nucl Med. 2014;55:1856–1863. doi: 10.2967/jnumed.114.144881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Beaino W, Nedrow JR, Anderson CJ. Evaluation of (68)Ga- and (177)Lu-DOTA-PEG4-LLP2A for VLA-4-targeted PET imaging and treatment of metastatic melanoma. Mol Pharm. 2015;12:1929–1938. doi: 10.1021/mp5006917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Denardo SJ, Liu R, Albrecht H, Natarajan A, Sutcliffe JL, Anderson C, Peng L, Ferdani R, Cherry SR, Lam KS. 111In-LLP2A-DOTA polyethylene glycol-targeting {alpha}4{beta}1 integrin: comparative pharmacokinetics for imaging and therapy of lymphoid malignancies. J Nucl Med. 2009;50:625–634. doi: 10.2967/jnumed.108.056903. [DOI] [PubMed] [Google Scholar]
- 150.Peng L, Liu R, Andrei M, Xiao W, Lam KS. In vivo optical imaging of human lymphoma xenograft using a library-derived peptidomimetic against alpha4beta1 integrin. Mol Cancer Ther. 2008;7:432–437. doi: 10.1158/1535-7163.MCT-07-0575. [DOI] [PubMed] [Google Scholar]
- 151.Shokeen M, Zheleznyak A, Wilson JM, Jiang M, Liu R, Ferdani R, Lam KS, Schwarz JK, Anderson CJ. Molecular imaging of very late antigen-4 (alpha4beta1 integrin) in the premetastatic niche. J Nucl Med. 2012;53:779–786. doi: 10.2967/jnumed.111.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Soodgupta D, Hurchla MA, Jiang M, Zheleznyak A, Weilbaecher KN, Anderson CJ, Tomasson MH, Shokeen M. Very late antigen-4 (alpha(4)beta(1) integrin) targeted PET imaging of multiple myeloma. PLoS One. 2013;8:e55841. doi: 10.1371/journal.pone.0055841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zwingenberger AL, Kent MS, Liu R, Kukis DL, Wisner ER, DeNardo SJ, Taylor SL, Chen X, Lam KS. In-vivo biodistribution and safety of 99mTc-LLP2A-HYNIC in canine non-Hodgkin lymphoma. PLoS One. 2012;7:e34404. doi: 10.1371/journal.pone.0034404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zwingenberger AL, Kent MS, Shi C, Taylor SL, Chen X, Lam KS. Affinity of the alpha4-beta1 integrin-targeting peptide LLP2A to canine lymphoma. Vet Immunol Immunopathol. 2012;145:298–304. doi: 10.1016/j.vetimm.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Xiao W, Wang Y, Lau EY, Luo J, Yao N, Shi C, Meza L, Tseng H, Maeda Y, Kumaresan P, Liu R, Lightstone FC, Takada Y, Lam KS. The use of one-bead one-compound combinatorial library technology to discover high-affinity alphavbeta3 integrin and cancer targeting arginine-glycine-aspartic acid ligands with a built-in handle. Mol Cancer Ther. 2010;9:2714–2723. doi: 10.1158/1535-7163.MCT-10-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Paez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Vinals F, Inoue M, Bergers G, Hanahan D, Casanovas O. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell. 2009;15:220–231. doi: 10.1016/j.ccr.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell. 2009;15:232–239. doi: 10.1016/j.ccr.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kiessling F, Huppert J, Zhang C, Jayapaul J, Zwick S, Woenne EC, Mueller MM, Zentgraf H, Eisenhut M, Addadi Y, Neeman M, Semmler W. RGD-labeled USPIO inhibits adhesion and endocytotic activity of alpha v beta3-integrin-expressing glioma cells and only accumulates in the vascular tumor compartment. Radiology. 2009;253:462–469. doi: 10.1148/radiol.2532081815. [DOI] [PubMed] [Google Scholar]
- 159.Reynolds AR, Hart IR, Watson AR, Welti JC, Silva RG, Robinson SD, Da Violante G, Gourlaouen M, Salih M, Jones MC, Jones DT, Saunders G, Kostourou V, Perron-Sierra F, Norman JC, Tucker GC, Hodivala-Dilke KM. Stimulation of tumor growth and angiogenesis by low concentrations of RGD-mimetic integrin inhibitors. Nat Med. 2009;15:392–400. doi: 10.1038/nm.1941. [DOI] [PubMed] [Google Scholar]
- 160.Cho CF, Amadei GA, Breadner D, Luyt LG, Lewis JD. Discovery of novel integrin ligands from combinatorial libraries using a multiplex “beads on a bead” approach. Nano Lett. 2012;12:5957–5965. doi: 10.1021/nl3034043. [DOI] [PubMed] [Google Scholar]
- 161.Wong AH, Zhou D, Rini JM. The X-ray crystal structure of human aminopepti-dase N reveals a novel dimer and the basis for peptide processing. J Biol Chem. 2012;287:36804–36813. doi: 10.1074/jbc.M112.398842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang W, Wei Z, Zhang D, Ma H, Wang Z, Bu X, Li M, Geng L, Lausted C, Hood L, Fang Q, Wang H, Hu Z. Rapid screening of peptide probes through in situ single-bead sequencing microarray. Anal Chem. 2014;86:11854–11859. doi: 10.1021/ac503454z. [DOI] [PubMed] [Google Scholar]
- 163.Ding H, Prodinger WM, Kopecek J. Two-step fluorescence screening of CD21-binding peptides with one-bead one-compound library and investigation of binding properties of N-(2-hydroxypropyl)methacrylamide copolymer–peptide conjugates. Biomacromolecules. 2006;7:3037–3046. doi: 10.1021/bm060508f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Winssinger N, Damoiseaux R, Tully DC, Geierstanger BH, Burdick K, Harris JL. PNA-encoded protease substrate microarrays. Chem Biol. 2004;11:1351–1360. doi: 10.1016/j.chembiol.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 165.Zambaldo C, Barluenga S, Winssinger N. PNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:8–15. doi: 10.1016/j.cbpa.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 166.Svensen N, Diaz-Mochon JJ, Bradley M. Decoding a PNA encoded peptide library by PCR: the discovery of new cell surface receptor ligands. Chem Biol. 2011;18:1284–1289. doi: 10.1016/j.chembiol.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 167.Svensen N, Diaz-Mochon JJ, Bradley M. Encoded peptide libraries and the discovery of new cell binding ligands. Chem Commun (Camb) 2011;47:7638–7640. doi: 10.1039/c1cc11668a. [DOI] [PubMed] [Google Scholar]
- 168.Thapa P, Zhang RY, Menon V, Bingham JP. Native chemical ligation: a boon to peptide chemistry. Molecules. 2014;19:14461–14483. doi: 10.3390/molecules190914461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xu Q, Lam KS. Protein and chemical microarrays—powerful tools for proteomics. J Biomed Biotechnol. 2003;2003:257–266. doi: 10.1155/S1110724303209220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Ding Y, Li J, Xiao W, Xiao K, Lee J, Bhardwaj U, Zhu Z, Digiglio P, Yang G, Lam KS, Pan T. Microfluidic-enabled print-to-screen platform for high-throughput screening of combinatorial chemotherapy. Anal Chem. 2015;87:10166–10171. doi: 10.1021/acs.analchem.5b00826. [DOI] [PubMed] [Google Scholar]
- 171.Ding Y, Huang E, Lam KS, Pan T. Microfluidic impact printer with interchangeable cartridges for versatile non-contact multiplexed micropatterning. Lab Chip. 2013;13:1902–1910. doi: 10.1039/c3lc41372a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Marr A, Nissen F, Maisch D, Altmann A, Rana S, Debus J, Huber PE, Haberkorn U, Askoxylakis V. Peptide arrays for development of PDGFRbeta affine molecules. Mol Imaging Biol. 2013;15:391–400. doi: 10.1007/s11307-013-0616-0. [DOI] [PubMed] [Google Scholar]
- 173.Guan M, Yao W, Liu R, Lam KS, Nolta J, Jia J, Panganiban B, Meng L, Zhou P, Shahnazari M, Ritchie RO, Lane NE. Directing mesenchymal stem cells to bone to augment bone formation and increase bone mass. Nat Med. 2012;18:456–462. doi: 10.1038/nm.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhu D, Qin Y, Wang J, Zhang L, Zou S, Zhu X, Zhu L. Novel glypican-3-binding peptide for in vivo hepatocellular carcinoma fluorescent imaging. Bioconjug Chem. 2016 doi: 10.1021/acs.bioconjchem.6b00030. [DOI] [PubMed] [Google Scholar]
- 175.Chang HN, Liu BY, Qi YK, Zhou Y, Chen YP, Pan KM, Li WW, Zhou XM, Ma WW, Fu CY, Qi YM, Liu L, Gao YF. Blocking of the PD-1/PD-L1 interaction by a D-peptide antagonist for cancer immunotherapy. Angew Chem Int Ed Eng. 2015;54:11760–11764. doi: 10.1002/anie.201506225. [DOI] [PubMed] [Google Scholar]
- 176.Bertoldo D, Khan MM, Dessen P, Held W, Huelsken J, Heinis C. Phage selection of peptide macrocycles against beta-catenin to interfere with Wnt signaling. ChemMedChem. 2016 doi: 10.1002/cmdc.201500557. [DOI] [PubMed] [Google Scholar]
- 177.Cho JH, Ha NR, Koh SH, Yoon MY. Design of a PKCdelta-specific small peptide as a theragnostic agent for glioblastoma. Anal Biochem. 2016;496:63–70. doi: 10.1016/j.ab.2015.12.010. [DOI] [PubMed] [Google Scholar]
- 178.Qin X, Zhang C, Xue X, Li M, Li W, Hao Q, Zhang Y, Yan Z, Wei Z. Identification of a novel peptide ligand of human transferrin receptor 1 for targeted tumor delivery drug. Protein Pept Lett. 2013;20:96–101. [PubMed] [Google Scholar]
- 179.Mai J, Song S, Rui M, Liu D, Ding Q, Peng J, Xu Y. A synthetic peptide mediated active targeting of cisplatin liposomes to Tie2 expressing cells. J Control Release. 2009;139:174–181. doi: 10.1016/j.jconrel.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 180.Ding H, Prodinger WM, Kopecek J. Identification of CD21-binding peptides with phage display and investigation of binding properties of HPMA copolymer–peptide conjugates. Bioconjug Chem. 2006;17:514–523. doi: 10.1021/bc0503162. [DOI] [PubMed] [Google Scholar]
- 181.El-Mousawi M, Tchistiakova L, Yurchenko L, Pietrzynski G, Moreno M, Stanimirovic D, Ahmad D, Alakhov V. A vascular endothelial growth factor high affinity receptor 1-specific peptide with antiangiogenic activity identified using a phage display peptide library. J Biol Chem. 2003;278:46681–46691. doi: 10.1074/jbc.M308681200. [DOI] [PubMed] [Google Scholar]
- 182.Naiyer MM, Saha S, Hemke V, Roy S, Singh S, Musti KV, Saha B. Identification and characterization of a human IL-10 receptor antagonist. Hum Immunol. 2013;74:28–31. doi: 10.1016/j.humimm.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 183.Song S, Liu D, Peng J, Sun Y, Li Z, Gu JR, Xu Y. Peptide ligand-mediated liposome distribution and targeting to EGFR expressing tumor in vivo. Int J Pharm. 2008;363:155–161. doi: 10.1016/j.ijpharm.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 184.Dejesus OT. Synthesis of [64Cu]Cu-NOTA-Bn-GE11 for PET imaging of EGFR-rich tumors. Curr Radiopharm. 2012;5:15–18. doi: 10.2174/1874471011205010015. [DOI] [PubMed] [Google Scholar]
- 185.Cheng Y, Meyers JD, Agnes RS, Doane TL, Kenney ME, Broome AM, Burda C, Basilion JP. Addressing brain tumors with targeted gold nanoparticles: a new gold standard for hydrophobic drug delivery? Small. 2011;7:2301–2306. doi: 10.1002/smll.201100628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Kohno M, Horibe T, Haramoto M, Yano Y, Ohara K, Nakajima O, Matsuzaki K, Kawakami K. A novel hybrid peptide targeting EGFR-expressing cancers. Eur J Cancer. 2011;47:773–783. doi: 10.1016/j.ejca.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 187.Abourbeh G, Shir A, Mishani E, Ogris M, Rodl W, Wagner E, Levitzki A. PolyIC GE11 polyplex inhibits EGFR-overexpressing tumors. IUBMB Life. 2012;64:324–330. doi: 10.1002/iub.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Schafer A, Pahnke A, Schaffert D, van Weerden WM, de Ridder CM, Rodl W, Vetter A, Spitzweg C, Kraaij R, Wagner E, Ogris M. Disconnecting the yin and yang relation of epidermal growth factor receptor (EGFR)-mediated delivery: a fully synthetic, EGFR-targeted gene transfer system avoiding receptor activation. Hum Gene Ther. 2011;22:1463–1473. doi: 10.1089/hum.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, Fujita K, Mizutani T, Ohgi T, Ochiya T, Gotoh N, Kuroda M. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Navari M, Zare M, Javanmardi M, Asadi-Ghalehni M, Modjtahedi H, Rasaee MJ. Epitope mapping of epidermal growth factor receptor (EGFR) monoclonal antibody and induction of growth-inhibitory polyclonal antibodies by vaccination with EGFR mimotope. Immunopharmacol Immunotoxicol. 2014;36:309–315. doi: 10.3109/08923973.2014.945127. [DOI] [PubMed] [Google Scholar]
- 191.Zhou J, Joshi BP, Duan X, Pant A, Qiu Z, Kuick R, Owens SR, Wang TD. EGFR overexpressed in colonic neoplasia can be detected on wide-field endoscopic imaging. Clin Transl Gastroenterol. 2015;6:e101. doi: 10.1038/ctg.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Wang W, Chen X, Li T, Li Y, Wang R, He D, Luo W, Li X, Wu X. Screening a phage display library for a novel FGF8b-binding peptide with anti-tumor effect on prostate cancer. Exp Cell Res. 2013;319:1156–1164. doi: 10.1016/j.yexcr.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 193.Dai X, Cai C, Xiao F, Xiong Y, Huang Y, Zhang Q, Xiang Q, Lou G, Lian M, Su Z, Zheng Q. Identification of a novel aFGF-binding peptide with anti-tumor effect on breast cancer from phage display library. Biochem Biophys Res Commun. 2014;445:795–801. doi: 10.1016/j.bbrc.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 194.Wu XP, Yan QX, Huang YD, Huang HX, Su ZJ, Xiao J, Zeng YY, Wang Y, Nie CJ, Yang YG, Li XK. Isolation of a novel basic FGF-binding peptide with potent antiangiogenetic activity. J Cell Mol Med. 2010;14:351–356. doi: 10.1111/j.1582-4934.2008.00506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Su JL, Lai KP, Chen CA, Yang CY, Chen PS, Chang CC, Chou CH, Hu CL, Kuo ML, Hsieh CY, Wei LH. A novel peptide specifically binding to interleukin-6 receptor (gp80) inhibits angiogenesis and tumor growth. Cancer Res. 2005;65:4827–4835. doi: 10.1158/0008-5472.CAN-05-0188. [DOI] [PubMed] [Google Scholar]
- 196.Koivunen E, Gay DA, Ruoslahti E. Selection of peptides binding to the alpha 5 beta 1 integrin from phage display library. J Biol Chem. 1993;268:20205–20210. [PubMed] [Google Scholar]
- 197.Koivunen E, Wang B, Ruoslahti E. Isolation of a highly specific ligand for the alpha 5 beta 1 integrin from a phage display library. J Cell Biol. 1994;124:373–380. doi: 10.1083/jcb.124.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Murayama O, Nishida H, Sekiguchi K. Novel peptide ligands for integrin alpha 6 beta 1 selected from a phage display library. J Biochem (Tokyo) 1996;120:445–451. doi: 10.1093/oxfordjournals.jbchem.a021431. [DOI] [PubMed] [Google Scholar]
- 199.Arap W, Pasqualini R, Ruoslahti E. Cancer treatment by targeted drug delivery to tumor vasculature in a mouse model. Science. 1998;279:377–380. doi: 10.1126/science.279.5349.377. [DOI] [PubMed] [Google Scholar]
- 200.Line BR, Mitra A, Nan A, Ghandehari H. Targeting tumor angiogenesis: comparison of peptide and polymer–peptide conjugates. J Nucl Med. 2005;46:1552–1560. [PubMed] [Google Scholar]
- 201.Hajitou A, Trepel M, Lilley CE, Soghomonyan S, Alauddin MM, Marini FC, 3rd, Restel BH, Ozawa MG, Moya CA, Rangel R, Sun Y, Zaoui K, Schmidt M, von Kalle C, Weitzman MD, Gelovani JG, Pasqualini R, Arap W. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 202.Wang H, Chen K, Cai W, Li Z, He L, Kashefi A, Chen X. Integrin-targeted imaging and therapy with RGD4C–TNF fusion protein. Mol Cancer Ther. 2008;7:1044–1053. doi: 10.1158/1535-7163.MCT-07-2084. [DOI] [PubMed] [Google Scholar]
- 203.Fueyo J, Alemany R, Gomez-Manzano C, Fuller GN, Khan A, Conrad CA, Liu TJ, Jiang H, Lemoine MG, Suzuki K, Sawaya R, Curiel DT, Yung WK, Lang FF. Pre-clinical characterization of the antiglioma activity of a tropism-enhanced adenovi-rus targeted to the retinoblastoma pathway. J Natl Cancer Inst. 2003;95:652–660. doi: 10.1093/jnci/95.9.652. [DOI] [PubMed] [Google Scholar]
- 204.Chen K, Chen X. Integrin targeted delivery of chemotherapeutics. Theranostics. 2011;1:189–200. doi: 10.7150/thno/v01p0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Pameijer CR, Navanjo A, Meechoovet B, Wagner JR, Aguilar B, Wright CL, Chang WC, Brown CE, Jensen MC. Conversion of a tumor-binding peptide identified by phage display to a functional chimeric T cell antigen receptor. Cancer Gene Ther. 2007;14:91–97. doi: 10.1038/sj.cgt.7700993. [DOI] [PubMed] [Google Scholar]
- 206.Koivunen E, Arap W, Valtanen H, Rainisalo A, Medina OP, Heikkila P, Kantor C, Gahmberg CG, Salo T, Konttinen YT, Sorsa T, Ruoslahti E, Pasqualini R. Tumor targeting with a selective gelatinase inhibitor. Nat Biotechnol. 1999;17:768–774. doi: 10.1038/11703. [DOI] [PubMed] [Google Scholar]
- 207.Medina OP, Kairemo K, Valtanen H, Kangasniemi A, Kaukinen S, Ahonen I, Permi P, Annila A, Sneck M, Holopainen JM, Karonen SL, Kinnunen PK, Koivunen E. Radionuclide imaging of tumor xenografts in mice using a gelatinase-targeting peptide. Anticancer Res. 2005;25:33–42. [PubMed] [Google Scholar]
- 208.Sprague JE, Li WP, Liang K, Achilefu S, Anderson CJ. In vitro and in vivo investigation of matrix metalloproteinase expression in metastatic tumor models. Nucl Med Biol. 2006;33:227–237. doi: 10.1016/j.nucmedbio.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 209.Robinson P, Stuber D, Deryckere F, Tedbury P, Lagrange M, Orfanoudakis G. Identification using phage display of peptides promoting targeting and internaliza-tion into HPV-transformed cell lines. J Mol Recognit. 2005;18:175–182. doi: 10.1002/jmr.723. [DOI] [PubMed] [Google Scholar]
- 210.Karmali PP, Kotamraju VR, Kastantin M, Black M, Missirlis D, Tirrell M, Ruoslahti E. Targeting of albumin-embedded paclitaxel nanoparticles to tumors. Nanomedicine. 2009;5:73–82. doi: 10.1016/j.nano.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zhang F, Niu G, Lin X, Jacobson O, Ma Y, Eden HS, He Y, Lu G, Chen X. Imaging tumor-induced sentinel lymph node lymphangiogenesis with LyP-1 peptide. Amino Acids. 2012;42:2343–2351. doi: 10.1007/s00726-011-0976-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Medina OP, Soderlund T, Laakkonen LJ, Tuominen EK, Koivunen E, Kinnunen PK. Binding of novel peptide inhibitors of type IV collagenases to phospho-lipid membranes and use in liposome targeting to tumor cells in vitro. Cancer Res. 2001;61:3978–3985. [PubMed] [Google Scholar]
- 213.Penate Medina O, Haikola M, Tahtinen M, Simpura I, Kaukinen S, Valtanen H, Zhu Y, Kuosmanen S, Cao W, Reunanen J, Nurminen T, Saris PE, Smith-Jones P, Bradbury M, Larson S, Kairemo K. Liposomal tumor targeting in drug delivery utilizing MMP-2- and MMP-9-binding ligands. J Drug Deliv. 2011;2011:160515. doi: 10.1155/2011/160515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cai J, Wei R, Cheng J. Preparation and characterization of a novel chimeric protein VEGI-CTT in Escherichia coli. J Biomed Biotechnol. 2008;2008:564969. doi: 10.1155/2008/564969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zou Y, Chen Y, Jiang Y, Gao J, Gu J. Targeting matrix metalloproteinases and en-dothelial cells with a fusion peptide against tumor. Cancer Res. 2007;67:7295–7300. doi: 10.1158/0008-5472.CAN-06-3920. [DOI] [PubMed] [Google Scholar]
- 216.Heikkila P, Suojanen J, Pirila E, Vaananen A, Koivunen E, Sorsa T, Salo T. Human tongue carcinoma growth is inhibited by selective antigelatinolytic peptides. Int J Cancer. 2006;118:2202–2209. doi: 10.1002/ijc.21540. [DOI] [PubMed] [Google Scholar]
- 217.Sun JM, Zhang C, Li XN. Initial screening of binding-peptide of the cell surface marker CD133 of cancer stem cells. Beijing Da Xue Xue Bao. 2008;40:476–479. [PubMed] [Google Scholar]
- 218.Devemy E, Blaschuk OW. Identification of a novel N-cadherin antagonist. Peptides. 2008;29:1853–1861. doi: 10.1016/j.peptides.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 219.Devemy E, Blaschuk OW. Identification of a novel dual E- and N-cadherin antagonist. Peptides. 2009;30:1539–1547. doi: 10.1016/j.peptides.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 220.Aggarwal S, Singh P, Topaloglu O, Isaacs JT, Denmeade SR. A dimeric peptide that binds selectively to prostate-specific membrane antigen and inhibits its enzymatic activity. Cancer Res. 2006;66:9171–9177. doi: 10.1158/0008-5472.CAN-06-1520. [DOI] [PubMed] [Google Scholar]
- 221.Shi LF, Wu Y, Li CY. Identification of high-affinity VEGFR3-binding peptides through a phage-displayed random peptide library. J Gynecol Oncol. 2015;26:327–335. doi: 10.3802/jgo.2015.26.4.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yang L, Jiang H, Shi B, Wang H, Li J, Wang H, Yao M, Li Z. Identification and characterization of Ch806 mimotopes. Cancer Immunol Immunother. 2010;59:1481–1487. doi: 10.1007/s00262-010-0872-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Askoxylakis V, Garcia-Boy R, Rana S, Kramer S, Hebling U, Mier W, Altmann A, Markert A, Debus J, Haberkorn U. A new peptide ligand for targeting human carbonic anhydrase IX, identified through the phage display technology. PLoS One. 2010;5:e15962. doi: 10.1371/journal.pone.0015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Koolpe M, Dail M, Pasquale EB. An ephrin mimetic peptide that selectively targets the EphA2 receptor. J Biol Chem. 2002;277:46974–46979. doi: 10.1074/jbc.M208495200. [DOI] [PubMed] [Google Scholar]
- 225.Blackburn WH, Dickerson EB, Smith MH, McDonald JF, Lyon LA. Peptide-functionalized nanogels for targeted siRNA delivery. Bioconjug Chem. 2009;20:960–968. doi: 10.1021/bc800547c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.van Geer MA, Bakker CT, Koizumi N, Mizuguchi H, Wesseling JG, Oude Elferink RP, Bosma PJ. Ephrin A2 receptor targeting does not increase adenoviral pancreatic cancer transduction in vivo. World J Gastroenterol. 2009;15:2754–2762. doi: 10.3748/wjg.15.2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang S, Noberini R, Stebbins JL, Das S, Zhang Z, Wu B, Mitra S, Billet S, Fernandez A, Bhowmick NA, Kitada S, Pasquale EB, Fisher PB, Pellecchia M. Targeted delivery of paclitaxel to EphA2-expressing cancer cells. Clin Cancer Res. 2013;19:128–137. doi: 10.1158/1078-0432.CCR-12-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Scarberry KE, Dickerson EB, McDonald JF, Zhang ZJ. Magnetic nanoparticle– peptide conjugates for in vitro and in vivo targeting and extraction of cancer cells. J Am Chem Soc. 2008;130:10258–10262. doi: 10.1021/ja801969b. [DOI] [PubMed] [Google Scholar]
- 229.Scarberry KE, Mezencev R, McDonald JF. Targeted removal of migratory tumor cells by functionalized magnetic nanoparticles impedes metastasis and tumor progression. Nanomedicine (London) 2011;6:69–78. doi: 10.2217/nnm.10.103. [DOI] [PubMed] [Google Scholar]
- 230.Koolpe M, Burgess R, Dail M, Pasquale EB. EphB receptor-binding peptides identified by phage display enable design of an antagonist with ephrin-like affinity. J Biol Chem. 2005;280:17301–17311. doi: 10.1074/jbc.M500363200. [DOI] [PubMed] [Google Scholar]
- 231.Xiong C, Huang M, Zhang R, Song S, Lu W, Flores L, 2nd, Gelovani J, Li C. In vivo small-animal PET/CT of EphB4 receptors using 64Cu-labeled peptide. J Nucl Med. 2011;52:241–248. doi: 10.2967/jnumed.110.081943. [DOI] [PubMed] [Google Scholar]
- 232.You J, Zhang R, Xiong C, Zhong M, Melancon M, Gupta S, Nick AM, Sood AK, Li C. Effective photothermal chemotherapy using doxorubicin-loaded gold nano-spheres that target EphB4 receptors in tumors. Cancer Res. 2012;72:4777–4786. doi: 10.1158/0008-5472.CAN-12-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Thapa N, Kim S, So IS, Lee BH, Kwon IC, Choi K, Kim IS. Discovery of a phosphatidylserine-recognizing peptide and its utility in molecular imaging of tumour apoptosis. J Cell Mol Med. 2008;12:1649–1660. doi: 10.1111/j.1582-4934.2008.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Diderich P, Heinis C. Phage selection of bicyclic peptides binding Her2. Tetrahedron. 2014;70:7733–7739. [Google Scholar]
- 235.Witsch EJ, Mahlknecht G, Wakim J, Sertchook R, Bublil E, Yarden Y, Sela M. Generation and characterization of peptide mimotopes specific for anti ErbB-2 monoclonal antibodies. Int Immunol. 2011;23:391–403. doi: 10.1093/intimm/dxr028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Karasseva NG, Glinsky VV, Chen NX, Komatireddy R, Quinn TP. Identification and characterization of peptides that bind human ErbB-2 selected from a bacteri-ophage display library. J Protein Chem. 2002;21:287–296. doi: 10.1023/a:1019749504418. [DOI] [PubMed] [Google Scholar]
- 237.Kumar SR, Quinn TP, Deutscher SL. Evaluation of an 111In-radiolabeled peptide as a targeting and imaging agent for ErbB-2 receptor expressing breast carcinomas. Clin Cancer Res. 2007;13:6070–6079. doi: 10.1158/1078-0432.CCR-07-0160. [DOI] [PubMed] [Google Scholar]
- 238.Deutscher SL, Figueroa SD, Kumar SR. In-labeled KCCYSL peptide as an imaging probe for ErbB-2-expressing ovarian carcinomas. J Label Compd Radiopharm. 2009;52:583–590. doi: 10.1002/jlcr.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kumar SR, Gallazzi FA, Ferdani R, Anderson CJ, Quinn TP, Deutscher SL. In vitro and in vivo evaluation of (6) (4)Cu-radiolabeled KCCYSL peptides for targeting epidermal growth factor receptor-2 in breast carcinomas. Cancer Biother Radiopharm. 2010;25:693–703. doi: 10.1089/cbr.2010.0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Houimel M, Schneider P, Terskikh A, Mach JP. Selection of peptides and synthesis of pentameric peptabody molecules reacting specifically with ErbB-2 receptor. Int J Cancer. 2001;92:748–755. doi: 10.1002/1097-0215(20010601)92:5<748::aid-ijc1258>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 241.Pero SC, Shukla GS, Armstrong AL, Peterson D, Fuller SP, Godin K, Kingsley-Richards SL, Weaver DL, Bond J, Krag DN. Identification of a small peptide that inhibits the phosphorylation of ErbB2 and proliferation of ErbB2 overexpressing breast cancer cells. Int J Cancer. 2004;111:951–960. doi: 10.1002/ijc.20306. [DOI] [PubMed] [Google Scholar]
- 242.Shukla GS, Krag DN. Cancer cell-specific internalizing ligands from phage displayed beta-lactamase–peptide fusion libraries. Protein Eng Des Sel. 2010;23:431–440. doi: 10.1093/protein/gzq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Rusckowski M, Gupta S, Liu G, Dou S, Hnatowich DJ. Evidence of specificity of radiolabeled phage display peptides for the TAG-72 antigen. Cancer Biother Radiopharm. 2007;22:564–572. doi: 10.1089/cbr.2006.307. [DOI] [PubMed] [Google Scholar]
- 244.Chen L, Wang Y, Cheng D, Dou S, Liu X, Liu G, Hnatowich DJ, Rusckowski M. Comparing two TAG-72 binding peptides previously identified by phage display as potential imaging agents. Nucl Med Commun. 2011;32:920–924. doi: 10.1097/MNM.0b013e328348fc64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Xiao N, Cheng D, Wang Y, Chen L, Liu X, Dou S, Liu G, Liang M, Hnatowich DJ, Rusckowski M. Identification of a high affinity TAG-72 binding peptide by phage display selection. Cancer Biol Ther. 2011;11:22–31. doi: 10.4161/cbt.11.1.13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chen L, Wang Y, Liu X, Dou S, Liu G, Hnatowich DJ, Rusckowski M. A new TAG-72 cancer marker peptide identified by phage display. Cancer Lett. 2008;272:122–132. doi: 10.1016/j.canlet.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Zou J, Glinsky VV, Landon LA, Matthews L, Deutscher SL. Peptides specific to the galectin-3 carbohydrate recognition domain inhibit metastasis-associated cancer cell adhesion. Carcinogenesis. 2005;26:309–318. doi: 10.1093/carcin/bgh329. [DOI] [PubMed] [Google Scholar]
- 248.Kumar SR, Deutscher SL. 111In-labeled galectin-3-targeting peptide as a SPECT agent for imaging breast tumors. J Nucl Med. 2008;49:796–803. doi: 10.2967/jnumed.107.048751. [DOI] [PubMed] [Google Scholar]
- 249.Deutscher SL, Figueroa SD, Kumar SR. Tumor targeting and SPECT imaging properties of an (111)In-labeled galectin-3 binding peptide in prostate carcinoma. Nucl Med Biol. 2009;36:137–146. doi: 10.1016/j.nucmedbio.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 250.Landon LA, Peletskaya EN, Glinsky VV, Karasseva N, Quinn TP, Deutscher SL. Combinatorial evolution of high-affinity peptides that bind to the Thomsen– Friedenreich carcinoma antigen. J Protein Chem. 2003;22:193–204. doi: 10.1023/a:1023483232397. [DOI] [PubMed] [Google Scholar]
- 251.Landon LA, Zou J, Deutscher SL. Effective combinatorial strategy to increase affinity of carbohydrate binding by peptides. Mol Divers. 2004;8:35–50. doi: 10.1023/b:modi.0000006897.40575.41. [DOI] [PubMed] [Google Scholar]
- 252.Kumar SR, Gallazzi FA, Quinn TP, Deutscher SL. (64)Cu-labeled peptide for PET of breast carcinomas expressing the Thomsen–Friedenreich carbohydrate antigen. J Nucl Med. 2011;52:1819–1826. doi: 10.2967/jnumed.111.093716. [DOI] [PubMed] [Google Scholar]
- 253.Peletskaya EN, Glinsky VV, Glinsky GV, Deutscher SL, Quinn TP. Characterization of peptides that bind the tumor-associated Thomsen–Friedenreich antigen selected from bacteriophage display libraries. J Mol Biol. 1997;270:374–384. doi: 10.1006/jmbi.1997.1107. [DOI] [PubMed] [Google Scholar]
- 254.Pilch J, Brown DM, Komatsu M, Jarvinen TA, Yang M, Peters D, Hoffman RM, Ruoslahti E. Peptides selected for binding to clotted plasma accumulate in tumor stroma and wounds. Proc Natl Acad Sci U S A. 2006;103:2800–2804. doi: 10.1073/pnas.0511219103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Ballinger MD, Shyamala V, Forrest LD, Deuter-Reinhard M, Doyle LV, Wang JX, Panganiban-Lustan L, Stratton JR, Apell G, Winter JA, Doyle MV, Rosenberg S, Kavanaugh WM. Semirational design of a potent, artificial agonist of fibroblast growth factor receptors. Nat Biotechnol. 1999;17:1199–1204. doi: 10.1038/70746. [DOI] [PubMed] [Google Scholar]
- 256.Maruta F, Parker AL, Fisher KD, Hallissey MT, Ismail T, Rowlands DC, Chandler LA, Kerr DJ, Seymour LW. Identification of FGF receptor-binding peptides for cancer gene therapy. Cancer Gene Ther. 2002;9:543–552. doi: 10.1038/sj.cgt.7700470. [DOI] [PubMed] [Google Scholar]
- 257.McConnell SJ, Thon VJ, Spinella DG. Isolation of fibroblast growth factor receptor binding sequences using evolved phage display libraries. Comb Chem High Throughput Screen. 1999;2:155–163. [PubMed] [Google Scholar]
- 258.Fukuda MN, Ohyama C, Lowitz K, Matsuo O, Pasqualini R, Ruoslahti E, Fukuda M. A peptide mimic of E-selectin ligand inhibits sialyl Lewis X-dependent lung colonization of tumor cells. Cancer Res. 2000;60:450–456. [PubMed] [Google Scholar]
- 259.Mueller J, Gaertner FC, Blechert B, Janssen KP, Essler M. Targeting of tumor blood vessels: a phage-displayed tumor-homing peptide specifically binds to matrix metalloproteinase-2-processed collagen IV and blocks angiogenesis in vivo. Mol Cancer Res. 2009;7:1078–1085. doi: 10.1158/1541-7786.MCR-08-0538. [DOI] [PubMed] [Google Scholar]
- 260.Wu P, Leinonen J, Koivunen E, Lankinen H, Stenman UH. Identification of novel prostate-specific antigen-binding peptides modulating its enzyme activity. Eur J Biochem. 2000;267:6212–6220. doi: 10.1046/j.1432-1327.2000.01696.x. [DOI] [PubMed] [Google Scholar]
- 261.Urech-Varenne C, Radtke F, Heinis C. Phage selection of bicyclic peptide ligands of the Notch1 receptor. ChemMedChem. 2015;10:1754–1761. doi: 10.1002/cmdc.201500261. [DOI] [PubMed] [Google Scholar]
- 262.Zhang D, Jia H, Li W, Hou Y, Lu S, He S. Screening and identification of a phage display derived peptide that specifically binds to the CD44 protein region encoded by variable exons. J Biomol Screen. 2016;21:44–53. doi: 10.1177/1087057115608604. [DOI] [PubMed] [Google Scholar]
- 263.Zhang D, Jia H, Wang Y, Li WM, Hou YC, Yin SW, Wang TD, He SX, Lu SY. A CD44 specific peptide developed by phage display for targeting gastric cancer. Biotechnol Lett. 2015;37:2311–2320. doi: 10.1007/s10529-015-1896-z. [DOI] [PubMed] [Google Scholar]
- 264.Wang W, Chen T, Li H, Chen Y, Wu Z, Feng T, Zhang X, Zhong Q, Zhong Q, Li G, Guo L, Zhou L, Zhou J. Screening a novel FGF3 antagonist peptide with anti-tumor effects on breast cancer from a phage display library. Mol Med Rep. 2015;12:7051–7058. doi: 10.3892/mmr.2015.4248. [DOI] [PubMed] [Google Scholar]
- 265.Han Z, Zhou Z, Shi X, Wang J, Wu X, Sun D, Chen Y, Zhu H, Magi-Galluzzi C, Lu ZR. EDB fibronectin specific peptide for prostate cancer targeting. Bioconjug Chem. 2015;26:830–838. doi: 10.1021/acs.bioconjchem.5b00178. [DOI] [PubMed] [Google Scholar]
- 266.He XQ, Guan J, Liu F, Li J, He MR. Identification of the sAPRIL binding peptide and its growth inhibition effects in the colorectal cancer cells. PLoS One. 2015;10:e0120564. doi: 10.1371/journal.pone.0120564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Khemthongcharoen N, Ruangpracha A, Sarapukdee P, Rattanavarin S, Jolivot R, Jarujareet U, Plaimas K, Bhattarakosol P, Patumraj S, Piyawattanametha W. Novel p16 binding peptide development for p16-overexpressing cancer cell detection using phage display. J Pept Sci. 2015;21:265–273. doi: 10.1002/psc.2726. [DOI] [PubMed] [Google Scholar]
- 268.Bose D, Nahar S, Rai MK, Ray A, Chakraborty K, Maiti S. Selective inhibition of miR-21 by phage display screened peptide. Nucleic Acids Res. 2015;43:4342–4352. doi: 10.1093/nar/gkv185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wang XF, Birringer M, Dong LF, Veprek P, Low P, Swettenham E, Stantic M, Yuan LH, Zobalova R, Wu K, Ledvina M, Ralph SJ, Neuzil J. A peptide conjugate of vitamin E succinate targets breast cancer cells with high ErbB2 expression. Cancer Res. 2007;67:3337–3344. doi: 10.1158/0008-5472.CAN-06-2480. [DOI] [PubMed] [Google Scholar]
- 270.Shadidi M, Sioud M. Identification of novel carrier peptides for the specific delivery of therapeutics into cancer cells. FASEB J. 2003;17:256–258. doi: 10.1096/fj.02-0280fje. [DOI] [PubMed] [Google Scholar]
- 271.Luo H, Yang J, Jin H, Huang C, Fu J, Yang F, Gong H, Zeng S, Luo Q, Zhang Z. Tet-rameric far-red fluorescent protein as a scaffold to assemble an octavalent peptide nanoprobe for enhanced tumor targeting and intracellular uptake in vivo. FASEB J. 2011;25:1865–1873. doi: 10.1096/fj.10-174318. [DOI] [PubMed] [Google Scholar]
- 272.Jie LY, Cai LL, Wang LJ, Ying XY, Yu RS, Zhang MM, Du YZ. Actively-targeted LTVSPWY peptide-modified magnetic nanoparticles for tumor imaging. Int J Nanomedicine. 2012;7:3981–3989. doi: 10.2147/IJN.S33593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Orban E, Manea M, Marquadt A, Banoczi Z, Csik G, Fellinger E, Bosze S, Hudecz F. A new daunomycin–peptide conjugate: synthesis, characterization and the effect on the protein expression profile of HL-60 cells in vitro. Bioconjug Chem. 2011;22:2154–2165. doi: 10.1021/bc2004236. [DOI] [PubMed] [Google Scholar]
- 274.Wu CH, Kuo YH, Hong RL, Wu HC. alpha-Enolase-binding peptide enhances drug delivery efficiency and therapeutic efficacy against colorectal cancer. Sci Transl Med. 2015;7:290ra291. doi: 10.1126/scitranslmed.aaa9391. [DOI] [PubMed] [Google Scholar]
- 275.Jeong MH, Kim K, Kim EM, Cheong SJ, Lee CM, Jeong HJ, Kim DW, Lim ST, Sohn MH, Chung J. In vivo and in vitro evaluation of Cy5.5 conjugated epidermal growth factor receptor binding peptide. Nucl Med Biol. 2012;39:805–812. doi: 10.1016/j.nucmedbio.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 276.Staquicini FI, Tandle A, Libutti SK, Sun J, Zigler M, Bar-Eli M, Aliperti F, Perez EC, Gershenwald JE, Mariano M, Pasqualini R, Arap W, Lopes JD. A subset of host B lymphocytes controls melanoma metastasis through a melanoma cell adhesion molecule/MUC18-dependent interaction: evidence from mice and humans. Cancer Res. 2008;68:8419–8428. doi: 10.1158/0008-5472.CAN-08-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Wang T, D'Souza GG, Bedi D, Fagbohun OA, Potturi LP, Papahadjopoulos-Sternberg B, Petrenko VA, Torchilin VP. Enhanced binding and killing of target tumor cells by drug-loaded liposomes modified with tumor-specific phage fusion coat protein. Nanomedicine (London) 2010;5:563–574. doi: 10.2217/nnm.10.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Fagbohun OA, Bedi D, Grabchenko NI, Deinnocentes PA, Bird RC, Petrenko VA. Landscape phages and their fusion proteins targeted to breast cancer cells. Protein Eng Des Sel. 2012;25:271–283. doi: 10.1093/protein/gzs013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Wang T, Petrenko VA, Torchilin VP. Paclitaxel-loaded polymeric micelles modified with MCF-7 cell-specific phage protein: enhanced binding to target cancer cells and increased cytotoxicity. Mol Pharm. 2010;7:1007–1014. doi: 10.1021/mp1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wu C, Lo SL, Boulaire J, Hong ML, Beh HM, Leung DS, Wang S. A peptide-based carrier for intracellular delivery of proteins into malignant glial cells in vitro. J Control Release. 2008;130:140–145. doi: 10.1016/j.jconrel.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 281.Wang C, Ning L, Wang H, Lu Z, Li X, Fan X, Wang X, Liu Y. A peptide-mediated targeting gene delivery system for malignant glioma cells. Int J Nanomedicine. 2013;8:3631–3640. doi: 10.2147/IJN.S44990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Kim JW, Kane JR, Young JS, Chang AL, Kanojia D, Morshed RA, Miska J, Ahmed AU, Balyasnikova IV, Han Y, Zhang L, Curiel DT, Lesniak MS. A genetically modified adenoviral vector with a phage display-derived peptide incorporated into fiber fibritin chimera prolongs survival in experimental glioma. Hum Gene Ther. 2015;26:635–646. doi: 10.1089/hum.2015.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Beck S, Jin X, Yin J, Kim SH, Lee NK, Oh SY, Jin X, Kim MK, Kim EB, Son JS, Kim SC, Nam DH, Kim SH, Kang SK, Kim H, Choi YJ. Identification of a peptide that interacts with Nestin protein expressed in brain cancer stem cells. Biomaterials. 2011;32:8518–8528. doi: 10.1016/j.biomaterials.2011.07.048. [DOI] [PubMed] [Google Scholar]
- 284.Matsuo AL, Tanaka AS, Juliano MA, Rodrigues EG, Travassos LR. A novel melanoma-targeting peptide screened by phage display exhibits antitumor activity. J Mol Med (Berl) 2010;88:1255–1264. doi: 10.1007/s00109-010-0671-9. [DOI] [PubMed] [Google Scholar]
- 285.Kelly K, Alencar H, Funovics M, Mahmood U, Weissleder R. Detection of invasive colon cancer using a novel, targeted, library-derived fluorescent peptide. Cancer Res. 2004;64:6247–6251. doi: 10.1158/0008-5472.CAN-04-0817. [DOI] [PubMed] [Google Scholar]
- 286.Kelly KA, Jones DA. Isolation of a colon tumor specific binding peptide using phage display selection. Neoplasia. 2003;5:437–444. doi: 10.1016/s1476-5586(03)80046-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Huang G, Zhang C, Li S, Khemtong C, Yang SG, Tian R, Minna JD, Brown KC, Gao J. A novel strategy for surface modification of superparamagnetic iron oxide nanoparticles for lung cancer imaging. J Mater Chem. 2009;19:6367–6372. doi: 10.1039/b902358e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zhou X, Chang YC, Oyama T, McGuire MJ, Brown KC. Cell-specific delivery of a chemotherapeutic to lung cancer cells. J Am Chem Soc. 2004;126:15656–15657. doi: 10.1021/ja0446496. [DOI] [PubMed] [Google Scholar]
- 289.Li S, McGuire MJ, Lin M, Liu YH, Oyama T, Sun X, Brown KC. Synthesis and characterization of a high-affinity {alpha}v{beta}6-specific ligand for in vitro and in vivo applications. Mol Cancer Ther. 2009;8:1239–1249. doi: 10.1158/1535-7163.MCT-08-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Li S, Gray BP, McGuire MJ, Brown KC. Synthesis and biological evaluation of a peptide–paclitaxel conjugate which targets the integrin alphavbeta(6) Bioorg Med Chem. 2011;19:5480–5489. doi: 10.1016/j.bmc.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Guan H, McGuire MJ, Li S, Brown KC. Peptide-targeted polyglutamic acid doxo-rubicin conjugates for the treatment of alpha(v)beta(6)-positive cancers. Bioconjug Chem. 2008;19:1813–1821. doi: 10.1021/bc800154f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Guthi JS, Yang SG, Huang G, Li S, Khemtong C, Kessinger CW, Peyton M, Minna JD, Brown KC, Gao J. MRI-visible micellar nanomedicine for targeted drug delivery to lung cancer cells. Mol Pharm. 2010;7:32–40. doi: 10.1021/mp9001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Gray BP, Li S, Brown KC. From phage display to nanoparticle delivery: functionalizing liposomes with multivalent peptides improves targeting to a cancer biomarker. Bioconjug Chem. 2013;24:85–96. doi: 10.1021/bc300498d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Tang B, Li Z, Huang D, Zheng L, Li Q. Screening of a specific peptide binding to VPAC1 receptor from a phage display peptide library. PLoS One. 2013;8:e54264. doi: 10.1371/journal.pone.0054264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Choi JH, Lee WK, Han SH, Ha S, Ahn SM, Kang JS, Choi YJ, Yun CH. Identification and characterization of nonapeptide targeting a human B cell lymphoma, Raji. Int Immunopharmacol. 2008;8:852–858. doi: 10.1016/j.intimp.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 296.Sun X, Niu G, Yan Y, Yang M, Chen K, Ma Y, Chan N, Shen B, Chen X. Phage display-derived peptides for osteosarcoma imaging. Clin Cancer Res. 2010;16:4268–4277. doi: 10.1158/1078-0432.CCR-10-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Wang D, Li W, Zhang H, Mao Q, Xia H. A targeting peptide improves adenovirus-mediated transduction of a glioblastoma cell line. Oncol Rep. 2014;31:2093–2098. doi: 10.3892/or.2014.3065. [DOI] [PubMed] [Google Scholar]
- 298.Kim Y, Lillo AM, Steiniger SC, Liu Y, Ballatore C, Anichini A, Mortarini R, Kaufmann GF, Zhou B, Felding-Habermann B, Janda KD. Targeting heat shock proteins on cancer cells: selection, characterization, and cell-penetrating properties of a peptidic GRP78 ligand. Biochemistry. 2006;45:9434–9444. doi: 10.1021/bi060264j. [DOI] [PubMed] [Google Scholar]
- 299.Yoneda Y, Steiniger SC, Capkova K, Mee JM, Liu Y, Kaufmann GF, Janda KD. A cell-penetrating peptidic GRP78 ligand for tumor cell-specific prodrug therapy. Bioorg Med Chem Lett. 2008;18:1632–1636. doi: 10.1016/j.bmcl.2008.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Kang J, Zhao G, Lin T, Tang S, Xu G, Hu S, Bi Q, Guo C, Sun L, Han S, Xu Q, Nie Y, Wang B, Liang S, Ding J, Wu K. A peptide derived from phage display library exhibits anti-tumor activity by targeting GRP78 in gastric cancer multidrug resistance cells. Cancer Lett. 2013;339:247–259. doi: 10.1016/j.canlet.2013.06.016. [DOI] [PubMed] [Google Scholar]
- 301.Passarella RJ, Spratt DE, van der Ende AE, Phillips JG, Wu H, Sathiyakumar V, Zhou L, Hallahan DE, Harth E, Diaz R. Targeted nanoparticles that deliver a sustained, specific release of Paclitaxel to irradiated tumors. Cancer Res. 2010;70:4550–4559. doi: 10.1158/0008-5472.CAN-10-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Essler M, Ruoslahti E. Molecular specialization of breast vasculature: a breast-homing phage-displayed peptide binds to aminopeptidase P in breast vasculature. Proc Natl Acad Sci U S A. 2002;99:2252–2257. doi: 10.1073/pnas.251687998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Arap W, Kolonin MG, Trepel M, Lahdenranta J, Cardo-Vila M, Giordano RJ, Mintz PJ, Ardelt PU, Yao VJ, Vidal CI, Chen L, Flamm A, Valtanen H, Weavind LM, Hicks ME, Pollock RE, Botz GH, Bucana CD, Koivunen E, Cahill D, Troncoso P, Baggerly KA, Pentz RD, Do KA, Logothetis CJ, Pasqualini R. Steps toward mapping the human vasculature by phage display. Nat Med. 2002;8:121–127. doi: 10.1038/nm0202-121. [DOI] [PubMed] [Google Scholar]
- 304.Wang W, Ke S, Kwon S, Yallampalli S, Cameron AG, Adams KE, Mawad ME, Sevick-Muraca EM. A new optical and nuclear dual-labeled imaging agent targeting interleukin 11 receptor alpha-chain. Bioconjug Chem. 2007;18:397–402. doi: 10.1021/bc0602679. [DOI] [PubMed] [Google Scholar]
- 305.Zurita AJ, Troncoso P, Cardo-Vila M, Logothetis CJ, Pasqualini R, Arap W. Combinatorial screenings in patients: the interleukin-11 receptor alpha as a candidate target in the progression of human prostate cancer. Cancer Res. 2004;64:435–439. doi: 10.1158/0008-5472.can-03-2675. [DOI] [PubMed] [Google Scholar]
- 306.Joyce JA, Laakkonen P, Bernasconi M, Bergers G, Ruoslahti E, Hanahan D. Stage-specific vascular markers revealed by phage display in a mouse model of pancreatic islet tumorigenesis. Cancer Cell. 2003;4:393–403. doi: 10.1016/s1535-6108(03)00271-x. [DOI] [PubMed] [Google Scholar]
- 307.Hamzah J, Nelson D, Moldenhauer G, Arnold B, Hammerling GJ, Ganss R. Vascular targeting of anti-CD40 antibodies and IL-2 into autochthonous tumors enhances immunotherapy in mice. J Clin Invest. 2008;118:1691–1699. doi: 10.1172/JCI33201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Pasqualini R, Koivunen E, Kain R, Lahdenranta J, Sakamoto M, Stryhn A, Ashmun RA, Shapiro LH, Arap W, Ruoslahti E. Aminopeptidase N is a receptor for tumor-homing peptides and a target for inhibiting angiogenesis. Cancer Res. 2000;60:722–727. [PMC free article] [PubMed] [Google Scholar]
- 309.Corti A, Curnis F, Arap W, Pasqualini R. The neovasculature homing motif NGR: more than meets the eye. Blood. 2008;112:2628–2635. doi: 10.1182/blood-2008-04-150862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Pastorino F, Brignole C, Marimpietri D, Cilli M, Gambini C, Ribatti D, Longhi R, Allen TM, Corti A, Ponzoni M. Vascular damage and anti-angiogenic effects of tumor vessel-targeted liposomal chemotherapy. Cancer Res. 2003;63:7400–7409. [PubMed] [Google Scholar]
- 311.Pastorino F, Di Paolo D, Piccardi F, Nico B, Ribatti D, Daga A, Baio G, Neumaier CE, Brignole C, Loi M, Marimpietri D, Pagnan G, Cilli M, Lepekhin EA, Garde SV, Longhi R, Corti A, Allen TM, Wu JJ, Ponzoni M. Enhanced antitumor efficacy of clinical-grade vasculature-targeted liposomal doxorubicin. Clin Cancer Res. 2008;14:7320–7329. doi: 10.1158/1078-0432.CCR-08-0804. [DOI] [PubMed] [Google Scholar]
- 312.Curnis F, Sacchi A, Borgna L, Magni F, Gasparri A, Corti A. Enhancement of tumor necrosis factor alpha antitumor immunotherapeutic properties by targeted delivery to aminopeptidase N (CD13) Nat Biotechnol. 2000;18:1185–1190. doi: 10.1038/81183. [DOI] [PubMed] [Google Scholar]
- 313.Curnis F, Sacchi A, Corti A. Improving chemotherapeutic drug penetration in tu-morsby vascular targeting and barrier alteration. J Clin Invest. 2002;110:475–482. doi: 10.1172/JCI15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Corti A, Pastorino F, Curnis F, Arap W, Ponzoni M, Pasqualini R. Targeted drug delivery and penetration into solid tumors. Med Res Rev. 2012;32:1078–1091. doi: 10.1002/med.20238. [DOI] [PubMed] [Google Scholar]
- 315.Meng J, Yan Z, Wu J, Li L, Xue X, Li M, Li W, Hao Q, Wan Y, Qin X, Zhang C, You Y, Han W, Zhang Y. High-yield expression, purification and characterization of tumor-targeted IFN-alpha2a. Cytotherapy. 2007;9:60–68. doi: 10.1080/14653240601094322. [DOI] [PubMed] [Google Scholar]
- 316.Zhang B, Gao B, Dong S, Zhang Y, Wu Y. Anti-tumor efficacy and pre-clinical im-munogenicity of IFNalpha2a-NGR. Regul Toxicol Pharmacol. 2011;60:73–78. doi: 10.1016/j.yrtph.2011.02.007. [DOI] [PubMed] [Google Scholar]
- 317.Meng J, Ma N, Yan Z, Han W, Zhang Y. NGR enhanced the anti-angiogenic activity of tum-5. J Biochem. 2006;140:299–304. doi: 10.1093/jb/mvj152. [DOI] [PubMed] [Google Scholar]
- 318.Fogal V, Zhang L, Krajewski S, Ruoslahti E. Mitochondrial/cell-surface protein p32/gC1qR as a molecular target in tumor cells and tumor stroma. Cancer Res. 2008;68:7210–7218. doi: 10.1158/0008-5472.CAN-07-6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E. A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nat Med. 2002;8:751–755. doi: 10.1038/nm720. [DOI] [PubMed] [Google Scholar]
- 320.Herringson TP, Altin JG. Effective tumor targeting and enhanced anti-tumor effect of liposomes engrafted with peptides specific for tumor lymphatics and vascu-lature. Int J Pharm. 2011;411:206–214. doi: 10.1016/j.ijpharm.2011.03.044. [DOI] [PubMed] [Google Scholar]
- 321.Park JH, von Maltzahn G, Xu MJ, Fogal V, Kotamraju VR, Ruoslahti E, Bhatia SN, Sailor MJ. Cooperative nanomaterial system to sensitize, target, and treat tumors. Proc Natl Acad Sci U S A. 2010;107:981–986. doi: 10.1073/pnas.0909565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Staquicini FI, Ozawa MG, Moya CA, Driessen WH, Barbu EM, Nishimori H, Soghomonyan S, Flores LG, 2nd, Liang X, Paolillo V, Alauddin MM, Basilion JP, Furnari FB, Bogler O, Lang FF, Aldape KD, Fuller GN, Hook M, Gelovani JG, Sidman RL, Cavenee WK, Pasqualini R, Arap W. Systemic combinatorial peptide selection yields a non-canonical iron-mimicry mechanism for targeting tumors in a mouse model of human glioblastoma. J Clin Invest. 2011;121:161–173. doi: 10.1172/JCI44798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Laakkonen P, Akerman ME, Biliran H, Yang M, Ferrer F, Karpanen T, Hoffman RM, Ruoslahti E. Antitumor activity of a homing peptide that targets tumor lymphatics and tumor cells. Proc Natl Acad Sci U S A. 2004;101:9381–9386. doi: 10.1073/pnas.0403317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Hariri G, Yan H, Wang H, Han Z, Hallahan DE. Radiation-guided drug delivery to mouse models of lung cancer. Clin Cancer Res. 2010;16:4968–4977. doi: 10.1158/1078-0432.CCR-10-0969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Han Z, Fu A, Wang H, Diaz R, Geng L, Onishko H, Hallahan DE. Noninvasive assessment of cancer response to therapy. Nat Med. 2008;14:343–349. doi: 10.1038/nm1691. [DOI] [PubMed] [Google Scholar]
- 326.Wang H, Yan H, Fu A, Han M, Hallahan D, Han Z. TIP-1 translocation onto the cell plasma membrane is a molecular biomarker of tumor response to ionizing radiation. PLoS One. 2010;5:e12051. doi: 10.1371/journal.pone.0012051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Lowery A, Onishko H, Hallahan DE, Han Z. Tumor-targeted delivery of liposome-encapsulated doxorubicin by use of a peptide that selectively binds to irradiated tumors. J Control Release. 2011;150:117–124. doi: 10.1016/j.jconrel.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Hariri G, Wellons MS, Morris WH, 3rd, Lukehart CM, Hallahan DE. Multifunctional FePt nanoparticles for radiation-guided targeting and imaging of cancer. Ann Biomed Eng. 2011;39:946–952. doi: 10.1007/s10439-010-0219-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Hallahan D, Geng L, Qu S, Scarfone C, Giorgio T, Donnelly E, Gao X, Clanton J. Integrin-mediated targeting of drug delivery to irradiated tumor blood vessels. Cancer Cell. 2003;3:63–74. doi: 10.1016/s1535-6108(02)00238-6. [DOI] [PubMed] [Google Scholar]
- 330.Akita N, Maruta F, Seymour LW, Kerr DJ, Parker AL, Asai T, Oku N, Nakayama J, Miyagawa S. Identification of oligopeptides binding to peritoneal tumors of gastric cancer. Cancer Sci. 2006;97:1075–1081. doi: 10.1111/j.1349-7006.2006.00291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Agemy L, Friedmann-Morvinski D, Kotamraju VR, Roth L, Sugahara KN, Girard OM, Mattrey RF, Verma IM, Ruoslahti E. Targeted nanoparticle enhanced proapoptotic peptide as potential therapy for glioblastoma. Proc Natl Acad Sci U S A. 2011;108:17450–17455. doi: 10.1073/pnas.1114518108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Greenwald DR, Ruoslahti E. Coadministration of a tumor-penetrating peptide enhances the efficacy of cancer drugs. Science. 2010;328:1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Yan Z, Yang Y, Wei X, Zhong J, Wei D, Liu L, Xie C, Wang F, Zhang L, Lu W, He D. Tumor-penetrating peptide mediation: an effective strategy for improving the transport of liposomes in tumor tissue. Mol Pharm. 2014;11:218–225. doi: 10.1021/mp400393a. [DOI] [PubMed] [Google Scholar]
- 334.Mintz PJ, Cardo-Vila M, Ozawa MG, Hajitou A, Rangel R, Guzman-Rojas L, Christianson DR, Arap MA, Giordano RJ, Souza GR, Easley J, Salameh A, Oliviero S, Brentani RR, Koivunen E, Arap W, Pasqualini R. An unrecognized extracellular function for an intracellular adapter protein released from the cytoplasm into the tumor microenvironment. Proc Natl Acad Sci U S A. 2009;106:2182–2187. doi: 10.1073/pnas.0807543105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Kelly KA, Bardeesy N, Anbazhagan R, Gurumurthy S, Berger J, Alencar H, Depinho RA, Mahmood U, Weissleder R. Targeted nanoparticles for imaging incipient pancreatic ductal adenocarcinoma. PLoS Med. 2008;5:e85. doi: 10.1371/journal.pmed.0050085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Bausch D, Thomas S, Mino-Kenudson M, Fernandezdel CC, Bauer TW, Williams M, Warshaw AL, Thayer SP, Kelly KA. Plectin-1 as a novel biomarker for pancreatic cancer. Clin Cancer Res. 2011;17:302–309. doi: 10.1158/1078-0432.CCR-10-0999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Foight GW, Ryan JA, Gulla SV, Letai A, Keating AE. Designed BH3 peptides with high affinity and specificity for targeting Mcl-1 in cells. ACS Chem Biol. 2014;9:1962–1968. doi: 10.1021/cb500340w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Dutta S, Gulla S, Chen TS, Fire E, Grant RA, Keating AE. Determinants of BH3 binding specificity for Mcl-1 versus BcI-x(L) J Mol Biol. 2010;398:747–762. doi: 10.1016/j.jmb.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Kimura RH, Levin AM, Cochran FV, Cochran JR. Engineered cystine knot pep-tides that bind alphavbeta3, alphavbeta5, and alpha5beta1 integrins with low-nanomolar affinity. Proteins. 2009;77:359–369. doi: 10.1002/prot.22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Li WH, Lei P, Yu B, Wu S, Peng JL, Zhao XP, Zhu HF, Kirschfink M, Shen GX. Screening and identification of a novel target specific for hepatoma cell line HepG2 from the FliTrx bacterial peptide library. Acta Biochim Biophys Sin. 2008;40:443–451. doi: 10.1111/j.1745-7270.2008.00412.x. [DOI] [PubMed] [Google Scholar]
- 341.Zitzmann S, Kramer S, Mier W, Mahmut M, Fleig J, Altmann A, Eisenhut M, Haberkorn U. Identification of a new prostate-specific cyclic peptide with the bacterial FliTrx system. J Nucl Med. 2005;46:782–785. [PubMed] [Google Scholar]
- 342.Yang W, Luo D, Wang S, Wang R, Chen R, Liu Y, Zhu T, Ma X, Liu R, Xu G, Meng L, Lu Y, Zhou J, Ma D. TMTP1, a novel tumor-homing peptide specifically targeting metastasis. Clin Cancer Res. 2008;14:5494–5502. doi: 10.1158/1078-0432.CCR-08-0233. [DOI] [PubMed] [Google Scholar]
- 343.Dane KY, Chan LA, Rice JJ, Daugherty PS. Isolation of cell specific peptide li-gands using fluorescent bacterial display libraries. J Immunol Methods. 2006;309:120–129. doi: 10.1016/j.jim.2005.11.021. [DOI] [PubMed] [Google Scholar]
- 344.Dane KY, Gottstein C, Daugherty PS. Cell surface profiling with peptide libraries yields ligand arrays that classify breast tumor subtypes. Mol Cancer Ther. 2009;8:1312–1318. doi: 10.1158/1535-7163.MCT-08-1105. [DOI] [PubMed] [Google Scholar]
- 345.Brown CK, Modzelewski RA, Johnson CS, Wong MK. A novel approach for the identification of unique tumor vasculature binding peptides using an E. coli peptide display library. Ann Surg Oncol. 2000;7:743–749. doi: 10.1007/s10434-000-0743-0. [DOI] [PubMed] [Google Scholar]
- 346.Dong B, Wang AX, Yuan LH, Chen LS, Pu KF, Duan W, Yan XY, Zhu YM. Peptide-fluorescent bacteria complex as luminescent reagents for cancer diagnosis. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Hayashi Y, Morimoto J, Suga H. In vitro selection of anti-Akt2 thioether-macrocyclic peptides leading to isoform-selective inhibitors. ACS Chem Biol. 2012;7:607–613. doi: 10.1021/cb200388k. [DOI] [PubMed] [Google Scholar]
- 348.Morimoto J, Hayashi Y, Suga H. Discovery of macrocyclic peptides armed with a mechanism-based warhead: isoform-selective inhibition of human deacetylase SIRT2. Angew Chem Int Ed Eng. 2012;51:3423–3427. doi: 10.1002/anie.201108118. [DOI] [PubMed] [Google Scholar]
- 349.Lau D, Guo L, Liu R, Marik J, Lam K. Peptide ligands targeting integrin alpha3beta1 in non-small cell lung cancer. Lung Cancer. 2006;52:291–297. doi: 10.1016/j.lungcan.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 350.Aina OH, Marik J, Liu R, Lau DH, Lam KS. Identification of novel targeting pep-tides for human ovarian cancer cells using “one-bead one-compound” combinatorial libraries. Mol Cancer Ther. 2005;4:806–813. doi: 10.1158/1535-7163.MCT-05-0029. [DOI] [PubMed] [Google Scholar]
- 351.Lau DH, Guo L, Liu R, Song A, Shao C, Lam KS. Identifying peptide ligands for cell surface receptors using cell-growth-on-bead assay and one-bead one-compound combinatorial library. Biotechnol Lett. 2002;24:497–500. [Google Scholar]
- 352.DeRoock IB, Pennington ME, Sroka TC, Lam KS, Bowden GT, Bair EL, Cress AE. Synthetic peptides inhibit adhesion of human tumor cells to extracellular matrix proteins. Cancer Res. 2001;1:3308–3313. [PubMed] [Google Scholar]
- 353.Nair RR, Emmons MF, Cress AE, Argilagos RF, Lam K, Kerr WT, Wang HG, Dalton WS, Hazlehurst LA. HYD1-induced increase in reactive oxygen species leads to autophagy and necrotic cell death in multiple myeloma cells. Mol Cancer Ther. 2009;8:2441–2451. doi: 10.1158/1535-7163.MCT-09-0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Pennington ME, Lam KS, Cress AE. The use of a combinatorial library method to isolate human tumor cell adhesion peptides. Mol Divers. 1996;2:19–28. doi: 10.1007/BF01718696. [DOI] [PubMed] [Google Scholar]
- 355.Aggarwal S, Janssen S, Wadkins RM, Harden JL, Denmeade SR. A combinatorial approach to the selective capture of circulating malignant epithelial cells by peptide ligands. Biomaterials. 2005;26:6077–6086. doi: 10.1016/j.biomaterials.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 356.Lam KS, Lou Q, Zhao ZG, Smith J, Chen ML, Pleshko E, Salmon SE. Idiotype specific peptides bind to the surface immunoglobulins of two murine B-cell lymphoma lines, inducing signal transduction. Biomed Pept Proteins Nucleic Acids. 1995;1:205–210. [PubMed] [Google Scholar]
- 357.Mikawa M, Wang H, Guo L, Liu R, Marik J, Takada Y, Lam K, Lau D. Novel peptide ligands for integrin alpha 4 beta 1 overexpressed in cancer cells. Mol Cancer Ther. 2004;3:1329–1334. [PubMed] [Google Scholar]
- 358.Liu R, Peng L, Han HJ, Lam KS. Structure-activity relationship studies of a series of peptidomimetic ligands for alpha(4)beta(1) integrin on Jurkat T-leukemia cells. Biopolymers. 2006;84:595–604. doi: 10.1002/bip.20588. [DOI] [PubMed] [Google Scholar]
- 359.Zhang H, Aina OH, Lam KS, de Vere White R, Evans C, Henderson P, Lara PN, Wang X, Bassuk JA, Pan CX. Identification of a bladder cancer-specific ligand using a combinatorial chemistry approach. Urol Oncol. 2012;30:635–645. doi: 10.1016/j.urolonc.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Lin TY, Zhang H, Wang S, Xie L, Li B, Rodriguez CO, Jr, de Vere White R, Pan CX. Targeting canine bladder transitional cell carcinoma with a human bladder cancer-specific ligand. Mol Cancer. 2011;10:9. doi: 10.1186/1476-4598-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.TY Lin, Zhang H, Luo J, Li Y, Gao T, Lara PN, Jr, de Vere White R, Lam KS, Pan CX. Multifunctional targeting micelle nanocarriers with both imaging and therapeutic potential for bladder cancer. Int J. 2012;7:2793–2804. doi: 10.2147/IJN.S27734. [DOI] [PMC free article] [PubMed] [Google Scholar]


